Abstract
InhA, the primary target for the first line anti-tuberculosis drug isoniazid, is a key enzyme of the fatty-acid synthase II system involved in mycolic acid biosynthesis in Mycobacterium tuberculosis. In this study, we show that InhA is a substrate for mycobacterial serine/threonine protein kinases. Using a novel approach to validate phosphorylation of a substrate by multiple kinases in a surrogate host (Escherichia coli), we have demonstrated efficient phosphorylation of InhA by PknA, PknB, and PknH, and to a lower extent by PknF. Additionally, the sites targeted by PknA/PknB have been identified and shown to be predominantly located at the C terminus of InhA. Results demonstrate in vivo phosphorylation of InhA in mycobacteria and validate Thr-266 as one of the key sites of phosphorylation. Significantly, our studies reveal that the phosphorylation of InhA by kinases modulates its biochemical activity, with phosphorylation resulting in decreased enzymatic activity. Co-expression of kinase and InhA alters the growth dynamics of Mycobacterium smegmatis, suggesting that InhA phosphorylation in vivo is an important event in regulating its activity. An InhA-T266E mutant, which mimics constitutive phosphorylation, is unable to rescue an M. smegmatis conditional inhA gene replacement mutant, emphasizing the critical role of Thr-266 in mediating post-translational regulation of InhA activity. The involvement of various serine/threonine kinases in modulating the activity of a number of enzymes of the mycolic acid synthesis pathway, including InhA, accentuates the intricacies of mycobacterial signaling networks in parallel with the changing environment.
Keywords: Bacterial Protein Kinases, Bacterial Signal Transduction, Enzyme Catalysis, Protein Phosphorylation, Reductase, InhA, Mycobacteria, Mycolic Acid, Substrate
Introduction
The characteristic nature of the cell envelope of Mycobacterium tuberculosis is linked to its pathogenicity. The thick layer of lipids on the outer surface of mycobacteria is protective in nature, and mycolic acids comprise the bulk of this layer. Mycolic acids also constitute structural components of the cell wall and envelope (1, 2). Mycobacteria are unique in having two fatty-acid synthase (FAS)5 systems, FAS-I and FAS-II, and both of these pathways are involved in the synthesis of mycolic acids. Eukaryotic-like FAS-I is a single multidomain enzyme, whereas FAS-II includes discrete monofunctional enzymes that carry out the various sequential steps involved in the process of synthesis (3). The FAS-I system is responsible for the de novo fatty acid biosynthesis (4), producing C20- and C26-S-enzyme derivatives that are converted to the respective coenzyme A (CoA) forms and then released. The C20 fatty acid is the probable starting point for the FAS-II system (2), and the C26 fatty acid is incorporated as the α-branch of mycolic acid (5).
FabH (β-ketoacyl-ACP synthase III) forms the link between the two FAS systems by condensing the long chain acyl-CoA products (produced by FAS-I) and malonyl-ACP (produced from malonyl-CoA by the action of FabD (malonyl-CoA:ACP transacylase) (6)). The β-ketoacyl-S-ACP product (7) thus formed is subjected to keto-reduction, dehydration, and enoyl reduction consecutively, reactions catalyzed by MabA (β-ketoacyl-ACP reductase), β-hydroxyl-ACP dehydratase complex (Rv0636 + Rv0635, 0637), and InhA (enoyl-ACP reductase), respectively (2, 8). The ACP-bound acyl chain thus becomes two carbon units longer than the starting chain and now undergoes repetitive cycles of reduction, with the exception that the ketosynthases KasA or KasB substitute for FabH. InhA, the 2-trans-enoyl-ACP reductase of the FAS-II system (9), was first identified as the gene encoding a target of the drugs isoniazid and ethionamide, following the characterization of genetic variants that conferred resistance to these drugs (10). Biochemical analysis of InhA revealed that InhA catalyzes NADH-specific reduction of 2-trans-enoyl-ACP with preference for long chain substrates (C12–C24) (11). InhA is also essential for the survival of mycobacteria; inactivation of InhA affects mycobacteria acutely, causing accumulation of saturated fatty acids, the end products of FAS-I (12), “blebbing” of the cell wall, and consequent cell death by lysis (12, 13).
Protein phosphorylation is a principal mechanism by which extracellular signals are transduced into cellular responses. M. tuberculosis genome encodes for 11 eukaryote-like serine/threonine protein kinases (STPKs) (14–17). The M. tuberculosis STPKs affect key mycobacterial processes. Signal transduction events mediated by PknA and PknB play an important role in determining cell shape, morphology, and possibly cell division (18); PknG (19–21) and PknH (22) influence M. tuberculosis virulence, adaptation, and growth within the host; and PknF affects cell division, growth rate, morphology, and glucose transport (23). The mycobacterial FAS-II multienzyme complex has been identified to be a target of M. tuberculosis STPKs, with substrates, including the malonyl-CoA:ACP transacylase FabD and the β-ketoacyl-ACP synthases KasA and KasB. These proteins are phosphorylated by various kinases in vitro; however, the phosphorylation is found to be most efficient with PknA. Interestingly, although phosphorylation has a negative impact on the activity of KasA, KasB activity is enhanced upon phosphorylation (24, 25). Recently, MabA, the β-ketoacyl-ACP reductase, has been demonstrated to be negatively regulated following phosphorylation (26). FabH (β-ketoacyl-ACP synthase III) has also been shown to be a substrate of several kinases, particularly PknA and PknF. Phosphorylation of FabH has an inhibitory effect on enzyme activity (27).
In this study, we have identified InhA to be a substrate of multiple mycobacterial STPKs. InhA phosphorylation mediated by the STPKs has been validated in vivo, using both Escherichia coli as a surrogate host as well as in Mycobacterium smegmatis. The putative phosphorylation sites have been identified. We demonstrate that the phosphorylation status of InhA is critical to the function and/or activity of InhA and alters the growth profile of mycobacteria. Most importantly, results from studies carried out with an inhA conditional gene replacement strain emphasize the crucial role of the phosphorylation status of InhA in mediating mycobacterial survival.
EXPERIMENTAL PROCEDURES
Reagents and Radioisotopes
Restriction/modification enzymes were obtained from New England Biolabs. Cloning and expression vectors pENTR/Directional TOPO cloning kit (Invitrogen), pMAL-c2X (New England Biolabs), pGEX-4T-2 (GE Healthcare), pQE2 (Qiagen), and pET-Duet1 (Novagen) were purchased from respective sources. pMV261-apra and pJAM2 (E. coli-Mycobacterium shuttle vectors) were kind gifts from Dr. William Jacobs and Dr. Tanya Parish, respectively. [γ-32P]ATP (6000 Ci/mmol) was purchased from PerkinElmer Life Sciences. Oligonucleotide primers and analytical grade chemicals were purchased from Sigma.
Generation of Plasmid Constructs
M. tuberculosis pknA and pknB (full length) were PCR-amplified, respectively, from BAC clone Rv13 (a kind gift from Prof. Stewart Cole, Pasteur Institute, France) using gene-specific primers and Phusion DNA polymerase (New England Biolabs). pknG (full length) and kinase domains, including the region up to transmembrane domains of pknD, pknE, pknF, pknH, pknI, pknJ, and pknL, were amplified from BAC clones Rv313 (pknG), Rv103 (pknD), Rv302 (pknE and pknF), Rv407 (pknH), Rv209 (pknI), Rv412 (pknJ), and Rv269 (pknL), respectively. pknA, pknD, pknE, pknF, pknG, and pknI amplicons were digested with EcoRI and HindIII and cloned into corresponding sites on pMAL-c2X (New England Biolabs). pknB, pknH, and pknL amplicons were digested with BamHI and HindIII and cloned into corresponding sites on pMAL-c2X vector. pknJ amplicon was digested with XbaI and HindIII and cloned into corresponding sites on pMAL-c2X vector. pMAL-PknK (full length) and pGEX-PknB were generated as described previously (28, 29). pJAM-PknA and pJAM-PknB were created by cloning the PCR amplicon obtained using the specific primers into BamHI/XbaI sites of pJAM2 vector (30).
inhA gene was amplified from H37Rv BAC clone Rv58 using gene-specific primers and cloned into pENTR/Directional TOPO cloning vector (Invitrogen). inhA was further subcloned into NdeI/NotI sites in pQE2 vector (Qiagen). The point mutants used in this study were generated by overlapping PCR mutagenesis using appropriate mutagenesis primers (details are available upon request). inhA gene was amplified using specific primers, and amplicon was cloned into BamHI/XbaI sites in MCS of pJAM2 to generate pJAM-InhA. To generate pMV261apra-InhA wild type and mutant constructs, the respective wild type and mutant pQE2-InhA constructs were used as templates. Primers used for the amplification were such that amplicon obtained included sequence for His tag at the N terminus. Amplicons obtained were cloned into PvuII/HindIII sites of pMV261-apra vector.
For the co-expression studies, the dual expression vector pET-Duet1 (Novagen) was used. The different MBP-tagged kinase constructs were digested with HindIII, and the overhangs generated were filled in using Klenow polymerase followed by heat inactivation and digestion with NdeI. These fragments were subcloned into MCS2 of pET-Duet1 at NdeI/EcoRV sites. InhA was cloned into the NotI site in the first MCS of pET-Duet1 vector. Strains and constructs used in this study are described in Table 1.
TABLE 1.
Strains and constructs used in this study
Expression and Purification of Recombinant Proteins
Plasmids overexpressing PknA, PknB, and InhA (wild type and mutants), or plasmids co-expressing a kinase and InhA, were transformed into E. coli BL21 (DE3) Codon Plus cells (Stratagene). Cultures were induced with 0.1 mm isopropyl 1-thio-β-d-galactopyranoside and further grown for 12–16 h at 18 °C. Cells were harvested and lysed by sonication in lysis buffer (20 mm Tris-HCl (pH 7.5), 200 mm NaCl, and 10 mm β-mercaptoethanol for MBP fusion proteins or phosphate-buffered saline (pH 7.4), 10 mm β-mercaptoethanol for His-tagged proteins). The cell lysates containing MBP or His fusion proteins were nutated with equilibrated amylose resin (New England Biolabs) or Ni-NTA-agarose (GE Healthcare) affinity resins, respectively. His-tagged proteins were eluted with lysis buffer containing 200–250 mm imidazole. For the elution of MBP-tagged proteins, 10–20 mm maltose was used. GST-PknB was purified as described earlier (29). Peak fractions were dialyzed against dialysis buffer (10 mm Tris-HCl (pH 7.4), 50 mm NaCl, and 20% glycerol or PBS containing 20% glycerol).
In Vitro Kinase Assay, Phosphoamino Acid Analysis, and Phosphopeptide Mapping
In vitro kinase assays were performed by incubating 1–5 pmol of PknA or PknB and 25–30 pmol of InhA (WT or mutants) as described earlier (29). Phosphoamino acid analysis and peptide mapping were carried out as described previously (29, 31).
Preparation of M. smegmatis Cell Lysates
Electrocompetent cells of M. smegmatis mc2155 were prepared, and 500 ng of plasmid DNA was electroporated as described previously (32). Large scale cultures were inoculated with 1% of primary culture and grown with shaking for 36–40 h. Cultures were harvested and lysed using a bead beater (BioSpec Products), and the lysate was clarified by centrifugation at 13,200 rpm for 50 min at 4 °C. The lysate was incubated with Ni-NTA affinity beads to allow for pulldown of His-tagged protein, and beads were washed with PBS and resuspended in 2× SDS sample buffer.
Two-dimensional Gel Electrophoresis
20 μg of InhA purified from E. coli strains co-expressing InhA and MBP/MBP-PknA/MBP-PknB were precipitated with 5 volumes of cold acetone. Precipitated proteins were resuspended in 300 μl of two-dimensional sample buffer (33). Aliquots of the samples were analyzed on SDS-PAGE to normalize the loading for the two-dimensional gels. Equal amounts (∼15 μg) of the samples were resolved by two-dimensional gel electrophoresis as described earlier (33). Gels were either silver-stained or transferred onto nitrocellulose membranes and probed with anti-InhA antibodies raised in mice. For evaluating the InhA and InhA-T266A mutant expressed in M. smegmatis by two-dimensional gels, ∼7.5 mg of lysates prepared from M. smegmatis strains harboring pMV261-InhA or pMV261-InhA-T266A or pJAM-InhA were used for pulldowns using Ni-NTA resin to isolate His-InhA, His-InhA-T266A, and InhA-His, respectively. Ni-NTA beads were resuspended in PBS containing 2% SDS to elute bound proteins. Proteins thus eluted were acetone-precipitated and resuspended in 300 μl of two-dimensional sample buffer. Aliquots of the samples were analyzed on SDS-PAGE, and ∼5 μg of His-InhA or His-InhA-T266A or InhA-His was resolved by two-dimensional gels followed by Western blotting using anti-InhA antibodies. To examine endogenous InhA, 300 μg of M. smegmatis mc2155 whole cell lysates were precipitated with acetone followed by two-dimensional gels and Western blotting using anti-InhA antibodies.
Enoyl-CoA Reductase Assay and Determination of Kinetic Parameters
2-trans-Octenoyl-CoA was prepared from trans-2-octenoic acid by the mixed anhydride method as reported previously (34). The sample was purified by reverse phase-HPLC (C18 250 × 4.60-mm column) using an acetonitrile/water/TFA (trifluoroacetic acid) solvent system (gradient, 4–72% B in 50 min; A, 0.1% TFA; B, acetonitrile containing 0.1% TFA; flow rate, 1 ml/min). The chromatogram was monitored at 210 and 265 nm. Fractions containing 2-trans-octenoyl-CoA were pooled, lyophilized, and stored at −20 °C till use. The chemical identity of the compound was confirmed by electrospray mass spectrometry.
InhA activity was assayed essentially as described previously (11), with minor modifications. Briefly, the reactions were carried out in a total volume of 50 μl in 30 mm PIPES buffer (pH 6.8) containing 500 μm 2-trans-octenoyl-CoA, 150 μm NADH, and 5–50 pmol of InhA enzyme (InhA or p-InhA, respectively). Time course assays were carried out, monitoring oxidation of NADH by measuring absorbance at 340 nm for 15 min, at 1-min intervals (GE Ultrospec 2100pro). NAD formed was calculated as ((A340 at 0 min − A340 at time t (1–15 min)/A340 at 0 min) × (NADH μm/pmol of InhA) and plotted as a function of time.
To determine the kinetic constants with respect to 2-trans-octenoyl-CoA, reactions were carried out in a 50-μl volume of 30 mm PIPES containing 150 μm NADH, 15 or 30 pmol of InhA or p-InhA, respectively, and varying concentrations of 2-trans-ocetenoyl-CoA. The reaction was performed for 15 min, and the absorbance was measured at 340 nm. NAD formed was calculated as ((initial A340 − final A340/initial A340) × (NADH μm/min/mg enzyme)). Km and Vmax values were determined by nonlinear regression analyses carried out with GraphPad Prism software. Data from two independent experiments were plotted, and results are presented as average values ± S.D. Similarly, kinetic constants were determined for NADH in 50-μl reactions containing 500 μm 2-trans-octenoyl-CoA, 15 or 30 pmol of InhA or p-InhA, respectively, and varying concentrations of NADH.
Growth Curve Analysis
M. smegmatis mc2155 strains, containing either pMV261apra or pMV261apra-InhA, were electroporated with pJAM-PknA (acetamidase-inducible). To carry out analysis of PknA, InhA, or PknA + InhA overexpression in mycobacteria, each strain was grown in 7H9-ADC without inducer to the exponential phase and re-inoculated into fresh medium (in the presence and absence of 0.2% acetamide) such that A600 was 0.02 for the secondary culture at time 0. Samples were withdrawn at different time points for A600 measurement. Studies of the growth pattern of InhA mutants were similarly carried out. M. smegmatis mc2155 strains electroporated with pMV261 vector only, pMV261-InhA, pMV261-InhA T266A, and pMV261-InhA T266E were used to overexpress the respective proteins. For analyzing effects of PknA co-expression with various InhA mutants, M. smegmatis mc2155 was electroporated with the integration-proficient construct pST-HI-acet-PknA. The resulting strain contains a single copy of pknA under the control of the acetamidase promoter. pMV261 vector, pMV261-InhA, pMV261-InhA T266A, and pMV261-InhA-T266E were electroporated into this strain. The resulting strains were grown in LB/Tween 80 without inducer to exponential phase and re-inoculated into fresh medium (in the presence and absence of 0.5% acetamide) such that A600 was 0.02 for the secondary culture at time 0. Samples were withdrawn at different time points for A600 measurement.
Assessment of in Vivo Functionality of InhA Phosphorylation Mutants
pMV261-InhA, pMV261-InhA-T266A, and pMV261-InhA-T266E were introduced into the M. smegmatis inhA conditional mutant strain mc24751 (13) by electroporation to generate the transformed strains, mc24751/pMV261-inhA, mc24751/pMV261-inhA-T266A, and mc24751/pMV261-inhA-T266E. The ability of the transforming inhA allele to rescue growth defects of the conditional mutant was assessed by growing the transformants in the presence or absence of acetamide (0.2% in tryptic soy broth) as described earlier (13).
CD Spectroscopic Analyses
Protein samples were prepared in PBS (pH 7.4). CD spectra were recorded on Jasco J-710 spectropolarimeter. Data were collected at 25 °C using 2-mm path length cell from 250 to 190 nm in 1-nm steps, at a scanning speed of 200 nm/min. Spectra presented are the averages of 20 scans. The results are reported as mean residue ellipticity expressed as degrees cm2 dmol−1.
Structural Modeling
Three-dimensional models of the InhA T266A and T266E mutants were created using PyMOL software (35). The Protein Data Bank code 1BVR of the published M. tuberculosis InhA structure (36) was used as the template for rendering these in silico structures.
RESULTS
InhA Is a Novel Target of M. tuberculosis STPKs
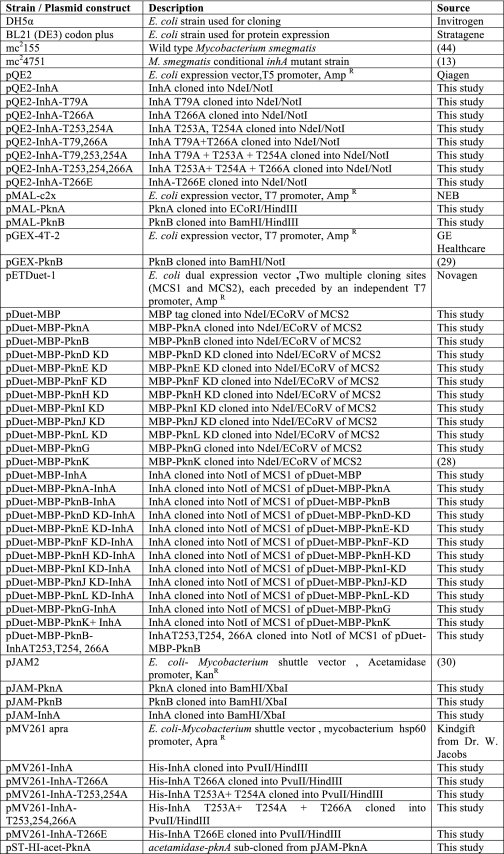
Kang et al. (18) have demonstrated that PknA and PknB show preferential phosphorylation of Thr residues that precede a Gln residue. Interestingly, several substrates of these kinases identified to date conform to this motif, including KasB of the FAS-II system (24). The presence of a TQ motif in InhA, and considering that several other enzymes of the FAS-II pathway were found to be substrates for PknA and/or PknB, we set out to explore whether InhA could also be a target for mycobacterial STPKs. Toward this, inhA, pknA, and pknB from M. tuberculosis were cloned and the respective proteins overexpressed in and purified from E. coli (Fig. 1A). In vitro kinase reactions carried out with recombinant InhA and PknA or PknB demonstrated that InhA was phosphorylated by both kinases (Fig. 1B). The InhA thus phosphorylated was subjected to phosphoamino acid and peptide mapping analyses. It was observed that InhA is phosphorylated specifically on threonine residues by PknA (Fig. 1C) and PknB (data not shown). The peptide map obtained was similar for InhA phosphorylated by PknA (Fig. 1D) and PknB (data not shown) and shows the presence of multiple spots, indicating that InhA is phosphorylated on multiple sites by these kinases in vitro.
FIGURE 1.
InhA is an in vitro substrate of PknA and PknB. A, E. coli BL21 (DE3) codon plus cells were transformed with pQE2-InhA, pMAL-PknA, and pGEX-PknB, respectively, and the corresponding His-InhA, MBP-PknA, and GST-PknB proteins were purified as described under “Experimental Procedures.” Bands corresponding to the purified proteins are indicated. B, in vitro kinase assays carried out using MBP-PknA (5 pmol) or GST-PknB (1 pmol) with InhA (30 pmol) in the presence of [γ-32P]ATP. Bands corresponding to autophosphorylated PknA or PknB and phosphorylated InhA are indicated by arrowheads. C, phosphoamino acid analysis of in vitro phosphorylated InhA. The panel shows the autoradiogram and the positions of pSer, pThr, and pTyr standards on the TLC plates are indicated (p = phosphorylated). D, two-dimensional peptide map of in vitro phosphorylated InhA autoradiogram is shown.
InhA Is Phosphorylated by Multiple STPKs
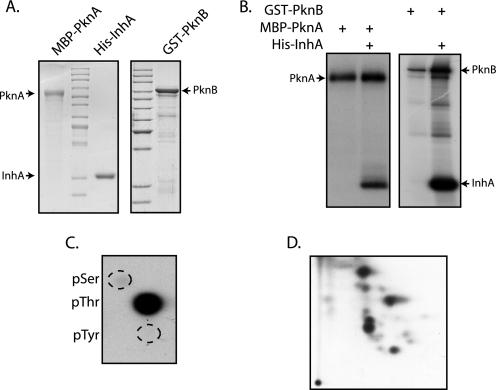
Many substrates identified thus far are phosphorylated by multiple M. tuberculosis kinases. To date, reports demonstrating phosphorylation of substrate by multiple kinases have been carried out in vitro using purified substrates and kinases. To investigate phosphorylation of InhA by M. tuberculosis serine/threonine kinases, we developed a convenient approach, utilizing E. coli as a surrogate host. pDuet-1 vector has two multiple cloning sites (MCS1 and MCS2) with independent T7 promoters, thus enabling co-expression of two proteins. We cloned either the full-length kinase (PknA, PknB, PknG, and PknK) or the kinase domains, including the region up to the transmembrane domain (kinase domain) (PknD, PknE, PknF, PknH, PknI, PknJ and PknL) in pMAL-c2x vector as described under “Experimental Procedures.” All 11 MBP-tagged kinases and the MBP tag were independently subcloned into the MCS2 of pDuet1 vector (Fig. 2A). As a first step, we determined the expression status of MBP and the MBP-tagged kinases. It is apparent from the results (Fig. 2B) that MBP and the MBP-tagged kinase expressed robustly. Restriction sites that are unique in MCS1 and can be used for substrate cloning are shown in Fig. 2A. Upon cloning, the substrate of interest into MCS1 of pDuet-MBP/MBP kinase constructs, one can express both the substrate and the kinase from a single construct in E. coli BL21 codon plus strain, thus providing an in vivo tool to assess phosphorylation of the substrate by corresponding kinase.
FIGURE 2.
pDuet-1 constructs designed for co-expression of MBP/MBP kinase and substrate. A, schematic representation of the MCS region of pDuet-MBP/MBP kinase constructs cloned in MCS2. Unique sites that can be used for cloning test substrate in MCS1 are shown. B, expression profiles of the various pDuet-MBP/MBP kinase constructs. Respective constructs were transformed in BL21 (DE3) BL21 codon plus cells, and corresponding cultures were induced. Lysates were probed with anti-MBP antibodies (New England Biolabs). KD refers to kinase domains, including the region up to transmembrane region.
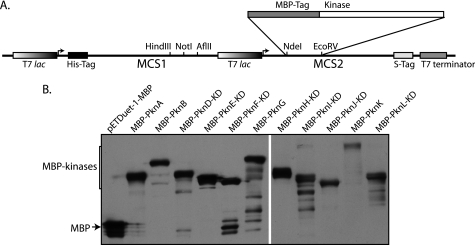
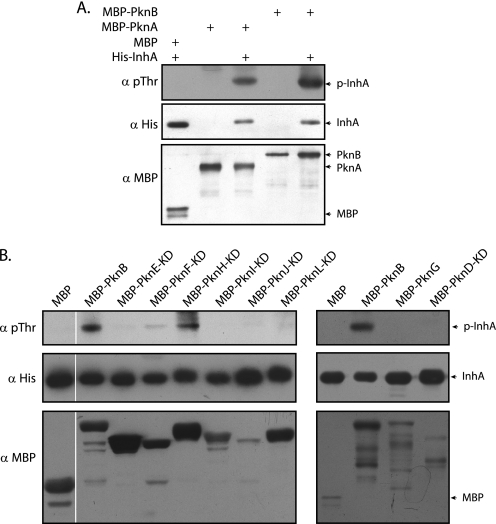
To validate the in vitro phosphorylation of InhA by PknA and PknB, InhA was co-expressed with MBP, MBP-PknA, or MBP-PknB, respectively, by means of the specific pDuet constructs. Phosphorylation status of InhA was determined by probing the His pulldowns with phosphothreonine antibodies (Fig. 3A). It was evident that both PknA and PknB phosphorylated InhA efficiently in vivo. To ascertain whether InhA is a target of any of the other STPKs, we performed an analogous experiment with the other kinases, with InhA co-expressed with MBP and MBP-PknB as negative and positive reference points, respectively (Fig. 3B). Our results showed that in addition to PknA and PknB, PknH and to an extent PknF also phosphorylated InhA. Together, these results indicate that InhA is a target substrate of multiple kinases that probably phosphorylate the protein on more than one threonine residue.
FIGURE 3.
InhA is targeted by multiple STPKs of M. tuberculosis. A, pDuet constructs with MBP, MBP-pknA, or MBP-pknB cloned in MCS2 with or without InhA cloned in MCS1 were transformed in BL21 (DE3) codon plus cells. Lysates and His pulldowns were probed with anti-Thr(P) (pThr) (Cell Signaling), anti-His (BD Biosciences), and anti-MBP (New England Biolabs) antibodies. B, pDuet-1 co-expression constructs containing MBP or kinases in MCS2 and InhA in MCS1 were subjected to Western blot analysis as above. KD refers to kinase domains, including the region up to transmembrane region. Top panel, Thr(P) blot; middle panel, His blot performed on His pulldowns; lower panel, MBP blot on lysates to show the expression of kinases.
InhA Is Phosphorylated in the C-terminal Region
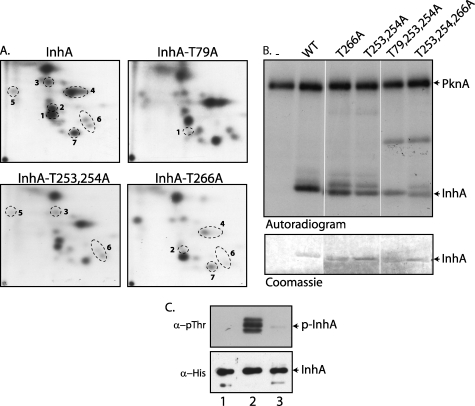
Primary sequence analysis of InhA showed the presence of only one “TQ” motif. The peptide map of InhA phosphorylated by PknA/PknB, however, indicated that there are multiple phosphorylation sites, with threonines specifically being targeted (Fig. 1, C and D). Analysis of the InhA primary sequence revealed presence of 13 threonine residues. We mutagenized all 13 potential phosphorylation sites individually or in combination (Thr-253 and Thr-254), from threonine to alanine residues, to determine which sites would modify the phosphopeptide maps. Nine of the twelve mutants had the same phosphopeptide pattern as the wild type InhA (data not shown). The peptide maps for the other three mutants, namely T79A, T266A, and T253A,T254A, were distinct from that of the wild type InhA. Spots that either disappeared or showed substantial decrease in intensity as compared with the wild type map are numbered from 1 to 7 (Fig. 4A). Because all the TLC plates were subjected to autoradiography for the same period of time, we correlated the disappearance and/or decreased intensity with the phosphorylation on a particular site. Peptide map of InhA-T79A showed absence of spot 1, indicating phosphorylation of InhA on the Thr-79 residue. Mutation of Thr-266 resulted in disappearance of spot 2 and decreased intensities of spots 4, 6, and 7. In the case of T253A,T254A mutant, we observed a decrease in the intensities of spots 3, 5, and 6. Maps of T253A and T254A single mutants were similar to the peptide map of InhA-T253A,T254A double mutant (data not shown). Theoretical tryptic digestion of InhA revealed that the C-terminal tryptic fragment has four threonine residues (Thr-241, Thr-253, Thr-254, and Thr-266). Mutation of one or the other threonines may have influence on the in vitro phosphorylation of neighboring threonine residues in the peptide. This may lead to further changes in the intensities of spots we detect on TLC. The complexity of the maps owing to a combination of multiple phosphorylation sites, more than one threonine residue on a tryptic peptide and/or the partial tryptic digestions did not permit complete assignment of all the spots.
FIGURE 4.
InhA is phosphorylated at its C terminus. A, in vitro phosphorylated InhA mutants were digested with trypsin, and the resulting phosphopeptides were mapped by two-dimensional resolution on thin layer chromatography. The spots undergoing a change in relative intensity in the InhA-mutant peptide maps compared with the wild type InhA map are numbered 1–7. B, comparative kinase assay of InhA wild type and various single and combination threonine point mutants. 5 pmol of MBP-PknA and 30 pmol of InhA or InhA mutants were used for the assays. Upper panel, autoradiogram; lower panel, Coomassie-stained protein bands of the InhA mutants. C, 4 μg of His-InhA or His-InhA- T253A,T254A,T266A (T253,254,266A) purified from E. coli co-expressing MBP or MBP-PknB were resolved, transferred, and probed with anti-phosphothreonine antibodies. Similarly, 500 ng of the proteins were resolved and probed using anti-His antibodies. Lane 1, His-InhA co-expressed with MBP; lane 2, His-InhA co-expressed with MBP-PknB; lane 3, InhA-T253,254,266A mutant co-expressed with MBP-PknB.
Next, generated combination threonine site mutants of InhA, and carried out comparative kinase assays with wild type and various InhA mutants (Fig. 4B). We observed a decrease in the extent of phosphorylation for most of the mutants when contrasted with the wild type InhA. Notably, the combination mutant in which threonine residues at positions 253, 254, and 266 are mutated to alanine concurrently (TriT: T253A,T254A,T266A mutant) showed significantly decreased phosphorylation (Fig. 4B). To substantiate these findings, we purified InhA from E. coli strains co-expressing either the MBP or MBP-PknB with InhA wild type or the TriT mutant and analyzed the phosphorylation status of InhA (Fig. 4C). It is apparent from the results that the triple mutant was negligibly phosphorylated by the kinase as compared with the wild type InhA. Taken together, our results suggest that Thr-79 is a minor phosphorylation site, whereas Thr-253, Thr-254, and Thr-266, located in the C-terminal region, are major targets of PknA/PknB-mediated phosphorylation events.
InhA Is Phosphorylated in Vivo
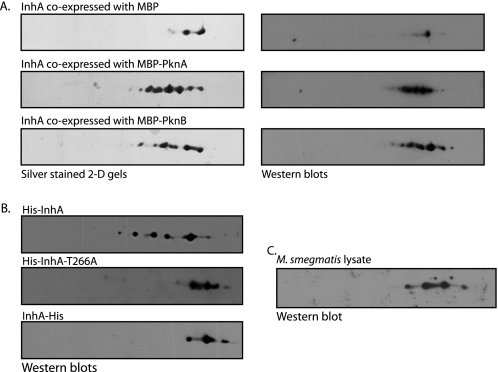
Phosphorylation is known to alter the pattern of protein migration in a two-dimensional gel, as negative charge on the phosphate group results in a shift toward the acidic end of the IPG strip. This property of a differential migration pattern of phosphorylated proteins was used to determine whether the various phosphorylated forms of InhA could be distinguished by two-dimensional gel electrophoresis. InhA was purified from E. coli co-expressing InhA and MBP, MBP-PknA, or MBP-PknB. These proteins were resolved in two dimensions, and the protein spots were visualized by silver staining and Western blotting with InhA-specific antibodies (Fig. 5A). Compared to InhA co-expressed with MBP, InhA co-expressed with either PknA or PknB showed additional spots, demonstrating phosphorylation of InhA by these kinases (Fig. 5A). The spot intensities as calculated by ImageJ software indicated that ∼50% of InhA protein was phosphorylated by PknA or PknB, respectively, under these conditions.
FIGURE 5.
InhA is phosphorylated in vivo, and Thr-266 is a major target site. A, His-InhA protein purified from E. coli following co-expression with MBP, MBP-PknA, or MBP-PknB was precipitated with 5 volumes of cold acetone. Approximately 15 μg of precipitated proteins were resolved on a 13-cm pH 4–7 linear gradient IPG strips (GE Healthcare) for 25,000 V-h. IPG strips were resolved in the second dimension on 10% SDS-polyacrylamide gels and were either visualized by silver staining or transferred to nitrocellulose membrane and subjected to Western blotting with anti-InhA antibodies raised in mice. B, two-dimensional gels of InhA protein isolated from M. smegmatis overexpressing InhA from pMV261-InhA, pMV261-InhA-T266A, or pJAM-InhA, respectively. Approximately 5 μg of the respective proteins were resolved as described. Western blots of the resolved proteins probed with anti-InhA antibodies are shown. C, 300 μg of M. smegmatis mc2155 whole cell lysate was precipitated with acetone, followed by resolution using two-dimensional gels as above, followed by Western blotting using anti-InhA antibodies.
Next, to ascertain whether InhA is indeed phosphorylated in mycobacteria, we transformed M. smegmatis with plasmid encoding M. tuberculosis inhA (pMV261-InhA), purified the recombinant InhA bearing N-terminal His tag, and analyzed the protein by two-dimensional gel electrophoresis. Western blot analysis of the two-dimensional gels showed the presence of multiple spots, suggesting phosphorylation of InhA in M. smegmatis (Fig. 5B, upper panel). Results in Fig. 4 demonstrated that InhA was mainly phosphorylated on Thr-253, Thr-254, and Thr-266. We generated pMV261-InhA-T266A, pMV261-InhA-T253A, T254A, and pMV261-InhA-TriT, plasmid-borne versions of the mutants, and transformed them into M. smegmatis. However, we could only detect expression of InhA and InhA-T266A mutant; the other mutants failed to express despite the sequence analysis of the clones showing no discrepancy. We resolved the His-InhA-T266A mutant isolated from M. smegmatis by two-dimensional gel electrophoresis. It is clear from our results (Fig. 5B, middle panel) that mutating the Thr-266 residue resulted in disappearance of spots. These results strongly suggest that Thr-266 is a bona fide phosphorylation site of InhA. Furthermore, because the phosphorylation of InhA occurs mainly at the C terminus, we reasoned that addition of a hexa-His tag at the C-terminal protein may interfere with the phosphorylation. Hence, we generated a pJAM-InhA construct with C-terminal His tag. Interestingly, when InhA-His protein purified from M. smegmatis transformed with pJAM-InhA was resolved by two-dimensional gels, Fig. 5B, bottom panel, we observed a pattern very similar to that observed for InhA-T266A. These results are in agreement with our earlier data that showed phosphorylation of InhA in the C-terminal region (Fig. 4). To verify that InhA is phosphorylated at endogenous expression levels, M. smegmatis whole cell lysate was resolved by two-dimensional gel electrophoresis, transferred to nitrocellulose membrane, and probed with anti-InhA antibodies. We observed multiple spots (Fig. 5C), suggesting that endogenous InhA is phosphorylated in mycobacteria. The number of spots detected was less than those observed for the His-InhA, most likely due to significantly lower amounts of the InhA protein present in the total lysates.
Phosphorylation of InhA Affects Its Biochemical Activity
InhA from M. tuberculosis has been shown to catalyze NADH-specific reduction of 2-trans-enoyl-CoA with preference for long chain substrates (C12–C24), with C16–C20 substrates being most efficiently reduced. A chain length of C8 was the minimum required to be a substrate for M. tuberculosis InhA (11). We synthesized 2-trans-octenoyl-CoA, as reported previously (34), for use in an in vitro enzyme assay. The experimental mass of the 2-trans-octenoyl-CoA was in accord with its calculated mass (891.29 Da), and the HPLC trace of the final product showed presence of a single peak (data not shown).
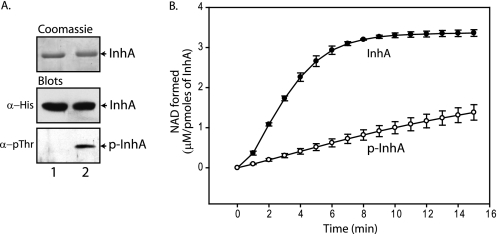
InhA co-expressed with MBP or MBP-PknB in E. coli was purified to represent unphosphorylated and phosphorylated forms of the protein, InhA and p-InhA, respectively. Phosphorylation of InhA by PknB was confirmed by Western blotting with anti-phosphothreonine antibodies (Fig. 6A). Activity of phosphorylated (p-InhA) and nonphosphorylated InhA was determined by measuring NADH oxidation over time. The NAD formed at each time point was calculated and plotted as a function of time for both the enzyme sets (Fig. 6B). We observed ∼5-fold reduction in the activity for p-InhA compared with nonphosphorylated InhA, suggesting negative modulation of InhA enzymatic activity upon phosphorylation. Furthermore, we determined the kinetic parameters for InhA and p-InhA for both the substrates 2-trans-octenoyl-CoA and NADH (Table 2). Specific activity of InhA was found to be ∼5-fold higher compared with p-InhA in agreement with our previous results (Fig. 6B). However, if the stoichiometry of the phosphorylation was taken into consideration (∼50% utilizing pDuet co-expression system), the difference in the specific activity would be ∼10-fold. We observed similar Km values for 2-trans-octenoyl-CoA for both the enzymes, although InhA seems to have higher affinity for NADH compared with p-InhA (Table 2).
FIGURE 6.
Phosphorylation of InhA affects its biochemical activity. A, InhA proteins used for enoyl-CoA reductase assay, visualized by Coomassie staining (top panel), His blot (middle panel), and phospho-blot (lower panel). Lane 1, InhA purified from E. coli co-expressing MBP tag; lane 2, InhA purified from E. coli co-expressing MBP-PknB that represents phosphorylated protein (p-InhA) (p = phosphorylated). B, graphical representation of time course assays of InhA (●) and p-InhA (○), carried out as detailed under “Experimental Procedures.” The y axis corresponds to NAD formed (μm/pmol of InhA) at each time point, and the x axis signifies time (min). Average values of three independent experiments are presented; the error bars represent the standard error.
TABLE 2.
Kinetic parameters of M. tuberculosis InhA and p-InhA
Experimental conditions and data analysis are described under “Experimental Procedures.” Data from two independent experiments are presented as average values ± S.D.
| InhA | p-InhA | |
|---|---|---|
| 2-trans-Octenoyl-CoA Km (μm) | 528 ± 16.7 | 511 ± 13 |
| NADH Km (μm) | 19.1 ± 5.4 | 57.8 ± 5.2 |
| Vmax (μm product/min/mg enzyme) | 15.3 ± 2.4 | 3.4 ± 0.8 |
Phosphorylation of InhA Impacts Growth of Mycobacterium
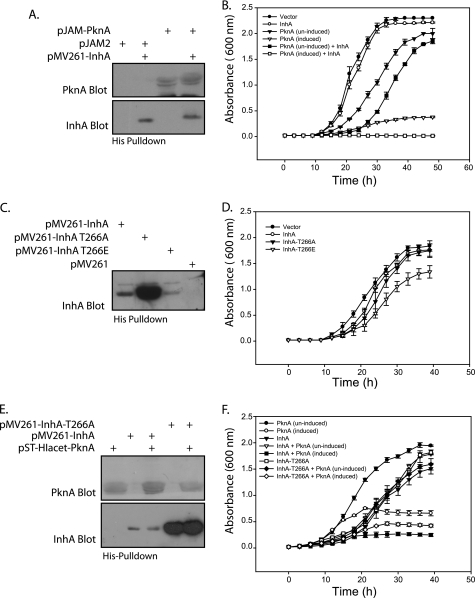
Loss of InhA activity leads to slowing of mycobacterial growth and subsequent lysis of cells (12, 13). Given the relatively lower activity of phosphorylated InhA as compared with the nonphosphorylated form in our enzyme assays (Fig. 6A), we sought to determine the effects of phosphorylation of InhA on the growth pattern of mycobacteria. For this purpose, M. smegmatis was transformed with plasmids that expressed either InhA (pMV261-InhA) or PknA (pJAM-PknA) or both. An acetamide-inducible promoter in pJAM-PknA allowed controllable expression of PknA. Western blot analysis demonstrated independent as well as co-expression of InhA and PknA (Fig. 7A). Similar efforts to co-express PknB and InhA in M. smegmatis were unsuccessful, although PknB expression by itself was quite stable. InhA overexpression alone had no discernible effects on the growth of M. smegmatis (Fig. 7B). However, we observed that growth of M. smegmatis expressing PknA was compromised even in the absence of inducer, likely due to basal expression from the acetamidase promoter. This was not surprising, as overexpression of PknA has been shown to affect the growth of the organism (18). While our results were compounded by the fact that PknA expression per se had a deleterious effect on mycobacterial growth (Fig. 7B) and (18), we did observe a further quantifiable decrease in the growth of cultures expressing both InhA and PknA, compared with the cultures expressing only PknA (Fig. 7B). This suggested that the PknA-mediated phosphorylation of InhA may play a role in modulating the growth pattern of the bacterium.
FIGURE 7.
Co-expression of PknA and InhA leads to diminished growth in mycobacteria. A, expression profile of M. smegmatis strains overexpressing pknA and/or inhA from pJAM-PknA and pMV261-InhA constructs, respectively. His pulldowns were subjected to Western blotting with anti-PknA (raised in rabbit) or anti-InhA antibodies (raised in mice). B, growth pattern analysis of M. smegmatis strains overexpressing pknA and/or inhA. InhA expression was constitutive, whereas PknA expression was under the control of acetamide-inducible promoter. Growth of strains containing vector control (pJAM2) only (●), vector control (pJAM2) and InhA (○), PknA only (▾), PknA in presence of acetamide (▿), PknA and InhA (■), and PknA and InhA in presence of acetamide (□) was monitored over a period of 48 h, every 3 h. A600 is plotted as a function of time. Average absorbance values at each time point from three independent experiments are plotted; the error bars represent the standard deviation of each data set. C, expression profile of M. smegmatis strains overexpressing InhA, InhA-T266A, or InhA-T266E from respective pMV261-InhA constructs. His pulldowns subjected to Western blotting with anti-InhA antibodies. D, growth curves of M. smegmatis strains overexpressing InhA, InhA-T266A, and InhA-T266E. Growth of strains containing vector control (pMV261 apra) only (●), InhA (○), InhA-T266A (▾), and InhA-T266E (▿) were monitored over a 40-h period at an interval of every 3 h. A600 is plotted as a function of time as above. E, expression profile of M. smegmatis strains overexpressing pknA and/or inhA mutants from pST-HI-acet-PknA (integrated single copy of PknA) and pMV261-InhA/pMV261-InhA-T266A/pMV261-InhA-T266E constructs, respectively. His pulldowns were subjected to Western blotting with anti-PknA or anti-InhA antibodies. F, growth pattern analysis of M. smegmatis strains overexpressing pknA and/or inhA/inhA T266A. InhA expression was constitutive, whereas PknA expression was under the control of acetamide-inducible promoter. Growth of strains containing PknA only (●), PknA in presence of acetamide (○) and InhA (▾), PknA and InhA (▿), and PknA and InhA in presence of acetamide (■), InhA-T266A (□), PknA and InhA-T266A (♦), and PknA and InhA-T266A in presence of acetamide (♢) was monitored over a period of 40 h. A600 is plotted as a function of time as above.
To investigate this hypothesis further, we created an InhA-T266E mutant. The glutamate residue would mimic a phosphorylated threonine residue, and hence this mutant was expected to function like InhA constitutively phosphorylated at the Thr-266 position. We studied the growth pattern of M. smegmatis strains transformed with plasmids for overexpression of InhA, InhA-T266A, and InhA-T266E, alongside a control strain. Surprisingly, the expression levels of these mutants varied drastically (Fig. 7C). We observed that M. smegmatis expressing the T266E mutant exhibited a slower growth rate compared with either InhA or InhA-T266A (Fig. 7D). Furthermore, we attempted to co-express the InhA, InhA-T266A, and InhA-T266E mutants, respectively, with PknA. For this purpose, the M. smegmatis strain containing the single integrated acetamidase-inducible copy of PknA was used as parent strain (PknA merodiploid). Unlike ectopically expressing pJAM-PknA, the single integrated copy of PknA serves to minimize the detrimental effects of PknA overexpression on growth. PknA merodiploid strain was electroporated with pMV261, pMV261-InhA, pMV261-InhA-T266A, and pMV261-InhA-T266E, respectively. Despite repeated attempts, we could not co-express InhA-T266E along with PknA. Analysis of growth patterns (Fig. 7F) showed that co-expression of InhA-T266A with PknA partially alleviates the diminution in growth compared with PknA-InhA co-expression (Fig. 7F). Inability of InhA-T266A mutant co-expression alongside PknA to completely revert the diminution of growth to only PknA overexpression levels could be due to the possibility that sites other than Thr-266, such as Thr-253 and Thr-254, may be still be targeted by PknA. Our results in Fig. 6B showed that phosphorylation of InhA led to decreased catalytic activity. Taken together, these results suggest that overexpressed PknA may be hyperphosphorylating InhA, which may result in decreased activity of InhA, and hence significantly compromised growth.
Phosphorylated State of InhA Is Functionally Inactive in Vivo
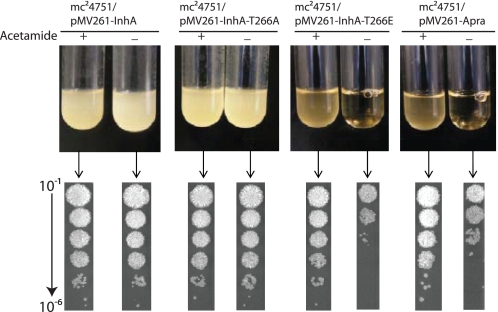
To examine the in vivo relevance of the observed differences, we tested the ability of the above described mutant alleles of inhA to rescue an M. smegmatis conditional inhA gene replacement mutant from death by lysis. The M. smegmatis mc24751 strain contains a deletion in the native inhA gene and has a recombinant, acetamide-inducible copy of inhA integrated into the chromosome (13). This strain grows normally in medium containing acetamide, whereas growth in medium devoid of acetamide results in loss of mycolic acid biosynthesis and subsequent cell lysis. The mutant alleles of inhA can thus be functionally analyzed in vivo by testing the ability of this strain transformed with plasmid-borne copies of the mutant alleles to survive (no lysis) in the medium devoid of acetamide. Strains of M. smegmatis mc24751 transformed with pMV261-InhA-T266A or pMV261-InhA-T266E were cultured in medium with or without acetamide as described previously (13). Strains containing vector only (pMV261apra) or native inhA (pMV261-InhA) were used as controls. As expected, although mc24751/pMV261apra lysed in medium without acetamide, mc24751 transformed with pMV261-InhA was able to grow equally well in medium with or without acetamide (Fig. 8). The strain mc24751/pMV261-InhA-T266A also grew normally in medium devoid of acetamide. On the other hand, mc24751/pMV261-inhA-T266E showed lysis when cultured in acetamide-free medium, thus indicating that the InhA-T266E mutant was unable to function efficiently in vivo. In addition, viable counts observed for vector control were ∼10-fold higher compared with mc24751/pMV261-inhA-T266E, thus suggesting that InhA-T266E may be behaving like a dominant negative form. These findings stress the importance of kinase-mediated phosphorylation of InhA.
FIGURE 8.
Assessment of the role of InhA phosphorylation in vivo. The top panel shows cultures of mc24751 strains transformed with different plasmids following 24 h of growth in TSB with or without acetamide. The lower panel corresponds to viable counts obtained from each tube, where 10-fold serial dilutions were spotted (5 μl) on TSB-agar plates containing acetamide.
Gross Conformation of InhA Is Unaffected by Mutations at Thr-266 Residue
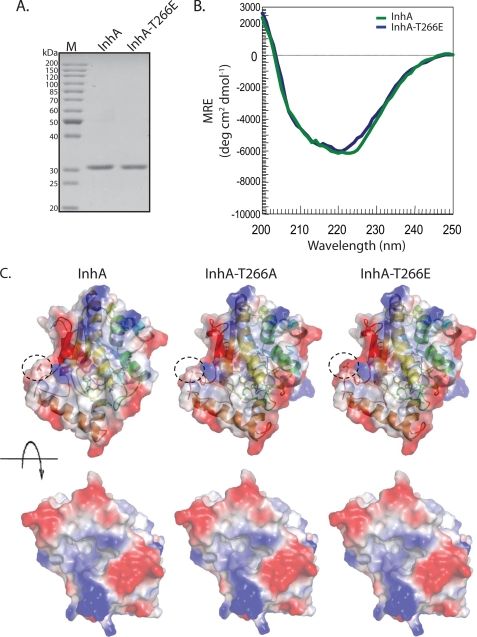
It is clear from our data that InhA-T266E mutant could not functionally complement WT-InhA. We investigated whether this could be a consequence of conformational aberrations caused by the InhA-T266E mutation. To verify that the overall conformational features of wild type are maintained in the mutant protein, we purified InhA and InhA-T266E mutants and analyzed them by CD spectroscopy (Fig. 9, A and B). The spectra of the two proteins in Fig. 9B are very similar suggesting that the T266E mutation does not perturb the gross folding of InhA. Additionally, in silico modeling of the three-dimensional structure of the InhA-T266E mutant using the published structure of M. tuberculosis WT InhA as a template revealed no apparent changes in the structure of the mutated protein or the substrate binding pocket (Fig. 9C). InhA is known to form homodimers. Hence, it is possible that T266E mutant may act as the dominant negative enzyme by dimerizing with wild type InhA. Combined with our kinetic data, we speculate that subtle conformational changes, if any, due to increase in charge because of phosphorylation might also modulate NADH binding and catalysis.
FIGURE 9.
Gross conformation of InhA is unaffected by mutations at Thr-266. A, 1 μg of purified InhA and InhA-T266E were resolved on 10% SDS-PAGE and visualized by Coomassie staining. B, CD spectra of InhA (green) and InhA-T266E (blue) are presented as a measure of mean residue ellipticity (MRE). C, in silico modeling of InhA mutants. The top panel represents superimposed ribbon diagrams and electrostatic surface potential maps (residue 266 is shown within a dotted circle). The bottom panel shows the opposite face of the protein where the substrate binding pocket can be visualized.
DISCUSSION
M. tuberculosis kinases have a plethora of substrates (16), including several key enzymes of the FAS system (24, 26, 27, 37). As the substrates identified to date in the FAS pathway are proficiently phosphorylated by PknA, we scrutinized the primary sequence of all the enzymes in the FAS pathway for the presence of the consensus “TQ” motif. Our analysis revealed that KasB, which was shown to be a substrate of these kinases (24), and InhA harbor this motif. The possibility of InhA also being a part of the signaling network of these kinases thus became a subject of interest to us. Our data illustrate robust phosphorylation of InhA by PknA and PknB, in agreement with the prediction of this enzyme being a target substrate (Fig. 1). InhA is an important component of the FAS-II pathway, the loss of which results in retarded growth and subsequent cell lysis (12, 13). Our in vitro InhA activity assay results suggested that phosphorylation of InhA results in decreased enzymatic activity (Fig. 6). Furthermore, in line with these results, we found that the strain expressing InhA-T266E has a delayed growth phenotype compared with the strain expressing the wild type form of the protein (Fig. 7). Most importantly, the InhA-T266E mutant failed to functionally complement the inhA conditional gene replacement mutant (Fig. 8) (13), implying phosphorylation-dependent modulation of InhA function. The fact that a number of enzymes involved mycolic acid biosynthesis pathway, including InhA, are regulated by serine/threonine protein kinase-mediated phosphorylation emphasizes the crucial role of the phosphorylation in mediating mycobacterial survival.
A complete understanding of phosphorylation events in a global context entails determining whether a substrate is part of a larger network involving multiple kinases. Thus far, in vitro kinase assays with purified M. tuberculosis kinases and the substrate in question have been employed by various groups to address this vital issue (24, 26, 27, 38, 39). We have devised an expedient method to assess the potential candidacy of protein substrates using all 11 M. tuberculosis STPKs (Fig. 2). This involves use of a dual expression vector pDuet-1. We have generated a series of 12 pDuet constructs harboring MBP kinase/MBP tag in MCS2; the putative substrate can be cloned in MCS1 (Fig. 2). The resulting plasmids are ideal for co-expression of the kinase-substrate pair, respectively, in the surrogate host E. coli. As the phosphorylation event takes place in vivo in the surrogate host, results obtained are free of the problems associated with in vitro reactions. This methodology provides an excellent in vivo tool, which can be used widely to assess phosphorylation of potential substrate by all 11 kinases of M. tuberculosis. Using this method, we have been able to demonstrate that PknA, PknB, and PknH efficiently target InhA, although PknF also targets it but to a lower extent (Fig. 3).
Identifying phosphorylation sites on a target substrate is a challenging task. We used a combination of peptide maps with mutant substrates and comparative kinase assays to determine the phosphorylation sites on InhA. A detailed scrutiny indicated that major sites of phosphorylation were present in the same tryptic peptide of InhA. Our inability to account for all the spots observed could be due to the presence of multiple phosphorylation sites on the same peptide (Fig. 4). In conjunction with kinase assays, we identified Thr-253, Thr-254, and Thr-266 to be the major targets for PknA/PknB-mediated phosphorylation and validated the same (Fig. 4). Two-dimensional gels have been used to confirm in vivo phosphorylation status of KasA, KasB, and FabH (24, 27). In agreement with our in vitro results, two-dimensional gel electrophoresis analysis of InhA co-expressed with PknA or PknB in E. coli demonstrated phosphorylation on multiple sites (Fig. 5). InhA isolated from M. smegmatis strain harboring pMV261-InhA plasmid appears to be effectively phosphorylated as seen by two-dimensional gel analysis. We also validated Thr-266 to be a major phosphorylation site in vivo in mycobacteria by means of the same approach (Fig. 5). Furthermore, we observed multiple spots when M. smegmatis lysates were probed with anti-InhA antibodies (Fig. 5), suggesting phosphorylation of InhA under endogenous expression conditions.
Phosphorylation-dephosphorylation events are a mode of regulating cellular functions. STPK-mediated phosphorylation events in M. tuberculosis are no exception to this paradigm. Several illustrations have been well documented in the literature (37). Phosphorylation events of the transcriptional regulator EmbR affect its ATPase activity and binding with DNA (40). Interactions of anti-anti-σ factor homolog Rv0516c with its anti-anti-anti-σ factor, and of Rv2175c, a DNA-binding protein of unknown function, with DNA, are abrogated upon phosphorylation by kinases (39, 41), whereas transcription factor VirS binds more strongly to its target promoter region (28). Functions of cell division proteins Wag31 and FtsZ are also modulated by phosphorylation (42, 43) The activity of enzymes like GlmU, KasA, FabH, and MabA is down-regulated upon phosphorylation, although the action of KasB in the phosphorylated form is enhanced (24, 26, 27, 29). Quemard et al. (11) in 1995 demonstrated activity of InhA by an indirect method in which the action of InhA is measured by monitoring NADH oxidation that occurs concurrently with substrate reduction. Upon performing time course assays and kinetics experiments with InhA and p-InhA, we observed a stark difference, with phosphorylation appearing to down-regulate the enzymatic efficiency of InhA (Fig. 6).
Because the phosphorylation of InhA results in decreased activity, we hypothesized that the overexpression of InhA and kinase may impact the growth of mycobacteria, due to hyperphosphorylation of InhA. PknA overexpression was demonstrated by to be detrimental to the growth (18). This is most likely due to hyperphosphorylation of a number of substrates, including InhA. We observed that overexpression of InhA and PknA significantly compromised the growth over and above the effects observed due to overexpression of PknA alone (Fig. 7). Despite our best efforts, we were unable to co-express PknB and InhA in M. smegmatis. It is possible that the deleterious effects on mycobacterial growth upon co-expression of PknB and InhA were so severe that cells overexpressing both proteins may not have survived. Furthermore, in line with these results, we found that the strain expressing InhA-T266E has a delayed growth phenotype compared with the strain expressing the wild type form of the protein (Fig. 7). Most importantly, InhA-T266E mutant failed to functionally complement the InhA conditional gene replacement mutant (13), strengthening our hypothesis (Fig. 8). In addition, co-expression of InhA-T266A mutant alongside PknA was found to partially alleviate the diminution in growth (Fig. 7), emphasizing the importance of phosphorylation at Thr-266. The fact that InhA-T266A mutant + PknA overexpression did not bring growth back to PknA-only overexpression levels suggests that, in addition to Thr-266, PknA also targets other sites such as Thr-253 and Thr-254 in vivo. InhA is shown to function as a dimer. Hence, it is likely that hyperphosphorylated InhA (when the kinase is overexpressed) is behaving like a dominant negative enzyme by titrating away unphosphorylated InhA.
Based on our data, we conclude that InhA is a target of multiple mycobacterial STPKs, and the resulting phosphorylations are instrumental in negatively modulating the growth of the bacterium by virtue of decreased activity of this enzyme. In depth studies by other groups have shown that other members of the FAS-II pathway, namely FabD, KasA, KasB, and MabA, as well as FabH that links the FAS-I and FAS-II pathways, are also regulated by phosphorylation attributed to these kinases (24, 26, 27). The identification of InhA as a novel substrate being phosphorylated by multiple STPKs, along with other FAS enzymes, is indicative of the various permutations and combinations by which the mycolic acid synthesis is regulated in vivo. A blend of multiple kinases, multiple substrates, and multiple levels of fine-tuning of enzymatic activity based on phosphorylation status puts forth an elegant and elaborate manner in which mycolic acid synthesis can be tightly restrained, in tandem with a multitude of intracellular and extracellular cues in the bacteria.
Acknowledgments
We thank Dr. Tanya Parish and Dr. Jacobs, Jr., for providing pJAM2 and pMV-261apra vectors, respectively.
This work was supported in part by the Department of Biotechnology, India (to V. K. N. and R. P. R.).
- FAS
- fatty-acid synthase
- MBP
- maltose-binding protein
- Ni-NTA
- nickel-nitrilotriacetic acid
- ACP
- acyl carrier protein
- STPK
- serine/threonine protein kinase.
REFERENCES
- 1.Barry C. E., 3rd, Lee R. E., Mdluli K., Sampson A. E., Schroeder B. G., Slayden R. A., Yuan Y. (1998) Prog. Lipid Res. 37, 143–179 [DOI] [PubMed] [Google Scholar]
- 2.Takayama K., Wang C., Besra G. S. (2005) Clin. Microbiol. Rev. 18, 81–101 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Bloch K. (1977) Adv. Enzymol. Relat. Areas Mol. Biol. 45, 1–84 [DOI] [PubMed] [Google Scholar]
- 4.Smith S., Witkowski A., Joshi A. K. (2003) Prog. Lipid Res. 42, 289–317 [DOI] [PubMed] [Google Scholar]
- 5.Asselineau J., Lederer E. (1950) Nature 166, 782–783 [DOI] [PubMed] [Google Scholar]
- 6.Kremer L., Nampoothiri K. M., Lesjean S., Dover L. G., Graham S., Betts J., Brennan P. J., Minnikin D. E., Locht C., Besra G. S. (2001) J. Biol. Chem. 276, 27967–27974 [DOI] [PubMed] [Google Scholar]
- 7.Choi K. H., Kremer L., Besra G. S., Rock C. O. (2000) J. Biol. Chem. 275, 28201–28207 [DOI] [PubMed] [Google Scholar]
- 8.Sacco E., Covarrubias A. S., O'Hare H. M., Carroll P., Eynard N., Jones T. A., Parish T., Daffé M., Bäckbro K., Quémard A. (2007) Proc. Natl. Acad. Sci. U.S.A. 104, 14628–14633 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Marrakchi H., Lanéelle G., Quémard A. (2000) Microbiology 146, 289–296 [DOI] [PubMed] [Google Scholar]
- 10.Banerjee A., Dubnau E., Quemard A., Balasubramanian V., Um K. S., Wilson T., Collins D., de Lisle G., Jacobs W. R., Jr. (1994) Science 263, 227–230 [DOI] [PubMed] [Google Scholar]
- 11.Quémard A., Sacchettini J. C., Dessen A., Vilcheze C., Bittman R., Jacobs W. R., Jr., Blanchard J. S. (1995) Biochemistry 34, 8235–8241 [DOI] [PubMed] [Google Scholar]
- 12.Vilchèze C., Morbidoni H. R., Weisbrod T. R., Iwamoto H., Kuo M., Sacchettini J. C., Jacobs W. R., Jr. (2000) J. Bacteriol. 182, 4059–4067 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Bhatt A., Kremer L., Dai A. Z., Sacchettini J. C., Jacobs W. R., Jr. (2005) J. Bacteriol. 187, 7596–7606 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Cole S. T., Brosch R., Parkhill J., Garnier T., Churcher C., Harris D., Gordon S. V., Eiglmeier K., Gas S., Barry C. E., 3rd, Tekaia F., Badcock K., Basham D., Brown D., Chillingworth T., Connor R., Davies R., Devlin K., Feltwell T., Gentles S., Hamlin N., Holroyd S., Hornsby T., Jagels K., Krogh A., McLean J., Moule S., Murphy L., Oliver K., Osborne J., Quail M. A., Rajandream M. A., Rogers J., Rutter S., Seeger K., Skelton J., Squares R., Squares S., Sulston J. E., Taylor K., Whitehead S., Barrell B. G. (1998) Nature 393, 537–544 [DOI] [PubMed] [Google Scholar]
- 15.Av-Gay Y., Everett M. (2000) Trends Microbiol. 8, 238–244 [DOI] [PubMed] [Google Scholar]
- 16.Chao J., Wong D., Zheng X., Poirier V., Bach H., Hmama Z., Av-Gay Y. (2010) Biochim. Biophys. Acta 1804, 620–627 [DOI] [PubMed] [Google Scholar]
- 17.Alber T. (2009) Curr. Opin. Struct. Biol. 19, 650–657 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Kang C. M., Abbott D. W., Park S. T., Dascher C. C., Cantley L. C., Husson R. N. (2005) Genes Dev. 19, 1692–1704 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Cowley S., Ko M., Pick N., Chow R., Downing K. J., Gordhan B. G., Betts J. C., Mizrahi V., Smith D. A., Stokes R. W., Av-Gay Y. (2004) Mol. Microbiol. 52, 1691–1702 [DOI] [PubMed] [Google Scholar]
- 20.Walburger A., Koul A., Ferrari G., Nguyen L., Prescianotto-Baschong C., Huygen K., Klebl B., Thompson C., Bacher G., Pieters J. (2004) Science 304, 1800–1804 [DOI] [PubMed] [Google Scholar]
- 21.Tiwari D., Singh R. K., Goswami K., Verma S. K., Prakash B., Nandicoori V. K. (2009) J. Biol. Chem. 284, 27467–27479 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Papavinasasundaram K. G., Chan B., Chung J. H., Colston M. J., Davis E. O., Av-Gay Y. (2005) J. Bacteriol. 187, 5751–5760 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Deol P., Vohra R., Saini A. K., Singh A., Chandra H., Chopra P., Das T. K., Tyagi A. K., Singh Y. (2005) J. Bacteriol. 187, 3415–3420 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Molle V., Brown A. K., Besra G. S., Cozzone A. J., Kremer L. (2006) J. Biol. Chem. 281, 30094–30103 [DOI] [PubMed] [Google Scholar]
- 25.Bhatt A., Molle V., Besra G. S., Jacobs W. R., Jr., Kremer L. (2007) Mol. Microbiol. 64, 1442–1454 [DOI] [PubMed] [Google Scholar]
- 26.Veyron-Churlet R., Zanella-Cléon I., Cohen-Gonsaud M., Molle V., Kremer L. (2010) J. Biol. Chem. 285, 12714–12725 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Veyron-Churlet R., Molle V., Taylor R. C., Brown A. K., Besra G. S., Zanella-Cléon I., Fütterer K., Kremer L. (2009) J. Biol. Chem. 284, 6414–6424 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Kumar P., Kumar D., Parikh A., Rananaware D., Gupta M., Singh Y., Nandicoori V. K. (2009) J. Biol. Chem. 284, 11090–11099 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Parikh A., Verma S. K., Khan S., Prakash B., Nandicoori V. K. (2009) J. Mol. Biol. 386, 451–464 [DOI] [PubMed] [Google Scholar]
- 30.Triccas J. A., Parish T., Britton W. J., Gicquel B. (1998) FEMS Microbiol. Lett. 167, 151–156 [DOI] [PubMed] [Google Scholar]
- 31.Boyle W. J., van der Geer P., Hunter T. (1991) Methods Enzymol. 201, 110–149 [DOI] [PubMed] [Google Scholar]
- 32.Parish T., Stoker N. G. (1998) Methods Mol. Biol. 101, 129–144 [DOI] [PubMed] [Google Scholar]
- 33.Eblen S. T., Kumar N. V., Shah K., Henderson M. J., Watts C. K., Shokat K. M., Weber M. J. (2003) J. Biol. Chem. 278, 14926–14935 [DOI] [PubMed] [Google Scholar]
- 34.He X., Alian A., Stroud R., Ortiz de Montellano P. R. (2006) J. Med. Chem. 49, 6308–6323 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.DeLano W. L. (2002) The PyMOL Molecular Graphies System, DeLano, Scienctific LLC, San Carlos, CA [Google Scholar]
- 36.Rozwarski D. A., Vilchèze C., Sugantino M., Bittman R., Sacchettini J. C. (1999) J. Biol. Chem. 274, 15582–15589 [DOI] [PubMed] [Google Scholar]
- 37.Molle V., Kremer L. (2010) Mol. Microbiol. 75, 1064–1077 [DOI] [PubMed] [Google Scholar]
- 38.Canova M. J., Kremer L., Molle V. (2009) J. Bacteriol. 191, 2876–2883 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Greenstein A. E., MacGurn J. A., Baer C. E., Falick A. M., Cox J. S., Alber T. (2007) PLoS Pathog. 3, e49. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Sharma K., Gupta M., Krupa A., Srinivasan N., Singh Y. (2006) FEBS J. 273, 2711–2721 [DOI] [PubMed] [Google Scholar]
- 41.Cohen-Gonsaud M., Barthe P., Canova M. J., Stagier-Simon C., Kremer L., Roumestand C., Molle V. (2009) J. Biol. Chem. 284, 19290–19300 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Kang C. M., Nyayapathy S., Lee J. Y., Suh J. W., Husson R. N. (2008) Microbiology 154, 725–735 [DOI] [PubMed] [Google Scholar]
- 43.Thakur M., Chakraborti P. K. (2006) J. Biol. Chem. 281, 40107–40113 [DOI] [PubMed] [Google Scholar]
- 44.Snapper S. B., Melton R. E., Mustafa S., Kieser T., Jacobs W. R., Jr. (1990) Mol. Microbiol. 4, 1911–1919 [DOI] [PubMed] [Google Scholar]