Abstract
Telomeres consisting of tandem guanine-rich repeats can form secondary DNA structures called G-quadruplexes that represent potential targets for DNA repair enzymes. While G-quadruplexes interfere with DNA synthesis in vitro, the impact of G-quadruplex formation on telomeric repeat replication in human cells is not clear. We investigated the mutagenicity of telomeric repeats as a function of G-quadruplex folding opportunity and thermal stability using a shuttle vector mutagenesis assay. Since single stranded DNA during lagging strand replication increases the opportunity for G-quadruplex folding, we tested vectors with G-rich sequences on the lagging versus the leading strand. Contrary to our prediction, vectors containing human [TTAGGG]10 repeats with a G-rich lagging strand were significantly less mutagenic than vectors with a G-rich leading strand, after replication in normal human cells. We show by UV melting experiments that G-quadruplexes from ciliates [TTGGGG]4 and [TTTTGGGG]4 are thermally more stable compared to human [TTAGGG]4. Consistent with this, replication of vectors with ciliate [TTGGGG]10 repeats yielded a 3-fold higher mutant rate compared to the human [TTAGGG]10 vectors. Furthermore, we observed significantly more mutagenic events in the ciliate repeats compared to the human repeats. Our data demonstrate that increased G-quadruplex opportunity (repeat orientation) in human telomeric repeats decreased mutagenicity, while increased thermal stability of telomeric G-qaudruplexes was associated with increased mutagenicity.
Keywords: telomere, G-quadruplex, genomic instability, DNA replication, mutagenesis
1. Introduction
Telomeres are nucleoprotein structures at chromosome ends that critically impact lifespan and health, as well as cell viability and genome stability [1-3]. Progress in recent years indicates that the inability to completely replicate chromosome ends is not the only source of telomere attrition, and that inappropriate processing by DNA repair enzymes or failures in telomere replication can cause rapid telomere loss (reviewed in [4]). Telomeres consist of an array of repeat sequences that interact with specific proteins to prevent the chromosome ends from being recognized as double strand breaks [5, 6]. Mammalian telomeres comprise of TTAGGG repeats, and human telomere lengths vary from 5 -15 kb and terminate in a 3′ ssDNA tail that is 50 -500 nt long [7]. The 3′ tails can invade preceding telomeric repeats to form a lariat like t-loop/D-loop structure that is further stabilized by the shelterin protein complex [8, 9]. Shelterin proteins TRF2 and TRF1 bind duplex telomeric DNA and POT1 binds to single strand TTAGGG repeats [10, 11], and together they recruit the remaining shelterin proteins TIN2, RAP1, and TPP1[4]. How these proteins influence the fundamental processes of DNA repair and replication in telomeric repeats has yet to be fully realized.
Cellular evidence indicates that telomeres are fraught with potential obstacles to DNA replication and require specific proteins to prevent stalling. In S. cerevisiae DNA replication fork stalling is greatly increased at telomeres in the absence of the Rrm3p helicase [12]. In S. pombe and humans the telomeric proteins Taz1 and TRF1, respectively, are required to prevent replication fork stalling at telomeres [13, 14]. The precise mechanism is not known, but some evidence suggests that TRF1 recruits helicases BLM and RTEL to dissociate alternate DNA structures [15]. The consequences of fork stalling in the telomeres can be loss of telomeric DNA or aberrant telomere structures including doublets that resemble broken telomeres [14-16]. Telomere doublets are induced by aphidicolin treatment which stalls replication forks and induces breaks at fragile sites [14]. The mechanistic models of mutagenesis in repetitive sequences involve stalling and/or dissociation of the DNA replication fork due to road blocks [17]. Studies in yeast and bacteria demonstrate that sites of stalled replication forks are susceptible to chromosomal breakage [12, 18, 19]. Thus, replication-mediated breaks in telomeres may represent an important source of telomeric loss.
Possible sources of replication fork stalling at telomeres include oxidative DNA damage which preferentially occurs at G runs [20], or alternate DNA structures including the t-loop/D-loop or G-quadruplex (G4) DNA which can form in ssDNA with tandem guanines. Telomeric DNA forms G4 structures spontaneously in vitro and in vivo [21-26], that block DNA polymerase progression in vitro [27]. G4 structures consist of planar arrays of quartets, and each quartet is formed by four guanines interacting through Hoogsteen base pairing [28] (Fig. 1A). The number of quartets in a quadruplex influences the stability of the structure and depends on the number of guanine residues [29]. The potential for G4 formation in the telomeres exists either in the 3′overhang, displaced DNA in the D-loop, or in the G-rich sequences present on the lagging strand. Okazaki fragment processing during lagging strand DNA synthesis is expected to produce transient regions of ssDNA, and G4 DNA folds in ssDNA regions [26, 30]. Cells deficient in the Werner syndrome protein (WRN), POT1 or FEN1 exhibit preferential loss of telomeres replicated from the G-rich lagging strand [15, 31, 32], suggesting these proteins may function in preventing and/or dissociating G4 structures. Furthermore, an agent that stabilizes G4 DNA induces defects in telomere replication and causes telomeric aberrations [33]. Whether G4 structures can interfere with telomere replication in normal cells has yet to be established.
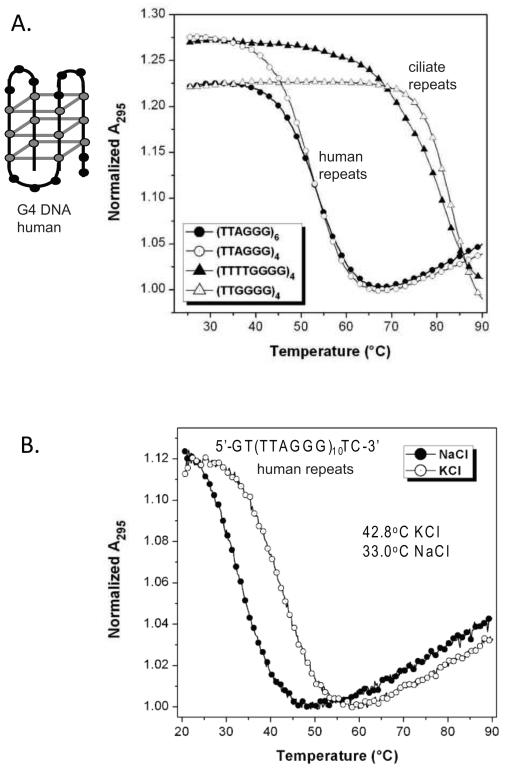
Fig. 1.
Ciliate telomeric repeats form G4 DNA structures of greater thermal stability compared to human telomeric repeats. (A) UV Melting curves showing intra-molecular G-quadruplex formation in telomeric ssDNA with flanking sequence. Melting curves for oligonucleotides GT-(TTAGGG)4-TC, GT-(TTAGGG)6-TC, GT-(TTGGGG)4-TC and GT-(TTTTGGGG)4-TC recorded at 295 nm in solutions containing 100 μM KCl. A schematic of one possible G4 conformation for human telomere repeats is shown. Grey balls indicates Gs, and grey lines indicate a quartet formed by base pairing between four Gs. (B) UV melting curves for the olignucleotide GT-(TTTAGGG)10-TC in two different salt solutions, 100 μM KCl and 100 μM NaCl.
Previous work indicates that sequences with the ability to form various alternate structures exhibit increased mutagenic potential (reviewed in [34]). In these studies shuttle vectors with mutation reporter genes have been invaluable. The insertion of sequences with the potential to form H-DNA and Z-DNA adjacent to a reporter gene induced breaks and large deletions in the shuttle vector after transfection into normal mammalian cells [35, 36]. The impact of G4 DNA on shuttle vector stability is unknown, but studies in yeast and worms suggest that G4 structures can be mutagenic. Loss of DOG-1 helicase in C. elegans leads to deletions in genes containing G-runs [37], and loss of Pif1 helicase in S. cerevisiea promotes instability in an artificial human G-rich minisatellite in the yeast genome [38]. However, the fidelity of telomeric repeat replication and the impact of G4 potential on the mutagenicity of telomeric repeats in human cells are largely unexamined.
Studies of ciliated protozoa provide evidence for G4 formation at telomeres and G4 resolution during replication. Ciliates contain a macronucleus consisting of up to 108 small DNA molecules that are terminate by telomeres consisting of about 20 bp of duplex DNA and a 16 nucleotide 3′ G-rich ssDNA tail (reviewed in [39]). This high concentration of telomeres allowed for the detection of G4 DNA by immuno-staining with antibodies raised against G4 structures [40]. DNA replication occurs exclusively in a distinct replication band [41] in which G4 DNA is not detected [40]. G4 formation is regulated by telomere binding proteins TEBP-α and TEBP-β [23, 25]. These studies suggest that G4 DNA is resolved during telomere replication in ciliates. In this study our goal was to test the mutagenic potential of telomeric repeat sequences and their ability to induce breaks and deletions upon replication in normal human cells, using a well established shuttle vector mutagenesis assay. We hypothesized that the mutagenicity of telomeric repeats correlates with G4 forming potential and thermal stability. To test this we examined various telomeric repeats that differ in G-quartet numbers and compared repeats with the G-rich sequence on the lagging strand versus the leading strand. We show that the ciliate repeats from T. thermophila (TTGGGG) and O. nova (TTTTGGGG) form more stable G4 DNA than human repeats (TTAGGG) in vitro. We demonstrate that while all of the vectors with various telomeric repeats exhibited low mutant rates after replication in human cells, the orientation of the human telomeric repeats (G-rich lagging versus leading strand) and the stability of the potential G4 structures significantly affected the vector mutant rates. We also observed an increase in mutagenic events in the ciliate telomeric repeats compared to the human repeats. However, in contrast to H-DNA and Z-DNA forming sequences, our data indicate that normal human cells possess the ability to effectively manage G4 forming sequences, particularly human telomeric repeats, during replication.
2. Materials and Methods
2.1. Reagents
Oligonucleotides containing telomeric repeat sequences and primers used in sequencing reactions were ordered from Integrated DNA technologies Inc. (Coralville, IA) (Supplemental Table S1). Restriction enzymes were purchased from New England Biolabs (Ipswich, MA). 5-fluoro-2′-deoxyuridine (FUdR) and chloramphenicol (chlor) were purchased from Sigma Chemical Co. (St.Louis, MO). Hygromycin and gentamycin were purchased from EMD Chemicals Inc (Gibbstown, NJ) and Fisher BioReagents respectively. Proteinase K and cell culture reagents RPMI-1640 and FBS were purchased from Invitrogen Corporation (Carlsbad, CA).
2.2. UV melting curves
All DNAs used in the UV melting curve experiments were purified by gel filtration chromatography, except for 5-GT-(TTAGGG)10-TC-3′, which was purified by denaturing polyacrylamide gel electrophoresis. DNA stock solutions were prepared in pure water and concentrations were determined by UV absorbance at 260 nm at 85°C on a Varian Cary 3 Bio spectrophotometer. At high temperature the bases are presumably unstacked, and the extinction coefficient can be calculated as the sum of the individual bases. The DNA base extinction coefficients were obtained from the literature [42]. Solutions containing 2.5 μM DNA in 10 mM Tris-HCl (pH 7), 100 mM KCl and 1.0 mM EDTA were prepared in 1 cm pathlength quartz cuvettes. Samples were placed in a Varian Cary 3 Bio spectrophotometer equipped with a thermoelectrically controlled multicell holder. The solutions were heated to 90°C and equilibrated for 5 min. Then a cooling gradient was applied at a rate of 1°C/min down to 25°C, when samples were equilibrated for 5 min before starting a heating gradient at the same rate up to 90°C. The absorbance at 295 nm was recorded as a function of temperature every 0.5°C. Melting temperatures were estimated by calculating the first derivative of the melting curve then determining the maximum value. Each melting curve was normalized by dividing the entire curve by the minimum absorbance values at 295 nm.
2.3. Cell culture
The non-tumorigenic human lymphoblastoid cell line LCL-721 was used in all experiments. These cells are EBV-transformed and were established from a clinically normal female donor. They were cultured in RPMI-1640 supplemented with 10% FBS and 50 μg/ml gentamycin as described [43].
2.4. Construction of shuttle vectors containing telomeric repeat sequences
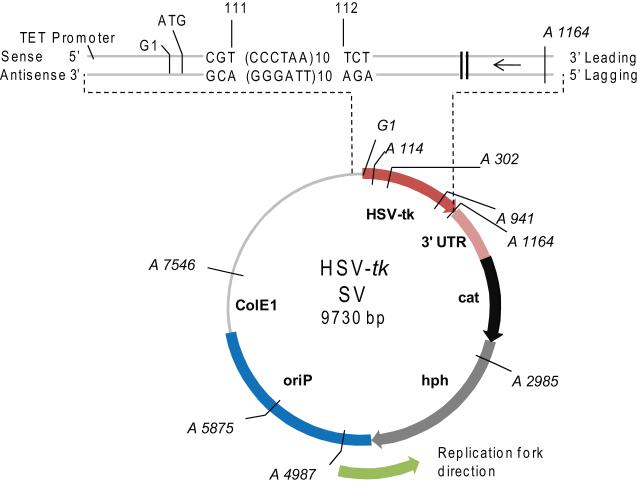
Vectors containing various telomeric repeat sequences within the 5′ coding region of the HSV-tk were constructed as previously described [44]. Briefly, oligonucleotides containing telomeric repeat sequences were annealed and cloned into the BsiWI and MluI sites of HSV-tk gene on the pGTK4 plasmid [45] (Supplemental Table S1). Telomeric repeats from human [TTAGGG]6, [TTAGGG]10, [CCCTAA]6, [CCCTAA]10 and ciliates [TTGGGG]10, [CCCCAA]10, [GGGGTTTT]5 and [CCCCAAAA]5 were inserted in-frame into the HSV-tk gene between bases 111 and 112 (Fig. 2). Only the sequence of the HSV-tk antisense strand will be referred to throughout the manuscript. To avoid introducing a rare codon that would lead to insufficient production of the HSV-tk reporter gene product, one of the telomere repeats of the [TTAGGG]6, [TTAGGG]10, and [TTGGGG]10 vectors was interrupted and the [GGGGTTTT]5 repeats began with a run of Gs rather than Ts (Supplemental Table S1; Figure 6). Vector names remained the same for simplicity. The HSV-tk gene was then subcloned into pND123 shuttle vector to generate the pJY parent control vector [43, 46] and the various telomeric repeat containing shuttle vectors. All vectors were introduced and amplified in the DH5α bacterial strain and the telomeric repeat sequence in each vector was confirmed by DNA sequence analysis.
Fig. 2.
pJY shuttle vector indicating the site of telomeric repeat insertion within the HSV-tk gene. The positions of the mutagenic reporter HSV-tk gene including the 3′UTR, EBV (oriP) origin of replication, ColE1 bacterial origin of replication, choramphenicol acetyltransferase gene (cat) for selection in bacteria, and hygromycin phosphoryltransferase gene (hph) for selection in human cells are indicated. The region from the Tet promoter to the 3′end of the HSV-tk gene is amplified to indicate insertion of the telomeric repeats in-frame between positions 111 and 112. Human repeats in the correct orientation are shown as an example. The arrow indicates the primary direction of replication in human cells and bacteria. The positions of restrictions sites used to screen for large deletions or rearrangements are shown; A = AvaI, and G = BglII.
Fig. 6.
Spectra of mutations arising within the telomeric repeats for the various shuttle vectors. The sequences of the various telomere repeats (upper case) with flanking HSV-tk coding sequence (lower case) are shown 3′ to 5′ for the lagging strand of the vector (see Fig. 2). Sequence alterations in the mutants are shown in bold. A line indicates deleted sequence. The clone from which the mutant was derived is shown, and the number (no.) next to the clone indicates the number of mutant isolates from the clone that exhibited the indicated mutation (See Supplemental Tables S6-S10). Arrow indicates the direction of the replication fork.
2.5. HSV-tk mutational analyses of telomeric repeat shuttle vectors
Vectors (10 μg) were electroporated into 107 LCL721 cells and plasmid-bearing cells were selected and cloned as previously described [44]. To test for the frequency of pre-existing mutations in the vectors generated spontaneously during vector propagation and selection in E. coli, background HSV-tk mutant frequencies of all vectors were calculated as previously described [45]. To avoid the selection and propagation of LCL721 clones that received a vector with a pre-exisiting mutation, cells were cloned at densities of 1-20 cells/well that were less than 0.1 × (1/μ), where μ is the background HSV-tk mutant frequency in E. coli [44]. Each clonal population was propagated for 26 – 29 generations in media containing 150 μg/ml hygromycin, after which an alkaline extraction method [43] was used to isolate shuttle vector DNA from 2 – 3 ×108 cells.
To determine HSV-tk mutant frequencies, the isolated shuttle vectors were transformed into FT334 bacteria (recA113, upp, tdk) followed by selective plating on Vogel Bonner Minimal Salts media (VBA) supplemented with 50 μg/ml chlor with and without 40 μM FUdR [47]. Chlor selects for plasmid-bearing bacteria and FUdR selects for bacteria containing shuttle vectors with a mutation that inactivates the HSV-tk gene product. The HSV-tk mutant frequency was determined as the number of FudR resistant colonies divided by the total number of plasmid-bearing colonies. HSV-tk mutant rates were calculated as the mutant frequency divided by the number of cell generations the clone was propagated to at the time of DNA isolation. Median mutant rates were determined for each telomeric shuttle vector and were analyzed by the non-parametric Mann Whitney test for pair-wise comparisons. Values determined to be outliers by the Grubb’s statistical test for outliers were excluded.
2.6. Generation of HSV-tk telomeric shuttle vector mutation spectra
Restriction enzyme digests and DNA sequencing were used to determine the types and locations of mutations in the mutant shuttle vectors isolated after replication in the human cells. For this, plasmids from 5-10 independent HSV-tk mutants from at least 3 clones for each shuttle vector were isolated and analyzed as described previously [44]. After electroporation of replicated shuttle vectors into FT334 cells, the bacteria were placed on ice and aliquoted into multiple tubes containing 1 ml VBA broth. After the 2h recovery period at 37°C, each culture was plated on selective media, and one FUdR-resistant mutant was isolated for plasmid purification and sequencing. This ensures that any mutational hotspots were not due to division of bacteria harboring HSV-tk mutant vectors during the 2 h recovery. To identify mutants with large deletions or rearrangements the plasmids were digested with AvaI and BglII restriction enzymes (Fig. 2). For this analysis each mutant obtained from a single clone was considered independent since the low resolution of this assay cannot distinguish between potential siblings and independent events. Sequence changes and mutations within the promoter and coding region of the HSV-tk gene, as well as the telomeric inserts, were determined by dideoxy DNA sequencing at ACGT Inc. (Wheeling, IL). DNA sequence analysis was done using Align-X software of Vector NTI Advance (Invitrogen Corporation). Although rare, some mutants from the same clone exhibited the identical mutation due to either that mutation occurring independently in different plasmids within a clone, or due to an early mutagenic event that was replicated multiple times producing mutant siblings. To maintain rigor and consistency, mutants from the same clone that exhibited the identical mutation were considered siblings and that mutation was scored once. The same mutations occurring in different clones were considered independent.
3. Results
3.1 Ciliate telomeric repeats form more stable G4 structures compared to human
Previous biophysical studies showed that O. nova telomeric (GGGGTTTT)3GGGG substrates formed significantly more stable G4 structures compared to human telomeric (GGGTTA)3GGG substrates [29]. We directly compared the G4 structure stability of the human, O.nova and T.thermophila telomeric repeats in the context of the shuttle vector flanking sequence in one orientation. The various telomeric repeats were inserted in-frame between positions 111 and 112 of the HSV-tk reporter gene cassette on the shuttle vector, as previously described for other repeat sequences [43, 44] (Fig. 2). G4 formation and relative thermal stability of the inserted sequences was measured by standard UV melting experiments. G4 structures exhibit hypochromic transitions as a function of temperature at 295 nm absorbance, which can serve as a signature for G4 formation [48, 49] (Supplemental Fig. S1). Melting curves for oligonucleotides GT-(TTAGGG)4-TC, GT-(TTAGGG)6-TC, GT-(TTGGGG)4-TC and GT-(TTTTGGGG)4-TC in 100 mM KCl (Fig. 1) yielded melting temperatures (Tm) of 52.5, 53.5, 83.0 and 81.0°C, respectively. Similar results were obtained with 100 mM NaCl, which also promotes G4 folding, although the G4 stabilities were decreased relative to 100 mM KCl (Supplemental Table S2). Although the O. nova G4 DNA exhibited large hysteresis between the heating and cooling curves, the melting curves and Tm values were not dependent on substrate concentration (Supplemental Fig. S2). This indicates that an intra-molecular G4 structure was formed but that the re-folding rate was slow relative to the cooling rate for the experiment. These data confirm that the telomeric repeats with flanking sequence can form intra-molecular G4 DNA, and that the ciliate G4 units with four quartets exhibit increased thermal stability compared to human G4 units with three quartets.
Next we asked whether sequences that can form two G4 units (at least 8 G-runs) fold into more stable structures compared to sequences that form a single G4 unit (< 8 G-runs). We compared the thermal stabilities of TC-(TTAGGG)6-GT to TC-(TTAGGG)10-GT, which represent the repeat number inserted into the shuttle vector (Figs. 1A and B). The Tm value for ten repeats at 42.8°C in KCl was 9.3-10.3°C lower than the Tm for the shorter oligonucleotides containing 4 or 6 repeats of the human telomeric sequence. Thus, the potential to form two G4 structures for TC-(TTAGGG)10-GT did not increase G4 stability. Importantly, these studies confirm that telomeric repeats of lengths that were inserted into the shuttle vector (6 and 10) can form uni-molecular G4 structures.
3.2. Experimental system for analysis of telomeric DNA mutagenesis
Deletions leading to telomere loss even in cells lacking the WRN helicase are rare but can still critically impact cell function [15, 50]. The shortest telomere, rather than average telomere length, determines cell survival and genome stability [51]. Therefore, we required a highly sensitive assay to detect the types of mutagenic events that impact telomere structure and function, namely deletions and rearrangements. The in vitro/ex vivo shuttle vector HSV-tk mutagenesis assay was chosen for its proven ability to detect rare spontaneous mutations (frequencies as low as 1×10−5) [43], and for the multiple unique advantages it offers for analyzing telomeric repeat replication. First, episomal vectors avoid complications of random insertion at sites of endogenous telomeres. Second, loss of telomeric DNA in the vector will not impact cell survival, unlike loss of endogenous telomeres. Third, oriP episomes are used to study human chromosome replication because they replicate once in S-phase, form chromatin structure, and segregate with sister chromatids [52, 53]. Lastly, the vectors have a defined EBV oriP replication origin and are replicated by the host proteins and the oriP binding protein EBNA-1 which lacks enzymatic activity [54, 55].
Replication of the shuttle vectors initiates at the oriP DS element and terminates at the 20 tandem FR repeats, which causes primarily unidirectional replication [56-62]. This affords the unique opportunity to examine the mutagenic potential of the TTAGGG (G-rich) sequence when replicated by lagging strand versus leading strand DNA synthesis. Single strand gaps during lagging strand replication are thought to permit G4 folding. The correct orientation on human chromosomes is the 5′-TTAGGG-3′ sequence on the lagging strand, therefore, we named all the vectors according to the lagging strand sequence. Since the replication fork starts at the 3′ end of the HSV-tk gene, the lagging strand is the antisense strand. For example, the [TTAGGG]10 vector has the correct repeat orientation with the G-rich sequence on the lagging strand (Fig. 2), and the [CCCTAA]10 vector has the reverse orientation with the G-rich sequence on the leading strand. The assay will detect any mutation that inactivates the HSV-tk gene product, thus, mutation frequencies and specificities in the coding region serve as an internal control. Importantly, we can detect potential deletions or rearrangements induced by the presence of the telomeric repeats because flanking HSV-tk sequence is also affected. The assay is designed to detect events that are most likely to impact the telomere integrity, rather than minor alterations in repeat number.
3.3. Stability of telomeric vectors in bacteria
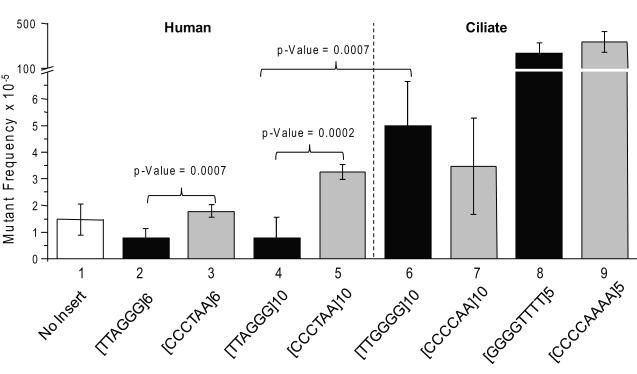
We first examined the stability of the telomeric repeat vectors in E.coli by determining the mutant frequency for each compared to the control vector which lacks telomeric repeats. The shuttle vectors have the bacterial replication origin derived from pBR322 which is also unidirectional (Fig. 2 arrow) [63] in the same direction as oriP. The vectors containing the human telomeric repeats were stable in bacteria and yielded HSV-tk mutant frequencies that were similar to the control vector (Fig. 3, bars 1-5). Increasing the repeat number from six to ten did not significantly impact the vector mutant frequency (Fig. 3 compare bars 2 and 4; 3 and 5). Surprisingly, the [TTAGGG]10 vector with the G-rich lagging strand orientation was highly stable and yielded an average mutant frequency of 0.77×10−5 that was 4-fold lower than the mutant frequency obtained for G-rich leading strand [CCCTAA]10 vector (3.2×10−5). The difference was statistically significant whether there were six repeats (p-value = 0.0007) or ten repeats in the vectors (p-value= 0.0002). Therefore, the orientation of the human telomeric repeats influences the stability of the vectors in E. coli.
Fig. 3.
HSV-tk mutant frequencies of telomeric shuttle vectors in recA- E. coli. Mutant frequencies are based on 1.3×104 to 1.7×106 chloramphenicol-resistant bacteria colonies. The sequence of the telomeric repeats on the antisense (lagging) strand is indicated. Black bars indicate that the G-rich strand of telomeric repeats in the vector is replicated as the lagging strand (correct orientation), and the grey bars indicate replication as the leading strand (reverse oriention). Values represent the mean and standard deviations from at least 5 independent experiments.
Next we examined the stability of the vectors with ciliate telomeric repeats. In contrast to the human repeats, the mutant frequencies obtained for the ciliate telomeric vectors were statistically similar regardless of repeat orientation (G-rich lagging vs. G-rich leading strand) (Fig. 3, compare bars 6 to 7 and 8 to 9). However, the T.thermophila [TTGGGG]10 vector with the G-rich lagging strand sequence yielded a mutant frequency of 5.0 ×10−5 that was significantly higher than the human [TTAGGG]10 vector with the G-rich lagging strand (0.77×10−5) (Fig. 3 compared bars 4 and 6, p-value = 0.0007). These repeats differ by one base. Interestingly, when the G-rich sequence was on the leading strand, the mutant frequencies for the ciliate [CCCCAA]10 and human [CCCTAA]10vectors were not statistically different (Fig. 3). Thus, T.thermophila telomeric vectors were less stable than human telomeric vectors only when the repeats were in the G-rich lagging strand orientation.
The vectors containing the O.nova [GGGGTTTT]5 and [CCCCAAAA]5 telomeric inserts showed a >100-fold increase in mutant frequency over the control vector (Fig. 3). Since the O. nova repeats are multiples of 8, changes in repeat number alter the reading frame and inactivate the HSV-tk gene. To determine if this was the reason for the dramatic increase in mutant frequency we sequenced the mutants. Most (14/15) of the [GGGGTTTT]5 vector mutants had alterations in the telomeric repeat number (+1 repeat, 2 events; − 1 repeat, 12 events). Therefore, the O.nova vector affords us the opportunity to examine alterations in repeat number in telomeric repeats that share similar properties as human repeats (i.e. tandem G-runs).
3.4. Replication of telomeric vectors in human cells
The vectors with telomeric inserts were introduced into LCL721 human lymphoblastoid cells by electroporation, and plasmid bearing clones were expanded for 25 to 29 population doublings [44]. Mutant rates were calculated from 4 – 7 clones from each independent experiment. Shuttle vector DNA was extracted and transformed into FT334 bacteria and mutant frequencies were calculated and converted to mutant rates (see Materials and Methods). The median mutant rate for the control vector was calculated as 3.3 × 10−6 from a total of 12 clones isolated from two independent experiments. Importantly, the median mutant rates from each experiment were very similar (4.4×10−6 and 3.4×10−6, Supplemental Table S3) and were consistent with previous studies (3.6 × 10−6 [44]).
3.4.1. Stability of human telomeric repeat vectors in human cells
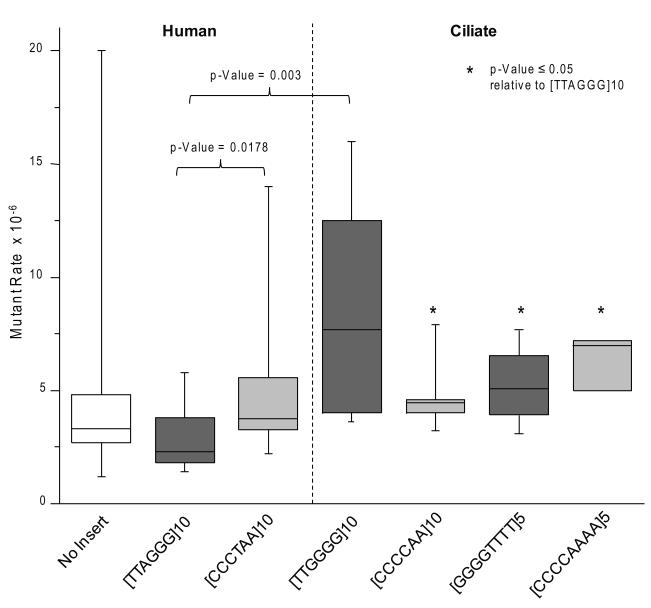
Next we examined the mutagenic potential of vectors with human telomeric repeats after replication in human LCL721 cells. The human telomeric repeat shuttle vectors were highly stable and yielded median mutant rates that were not statistically different from the control vector (Fig. 4 and Supplemental Tables S3 and S5). The data are presented in Fig. 4 as a box plot in which each box represents the interquartile range (25th to 75th percentiles), horizontal bars in the box represent the medians, and the vertical bars represent the maximal and minimal values. When the G-rich sequence was on the lagging strand the vector median mutant rate was 2.3×10−6 ([TTAGGG]10 vector), which was significantly lower than the mutant rate obtained for the G-rich leading strand vector ([CCCTAA]10 vector = 3.8×10−6)(p-value = 0.0178, Mann Whitney) (Fig. 4 and Supplemental Table S5). Importantly, the G-rich lagging strand [TTAGGG]10 vector has the repeats in the correct orientation as they occur at chromosomal telomeres in humans. Similar to results in bacteria, the human telomeric repeat sequences are highly stable in human LCL721 cells, and the repeat orientation significantly impacts the overall median mutant rates of the vectors.
Fig. 4.
Median mutant rates of telomeric shuttle vectors in LCL721 human cells. Clonal populations of human cells bearing various shuttle vectors were expanded for 26 to 29 generations. Shuttle vector DNA was isolated from each clone and the HSV-tk mutant frequency was determined as described in Materials and Methods. The mutant rate was calculated by dividing the mutant frequency by the number of generations the clone was expanded to at the time of shuttle vector isolation (see also Supplemental Tables S3 and S4). Each box represents the interquartile range (25th to 75th percentiles). The horizontal bars in the boxes represent the medians. The vertical bars represent the range of values included in the statistics, from the maximal to the minimal value. The following outliers were excluded: 27 [no insert control]; 71 [CCCTAA]10; and 38 [CCCCAAAA]5. * indicates p≤ 0.05 relative to [TTAGGG]10 (Mann-Whitney rank sum test (two-sided)).
3.4.2. Stability of vectors with human telomere repeats compared to ciliate repeats
Based on the model that G4 DNA in telomeric sequences may lead to replication fork demise, we predicted that telomeric sequences that form more stable G4 DNA would have a greater destabilizing effect on the vectors, compared to the human repeats. Ciliate telomeric repeats (T.thermophila and O.nova) form G4 structures of greater thermal stability than human sequences (Fig. 1). We compared T.thermophila repeats to human repeats since both are multiples of six. Vectors with the correct G-rich lagging strand orientation yielded a median mutant rate for T.thermophila [TTGGGG]10 vector that was 3.3-fold higher than the mutant rate obtained for the human [TTAGGG]10 vector; 7.7×10−6 and 2.3×10−6, respectively (Fig. 4). This difference was statistically significant (p-value = 0.003) (Supplemental Tables S4-S5). In contrast, the vectors with the G-rich leading strand yielded very similar median mutant rates; 4.4×10−6 for the T.thermopila [CCCCTT]10 vector, compared to 3.8×10−6 for the human [CCCTAA]10 vector (Fig. 4, Supplemental Tables S3-S5). G4 units are expected to fold more frequently in the lagging strand, compared to leading strand, and these data suggest that the T.thermopila G4 units may impede replication more than human G4 units. Consistent with this, we observed that the vector mutant rate was nearly 2-fold higher when the T.thermopila G-rich sequence was on the lagging strand ([TTGGGG]10 vector), compared to its presence on the leading strand ([CCCCAA]10 vector) (Fig. 4, Supplemental Table S4). However this difference was not statistically significant, perhaps due to the strikingly large spread of values obtained for the [TTGGGG]10 vector (Fig. 4). In summary, vectors with the correct human repeat orientation (G-rich lagging) were more stable than vectors with T.thermopila repeats. The orientation of the telomeric repeats for both human and T.thermopila influences the vector mutant rate, but in opposite directions.
3.4.3. Vectors with O.nova repeats are more stable in human cells compared to bacteria
To determine whether tandem telomeric repeats are susceptible to the same types of repeat alterations that occur in other simple tandem repeat (STR) sequences [43], we took advantage of the O.nova (GGGGTTTT) sequence. Alterations in O.nova repeat number inactivate the HSV-tk gene (Fig. 2). These repeats form stable G4 structures (Fig. 1), and resemble human repeats in that they consist of alternating pyrimine and purine runs and the G-rich sequence is present on the lagging strand [64]. Thus, we asked whether these characteristics of telomeric repeats make them susceptible to alterations in repeat number. Replication of the G-rich lagging strand [GGGGTTTT]5 vector yielded a mediate mutant rate of 5.1×10−6 that was slightly lower than the rate for the vector with the G-rich leading strand ([CCCCAAAA]5 vector = 7.1×10−6) (Fig. 4, Supplemental Tables S4-S5). However, this difference was not significant, and neither vector yielded a mutant rate that was statistically different from the control (Supplemental Table S5). This is in stark contrast to the 100- and 300-fold increase in mutant frequencies observed for these vectors in bacteria, compared to controls (Fig. 3). Thus, while vectors containing O.nova repeats are highly unstable in E. coli, they exhibit similar stabilities as a vector lacking these repeats after replication in LCL721 human cells.
3.5. Insertion of telomeric repeats decreases the frequencies of vector alterations
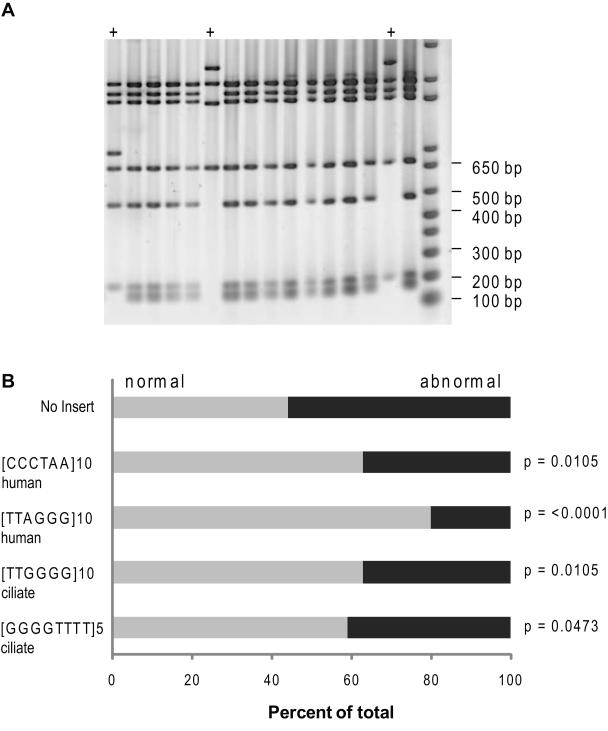
Next we asked whether the mutation types and frequencies arising within the shuttle vectors were influenced by the various telomeric repeats. We isolated 5 to 10 independent mutants from about 3 clones for each shuttle vector for mutational analysis (see Supplemental Tables S3-S4 for clones). Previous reports indicate that H-DNA and Z-DNA forming sequences increase the frequency of large deletions and rearrangements within a shuttle vector [35, 36]. To test whether this was true for the telomeric G4 forming sequences, the mutant vectors were digested with specific restriction enzymes to test the integrity of the shuttle vector as described previously [43] (Fig. 2). The proportion of mutant vectors harboring the human G-rich lagging ([TTAGGG]10) or leading ([CCCTAA]10) strand repeats that exhibited abnormal digest patterns was significantly lower at 20% or 37%, respectively, compared to the control vector (56%) (p-values were <0.0001 or 0.01, respectively, Fisher’s exact test) (Fig. 5). Similar results were observed for the vector mutants bearing ciliate telomeric repeats [TTGGGG]10 and [GGGGTTTTT]5 (p-values = 0.01 and 0.047, respectively; Fisher’s exact test). Since the orientation of the ciliate repeats did not significantly alter the overall mutant rates (Fig. 4), we only analyzed mutants of the vectors with the correct G-rich lagging strand orientation. In summary, the insertion of telomeric repeats decreased the frequency of shuttle vector alterations (i.e. large deletions and rearrangements induced throughout the vectors), and the human repeats in the correct G-rich lagging strand orientation yielded the most significant decrease.
Fig. 5.
Proportion of mutants with large deletions and alterations throughout the shuttle vector. Shuttle vector DNA from 5 to 10 mutants isolated from approximately 3 clones for each shuttle vector was digested with AvaI and BglII restriction enzymes for 2 hr at 37°C. Reactions were run on an 0.8% agarose gel. A representative image is shown in panel A. The + indicates an abnormal digest pattern. B, the percent of mutant DNA exhibiting a wild type digest pattern (grey bars) or an abnormal digest pattern (black bars) was calculated as a function of total mutant vectors that were digested. All mutants isolated from the same clone were considered independent (see Materials and Methods). p-values calculated from Fisher’s exact test.
Deletions that span the telomeric repeats could have arisen from breaks induced by the G4 forming sequences. To determine the frequencies of defined deletions and rearrangements that involved loss of the telomeric repeats we sequenced the entire 1110-bp HSV-tk gene and upstream promoter region. The median mutation rate for defined deletions/rearrangements occurring within the HSV-tk gene was calculated by multiplying the proportion of these events by the total median mutant rate (Fig. 4 and Table 1). The deletion/rearrangement mutation rates for human telomeric vectors were not greatly influenced by repeat orientation and were decreased approximately 2- to 3- fold (5.6×10−7 and 7.6×10−7), compared to the control and T.thermophila vectors (13×10−7 and 18×10−7, respectively) (Table 1). Vectors with the O.nova repeats exhibited the lowest rates at 1.5×10−7. However, there was no significant bias for deletions/rearrangements involving loss of the telomeric repeats for any of the vectors (Table 1). The proportions of events involving loss of the telomeric repeats were similar to the proportion of events that spanned the repeat insertion site (111-112 bps) for the control vector (Fisher’s exact test) (Table 1). This indicates that the presence of telomeric repeats did not destabilize the insertion region in the HSV-tk gene (Fig. 2).
Table 1.
Sequenced deletions and rearrangements arising in the HSV-tk gene that involve the telomeric repeat region after replication in human cells
| Mutation Class | Telomeric Insertsa | ||||
|---|---|---|---|---|---|
| No insert | [CCCTAA]10 | [TTAGGG]10 | [TTGGGG]10 | [GGGGTTTT]5 | |
| Deletions and Rearrangements | |||||
| Median mutation rate | 13×10−7 | 5.6×10−7 | 7.6×10−7 | 18×10−7 | 1.5×10−7 |
| Proportion of total mutations | 0.39 | 0.15 | 0.33 | 0.24 | 0.030 |
| Number | 13 | 6 | 10 | 9 | 1 |
| Loss of repeats (proportion)b | 6 (0.46)c | 3 (0.50) | 5 (0.50) | 4 (0.44) | 0 |
| Retain repeats (proportion)b | 7 (0.54) | 3 (0.50) | 5 (0.50) | 5 (0.56) | 1 |
Values were derived from sequenced HSV-tk mutants in Supplemental Tables S6-S10. Small deletio ns (≤ 12 bp) were excluded. Identical mutants from a single clone were counted once to avoid including potential siblings.
DNA sequence of the lagging strand of replication
The proportion is based on the number of deletions and rearrangements from line 3.
Deletions or rearrangements that involved loss of base pairs 111 and 112. This is the region in which telomeric repeats were inserted for the other vectors (see Figure 2).
3.6. Ciliate telomeric repeats exhibit higher mutation frequencies than human repeats
Next we compared the rates of mutations occurring within the various telomeric repeats. Mutational events that affected only the HSV-tk coding sequence served as an internal control, and included base substitutions, frameshifts and small deletions (Table 2). The proportions of HSV-tk coding sequence mutations were similar among the various vectors (Table 2). Furthermore, the spectra of mutations in the HSV-tk coding sequence were all dominated by base substitutions, and the distribution of mutations was not altered by the telomeric repeats in an obvious manner (Supplemental Tables S6-10). However, we observed a significantly higher proportion of mutations within the ciliate [TTGGGG]10 and [GGGGTTTT]10 repeats compared to the human [TTAGGG]10 repeats (p-value = 0.0309 and 0.0309, respectively, Fisher’s exact test). This translated to a near 10-fold increase in median mutation rate for the T.thermophila repeats (14×10−7) compared to human repeats in the same orientation (1.5×10−7) (Table 2). The occurrence of telomeric mutations in the human repeats was rare and was not significantly influenced by repeat orientation. The mutations detected were alterations in repeat number that were accompanied by mutations elsewhere in the HSV-tk gene, and a frameshift within one telomere repeat (Fig. 6 and Supplemental Tables S6-7). The mutations in the T.thermophila repeats also included alterations in repeat number, but more strikingly, we observed a total of 8 mutants containing large deletions with an endpoint in the [TTGGGG]10 repeats (Fig. 6). Since six of these mutants were isolated from the same clone (C) they could potentially be siblings and were scored once (Tables 1-2), but it is worth noting that siblings were rare (Supplemental Tables S6-10). These data indicate that ciliate [TTGGGG] repeats induce deletions likely due to breaks within the repeats.
Table 2.
Defined mutational events within the telomeric repeats and HSV-tk coding regions after replication in LCL721 human cells
| Mutation Class | Telomeric Inserts |
||||
|---|---|---|---|---|---|
| No insert | [CCCTAA]10 | [TTAGGG]10 | [TTGGGG]10 | [GGGGTTTT]5 | |
| Telomeric Sequence a | |||||
| Median mutation rate | n/a | 1.0×10−7 | 1.5×10−7 | 14×10−7 | 9.2×10−7 |
| Number | n/a | 1 | 2 | 7 | 6 |
| Proportion of total | n/a | 0.025 | 0.067 | 0.18 | 0.18 |
| HSV-tk Coding Region b | |||||
| Median mutation rate | 20×10−7 | 31×10−7 | 15×10−7 | 51×10−7 | 40×10−7 |
| Number | 20 | 33 | 19 | 25 | 26 |
| Proportion of total | 0.61 | 0.83 | 0.63 | 0.66 | 0.79 |
| Total Mutations | 33 | 41 | 30 | 38 | 33 |
Values were derived from sequenced HSV-tk mutants in Supplemental Tables S6 – S10. Identical mutants from a single clone were counted once to avoid including potential siblings
Includes large deletions with a border in the telomeric repeats
Includes base substitutions, frameshifts and small deletions < 12 bp confined to the HSV-tk coding sequence.
The O.nova [GGGGTTTT]5 vectors exhibited an increase in median telomeric mutation rate (9.2×10−7), compared to vectors with human repeats (1.5×10−7), due to the potential for alterations in repeat number to shift the HSV-tk reading frame (Table 2). Consistent with this, all of the sequenced telomeric mutations involved repeat alterations, and the loss of telomeric repeat units occurred more frequently compared to repeat additions (Fig. 6). Some mutants from the same clone showed the identical mutation and were considered to be siblings (Table 2). But we cannot rule out the possibility that these were independent events, especially considering that similar mutations were observed in multiple distinct clones (Fig. 6). However, none of sequenced O.nova mutants exhibited large deletions with endpoints in the [GGGGTTTT]5 region, in contrast to the T.thermophila repeats. Thus, the mutation spectra revealed that the T.thermopila and O.nova repeats yielded higher telomeric mutation rates compared to the human repeats due to distinctly different mechanisms.
4. Discussion
In this study we measured the mutagenic potential of various telomeric repeats in E. coli and clonal populations of human lymphoblastoid cells as a function of repeat orientation and G-quadruplex thermal stability. To our knowledge this is the first report of spontaneous mutation rates of telomeric repeat sequences using a shuttle vector mutagenesis assay. This highly sensitive assay allowed us to quantitate rare and independent mutagenic events that are expected to impact telomere integrity and function, namely large deletions and rearrangements. Importantly, the unidirectional replication origins afforded a unique opportunity to compare leading versus lagging strand replication of the G-rich sequences which possess quadruplex forming potential. As expected we observed that molecules with telomeric repeats from ciliates formed G4 structures with higher thermal stability compared to human repeats in vitro. Consistent with this, shuttle vectors with T.thermophila [TTGGGG]10 repeats exhibited greater mutant frequencies compared to vectors with human [TTAGGG]10 repeats in both bacteria and human cells. This translated to a near 10-fold higher rate of mutations within the T.thermophila repeats compared to human repeats (Table 2). Interestingly, the vectors with the human repeats in the correct orientation (lagging G-rich strand) were highly stable, and exhibited significantly lower mutant frequencies compared to vectors with the reverse repeat orientation (leading G-rich strand) for both bacteria and human cells. In general, we observed that the mutagenic potential of various telomeric repeats is relatively low in human cells, but is influenced by repeat orientation and thermal stability of the folded G4 DNA.
Replication of G-rich sequences as the lagging strand template mimics the process of telomere replication on human chromosomes. The result that the human telomeric repeat vector with the G-rich lagging strand sequence yielded significantly lower mutant frequencies compared to the vector with G-rich leading strand sequence, in both human cells and bacteria, was unexpected (Figs. 2-3). Transient ssDNA in Okazaki fragment processing is predicted to permit G4 folding, and is thought to explain the preferential loss of telomeres replicated from the G-rich lagging strand in cells defective for WRN helicase, FEN1, and POT1 proteins [15, 31, 32, 65]. G4 impediments to replication were also attributed to the deletions and rearrangements in G-rich repeat sequences in S.cerevisiea lacking Pif1 helicase [38] and C.elegans lacking the FANCJ helicase homolog Dog-1[37]. One possible explanation for our results is that quadruplexes did not form, or did not fold more frequently when the G-rich runs were present on the lagging strand versus the leading strand. We believe the former is unlikely because TTAGGG repeats were observed to form G4 DNA on plasmids in vivo during transcription [26]. Another possibility is that regulated G4 DNA folding is favorable for replication of human telomeric repeats rather than detrimental. A recent study reported a positive role for G4 DNA in the maintenance of telomere lengths, and suggested that intra-molecular G4 DNA promotes telomerase translocation in S.cerevisiea [66]. We propose that G4 DNA may recruit helicases such as the RecQ helicases WRN and BLM or FANCJ in human cells, and RecQ in bacteria, which all exhibit high affinity for, and unwind G4 structures in vitro [67-69]. These enzymes are known to act in pathways that prevent replication fork demise, restore stalled replication forks, and promote genomic stability [70]. Consistent with this, the presence of TTAGGG repeats on the lagging strand had the greatest stabilizing effect on the shuttle vectors (Figs. 2-3, 5), and offered the potential for G4 folding during Okazaki fragment processing.
Our studies indicate that the thermal stability of G4 structures also influences the mutagenic potential of G4 forming sequences. The higher thermal stability of the ciliate repeat G4 DNA, compared to human repeat G4 DNA (Fig. 1), agrees with previous work and is due to the extra guanine tetrad in the ciliate repeats [29, 71]. Even in the presence of a complementary strand the T.thermophila (TTGGGG)4 strand predominately formed G4 DNA, whereas the human (TTAGGG)4 strand existed in a mixture of G4 and duplex DNA [71]. This correlated with a higher mutant frequency for vectors with the TTGGGG lagging strand repeats, compared to vectors with this sequence on the leading strand, and vectors with the human TTAGGG lagging strand repeats in both bacteria and humans (Figs. 3-4). Thus, in the case of the T.thermophila repeats, the expected higher G4 potential when the G-rich sequence was on the lagging strand (correct orientation), compared to leading strand (reverse orientation), was detrimental rather than beneficial unlike the human repeats. The G4 forming sequence not only determines thermal stability but also topology (i.e. parallel vs. anti-parallel, loop size between tetrads, arrangement) [72], therefore, it was proposed that various helicases exhibit differing efficiencies of G4 unwinding depending on the G4 structure [38]. Humans and E.coli may possess an abundance of helicases, such as RecQ helicases, that are well suited for disrupting human telomeric G4 DNA, compared to other types of G4 sequence structures. Recruitment of telomeric proteins TRF1 and TRF2 to the human repeats may further enhance their stability within the shuttle vector. In human cells, we observed a higher rate of mutations within the T.thermophila TTGGGG repeats compared to human repeats in either orientation, and we observed large deletions with endpoints in the TTGGGG repeats only (Fig. 6). These deletions suggest the T.thermophila repeats may have impeded replication fork progression. Chromatin immuno-precipitation studies showed that TRF1 and TRF2 proteins bind the TTAGGGTTA site in the oriP in human cells [44, 73], indicating that the human telomeric sequence we inserted into the shuttle vectors should also bind telomeric proteins. This raises the possibility that replication might have initiated within the telomeric repeats, but this is unlikely because only oriP can support plasmid establishment and long term replication in clonal expansion [74]. TRF1 and TRF2 interact with G quadruplex resolving helicases including WRN and BLM [75, 76], and may enhance recruitment of these helicases to the shuttle vectors with human telomeric repeats. Recent work indicates that TRF1 is required to prevent replication fork stalling at human telomeric repeats [14]. It is important to note that the structure of the TRF1 and TRF2 proteins with respect to the progressing replication fork will be influenced by the orientation of their TTAGGG binding sequences, and this might also influence replication fidelity.
Since O.nova telomeric repeats are multiples of 8, we could directly compare the stability of sequences with telomeric character and G-quadruplex forming potential, to Short Tandem Repeat (STR) sequences elsewhere in the genome. Alterations in repeat number shift the HSV-tk reading frame. Surprisingly, the vectors with O.nova (GGGGTTTT)5 repeats in either orientation were highly unstable in E. coli and exhibited a near 100- to 300-fold increase in mutant frequency compared to controls due to changes in repeat number (Fig. 3 and data not shown). In stark contrast, these repeats were stably replicated in human LCL721 cells (Fig. 4), and while some mutants also exhibited alterations in repeat number the frequencies were very low (Table 2 and Fig. 6). The human and bacterial replicative polymerases may differ in their tendency to slip in regions with high G-C content. However, our data indicate that in normal human cells the G4 forming potential of repeat sequences is unlikely to promote DNA polymerase slippage and primer/template misalignments that could lead to alterations in repeat number. Consistent with this, the vectors with O.nova repeats in either orientation were more stable and exhibited lower mutant rates (5.1×10−6 and 7.1×10−6) than the STR sequences [TTTC/AAAG]9 and [TCTA/AGAT]9 (38×10−6 and 58×10−6) reported earlier using our system in human cells [44]. While the G4 structures from O.nova repeats exhibited similar thermal stabilities as the G4 units from T.thermophila, large deletions were only observed in the T.thermophila repeats (Fig. 6). This suggests that the T.thermophila repeats likely induced replication fork stalling. RecQ helicases have been found to efficiently disrupt O.nova telomeric G4 DNA [69], and the larger (TTTT) loop between G-tetrads compared to the (TT) loop for T.thermophila influences the G4 topology [77]. Thus, not all G4 forming sequences exhibit the same mutagenic potential and mutation specificities.
While differences were observed among the various telomeric repeats, all of the telomeric vectors exhibited relatively low mutant rates after replication in human cells. One limitation of the assay is that the vectors contain only 5 to 10 telomeric repeats, whereas upwards of 800 repeats exist in chromosomal ends [7]. However, the shuttle vector with 10 telomeric repeats approaches the average length of an Okazaki fragment during DNA replication (~140 nt) [78], and thus, mimics the span of ssDNA arising during normal telomere replication that could form G4 DNA. Furthermore, previous reports indicate that a greater number of telomeric repeats does not correlate with more G4 DNA units [79]. In contrast to the G4 forming telomeric repeats, the insertion of sequences of similar length (25-32 bp) that can form alternate non-B structures (H-DNA and Z-DNA), led to a significant increase in deletions [35, 36]. These data suggest that some H-DNA and Z-DNA structures may impede replication or provoke breakage [35, 36], in stark contrast to the telomeric G4 sequences. Our results are consistent with studies that indicate natural interstitial telomeric sequences of perfect repeats in human cells are not prone to breakage [80].
In summary, our experiments indicate that human and ciliate telomeric repeats are stably replicated in normal human somatic non-tumorigenic cells, and that thermal stability and topology of potential G4 structures and repeat orientation influence the mutagenic potential. Our studies suggest that normal human cells have evolved mechanisms, including a plethora of helicases, to facilitate replication through G4 forming telomeric sequences. Consistent with this, the vectors with human repeats in the correct orientation exhibited significantly lower mutant rates than the control and other telomeric vectors (Fig. 4), and the lowest proportion of vector alterations (Fig. 5). These results suggest that human cells have evolved mechanisms for optimizing replication specifically of human telomeres. This may partly explain why telomeres in normal cells are stably replicated despite the tracts of repetitive sequences that can form alternate structures. The mutagenesis assay used in this study now provides a highly sensitive approach for elucidating the roles of various proteins in promoting faithful telomere replication and telomere stability, such as various DNA helicases.
Supplementary Material
Acknowledgements
This work was supported by NIH grants [RO1 ES0515052 (P.L.O.); RO1 CA100060 ( K.A.E.)], the Ellison Medical Foundation (P.L.O.), and the Jake Gittlen Cancer Research Foundation (K.A.E.). We thank members of the Opresko and Eckert lab for critical reading of the manuscript, and Gregory Sowd and Suzanne Hile for technical support and assistance.
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
References
- [1].Palm W, de Lange T. How Shelterin Protects Mammalian Telomeres. Annual Review of Genetics. 2008;42:301–334. doi: 10.1146/annurev.genet.41.110306.130350. [DOI] [PubMed] [Google Scholar]
- [2].Blasco MA. Telomeres and human disease: ageing, cancer and beyond. Nat Rev Genet. 2005;6:611–622. doi: 10.1038/nrg1656. [DOI] [PubMed] [Google Scholar]
- [3].Armanios M. Syndromes of Telomere Shortening. Annual Review of Genomics and Human Genetics. 2009;10:45–61. doi: 10.1146/annurev-genom-082908-150046. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [4].de Lange T. Shelterin: the protein complex that shapes and safeguards human telomeres. Genes Dev. 2005;19:2100–2110. doi: 10.1101/gad.1346005. [DOI] [PubMed] [Google Scholar]
- [5].di Fagagna F. d’Adda, Reaper PM, Clay-Farrace L, Fiegler H, Carr P, Von Zglinicki T, Saretzki G, Carter NP, Jackson SP. A DNA damage checkpoint response in telomere-initiated senescence. Nature. 2003;426:194–198. doi: 10.1038/nature02118. [DOI] [PubMed] [Google Scholar]
- [6].Takai H, Smogorzewska A, de Lange T. DNA damage foci at dysfunctional telomeres. Curr Biol. 2003;13:1549–1556. doi: 10.1016/s0960-9822(03)00542-6. [DOI] [PubMed] [Google Scholar]
- [7].Riethman H. Human telomere structure and biology. Annu Rev Genomics Hum Genet. 2008;9:1–19. doi: 10.1146/annurev.genom.8.021506.172017. [DOI] [PubMed] [Google Scholar]
- [8].Griffith JD, Comeau L, Rosenfield S, Stansel RM, Bianchi A, Moss H, de Lange T. Mammalian telomeres end in a large duplex loop. Cell. 1999;97:503–514. doi: 10.1016/s0092-8674(00)80760-6. [DOI] [PubMed] [Google Scholar]
- [9].Stansel RM, de Lange T, Griffith JD. T-loop assembly in vitro involves binding of TRF2 near the 3′ telomeric overhang. EMBO J. 2001;20:5532–5540. doi: 10.1093/emboj/20.19.5532. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [10].Broccoli D, Smogorzewska A, Chong L, de Lange T. Human telomeres contain two distinct Myb-related proteins, TRF1 and TRF2. Nat Genet. 1997;17:231–235. doi: 10.1038/ng1097-231. [DOI] [PubMed] [Google Scholar]
- [11].Lei M, Podell ER, Cech TR. Structure of human POT1 bound to telomeric single-stranded DNA provides a model for chromosome end-protection. Nat Struct Mol Biol. 2004;11:1223–1229. doi: 10.1038/nsmb867. [DOI] [PubMed] [Google Scholar]
- [12].Ivessa AS, Zakian VA. To fire or not to fire: origin activation in Saccharomyces cerevisiae ribosomal DNA. Genes Dev. 2002;16:2459–2464. doi: 10.1101/gad.1033702. [DOI] [PubMed] [Google Scholar]
- [13].Miller KM, Rog O, Cooper JP. Semi-conservative DNA replication through telomeres requires Taz1. Nature. 2006;440:824–828. doi: 10.1038/nature04638. [DOI] [PubMed] [Google Scholar]
- [14].Sfeir A, Kosiyatrakul ST, Hockemeyer D, MacRae SL, Karlseder J, Schildkraut CL, de Lange T. Mammalian telomeres resemble fragile sites and require TRF1 for efficient replication. Cell. 2009;138:90–103. doi: 10.1016/j.cell.2009.06.021. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [15].Crabbe L, Verdun RE, Haggblom CI, Karlseder J. Defective telomere lagging strand synthesis in cells lacking WRN helicase activity. Science. 2004;306:1951–1953. doi: 10.1126/science.1103619. [DOI] [PubMed] [Google Scholar]
- [16].Martinez P, Thanasoula M, Munoz P, Liao C, Tejera A, McNees C, Flores JM, Fernandez-Capetillo O, Tarsounas M, Blasco MA. Increased telomere fragility and fusions resulting from TRF1 deficiency lead to degenerative pathologies and increased cancer in mice. Genes Dev. 2009;23:2060–2075. doi: 10.1101/gad.543509. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [17].Hile SE, Eckert KA. Positive correlation between DNA polymerase alpha-primase pausing and mutagenesis within polypyrimidine/polypurine microsatellite sequences. J Mol Biol. 2004;335:745–759. doi: 10.1016/j.jmb.2003.10.075. [DOI] [PubMed] [Google Scholar]
- [18].Cha RS, Kleckner N. ATR homolog Mec1 promotes fork progression, thus averting breaks in replication slow zones. Science. 2002;297:602–606. doi: 10.1126/science.1071398. [DOI] [PubMed] [Google Scholar]
- [19].Courcelle J, Donaldson JR, Chow KH, Courcelle CT. DNA damage-induced replication fork regression and processing in Escherichia coli. Science. 2003;299:1064–1067. doi: 10.1126/science.1081328. [DOI] [PubMed] [Google Scholar]
- [20].Petersen S, Saretzki G, von Zglinicki T. Preferential accumulation of single-stranded regions in telomeres of human fibroblasts. Exp Cell Res. 1998;239:152–160. doi: 10.1006/excr.1997.3893. [DOI] [PubMed] [Google Scholar]
- [21].Luu KN, Phan AT, Kuryavyi V, Lacroix L, Patel DJ. Structure of the Human Telomere in K+ Solution: An Intramolecular (3 + 1) G-Quadruplex Scaffold. Journal of the American Chemical Society. 2006;128:9963–9970. doi: 10.1021/ja062791w. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [22].Zahler AM, Williamson JR, Cech TR, Prescott DM. Inhibition of telomerase by G-quartet DMA structures. Nature. 1991;350:718–720. doi: 10.1038/350718a0. [DOI] [PubMed] [Google Scholar]
- [23].Paeschke K, Simonsson T, Postberg J, Rhodes D, Lipps HJ. Telomere end-binding proteins control the formation of G-quadruplex DNA structures in vivo. Nat Struct Mol Biol. 2005;12:847–854. doi: 10.1038/nsmb982. [DOI] [PubMed] [Google Scholar]
- [24].Yang Q, Xiang J, Yang S, Zhou Q, Li Q, Tang Y, Xu G. Verification of specific G-quadruplex structure by using a novel cyanine dye supramolecular assembly: I. recognizing mixed G-quadruplex in human telomeres. Chem Commun (Camb) 2009:1103–1105. doi: 10.1039/b820101c. [DOI] [PubMed] [Google Scholar]
- [25].Paeschke K, Juranek S, Simonsson T, Hempel A, Rhodes D, Lipps HJ. Telomerase recruitment by the telomere end binding protein-beta facilitates G-quadruplex DNA unfolding in ciliates. Nat Struct Mol Biol. 2008;15:598–604. doi: 10.1038/nsmb.1422. [DOI] [PubMed] [Google Scholar]
- [26].Duquette ML, Handa P, Vincent JA, Taylor AF, Maizels N. Intracellular transcription of G-rich DNAs induces formation of G-loops, novel structures containing G4 DNA. Genes Dev. 2004;18:1618–1629. doi: 10.1101/gad.1200804. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [27].Kamath-Loeb AS, Loeb LA, Johansson E, Burgers PM, Fry M. Interactions between the Werner syndrome helicase and DNA polymerase delta specifically facilitate copying of tetraplex and hairpin structures of the d(CGG)n trinucleotide repeat sequence. J Biol Chem. 2001;276:16439–16446. doi: 10.1074/jbc.M100253200. [DOI] [PubMed] [Google Scholar]
- [28].Maizels N. Dynamic roles for G4 DNA in the biology of eukaryotic cells. Nat Struct Mol Biol. 2006;13:1055–1059. doi: 10.1038/nsmb1171. [DOI] [PubMed] [Google Scholar]
- [29].Lee JY, Yoon J, Kihm HW, Kim DS. Structural diversity and extreme stability of unimolecular Oxytricha nova telomeric G-quadruplex. Biochemistry. 2008;47:3389–3396. doi: 10.1021/bi702013d. [DOI] [PubMed] [Google Scholar]
- [30].Neaves KJ, Huppert JL, Henderson RM, Edwardson JM. Direct visualization of G-quadruplexes in DNA using atomic force microscopy. Nucleic Acids Res. 2009;37:6269–6275. doi: 10.1093/nar/gkp679. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [31].Wu L, Multani AS, He H, Cosme-Blanco W, Deng Y, Deng JM, Bachilo O, Pathak S, Tahara H, Bailey SM, Behringer RR, Chang S. Pot1 deficiency initiates DNA damage checkpoint activation and aberrant homologous recombination at telomeres. Cell. 2006;126:49–62. doi: 10.1016/j.cell.2006.05.037. [DOI] [PubMed] [Google Scholar]
- [32].Saharia A, Guittat L, Crocker S, Lim A, Steffen M, Kulkarni S, Stewart SA. Flap endonuclease 1 contributes to telomere stability. Curr Biol. 2008;18:496–500. doi: 10.1016/j.cub.2008.02.071. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [33].Rizzo A, Salvati E, Porru M, D’Angelo C, Stevens MF, D’Incalci M, Leonetti C, Gilson E, Zupi G, Biroccio A. Stabilization of quadruplex DNA perturbs telomere replication leading to the activation of an ATR-dependent ATM signaling pathway. Nucleic Acids Res. 2009;37:5353–5364. doi: 10.1093/nar/gkp582. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [34].Wang G, Vasquez KM. Non-B DNA structure-induced genetic instability. Mutation Research/Fundamental and Molecular Mechanisms of Mutagenesis. 2006;598:103–119. doi: 10.1016/j.mrfmmm.2006.01.019. [DOI] [PubMed] [Google Scholar]
- [35].Wang G, Christensen LA, Vasquez KM. Z-DNA-forming sequences generate large-scale deletions in mammalian cells. Proceedings of the National Academy of Sciences of the United States of America. 2006;103:2677–2682. doi: 10.1073/pnas.0511084103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [36].Wang G, Vasquez KM. Naturally occurring H-DNA-forming sequences are mutagenic in mammalian cells. Proceedings of the National Academy of Sciences of the United States of America. 2004;101:13448–13453. doi: 10.1073/pnas.0405116101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [37].Cheung I, Schertzer M, Rose A, Lansdorp PM. Disruption of dog-1 in Caenorhabditis elegans triggers deletions upstream of guanine-rich DNA. Nat Genet. 2002;31:405–409. doi: 10.1038/ng928. [DOI] [PubMed] [Google Scholar]
- [38].Ribeyre C, Lopes J, Boule JB, Piazza A, Guedin A, Zakian VA, Mergny JL, Nicolas A. The yeast Pif1 helicase prevents genomic instability caused by G-quadruplex-forming CEB1 sequences in vivo. PLoS Genet. 2009;5:e1000475. doi: 10.1371/journal.pgen.1000475. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [39].Lipps HJ, Rhodes D. G-quadruplex structures: in vivo evidence and function. Trends in Cell Biology. 2009;19:414–422. doi: 10.1016/j.tcb.2009.05.002. [DOI] [PubMed] [Google Scholar]
- [40].Schaffitzel C, Berger I, Postberg J, Hanes J, Lipps HJ, Pluckthun A. In vitro generated antibodies specific for telomeric guanine-quadruplex DNA react with Stylonychia lemnae macronuclei. Proc Natl Acad Sci U S A. 2001;98:8572–8577. doi: 10.1073/pnas.141229498. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [41].Prescott DM. The DNA of ciliated protozoa. Microbiol Rev. 1994;58:233–267. doi: 10.1128/mr.58.2.233-267.1994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [42].Dawson RMC. Data for biochemical research. 3rd ed Clarendon Press; Oxford: 1986. [Google Scholar]
- [43].Hile SE, Yan G, Eckert KA. Somatic Mutation Rates and Specificities at TC/AG and GT/CA Microsatellite Sequences in Nontumorigenic Human Lymphoblastoid Cells. Cancer Res. 2000;60:1698–1703. [PubMed] [Google Scholar]
- [44].Eckert KA, Yan G, Hile SE. Mutation rate and specificity analysis of tetranucleotide microsatellite DNA alleles in somatic human cells. Mol Carcinog. 2002;34:140–150. doi: 10.1002/mc.10058. [DOI] [PubMed] [Google Scholar]
- [45].Eckert K, Hile S, Vargo P. Development and use of an in vitro HSV-tk forward mutation assay to study eukaryotic DNA polymerase processing of DNA alkyl lesions. Nucl. Acids Res. 1997;25:1450–1457. doi: 10.1093/nar/25.7.1450. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [46].Drinkwater NR, Klinedinst DK. Chemically induced mutagenesis in a shuttle vector with a low-background mutant frequency. Proc Natl Acad Sci U S A. 1986;83:3402–3406. doi: 10.1073/pnas.83.10.3402. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [47].Eckert KA, Drinkwater NR. recA-dependent and recA-independent N-ethyl-N-nitrosourea mutagenesis at a plasmid-encoded herpes simplex virus thymidine kinase gene in Escherichia coli. Mutation Research/Fundamental and Molecular Mechanisms of Mutagenesis. 1987;178:1–10. doi: 10.1016/0027-5107(87)90079-0. [DOI] [PubMed] [Google Scholar]
- [48].Mergny JL, Phan AT, Lacroix L. Following G-quartet formation by UV-spectroscopy. FEBS Lett. 1998;435:74–78. doi: 10.1016/s0014-5793(98)01043-6. [DOI] [PubMed] [Google Scholar]
- [49].Roy S, Tanious FA, Wilson WD, Ly DH, Armitage BA. High-affinity homologous peptide nucleic acid probes for targeting a quadruplex-forming sequence from a MYC promoter element. Biochemistry. 2007;46:10433–10443. doi: 10.1021/bi700854r. [DOI] [PubMed] [Google Scholar]
- [50].Bai Y, Murnane JP. Telomere instability in a human tumor cell line expressing a dominant-negative WRN protein. Hum Genet. 2003;113:337–347. doi: 10.1007/s00439-003-0972-y. [DOI] [PubMed] [Google Scholar]
- [51].Hemann MT, Strong MA, Hao LY, Greider CW. The shortest telomere, not average telomere length, is critical for cell viability and chromosome stability. Cell. 2001;107:67–77. doi: 10.1016/s0092-8674(01)00504-9. [DOI] [PubMed] [Google Scholar]
- [52].Sears J, Ujihara M, Wong S, Ott C, Middeldorp J, Aiyar A. The amino terminus of Epstein-Barr Virus (EBV) nuclear antigen 1 contains AT hooks that facilitate the replication and partitioning of latent EBV genomes by tethering them to cellular chromosomes. J Virol. 2004;78:11487–11505. doi: 10.1128/JVI.78.21.11487-11505.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [53].Sugden B. In the beginning: a viral origin exploits the cell. Trends Biochem Sci. 2002;27:1–3. doi: 10.1016/s0968-0004(01)02032-1. [DOI] [PubMed] [Google Scholar]
- [54].Dhar SK, Yoshida K, Machida Y, Khaira P, Chaudhuri B, Wohlschlegel JA, Leffak M, Yates J, Dutta A. Replication from oriP of Epstein-Barr virus requires human ORC and is inhibited by geminin. Cell. 2001;106:287–296. doi: 10.1016/s0092-8674(01)00458-5. [DOI] [PubMed] [Google Scholar]
- [55].Yates JL, Warren N, Sugden B. Stable replication of plasmids derived from Epstein-Barr virus in various mammalian cells. Nature. 1985;313:812–815. doi: 10.1038/313812a0. [DOI] [PubMed] [Google Scholar]
- [56].Lindner SE, Sugden B. The plasmid replicon of Epstein-Barr virus: mechanistic insights into efficient, licensed, extrachromosomal replication in human cells. Plasmid. 2007;58:1–12. doi: 10.1016/j.plasmid.2007.01.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [57].Ermakova OV, Frappier L, Schildkraut CL. Role of the EBNA-1 protein in pausing of replication forks in the Epstein-Barr virus genome. J Biol Chem. 1996;271:33009–33017. doi: 10.1074/jbc.271.51.33009. [DOI] [PubMed] [Google Scholar]
- [58].Wang J, Lindner SE, Leight ER, Sugden B. Essential elements of a licensed, mammalian plasmid origin of DNA synthesis. Mol Cell Biol. 2006;26:1124–1134. doi: 10.1128/MCB.26.3.1124-1134.2006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [59].Aiyar A, Aras S, Washington A, Singh G, Luftig RB. Epstein-Barr Nuclear Antigen 1 modulates replication of oriP-plasmids by impeding replication and transcription fork migration through the family of repeats. Virol J. 2009;6:29. doi: 10.1186/1743-422X-6-29. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [60].Gahn TA, Schildkraut CL. The Epstein-Barr virus origin of plasmid replication, oriP, contains both the initiation and termination sites of DNA replication. Cell. 1989;58:527–535. doi: 10.1016/0092-8674(89)90433-9. [DOI] [PubMed] [Google Scholar]
- [61].Platt TH, Tcherepanova IY, Schildkraut CL. Effect of number and position of EBNA-1 binding sites in Epstein-Barr virus oriP on the sites of initiation, barrier formation, and termination of replication. J Virol. 1993;67:1739–1745. doi: 10.1128/jvi.67.3.1739-1745.1993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [62].Dhar V, Schildkraut CL. Role of EBNA-1 in arresting replication forks at the Epstein-Barr virus oriP family of tandem repeats. Mol Cell Biol. 1991;11:6268–6278. doi: 10.1128/mcb.11.12.6268. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [63].Viguera E, Hernández P, Krimer DB, Boistov AS, Lurz R, Alonso JC, Schvartzman JB. The ColE1 Unidirectional Origin Acts as a Polar Replication Fork Pausing Site. Journal of Biological Chemistry. 1996;271:22414–22421. doi: 10.1074/jbc.271.37.22414. [DOI] [PubMed] [Google Scholar]
- [64].Gottschling DE, Zakian VA. Telomere proteins: specific recognition and protection of the natural termini of Oxytricha macronuclear DNA. Cell. 1986;47:195–205. doi: 10.1016/0092-8674(86)90442-3. [DOI] [PubMed] [Google Scholar]
- [65].Vallur AC, Maizels N. Distinct activities of exonuclease 1 and flap endonuclease 1 at telomeric g4 DNA. PLoS ONE. 5:e8908. doi: 10.1371/journal.pone.0008908. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [66].Zhang ML, Tong XJ, Fu XH, Zhou BO, Wang J, Liao XH, Li QJ, Shen N, Ding J, Zhou JQ. Yeast telomerase subunit Est1p has guanine quadruplex-promoting activity that is required for telomere elongation. Nat Struct Mol Biol. 17:202–209. doi: 10.1038/nsmb.1760. [DOI] [PubMed] [Google Scholar]
- [67].Wu X, Maizels N. Substrate-specific inhibition of RecQ helicase. Nucleic Acids Res. 2001;29:1765–1771. doi: 10.1093/nar/29.8.1765. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [68].Wu Y, Shin-ya K, Brosh RM., Jr. FANCJ Helicase Defective in Fanconia Anemia and Breast Cancer Unwinds G-Quadruplex DNA To Defend Genomic Stability. Mol. Cell. Biol. 2008;28:4116–4128. doi: 10.1128/MCB.02210-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [69].Mohaghegh P, Karow JK, Brosh RM, Jr, Bohr VA, Hickson ID. The Bloom’s and Werner’s syndrome proteins are DNA structure-specific helicases. Nucleic Acids Res. 2001;29:2843–2849. doi: 10.1093/nar/29.13.2843. Jr. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [70].Wu L, Hickson ID. DNA Helicases Required for Homologous Recombination and Repair of Damaged Replication Forks. Annual Review of Genetics. 2006;40:279–306. doi: 10.1146/annurev.genet.40.110405.090636. [DOI] [PubMed] [Google Scholar]
- [71].Risitano A, Fox KR. Stability of Intramolecular DNA Quadruplexes: Comparison with DNA Duplexesâ€. Biochemistry. 2003;42:6507–6513. doi: 10.1021/bi026997v. [DOI] [PubMed] [Google Scholar]
- [72].Burge S, Parkinson GN, Hazel P, Todd AK, Neidle S. Quadruplex DNA: sequence, topology and structure. Nucleic Acids Res. 2006;34:5402–5415. doi: 10.1093/nar/gkl655. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [73].Deng Z, Atanasiu C, Burg JS, Broccoli D, Lieberman PM. Telomere Repeat Binding Factors TRF1, TRF2, and hRAP1 Modulate Replication of Epstein-Barr Virus OriP. J. Virol. 2003;77:11992–12001. doi: 10.1128/JVI.77.22.11992-12001.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [74].Wang C-Y, Sugden B. Identifying a property of origins of DNA synthesis required to support plasmids stably in human cells. Proceedings of the National Academy of Sciences. 2008;105:9639–9644. doi: 10.1073/pnas.0801378105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [75].Opresko PL, von Kobbe C, Laine JP, Harrigan J, Hickson ID, Bohr VA. Telomere-binding protein TRF2 binds to and stimulates the Werner and Bloom syndrome helicases. J Biol Chem. 2002;277:41110–41119. doi: 10.1074/jbc.M205396200. [DOI] [PubMed] [Google Scholar]
- [76].Lillard-Wetherell K, Machwe A, Langland GT, Combs KA, Behbehani GK, Schonberg SA, German J, Turchi JJ, Orren DK, Groden J. Association and regulation of the BLM helicase by the telomere proteins TRF1 and TRF2. Hum Mol Genet. 2004;13:1919–1932. doi: 10.1093/hmg/ddh193. [DOI] [PubMed] [Google Scholar]
- [77].Tang J, Kan Z.-y., Yao Y, Wang Q, Hao Y.-h., Tan Z. G-quadruplex preferentially forms at the very 3′ end of vertebrate telomeric DNA. Nucl. Acids Res. 2008;36:1200–1208. doi: 10.1093/nar/gkm1137. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [78].Cleary JD, Nichol K, Wang Y-H, Pearson CE. Evidence of cis-acting factors in replication-mediated trinucleotide repeat instability in primate cells. Nat Genet. 2002;31:37–46. doi: 10.1038/ng870. [DOI] [PubMed] [Google Scholar]
- [79].Vorlickova M, Chladkova J, Kejnovska I, Fialova M, Kypr J. Guanine tetraplex topology of human telomere DNA is governed by the number of (TTAGGG) repeats. Nucleic Acids Res. 2005;33:5851–5860. doi: 10.1093/nar/gki898. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [80].Ruiz-Herrera A, Nergadze SG, Santagostino M, Giulotto E. Telomeric repeats far from the ends: mechanisms of origin and role in evolution. Cytogenet Genome Res. 2008;122:219–228. doi: 10.1159/000167807. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.