Abstract
Race-specific disease resistance in plants depends on the presence of resistance (R) genes. Most R genes encode NB-ARC-LRR proteins that carry a C-terminal leucine-rich repeat (LRR). Of the few proteins found to interact with the LRR domain, most have proposed (co)chaperone activity. Here, we report the identification of RSI2 (Required for Stability of I-2) as a protein that interacts with the LRR domain of the tomato R protein I-2. RSI2 belongs to the family of small heat shock proteins (sHSPs or HSP20s). HSP20s are ATP-independent chaperones that form oligomeric complexes with client proteins to prevent unfolding and subsequent aggregation. Silencing of RSI2-related HSP20s in Nicotiana benthamiana compromised the hypersensitive response that is normally induced by auto-active variants of I-2 and Mi-1, a second tomato R protein. As many HSP20s have chaperone properties, the involvement of RSI2 and other R protein (co)chaperones in I-2 and Mi-1 protein stability was examined. RSI2 silencing compromised the accumulation of full-length I-2 in planta, but did not affect Mi-1 levels. Silencing of heat shock protein 90 (HSP90) and SGT1 led to an almost complete loss of full-length I-2 accumulation and a reduction in Mi-1 protein levels. In contrast to SGT1 and HSP90, RSI2 silencing led to accumulation of I-2 breakdown products. This difference suggests that RSI2 and HSP90/SGT1 chaperone the I-2 protein using different molecular mechanisms. We conclude that I-2 protein function requires RSI2, either through direct interaction with, and stabilization of I-2 protein or by affecting signalling components involved in initiation of the hypersensitive response.
Keywords: HSP20, NB-LRR protein, alpha crystallin domain, hypersensitive response, resistasome, immunity
Introduction
Resistance (R) proteins in plants mediate recognition of specific pathogen-derived factors called Avirulence (Avr) proteins. Upon Avr perception, R proteins initiate defence responses that limit further pathogen ingress. These responses often result in macroscopically visible cell death, referred to as the hypersensitive response (HR).
The majority of R proteins are NB-ARC-LRR proteins, which contain a central nucleotide-binding and -hydrolysing domain (NB-ARC) and a C-terminal leucine-rich repeat (LRR) domain (Martin et al., 2003; van Ooijen et al., 2007; Tameling and Takken, 2008). The LRR mediates both intra- and intermolecular interactions (reviewed by Lukasik and Takken, 2009). Known intermolecular interactions include those with (co)chaperones, with the best studied being heat shock protein 90 (HSP90) that physically interacts with the LRRs of the R proteins I-2, RPM1, N and Rx (Hubert et al., 2003; Lu et al., 2003; Liu et al., 2004; de la Fuente van Bentem et al., 2005). HSP90 is a highly conserved ATP-dependent molecular chaperone that is responsible for the stability and function of a large number of proteins, collectively referred to as HSP90 client proteins (Pearl and Prodromou, 2006). The activity of HSP90 is regulated by interactions with co-chaperones (Pearl and Prodromou, 2006). Some of these co-chaperones also directly interact with HSP90 client proteins. For example, the co-chaperone protein phosphatase 5 (PP5) binds both the C-terminus of HSP90 and the LRR domain of R protein I-2 (de la Fuente van Bentem et al., 2005). Another HSP90-interacting co-chaperone, SGT1 (suppressor of the G2 allele of Skp1), has been shown to interact with the LRR of the barley resistance protein Mla-1 (Bieri et al., 2004). HSP90 and SGT1 have also been shown to interact with a third partner; RAR1 (Required for Mla12 Resistance) (Shirasu et al., 1999; Hubert et al., 2003; Takahashi et al., 2003; Bieri et al., 2004; Liu et al., 2004; Azevedo et al., 2006; Boter et al., 2007; Heise et al., 2007). The combined activities of the three (co)chaperones RAR1, SGT1 and HSP90 has been shown to be important for R protein stability and accumulation, and thus for R protein-mediated signalling responses (Zhang et al., 2004; Azevedo et al., 2006; Boter et al., 2007).
Another class of proteins with chaperone-associated functions are small heat shock proteins (sHSPs), or HSP20s, which range in size from 12 to 43 kDa. HSP20s are of variable sequence, and are characterized by a conserved domain of approximately 90 residues forming an α-crystallin domain (ACD) (Caspers et al., 1995). These proteins form large oligomers and perform their ATP-independent chaperone function in vitro by binding to (partially) denatured proteins (Lee et al., 1995; Helm et al., 1997; Kirschner et al., 2000). In vivo, HSP20s are believed to confer a protective function by preventing unfolding or disassembly of other proteins (van Montfort et al., 2001a). HSP20s probably maintain denatured proteins in a folding-competent state to allow subsequent ATP-dependent disaggregation by the HSP70/90 chaperone system (Kotak et al., 2007; Liberek et al., 2008).
Here, we describe the identification of RSI2 (Required for Stability of I-2), an HSP20 member that specifically interacts with the tomato (Solanum lycopersicum) R protein I-2. I-2 confers resistance to Fusarium oxysporum (Simons et al., 1998). In addition to analysing the physical interaction between I-2 and RSI2, we used a virus-induced gene silencing approach to analyse the functional involvement of RSI2 and other known (co)chaperones in the HR mediated by auto-active variants of I-2 and a second tomato R protein, Mi-1. Mi-1 belongs to a different subgroup of NB-ARC-LRR proteins (van Ooijen et al., 2007), and confers resistance to root-knot nematodes (Meloidogyne sp.), potato top aphid (Macrosiphum euphorbiae) and whitefly (Bemisia tabaci) (Vos et al., 1998). Furthermore, we analysed the effect of silencing of RSI2 and other (co)chaperones on I-2 and Mi-1 protein abundance and stability in planta.
Results
RSI2 interacts with the I-2 LRR domain in the yeast two-hybrid system
We reported previously the identification of RSI2 (originally referred to as an HSP17 protein) and PP5 as interactors of the I-2 LRR domain in a yeast two-hybrid screen (de la Fuente van Bentem et al., 2005). PP5 was identified using LRR1–29 as bait, whereas RSI2 was identified using a different bait, LRR12–29, which corresponds to amino acid residues 823–1250 (LRR annotation as described by de la Fuente van Bentem et al., 2005). Screening of 6 × 106 clones of a tomato cDNA library with bait LRR12–29 revealed two independent interacting clones. The two cDNA clones carried 733 and 742 bp inserts (Genbank accession number AY150040). The inserts are overlapping and differ only in the length of their 5′ UTRs. They encode a full-length HSP20, based on the presence of an α-crystallin domain (ACD) fused to an N-terminal domain, with a predicted mass of 17.8 kDa. Plants contain at least six subclasses of HSP20 proteins targeted to various cellular compartments (Waters and Vierling, 1999; Scharf et al., 2001). A phylogenetic tree derived from multiple sequence alignment of the ACDs of tomato, potato, pepper, rice and Arabidopsis HSP20 sequences clearly shows the various subfamilies (classification as described by Scharf et al., 2001), and places RSI2 in class I of the cytosolic HSP20s (Figure 1a, Figure S1 and Appendix S1). The structure of the class I wheat HSP20 (van Montfort et al., 2001a) reveals that the ACD adopts a β-sandwich fold consisting of two layers of five β-sheets flanked by helices and loops. A similar fold is predicted to be present in other HSP20s. Strikingly, we noted a strong paired-clustering of paralogues over orthologues for cytosolic class I proteins, and paralogues appear to have expanded in Solanaceae. This is suggestive of a rapid birth and death process, implying that this class of HSP20s (including RSI2) is under strong selection in the various hosts. Other classes of HSP20s targeted to other cellular compartments (e.g. mitochondria, peroxisomes, etc.) appear to have undergone less diversification.
Figure 1.
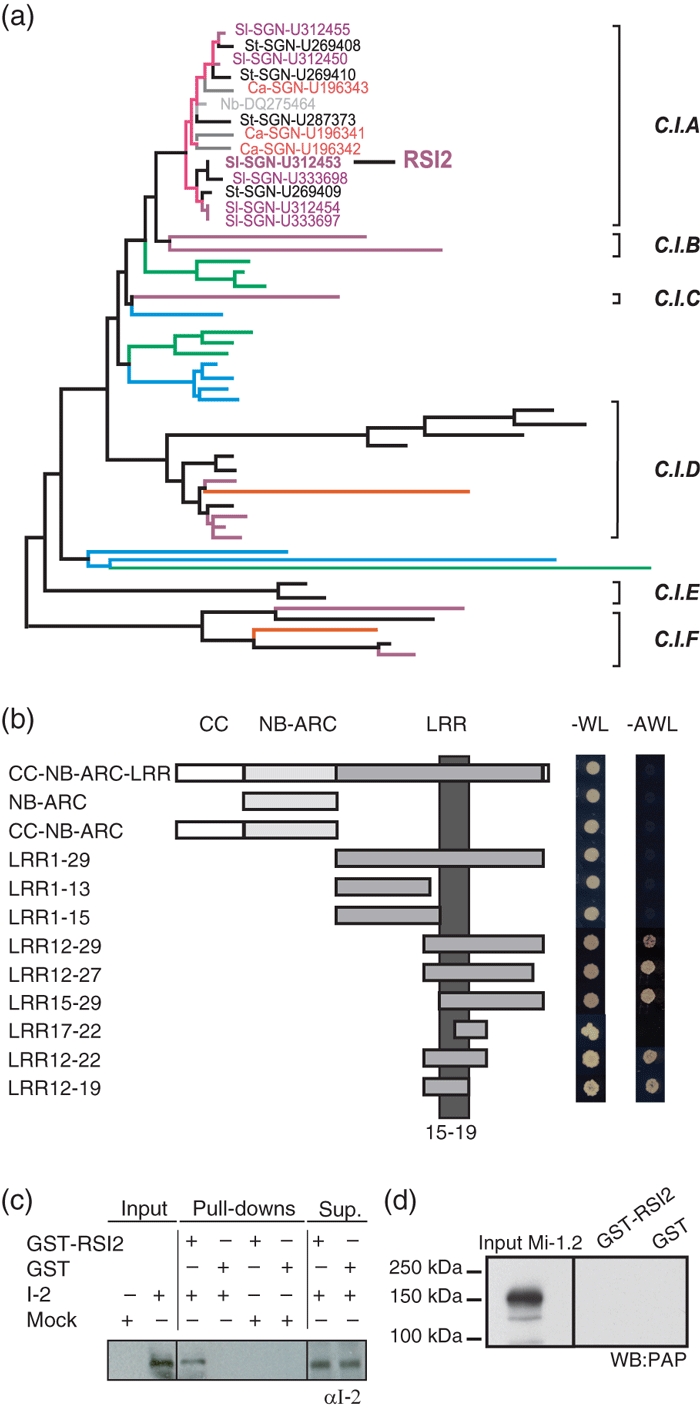
RSI2 interacts with I-2.(a) Evolutionary relationship of class I (C.I.) cytosolic HSP20s from tomato (Sl, purple), potato (St, black), Capsicum annuum (Ca, red) and N. benthamiana (Nb, grey). Six sub-clades can be distinguished (C.I.A to C.I.F). The tree contains 51 taxa, and is part of a larger tree containing 113 taxa (Figure S1).(b) Yeast two-hybrid assays showing interactions between RSI2 and truncated versions of I-2. The presence of bait and prey plasmids was confirmed by growth on minimal medium lacking tryptophan and leucine (–WL), and the interaction between bait and prey proteins was analysed by growth on minimal medium lacking tryptophan, leucine and adenine (–AWL). The dark grey bar highlights the I-2 region required for RSI2 interaction.(c) Western blot on total protein lysates from N. benthamiana leaves transiently expressing I-2 or mock-infiltrated and probed with I-2 antibody (αI-2). The presence of full-length R proteins in these extracts is shown in the left lanes. GST–RSI2 and GST proteins immobilized on glutathione Sepharose beads were incubated with these extracts as indicated (±). GST?–RSI2 and GST interacting proteins were subjected to SDS–PAGE and Western blot analysis to detect the presence of I-2. The supernatant remaining after pull-down was blotted to show that I-2 stability was unaffected by the various experimental conditions.(d) As (c), but using TAP-tagged Mi-1 and the PAP antibody to detect TAP-tagged Mi-1.
To assess the specificity of the I-2/HSP20 interaction, representative ACD members of class I were selected based on the phylogenetic tree (Figure S1). Full-length cDNAs were amplified from tomato EST sequences provided by the Kazusa DNA Research Institute (Kisarazu, Chiba, Japan). Two closely related homologues from class IA were selected (SL-SGN-U312450 and SL-SGN-U312454). One EST (SL-SGN-U316206) was also selected from class ID to represent a more distantly related homologue. The interaction of these homologues with I-2 LRR12–29 was analysed using yeast two-hybrid assays, and accumulation of the HSP20 proteins in yeast was verified by Western blot analysis (Figure S2b). Of the four homologues analysed, RSI2 was the only HSP20 that interacted with I-2 LRR12–29 (Figure S2a), which implies that the interaction between I-2 and RSI2 is specific.
To pinpoint the region of the I-2 protein responsible for the interaction with RSI2, various N- and C-terminal truncations of the I-2 protein were analysed for their interaction with RSI2 in yeast two-hybrid assays (Figure 1b). The minimal RSI2-interacting region of the I-2 LRR domain lies within LRR15–19, corresponding to amino acids 906–1015 (Figure 1b). Notably, the full-length I-2 protein and the full-length LRR domain (LRR1–29) did not interact with RSI2 when expressed in yeast (de la Fuente van Bentem et al., 2005).
GST–RSI2 fusion protein interacts with I-2 from plant protein extracts
We next investigated whether recombinant RSI2 associates with plant-produced I-2 in plant protein extracts. We performed pull-down experiments using Escherichia coli-produced GST–RSI2 and GST alone with non-tagged plant-produced I-2 rather than immunoprecipitation of proteins produced in planta, as this approach has previously been used successfully to reveal interactions between other members of an I-2 complex in plant protein extracts (de la Fuente van Bentem et al., 2005). The two GST bait proteins were affinity-purified and subsequently analysed by Coomassie staining of an SDS–PAGE gel. As shown in Figure S3, the proteins migrated according to their expected molecular weights (44 and 26 kDa), and little contamination of co-purifying protein was observed. Endogenous I-2 is expressed at very low levels in tomato and was undetectable in total protein leaf extracts using our affinity-purified I-2 antibody (Tameling et al., 2002), and labelling of I-2 with a peptide tag abolishes its biological activity (van Ooijen et al., 2008b). To achieve detectable expression levels, we produced full-length I-2 by transient transformation of Nicotiana benthamiana leaves using agroinfiltration. The N. benthamiana-produced full-length I-2 protein was readily detected in I-2-transformed plants, but not in mock-infiltrated plants (Figure 1c). Next, total protein extracts of N. benthamiana leaves infiltrated with either A. tumefaciens carrying I-2 constructs or with buffer were incubated with beads loaded with GST or GST–RSI2. The stability of the I-2 protein during the assay did not differ between the GST and GST–RSI2 samples, as shown by Western blot analysis of the supernatant fractions after GST pull-down. Moreover, I-2 was consistently co-purified with GST–RSI2, but not with the control containing GST alone (Figure 1c). The specific co-precipitation of I-2 with GST–RSI2 indicates that RSI2 interacts with the I-2 protein complex present in plant extracts.
We analysed the interaction of GST–RSI2 with the R protein Mi-1 in a similar manner. A tandem affinity purification (TAP)-tagged version of Mi-1 was used, as the polyclonal Mi-1 antibody is known to cross-react with the GST tag (van Ooijen et al., 2008b). The TAP tag does not appear to affect Mi-1 protein function, as the TAP-tagged auto-active mutant Mi-1H840A (van Ooijen et al., 2008b) induces an HR comparable to that of the non-tagged auto-active mutant in N. benthamiana (Figure S4). Full-length TAP-tagged Mi-1 can be readily detected on Western blot using PAP (peroxidise anti-peroxidase) antibody (Figure 1d). However, TAP-tagged Mi-1 did not co-precipitate with the GST–RSI2 fusion protein under the conditions used. These results indicate that, although RSI2 and I-2 can form a complex, RSI2 does not interact with the Mi-1 protein complex under the same conditions.
VIGS reveals a role for RSI2 in HR mediated by I-2 and Mi-1
R protein function depends on the activities of a number of chaperones or chaperone-associated proteins (de la Fuente van Bentem et al., 2005; Boter et al., 2007). To analyse whether RSI2 is also required for R protein function, we used virus-induced gene silencing (VIGS) of RSI2. For comparison, the (co)chaperones SGT1, RAR1, PP5 and HSP90 were included in these experiments. VIGS was induced using the tobacco rattle virus (TRV) silencing system, which was delivered by agroinfiltration of viral vector constructs into the leaflets of 2-week-old N. benthamiana plants (Ratcliff et al., 2001). To confirm onset and spread of silencing over time, phytoene desaturase (PDS) was used as a visible marker (Demmig-Adams and Adams, 1992). The nearly complete photobleaching that was consistently observed 3 weeks after agroinfiltration indicates extensive PDS silencing throughout these plants at this time point (Figure S5). To assess RSI2 silencing levels, specific primers were designed on the basis of the closest N. benthamiana homologue (shown in Figure 1b) present in the Institute for Genomic Research (TIGR) database (GenBank accession number DQ275464). In the sequenced region, this N. benthamiana gene is only 80% identical to RSI2, and is not predicted to be a silencing target (Xu et al., 2006). However, the relative expression level of this RSI2 homologue was reduced to 30% compared to the non-silenced controls in leaf extracts (Figure 2a). The presence of other, possibly even more closely related, unknown RSI2 genes in the N. benthamiana genome cannot be excluded, and silencing might therefore also target other genes encoding RSI2 homologues belonging to class IA. Efforts to analyse total class I protein levels by Western blotting of silenced plants were not successful because the affinity of several tested antibodies was insufficient to detect HSP20s in N. benthamiana protein extracts.
Figure 2.
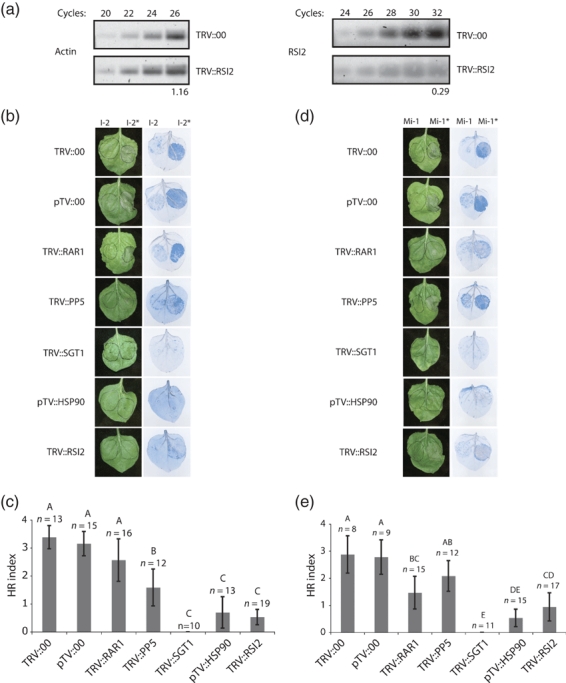
VIGS reveals a role for RSI2 in I-2- and Mi-1-mediated HR signaling.(a) Silencing efficiency of RSI2 was determined using semi-quantitative RT-PCR (right panel). Actin expression levels were measured as a control for equal cDNA quantity and quality (left panel). The number of PCR cycles is indicated at the top. The relative signal intensity compared to TRV::00 (indexed at 1) is indicated below.(b, d) Agroinfiltration of wild-type (left side of the leaf) or constitutively active mutants (right side of the leaf) of I-2 (b) and Mi-1 (d) into N. benthamiana 3 weeks after induction of silencing using the indicated TRV vectors. Photographs were taken 3 days after agroinfiltration, and representative leaves were stained using trypan blue to visualize cell death (right panels).(c, e) Severity of the HR upon I-2D495V (c) or Mi-1D841V (e) expression in silenced plants. HR was quantified visually on a scale from 0 (no symptoms) to 4 (full necrosis). Significant differences were determined by one-way anova (P < 0.05) and are indicated by different letters above the bars. The error bars represent the 95% confidence level; n is the number of plants analysed.
To assess I-2 function in the silenced plants, leaves were agroinfiltrated with constructs expressing wild-type I-2 (left side of the leaves) and the auto-active I-2D495V (I-2*) mutant (right side of the leaves) (Figure 2b,c). The level of cell death induced by I-2D495V was scored 3 days after agroinfiltration on a scale from 0 (absolutely no tissue collapse) to 4 (fully developed HR in the entire infiltrated region). The scale is shown in Figure S6. To enhance clarity of the HR, the infiltrated leaves were stained for cell death using trypan blue (Figure 2b).
Leaves of plants infected with the empty virus control showed a clear HR upon expression of I-2D495V, but not upon expression of wild-type I-2 (Figure 2b). This result showed that induction of the HR by I-2D495V was not compromised by infection with TRV. Silencing of the genes encoding the established R protein (co)chaperones HSP90 or SGT1 severely suppressed the HR triggered by I-2D495V. Similarly, no or only minor tissue collapse induced by I-2D495V was observed in the RSI2 silenced plants (0.5 ± 0.3) compared to control plants (3.4 ± 0.4) (Figure 2c). A one-way anova showed that the severity of HR symptoms in RSI2, HSP90 and SGT1 silenced plants was significantly reduced, reaching similar low levels, indicating a requirement for each of these genes for I-2-mediated HR. Silencing of PP5 only partially compromised I-2D495V-mediated HR, and RAR1 silencing did not significantly affect the HR (Figure 2b,c).
To determine whether RSI2 is also involved in the HR mediated by Mi-1, we assessed induction of the HR in silenced plants by the auto-active mutant Mi-1D841V (Mi-1*) (van Ooijen et al., 2008b). In plants infected with empty virus, a strong HR (2.9 ± 0.7) was induced by this mutant (Figure 2d, right side of the leaves) but not by the wild-type Mi-1 protein (left side of the leaves). In plants silenced for RSI2, the HR induced by Mi-1D841V was consistently and strongly compromised (0.9 ± 0.5) (Figure 2d,e). As with I-2D495, Mi-1D841V-mediated HR induction is also severely compromised upon SGT1 and HSP90 silencing. PP5 and RAR1 silencing also affected HR induction by Mi-1D841V, but to a lesser extent. These data conclusively show that the HR mediated by auto-active I-2 and Mi-1 is strongly reduced when expression of class IA HSP20s is diminished by gene silencing.
RSI2 silencing negatively affects I-2 protein accumulation
The suppressive effect of SGT1 gene silencing on N and Rx function is a result of compromised R protein stability, possibly due to reduced chaperone activity (Azevedo et al., 2006; Mestre and Baulcombe, 2006; Boter et al., 2007). HSP20s are also associated with chaperone functions in vitro (Nover and Scharf, 1997; Kirschner et al., 2000). To analyse whether the observed effects on the HR by silencing RSI2, SGT1, HSP90, PP5 or RAR1 can be attributed to reduced R protein stability, accumulation of I-2 and Mi-1 was analysed in the silenced plants.
To exclude the possibility that silencing of any of these genes adversely influences transgene expression via agroinfiltration per se, we agroinfiltrated a construct expressing TAP-tagged GUS protein. Expression levels of this control protein were not altered by silencing any of the indicated genes (Figure 3c), as shown by Western blotting using an antibody recognising the TAP tag.
Figure 3.
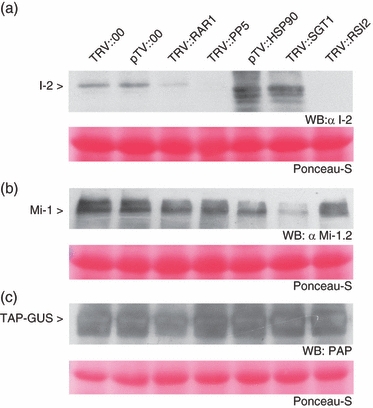
Expression levels of I-2, Mi-1 and GUS protein in silenced Nicotiana benthamiana leaves.N. benthamiana plants were silenced using the indicated TRV-based silencing constructs. Three weeks after induction of silencing, the upper leaves were agroinfiltrated with constructs expressing either I-2, Mi-1 or TAP-tagged GUS. One day after agroinfiltration, leaves were harvested and protein was extracted. Total protein (50 μg for I-2 and Mi-1 or 10 μg for TAP-tagged GUS) was loaded onto SDS–PAGE gels. Expression of I-2 (a), Mi-1 (b) or TAP-tagged GUS (c) was analysed by Western blotting using the antibody indicated. Equal loading was confirmed by Ponceau S staining of Rubisco (lower panels).
R protein accumulation in the silenced plants was assessed after agroinfiltration with I-2 and Mi-1 constructs. Compared to the vector controls, silencing of RAR1 and PP5 appeared to reduce I-2 protein levels (Figure 3a). Full-length I-2 protein was not detected at all in plants silenced for SGT1 and HSP90. However, strong accumulation of smaller proteins reacting with the I-2 antibody (putative I-2-derived degradation products) was observed. Silencing of RSI2 also abolished the accumulation of full-length I-2; however, this did not result in accumulation of smaller proteins that react with the I-2 antibody.
Mi-1 protein levels in RSI2, RAR1 or PP5 silenced plants were similar to those observed in empty virus controls (Figure 3b). Silencing of SGT1 led to a clear reduction of Mi-1 protein abundance, whereas HSP90 silencing only slightly affected Mi-1 protein accumulation. In neither case did we detect lower-molecular-weight products cross-reacting with the Mi-1 antibody. These data show that, although RSI2 is necessary for HR induction by auto-active Mi-1, silencing of RSI2 does not directly affect accumulation of the Mi-1 protein.
In conclusion, our data indicate that HSP90 and SGT1 are both required to maintain I-2 and Mi-1 integrity and stability. The functional involvement of RSI2 in I-2-mediated HR is associated with I-2 protein stabilization, but no effect on Mi-1 stability was observed.
Discussion
We here identify RSI2 as component of the I-2 protein complex, and show its requirement for the I-2- and Mi-1-mediated HR. We find that silencing of RSI2 severely reduces I-2 protein accumulation in planta (Figure 3), indicating a role for RSI2 in chaperoning I-2. As shown in Figure 1(a), RSI2 is a class I cytosolic HSP20 protein. Members of this class of HSP20s have been described to form large assemblies that prevent aggregation of unfolded client proteins (Lee et al., 1997; Lee and Vierling, 2000). HSP20 binding maintains these client proteins in a folding-competent state, allowing refolding by the ATP-dependent HSP70/HSP90 machinery (reviewed by Liberek et al., 2008). In the absence of chaperone activity, a client protein will aggregate and be degraded.
We also analysed the effect on I-2- and Mi-1-mediated HR after silencing other (co)chaperones (SGT1, HSP90, RAR1 and PP5) that interact with R proteins. In agreement with a previous study, silencing of SGT1, HSP90 or RAR1 reduced I-2-mediated HR. In that study, silencing of PP5 had no effect on I-2-mediated HR (de la Fuente van Bentem et al., 2005), whereas a relatively small, but significant, suppression of I-2-mediated HR was found in the present study. The difference between these studies might be due to enhanced efficiency of the Agrobacterium-mediated transformation procedure and higher silencing levels in our study due to use of the enhanced transformation procedures described by Fu et al., 2006. This is supported by the more extensive bleaching observed upon PDS silencing in our study (Figure S5) compared to the previous one (de la Fuente van Bentem et al., 2005).
We found that the HR mediated by an auto-active variant of Mi-1 is dependent on SGT1 and HSP90, which is consistent with the involvement of these proteins in Mi-1-mediated resistance towards whitefly (Bhattarai et al., 2007). RAR1 was found not to be involved in Mi-1-mediated whitefly resistance (Bhattarai et al., 2007), but we found that this protein is required for full HR induction by Mi-1. This difference may suggest that the HR is not required for whitefly resistance.
Class I HSP20s (including RSI2) are proposed to act sequentially to HSP90 to disaggregate misfolded client proteins (Liberek et al., 2008). Such a chaperone function for RSI2, i.e. maintaining misfolded I-2 in a soluble form to allow its refolding, agrees with the observed reduced accumulation of the full-length I-2 protein upon RSI2 or HSP90 silencing. Alternatively, RSI2 may function in direct conjunction with HSP90. The crystal structure of wheat HSP20 (van Montfort et al., 2001b) reveals that its fold closely resembles that of the CS (CHORD and SGT1) domain (Finn et al., 2006) found in SGT1 and p23, which are required for binding to HSP90 (Boter et al., 2007). p23 regulates human HSP90 activity by direct interaction with the ATP-bound active form of HSP90 (Johnson et al., 1994). The related folds of HSP20s and p23, together with the experimentally verified chaperoning activities of both, could indicate a similar biochemical function as chaperones and co-regulators of HSP90 activity (Bose et al., 1996). However, we were not able to detect an interaction between RSI2 and the I-2-interacting HSP90 isoform (de la Fuente van Bentem et al., 2005) using yeast two-hybrid assays (results not shown). Likewise, we were unable to establish the presence of a ternary complex of I-2, RSI2 and HSP90 in RSI2 pull-down assays (Figure 2) after probing the blot with an HSP90 antibody (data not shown). Future studies are required to determine whether RSI2 can function as a p23-like regulator of the HSP90/SGT1 machinery or whether it performs its chaperone functions independently of HSP90.
We currently cannot exclude the possibility that the interaction between I-2 and RSI2 is indirect and requires a bridging protein that is functionally conserved in yeast. Future experiments aimed to analyse the RSI2 and I-2 interaction in planta and to identify other proteins in this I-2/RSI2 complex using mass spectrometry might provide a partial answer this question. Co-expressing RSI2 and I-2 in N. benthamiana via agroinfiltration followed by co-immunoprecipitation could show co-existence of both partners in one complex, but this would not exclude the requirement for a bridging protein. The requirement for a bridging protein could be determined by analysing the dynamic composition of the I-2 protein complex immunoprecipitated from tomato under various infection states, but this strategy requires the development of I-2 antibodies suitable for immunoprecipitation or a protein tag that does not interfere with I-2 function. However, we consider a direct interaction more likely, as suggested in the current model for class I HSP20 function, which proposes direct interaction with client proteins through both the non-conserved N-terminal domain and the conserved ACD (Basha et al., 2006). The strong conservation in the ACD suggests that this region is essential for chaperone activity, whereas the variable N-terminus may be involved in client protein recognition specificity. The RSI2-interacting region was mapped to LRR15–19 of I-2 (Figure 1). This region differs from that required for interaction with HSP90 (LRR1–11), but overlaps with the PP5-interacting region (LRR12–22) (de la Fuente van Bentem et al., 2005), suggesting that these proteins might compete for binding. LRR1–29 and full-length I-2 contain the RSI2 interaction surface, but no interaction was observed in the yeast two-hybrid assay. Similarly, the PP5/I-2 interaction in yeast was observed with truncated versions of I-2 but not with the full-length protein (de la Fuente van Bentem et al., 2005). Possibly, folding of extended LRR domains in yeast differs from that in the plant background, disrupting the interaction surface.
We did not observe an interaction between RSI2 and Mi-1 in pull-down assays (Figure 1), and accumulation of Mi-1 expression in planta was not affected by RSI2 silencing (Figure 3), implying that Mi-1 is not itself a RSI2 client protein. However, Mi-1-mediated HR was compromised by RSI2 silencing (Figure 2). Possible explanations are that Mi-1 interacts with an RSI2 homologue that is silenced in these plants, or that RSI2 is required for the function of a protein downstream of Mi-1. A possible downstream candidate is the NB-ARC-LRR protein NRC1, which is involved in signalling mediated by many resistance proteins including Mi-1 (Gabriëls et al., 2007). Further studies are required to determine whether NRC1 is indeed an RSI2 client protein.
This report links an HSP20 to accumulation and function of R proteins. Another HSP20 (Nt-sHSP) has previously been linked to disease resistance in plants (Maimbo et al., 2007). In NtsHSP silenced plants, disease symptoms triggered by Ralstonia solanacearum are enhanced and expression of defence-related marker genes is compromised. It remains to be investigated, however, whether this HSP20 is also involved in R protein-mediated resistance, as the HR triggered by a pathogen or the elicitor INF1 was unaffected in these silenced lines (Maimbo et al., 2007). The phylogenetic tree in Figure S1 shows that Nt-sHSP is distantly related to RSI2, and further shows that plants possess a large number of highly diverse HSP20s with various subcellular localizations. RSI2 is member of a relatively large cluster of paralogues specific to species of the Solanaceae family (tomato, potato, Capsicum and Nicotiana benthamiana). The closest non-Solanaceae family member in this branch (from Arabidopsis thaliana) is relatively distant. A similar clustering of orthologues is seen in other branches of class I, implying that there is strong selection pressure for rapid diversification of the class I HSP20s. It is tempting to speculate that this reflects co-evolution of class I HSP20s with their (rapidly evolving) client proteins. Prime candidates for such client proteins are the R proteins, as these are rapidly evolving and the spectrum of NB-LRR subfamilies differs greatly between plant families (McHale et al., 2006). Co-evolution between specific HSP20s and R proteins is supported by the observed specificity of I-2 for RSI2 reported here. First, we did not retrieve other HSP20 orthologues when we screened a tomato cDNA library (de la Fuente van Bentem et al., 2005). Second, we found that closely related tomato HSP20 homologues did not interact with I-2 in the yeast two-hybrid assay (Figure S2).
A specific role for RSI2-like chaperones in maintaining R protein stability implies that R proteins are generally unstable and prone to inactivation. Support for this idea is the loss of function observed for many R proteins, including Mi-1 (Dropkin, 1969), at elevated temperatures.
Silencing of HSP90 or SGT1 also suppresses the ability of auto-active mutants of Mi-1 and I-2 to trigger an HR. The interaction between SGT1 and HSP90 is necessary for SGT1 to fulfil its function in resistance mediated by the R protein Rx (Boter et al., 2007). SGT1 is linked to protein degradation by the ubiquitin/26S proteasome pathway, as it is important for the function of several SCF (for SKP1/CULLIN1/F-box protein) complexes (Kitagawa et al., 1999; Azevedo et al., 2002; Gray et al., 2003). Silencing of the genes encoding SGT1 or its interaction partner HSP90 affected accumulation of full-length I-2 protein, possibly by reduced chaperoning activity. At the same time, the cell may not be able to completely degrade the (partially) unfolded I-2 products because a link to the 26S proteasome is broken by SGT1 silencing. This may explain the accumulation of intermediate degradation products upon HSP90 or SGT1 silencing (Figure 3). Such accumulation was not observed upon RSI2 silencing, which indicates that RSI2 is involved in stabilizing the I-2 protein but not in its elimination by the 26S proteasome.
To summarize, we report an HSP20 that is a part of the I-2 protein complex in vitro, and is required for the HR mediated by auto-active mutants of I-2 and Mi-1 in planta. We also demonstrate that RSI2, SGT1 and HSP90 are required for I-2 accumulation. We can thus add RSI2 to the list of chaperones that not only interact with NB-ARC-LRR R proteins but are also required for their function.
Experimental procedures
Yeast two-hybrid assays and protein isolation from yeast
Construction of a Fusarium-infected tomato cDNA interaction library and the yeast two-hybrid screening method, including the I-2 baits used in this study, have been described previously (de la Fuente van Bentem et al., 2005). The host strain PJ69-4a was transformed with bait (pAS2-1) and prey (pACT2) constructs (Clontech, http://www.clontech.com/) and the presence of both vectors was selected for on minimal medium (MM) plates lacking tryptophan and leucine (–WL). Droplets of a cell dilution corresponding to 104 colony-forming units were spotted on minimal medium plates that also lacked adenine (–AWL) to analyse transcriptional activation of the marker genes that indicate interaction between the bait and prey proteins. To test interaction of the HSP20 orthologues, dilutions corresponding to 107, 106 and 105 colony-forming units were used.
To confirm expression of the HSP20 prey proteins, transformed yeast was grown for 5 days in selective minimal medium (–WL). This culture was used to inoculate 25 ml of YPAD medium (20 g/l Peptone, 20 g/l Dextrose, 10 g/l Yeast extract, 60 mg/l Adenine hemisulphate), and grown for an additional 24 h. Cells were collected and proteins were extracted as described in the Clontech yeast protocol book (http://www.clontech.com/images/pt/PT3024-1.pdf) by adding 100 μl of cracking buffer [8 m urea, 5% w/v SDS, 40 mm Tris/HCl pH 6.8, 0.1 mm EDTA, 0.4 mg/ml bromophenol blue, 2 mm PMSF and 1× protease inhibitor cocktail (Roche, http://www.roche.com)] per 7.5 OD units (600 nm) of yeast cells. Collected cells were broken using glass beads in a Fastprep (2 × 45 sec) (Q-Bio Gene, http://www.qbiogene.com). Ten microlitre aliquots of the resulting protein fractions (supernatant) were loaded on SDS–PAGE gels for immunoblotting. The blots were probed using horseradish peroxidase-conjugated anti-HA antibody (monoclonal 12CA5; Roche) at a 1:1000 dilution, and luminescence was used to visualize the HA-tagged proteins.
Phylogenetic tree and sequence alignments
SGN sequence data were downloaded from the Unigene (contigs) family 109 HSP20 with 118 family members (http://www.sgn.cornell.edu). SGN contigs were checked for sequencing errors and corrected when possible. The variable N/C-terminal domains flanking the ACD were trimmed even though they contain areas of conservation in sub-clades. Contigs that did not cover the entire HSP20 core were excluded from further analysis. The Arabidopsis and rice sequences were obtained based on cut-off assignment of the presence of the HSP20 protein domain (Pfam HMM model). The evolutionary history was inferred using the minimum evolution method. The distances were computed using the Poisson correction method, and are given as the number of amino acid substitutions per site. The minimum evolution tree was searched using the close neighbour interchange (CNI) algorithm at a search level of 1. Positions containing alignment gaps and missing data were eliminated in pairwise sequence comparisons. There were a total of 194 positions in the final dataset. Phylogenetic analyses were performed using MEGA4 (Tamura et al., 2007). Clade annotations were as described previously (Scharf et al., 2001).
Vector construction
The binary vectors carrying wild-type I-2, I-2D495V, wild-type Mi-1 and Mi-1D841V have been described previously (van Ooijen et al., 2008b). For construction of the TAP-tagged Mi-1 construct, the coding sequence was amplified by PCR on pSE23 (Gabriëls et al., 2007) using primers 5′-AAAAAGCAGGCTCTATGGAAAAACGAAAAGATATT-3′ and 5′-AGAAAGCTGGGTTCTTAAATAAGGGGATATTCTTCTG-3′. Gateway attB sequences were added by adapter PCR using the primers 5′-GGGGACAAGTTTGTACAAAAAAGCAGGCT-3′ and 5′-GGGGACCACTTTGTACAAGAAAGCTGGGT-3′. The resulting PCR products were transferred directly to binary vector CTAPi (Rohila et al., 2004) using the Gateway one-tube protocol (Invitrogen, http://www.invitrogen.com/). To create a TAP-tagged auto-active Mi-1 mutant, the Bsp119I/BcuI fragment of a tagged wild-type clone was ligated in the corresponding sites of the non-tagged Mi-1 H840A construct, thereby removing the stop codon and allowing a translational fusion to the tag (van Ooijen et al., 2008a). A TAP–GUS fusion control was generated by a Gateway LR reaction using the GUS control plasmid included in the LR kit (Invitrogen) and the binary vector NTAPi (Rohila et al., 2004).
RSI2 silencing was performed using the tobacco rattle virus (TRV) system by cloning an EcoRI/XhoI fragment from the cDNA pACT2 library clone (de la Fuente van Bentem et al., 2005) into pYL156 (Liu et al., 2002). An EcoRI/SalI fragment from the pAS2-1 bait vector carrying the tomato PP5 tetratricopeptide-repeat (TPR) domain (de la Fuente van Bentem et al., 2005) was ligated into pYL156 digested with EcoRI/XhoI. A BamHI/SalI fragment from the SGT1 silencing vector used by Peart et al. (2002) was ligated into pYL156 digested with BamHI/XhoI. RAR1 was silenced using the silencing vector described by Liu et al. (2002). HSP90 silencing was performed using the TRV silencing vector described by Ratcliff et al. (2001) and the RNA2 clone described by (de la Fuente van Bentem et al. (2005).
pGEX-RSI2 was generated by cloning the full-length RSI2 sequence from the pACTII interaction clone (de la Fuente van Bentem et al., 2005) into pGEX-KG (Guan and Dixon, 1991) digested with NcoI/XhoI. All clones were validated by sequencing.
Full-length cDNAs of three selected class I HSP20s were PCR-amplified from EST clones (National BioResource Project of Japan) using primers containing either an EcoRI or XhoI site (underlined) for directional cloning into pACT2. EST clone FC02DH06 (SGN-U312450) was amplified using primers 5′-GGAATTCAAATGTCACTGATCCCAAGAATC-3′ and 5′-GGCTCGAGTTAACCAGAGATCTCAATGGA-3′, clone FB14BB10 (SGN-U312454) was amplified using primers 5′-GGAATTCAAATGTCTCTGATCCCAAGAATT-3′ and 5′-GGCTCGAGTTAACCAGAAATCTCAATGGA, and clone FC08BB05 (SGN-U316206) was amplified using primers 5′-GGAATTCAAATGTCTCTGATTCCAAGCTTC-3′ and 5′-GGCTCGAGTTAACCAGAGATGTCAATTGCC-3′. These clones have been described previously (Tsugane et al., 2005). All clones were sequenced to verify the presence of the correct insertion sequence.
GST–RSI2 production and pull-down
Escherichia coli strain BL21 (DE3) was transformed with empty pGEX-KG vector and with pGEX-KG containing full-length RSI2. Protein expression was induced by incubation at room temperature for 5 h in the presence of 1.5 mm IPTG. Pelleted cells (13 000 g) were frozen, and then thawed by resuspending in 10 ml ice-cold PBS, pH 7.4, supplemented with 1× protein inhibitor cocktail (Roche). Lysozyme (1 mg/ml) was added, and the suspension was rotated at 4°C for 30 min, followed by addition of 0.5% v/v Triton X-100 and incubation for a further 30 min. The cell mixture was sonicated, and cell debris was removed by centrifugation at 18 000 g at 4°C. To 1 ml of crude extract of induced E. coli cells, 200 μl of 50% pre-equilibrated glutathione bead slurry (GE Healthcare, http://www.gehealthcare.com) was added, and capture of GST-tagged proteins was performed with rotation at 4°C for 1.5 h. Pelleted beads (100 g) were rinsed four times with 0.5 ml ice-cold PBS, pH 7.4, supplemented with Complete protease inhibitors (Roche). Total protein was extracted from N. benthamiana tissue in which either I-2 or TAP-tagged Mi-1 proteins were expressed by Agrobacterium transformation. Total plant protein (10 mg) was supplemented with 0.1% v/v NP-40 to obtain an interaction buffer, and added to immobilized GST–RSI2 or GST protein alone. Equal loading of GST–RSI2 and GST (10 μg per pull-down) was confirmed using Bradford protein quantification. The final interaction buffer composition was 25 mm Tris pH 8, 1 mm EDTA, 150 mm NaCl, 5 mm DTT, 0.2% v/v NP-40, 1 x Complete protease inhibitor cocktail (Roche) in a total volume of 1.5 ml. This mixture was incubated with rotation overnight at 4°C to allow binding. The beads were pelleted (100 g) and rinsed five times with ice-cold interaction buffer. Proteins were eluted by addition of Laemmli sample buffer to the pelleted beads and boiling for 5 min. Blotting and immunodetection were performed as described previously (van Ooijen et al., 2008b).
Agrobacterium-mediated transformation and VIGS
Agrobacterium tumefaciens strain GV3101(pMP90) (Koncz and Schell, 1986) was transformed with the indicated vectors and infiltrated as described previously (van Ooijen et al., 2008b). Agroinfiltrations were performed in the laboratory (no direct sunlight, 20°C). For infiltration of Agrobacterium carrying TRV-based silencing constructs (Ratcliff et al., 2001; Liu et al., 2002), 2-week-old N. benthamiana leaves were used. Silencing was performed using the indicated RNA1 and RNA2 constructs, infiltrated at an OD600 of 1. Empty RNA2 vectors and their corresponding RNA1 constructs were used as negative controls. Plants were kept in the laboratory for 3 days and then transferred back to the greenhouse.
Agrobacterium-mediated expression of the R proteins was performed using a bacterial suspension with an OD600 of 1 in the laboratory using 3-week-old N. benthamiana plants or 2.5 weeks after silencing. HR was quantified on a scale from 0 to 4 as shown in Figure S6, and cell death was visualized using trypan blue staining as described previously (van Ooijen et al., 2008b).
For analysis of protein expression levels in silenced plants, three infiltrated leaves from three independent silenced plants (total nine) were harvested and pooled 24 h after agroinfiltration. Protein extraction was performed as described previously (van Ooijen et al., 2008b).
RT-PCR
Total RNA was extracted using Trizol (Invitrogen). cDNA was amplified from 1 μg RNA using SuperscriptI II (Invitrogen) as described by the manufacturer. RSI2 fragments were amplified using the primers 5′-CTGAAGCACATGTGTTTAAGGCC-3′ and 5′-CTTGACATCAGGCTTCTTCAC-3′. The resolving agarose gel was stained using SYBR Green (Invitrogen) and scanned using a STORM phosphoimager (Amersham Bioscience, http://www5.amershambiosciences.com/). The signal intensities were measured using ImageQuant (GE Healthcare) and corrected for background signals.
One-way anova
For statistical analysis of HR induction by auto-active R protein mutants on silenced plants, HR was visually scored 3 (I-2) or 4 (Mi-1) days after agroinfiltration as described above. Datasets from three independent silencing experiments were scored, totalling the number of replicates indicated in Figure 2. A one-way anova was performed on these data using a significance interval of 95%. Error bars represent a 95% confidence level calculated using Excel.
Acknowledgments
We thank J. van de Hove, L. Ma, W. Roels, K.-J. de Vries, Marianne de Vroomen and P. Mak for their help with the yeast two-hybrid assays, vector construction or testing HSP20 antibodies. The greenhouse staff are acknowledged for plant care, D. Baulcombe (Department of Plant Sciences, University of Cambridge, UK) and S. Dinesh-Kumar (Department of Molecular, Cellular, and Developmental Biology, Yale University, USA) for providing TRV silencing constructs, and the National Bioresource Project (Ministry of Education, Culture, Sport, Science and Technology, Japan) for the EST clones containing HSP20. The authors would like to thank the reviewers, the editor, and M. Rep for critical review of the manuscript and providing helpful comments. G.v.O. was supported by the Netherlands Research Council for Earth and Life Sciences, with financial aid from the Netherlands Organization for Scientific Research (grant number 813.06.005), and by the Centre for BioSystems Genomics, The Netherlands. H.v.d.B. was supported by the Netherlands Organization for Scientific Research (VENI grant number 863.04.018).
The Genbank accession number for the RSI2 sequence is AY150040.
SUPPORTING INFORMATION
Additional Supporting Information may be found in the online version of this article:
Figure S1. Evolutionary relationships of 113 HSP20 taxa.
Figure S2. Yeast two-hybrid interactions between I-2 and various class IA HSP20 proteins.
Figure S3. Coomassie-stained SDS–PAGE gel showing purified GST–RSI2 and GST proteins produced in Escherichia coli.
Figure S4. C-terminally TAP-tagged Mi-1H840 induces the HR.
Figure S5. N. benthamiana plants silenced for phytoene desaturase.
Figure S6. HR intensity scores.
Appendix S1. Clustal W (1.8) multiple sequence alignment of the core HSP20 domain used for the phylogeny analysis.
Please note: As a service to our authors and readers, this journal provides supporting information supplied by the authors. Such materials are peer-reviewed and may be re-organized for online delivery, but are not copy-edited or typeset. Technical support issues arising from supporting information (other than missing files) should be addressed to the authors.
References
- Azevedo C, Sadanandom A, Kitagawa K, Freialdenhoven A, Shirasu K, Schulze-Lefert P. The RAR1 interactor SGT1, an essential component of R gene-triggered disease resistance. Science. 2002;295:2073–2076. doi: 10.1126/science.1067554. [DOI] [PubMed] [Google Scholar]
- Azevedo C, Betsuyaku S, Peart J, Takahashi A, Noel L, Sadanandom A, Casais C, Parker J, Shirasu K. Role of SGT1 in resistance protein accumulation in plant immunity. EMBO J. 2006;25:2007–2016. doi: 10.1038/sj.emboj.7601084. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Basha E, Friedrich KL, Vierling E. The N-terminal arm of small heat shock proteins is important for both chaperone activity and substrate specificity. J. Biol. Chem. 2006;281:39943–39952. doi: 10.1074/jbc.M607677200. [DOI] [PubMed] [Google Scholar]
- Bhattarai KK, Li Q, Liu Y, Dinesh-Kumar SP, Kaloshian I. The MI-1-mediated pest resistance requires HSP90 and Sgt1. Plant Physiol. 2007;144:312–323. doi: 10.1104/pp.107.097246. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bieri S, Mauch S, Shen QH, et al. RAR1 positively controls steady state levels of barley MLA resistance proteins and enables sufficient MLA6 accumulation for effective resistance. Plant Cell. 2004;16:3480–3495. doi: 10.1105/tpc.104.026682. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bose S, Weikl T, Bugl H, Buchner J. Chaperone function of HSP90-associated proteins. Science. 1996;274:1715–1717. doi: 10.1126/science.274.5293.1715. [DOI] [PubMed] [Google Scholar]
- Boter M, Amigues B, Peart J, et al. Structural and functional analysis of SGT1 reveals that its interaction with HSP90 is required for the accumulation of Rx, an R protein involved in plant immunity. Plant Cell. 2007;19:3791–3804. doi: 10.1105/tpc.107.050427. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Caspers GJ, Leunissen JA, de Jong WW. The expanding small heat-shock protein family, and structure predictions of the conserved ‘alpha-crystallin domain’. J. Mol. Evol. 1995;40:238–248. doi: 10.1007/BF00163229. [DOI] [PubMed] [Google Scholar]
- Demmig-Adams B, Adams WW. Photoprotection and other responses of plants to high light stress. Annu. Rev. Plant Physiol. Plant Mol. Biol. 1992;43:599–626. [Google Scholar]
- Dropkin VH. The necrotic reaction of tomatoes and other hosts resistant to Meloidogyne: reversal by temperature. Phytopathology. 1969;59:1632–1637. [Google Scholar]
- Finn RD, Mistry J, Schuster-Bockler B, et al. Pfam: clans, web tools and services. Nucleic Acids Res. 2006;34:D247–D251. doi: 10.1093/nar/gkj149. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fu DQ, Zhu BZ, Zhu HL, Zhang HX, Xie YH, Jiang WB, Zhao XD, Luo KB. Enhancement of virus-induced gene silencing in tomato by low temperature and low humidity. Mol. Cells. 2006;21:153–160. [PubMed] [Google Scholar]
- de la Fuente van Bentem S, Vossen JH, de Vries KJ, van Wees S, Tameling WIL, Dekker HL, de Koster CG, Haring MA, Takken FLW, Cornelissen BJC. Heat shock protein 90 and its co-chaperone protein phosphatase 5 interact with distinct regions of the tomato I-2 disease resistance protein. Plant J. 2005;43:284–298. doi: 10.1111/j.1365-313X.2005.02450.x. [DOI] [PubMed] [Google Scholar]
- Gabriëls SHEJ, Vossen JH, Ekengren SK, et al. An NB-LRR protein required for HR signalling mediated by both extra- and intracellular resistance proteins. Plant J. 2007;50:14–28. doi: 10.1111/j.1365-313X.2007.03027.x. [DOI] [PubMed] [Google Scholar]
- Gray WM, Muskett PR, Chuang HW, Parker JE. Arabidopsis SGT1b is required for SCF(TIR1)-mediated auxin response. Plant Cell. 2003;15:1310–1319. doi: 10.1105/tpc.010884. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Guan KL, Dixon JE. Eukaryotic proteins expressed in Escherichia coli: an improved thrombin cleavage and purification procedure of fusion proteins with glutathione S-transferase. Anal. Biochem. 1991;192:262–267. doi: 10.1016/0003-2697(91)90534-z. [DOI] [PubMed] [Google Scholar]
- Heise CT, Le Duff CS, Boter M, et al. Biochemical characterization of RAR1 cysteine- and histidine-rich domains (CHORDs): a novel class of zinc-dependent protein–protein interaction modules. Biochemistry. 2007;46:1612–1623. doi: 10.1021/bi062174k. [DOI] [PubMed] [Google Scholar]
- Helm KW, Lee GJ, Vierling E. Expression and native structure of cytosolic class II small heat-shock proteins. Plant Physiol. 1997;114:1477–1485. doi: 10.1104/pp.114.4.1477. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hubert DA, Tornero P, Belkhadir Y, Krishna P, Takahashi A, Shirasu K, Dangl JL. Cytosolic HSP90 associates with and modulates the Arabidopsis RPM1 disease resistance protein. EMBO J. 2003;22:5679–5689. doi: 10.1093/emboj/cdg547. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Johnson JL, Beito TG, Krco CJ, Toft DO. Characterization of a novel 23-kilodalton protein of unactive progesterone receptor complexes. Mol. Cell. Biol. 1994;14:1956–1963. doi: 10.1128/mcb.14.3.1956. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kirschner M, Winkelhaus S, Thierfelder JM, Nover L. Transient expression and heat-stress-induced co-aggregation of endogenous and heterologous small heat-stress proteins in tobacco protoplasts. Plant J. 2000;24:397–411. doi: 10.1046/j.1365-313x.2000.00887.x. [DOI] [PubMed] [Google Scholar]
- Kitagawa K, Skowyra D, Elledge SJ, Harper JW, Hieter P. SGT1 encodes an essential component of the yeast kinetochore assembly pathway and a novel subunit of the SCF ubiquitin ligase complex. Mol. Cell. 1999;4:21–33. doi: 10.1016/s1097-2765(00)80184-7. [DOI] [PubMed] [Google Scholar]
- Koncz C, Schell J. The promoter of Tl-DNA gene 5 controls the tissue-specific expression of chimaeric genes carried by a novel type of Agrobacterium binary vector. Mol. Gen. Genet. 1986;204:383–396. [Google Scholar]
- Kotak S, Larkindale J, Lee U, von Koskull-Doring P, Vierling E, Scharf KD. Complexity of the heat stress response in plants. Curr. Opin. Plant Biol. 2007;10:310–316. doi: 10.1016/j.pbi.2007.04.011. [DOI] [PubMed] [Google Scholar]
- Lee GJ, Vierling E. A small heat shock protein cooperates with heat shock protein 70 systems to reactivate a heat-denatured protein. Plant Physiol. 2000;122:189–198. doi: 10.1104/pp.122.1.189. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lee GJ, Pokala N, Vierling E. Structure and in vitro molecular chaperone activity of cytosolic small heat shock proteins from pea. J. Biol. Chem. 1995;270:10432–10438. doi: 10.1074/jbc.270.18.10432. [DOI] [PubMed] [Google Scholar]
- Lee GJ, Roseman AM, Saibil HR, Vierling E. A small heat shock protein stably binds heat-denatured model substrates and can maintain a substrate in a folding-competent state. EMBO J. 1997;16:659–671. doi: 10.1093/emboj/16.3.659. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liberek K, Lewandowska A, Zietkiewicz S. Chaperones in control of protein disaggregation. EMBO J. 2008;27:328–335. doi: 10.1038/sj.emboj.7601970. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu Y, Schiff M, Serino G, Deng XW, Dinesh-Kumar SP. Role of SCF ubiquitin-ligase and the COP9 signalosome in the N gene-mediated resistance response to Tobacco mosaic virus. Plant Cell. 2002;14:1483–1496. doi: 10.1105/tpc.002493. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu Y, Burch-Smith T, Schiff M, Feng S, Dinesh-Kumar SP. Molecular chaperone HSP90 associates with resistance protein N and its signaling proteins SGT1 and Rar1 to modulate an innate immune response in plants. J. Biol. Chem. 2004;279:2101–2108. doi: 10.1074/jbc.M310029200. [DOI] [PubMed] [Google Scholar]
- Lu R, Malcuit I, Moffett P, Ruiz MT, Peart J, Wu AJ, Rathjen JP, Bendahmane A, Day L, Baulcombe DC. High throughput virus-induced gene silencing implicates heat shock protein 90 in plant disease resistance. EMBO J. 2003;22:5690–5699. doi: 10.1093/emboj/cdg546. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lukasik E, Takken FLW. STANDing strong, resistance proteins instigators of plant defence. Curr. Opin. Plant Biol. 2009;12:427–436. doi: 10.1016/j.pbi.2009.03.001. [DOI] [PubMed] [Google Scholar]
- Maimbo M, Ohnishi K, Hikichi Y, Yoshioka H, Kiba A. Induction of a small heat shock protein and its functional roles in Nicotiana plants in the defense response against Ralstonia solanacearum. Plant Physiol. 2007;145:1588–1599. doi: 10.1104/pp.107.105353. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Martin GB, Bogdanove AJ, Sessa G. Understanding the functions of plant disease resistance proteins. Annu. Rev. Plant Biol. 2003;54:23–61. doi: 10.1146/annurev.arplant.54.031902.135035. [DOI] [PubMed] [Google Scholar]
- McHale L, Tan X, Koehl P, Michelmore RW. Plant NBS-LRR proteins: adaptable guards. Genome Biol. 2006;7:212. doi: 10.1186/gb-2006-7-4-212. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mestre P, Baulcombe DC. Elicitor-mediated oligomerization of the tobacco N disease resistance protein. Plant Cell. 2006;18:491–501. doi: 10.1105/tpc.105.037234. [DOI] [PMC free article] [PubMed] [Google Scholar]
- van Montfort R, Slingsby C, Vierling E. Structure and function of the small heat shock protein/alpha-crystallin family of molecular chaperones. Adv. Protein Chem. 2001a;59:105–156. doi: 10.1016/s0065-3233(01)59004-x. [DOI] [PubMed] [Google Scholar]
- van Montfort RL, Basha E, Friedrich KL, Slingsby C, Vierling E. Crystal structure and assembly of a eukaryotic small heat shock protein. Nat. Struct. Biol. 2001b;8:1025–1030. doi: 10.1038/nsb722. [DOI] [PubMed] [Google Scholar]
- Nover L, Scharf KD. Heat stress proteins and transcription factors. Cell. Mol. Life Sci. 1997;53:80–103. doi: 10.1007/PL00000583. [DOI] [PMC free article] [PubMed] [Google Scholar]
- van Ooijen G, van den Burg HA, Cornelissen BJC, Takken FLW. Structure and function of resistance proteins in solanaceous plants. Annu. Rev. Phytopathol. 2007;45:43–72. doi: 10.1146/annurev.phyto.45.062806.094430. [DOI] [PubMed] [Google Scholar]
- van Ooijen G, Mayr G, Albrecht M, Cornelissen BJC, Takken FLW. Transcomplementation, but not physical association of the CC-NB-ARC and LRR domains of tomato R protein Mi-1.2 is altered by mutations in the ARC2 subdomain. Mol. Plant. 2008a;1:401–410. doi: 10.1093/mp/ssn009. [DOI] [PubMed] [Google Scholar]
- van Ooijen G, Mayr G, Kasiem MM, Albrecht M, Cornelissen BJ, Takken FL. Structure–function analysis of the NB-ARC domain of plant disease resistance proteins. J. Exp. Bot. 2008b;59:1383–1397. doi: 10.1093/jxb/ern045. [DOI] [PubMed] [Google Scholar]
- Pearl LH, Prodromou C. Structure and mechanism of the HSP90 molecular chaperone machinery. Annu. Rev. Biochem. 2006;75:271–294. doi: 10.1146/annurev.biochem.75.103004.142738. [DOI] [PubMed] [Google Scholar]
- Peart JR, Lu R, Sadanandom A, et al. Ubiquitin ligase-associated protein SGT1 is required for host and nonhost disease resistance in plants. Proc. Natl Acad. Sci. USA. 2002;99:10865–10869. doi: 10.1073/pnas.152330599. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ratcliff F, Martin-Hernandez AM, Baulcombe DC. Tobacco rattle virus as a vector for analysis of gene function by silencing. Plant J. 2001;25:237–245. doi: 10.1046/j.0960-7412.2000.00942.x. [DOI] [PubMed] [Google Scholar]
- Rohila JS, Chen M, Cerny R, Fromm ME. Improved tandem affinity purification tag and methods for isolation of protein heterocomplexes from plants. Plant J. 2004;38:172–181. doi: 10.1111/j.1365-313X.2004.02031.x. [DOI] [PubMed] [Google Scholar]
- Scharf KD, Siddique M, Vierling E. The expanding family of Arabidopsis thaliana small heat stress proteins and a new family of proteins containing alpha-crystallin domains (Acd proteins) Cell Stress Chaperones. 2001;6:225–237. doi: 10.1379/1466-1268(2001)006<0225:tefoat>2.0.co;2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shirasu K, Lahaye T, Tan MW, Zhou F, Azevedo C, Schulze-Lefert P. A novel class of eukaryotic zinc-binding proteins is required for disease resistance signaling in barley and development in C. elegans. Cell. 1999;99:355–366. doi: 10.1016/s0092-8674(00)81522-6. [DOI] [PubMed] [Google Scholar]
- Simons G, Groenendijk J, Wijbrandi J, et al. Dissection of the Fusarium I2 gene cluster in tomato reveals six homologs and one active gene copy. Plant Cell. 1998;10:1055–1068. doi: 10.1105/tpc.10.6.1055. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Takahashi A, Casais C, Ichimura K, Shirasu K. HSP90 interacts with RAR1 and SGT1 and is essential for RPS2-mediated disease resistance in Arabidopsis. Proc. Natl Acad. Sci. USA. 2003;100:11777–11782. doi: 10.1073/pnas.2033934100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tameling WIL, Takken FLW. Resistance proteins: scouts of the plant innate immune system. Eur. J. Plant Pathol. 2008;121:243–255. [Google Scholar]
- Tameling WIL, Elzinga SD, Darmin PS, Vossen JH, Takken FLW, Haring MA, Cornelissen BJC. The tomato R gene products I-2 and Mi-1 are functional ATP binding proteins with ATPase activity. Plant Cell. 2002;14:2929–2939. doi: 10.1105/tpc.005793. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tamura K, Dudley J, Nei M, Kumar S. MEGA4: Molecular Evolutionary Genetics Analysis (MEGA) software version 4.0. Mol. Biol. Evol. 2007;24:1596–1599. doi: 10.1093/molbev/msm092. [DOI] [PubMed] [Google Scholar]
- Tsugane T, Watanabe M, Yano K, Sakurai N, Suzuki H, Shibata D. Expressed sequence tags of full-length cDNA clones from the miniature tomato (Lycopersicon escumlentum) cultivar Micro-Tom. Plant Biotechnol. 2005;22:161–165. [Google Scholar]
- Vos P, Simons G, Jesse T, et al. The tomato Mi-1 gene confers resistance to both root-knot nematodes and potato aphids. Nat. Biotechnol. 1998;16:1365–1369. doi: 10.1038/4350. [DOI] [PubMed] [Google Scholar]
- Waters ER, Vierling E. The diversification of plant cytosolic small heat shock proteins preceded the divergence of mosses. Mol. Biol. Evol. 1999;16:127–139. doi: 10.1093/oxfordjournals.molbev.a026033. [DOI] [PubMed] [Google Scholar]
- Xu P, Zhang Y, Kang L, Roossinck MJ, Mysore KS. Computational estimation and experimental verification of off-target silencing during posttranscriptional gene silencing in plants. Plant Physiol. 2006;142:429–440. doi: 10.1104/pp.106.083295. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang Y, Dorey S, Swiderski M, Jones JDG. Expression of RPS4 in tobacco induces an AvrRps4-independent HR that requires EDS1, SGT1 and HSP90. Plant J. 2004;40:213–224. doi: 10.1111/j.1365-313X.2004.02201.x. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.


