Abstract
Plastids contain sigma factors, i.e. gene-regulatory proteins for promoter binding and transcription initiation. Despite the physical and functional similarity shared with their prokaryotic counterparts, the plant sigma factors have distinguishing features: most notably the existence of a variable extra sequence comprising their N-terminal portions. This distinct architecture is reflected by functional differences, including phosphorylation control by organellar protein kinase(s) closely related to nucleocytosolic, rather than bacterial-type, enzymes. In particular, cpCK2, a nuclear-coded plastid-targeted casein kinase 2, has been implicated as a key component in plant sigma factor phosphorylation and transcriptional regulation (Eur. J. Biochem. 269, 2002, 3329; Planta, 219, 2004, 298). Although this notion is based mainly on biochemical evidence and in vitro systems, the recent availability of Arabidopsis sigma knock-out lines for complementation by intact and mutant sigma cDNAs has opened up new strategies for the study of transcription regulatory mechanisms in vivo. Using Arabidopsis sigma factor 6 (AtSIG6) as a paradigm, we present data suggesting that: (i) this factor is a substrate for regulatory phosphorylation by cpCK2 both in vitro and in vivo; (ii) cpCK2 phosphorylation of SIG6 occurs at multiple sites, which can widely differ in their effect on the visual and/or molecular phenotype; (iii) in vivo usage of the perhaps most critical cpCK2 site defined by Ser174 requires (pre-)phosphorylation at the n + 3 serine residue Ser177, pointing to ‘pathfinder’ kinase activity capable of generating a functional cpCK2 substrate site.
Keywords: plant sigma factor, plastid transcription kinase, multisite phosphorylation control, chloroplast transcription, Arabidopsis phenotype, point mutants
Introduction
Plant cells are tripartite genetic systems consisting of three transcriptionally active compartments, i.e. the nucleus, the mitochrondria and the plastids. The latter contain two principally different forms of RNA polymerases for transcription of a full complement of organellar genes in normal (wild-type) plants. Nucleus-encoded polymerase (NEP) is a single-subunit enzyme closely related to those of T7/T3 phages and mitochondria. In contrast, plastid-encoded polymerase (PEP) is a multi-subunit bacterial-type enzyme with α-, β- and β′-equivalent core subunits that are encoded by plastid genes (Maliga, 1998; Hess and Börner, 1999). It has become clear, however, that the core polypeptides are embedded into a much larger functional complex, made up of nucleus-encoded polypeptides, most of which seem to represent chloroplast versions of ‘eukaryotic’ nucleo/cytosolic proteins (Pfannschmidt et al., 2000; Suzuki et al., 2004; Pfalz et al., 2006).
An intriguing feature of PEP transcription is the involvement of typical ‘prokaryotic’, yet nucleus-encoded, sigma initiation factors (for review see e.g. Gross et al., 1998; Burgess and Anthony, 2001; Helmann, 2009). Plastids usually contain a small set of these factors, each of which reveals the principal sigma domains within its conserved C-terminal region (CR) (Campbell et al., 2002; Murakami et al., 2002). Unlike many bacterial sigma factors, however, the plant factors have a highly variable unconserved region (UCR) comprising their N-terminal portions (Shiina et al., 2005; Lysenko, 2007). Although the role of UCR has long remained enigmatic, recent work suggests that it is critically involved in specifying the visual and molecular phenotype (Kubota et al., 2007; Schweer et al., 2009).
Arabidopsis thaliana contains a family of six sigma genes, named AtSig1–AtSig6 (Isono et al., 1997; Tanaka et al., 1997; Fujiwara et al., 2000; Hakimi et al., 2000). Two lines of evidence have led to an assignment of the gene products as true sigma factors: (i) in vitro transcription and DNA-binding experiments using the bacterially expressed recombinant factors and Escherichia coli core RNA polymerase; and (ii) sigma knock-out and antisense plants as tools that allow us to establish causal relationships between sigma genes and phenotypic traits in vivo (Hanaoka et al., 2003; Privat et al., 2003).
Despite general agreement on the existence of ‘true’ sigma factors in plants, it has turned out to be more difficult than anticipated to assign each individual factor a well-defined role in gene (promoter)-specific transcription, with noticeable consequences for plant development and function. This is in part because of a certain level of functional overlap between factors, which helps maintain the overall transcription program, even in adverse situations (Kanamaru and Tanaka, 2004; Lysenko, 2007). Another, related, reason is that sigma factor steady-state levels do not necessarily correlate with the magnitude of downstream effects in plastid gene expression. In part, this can be assigned to reversible protein modification, such as phosphorylation, which results in altered promoter binding, and thus greater flexibility of transcription (Tiller and Link, 1993).
Available evidence (Baginsky et al., 1997, 1999; Baena-Gonzalez et al., 2001) suggests that a PEP-associated Ser/Thr protein kinase, termed plastid transcription kinase (PTK), is a major player in plastid sigma factor phosphorylation. Cloning, sequencing and functional characterization revealed that the catalytically active component is a nucleus-encoded and chloroplast-targeted protein closely related to the α-subunit of nucleocytosolic casein kinase 2 (CK2) (Pinna, 1990), which was hence named cpCK2 (Ogrzewalla et al., 2002). Subsequent work established the presence of a single gene for cpCK2 in a number of plant species, including Arabidopsis (Loschelder et al., 2004; Salinas et al., 2006). Nevertheless, despite identification and characterization of the transcription kinase itself, the extent to which phosphorylation control is responsible for the activation or inactivation of plant sigma factors in vivo has not yet been reported.
To help clarify this question, we focused our attention on one of the Arabidopsis sigma factors, AtSIG6, for which mutant lines with readily discernible phenotypes are available (Ishizaki et al., 2005; Loschelder et al., 2006; Schweer et al., 2006, 2009; Coll et al., 2009). We asked whether AtSIG6 could serve as a PTK/cpCK2 target and, if so, which phosphorylation site(s) might be functionally relevant in Arabidopsis in vivo compared with the bacterially expressed recombinant protein in vitro. Using site-directed mutagenesis and retransformation of an AtSig6 knock-out line, we present evidence suggesting phosphorylation control of SIG6 activity by cpCK2 and probably one other protein kinase.
Results
Localization and selection of putative PTK/cpCK2 phosphorylation sites on sigma factor AtSIG6 using prediction tools and sequence alignments
In vitro experiments with authentic chloroplast sigma factors had provided initial clues suggesting that their activity depends on phosphorylation state (Tiller and Link, 1993). In addition, recombinant sigma factor SaSIG1 from mustard was shown to be a cpCK2 substrate (Ogrzewalla et al., 2002), although the functional consequences were not investigated. It thus remained to be established if (cpCK2) phosphorylation might have a regulatory effect on plant sigma factors, whether or not such a mechanism plays a role both in vitro and in vivo, and where exactly the relevant sites might be located on the substrate protein(s). To gain information whether AtSIG6 can be a potential substrate for CK2 phosphorylation, we therefore searched for putative sites in the derived protein sequence.
Consensus phosphorylation site motifs for (nucleocytosolic) CK2 often conform to the sequence motifs S*/T*xxEx and S*/T*xxDx, respectively (Pinna, 1990; Meggio and Pinna, 2003) (Table 1, bottom). In addition, as an alternative to the acidic residues aspartate or glutamate, a serine at the n + 3 position can help create a CK2 substrate site if it is converted to phosphoserine by another protein kinase (Roach, 1991; Meggio and Pinna, 2003). The plastid transcription kinase PTK (cpCK2) is highly homologous to, and shares principal enzymatic properties with, the nucleocytosolic members of the CK2 family (Ogrzewalla et al., 2002; Loschelder et al., 2004), suggesting that prediction tools suitable for CK2 could also provide valid answers with regard to cpCK2 phosphorylation site(s) on AtSIG6.
Table 1.
Prediction of putative casein kinase 2 (CK2) phosphorylation sites in AtSIG6
| Position in consensus motif | ||||||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Position | Aminoacid | NetPhos 2.0 | NetPhosK 1.0 | KinasePhos 2.0 | disphos | gps | PREDPhospho | ScanSite | −2 | −1 | * | +1 | +2 | +3 | +4 | Homology |
| 26 | Serine | * | * | Y | S | S | P | S | S | V | ||||||
| 94 | Serine | * | * | * | * | * | * | L | V | S | S | R | E | D | ||
| 95 | Serine | * | * | * | * | * | * | V | S | S | R | E | D | E | ||
| 149 | Serine | * | A | L | S | A | S | K | Q | |||||||
| 174 | Serine | * | * | * | * | S | L | S | T | S | S | S | AtSIG2 | |||
| 176 | Serine | * | * | * | S | T | S | S | S | M | S | |||||
| 180 | Serine | * | * | * | * | * | S | M | S | L | P | E | K | AtSIG5 | ||
| 206 | Serine | * | P | K | S | N | D | V | D | |||||||
| 244 | Threonine | * | P | E | T | K | Q | L | L | |||||||
| 249 | Threonine | * | L | L | T | A | K | E | E | AtSIG1-5 | ||||||
| 282 | Threonine | * | E | P | T | I | G | E | W | |||||||
| 306 | Serine | * | * | G | R | S | S | R | E | K | ||||||
| 411 | Serine | * | * | * | R | P | S | K | E | E | L | AtSIG4, 5 | ||||
| 423 | Threonine | * | V | S | T | E | K | L | D | |||||||
| 445 | Serine | * | * | I | W | S | D | Q | D | T | ||||||
| 450 | Threonine | * | * | D | T | T | F | Q | E | I | AtSIG4 | |||||
| 458 | Serine | * | * | * | * | P | D | S | G | I | E | T | AtSIG1, 3, 5 | |||
| 462 | Threonine | * | I | E | T | P | T | M | S | AtSIG2, 3, 4 | ||||||
| 484 | Serine | * | * | * | V | L | S | P | K | E | R | AtSIG4 | ||||
| 504 | Serine | * | * | * | Q | R | S | L | S | E | I | AtSIG1-5 | ||||
| CK2 consensus substrate site | S/T | x | x | E/D | x | |||||||||||
Prediction tools NetPhos 2.0 (Blom et al., 1999), NetPhosK 1.0 (Blom et al., 2004), KinasePhos 2.0 (Wong et al., 2007), disphos (Diella et al., 2004), gps (Xue et al., 2005), PREDPhospho (Kim et al., 2004) and ScanSite (Obenauer et al., 2003) were used, leading to the detection of putative sites (+). Those selected for subsequent experiments are set in bold. The phosphoacceptor residue (*) and the acidic residue at the n + 3 position are marked in bold. Arabidopsis sigma factors showing regional similarity with AtSIG6 CK2 sites are indicated in the last column (Figure S2 for detailed positions). Bottom: consensus sequence of CK2 phosphorylation sites (Meggio and Pinna, 2003).
Indeed, using NetPhos 2.0 (Blom et al., 1999), disphos 1.3 (Diella et al., 2004), KinasePhos 2.0 (Wong et al., 2007), NetPhosK 1.0 (Blom et al., 2004), gps (Xue et al., 2005), PREDPhospho (Kim et al., 2004) and ScanSite (Obenauer et al., 2003), a common picture emerges, suggesting a limited number of putative cpCK2 sites (Table 1). Based on their positions, these putative phosphorylation sites can be divided into ‘general’ and ‘unique’ sites, the former (those ranging from T244 to S504) being located within the CR, and the latter (from S26 to S206) being located within the UCR of the protein (Figure 1; Table 1).
Figure 1.
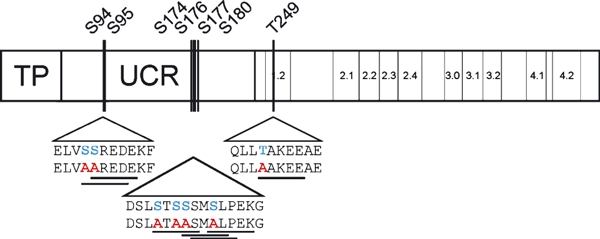
Positions of functionally tested cpCK2 phosphorylation sites on AtSIG6.Scheme showing the principal sigma factor architecture, with the conserved region containing subregions 1.2–4.2 on the right as well as the unconserved region (UCR) and the transit peptide sequence (TP) on the left. The sequences of the analysed substrate sites are enlarged below the triangles: wild type (upper row) and mutant (lower row). Original S or T residues that can function as phosphate acceptors are marked in blue, the exchanges are drawn in red and the entire cpCK2 site is underlined.
The ‘general’ site at T249 maps to the conserved sigma subregion 1.2 (Figure 1), which is known to be involved in core binding (Baldwin and Dombroski, 2001) and recognition control of the –10 promoter element (Zenkin et al., 2007) in bacterial systems. Although only one of the seven prediction tools (NetPhosK 1.0; Table 1) gave an acceptably high score for this site, sequence alignments showed a putative CK2 site at equivalent positions in all Arabidopsis sigma factors (Figure 2b), as well as sigma factors from other plant species, e.g. maize ZmSIG6 (Beardslee et al., 2002), Chlamydomonas RpoD (Carter et al., 2004) and Physcomitrella PpSIG2 (Hara et al., 2001) (Figure S1). A motif that would conform to the CK2 consensus site is noticeable even in bacterial sigma factors (Gruber and Gross, 2003), despite the lack of evidence for this kinase class in prokaryotes. Nevertheless, this ‘general’ site was included in subsequent analyses to allow for comparison with the ‘unique’ sites, i.e. potential CK2 substrate sites that are located within the UCR, and appear to be AtSIG6-specific.
Figure 2.

Alignment of analysed cpCK2 substrate motifs.The phosphoacceptor Ser or Thr residues are marked in red, and the acidic residues at n + 3 are marked in grey.(a) Multiple alignment of the eight cpCK2 substrate sites in AtSIG6 shown in Figure 1.(b) Alignment of the ‘general’ substrate site T249 from AtSIG6 with motifs from the conserved region of the other Arabidopsis sigma factors, AtSIG1– AtSIG5.
Of the latter, those at S94 and S95 were detected by almost all prediction tools, and the assigned score values were in the top segment of all sites investigated (Figure 1; Table 1). Sequence alignment with the other Arabidopsis sigma factors did not reveal any appreciable similarity to AtSIG6 around S94/S95 (Figure S2). A somewhat similar situation applies to the region encompassing S174, S176 and S180, all of which are potential CK2 sites predicted by several (between three and five) programs. With one possible exception (residues reminiscent of S174 are located at positions T141 in AtSIG2), none of these sites is conserved in other Arabidopsis sigma factors (Figure S2).
Phosphorylation control of recombinant AtSIG6 by PTK/cpCK2 in vitro
To gain information on sigma factor activity in response to phosphorylation state, electrophoretic mobility shift assay (EMSA) DNA binding experiments were carried out using bacterially expressed AtSIG6 in combination with E. coli core RNA polymerase and a cloned DNA fragment that carries the Arabidopsis chloroplast atpB PEP promoter (Figure 3). The recombinant sigma factor was either used without (SIG6) or with prior phosphorylation by recombinant cpCK2 (SIG6-P).
Figure 3.
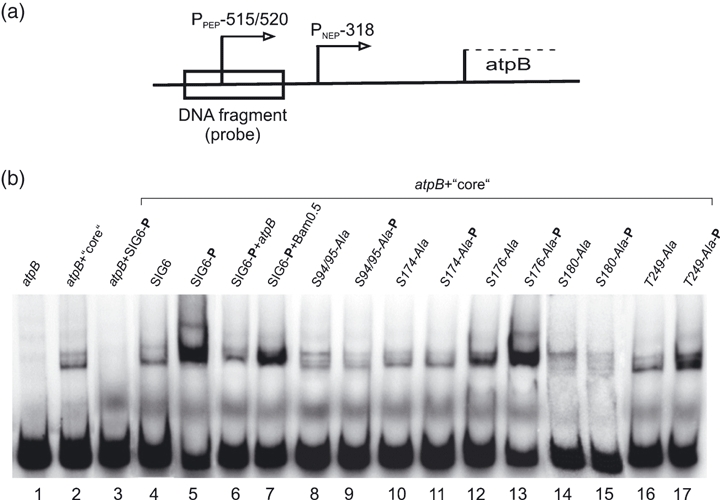
Phosphorylation-dependent DNA binding of recombinant AtSIG6 in vitro.(a) Map position of the DNA fragment used as a probe. The 424-bp fragment carries the atpB PEP-515/-520 promoter (Pfalz et al., 2006) and a 154-bp section downstream of the transcription start site.(b) EMSA experiments using promoter fragment, Escherichia coli core polymerase and recombinant sigma proteins, with or without prior phosphorylation by recombinant cpCK2. The free radiolabelled DNA fragment migrates at the bottom (lane 1; atpB). Core polymerase alone (lane 2) but not AtSIG6 alone (lane 3) is able to bind to the promoter fragment. Complete reactions containing probe, core enzyme and either phosphorylated or mock-phosphorylated sigma proteins are shown in lanes 4–17 (atpB + core). Lanes 6 and 7 show competition with a twofold molar excess of unlabelled promoter fragment (atpB), but not with a promoter-less fragment Bam0.5 at 10-fold excess. Like AtSIG6 itself, none of the mutagenized derivatives show sigma activity in the unphosporylated form (lanes 8, 10, 12, 14 and 16).
Following incubation of either unphosphorylated (not shown) or phosphorylated AtSIG6 alone (lane 3) in the presence of the labelled probe, but without core enzyme, the only detectable signal is at the position of the free probe (bottom band). The virtual absence of any additional band with retarded mobility indicates that the factor lacks DNA binding activity on its own. E. coli core enzyme alone in the absence of AtSIG6 (lane 2) resulted in a small but significant portion of labelled material to a shifted position. The core enzyme preparation (Epicentre, http://www.epibio.com) did not contain detectable levels of bacterial sigma factor(s), and the shifted material was thus taken into account as the baseline in subsequent experiments.
The full system, i.e. the labelled probe in the presence of both AtSIG6 and the core enzyme (lane 4), gave shifted signals at the same position and strength as seen for the core alone (lane 2), suggesting that unphosphorylated AtSIG6 has little if any sigma activity in this assay. However, when the phosphorylated recombinant factor was used (SIG6-P, lane 5), this led to a considerable increase in intensity, along with an additional small shift in migration position of the DNA–protein complex. Carrying out the same reaction with an added twofold molar excess of unlabelled atpB promoter fragment as a competitor (lane 6), the intensity of shifted radioactive material was largely reduced. In contrast, a promoter-less fragment that was tested as a non-specific competitor did not negatively affect the binding signal (Bam0.5; lane 7) (Homann and Link, 2003). Together, this suggests that AtSIG6 indeed confers specific and efficient promoter binding to the polymerase, yet only in its phosphorylated form.
To investigate this further, we next constructed and tested mutant versions of the AtSIG6 protein that contained altered residues at putative cpCK2 substrate sites (Figure 2a). As is evident from Figure 3(b), neither the protein containing Ser → Ala exchanges at positions S94/95 (lanes 8 and 9), nor the one with an exchange at position S174 (lanes 10 and 11), revealed any significant SIG6-mediated DNA binding activity in the complete EMSA reaction, regardless of phosphorylation state. In contrast, the mutant version with a Ser → Ala exchange at S176 (lanes 12 and 13) showed a strong increase in DNA-binding activity upon phosphorylation, comparable with the effect observed for the non-mutagenized AtSIG6 protein (lanes 4 and 5). This suggests that S94/95 as well as S174, but not S176, are critical positions for phosphorylation-dependent activation of AtSIG6 in vitro.
A Thr → Ala exchange at the ‘general’ position T249 results in a mutant protein that is still active in its phosphorylated state (lanes 16 and 17), although perhaps to a somewhat lesser extent than the wild-type factor (lanes 4 and 5). This would argue against a significant regulatory effect of phosphorylation at this site in the in vitro system, at least for the atpB promoter.
Construction and phenotypic analysis of Arabidopsis AtSig6 mutant lines containing altered phosphorylation sites
Having located putative cpCK2 phosphorylation sites on AtSIG6 (Figures 1 and 2), and demonstrated phosphorylation-dependent activation of the factor in vitro (Figure 3b), we next asked which of these sites might be functionally relevant in Arabidopsis in vivo. To clarify this, we constructed full-length AtSIG6 cDNAs including the transit peptide region, either wildtype or mutant sequences, with amino acid exchanges at selected positions (Figures 1 and 2a). Following mobilization to a binary vector and retransformation of the Arabidopsis sig6-2 knock-out, the progeny were analysed both for their visual (Figures 4 and S3) and molecular phenotypes (Figure 5).
Figure 4.

Phenotypic features of Arabidopsis sig6 mutant lines carrying altered phosphorylation sites.Plates show 14-day-old plantlets representing the wild type (WT), the parental sig6-2 knock-out line and the retransformed lines generated by cDNAs for either authentic AtSIG6 (sig6-com) or AtSIG6 derivatives, with exchange of residues 94, 95, 174, 176, 177, 180 or 249, as indicated. Several of the latter lines resemble the wild type in growth and pigmentation (S94-Ala, S95-Ala, S176-Ala, S180-Ala and T249-Ala), whereas others reveal a chlorophyll-deficient phenotype (S94/95-Ala, S174-Ala, S174-Gln, S177-Ala and S177-Asp) comparable with that of the parental sig6-2 knock-out (sig6-2). S, serine site; T, threonine site.
Figure 5.
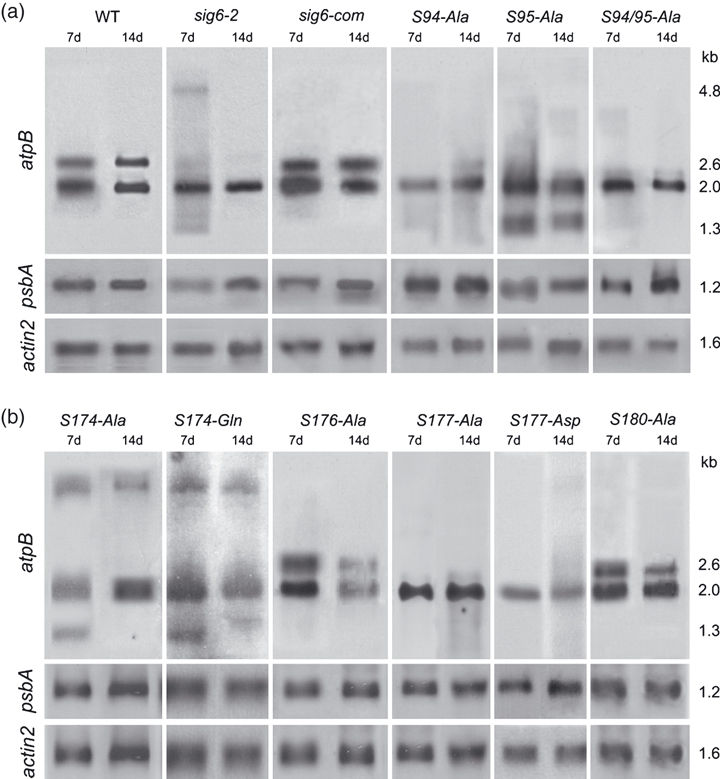
Site-directed phosphomutants are differentially affected in plastid gene expression. Northern blot analyses were carried out with total cellular RNA (1 μg each) from the wild type (WT), the AtSig6 knock-out line (sig6-2) and the retransformed lines, including those containing fully functional AtSIG6 cDNA (sig6-com) and various point mutant derivatives. Hybridization was performed using DIG-labelled RNA probes for atpB, psbA and a nuclear control gene (actin2), with RNA from 7-day-old seedlings (7d) and 14-day-old plantlets (14d). Transcript sizes (kb) are indicated in the right margin.
To facilitate the identification of significant phenotypic traits, mutant lines were studied at two developmental stages, i.e. 7-day-old seedlings (Figure S3) and 14-day-old plantlets (Figure 4). They were compared with the wild type and the parental sig6-2 knock-out line (Figures 4 and S3, first row), with the latter showing the previously described phenotype of white cotyledons but green true leaves (Loschelder et al., 2006; Schweer et al., 2006, 2009).
A number of phosphorylation site mutants that were tested exhibit the fully green wild-type phenotype: S94-Ala and S95-Ala (Figures 4 and S3, second row) as well as S176-Ala, S180-Ala and T249-Ala (fourth row). None of these lines showed gross deviation in growth parameters (size/shape) and pigmentation. However, in several other lines dramatic defects in pigmentation were found. These included the S94/95-Ala double exchange mutant (second row), as well as the mutant lines obtained after exchange at position S174 (S174-Ala and S174-Gln) or position S177 (S177-Ala and S177-Asp) (third row).
Within this chlorophyll-defective group of mutants, both the severity and the temporal mode of the pigment deficiency varied considerably. The most dramatic situation is found in the case of the S94/95 double mutant, which not only revealed white cotyledons (Figure S3) but also achlorophyllous primary leaves (Figure 4), leading to lethality at this stage. Not quite as extreme, but still more severe than the parental sig6-2 knock-out, the S174 and S177 mutants develop white and yellowish cotyledons, respectively, and their first true leaves also remain pigment deficient. Although pigmentation gradually recovers to wild-type levels, both the S174 and S177 mutants remain compromised, as indicated by their delayed growth.
Plastid gene expression in AtSig6 phosphorylation site mutants
Northern blot hybridization has previously proved useful as a rapid diagnostic means to define SIG6-related organellar gene expression states in wild-type and AtSig6 mutant lines (Loschelder et al., 2006; Schweer et al., 2006, 2009). Figure 5 shows a comparative analysis of phosphorylation site mutants following hybridization of total RNA samples (day 7 and 14) with either a chloroplast atpB or psbA probe, or with a nuclear actin2 gene probe. Data obtained with the wild type, the parental sig6-2 knock-out line, and the knock-out line retransformed and complemented with SIG6 cDNA were included as controls, to which the various phosporylation site mutants were compared.
In brief, with the atpB probe, the wild-type RNA sample reveals the two major atpB/E transcripts at 2.6 (SIG6/PEP-driven) and 2.0 kb (NEP-driven) (Schweer et al., 2006). The knock-out mutant lacks the 2.6-kb transcript but shows the smaller 2.0-kb species and a transient 4.8-kb transcript. Following the retransformation of the sig6-2 knock-out with SIG6 cDNA (sig6-com), the wild-type pattern is restored, i.e. both the 2.6- and 2.0-kb transcripts but not the 4.8-kb transcript are visible.
When RNA samples from the SIG6 phosphorylation site mutants were analysed, the hybridization patterns obtained were as follows: (i) The 2.0-kb (NEP) transcript is detectable in all lines; (ii) the larger 2.6-kb (SIG6/PEP) transcript is present at wild-type levels in both the S176 and the S180 seedlings, but is highly reduced or virtually absent in the other retransformed mutant lines; (iii) the 4.8-kb transcript is only detected in the S174 lines.
Based on these diagnostic transcript patterns, it seems that Ser174 is absolutely required for SIG6 activity in driving atpB gene expression in vivo. A noticeable, but less pronounced, effect is also obvious for Ser94 and Ser95, as well as for the ‘general’ site T249-Ala, whereas Ser176 and Ser180 do not seem to be required for activity.
Also shown in Figure 5 are the results of northern blot analyses with a psbA probe. The single 1.2-kb transcript is present in about equal relative quantities in wild-type RNA from 7-day-old seedlings and 14-day-old plantlets. In the sig6-2 knock-out mutant the signal intensity is reduced only at the earlier stage, and its intensity is restored to wild-type levels following retransformation of the knock-out with SIG6 cDNA (Loschelder et al., 2006). All phosphorylation site mutants show comparable transcript levels at both time points, i.e. they resemble the wild type rather than the parental knock-out with regard to psbA gene expression. Finally, the results obtained with an actin2 gene probe establish that all mutant lines do not show significant deviation from the wild-type RNA expression levels of this nuclear control gene.
Discussion
Here we have investigated putative cpCK2 phosphorylation sites in Arabidopsis sigma factor AtSIG6, and we have tested these sites for regulatory function. Together, the EMSA in vitro experiments (Figure 3) and the mutational analysis in vivo (Figures 4 and 5) support the notion that AtSIG6 is a cpCK2 substrate that responds to phosphorylation, resulting in altered DNA binding activity in vitro and changes in plastid gene expression patterns in vivo. Furthermore, based on the observation that atpB and psbA gene expression is differentially affected (Figure 5), the results obtained with the Arabidopsis AtSig6 mutants suggest apparent promoter specificity of the phosphorylation control.
As depicted in the model shown in Figure 6(a) for atpB transcription, AtSIG6 is able to confer productive binding to, and initiation from, the PEP promoter only in its phosphorylated state. The most critical phosphorylation sites seem to be those at S94/95 and S174, as inferred from the data indicating that mutational changes at these sites lead to dramatic alterations in phenotype (Figure 4) and plastid gene expression at the atpB PEP promoter in vivo (Figure 5). Interestingly, none of the analysed mutant lines show any appreciable effect on psbA gene expression in vivo (Figure 5). Furthermore, EMSA control experiments using the psbA promoter (Figure S5) did not reveal differences in binding activity of the phosphorylated versus unphosphorylated forms of AtSIG6. Together, this could mean that binding and initiation at the psbA promoter (Figure 6b) by AtSIG6 may be regulated differently compared with the atpB promoter (Figure 6a).
Figure 6.
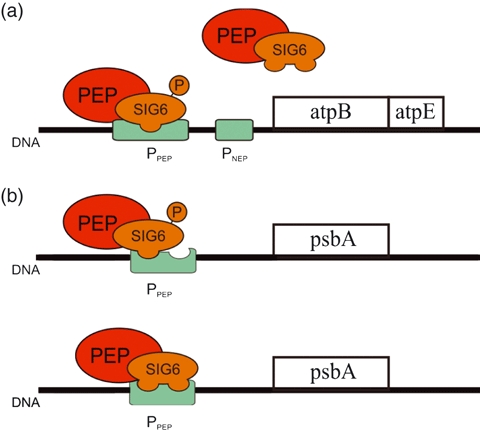
Model illustrating regulation by AtSIG6 phosphorylation at two different chloroplast promoters.(a) Transcription of the atpB/E operon is driven both from an NEP and a PEP promoter. The latter is recognized by the PEP/SIG6 complex, yet only if the sigma factor is phosphorylated at defined regulatory site(s).(b) In contrast, there is no apparent effect of SIG6 phosphorylation state on transcription at the psbA PEP promoter. The more complex architecture of this promoter (Figure S4 could be part of a scenario in which SIG6 is capable of binding at various phosphorylation sites, perhaps even further complicated by functional overlap with other sigma factors.Colour code: green, promoters; red, PEP; orange, AtSIG6.
This is likely to reflect differences in promoter architecture, including the presence of the TATA-box-like element and extended –10 element (Browning and Busby, 2004) within the psbA but not atpB promoter (for a review, see e.g. Liere and Maliga, 2001; Swiatecka-Hagenbruch et al., 2007). It could thus be envisaged that more than one choice exists for the efficient three-dimensional arrangement of the AtSIG6-PEP initiation complex at the psbA promoter. Furthermore, the psbA promoter is known to be recognized by more than one member of the sigma factor family (Hanaoka et al., 2003; Privat et al., 2003). Although psbA transcript levels are downregulated in AtSig2, AtSig5 and AtSig6 knock-out mutants, at least during certain developmental stages, the mRNA is never completely absent, which is consistent with the view that multiple sigma factors can initiate transcription from this promoter (Figure S4). In contrast, at the atpB promoter AtSIG6 does not seem to act in a redundant overlapping manner with other sigma factors (Schweer et al., 2006), and thus mutational defects would be readily detectable in gene expression patterns from the atpB/E transcription unit (Schweer et al., 2009).
Our present work establishes that only some of the putative cpCK2 phosphorylation sites on AtSIG6 are indeed utilized for regulation in vitro and/or in vivo. These include the two clusters of closely spaced sites S94/S95 and S174, S177 and S180.
The putative S94 and S95 sites each had high prediction scores with most of the programs used (Table 1). The positions n + 3 and n + 4, in the case of S94, and n + 2 through n + 4, in the case of S95, represent acidic residues in accordance with the CK2 consensus substrate site (Pinna, 1990). The exchange of Ser94 or Ser95 with alanine in each case affects the central phosphoacceptor residue of the consensus substrate site, which results in dramatic losses of the 2.6-kb atpB transcript originating from the atpB PEP promoter (Figure 5). Interestingly, however, both phosphorylation site mutants have a fully green phenotype resembling that of the wild type. Only if both Ser94 and Ser95 are mutated simultaneously (S94/S95 double mutant) is a chlorophyll-defective phenotype (Figure 4), as well as complete absence of the 2.6-kb atpB transcript (Figure 5), observed.
Consistent with these in vivo data, the EMSA experiments (Figure 3b) demonstrate that upon conversion of both Ser94 and Ser95 to Ala, the resulting mutant factor is no longer able to confer promoter-specific DNA binding in a phosphorylation-dependent manner in vitro. It is likely that Ser94 and Ser95 can both be used as phosphoacceptor residues in Arabidopsis, which may be an evolutionary safeguard to help warrant phosphorylation in this critical region of AtSIG6. Furthermore, it cannot be excluded that S94 and S95 are used alternatively during different developmental stages, and in different environmental situations. This idea is supported by the findings that the S94-Ala line showed highly diminished but still detectable levels of the 2.6-kb PEP transcript at 14 days, with a complete absence at 7 days, whereas the converse was true for S95-Ala (Figure 5).
The second region with clustered motifs for CK2 phosphorylation sites spans five serine residues between position 174 and 180 (Figure 1). Among these, only Ser174 and Ser177, but not Ser176 and Ser180, showed significant phenotypic effects in vivo upon conversion to other residues (Figures 4, 5 and S3). In addition, the Ser residues at 174 and 180, but not 176, appeared to be critical for phosphorylation-dependent DNA-binding activity in vitro (Figure 3b). It is notable that S174 is a ‘non-consensus’ CK2 substrate site because of the absence of an acidic (Asp or Glu) residue at position n + 3 (Pinna, 1990). However, this could be overcome by phosphorylation of the Ser at this position (Roach, 1991), which is a situation not rarely encountered in CK2 sites in other systems (Meggio and Pinna, 2003). Pre-phosphorylation of the n + 3 serine can be considered as a mechanism that ‘opens’ the CK2 site for phosphorylation in a temporal and/or spatial context of a kinase cascade. It should be noted, however, that a mimic phosphorylation experiment by Ser → Asp exchange at position S177 led to a chlorophyll-defective phenotype (Figure 4), and to a loss of the 2.6-kb atpB PEP transcript (Figure 5). Thus, the conclusion is that an ‘always on’ state at this position affects the SIG6 in vivo activity and the seedling phenotype in a negative, rather than positive, manner.
How could pre-phosphorylation at the n + 3 residue of the S174 site be envisaged? In principle, cpCK2 itself could qualify as a ‘pathfinder’ kinase, taking advantage of the two closely-spaced serine residues Ser177 and Ser180. Phosphorylation at Ser180 (the n + 3 position for Ser177) would ‘open’ Ser177, which can then serve as the n + 3 position for Ser174. This explanation, which is solely based on our current knowledge of consensus CK2 substrate sites, is fully consistent with the data obtained in the in vitro DNA binding assays (Figure 3b). It is also consistent with most of the in vivo RNA data (Figure 5), except those for line S180-Ala. As the latter is defective at Ser180, cpCK2 should not be able to ‘open’ Ser177, or ultimately Ser174. Nevertheless, the S180-Ala line reveals the 2.6-kb PEP transcript, despite its defective Ser180 site, which could mean that Ser177 is pre-phosphorylated by an alternative kinase in vivo.
Screening for other potential kinases that might be able to recognize Ser177 suggested ribosomal S6 kinase (RSK; group 4.2.6 according to PlantsP, http://plantsp.genomics.purdue.edu/html/families.html) as a possible candidate. In recent localization analyses, a member of the RSK family, PK-like protein (At3g44610), was identified as a plastid kinase (Kleffmann et al., 2004; Schliebner et al., 2008). Another kinase with reasonable score is CK1, a nucleocytosolic enzyme with functions in intracellular protein targeting (Marin et al., 2003). It is known that cytosolic phosphorylation can have an impact on chloroplast import (Martin et al., 2006) and transcription (Christopher et al., 1997). It would be interesting to investigate Arabidopsis kinase mutants to further test a regulatory role on sigma factor pre-phosphorylation.
PSIPRED (http://bioinf.cs.ucl.ac.uk/psipred; Jones, 1999) was used to distinguish between phosphorylation and alterations in secondary structure as a result of amino acid exchange. This tool suggested that there were no conformational differences in AtSIG6 compared with the site-directed mutant versions studied here. Furthermore, we experimentally addressed this question by the exchange of Ser for more than one alternative residue. For instance, as is evident from Figures 4 and S3 (third row), both Gln and Ala gave comparable results in the case of the S174 phosphorylation site.
The large-scale involvement of cpCK2 in generating the chloroplast phosphoproteome has recently been highlighted (Reiland et al., 2009). Of the almost 200 detectable chloroplast phosphoproteins, many seem to reflect CK2 activity. These results underline a central role of cpCK2 in controlling the crosstalk between chloroplast gene expression and general metabolism. Other reports have previously pinpointed the role of protein kinase(s) in the regulation and maintenance of the chloroplast transcription apparatus (Kim et al., 1999; Baena-Gonzalez et al., 2001; Jeong et al., 2004; Puthiyaveetil et al., 2008; Steiner et al., 2009). The critical role of nucleocytosolic CK2 in plant development and function has recently been established by using dominant-negative mutant lines (Moreno-Romero et al., 2008). Chloroplast protein kinases with key functions in organellar signalling have been characterized using molecular genetic techniques (Depege et al., 2003; Bonardi et al., 2005; Vainonen et al., 2005), whereas this is yet to be achieved for cpCK2. By addressing the question of how cpCK2 phosphorylation controls the activity of plastid sigma factors, our data reported here can help gain further insight into the role of this important signalling kinase in plants.
Experimental procedures
Plant growth conditions, harvesting and RNA extraction
Arabidopsis thaliana knock-out line sig6-2 was obtained from the GABI-Kat mutant collection at the Max-Planck Institute fuer Zuechtungsforschung (GABI-Kat identifier 242G06; http://www.gabi-kat.de). It has a single T-DNA insertion in exon 5 of the AtSig6 gene (Rosso et al., 2003; Loschelder et al., 2006). Wild-type, sig6-2 and retransformed sig6-2 seedlings (all A. thaliana ecotype Columbia) were grown on MS agar medium containing 0.4% (w/v) Gelrite and 1% (w/v) sucrose (Sigma-Aldrich, http://www.sigmaaldrich.com). Plates were maintained at 24°C under 8-h short-day conditions at a photofluence rate of 60 μmol m−2 sec−1. Cotyledons (day 7) and leaves (day 14) were collected, frozen and powdered in liquid nitrogen. Total RNA was prepared as decribed by Chomczynski and Sacchi (1987).
Re-transformation of the sig6-2 knock-out line
The full-length AtSIG6 cDNA including the transit peptide was mutagenized by using the QuikChange II site-directed mutagenesis kit (Stratagene, http://www.stratagene.com) and the oligonucleotides listed in Table S1. Products were cloned behind the CaMV 35S promoter of the binary vector pBINAR (Höfgen and Willmitzer, 1990). Each 35S promoter::cDNA construct was introduced into Rhizobium radiobacter (Agrobacterium tumefaciens) strain GV3101, and then transformed into the sig6-2 line by floral dip (Clough and Bent, 1998). Complemented T1 plants were selected by kanamycin resistance, followed by Southern blot analysis and PCR with primers for the resistance gene (npt1 and npt2) as well as those from within the AtSig6 coding region (UKSIG6-RP and UKSIG6-LP) (Loschelder et al., 2006). For each mutation, at least three independent lines were maintained and re-tested twice, except for the lethal double-mutant S94/95-Ala (six independent lines were tested once).
Northern blot analysis
Gene-specific RNA probes (Loschelder et al., 2006) were obtained by in vitro transcription of DNA regions cloned in pGEM-T Easy (Promega, http://www.promega.com). The linearized plasmids were transcribed by SP6 or T7 RNA polymerase in the presence of DIG-11-UTP (Roche, http://www.roche.com). Plant total RNA (1 μg) was gel-fractionated, transferred to positively charged nylon membrane (Roche) and hybridized with the DIG-labelled probe, followed by chemiluminescence detection (Schweer et al., 2006).
Recombinant proteins and phosphorylation
AtSIG6 cDNA or mutagenized derivatives thereof were amplified using the forward primer Sig6-TP (Table S1), which prevents the synthesis of the transit peptide region. PCR products were each cloned into pMAL-c2x (NEB, http://www.neb.com), and recombinant proteins were purified on amylose resin according to the pMAL manual. Purified proteins (15 μg) were phosphorylated in 50-μl reactions containing 20 mm Tris-HCl, pH 7.5, 50 mm KCl, 10 mm MgCl2, 0.1 mm ATP und 15 μg cpCK2 at 30°C for 30 min, as described by Ogrzewalla et al. (2002). Mock phosphorylation was carried out under identical conditions, without ATP. The recombinant kinase had less than 1/10th of the activity of a native commercial CK2 preparation (NEB), but revealed the typical CK2-type features, including ATP/GTP usage, acidic substrate preference and heparin sensitivity (Baginsky et al., 1997, 1999; Ogrzewalla et al., 2002). SIG6 and its mutant derivatives are cpCK2 substrates in vitro (H. Türkeri and G. Link, unpublished data).
EMSA
EMSA reactions (Fried and Crothers, 1981; Garner and Revzin, 1981) containing 30 pmol sigma protein, 5.7 pmol E. coli core RNA polymerase (Epicentre, http://www.epicentre.com), 5 ng 32P-labelled double-stranded probe (Figure 3a) and 3 μg poly[d(I-C)] were incubated in 50 μl binding buffer at 25°C for 15 min (Homann and Link, 2003). The probe was prepared by PCR-based cloning of a 424-bp fragment (positions 54 534–54 958 on chloroplast DNA circle; AP000423) that carries the Arabidopsis atpB PEP promoter, followed by cloning into pGEM T-Easy (Promega). After cutting out the fragment with BamHI/SalI (Promega), it was electrophoretically purified, eluted, and end-labelled using [γ-32P]ATP and polynucleotide kinase (NEB). The same fragment, but unlabelled, was used as a specific competitor. Bam0.5, a 500-bp DNA fragment from within the trnK intron (Homann and Link, 2003), served as a non-specific competitor. Competitors were used at up to 20-fold molar excess, and were added prior to the labelled probe. DNA–protein complex formation was analysed in triplicate on native 4% (w/v) polyacrylamide gels (37.5:1 acrylamide:bisacrylamide) in TBE buffer (50 mm Tris, 45 mm boric acid, 0.5 mm EDTA, pH 8.3). After drying, gels were analysed using an FLA-3000 phosphoimager (Fuji, http://www.fujifilm.com).
Acknowledgments
We would like to thank Professor B. Weisshaar, University of Bielefeld, and the GABI-Kat team at the Max-Planck-Institute fuer Zuechtungsforschung, Cologne, for the generous supply of the sig6-2 mutant line. This work was funded by the Deutsche Forschungsgemeinschaft (SFB 480/B7).
Supporting Information
Additional Supporting Information may be found in the online version of this article:
Figure S1. Alignment of conserved sigma factor subregion 1.2 in various plant species, showing the distribution of putative CK2 substrate motifs.
Figure S2. Alignment of the six Arabidopsis sigma factors reveals the presence or absence of homologous phosphorylation sites in CR and UCR, respectively.
Figure S3. Phenotype of 7-day-old seedlings from Arabidopsis wild type and SIG6 mutant lines.
Figure S4. Differences in chloroplast psbA and atpB PEP promoter architecture.
Figure S5. EMSA with AtSIG6 and its mutant derivatives using the psbA promoter.
Table S1. Oligonucleotides used for site-directed mutagenesis and cloning of AtSIG6 cDNA constructs as well as the EMSA probe fragment carrying the atpB PEP promoter.
Please note: As a service to our authors and readers, this journal provides supporting information supplied by the authors. Such materials are peer-reviewed and may be re-organized for online delivery, but are not copy-edited or typeset. Technical support issues arising from supporting information (other than missing files) should be addressed to the authors.
References
- Baena-Gonzalez E, Baginsky S, Mulo P, Summer H, Aro EM, Link G. Chloroplast transcription at different light intensities. Glutathione-mediated phosphorylation of the major RNA polymerase involved in redox-regulated organellar gene expression. Plant Physiol. 2001;127(3):1044–1052. doi: 10.1104/pp.010168. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Baginsky S, Tiller K, Link G. Transcription factor phosphorylation by a protein kinase associated with chloroplast RNA polymerase from mustard (Sinapis alba) Plant Mol. Biol. 1997;34(2):181–189. doi: 10.1023/a:1005802909902. [DOI] [PubMed] [Google Scholar]
- Baginsky S, Tiller K, Pfannschmidt T, Link G. PTK, the chloroplast RNA polymerase-associated protein kinase from mustard (Sinapis alba), mediates redox control of plastid in vitro transcription. Plant Mol. Biol. 1999;39(5):1013–1023. doi: 10.1023/a:1006177807844. [DOI] [PubMed] [Google Scholar]
- Baldwin NE, Dombroski AJ. Isolation and characterization of mutations in region 1.2 of Escherichia coli sigma70. Mol. Microbiol. 2001;42(2):427–437. doi: 10.1046/j.1365-2958.2001.02642.x. [DOI] [PubMed] [Google Scholar]
- Beardslee TA, Roy-Chowdhury S, Jaiswal P, Buhot L, Lerbs-Mache S, Stern DB, Allison LA. A nucleus-encoded maize protein with sigma factor activity accumulates in mitochondria and chloroplasts. Plant J. 2002;31(2):199–209. doi: 10.1046/j.1365-313x.2002.01344.x. [DOI] [PubMed] [Google Scholar]
- Blom N, Gammeltoft S, Brunak S. Sequence and structure-based prediction of eukaryotic protein phosphorylation sites. J. Mol. Biol. 1999;294(5):1351–1362. doi: 10.1006/jmbi.1999.3310. [DOI] [PubMed] [Google Scholar]
- Blom N, Sicheritz-Ponten T, Gupta R, Gammeltoft S, Brunak S. Prediction of post-translational glycosylation and phosphorylation of proteins from the amino acid sequence. Proteomics. 2004;4(6):1633–1649. doi: 10.1002/pmic.200300771. [DOI] [PubMed] [Google Scholar]
- Bonardi V, Pesaresi P, Becker T, Schleiff E, Wagner R, Pfannschmidt T, Jahns P, Leister D. Photosystem II core phosphorylation and photosynthetic acclimation require two different protein kinases. Nature. 2005;437(7062):1179–1182. doi: 10.1038/nature04016. [DOI] [PubMed] [Google Scholar]
- Browning DF, Busby SJ. The regulation of bacterial transcription initiation. Nat. Rev. Microbiol. 2004;2(1):57–65. doi: 10.1038/nrmicro787. [DOI] [PubMed] [Google Scholar]
- Burgess RR, Anthony L. How sigma docks to RNA polymerase and what sigma does. Curr. Opin. Microbiol. 2001;4(2):126–131. doi: 10.1016/s1369-5274(00)00177-6. [DOI] [PubMed] [Google Scholar]
- Campbell EA, Muzzin O, Chlenov M, Sun JL, Olson CA, Weinman O, Trester-Zedlitz ML, Darst SA. Structure of the bacterial RNA polymerase promoter specificity sigma subunit. Mol. Cell. 2002;9(3):527–539. doi: 10.1016/s1097-2765(02)00470-7. [DOI] [PubMed] [Google Scholar]
- Carter ML, Smith AC, Kobayashi H, Purton S, Herrin DL. Structure, circadian regulation and bioinformatic analysis of the unique sigma factor gene in Chlamydomonas reinhardtii. Photosynth. Res. 2004;82(3):339–349. doi: 10.1007/s11120-004-4213-6. [DOI] [PubMed] [Google Scholar]
- Chomczynski P, Sacchi N. Single-step method of RNA isolation by acid guanidinium thiocyanate-phenol-chloroform extraction. Anal. Biochem. 1987;162(1):156–159. doi: 10.1006/abio.1987.9999. [DOI] [PubMed] [Google Scholar]
- Christopher DA, Li X, Kim M, Mullet JE. Involvement of protein kinase and extraplastidic serine/threonine protein phosphatases in signalling pathways regulating plastid transcription and the psbD blue light-responsive promoter in barley. Plant Physiol. 1997;113(4):1273–1282. doi: 10.1104/pp.113.4.1273. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Clough SJ, Bent AF. Floral dip: a simplified method for Agrobacterium-mediated transformation of Arabidopsis thaliana. Plant J. 1998;16(6):735–743. doi: 10.1046/j.1365-313x.1998.00343.x. [DOI] [PubMed] [Google Scholar]
- Coll NS, Danon A, Meurer J, Cho WK, Apel K. Characterization of soldat8, a suppressor of singlet oxygen-induced cell death in Arabidopsis seedlings. Plant Cell Physiol. 2009;50(4):707–718. doi: 10.1093/pcp/pcp036. [DOI] [PubMed] [Google Scholar]
- Depege N, Bellafiore S, Rochaix JD. Role of chloroplast protein kinase Stt7 in LHCII phosphorylation and state transition in Chlamydomonas. Science. 2003;299(5612):1572–1575. doi: 10.1126/science.1081397. [DOI] [PubMed] [Google Scholar]
- Diella F, Cameron S, Gemund C, Linding R, Via A, Kuster B, Sicheritz-Ponten T, Blom N, Gibson TJ. Phospho.ELM: a database of experimentally verified phosphorylation sites in eukaryotic proteins. BMC Bioinformatics. 2004;5:79. doi: 10.1186/1471-2105-5-79. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fried M, Crothers DM. Equilibria and kinetics of lac repressor-operator interactions by polyacrylamide gel electrophoresis. Nucleic Acids Res. 1981;9(23):6505–6525. doi: 10.1093/nar/9.23.6505. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fujiwara M, Nagashima A, Kanamaru K, Tanaka K, Takahashi H. Three new nuclear genes, sigD, sigE and sigF, encoding putative plastid RNA polymerase sigma factors in Aarabidopsis thaliana. FEBS Lett. 2000;481(1):47–52. doi: 10.1016/s0014-5793(00)01965-7. [DOI] [PubMed] [Google Scholar]
- Garner MM, Revzin A. A gel electrophoresis method for quantifying the binding of proteins to specific DNA regions: application to components of the Escherichia coli lactose operon regulatory system. Nucleic Acids Res. 1981;9(13):3047–3060. doi: 10.1093/nar/9.13.3047. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gross CA, Chan C, Dombroski A, Gruber T, Sharp M, Tupy J, Young B. The functional and regulatory roles of sigma factors in transcription. Cold Spring Harb. Symp. Quant. Biol. 1998;63:141–155. doi: 10.1101/sqb.1998.63.141. [DOI] [PubMed] [Google Scholar]
- Gruber TM, Gross CA. Multiple sigma subunits and the partitioning of bacterial transcription space. Annu. Rev. Microbiol. 2003;57:441–466. doi: 10.1146/annurev.micro.57.030502.090913. [DOI] [PubMed] [Google Scholar]
- Hakimi MA, Privat I, Valay JG, Lerbs-Mache S. Evolutionary conservation of C-terminal domains of primary sigma(70)-type transcription factors between plants and bacteria. J. Biol. Chem. 2000;275(13):9215–9221. doi: 10.1074/jbc.275.13.9215. [DOI] [PubMed] [Google Scholar]
- Hanaoka M, Kanamaru K, Takahashi H, Tanaka K. Molecular genetic analysis of chloroplast gene promoters dependent on SIG2, a nucleus-encoded sigma factor for the plastid-encoded RNA polymerase, in Arabidopsis thaliana. Nucleic Acids Res. 2003;31(24):7090–7098. doi: 10.1093/nar/gkg935. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hara K, Morita M, Takahashi R, Sugita M, Kato S, Aoki S. Characterization of two genes, Sig1 and Sig2, encoding distinct plastid sigma factors in the moss Physcomitrella patens: phylogenetic relationships to plastid sigma factors in higher plants. FEBS Lett. 2001;499(1–2):87–91. doi: 10.1016/s0014-5793(01)02530-3. [DOI] [PubMed] [Google Scholar]
- Helmann JD. RNA polymerase: a nexus of gene regulation. Methods. 2009;47(1):1–5. doi: 10.1016/j.ymeth.2008.12.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hess WR, Börner T. Organellar RNA polymerases of higher plants. Int. Rev. Cytol. 1999;190:1–59. doi: 10.1016/s0074-7696(08)62145-2. [DOI] [PubMed] [Google Scholar]
- Höfgen R, Willmitzer L. Biochemical and genetic analysis of different patatin isoforms expressed in various organs of potato (Solanum tuberosum) Plant Sci. 1990;66:221–230. [Google Scholar]
- Homann A, Link G. DNA-binding and transcription characteristics of three cloned sigma factors from mustard (Sinapis alba L.) suggest overlapping and distinct roles in plastid gene expression. Eur. J. Biochem. 2003;270(6):1288–1300. doi: 10.1046/j.1432-1033.2003.03494.x. [DOI] [PubMed] [Google Scholar]
- Ishizaki Y, Tsunoyama Y, Hatano K, Ando K, Kato K, Shinmyo A, Kobori M, Takeba G, Nakahira Y, Shiina T. A nuclear-encoded sigma factor, Arabidopsis SIG6, recognizes sigma-70 type chloroplast promoters and regulates early chloroplast development in cotyledons. Plant J. 2005;42(2):133–144. doi: 10.1111/j.1365-313X.2005.02362.x. [DOI] [PubMed] [Google Scholar]
- Isono K, Shimizu M, Yoshimoto K, Niwa Y, Satoh K, Yokota A, Kobayashi H. Leaf-specifically expressed genes for polypeptides destined for chloroplasts with domains of sigma70 factors of bacterial RNA polymerases in Arabidopsis thaliana. Proc. Natl Acad. Sci. USA. 1997;94(26):14948–14953. doi: 10.1073/pnas.94.26.14948. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jeong SY, Peffer N, Meier I. Phosphorylation by protein kinase CKII modulates the DNA-binding activity of a chloroplast nucleoid-associated protein. Planta. 2004;219(2):298–302. doi: 10.1007/s00425-004-1215-8. [DOI] [PubMed] [Google Scholar]
- Jones DT. Protein secondary structure prediction based on position-specific scoring matrices. J. Mol. Biol. 1999;292(2):195–202. doi: 10.1006/jmbi.1999.3091. [DOI] [PubMed] [Google Scholar]
- Kanamaru K, Tanaka K. Roles of chloroplast RNA polymerase sigma factors in chloroplast development and stress response in higher plants. Biosci. Biotechnol. Biochem. 2004;68(11):2215–2223. doi: 10.1271/bbb.68.2215. [DOI] [PubMed] [Google Scholar]
- Kim M, Christopher DA, Mullet JE. ADP-Dependent phosphorylation regulates association of a DNA-binding complex with the barley chloroplast psbD blue-light-responsive promoter. Plant Physiol. 1999;119(2):663–670. doi: 10.1104/pp.119.2.663. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kim JH, Lee J, Oh B, Kimm K, Koh I. Prediction of phosphorylation sites using SVMs. Bioinformatics. 2004;20(17):3179–3184. doi: 10.1093/bioinformatics/bth382. [DOI] [PubMed] [Google Scholar]
- Kleffmann T, Russenberger D, von ZA, Christopher W, Sjolander K, Gruissem W, Baginsky S. The Arabidopsis thaliana chloroplast proteome reveals pathway abundance and novel protein functions. Curr. Biol. 2004;14(5):354–362. doi: 10.1016/j.cub.2004.02.039. [DOI] [PubMed] [Google Scholar]
- Kubota Y, Miyao A, Hirochika H, Tozawa Y, Yasuda H, Tsunoyama Y, Niwa Y, Imamura S, Shirai M, Asayama M. Two novel nuclear genes, OsSIG5 and OsSIG6, encoding potential plastid sigma factors of RNA polymerase in rice: tissue-specific and light-responsive gene expression. Plant Cell Physiol. 2007;48(1):186–192. doi: 10.1093/pcp/pcl050. [DOI] [PubMed] [Google Scholar]
- Liere K, Maliga P. Plastid RNA polymerases in higher plants. In: Aro E-M, Andersson B, editors. Regulation of Photosynthesis. Dordrecht, The Netherlands: Kluwer Academic Publishers; 2001. pp. 29–49. [Google Scholar]
- Loschelder H, Homann A, Ogrzewalla K, Link G. Proteomics-based sequence analysis of plant gene expression – the chloroplast transcription apparatus. Phytochemistry. 2004;65(12):1785–1793. doi: 10.1016/j.phytochem.2004.04.034. [DOI] [PubMed] [Google Scholar]
- Loschelder H, Schweer J, Link B, Link G. Dual temporal role of plastid sigma factor 6 in Arabidopsis development. Plant Physiol. 2006;142(2):642–650. doi: 10.1104/pp.106.085878. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lysenko EA. Plant sigma factors and their role in plastid transcription. Plant Cell Rep. 2007;26(7):845–859. doi: 10.1007/s00299-007-0318-7. [DOI] [PubMed] [Google Scholar]
- Maliga P. Two plastid RNA polymerases of higher plants: an evolving story. Trends Plant Sci. 1998;3(1):4–6. [Google Scholar]
- Marin O, Bustos VH, Cesaro L, Meggio F, Pagano MA, Antonelli M, Allende CC, Pinna LA, Allende JE. A noncanonical sequence phosphorylated by casein kinase 1 in beta-catenin may play a role in casein kinase 1 targeting of important signaling proteins. Proc. Natl Acad. Sci. USA. 2003;100(18):10193–10200. doi: 10.1073/pnas.1733909100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Martin T, Sharma R, Sippel C, Waegemann K, Soll J, Vothknecht UC. A protein kinase family in Arabidopsis phosphorylates chloroplast precursor proteins. J. Biol. Chem. 2006;281(52):40216–40223. doi: 10.1074/jbc.M606580200. [DOI] [PubMed] [Google Scholar]
- Meggio F, Pinna LA. One-thousand-and-one substrates of protein kinase CK2? FASEB J. 2003;17(3):349–368. doi: 10.1096/fj.02-0473rev. [DOI] [PubMed] [Google Scholar]
- Moreno-Romero J, Espunya MC, Platara M, Arino J, Martinez MC. A role for protein kinase CK2 in plant development: evidence obtained using a dominant-negative mutant. Plant J. 2008;55(1):118–130. doi: 10.1111/j.1365-313X.2008.03494.x. [DOI] [PubMed] [Google Scholar]
- Murakami KS, Masuda S, Campbell EA, Muzzin O, Darst SA. Structural basis of transcription initiation: an RNA polymerase holoenzyme-DNA complex. Science. 2002;296(5571):1285–1290. doi: 10.1126/science.1069595. [DOI] [PubMed] [Google Scholar]
- Obenauer JC, Cantley LC, Yaffe MB. Scansite 2.0: proteome-wide prediction of cell signaling interactions using short sequence motifs. Nucleic Acids Res. 2003;31(13):3635–3641. doi: 10.1093/nar/gkg584. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ogrzewalla K, Piotrowski M, Reinbothe S, Link G. The plastid transcription kinase from mustard (Sinapis alba L.). A nuclear-encoded CK2-type chloroplast enzyme with redox-sensitive function. Eur. J. Biochem. 2002;269(13):3329–3337. [PubMed] [Google Scholar]
- Pfalz J, Liere K, Kandlbinder A, Dietz KJ, Oelmüller R. pTAC2, -6, and -12 are components of the transcriptionally active plastid chromosome that are required for plastid gene expression. Plant Cell. 2006;18(1):176–197. doi: 10.1105/tpc.105.036392. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pfannschmidt T, Ogrzewalla K, Baginsky S, Sickmann A, Meyer HE, Link G. The multisubunit chloroplast RNA polymerase A from mustard (Sinapis alba L.). Integration of a prokaryotic core into a larger complex with organelle-specific functions. Eur. J. Biochem. 2000;267(1):253–261. doi: 10.1046/j.1432-1327.2000.00991.x. [DOI] [PubMed] [Google Scholar]
- Pinna LA. Casein kinase 2: an ‘eminence grise’ in cellular regulation? Biochim. Biophys. Acta. 1990;1054(3):267–284. doi: 10.1016/0167-4889(90)90098-x. [DOI] [PubMed] [Google Scholar]
- Privat I, Hakimi MA, Buhot L, Favory JJ, Mache-Lerbs S. Characterization of Arabidopsis plastid sigma-like transcription factors SIG1, SIG2 and SIG3. Plant Mol. Biol. 2003;51(3):385–399. doi: 10.1023/a:1022095017355. [DOI] [PubMed] [Google Scholar]
- Puthiyaveetil S, Kavanagh TA, Cain P, Sullivan JA, Newell CA, Gray JC, Robinson C, van der Giezen M, Rogers MB, Allen JF. The ancestral symbiont sensor kinase CSK links photosynthesis with gene expression in chloroplasts. Proc. Natl Acad. Sci. USA. 2008;105(29):10061–10066. doi: 10.1073/pnas.0803928105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Reiland S, Messerli G, Baerenfaller K, Gerrits B, Endler A, Grossmann J, Gruissem W, Baginsky S. Large-scale Arabidopsis phosphoproteome profiling reveals novel chloroplast kinase substrates and phosphorylation networks. Plant Physiol. 2009;150(2):889–903. doi: 10.1104/pp.109.138677. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Roach PJ. Multisite and hierarchal protein phosphorylation. J. Biol. Chem. 1991;266(22):14139–14142. [PubMed] [Google Scholar]
- Rosso MG, Li Y, Strizhov N, Reiss B, Dekker K, Weisshaar B. An Arabidopsis thaliana T-DNA mutagenized population (GABI-Kat) for flanking sequence tag-based reverse genetics. Plant Mol. Biol. 2003;53(1–2):247–259. doi: 10.1023/B:PLAN.0000009297.37235.4a. [DOI] [PubMed] [Google Scholar]
- Salinas P, Fuentes D, Vidal E, Jordana X, Echeverria M, Holuigue L. An extensive survey of CK2 alpha and beta subunits in Arabidopsis: multiple isoforms exhibit differential subcellular localization. Plant Cell Physiol. 2006;47(9):1295–1308. doi: 10.1093/pcp/pcj100. [DOI] [PubMed] [Google Scholar]
- Schliebner I, Pribil M, Zuhlke J, Dietzmann A, Leister D. A survey of chloroplast protein kinases and phosphatases in Arabidopsis thaliana. Curr. Genomics. 2008;9(3):184–190. doi: 10.2174/138920208784340740. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schweer J, Loschelder H, Link G. A promoter switch that can rescue a plant sigma factor mutant. FEBS Lett. 2006;580(28–29):6617–6622. doi: 10.1016/j.febslet.2006.11.010. [DOI] [PubMed] [Google Scholar]
- Schweer J, Geimer S, Meurer J, Link G. Arabidopsis mutants carrying chimeric sigma factor genes reveal regulatory determinants for plastid gene expression. Plant Cell Physiol. 2009;50(7):1382–1386. doi: 10.1093/pcp/pcp069. [DOI] [PubMed] [Google Scholar]
- Shiina T, Tsunoyama Y, Nakahira Y, Khan MS. Plastid RNA polymerases, promoters, and transcription regulators in higher plants. Int. Rev. Cytol. 2005;244:1–68. doi: 10.1016/S0074-7696(05)44001-2. [DOI] [PubMed] [Google Scholar]
- Steiner S, Dietzel L, Schroter Y, Fey V, Wagner R, Pfannschmidt T. The role of phosphorylation in redox regulation of photosynthesis genes psaA and psbA during photosynthetic acclimation of mustard. Mol Plant. 2009;2(3):416–429. doi: 10.1093/mp/ssp007. [DOI] [PubMed] [Google Scholar]
- Suzuki JY, Ytterberg AJ, Beardslee TA, Allison LA, Wijk KJ, Maliga P. Affinity purification of the tobacco plastid RNA polymerase and in vitro reconstitution of the holoenzyme. Plant J. 2004;40(1):164–172. doi: 10.1111/j.1365-313X.2004.02195.x. [DOI] [PubMed] [Google Scholar]
- Swiatecka-Hagenbruch M, Liere K, Borner T. High diversity of plastidial promoters in Arabidopsis thaliana. Mol. Genet. Genomics. 2007;277(6):725–734. doi: 10.1007/s00438-007-0222-4. [DOI] [PubMed] [Google Scholar]
- Tanaka K, Tozawa Y, Mochizuki N, Shinozaki K, Nagatani A, Wakasa K, Takahashi H. Characterization of three cDNA species encoding plastid RNA polymerase sigma factors in Arabidopsis thaliana: evidence for the sigma factor heterogeneity in higher plant plastids. FEBS Lett. 1997;413(2):309–313. doi: 10.1016/s0014-5793(97)00906-x. [DOI] [PubMed] [Google Scholar]
- Tiller K, Link G. Phosphorylation and dephosphorylation affect functional characteristics of chloroplast and etioplast transcription systems from mustard (Sinapis alba L.) EMBO J. 1993;12(5):1745–1753. doi: 10.1002/j.1460-2075.1993.tb05822.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vainonen JP, Hansson M, Vener AV. STN8 protein kinase in Arabidopsis thaliana is specific in phosphorylation of photosystem II core proteins. J. Biol. Chem. 2005;280(39):33679–33686. doi: 10.1074/jbc.M505729200. [DOI] [PubMed] [Google Scholar]
- Wong YH, Lee TY, Liang HK, Huang CM, Wang TY, Yang YH, Chu CH, Huang HD, Ko MT, Hwang JK. KinasePhos 2.0: a web server for identifying protein kinase-specific phosphorylation sites based on sequences and coupling patterns. Nucleic Acids Res. 2007;35(Web Server issue):W588–W594. doi: 10.1093/nar/gkm322. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xue Y, Zhou F, Zhu M, Ahmed K, Chen G, Yao X. GPS: a comprehensive www server for phosphorylation sites prediction. Nucleic Acids Res. 2005;33(Web Server issue):W184–W187. doi: 10.1093/nar/gki393. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zenkin N, Kulbachinskiy A, Yuzenkova Y, Mustaev A, Bass I, Severinov K, Brodolin K. Region 1.2 of the RNA polymerase sigma subunit controls recognition of the -10 promoter element. EMBO J. 2007;26(4):955–964. doi: 10.1038/sj.emboj.7601555. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.


