Abstract
FAN (factor associated with neutral sphingomyelinase [N-SMase] activation) exhibits striking structural homologies to Lyst (lysosomal trafficking regulator), a BEACH protein whose inactivation causes formation of giant lysosomes/Chediak-Higashi syndrome. Here, we show that cells lacking FAN show a statistically significant increase in lysosome size (although less pronounced as Lyst), pointing to previously unrecognized functions of FAN in regulation of the lysosomal compartment. Since FAN regulates activation of N-SMase in complex with receptor for activated C-kinase (RACK)1, a scaffolding protein that recruits and stabilizes activated protein kinase C (PKC) isotypes at cellular membranes, and since an abnormal (calpain-mediated) downregulation/membrane recruitment of PKC has been linked to the defects observed in Lyst-deficient cells, we assessed whether PKC is also of relevance in FAN signaling. Our results demonstrate that activation of PKC is not required for regulation of NSMase by FAN/RACK1. Conversely, activation of PKC and recruitment/stabilization by RACK1 occurs uniformly in the presence or absence of FAN (and equally, Lyst). Furthermore, regulation of lysosome size by FAN is not coupled to an abnormal downregulation/membrane recruitment of PKC by calpain. Identical results were obtained for Lyst, questioning the previously reported relevance of PKC for formation of giant lysosomes and in Chediak-Higashi syndrome. In summary, FAN mediates activation of N-SMase as well as regulation of lysosome size by signaling pathways that operate independent from activation/membrane recruitment of PKC.
Keywords: FAN, N-SMase, TNF, Lyst, RACK1, PKC, lysosome, vesicular trafficking, WD repeat proteins, BEACH proteins
INTRODUCTION
We have previously identified the protein FAN (factor associated with neutral sphingomyelinase activation) as an essential component in the activation of neutral sphingomyelinase (N-SMase, an enzyme that generates the lipid second messenger ceramide from sphingomyelin at the cell membrane and whose physiological role is just beginning to emerge [1, 2]) by the 55 kDa tumor necrosis factor (TNF) receptor (TNF-R55) [3]. Structurally, FAN is characterized by the presence of five C-terminal WD repeats, classifying FAN as a member of the ancient WD (or WD40) repeat protein family [3]. WD repeat proteins contribute to cellular functions ranging from signal transduction to cell cycle control and are conserved across eukaryotes as well as prokaryotes [4]. WD repeats consist of a conserved core of 23-41 amino acids, usually initiated by Gly-His (GH) and terminated by Trp-Asp (WD), and are found between four and 16 times in individual proteins [4]. They can fold into a propeller-like structure that serves as a scaffold for interaction with other proteins [4]. Accordingly, we have demonstrated that the WD repeats of FAN are required for binding to TNF-R55 and for subsequent activation of N-SMase [3]. A second conserved homology domain of FAN, the BEACH (for beige and CHS) domain is located N-terminal of the WD repeats and was originally described in the mouse WD repeat protein Beige (also known as Lyst, lysosomal trafficking regulator) and its human homologue, the CHS protein [5]. The structural architecture shared by FAN and Lyst is also found in an increasing number of other proteins that together constitute the emerging subfamily of BEACH proteins. Many BEACH proteins are very large (> 2000 amino acid residues) and have putative functions in vesicular transport or membrane dynamics, such as neurobeachin, a protein which is essential for neuronal synaptic transmission [6], the LvsA protein, governing membrane processing and cytokinesis in Dictyostelium [7], or LBA, a protein closely related to neurobeachin which has a possible function in polarized vesicle trafficking [8]. Analysis of the crystal structure of the BEACH domain from human neurobeachin revealed that it interacts with a weakly conserved pleckstrin-homology (PH) domain just before the BEACH domain. Functional studies with FAN have furthermore demonstrated that both the PH and the BEACH domains are required for signal transduction from TNF-R55, suggesting that the PH and BEACH domains may function as a single unit [9]. However, the exact molecular function of the BEACH domain is currently unknown. Similarly, the molecular pathways by which BEACH proteins affect vesicle formation and trafficking are largely undefined. Lyst/CHS, the most well characterized member of the BEACH protein family, is inactivated in patients with Chediak-Higashi syndrome, which suffer from hypopigmentation, severe immunological deficiency, neuronal abnormalities and a bleeding tendency [5]. Similar symptoms are seen in the beige mouse, the corresponding mouse model. At the cellular level, the disease is characterized by the occurrence of giant intracellular vesicles and by protein sorting defects into these organelles, most likely due to defects in the vesicular transport to and from the lysosome and late endosome [5, 10]. The molecular basis for this disease are mutations within Lyst/CHS that lead to a truncated protein. It is, however, unknown how Lyst/CHS exerts its normal function and how it links to vesicular transport [5]. Based on a yeast two hybrid screen showing that Lyst/CHS interacts with proteins important in vesicular transport and signal transduction, it was suggested that Lyst/CHS may function as an adapter protein that juxtaposes proteins mediating intracellular membrane fusion reactions [11]. Independently, an enhanced proteolysis of conventional protein kinase C (cPKC) isotypes by the thiol protease calpain, resulting in a disturbed membrane recruitment/activation of cPKC, has been implicated in the defects seen in cells lacking Lyst/CHS [12, 13]. The putative functions of FAN are likewise only marginally understood. Aside from its essential role in N-SMase activation [3, 14] (which is most likely required for the described effects of FAN on cutaneous barrier disruption [14] and apoptosis [15-19]), FAN may exert additional functions in actin reorganization in macrophages [20] and in control of lysosomal permeability [21]. However, the molecular mechanisms by which FAN participates in these functions are currently unknown. In a previous study, we have shown that the adapter protein RACK1 (receptor for activated C-kinase 1) is one of the components in the signaling pathways of FAN. We have demonstrated that RACK1 forms a complex with FAN, and that this interaction modulates the activation of N-SMase by TNF-R55 [22]. Similar to FAN, RACK1 also belongs to the family of WD repeat proteins, carrying seven individual WD repeats. RACK1 is highly conserved from Chlamydomonas to human [23], indicating that its function was established before the evolutionary divergence of plants and animals. RACK1 is a scaffolding protein that is involved in the recruitment, assembly and regulation of multiple different signaling molecules. These molecules interact with several independent protein binding sites located on the individual WD repeats of RACK1 [24], e. g. FAN binds to a region comprising at least parts of WD repeats V to VII of RACK1 [22]. Since RACK1 is also a constituent of the eukaryotic ribosome that regulates translation, it has been speculated that RACK1 may promote the recruitment of ribosomes to cellular sites where translation is required [24]. Originally, RACK1 has been isolated as a “receptor for activated C-kinase 1,” i. e. a protein that binds to activated PKC isotypes, facilitates their translocation to cellular membranes, and simultaneously prevents their premature degradation [25]. We therefore speculated that RACK1 might participate in the cellular functions of FAN by recruiting activated PKC into the vicinity of FAN. A putative role of PKC in the signaling pathways of FAN appeared further likely because of the shared structural architecture of FAN and Lyst (suggesting functional overlaps) in combination with the proposed implication of PKC in the regulation of vesicular transport through Lyst. In this study, we therefore investigated the interconnections between FAN, RACK1, PKC and vesicle formation in more detail.
MATERIALS AND METHODS
Reagents
Antibodies specific for PKCα (sc-208, rabbit IgG), PKCβI (sc-8049, mouse IgG1), PKCβII (sc-210, rabbit IgG) and conventional PKC isotypes (panPKC, sc-17769, mouse IgG2a) were purchased from Santa Cruz, Heidelberg, Germany. The RACK1-specific antibody (610168, mouse IgM) was provided by BD Biosciences, Heidelberg, Germany. Highly purified human recombinant TNF (hTNF) was supplied by BASF Bioresearch, Ludwigshafen, Germany. Phorbol 12-myristate 13-acetate (PMA) and chelerythrine were obtained from Sigma, Deisenhofen, Germany, Gö 6976, (2S, 3S)-trans-epoxyscuccinyl-L-leucylamido-3-methylbutane ethyl ester (E-64d), calpastatin and calpeptin were purchased from Calbiochem GmbH, Bad Soden, Germany.
Cell Culture
Embryonic Fibroblasts (EF) from wildtype or FAN-deficient mice as well as Lyst-deficient MCHSF2, C572CF wildtype control and Big24r (Lyst-deficient fibroblasts expressing a yeast artificial chromosome (YAC) complementing the Lyst-deficiency) fibroblasts have been described previously [14, 26-29]. Cells were maintained in DMEM supplemented with 10 % v/v FCS, 2 mM glutamine and 50 μg/ml each of streptomycin and penicillin in a humidified incubator containing 5 % w/v CO2. For Big24r fibroblasts, 1 mg/ml Geneticin (Invitrogen, Karlsruhe, Germany) was added to the culture medium to maintain the YAC. Primary lung fibroblasts from Lyst-deficient C57BL/6J-bgJ and wildtype C57BL/6 mice (Charles River Laboratories, Sulzfeld, Germany) were generated essentially as described [30], except that cells were cultivated in DMEM supplemented with 20 % v/v FCS. Primary human wildtype (GM08148) and Lyst-deficient (GM02075) fibroblasts were purchased from the Coriell Institute for Medical Research (Camden, NJ, USA) and maintained in DMEM supplemented with 15 % v/v FCS.
Immunofluorescence And Analysis of Lysosome Size
4 × 104 cells were seeded on poly-L-lysine-coated glass coverslips and grown over night at 37 °C. To visualize lysosomes, cells were incubated with 100 nM LysoTracker Red (Invitrogen) for 1 h at 37 °C, washed twice with PBS and fixed in 4% w/v paraformaldehyde for 30 min at room temperature before being analyzed using a Zeiss LSM 510 confocal laserscanning microscope (Zeiss, Oberkochen, Germany). Final digital images were processed by applying the auto-contrast function of Adobe Photoshop CS (Adobe Systems, Mountainview, CA, USA). For quantification of lysosome size, cell division was synchronized by starving the cells in medium containing 0.1% v/v FCS over night followed by recovery in normal medium before further treatment. Micrographs were generated by confocal laserscanning microscopy using constant magnification (400 ×) and stack size. The size of individual lysosomes was quantified from the obtained micrographs using the program ImageJ (Wayne Rasband, National Institutes of Health, Bethesda, MD, USA).
Preparation of Cytosolic and Membrane Fractions
To distinguish membrane-associated and cytosolic PKC, cells were washed twice in ice-cold PBS and lysed in homogenization buffer (20 mM Tris pH 7.5, 250 mM sucrose, 2 mM EDTA, 5 mM EGTA, 2 mM PMSF, 50 mM β-mercaptoethanol) by three rounds of sonication (10 s each; SONIFIER 250, Branson Ultrasonics, Danbury, CT, USA). After centrifugation (100000 × g, 1 h, 4 °C), the supernatant representing the cytosolic fraction was removed. The precipitate was resuspended in homogenization buffer containing 0.5 % v/v Triton X-100 and centrifuged again (40000 × g, 15 min, 4 °C) to obtain the membrane fraction (contained in the supernatant).
Immunoblots
Equal amounts of protein per lane were resolved by electrophoresis on SDS polyacrylamide gels (SDS-PAGE). After electrophoretic transfer to nitrocellulose, reactive proteins were detected using specific antibodies and the ECL detection kit (Amersham Biosciences, Freiburg, Germany).
Immunoprecipitation
Immunoprecipitation was performed over night on ice using 1 μg of antibody (sc-210 or sc-17769) and 500 μg of protein in homogenization buffer containing 0.25 % v/v Nonidet P-40. Immunecomplexes were collected by a 1 hour incubation with γ-bind-sepharose (Amersham Biosciences) and subsequent washing of the immunecomplexes for three times in cold homogenization buffer. The precipitates were resuspended in 25 μl of homogenization buffer and either analyzed in Western blots or assayed for PKCβII activity as described below, except that the ion exchange chromatography step was omitted.
PKC Assays
Cytosolic and membrane fractions were subjected to ion exchange chromatography to separate PKC from other protein kinases essentially as described [31]. Briefly, HiTrap DEAE FF columns (Amersham Biosciences) were equilibrated with a buffer containing 20 mM Tris HCl pH 7.5, 1 mM EDTA, 1 mM EGTA, 2 mM PMSF, 50 mM β-mercaptoethanol, once with 1 M NaCl, and again with equilibration buffer before loading of the samples. The columns were then washed again with equilibration buffer and eluted sequentially with 0.2, 0.4 and 0.6 M NaCl. PKC activity of the obtained fractions was assayed by determining the incorporation of 32P into a PKC specific substrate peptide using the Protein Kinase C Enzyme Biotrack Assay System (Amersham Biosciences) following the recommendations of the manufacturer. 32P-incorporation was quantified in a scintillation counter (Beckman Coulter, Krefeld, Germany). Since initial experiments demonstrated that PKC activity eluted preferentially at 0.2 M NaCl (data not shown), this fraction was routinely used in subsequent experiments.
Assays for Calpain Activity
Calpain activity was measured using the Fluorogenic Calpain Activity Kit (Calbiochem) following the recommendations of the manufacturer. Human calpain 1 (provided with the kit) served as a positive control for the assay. The release of 7-amino-4-methylcoumarin (amc) from the synthetic substrate Suc-LLVY-amc after digestion by calpains was measured as emission at 460 nm upon excitation at 355 nm using a Fluoroskan II fluorimeter equipped with a thermostated plate reader (Labsystems, Egelsbach, Germany).
RESULTS AND DISCUSSION
Impact of FAN on lysosome size
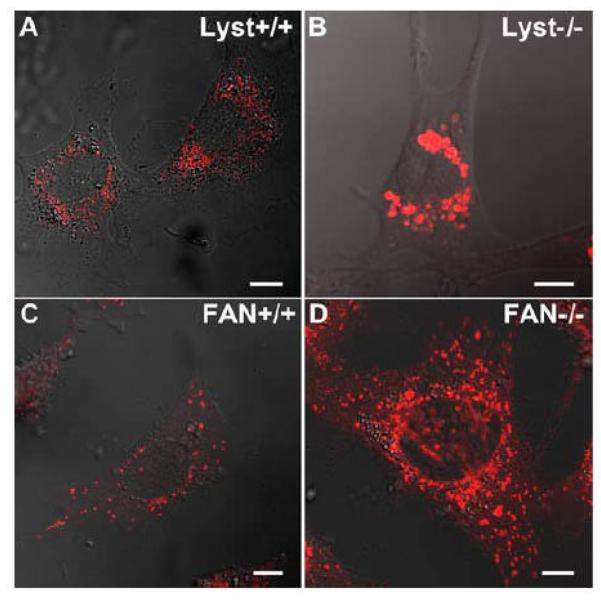
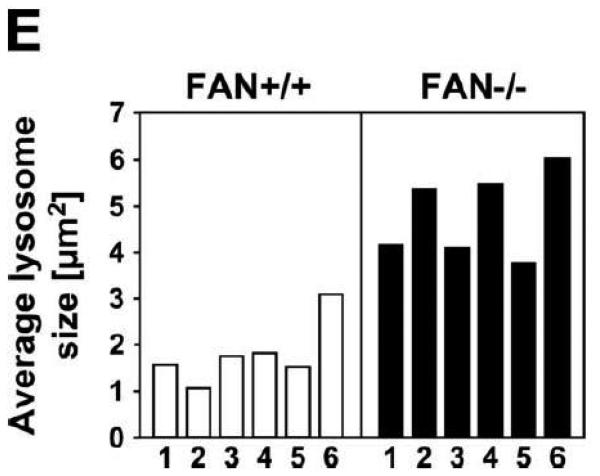
An immediately evident hallmark of Lyst-deficient cells is the presence of giant lysosomes with a clustered perinuclear distribution, whereas wildtype cells display small lysosomes that are dispersed throughout the cytoplasm [27, 32] (Fig. 1A, B). Given the striking structural similarity of FAN and Lyst, we were interested whether loss of FAN might likewise affect the lysosomal compartment. Therefore, FAN+/+ and FAN−/− EF were stained with LysoTracker Red, a red-fluorescent dye that stains acidic compartments in live cells. As shown in Fig. 1C, D, morphological inspection of FAN−/− EF suggested a slightly larger lysosome size when compared to FAN+/+ EF, although the difference was not as prominent as in Lyst−/− vs. Lyst+/+ fibroblasts. To substantiate this impression, we performed a quantitative analysis of lysosome size in FAN+/+ and FAN−/− EF. As it has been described for Lyst-deficient cells that lysosome size may vary within the cell cycle [33], we synchronized FAN+/+ and FAN−/− EF in their cell cycle by serum starvation for these experiments. Despite overlaps in the size of individual lysosomes (e. g. occasional large lysosomes in FAN+/+ EF or smaller lysosomes in FAN−/− EF; Fig. 1C, D), quantification of >5000 lysosomes in four independent experiments indicated that the average lysosome size in FAN−/− EF is consistently above that of FAN+/+ EF. Results from one representative experiment are shown in Fig. 1E, similar results were obtained in the other three experiments (data not shown). Analysis of variance confirmed that the difference in average lysosome size of FAN+/+ and FAN−/− cells was significant (p = 0.000067, 0.00048, 0.0032 and 0.0018 for each of the four experiments). Although we initially suspected that FAN−/− EF contained not only larger, but also more lysosomes than FAN+/+ EF (Fig. 1C, D), we however did not find a dependence on the presence or absence of FAN when we counted lysosome numbers in a larger number of individual FAN+/+ vs. FAN−/− EF cells. Overall, the average number of lysosomes per cell was comparable in FAN+/+ vs. FAN−/− EF (data not shown).
Figure 1.
Lysosome size in FAN- and Lyst-deficient cells and their wildtype counterparts. Lysosomes of Lyst+/+ (A) and Lyst−/− (B) fibroblasts as well as FAN+/+ (C) and FAN−/− EF (D) were stained with LysoTracker Red and visualized by confocal laserscanning microscopy. The results shown are representative for several experiments (n>5). Bar, 10 μm. (E) The average lysosome size of 6 individual FAN+/+ and FAN−/− EF cells was determined as described under “Materials And Methods. One out of four experiments is shown.
Our results implicate that in addition to Lyst, FAN likewise participates in signaling pathways whose disturbance has effects on the regulation of lysosome size (but not lysosome number). In line, other members of the BEACH protein family such as LvsA, LvsB, neurobeachin and LBA have also been implicated in the regulation of vesicular trafficking [6-8, 34]. However, deficiency of FAN does not affect lysosome size as dramatically as loss of Lyst (Fig. 1A-D). Moreover, mice deficient for FAN do not show overt abnormalities, especially no defects indicative for Chediak-Higashi syndrome [14]. Nevertheless, these mice display a delayed kinetics of skin recovery after cutaneous barrier disruption [14], a process that requires membrane remodeling and secretion of vesicles. Therefore, FAN may regulate signaling pathways that either overlap only partially with those of Lyst or which can largely be complemented by Lyst or other members of the BEACH protein family. However, the exact nature of these signaling pathways is still largely enigmatic. As one possibility, Werneburg and coworkers have recently put forward a function of FAN in the regulation of lysosomal permeability [21]. Alternatively, one might speculate that FAN could affect vesicular trafficking through its function as an essential activator of N-SMase [3]. Activation of SMases may have profound effects on the fluidity and even the curvature of the plasma membrane. Zha and coworkers have shown that upon addition of SMase, the surface of affected cells invaginated and formed small vesicles in the absence of any coat proteins [35]. It has been speculated that an involvement, directly or indirectly, in the modulation of local membrane lipid composition may be a common denominator of neurobeachin, Lyst, LvsA, and FAN [36]. Furthermore, it has been proposed that proteins with a structure similar to FAN may participate in membrane remodeling and/or vesicular trafficking by regulating the activity of SMases through their BEACH domain [7]. Therefore, BEACH proteins might control vesicle formation and dynamic remodeling of cell membranes through the regulation of membrane curvature by activation of SMases.
Impact of FAN/RACK1 on TNF-dependent membrane translocation/activation of PKC
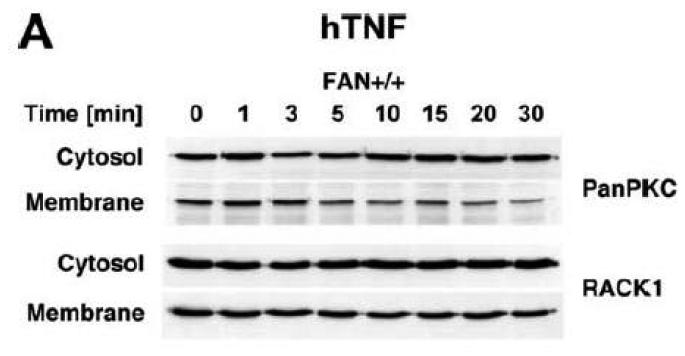
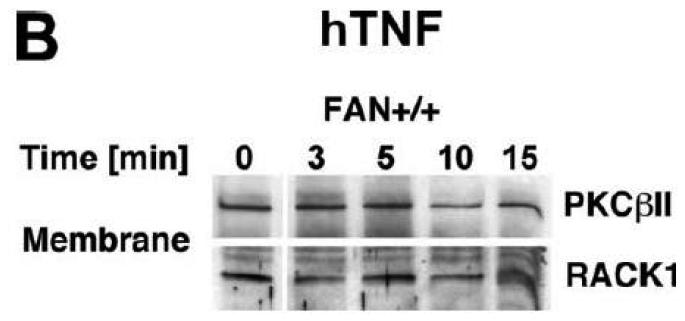
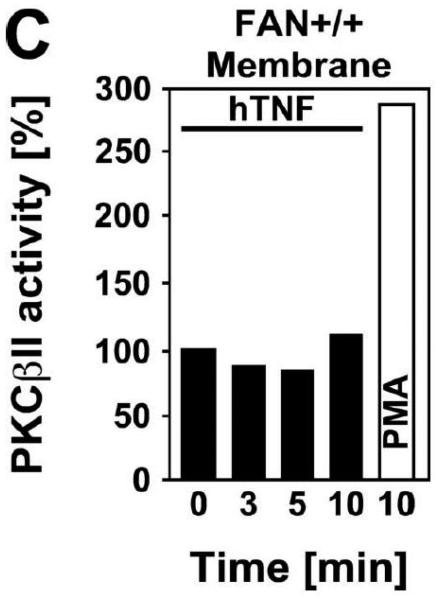
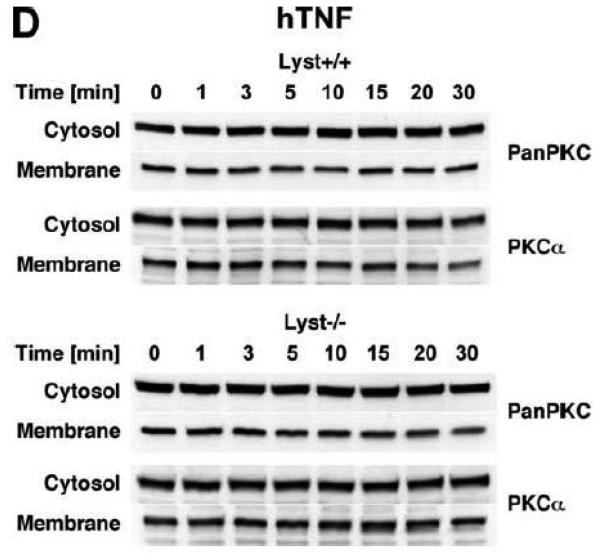
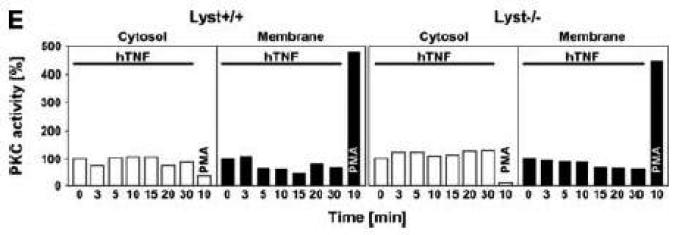
Subsequently, we wanted to determine the role of FAN and RACK1 in TNF-R55-induced activation of PKC. Since TNF can act as a potent inducer of PKC activation and translocation [31], activation of PKC by TNF may depend on formation of a complex consisting of TNFR55, FAN and RACK1. Therefore, we analyzed TNF-induced membrane translocation of PKC in FAN+/+ EF that harbor functional copies of TNF-R55, FAN and RACK1. As shown in Fig. 2A, TNF did not induce a detectable translocation of cPKC isotypes from the cytosolic to the membrane fraction in these cells. Moreover, we did not observe any TNF-induced changes in the distribution of RACK1 between the cytosolic and membrane fraction resulting from cotranslocation with activated PKC (Fig. 2A). Since RACK1 has been described as an anchoring protein with preference for activated PKCβII [37], we immunoprecipitated PKCβII from TNF-stimulated FAN+/+ EF to allow for a more specific detection. Again, as shown in Fig. 2B, neither the amount of immunoprecipitated PKCβII nor the quantity of coimmunoprecipitating RACK1 contained in the membrane fraction did show an increase in response to TNF-treatment. In corresponding measurements of PKCβII activity in the immunoprecipitates, we likewise did not observe an increase in response to TNF (whereas the phorbol ester PMA readily stimulated the activity of PKCβII; Fig. 2C). Similar results were obtained when we measured PKCβII activity in immunoprecipitates from TNF-stimulated FAN−/− EF (data not shown). As an additional control, we examined membrane translocation and activation of PKC in fibroblasts from wildtype (Lyst+/+) and Lyst-deficient (Lyst−/−) mice. Consistent with the observations made in FAN+/+ EF, TNF did not induce translocation of cPKC from the cytosolic to the membrane fraction. Identical results were obtained when we analyzed membrane translocation individually for PKC (Fig. 2D). Furthermore, when we measured cPKC activity directly in Lyst+/+ or Lyst−/− fibroblasts, treatment with TNF did not lead to a relevant increase of activity in the membrane fraction nor to a decrease in the cytosolic fraction (in contrast to stimulation with PMA used as a positive control; Fig. 2E). In addition, TNF did not increase the activity of PKCβII immunoprecipitated from membrane fractions of Lyst+/+ and Lyst−/− fibroblasts (data not shown).
Figure 2.
TNF does not induce membrane translocation or activation of PKC isotypes in fibroblasts. (A) EF from mice harboring a functional FAN protein (FAN+/+) were treated with 100 ng/ml hTNF for the indicated times. Cytosolic and membrane fractions were analyzed for the presence of cPKC isotypes (using an antibody simultaneously recognizing all cPKC isotypes; “panPKC”) or for presence of RACK1. Detection of β-actin was used as a loading control (data not shown). (B) FAN+/+ EF were treated with 50 ng/ml hTNF for the indicated times and PKCβII was immunoprecipitated from the membrane fraction. Subsequently, RACK1 coimmunoprecipitating with PKCβII and (for control of the immunoprecipitation reaction) PKCβII itself were detected by Western blot. (C) PKCβII activity was measured in immunoprecipitates from FAN+/+ EF that were generated as in panel (B). PKCβII activity measured in immunoprecipitates from FAN+/+ EF after treatment with 0.1 μM PMA for 10 min is shown for comparison. One out of two experiments with similar results is shown. (D) Fibroblasts from wildtype (Lyst+/+) and from Lyst-deficient (Lyst−/−) mice were treated and analyzed for membrane translocation of cPKC and PKCα by Western blot as in panel (A). Detection of β-actin was used as a loading control (data not shown). For detection of PKCα, membranes probed with panPKC were reprobed with an antibody specific for PKCα. Prior to reprobing, any residual peroxidase activity was eliminated by incubating the membranes in 15 % v/v H202. Furthermore, the utilized pan PKC and PKCα antibodies were generated in different species, thus ensuring the specificity of detection. (E), Cytosolic and membrane fractions from Lyst+/+ and Lyst−/− fibroblasts were assayed for PKC activity after stimulation with 50 ng/ml hTNF for the indicated times. Activity is shown relative to unstimulated cells. Treatment with 0.1 μM PMA for 10 min is shown for comparison.
Collectively, the above results indicate that in fibroblasts, TNF does not induce membrane translocation or activation of cPKC isotypes. In support of this assumption, Kellerer and coworkers have shown that in rat-1 fibroblasts, TNF induces a rapid translocation of PKCε, while no effect occurred on other isotypes [38]. Likewise, we have previously demonstrated that PKC is activated by TNF in human K562 erythroleukemia and in human leukemic Jurkat T cells, but not in the human fibroblast cell line CCD18 [31], suggesting that PKC activation may play a major role in TNF signal transduction in some, but not all target cells. Therefore, our results suggest that in the examined fibroblast cell lines, the signaling pathways of TNF and PKC are not linked to each other and that the previously reported regulation of N-SMase by FAN and RACK1 in response to TNF [3, 14, 22] occurs without involvement of PKC in these cells.
RACK1 functions as a “receptor for activated C-kinase” in membrane translocation/activation of PKC isotypes independent from a recruitment to FAN or Lyst
Subsequently, we examined whether FAN and RACK1 participate in PKC activation through signaling pathways independent from TNF. The striking structural similarity of FAN to Lyst as well as to other members of the BEACH protein subfamily [39] implicates potential overlaps in the biochemical or cellular functions of these proteins. Of note, a rapid downregulation of phorbol ester-induced PKC activity has been implicated in the manifestation of giant lysosomes in Lyst-deficient cells [12, 13]. This abnormal downregulation of membrane-associated PKC activity may be due to the absence of an anchoring protein that stabilizes activated PKC at the membrane. Since this is exactly the function that is executed by RACK1, we speculated that phorbol ester-induced activation of PKC might require a functional complex of RACK1 and a BEACH protein such as FAN or Lyst for maintaining and stabilizing PKC activity at the cell membrane.
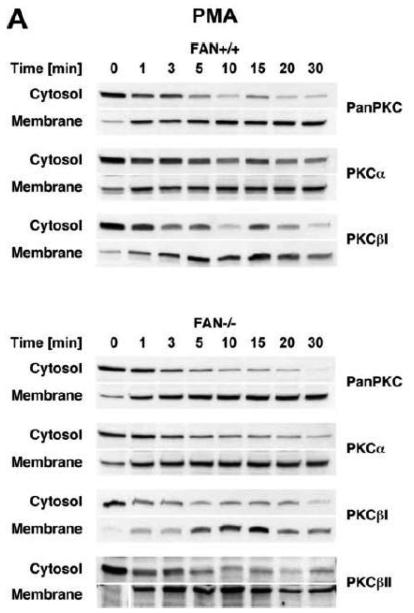
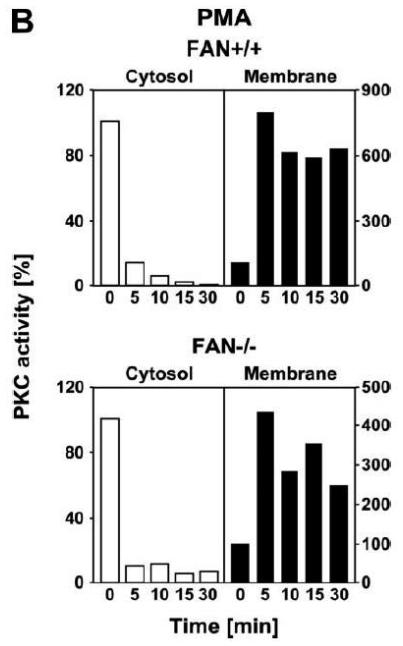
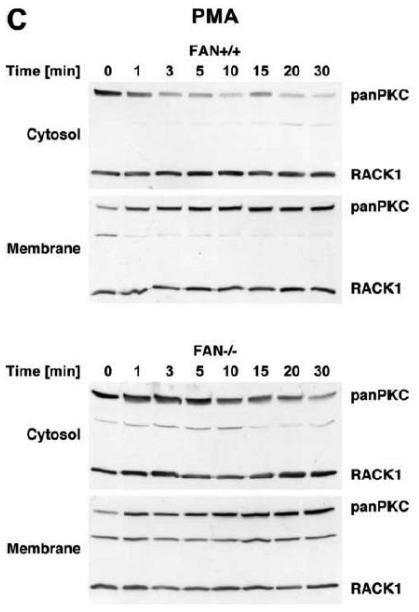
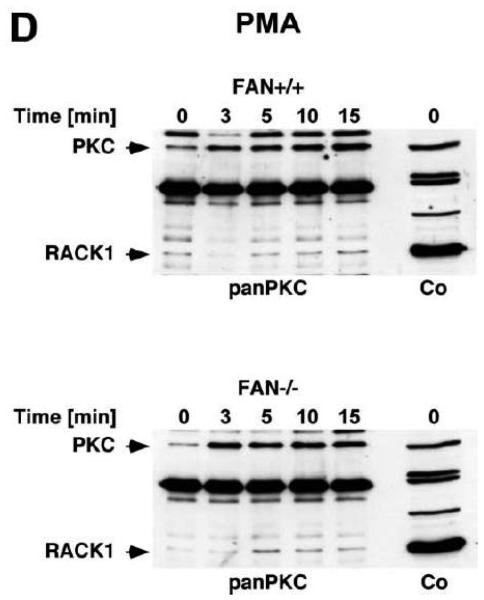
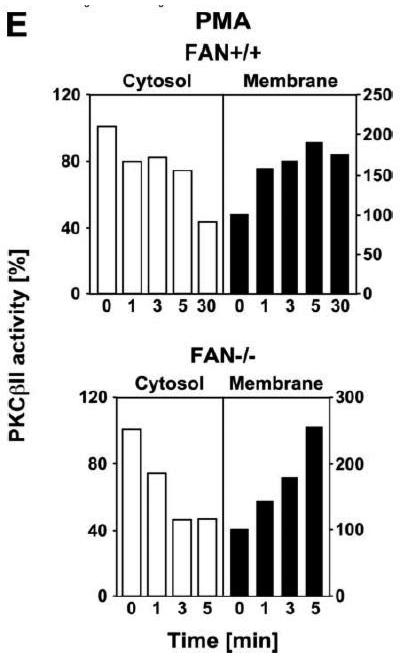
We therefore compared the translocation of cPKC isotypes in response to PMA from the cytosolic to the membrane fraction in EF from FAN+/+ and FAN−/−the cytosolic into the membrane fraction in FAN+/+ EF, indicative for enzymatic activation mice. As shown in Fig. 3A, treatment with PMA induced a rapid translocation (within 1 min) of cPKC isotypes from of cPKC. These results were additionally confirmed for the individual isotypes PKCα and PKCβI. When we analyzed membrane translocation in FAN−/− EF, we observed that cPKC, PKCα, PKCβI and the preferentially RACK1-binding isotype PKCβII translocated in an identical pattern. In assays that measured the enzymatic activity of PKC rather than protein translocation, treatment with PMA led to a clear increase of activity in the membrane fractions, and to a similarly pronounced decrease of activity in the cytosolic fractions of both FAN+/+ and FAN−/− EF (Fig. 3B), showing a kinetics corresponding to the translocation observed in Western blots. Therefore, we next investigated whether a cotranslocation of RACK1 and PKC from the cytosolic to the membrane fraction was detectable and whether the presence or absence of FAN was relevant for this translocation. As demonstrated in Fig. 3C, in contrast to the clearly visible translocation of cPKC isotypes, the total amount of RACK1 in the cytosolic and membrane fractions of FAN+/+ and FAN−/− EF did not change in response to PMA. However, RACK1 is abundantly expressed in both the cytosolic and membrane fractions of the analyzed cells, and thus, the total amount of RACK1 present in these fractions may interfere with detection of the fraction of RACK1 that associates and cotranslocates with specific PKC isotypes. Therefore, we analyzed RACK1 specifically coimmunoprecipitating with cPKCs. As shown in Fig. 3D, PMA induced a time-dependent accumulation of cPKC in the membrane fractions of both FAN+/+ and FAN−/− EF as well as an increase in the amount of cPKC-associated RACK1 in the membrane fraction of both cell lines. In a separate experiment, the activity of immunoprecipitated PKCβII (the preferentially RACK1-binding isotype) from PMA-treated FAN+/+ as well as FAN−/− EF showed a clear increase in the membrane fractions and an equivalent decrease in the cytosolic fractions (Fig. 3E), again corresponding to the translocation data obtained in Western blots. In summary, the above data clearly demonstrate that PMA-induced membrane translocation and enzymatic activation of PKC isotypes likewise occurs in FAN-expressing and FAN deficient EF, indicating that PKC activation by PMA does not depend on the presence of FAN. Apparently, RACK1 is able to recruit PKC isotypes (such as PKCβII) to the plasma membrane independent of its ability to form a complex with FAN.
Figure 3.
Membrane translocation and activation of PKC isotypes does not depend on recruitment of RACK1 to FAN. (A) FAN+/+ and FAN−/− EF were treated with 0.1 μM PMA for the indicated times before cytosolic and membrane fractions were assayed for the presence of cPKC isotypes (panPKC) or for individual PKCs α, βI, or βII by Western blot. Detection of β-actin was used as a loading control (data not shown). For detection of PKCα, membranes were reprobed as described in Fig. 2D. The results shown are representative of a total of three independent experiments. (B) Alternatively, enzymatic activity of PKC was measured in cytosolic and membrane preparations following treatment with PMA. Similar activation kinetics (albeit with a certain variation in the activation maximum) of FAN+/+ and FAN−/− EF were found in repeated experiments (n=4 for FAN+/+, n=9 for FAN−/− EF). (C), FAN+/+ and FAN−/− EF were treated with PMA as above, and both cPKC isotypes and RACK1 were simultaneously detected by Western blot (n=2). The additional band migrating between cPKC and RACK1 is due to a nonspecific reactivity of the utilized RACK1 antibody. (D) FAN+/+ and FAN−/− EF were either left untreated or stimulated with 0.1 μM PMA for the indicated times before immunoprecipitations were performed from membrane fractions using an antibody specific for cPKC (“panPKC”). Precipitated cPKC isotypes and coimmunoprecipitating RACK1 were detected simultaneously in Western blots using a mixture of RACK1 and panPKC antibody. Co: a lysate from 293 cells abundantly expressing cPKC and RACK1 was loaded in a separate lane as positive control for immunodetection/electrophoretic migration of cPKC and RACK1. (E) FAN+/+ and FAN−/− EF were treated with 0.1 μM PMA for the indicated times and PKCβII activity was measured after immunoprecipitation of the enzyme from cytosolic and membrane fractions (n=2). Activity is shown relative to untreated cells.
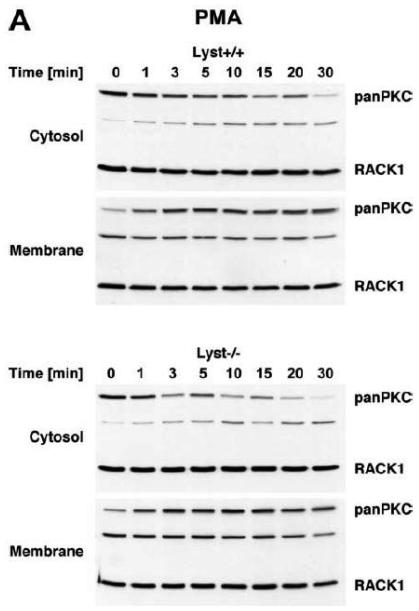
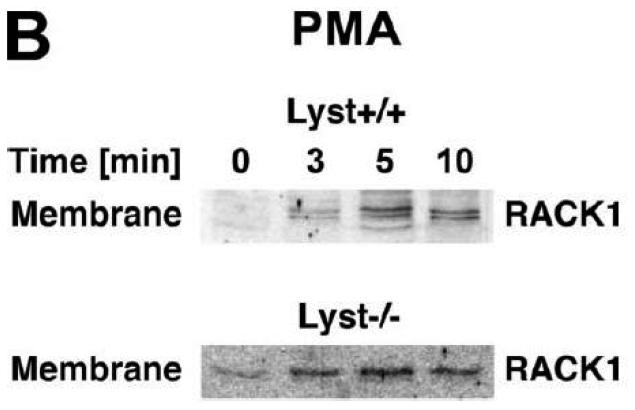
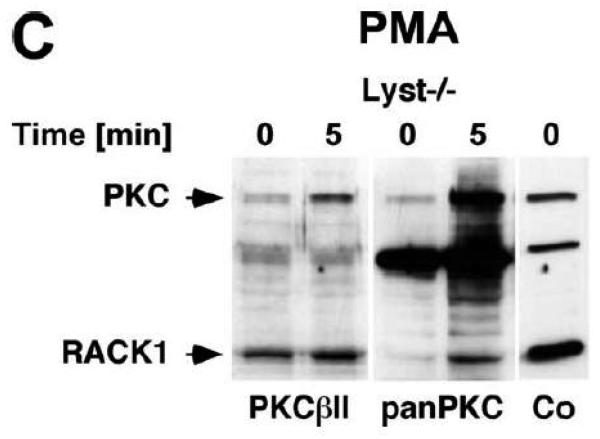
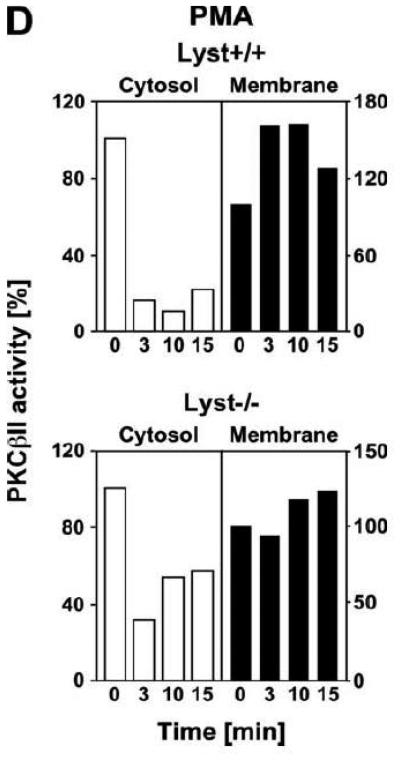
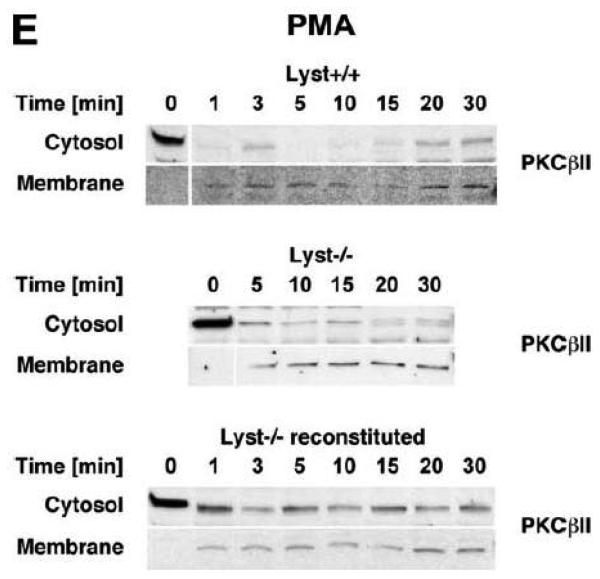
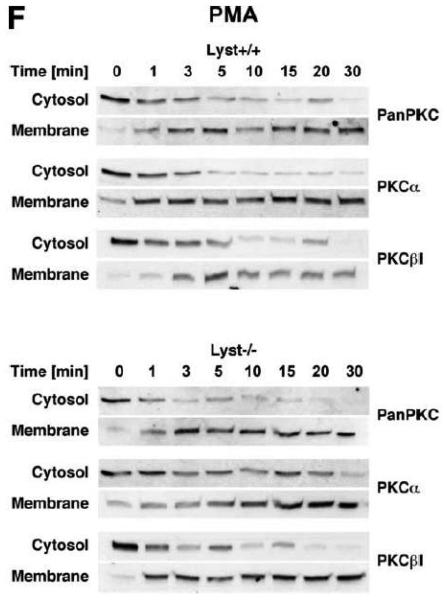
To explore the option that RACK1 requires interaction with a BEACH protein other than FAN for efficient activation and recruitment of PKC isotypes to the plasma membrane, we analyzed Lyst−/− fibroblasts and their wildtype counterparts. We considered Lyst as a potential candidate for interaction with RACK1 since a disturbed activation of PKC in response to PMA has been described in Lyst-deficient cells [12]. This disturbed activation may result from an inability of RACK1 to properly translocate, anchor and stabilize PKC at the plasma membrane when Lyst is missing. However, when we treated Lyst+/+ or Lyst−/− fibroblasts with PMA, we observed a clear cytosol-to-membrane translocation for cPKC in both cell lines, regardless whether Lyst was missing or not (“panPKC” in Fig. 4A). Similar to the results seen in FAN+/+ and FAN−/− EF, no alteration in the distribution of total cellular RACK1 was detectable (“RACK1” in Fig. 4A), again most likely due to the association/cotranslocation of only a minor fraction of total cellular RACK1 with specific PKC isotypes during activation. Therefore, we performed coimmunoprecipitations to assess the amount of RACK1 that specifically associates with PKCβII in Lyst+/+ as well as in Lyst−/− fibroblasts. As shown in Fig. 4B, a time-dependent increase of RACK1 coimmunoprecipitating with PKCβII was detectable in the membrane fraction of Lyst+/+ fibroblasts, but likewise in Lyst−/− cells, suggesting that RACK1 can bind to PKCβII and mediate its translocation to the plasma membrane without requirement for Lyst. This result was additionally corroborated in Lyst−/− fibroblasts by analysis of RACK1 coimmunoprecipitating specifically with PKCβII or with the PKC fraction bound by an antibody against all cPKCs (“panPKC”). In both cases, treatment with PMA induced a translocation of both the respective PKC and of PKC-associated RACK1 into the membrane fraction although Lyst was absent in these cells (Fig. 4C). Likewise, PMA induced a decrease of PKCβII activity in immunoprecipitates from the cytosolic fractions and a concurrent increase in the membrane fractions of fibroblasts, regardless whether Lyst was present or not (Fig. 4D). We further validated these results by conventional Western blot of PKCβII membrane translocation. In both Lyst+/+ as well as Lyst−/− fibroblasts, PMA induced a clear redistribution of PKCβII protein from the cytosolic to the membrane fraction. Identical results were obtained in Lyst−/− fibroblasts that are functionally reconstituted by a YAC complementing their Lyst-deficiency [29] (Fig. 4E). Therefore, the presence or absence of Lyst does not affect the ability of RACK1 to recruit and activate PKCβII in murine fibroblasts. In support of this result, RACK1 did not interact with Lyst in yeast two-hybrid studies utilizing a construct encoding the C-terminal portion of Lyst comprising the WD repeats and parts of the BEACH domain (BPM-1, [29]; data not shown).
Figure 4.
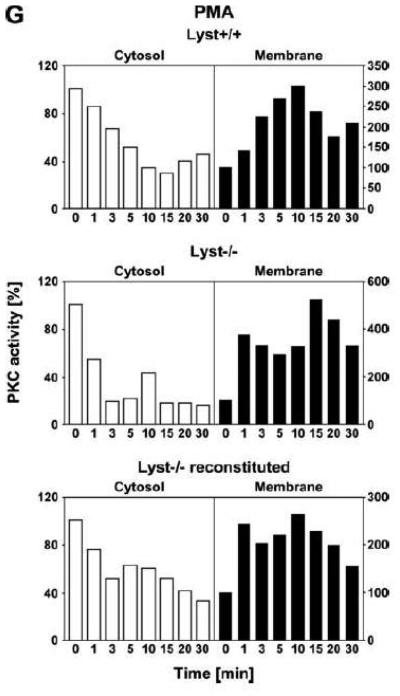
Function of RACK1 as a “receptor for activated C-kinase 1” is independent from an interaction with Lyst. (A) Fibroblasts expressing (+/+) or deficient for Lyst (−/−) were stimulated with 0.1 μM PMA for 0 to 30 min. Subsequently, cytosolic and membrane fractions were simultaneously analyzed for cPKC isotypes (panPKC) or for RACK1 (n=2). The additional band migrating between cPKC and RACK1 is due to a nonspecific reactivity of the utilized RACK1 antibody. (B) Lyst+/+ and Lyst−/− fibroblasts were treated with 0.1 μM PMA for the indicated times before PKCβII was immunoprecipitated from the membrane fraction and coimmunoprecipitating RACK1 was detected by Western blot. The double bands detected by the RACK1 antibody were routinely observed in gel electrophoretic separations with high resolution and most likely represent differentially modified/phosphorylated forms of RACK1 (phosphorylation of RACK1 by the kinase Src has been described [40]). (C) Fibroblasts deficient for Lyst were either left untreated or stimulated with 0.1 μM PMA for 5 min. PKCβII or cPKC isotypes were immunoprecipitated and detected together with coimmunoprecipitated RACK1 by Western blot as described in the legend to Fig. 3D. (D) Lyst+/+ and Lyst−/− fibroblasts were treated with 0.1 μM PMA as indicated and the enzymatic activity of PKCβII was determined in immunoprecipitates from cytosolic and membrane fractions (n=2). Activity is shown relative to untreated cells. (E) Translocation of PKCβII from the cytosolic to the membrane fraction was analyzed by Western blot in Lyst+/+, Lyst−/− and Lyst−/− fibroblasts carrying a YAC that reconstitutes the function of Lyst (Big24r cells). Prior to analysis, cells were stimulated with 0.1 μM PMA for the indicated times. (F) Lyst+/+ and Lyst−/− fibroblasts were stimulated as in panel (A) before cytosolic and membrane fractions were analyzed for the presence of cPKC isotypes (panPKC) or for individual PKCs α and β1 by Western blot (n=2). Detection of β-actin was used as a loading control (data not shown). For detection of PKCα, membranes were reprobed as described in the legend to Fig 2D. (G), Enzymatic activity of PKC was determined in the cytosolic and membrane fractions from Lyst+/+, Lyst−/− and Lyst-reconstituted Lyst−/− fibroblasts after stimulation with 0.1 μM PMA for the indicated times. Activity is shown relative to unstimulated cells. Similar results were obtained in multiple experiments (Lyst+/+: n=2, Lyst−/−: n=5, Lyst−/− reconstituted: n=3).
The regular translocation/activation of PKCβII in Lyst-deficient fibroblasts seen in our experiments in response to PMA is inconsistent with the disturbed activation of PKC that has been described for and been implicated in the manifestation of giant lysosomes in Lyst-deficient cells [12, 13]. Therefore, we extended our previous analyses and additionally compared cytosol-to-membrane translocation of individual cPKC isotypes, PKCα and PKCβI in Lyst+/+ and Lyst−/− fibroblasts. As shown in Fig. 4F, treatment of Lyst-deficient fibroblasts with PMA did not induce a translocation pattern of cPKC, PKCα, or PKCβI that was any different from the pattern seen in Lyst+/+ cells, from the pattern shown for PKCβII in Lyst+/+ or Lyst-reconstituted Lyst−/− Big24r cells (Fig. 4E), or from the pattern seen in FAN+/+ or FAN−/− EF (Fig. 3A). Moreover, in measurements of enzymatic activity, we did not observe a rapid downregulation of activated PKC in the membrane fraction of Lyst−/− fibroblasts. In contrast to the described almost complete reduction of activity within 5 min [12], we consistently measured an increase of activity in the membrane fraction as well as a corresponding decrease in the cytosolic fraction that followed comparable patterns in Lyst−/−, Lyst+/+ and Lyst-reconstituted Lyst−/− fibroblasts (Fig. 4G) and which again corresponded to the translocation patterns seen in Western blots. In conclusion, the Lyst-deficient murine fibroblasts examined here do not show a rapid downregulation or abnormal activation of PKC and the formation of enlarged lysosomes in these cells is most likely not linked to defects in membrane recruitment/activation of PKC.
Overall, these data clearly indicate that RACK1 is able to bind to cPKC isotypes in the presence or absence of either Lyst or FAN, strongly suggesting that association of RACK1 to BEACH proteins is not required for its function as a “receptor for activated C-kinase” (i. e. RACK1-mediated recruitment of activated cPKC isotypes to the plasma membrane and stabilization of the activated enzyme from premature degradation).
The role of FAN in regulation of lysosome size is not coupled to an enhanced proteolysis of PKC
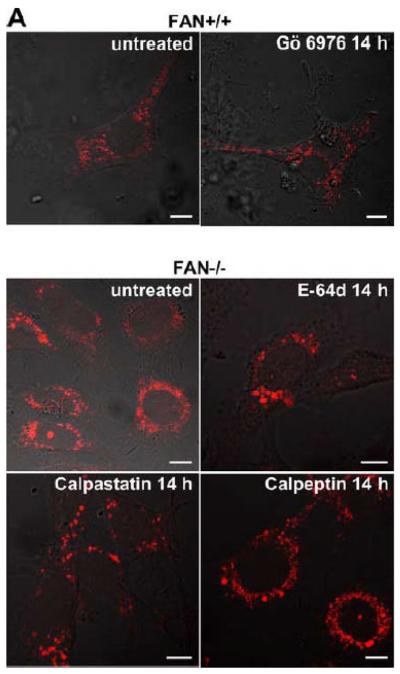
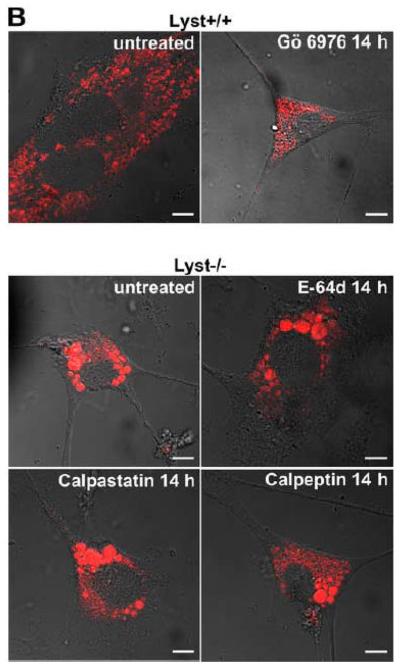
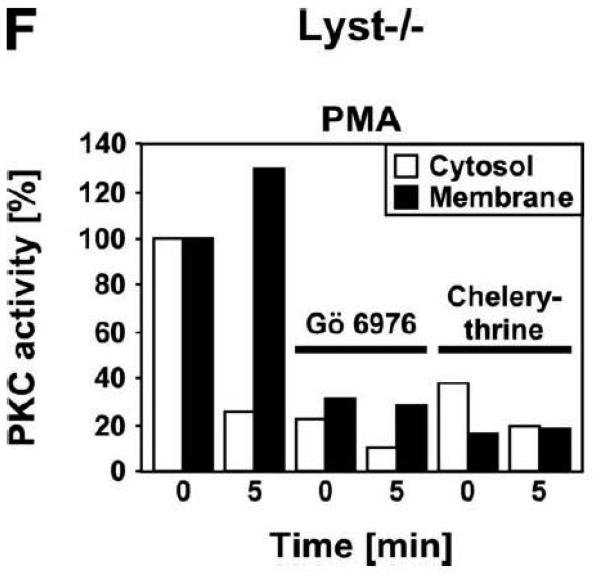
In Lyst-deficient cells, the formation of giant lysosomes has been linked to an enhanced proteolysis of PKC by the thiol proteinase calpain, resulting in an abnormal downregulation/membrane recruitment of PKC. Blockage of calpain by E-64d, a broad spectrum cysteine protease inhibitor, has been described not only to restore PKC activity, but also to prevent giant granule formation in Lyst-deficient cells. Likewise, PKC inhibitors have been described to promote giant granule formation in wildtype cells [13]. To evaluate whether these mechanisms were also of importance for the effects of FAN on lysosome size, we incubated FAN+/+ EF with Gö 6976, an inhibitor of cPKC. In parallel, FAN−/− EF were treated with the calpain inhibitors E-64d, calpastatin, or calpeptin. As shown in Fig. 5A, treatment with Gö 6976 did not induce a detectable enlargement of lysosomes in FAN+/+ EF, nor could we observe a noticeable reduction of lysosome size in FAN−/− EF that had been treated with calpain inhibitors. Likewise, in a reverse experiment, no effect of Gö 6976 on FAN−/− EF or of calpastatin on FAN+/+ EF was detectable (data not shown).
Figure 5.
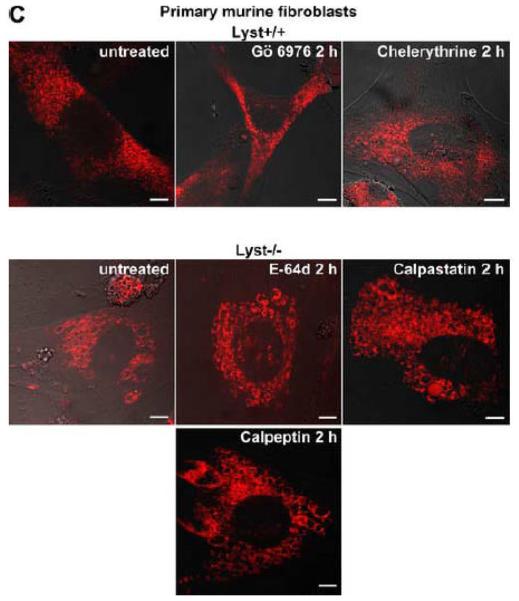
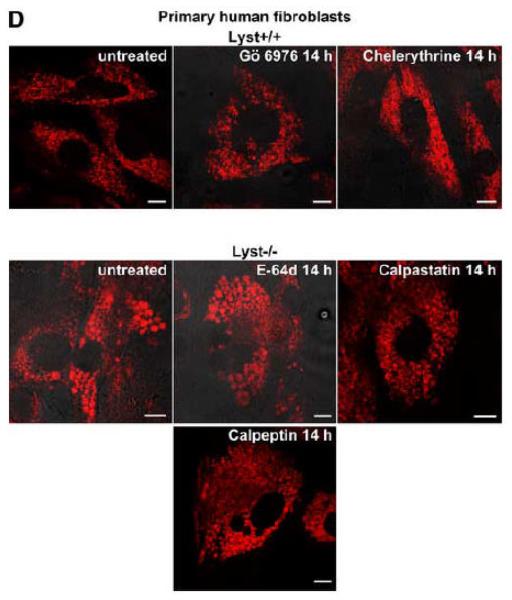
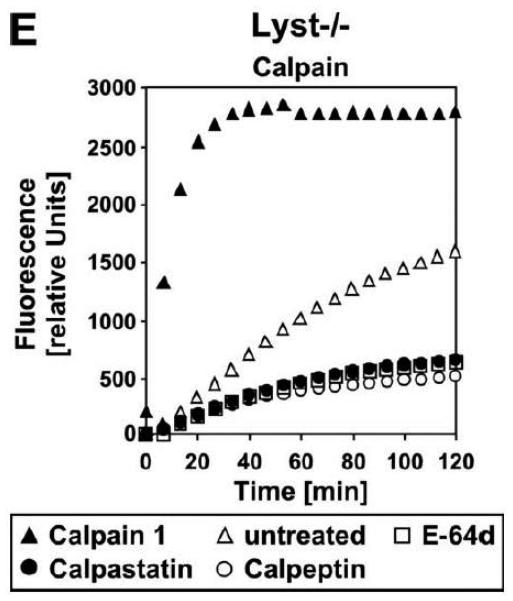
Lysosome size in wildtype fibroblasts or fibroblasts deficient for FAN or Lyst is not altered by inhibitors of PKC or of calpain-mediated PKC-proteolysis. FAN+/+ and FAN−/− EF (A), Lyst−/− and Lyst−/− fibroblasts (C572CF, MCHSF2; B), Lyst+/+ and Lyst−/− primary murine lung fibroblasts (C) and Lyst+/+ and Lyst−/− primary human fibroblasts (D) were either left untreated or stimulated with the PKC-inhibitors Gö 6976 (1 μM) or chelerythrine (1 μM) or with the calpain inhibitors E-64d (1 μg/ml), calpastatin (10 μM) or calpeptin (10 μM) for the indicated times. Following treatment, cells were stained with LysoTracker Red and visualized by confocal laserscanning microscopy. Bar, 10 μm. The circular staining seen in some panels (C, D) may be due to the presence of internal vesicles in these lysosomes that have not been broken down and which do not have the low pH necessary for staining with LysoTracker Red. (E) Lyst−/− fibroblasts (MCHSF2) were left untreated or preincubated for 14 h with 1 μg/ml E-64d, or 10 μM calpastatin or calpeptin. Subsequently, activity of cytoplasmic calpains was determined by measuring the digestion the fluorogenic substrate Suc-LLVY-amc over 120 minutes. Purified human calpain 1 served as a positive control. (F) Lyst+/+ fibroblasts (C572CF) were left untreated or preincubated for 2 h with 1 μM Gö 6976 or 1 μM chelerythrine before being stimulated with 0.1 μM PMA or not. Subsequently, PKC activity was measured in the cytosolic and membrane fractions. (G) Lyst−/− fibroblasts (MCHSF2) were left untreated or preincubated with 1 μg/ml E-64d, or 10 μM calpastatin or calpeptin for 2 h. Cells were then treated with 0.1 μM PMA or not and PKC activity of cytosolic and membrane fractions was determined. PKC activity in cells without PMA stimulation was not altered by preincubation with E-64d, calpastatin or calpeptin. Similar results were obtained in two other experiments (data not shown). (H) In parallel, cytosolic and membrane fractions were assayed for the presence of cPKC isotypes (panPKC) Western blot (PMA-treated fractions are shown from two independent stimulations).
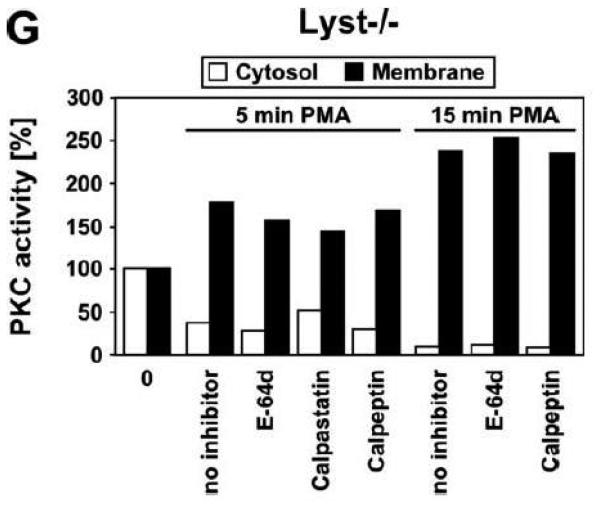
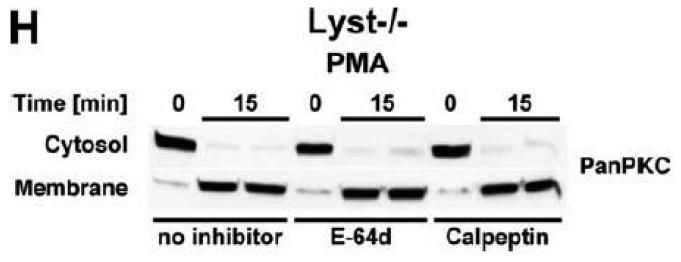
As additional controls, we analyzed the impact of PKC inhibition by Gö 6976 on Lyst+/+ fibroblasts as well as the effects of the calpain inhibitors E-64d, calpastatin or calpeptin in Lyst−/− fibroblasts (Fig. 5B). To exclude artifacts originating from immortalization of the above cell lines, we repeated these experiments in primary murine (Fig. 5C) and primary human (Fig. 5D) Lyst+/+ and Lyst−/− fibroblasts, additionally employing the PKC inhibitor chelerythrine. In all cases, inhibition of PKC uniformly did not enlarge lysosome size in wildtype fibroblasts, nor did inhibition of calpain lead to a shrinking of giant lysosomes in Lyst-deficient fibroblasts (Fig. 5B-D). We verified the integrity of E-64d, calpastatin and calpeptin in enzymatic assays where an unambiguous inhibition of calpain activity by all three inhibitors was observed in Lyst−/− fibroblasts (Fig. 5E). Likewise, the PKC inhibitors Gö 6976 and chelerythrine clearly reduced basal PKC activity and inhibited stimulation of PKC by PMA in Lyst+/+ fibroblasts (Fig. 5F). Thus, although these inhibitors were functional, they did not affect lysosome size, arguing that alterations in lysosome size in Lyst- and FAN-deficient cells are not coupled to an enhanced proteolysis of PKC by calpain. This was further corroborated in an experiment in which we directly measured membrane translocation and activation of PKC in the presence of calpain inhibitors. Preincubation of Lyst−/− fibroblasts with E-64d, calpeptin or calpastatin neither increased membrane-associated PKC activity (Fig. 5G), nor did preincubation with E-64d or calpeptin alter membrane translocation of PKC (Fig. 5H). This result is in concordance with our observation that Lyst−/− fibroblasts do not show a previously reported [12] rapid downregulation of PKC (Fig. 4) which therefore also cannot be restored by inhibition of calpains. It is also in concordance with the lacking effect of these inhibitors on lysosome size (Fig. 5A-D). These results clearly demonstrate that the deregulation of lysosome size in cells lacking the BEACH proteins FAN or Lyst is not due to an enhanced proteolysis or abnormal downregulation/membrane translocation of PKC.
Taken together, our data suggest that FAN exerts its cellular functions (such as activation of N-SMase and regulation of lysosome size) independently from the signaling pathways that lead to membrane translocation/activation of PKC. Although a role of PKC in the signaling cascades triggered by FAN would have been plausible in a model in which RACK1 acts as an interaction partner of FAN and concurrently recruits activated PKC into the vicinity of FAN, our data clearly demonstrate that the respective signaling pathways do not depend on each other. While FAN is essential for activation of N-SMase by TNF in EF [14], this occurs in the complete absence of membrane translocation/activation of PKC isotypes. Likewise, phorbol ester-induced membrane translocation/activation of PKC isotypes in EF lacking FAN is undisturbed and indistinguishable from the response seen in wildtype cells. Similarly, the function of RACK1 as a “receptor for activated C-kinase” is not affected by presence of absence of FAN, suggesting that RACK1 has distinct functions in the signaling pathways of FAN and of PKC. Finally, lysosome size is increased in FAN-deficient EF but unaffected by inhibitors of PKC or calpain. Of note, in the course of our experiments, we found that fibroblasts deficient for Lyst likewise show a perfectly normal activation/membrane translocation and binding of RACK1 to PKC isotypes, as well as an unaltered lysosome size in response to inhibition of PKC or calpain. This suggests that the causal role of a calpain-mediated abnormal downregulation/membrane recruitment of PKC isotypes in the formation of giant lysosomes that was previously reported for Lyst-deficient fibroblasts [13] may not be universal. However, we currently cannot rule out that downregulation/membrane recruitment of PKC isotypes affects lysosome formation in other Lyst-deficient cells types, such as macrophages and polymorphonuclear leukocytes [12].
In this study, we have shown that FAN has a novel, previously unrecognized function in the regulation of lysosome size, in addition to N-SMase activation [3, 14], cutaneous barrier repair [14], and control of lysosomal permeability [21]. However, PKC is apparently not critically involved in these signaling pathways. The identification and characterization of additional components of these pathways (e. g. novel interaction partners of the FAN/RACK1 complex) will provide further insight and is a current subject of investigation.
ACKNOWLEDGMENTS
This work was supported by grants of the Research Commission of the Medical Faculty and the Hensel-Stiftung, Kiel, to D. A. and grant NIH RO1 HL26922 to J. K.
Abbreviations
- amc
7-amino-4methylcoumarin
- BEACH
beige and CHS
- E-64d
(2S, 3S)-trans-epoxyscuccinyl-L-leucylamido-3-methylbutane ethyl ester
- EF
embryonic fibroblasts
- FAN
factor associated with neutral sphingomyelinase activation
- hTNF
human recombinant TNF
- Lyst
lysosomal trafficking regulator
- N-SMase
neutral sphingomyelinase
- PH
pleckstrin-homology
- PKC
protein kinase C
- PMA
phorbol 12-myristate 13-acetate
- RACK1
receptor for activated C-kinase 1
- TNF
tumor necrosis factor
- TNF-R55
55 kDa TNF receptor
- YAC
yeast artificial chromosome
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
REFERENCES
- 1.Clarke CJ, Hannun YA. Neutral sphingomyelinases and nSMase2: Bridging the gaps. Biochim. Biophys. Acta. 2006;1758:1893–1901. doi: 10.1016/j.bbamem.2006.06.025. [DOI] [PubMed] [Google Scholar]
- 2.Clarke CJ, Snook CF, Tani M, Matmati N, Marchesini N, Hannun YA. The extended family of neutral sphingomyelinases. Biochemistry. 2006;45:11247–11256. doi: 10.1021/bi061307z. [DOI] [PubMed] [Google Scholar]
- 3.Adam-Klages S, Adam D, Wiegmann K, Struve S, Kolanus W, Schneider-Mergener J, Krönke M. FAN, a novel WD-repeat protein, couples the p55 TNF-receptor to neutral sphingomyelinase. Cell. 1996;86:937–947. doi: 10.1016/s0092-8674(00)80169-5. [DOI] [PubMed] [Google Scholar]
- 4.Li D, Roberts R. WD-repeat proteins: structure characteristics, biological function, and their involvement in human diseases. Cell. Mol. Life Sci. 2001;58:2085–2097. doi: 10.1007/PL00000838. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Nagle DL, Karim MA, Woolf EA, Holmgren L, Bork P, Misumi DJ, McGrail SH, Dussault BJ, Perou CM, Boissy RE, Duyk GM, Spritz RA, Moore KJ. Identification and mutation analysis of the complete gene for Chediak-Higashi syndrome. Nat. Genet. 1996;14:307–311. doi: 10.1038/ng1196-307. [DOI] [PubMed] [Google Scholar]
- 6.Su Y, Balice-Gordon RJ, Hess DM, Landsman DS, Minarcik J, Golden J, Hurwitz I, Liebhaber SA, Cooke NE. Neurobeachin is essential for neuromuscular synaptic transmission. J. Neurosci. 2004;24:3627–3636. doi: 10.1523/JNEUROSCI.4644-03.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Kwak E, Gerald N, Larochelle DA, Vithalani KK, Niswonger ML, Maready M, De Lozanne A. LvsA, a protein related to the mouse beige protein, is required for cytokinesis in Dictyostelium. Mol. Biol. Cell. 1999;10:4429–4439. doi: 10.1091/mbc.10.12.4429. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Wang JW, Howson J, Haller E, Kerr WG. Identification of a novel lipopolysaccharide-inducible gene with key features of both a kinase anchor proteins and chs1/beige proteins. J. Immunol. 2001;166:4586–4595. doi: 10.4049/jimmunol.166.7.4586. [DOI] [PubMed] [Google Scholar]
- 9.Jogl G, Shen Y, Gebauer D, Li J, Wiegmann K, Kashkar H, Krönke M, Tong L. Crystal structure of the BEACH domain reveals an unusual fold and extensive association with a novel PH domain. EMBO J. 2002;21:4785–4795. doi: 10.1093/emboj/cdf502. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Barbosa MD, Nguyen QA, Tchernev VT, Ashley JA, Detter JC, Blaydes SM, Brandt SJ, Chotai D, Hodgman C, Solari RC, Lovett M, Kingsmore SF. Identification of the homologous beige and Chediak-Higashi syndrome genes [published erratum appears in Nature 1997 Jan 2;385(6611):97] Nature. 1996;382:262–265. doi: 10.1038/382262a0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Tchernev VT, Mansfield TA, Giot L, Kumar AM, Nandabalan K, Li Y, Mishra VS, Detter JC, Rothberg JM, Wallace MR, Southwick FS, Kingsmore SF. The Chediak-Higashi protein interacts with SNARE complex and signal transduction proteins. Mol. Med. 2002;8:56–64. [PMC free article] [PubMed] [Google Scholar]
- 12.Ito M, Tanabe F, Takami Y, Sato A, Shigeta S. Rapid down-regulation of protein kinase C in (Chediak-Higashi syndrome) beige mouse by phorbol ester. Biochem. Biophys. Res. Commun. 1988;153:648–656. doi: 10.1016/s0006-291x(88)81144-6. [DOI] [PubMed] [Google Scholar]
- 13.Tanabe F, Cui SH, Ito M. Abnormal down-regulation of PKC is responsible for giant granule formation in fibroblasts from CHS (Beige) mice - a thiol proteinase inhibitor, E-64-d, prevents giant granule formation in beige fibroblasts. J. Leukoc. Biol. 2000;67:749–755. doi: 10.1002/jlb.67.5.749. [DOI] [PubMed] [Google Scholar]
- 14.Kreder D, Krut O, Adam-Klages S, Wiegmann K, Scherer G, Plitz T, Jensen JM, Proksch E, Steinmann J, Pfeffer K, Krönke M. Impaired neutral sphingomyelinase activation and cutaneous barrier repair in FAN-deficient mice. EMBO J. 1999;18:2472–2479. doi: 10.1093/emboj/18.9.2472. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Ségui B, Andrieu-Abadie N, Adam-Klages S, Meilhac O, Kreder D, Garcia V, Bruno AP, Jaffrézou JP, Salvayre R, Krönke M, Levade T. CD40 signals apoptosis through FAN-regulated activation of the sphingomyelin-ceramide pathway. J. Biol. Chem. 1999;274:37251–37258. doi: 10.1074/jbc.274.52.37251. [DOI] [PubMed] [Google Scholar]
- 16.Ségui B, Cuvillier O, Adam-Klages S, Garcia V, Malagarie-Cazenave S, Lévêque S, Caspar-Bauguil S, Coudert J, Salvayre R, Krönke M, Levade T. Involvement of FAN in TNF-induced apoptosis. J. Clin. Invest. 2001;108:143–151. doi: 10.1172/JCI11498. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Malagarie-Cazenave S, Ségui B, Lévêque S, Garcia V, Carpentier S, Altié MF, Brouchet A, Gouazé V, Andrieu-Abadie N, Barreira Y, Benoist H, Levade T. Role of FAN in tumor necrosis factor-alpha and lipopolysaccharide-induced interleukin-6 secretion and lethality in D-galactosamine-sensitized mice. J. Biol. Chem. 2004;279:18648–18655. doi: 10.1074/jbc.M314294200. [DOI] [PubMed] [Google Scholar]
- 18.O'Brien NW, Gellings NM, Guo M, Barlow SB, Glembotski CC, Sabbadini RA. Factor associated with neutral sphingomyelinase activation and its role in cardiac cell death. Circ. Res. 2003;92:589–591. doi: 10.1161/01.RES.0000066290.29715.67. [DOI] [PubMed] [Google Scholar]
- 19.Neumeyer J, Hallas C, Merkel O, Winoto-Morbach S, Jakob M, Thon L, Adam D, Schneider-Brachert W, Schütze S. TNF-receptor I defective in internalization allows for cell death through activation of neutral sphingomyelinase. Exp. Cell. Res. 2006;312:2142–2153. doi: 10.1016/j.yexcr.2006.03.014. [DOI] [PubMed] [Google Scholar]
- 20.Peppelenbosch M, Boone E, Jones GE, van Deventer SJ, Haegeman G, Fiers W, Grooten J, Ridley AJ. Multiple signal transduction pathways regulate TNF-induced actin reorganization in macrophages: inhibition of Cdc42-mediated filopodium formation by TNF. J. Immunol. 1999;162:837–845. [PubMed] [Google Scholar]
- 21.Werneburg N, Guicciardi ME, Yin XM, Gores GJ. TNF-alpha-mediated lysosomal permeabilization is FAN and caspase 8/Bid dependent. Am. J. Physiol. Gastrointest. Liver Physiol. 2004;287:G436–443. doi: 10.1152/ajpgi.00019.2004. [DOI] [PubMed] [Google Scholar]
- 22.Tcherkasowa AE, Adam-Klages S, Kruse ML, Wiegmann K, Mathieu S, Kolanus W, Krönke M, Adam D. Interaction with factor associated with neutral sphingomyelinase activation, a WD motif-containing protein, identifies receptor for activated C-kinase 1 as a novel component of the signaling pathways of the p55 TNF receptor. J. Immunol. 2002;169:5161–5170. doi: 10.4049/jimmunol.169.9.5161. [DOI] [PubMed] [Google Scholar]
- 23.Neer EJ, Smith TF. G protein heterodimers: New structures propel new questions. Cell. 1996;84:175–178. doi: 10.1016/s0092-8674(00)80969-1. [DOI] [PubMed] [Google Scholar]
- 24.Nilsson J, Sengupta J, Frank J, Nissen P. Regulation of eukaryotic translation by the RACK1 protein: a platform for signalling molecules on the ribosome. EMBO Rep. 2004;5:1137–1141. doi: 10.1038/sj.embor.7400291. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Mochly-Rosen D, Gordon AS. Anchoring proteins for protein kinase C: a means for isozyme selectivity. FASEB J. 1998;12:35–42. [PubMed] [Google Scholar]
- 26.Lüschen S, Adam D, Ussat S, Kreder D, Schneider-Brachert W, Krönke M, Adam-Klages S. Activation of ERK1/2 and cPLA(2) by the p55 TNF receptor occurs independently of FAN. Biochem. Biophys. Res. Commun. 2000;274:506–512. doi: 10.1006/bbrc.2000.3173. [DOI] [PubMed] [Google Scholar]
- 27.Perou CM, Justice MJ, Pryor RJ, Kaplan J. Complementation of the beige mutation in cultured cells by episomally replicating murine yeast artificial chromosomes. Proc. Natl. Acad. Sci. U. S. A. 1996;93:5905–5909. doi: 10.1073/pnas.93.12.5905. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Perou CM, Moore KJ, Nagle DL, Misumi DJ, Woolf EA, McGrail SH, Holmgren L, Brody TH, Dussault BJ, Jr., Monroe CA, Duyk GM, Pryor RJ, Li L, Justice MJ, Kaplan J. Identification of the murine beige gene by YAC complementation and positional cloning. Nat. Genet. 1996;13:303–308. doi: 10.1038/ng0796-303. [DOI] [PubMed] [Google Scholar]
- 29.Ward DM, Shiflett SL, Huynh D, Vaughn MB, Prestwich G, Kaplan J. Use of expression constructs to dissect the functional domains of the CHS/Beige protein: Identification of multiple phenotypes. Traffic. 2003;4:403–415. doi: 10.1034/j.1600-0854.2003.00093.x. [DOI] [PubMed] [Google Scholar]
- 30.Grassmé H, Kirschnek S, Riethmueller J, Riehle A, von Kurthy G, Lang F, Weller M, Gulbins E. CD95/CD95 ligand interactions on epithelial cells in host defense to pseudomonas aeruginosa. Science. 2000;290:527–530. doi: 10.1126/science.290.5491.527. [DOI] [PubMed] [Google Scholar]
- 31.Schütze S, Nottrott S, Pfizenmaier K, Krönke M. Tumor necrosis factor signal transduction - cell-type specific activation and translocation of protein kinase C. J. Immunol. 1990;144:2604–2608. [PubMed] [Google Scholar]
- 32.Perou CM, Leslie JD, Green W, Li L, Ward DM, Kaplan J. The Beige/Chediak-Higashi syndrome gene encodes a widely expressed cytosolic protein. J. Biol. Chem. 1997;272:29790–29794. doi: 10.1074/jbc.272.47.29790. [DOI] [PubMed] [Google Scholar]
- 33.Lem L, Riethof DA, Scidmore-Carlson M, Griffiths GM, Hackstadt T, Brodsky FM. Enhanced interaction of HLA-DM with HLA-DR in enlarged vacuoles of hereditary and infectious lysosomal diseases. J. Immunol. 1999;162:523–532. [PubMed] [Google Scholar]
- 34.Harris E, Wang N, Wu W, Weatherford A, De Lozanne A, Cardelli J. Dictyostelium LvsB mutants model the lysosomal defects associated with Chediak-Higashi syndrome. Mol. Biol. Cell. 2002;13:656–669. doi: 10.1091/mbc.01-09-0454. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Zha XH, Pierini LM, Leopold PL, Skiba PJ, Tabas I, Maxfield FR. Sphingomyelinase treatment induces ATP-independent endocytosis. J. Cell. Biol. 1998;140:39–47. doi: 10.1083/jcb.140.1.39. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Wang XL, Herberg FW, Laue MM, Wüllner C, Hu B, Petrasch-Parwez E, Kilimann MW. Neurobeachin: A protein kinase A-anchoring, beige/Chediak-Higashi protein homolog implicated in neuronal membrane traffic. J. Neurosci. 2000;20:8551–8565. doi: 10.1523/JNEUROSCI.20-23-08551.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Schechtman D, Mochly-Rosen D. Adaptor proteins in protein kinase C-mediated signal transduction. Oncogene. 2001;20:6339–6347. doi: 10.1038/sj.onc.1204778. [DOI] [PubMed] [Google Scholar]
- 38.Kellerer M, Mushack J, Mischak H, Häring HU. Protein kinase C (PKC) epsilon enhances the inhibitory effect of TNF alpha on insulin signaling in HEK293 cells. FEBS Lett. 1997;418:119–122. doi: 10.1016/s0014-5793(97)01357-4. [DOI] [PubMed] [Google Scholar]
- 39.Wang N, Wu WI, DeLozanne A. BEACH family of proteins: Phylogenetic and functional analysis of six Dictyostelium BEACH proteins. J. Cell. Biochem. 2002;86:561–570. doi: 10.1002/jcb.10254. [DOI] [PubMed] [Google Scholar]
- 40.Chang BY, Harte RA, Cartwright CA. RACK1: a novel substrate for the Src protein-tyrosine kinase. Oncogene. 2002;21:7619–7629. doi: 10.1038/sj.onc.1206002. [DOI] [PubMed] [Google Scholar]