Abstract
Cervical cancer is the most common malignant disease responsible for the deaths of a large number of women in the developing world. Although certain strains of human papillomavirus (HPV) have been identified as the cause of this disease, events that lead to formation of malignant tumors are not fully clear. STAT3 is a major oncogenic transcription factor involved in the development and progression of a number of human tumors. However, the mechanisms that result in loss of control over STAT3 activity are not understood. Gene associated with Retinoid-Interferon-induced Mortality-19 (GRIM-19) is a tumor-suppressive protein identified using a genetic technique in the interferon/retinoid-induced cell death pathway. Here, we show that reduction in GRIM-19 protein levels occur in a number of primary human cervical cancers. Consequently, these tumors tend to express a high basal level of STAT3 and its downstream target genes. More importantly, using a surrogate model, we show that restoration of GRIM-19 levels reestablishes the control over STAT3-dependent gene expression and tumor growth in vivo. GRIM-19 suppressed the expression of tumor invasion- and angiogenesis-associated factors to limit tumor growth. This study identifies another major novel molecular pathway inactivated during the development of human cervical cancer.
Introduction
Cervical cancer is the most common gynecologic neoplasm affecting women in developing countries. It is estimated to be the cause of almost 260,000 deaths annually, of which about 80% occurred in developing countries (Castellsague 2008). Nearly all cases of cervical cancer are attributable to persistent infection by >100 known human papillomavirus (HPV) types, especially by HPV-16 and HPV-18, which cause ∼70% of all cervical cancers worldwide (Lowy and others 2008). It has been observed that the E6 and E7 oncoproteins of high-risk HPV play major roles in the development of cervical cancer (Schiffman and others 2007). These viral oncoproteins interfere with critical cell cycle regulators, proteins such as tumor suppressor (p53) and retinoblastoma (pRb), respectively. However, evidence indicates that HPV infection alone is insufficient to cause malignant changes. For example, the other host genetic changes and disorderly expression of various oncogenes and tumor suppressor genes are also important in the development of cervical cancer (Martin and others 2007).
In recent years, more attention has been paid to important signal transduction cascades, and it has become clear that the Signal Transducer and Activator of Transcription (STAT) proteins, in particular STAT3, may take a crucial part in malignant transformation (Bromberg and others 1998; Bromberg and others 1999). Although a constitutive activation of STAT3 is documented in a number of studies (Buettner and others 2002), it is unclear what other mechanisms, beyond oncogene-induced tyrosyl phosphorylation of STAT3, contribute to the promotion of tumor growth. In a previous report, we identified Gene associated with Retinoid-Interferon-induced Mortality-19 (GRIM-19), a novel gene product involved in IFN-β/RA-induced apoptosis, suppresses STAT3-induced gene expression (Zhang and others 2003). GRIM-19 expression has been found to be lost in renal cell cancers, certain prostate tumors, and colonic tumors (Alchanati and others 2006). GRIM-19 binds to STAT3 and inhibits its transcriptional activity (Lufei and others 2003). Because constitutively active STAT3 up-regulates growth-stimulating and antiapoptotic gene expression to promote oncogenesis, suppression of GRIM-19 may cause resistance to biological therapeutics. Chen and others reported elevated levels of phosphorylated STAT3 protein in cervical cancer cell lines and specimens (Chen and others 2007). The consequence of such a constitutive phosphorylation and the mechanisms that cause it are far from clear. In this study, we report a significant down-regulation of GRIM-19 protein levels in several primary human cervical cancers, which resulted in an increased expression of STAT3 and its downstream genes. We also show that restoration of GRIM-19 levels in a cervical cancer cell line restores growth suppression in vivo through a down-regulation of STAT3-induced gene expression.
Materials and Methods
Reagents
Lipofectamine™ 2000 (Invitrogen, Carlsbad, CA) was used for transfection. The kit for reverse transcription was from Promega and the PCR kit was from Takara Bio, Inc. Puromycin (BD Biosciences, Palo Alto, CA) was used to establish stable cell lines. MTT (Amresco, Solon, OH) was provided to assay proliferation. The primers used in the study were purchased from Shanghai Sangon Co., Ltd. GRIM-19 antibody was described previously (Hu and others 2002). STAT3 and pY-STAT3 antibodies were purchased from Cell Signaling Technology Inc. MMP-2, MMP-9, VEGF, β-actin, and Survivin antibodies were purchased from Santa Cruz Biotechnology Inc., Santa Cruz, CA. The secondary antibody kit and liquid 3,3′- diaminobenzidine substrate (DAB) were obtained from Beijing Zhongshan Biological Technology Co., Beijing, China. Recombinant human IFN-β was from Berlex Inc., Montville, NJ. Fresh stocks of RA (Sigma, St. Louis, MO) were prepared in ethanol and added to cultures under subdued light.
Cell culture
HeLa (human cervical cancer cell line) was grown in Dulbecco's modified Eagle's medium (Invitrogen Corporation, CA) supplemented with 10% fetal bovine serum, 100 U/mL penicillin, and 100 U/mL streptomycin.
Tumors
All cervical tissues were collected from individuals who underwent hysteromyomectomy between February 2007 and July 2008, with an informed consent of the patients under an institutionally approved protocol at the Anhui provincial hospital, Hefei, China. The ages of patients ranged from 35 to 59 years. Freshly excised tissues were flash frozen at −80°C for RNA and protein analyses. For pathologic diagnosis, a portion of the tissue was paraffin-embedded, cut into 5 to 7 μm thick sections. Clinical stages were determined by a certified gynecologic pathologist according to the International Federation of Gynecology Obstetrics (FIGO) staging system (or high-grade squamous intraepithelial lesions [HSIL] system). The 20 squamous epithelial carcinomas examined in these studies without distant metastases belonged to type Ib for 3 patients, IIa for 2 patients, IIb for 15 patients. Additional 15 normal cervical tissues, from women who underwent hysterectomy for reasons other than neoplasias of either cervix or endometrium, were collected and used as controls in this study. At least 2 different sections were prepared from each paraffin-embedded block and analyzed.
Immunohistochemical staining
The tissue sections were deparaffinized in xylene for 10 min, fixed using 100% ethanol for 5 min, and then rehydrated with graded ethanol. Endogenous peroxidase activity was quenched by incubation in 3% hydrogen peroxide in methanol for 10 min. The sections were washed twice with PBS, and incubated with the primary antibody (1:200 dilution) overnight at 4°C. They were rinsed twice with PBS and incubated with the corresponding secondary antibody for 30 min at 37°C. The sections were washed with PBS, and incubated for 1 min with high-sensitivity substrate (DAB). Finally, the slides were counterstained with 10% hematoxylin and photographs were captured using a microscope with a digital camera.
HPV DNA testing
DNA from normal cervical tissues and cervical cancer tissues were tested for the presence of high-risk HPV DNA by HC2 method as per the manufacturer's instructions (Digene Inc, Gaithersburg, MD). The probe cocktail is capable of detecting HPV types 16, 18, 31, 33, 35, 39, 45, 51, 52, 56, 58, 59, and 68, which are implicated in the initiation of cervical cancers. A positive cutoff point of 1.00 relative light unit per positive control (RLU/pc) was used to score the presence of HPV DNA in the samples. Each sample was tested in duplicate. Although it can detect HPV DNA in the sample, the HC2 test does not distinguish which HPV genotype(s) are present in them.
Plasmids and transfection
The pIRES-Puro2-Myc vector was generated by inserting an oligonucleotide coding for the Myc-tag into the Not I site of pIRES-Puro2 (Clontech, Inc., Mountain View, CA). Insertion of cDNAs of interest into the multiple cloning sites in frame results in an addition of a C-terminal Myc-tag to the expressed product. Since an internal ribosome entry site is located between the cDNA and Puromycin resistance gene, this ensures that all puromycin-resistant cell colonies express the transgene. pIRES-Puro2-GRIM-19-Myc contains a full-length ORF of human GRIM-19. HeLa cells were transfected with either the parent vector or pIRES-Puro2-GRIM-19-Myc into cells using Lipofectamine (Invitrogen, Carlsbad, CA). For stable expression of GRIM-19-Myc, transfectants were selected in growth medium containing 1.25 μg/mL puromycin. The established stable lines were designated as HeLa/Control and HeLa/GRIM-19, respectively.
Reverse transcription and quantitative PCR
Total RNA was extracted from tissues or cells and RNA (1 μg) was reverse-transcribed using random primers and Mu-MLV reverse transcriptase for 1 h at 37°C. The resultant cDNA was used as template in quantitative PCR employing SYBR chemistry (Sigma) using gene-specific primers (Supplementary Table 1; Supplementary materials are available online at http://www.liebertpub.com).
Immunoblotting
An equal amount of total protein was resolved on 10% SDS-PAGE. Proteins were transferred to PVDF membrane and probed with specific antibodies. Blots were developed using enhanced chemiluminescence reagent (Pierce, Thermo Scientific, UK).
MTT assay
Cells (2 × 103/well) were grown in a 96-well plate and incubated for indicated period times at 37°C. Cell growth was measured using color generation (MTT reduction) by adding 20 μL of MTT (5 mg/mL) into the culture medium. The cells were harvested and incubated with 100 μL of DMSO for 30 min and absorbance measured using a plate reader. All assays were performed in triplicate and the data were presented as mean ± SD.
Tumor xenograft growth
HeLa/GRIM-19, HeLa/Control (107) cells in 0.1 mL saline containing 50% Matrigel were implanted subcutaneously on the dorsal flank of 6-week-old female athymic nude mice (Beijing experimental animal center). Each group contained 8 mice. All mice were housed in a pathogen-free environment. Tumor growth was monitored by measuring tumor dimensions with a caliper and the volume was calculated using the formula V= (4/3) × πa2b, where 2a = minor axis, 2b = major axis of prolate spheroid (Ochsenbein and others 2001). At the end of the experiment, mice were euthanized and tumors collected. Tumor sections were processed for immunohistochemical and biochemical analyses.
Statistical analysis
The SPSS13.0 software was used for all statistical analyses. The significance of the differences between 2 groups was assessed by independent t-test. A P value of <0.05 was considered significant.
Results
Immunohistochemical analysis of GRIM-19 expression in cervical cancers
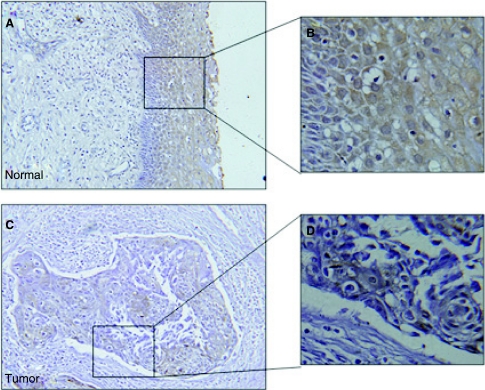
Since GRIM-19 was identified as a potential tumor suppressor in recent studies, we first checked the expression status of GRIM-19 in primary cervical cancers. Twenty paraffin-embedded cervical squamous carcinoma and 15 normal cervical tissues were evaluated for GRIM-19 expression using immunohistochemical analysis with a GRIM-19-specific monoclonal antibody. As shown in Figure 1, GRIM-19 was highly expressed in normal cervical epithelium (Fig. 1A). However, its expression was extremely reduced in cervical cancers (Fig. 1B). Similar pattern was found in 16 out of 20 primary tumors examined (Table 1).
FIG. 1.
Immunohistochemical analysis of normal cervical tissue and cervical cancer with gene associated with retinoid-interferon-induced mortality-19 (GRIM-19) antibodies. A representative picture of 15 independent normal samples and 20 tumor tissues are shown. An intense staining of GRIM-19 in normal cervical tissues is significantly suppressed in cervical tumors (200×). Photomicrographs on the right show a higher magnification (800×) of the boxed areas.
Table 1.
HPV, GRIM-19, and STAT3 Status in Normal Cervical and Cancerous Samples
| Tissue | Number of cases | HR-HPV-Positive cases | Low GRIM-19 expression with high STAT3 expression | GRIM-19 expression with moderate STAT3 expression |
|---|---|---|---|---|
| Normal cervix | 15 | 4a | 0 | 15 |
| Cervical cancer | 20 | 20 | 16* | 4 |
P value < 0.05 with respect to normal control.
No cervical hyperplasia. Asymptomatic infection.
GRIM-19 and STAT3 levels were determined by Western blot analysis.
HR-HPV (high-risk HPV) status was verified using a commercially available hybrid capture assay kit.
Since HPV types 16/18/31/33/35/39/45/51/52/56/58/59/6 8 are implicated in cervical carcinogenesis, we examined the HR-HPV infection in cervical tissues. The results showed that 4 cases from 15 normal cervical tissues were HR-HPV-positive; and all of cervical cancer tissues were HR-HPV-positive (Table 1). Pathological analysis did not suggest any detectable cervical cancer in the 4 normal samples. Therefore, they are considered as those who had an exposure to virus but were resolved at some point.
Down-regulation of GRIM-19 is associated with increased STAT3 activity in cervical cancer tissues
Because of the opposing effects of GRIM-19 and STAT3 on cell growth, we next examined if an inverse correlation existed between the expression of these proteins in primary tumors (n = 20) and normal tissues (n = 15). Tissue sections were screened for the expression of STAT3 and GRIM-19 using RT-PCR and Western blot analyses. Data from 4 representative samples are shown in Figure 2. In cervical tumor samples, significantly reduced expression of GRIM-19 was observed compared to normal cervical samples. Consistent with the reduction in GRIM-19 levels, expression of STAT3 target genes Cyclin B1, Bcl2-L1, and STAT3 was significantly up-regulated in the tumors compared to the normal controls (Fig. 2A). Reduction in GRIM-19 and up-regulation of STAT3 levels were confirmed by a Western blot analysis of the proteins extracted from cervical carcinomas and compared to normal controls (Fig. 2B). Normal cervical tissue showed a low STAT3 and generally high GRIM-19 levels. In contrast, cervical cancer tissues expressed significantly higher STAT3 and lower GRIM-19 than normal tissues (Table 1).
FIG. 2.
Down-regulation of GRIM-19 and up-regulation of STAT3 in cervical cancers. (A) RT-PCR analysis of the expression of GRIM-19, STAT3, Cyclin B1, and Bcl2-L1 in normal cervical tissue and cervical cancer tissue. (B) Equal amounts of total cellular extracts (50 μg) from the normal and tumor samples were resolved by SDS-PAGE and immunoblotted with anti-GRIM-19 and anti-STAT3 antibodies. Individual patient samples are indicated on top; N: normal; T: tumor.
GRIM-19 suppresses STAT3 and cell proliferation in HeLa cells
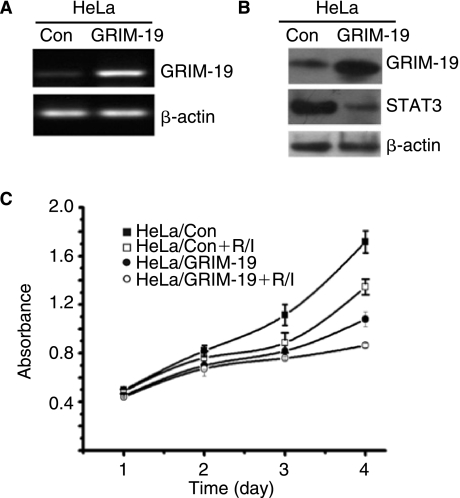
To test the biological relevance of GRIM-19 to cervical tumor cell growth, we established HeLa/GRIM-19 that expressed human GRIM-19 gene and a control line, HeLa/Control. HeLa cells express a low basal level of GRIM-19. The relative expression levels of GRIM-19 in these cell lines were confirmed using RT-PCR and Western blot analyses (Fig. 3A and B). Growth of HeLa/GRIM-19 cells, as indicated by MTT assay, was markedly (P < 0.05) inhibited on day 3 and day 4 (Fig. 3C), compared to the control cell line. Treatment of these cell lines with IFN-β/RA, as expected, suppressed cell growth further. Notably, a greater suppression of growth occurred in HeLa/GRIM-19 than in HeLa/Control. More importantly, the high expression of STAT3 found in HeLa/Control cells was significantly low in HeLa/GRIM-19 cells (Fig. 3B).
FIG. 3.
Overexpression of GRIM-19 suppresses HeLa cell proliferation. (A) RT-PCR analysis of the expression of GRIM-19 in HeLa/Control and HeLa/GRIM-19 cells. (B) Western blot analysis of the expression of GRIM-19 and STAT3 in HeLa/Control and HeLa/GRIM-19 cells. (C) An MTT assay was performed in the indicated cells. MTT reduction, due to cell growth, converts the colorless dye into a chromogen, whose levels were quantified at 570 nm using microplate reader. Each data point represents mean ± SE of 8 samples. Cells were treated with a combination of 1 μM retinoic acid and 1,000 U of interferon-β and were indicated with R/I.
GRIM-19 inhibits tumor growth by suppressing STAT3-regulated genes in vivo
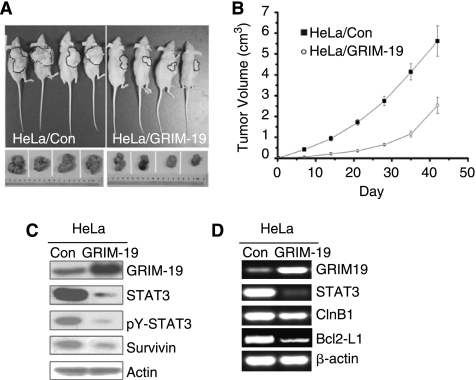
To evaluate the role of GRIM-19 in regulating tumor growth in vivo, we transplanted HeLa/Control and HeLa/GRIM-19 cells into athymic nude mice. Cells (107) were transplanted subcutaneously into the right flank (n = 8 per group) and tumor growth was monitored for 42 days (Fig. 4A and B). On day 42, mice were euthanized and tumors were collected. Tumor sizes in HeLa/GRIM-19 group were significantly smaller when compared with the HeLa/Control group (P < 0.05). Some representative mice bearing tumors and final tumor sizes are shown in Figure 4A. We next examined the levels of STAT3, pY-STAT3, and GRIM-19 in tumors derived from mice using RT-PCR or Western blot analyses (Fig. 4C and D). As expected, GRIM-19 expression levels were high in HeLa/GRIM-19 group. The levels of STAT3 and pY-STAT3 were strongly reduced in this group (Fig. 4C). We also examined the levels of STAT3, Cyclin B1, and Bcl2-L1 using RT-PCR analysis (Fig. 4D). The expression level of all these transcripts decreased in HeLa/GRIM-19 cells, compared to the HeLa/Control. In addition, expression of Survivin, an antiapoptotic protein regulated by STAT3, was strongly diminished in HeLa/GRIM-19 group (Fig. 4C). The inhibitory effect of GRIM-19 on tumor growth was further assessed by an immunohistochemical analysis of the expression of proliferation-associated antigen Ki67. As expected, the tumors expressing GRIM-19 had an extremely reduced expression of Ki67, compared to the control tumors (Supplementary Fig. 1).
FIG. 4.
Restoration of GRIM-19 inhibits the tumor growth. (A) Representative mice bearing HeLa/Control and HeLa/GRIM-19 tumors and the tumors developed in them are shown in the photographs. Tumors harvested on day 42 are shown. (B) Antitumor effects of GRIM-19 in vivo. Tumor volumes were measured over a period of 6 weeks. The tumor volume differences between the GRIM-19 and control group is highly significant (*P < 0.01). (C) Western blot analysis of the expression of GRIM-19, STAT3, p-STAT3, and Survivin in the tumor tissues. (D) RT-PCR analysis of the expression of GRIM-19 and STAT3, Cyclin B1, and Bcl2-L1 in the tumor tissues.
Down-regulation of angiogenesis- and tumor invasion-associated genes by GRIM-19
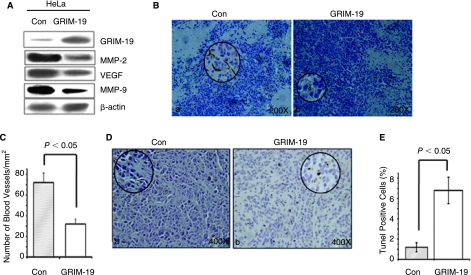
STAT3 is also known to up-regulate the expression of genes associated with angiogenesis and tumor invasion, such as the vascular endothelial growth factor (VEGF) and matrix metalloproteases (Dechow and others 2004; Xie and others 2004; Xie and others 2006; Wang and others 2007). We, therefore, examined if the tumors bearing GRIM-19 had reduced expression of these genes. Western blot analysis (Fig. 5A) revealed that the expression of GRIM-19 significantly suppressed the expression of VEGF (an angiogenic growth factor) and matrix metalloproteases, MMP-2 and MMP-9, involved in tumor invasion (Schaefer and others 2002; Wei and others 2003; Xu and others 2005).
FIG. 5.
GRIM-19 suppresses tumor angiogenesis and tumor invasion-associated gene expression. (A) Western blot analysis of the expression of tumor angiogenesis and metastasis-associated proteins (MMP-2 and MMP-9) and VEGF. A representative blot from several individual tumors (n = 4) is shown. (B) GRIM-19 suppresses tumor angiogenesis. Blood vessels were stained with CD31 antibodies. Note the loss of blood vessels in tumors with GRIM-19 (200×). Circled inserts show a magnified view (800×). Brown color deposits indicate positive staining for blood vessels. (C) Quantification of blood vessels in the tumors. Multiple regions (n = 5) from different tumors (n = 4) were quantified after staining with CD31. (D) TUNEL staining of tumors from HeLa/Control and HeLa/GRIM-19 (400×). Circled inserts show a magnified view (800×) of tumor apoptosis. (E) Quantification of apoptotic cells in tumors. Multiple regions (n = 5) from different tumors (n = 4) were quantified after TUNEL staining.
Suppression of angiogenesis was further ascertained with an additional experiment. Tumor sections were stained with an antibody against CD31, which detects new blood vessels (Charpin and others 2004). In agreement with the decline in VEGF expression, the GRIM-19-expressing tumors had a low number of CD31-positive blood vessels (Fig. 5B). A quantification of these data suggested that the tumors expressing GRIM-19 had significantly fewer (P < 0.05) vessels than the controls. The number of blood vessels in HeLa/Control tumors was 72 ± 9.3/mm2, while it was 32 ± 4.7/mm2 in HeLa/GRIM-19 tumors (Fig. 5C). Thus, inhibition of tumor growth directly correlated with reduced angiogenesis and increased apoptosis. Indeed, consistent with the suppression of angiogenic factors, significant number of apoptotic cells (6.8 ± 1.30%) were seen in the tumors expressing GRIM-19, compared to the controls (1.2 ± 0.45%), as revealed by TUNEL assay (Fig. 5D–E).
Discussion
Cervical cancer ranks among the top 3 cancers diagnosed in women worldwide, and is strongly associated with a subset of HPV infections (especially HPV-16 and HPV-18). The E6 and E7 proteins from the high-risk HPV types, such as HPV-16 and −18, bind to and degrade the tumor-suppressive proteins p53 and retinoblastoma proteins (Scheffner and others 1990; Scheffner and others 1992), respectively, in the pathogenesis of cervical cancer (Braun and others 2004). An ubiquitin ligase activity associated with the E6 protein targets p53 to degradation without causing an accumulation of mutations in the p53 gene (Huibregtse and others 1991). Although HPV infection is necessary, that by itself is insufficient to cause malignant transformation. Besides, persistence of HPV-16 or 18 in some women does not inevitably result in cancer (Santin and others 2005). While p53 and pRb inactivation likely cause early stages of benign transformation, they do not fully explain the processes leading to the development of malignant cervical cancer. Very little is known about other factors that affect growth and development of cervical cancer.
Earlier, we have described GRIM-19, a novel 16-kDa protein, involved in cellular apoptosis induced by IFN-β/RA (Hu and others 2002). GRIM-19 orthologs are present in most multicellular organisms. GRIM-19 is present in both cytoplasmic and nuclear compartments; however, the ratio of distribution seems to vary depending on the cell type. In the cytoplasm, GRIM-19 is found to be a part of the mitochondrial respiratory chain complex I (Fearnley and others 2001; Reeves and others 2007). This complex is vital for maximal ATP yield, and dysfunctions in this complex are related with neurodegenerative diseases (Lu and Cao 2008). A recent study has shown that a 2.1-kb non-coding RNA coded by the human cytomegalovirus binds to the complex I containing GRIM-19 and prevents apoptosis of virus-infected cells (Reeves and others 2007). In addition, GRIM-19 maps to a locus on human chromosome 19 that seems to be associated with prostate growth suppression and its expression is lost in some prostate tumors (Alchanati and others 2006; Papa and others 2007; Zhang and others 2008).
In an earlier study, we found that the vIRF1 oncoprotein of Kaposi's sarcoma-associated human herpes virus-8 binds to GRIM-19 and inhibits its proapoptotic action (Seo and others 2002). While investigating its effects, we wondered if other oncoprotein(s) from DNA viruses are also capable of binding to GRIM-19. We discovered that the E6 oncoprotein of HPV-16, but not that of HPV-6, binds to GRIM-19 in a preliminary coexpression experiment. However, we did not investigate if it had any biological or pathological relevance in those studies. Therefore, we investigated in this report if GRIM-19 status is affected in primary tumors. We found that its expression was suppressed in about ∼80% of advanced cervical cancers studied. Importantly, the expression levels of STAT3 and STAT3-inducible genes were elevated in these tumors. We also examined the HPV infection status in normal cervical tissues and cervical cancer tissues, the data showed that there was no evident correlation between HPV infection and levels of either GRIM-19 or STAT3. These observations are consistent with a previous report, which had indicated constitutive activation of STAT3 in most of the cervical cancer cell lines, including HeLa (HPV-18-positive), SiHa (HPV-16-positive), C33A (HPV-negative), HT-3 (HPV-negative) (Chen and others 2007).
Transcription factor STAT3 is necessary for a variety of activities in mammalian cells. STAT3 is also constitutively activated in a number of human cancers by autocrine growth factors or activated oncogenes (Frank 2007). Constitutive tyrosyl phosphorylation of STAT3, which is required for its DNA binding activity in many cases, is activated by a number of cellular oncogenes known to promote tumor growth and transform cells (Inghirami and others 2005). We and others have reported that GRIM-19 binds to STAT3 and inhibits its transcription-activating function (Lufei and others 2003; Zhang and others 2003). Inhibition of STAT3 by GRIM-19 appears to be an important step in suppressing oncogenesis (Kalakonda and others 2007), because a constitutively active STAT3 promotes antiapoptotic gene expression to support tumor survival (Bromberg and others 1999).
Since low expression of GRIM-19 occurs in renal, esophageal, prostate, and colonic tumors, and constitutive activation of STAT3 is known in cervical cancer (Chen and others 2007), we examined the status of GRIM-19 expression in cervical cancer tissues. The results from this study for the first time showed that GRIM-19 is suppressed in primary cervical cancers, with a corresponding increment in STAT3 activity. Current studies are similar to those observed with expression of GRIM-19, that is, reduced or totally suppressed, in a number of primary renal cell carcinomas and other tumors, which resulted in an up-regulation of STAT3-regulated genes in these tumors (Kalvakolanu 2004; Alchanati and others 2006). In an analogous manner, a cellular protein has been implicated in the inactivation of GRIM-19-dependent apoptosis in some esophageal cancers (Zhang and others 2004), although the effects on STAT3-dependent gene expression are unknown at this stage.
The biological relevance of our observations is further substantiated by a significant loss of growth of HeLa cell tumor in vitro and in vivo in presence of GRIM-19. Such loss of proliferation was associated with a decline in the steady-state levels of STAT3 and pY-STAT3 in these tumor cells. Besides, we detected a significantly low-level expression of Survivin in the tumor mass of HeLa/GRIM-19. Survivin is not only a target gene of STAT3 but also a member of the inhibitors of apoptosis family and has been implicated in both the regulation of cell division and suppression of apoptosis. Therefore, a loss of its expression might contribute to apoptosis of tumor cells (Mita and others 2008). Survivin is also an essential constituent of centromeres, required for mitotic division and chromosomal segregation (Speliotes and others 2000; Wheatley and others 2001; Lens and others 2003). We also found activated STAT3 was associated with increased expression of downstream antiapoptotic genes Bcl2-L1 and Cyclin B1 in these tissues, which can inhibit apoptosis and promote tumor cell proliferation, respectively. Interestingly, we find that the number of blood vessels in the tumors is significantly reduced in presence of GRIM-19 (Fig. 5). This effect appears to be due to a lower level expression of VEGF. Indeed, VEGF is one of the targets of STAT3 in other tumors (Schaefer and others 2002; Wei and others 2003; Xu and others 2005). These results are also consistent with previous reports in the literature that either IFNs (Sidky and Borden 1987; Dvorak and Gresser 1989) or IFN/RA combination suppress tumor-induced angiogenesis, especially by HPV-transformed cell lines (Majewski and others 1994). The antitumor effects of GRIM-19 was also seen in ME-180, another human cervical carcinoma cell line (data not shown), which is known to harbor an HPV type 39-like virus (Reuter and others 1991), suggesting that its effects appear to be global. We have now shown in other studies that GRIM-19 can suppress STAT3 in a number of tumor cell lines that were not infected by HPV (Alchanati and others 2006; Kalakonda and others 2007; Zhang and others 2008). Thus, there is no correlation between HPV status and GRIM-19's ability to inhibit STAT3.
As a result of down-regulation of the angiogenic factors, tumor cells seem to undergo apoptosis (Fig. 5). Thus, the tumor-suppressive function of GRIM-19 appears to be exerted in part through inactivation of STAT3. Lastly, loss of MMP-2 and MMP-9 expression might further inhibit the extravasation and invasive potential of tumor cells, therefore, limiting the spread of the tumor. It appears that restoration of GRIM-19 might be a new strategy for the treatment of cervical cancer. Consistent with these observations, a previous clinical study showed a strong tumor-suppressive activity of IFN-α/retinoic acid in patients with advanced squamous cell carcinomas (Lippman and others 1992). Additionally, our recent studies have shown that direct administration of GRIM-19 via expression plasmids into tumors growing in vivo could strongly suppress tumor growth (Zhang and others 2008). In addition, combination of GRIM-19 with shRNAs against STAT3 further robustly suppressed prostate tumor growth in vivo. Lastly, other IFN-regulated gene products, such as p56, appear to inhibit HPV growth by interfering with viral replication (Terenzi and others 2008). Thus, the IFNs seem to guard the host against development of virally induced tumors at multiple levels.
Supplementary Material
Acknowledgments
This work was supported by Anhui Provincial Science and Technology Development Foundation project, China (06013124B) and Anhui Provincial Natural Science Foundation project (#20090413117). W.H.X. is supported by National Natural Science Foundation of China (30771973, 30570704). D.V.K. is supported by US National Cancer Institute grants CA105005 and CA78282.
References
- Alchanati I. Nallar SC. Sun P. Gao L. Hu J. Stein A. Yakirevich E. Konforty D. Alroy I. Zhao X. Reddy SP. Resnick MB. Kalvakolanu DV. A proteomic analysis reveals the loss of expression of the cell death regulatory gene GRIM-19 in human renal cell carcinomas. Oncogene. 2006;25(54):7138–7147. doi: 10.1038/sj.onc.1209708. [DOI] [PubMed] [Google Scholar]
- Braun K. Ehemann V. Waldeck W. Pipkorn R. Corban-Wilhelm H. Jenne J. Gissmann L. Debus J. HPV18 E6 and E7 genes affect cell cycle, pRB and p53 of cervical tumor cells and represent prominent candidates for intervention by use peptide nucleic acids (PNAs) Cancer Lett. 2004;209(1):37–49. doi: 10.1016/j.canlet.2004.02.022. [DOI] [PubMed] [Google Scholar]
- Bromberg JF. Horvath CM. Besser D. Lathem WW. Darnell JE., Jr Stat3 activation is required for cellular transformation by v-src. Mol Cell Biol. 1998;18(5):2553–2558. doi: 10.1128/mcb.18.5.2553. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bromberg JF. Wrzeszczynska MH. Devgan G. Zhao Y. Pestell RG. Albanese C. Darnell JE., Jr Stat3 as an oncogene. Cell. 1999;98(3):295–303. doi: 10.1016/s0092-8674(00)81959-5. [DOI] [PubMed] [Google Scholar]
- Buettner R. Mora LB. Jove R. Activated STAT signaling in human tumors provides novel molecular targets for therapeutic intervention. Clin Cancer Res. 2002;8(4):945–954. [PubMed] [Google Scholar]
- Castellsague X. Natural history and epidemiology of HPV infection and cervical cancer. Gynecol Oncol. 2008;110(3 Suppl 2):S4–S7. doi: 10.1016/j.ygyno.2008.07.045. [DOI] [PubMed] [Google Scholar]
- Charpin C. Dales JP. Garcia S. Carpentier S. Djemli A. Andrac L. Lavaut MN. Allasia C. Bonnier P. Tumor neoangiogenesis by CD31 and CD105 expression evaluation in breast carcinoma tissue microarrays. Clin Cancer Res. 2004;10(17):5815–5819. doi: 10.1158/1078-0432.CCR-04-0021. [DOI] [PubMed] [Google Scholar]
- Chen CL. Hsieh FC. Lieblein JC. Brown J. Chan C. Wallace JA. Cheng G. Hall BM. Lin J. Stat3 activation in human endometrial and cervical cancers. Br J Cancer. 2007;96(4):591–599. doi: 10.1038/sj.bjc.6603597. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dechow TN. Pedranzini L. Leitch A. Leslie K. Gerald WL. Linkov I. Bromberg JF. Requirement of matrix metalloproteinase-9 for the transformation of human mammary epithelial cells by Stat3-C. Proc Natl Acad Sci USA. 2004;101(29):10602–10607. doi: 10.1073/pnas.0404100101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dvorak HF. Gresser I. Microvascular injury in pathogenesis of interferon-induced necrosis of subcutaneous tumors in mice. J Natl Cancer Inst. 1989;81(7):497–502. doi: 10.1093/jnci/81.7.497. [DOI] [PubMed] [Google Scholar]
- Fearnley IM. Carroll J. Shannon RJ. Runswick MJ. Walker JE. Hirst J. GRIM-19, a cell death regulatory gene product, is a sub-unit of bovine mitochondrial NADH:ubiquinone oxidoreductase (complex I) J Biol Chem. 2001;276(42):38345–38348. doi: 10.1074/jbc.C100444200. [DOI] [PubMed] [Google Scholar]
- Frank DA. STAT3 as a central mediator of neoplastic cellular transformation. Cancer Lett. 2007;251(2):199–210. doi: 10.1016/j.canlet.2006.10.017. [DOI] [PubMed] [Google Scholar]
- Hu J. Angell JE. Zhang J. Ma X. Seo T. Raha A. Hayashi J. Choe J. Kalvakolanu DV. Characterization of monoclonal antibodies against GRIM-19, a novel IFN-beta and retinoic acid-activated regulator of cell death. J Interferon Cytokine Res. 2002;22(10):1017–1026. doi: 10.1089/107999002760624242. [DOI] [PubMed] [Google Scholar]
- Huibregtse JM. Scheffner M. Howley PM. A cellular protein mediates association of p53 with the E6 oncoprotein of human papillomavirus types 16 or 18. EMBO J. 1991;10(13):4129–4135. doi: 10.1002/j.1460-2075.1991.tb04990.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Inghirami G. Chiarle R. Simmons WJ. Piva R. Schlessinger K. Levy DE. New and old functions of STAT3: a pivotal target for individualized treatment of cancer. Cell Cycle. 2005;4(9):1131–1133. doi: 10.4161/cc.4.9.1985. [DOI] [PubMed] [Google Scholar]
- Kalakonda S. Nallar SC. Lindner DJ. Hu J. Reddy SP. Kalvakolanu DV. Tumor-suppressive activity of the cell death activator GRIM-19 on a constitutively active signal transducer and activator of transcription 3. Cancer Res. 2007;67(13):6212–6220. doi: 10.1158/0008-5472.CAN-07-0031. [DOI] [PubMed] [Google Scholar]
- Kalvakolanu DV. The GRIMs: a new interface between cell death regulation and interferon/retinoid induced growth suppression. Cytokine Growth Factor Rev. 2004;15(2–3):169–194. doi: 10.1016/j.cytogfr.2004.01.002. [DOI] [PubMed] [Google Scholar]
- Lens SM. Wolthuis RM. Klompmaker R. Kauw J. Agami R. Brummelkamp T. Kops G. Medema RH. Survivin is required for a sustained spindle checkpoint arrest in response to lack of tension. EMBO J. 2003;22(12):2934–2947. doi: 10.1093/emboj/cdg307. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lippman SM. Kavanagh JJ. Paredes-Espinoza M. Delgadillo-Madrueno F. Paredes-Casillas P. Hong WK. Holdener E. Krakoff IH. 13-cis-retinoic acid plus interferon alpha-2a: highly active systemic therapy for squamous cell carcinoma of the cervix. J Natl Cancer Inst. 1992;84(4):241–245. doi: 10.1093/jnci/84.4.241. [DOI] [PubMed] [Google Scholar]
- Lowy DR. Solomon D. Hildesheim A. Schiller JT. Schiffman M. Human papillomavirus infection and the primary and secondary prevention of cervical cancer. Cancer. 2008;113(7 Suppl):1980–1993. doi: 10.1002/cncr.23704. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lu H. Cao X. GRIM-19 is essential for maintenance of mitochondrial membrane potential. Mol Biol Cell. 2008;19(5):1893–1902. doi: 10.1091/mbc.E07-07-0683. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lufei C. Ma J. Huang G. Zhang T. Novotny-Diermayr V. Ong CT. Cao X. GRIM-19, a death-regulatory gene product, suppresses Stat3 activity via functional interaction. EMBO J. 2003;22(6):1325–1335. doi: 10.1093/emboj/cdg135. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Majewski S. Szmurlo A. Marczak M. Jablonska S. Bollag W. Synergistic effect of retinoids and interferon alpha on tumor-induced angiogenesis: anti-angiogenic effect on HPV-harboring tumor-cell lines. Int J Cancer. 1994;57(1):81–85. doi: 10.1002/ijc.2910570115. [DOI] [PubMed] [Google Scholar]
- Martin CM. Kehoe L. Spillane CO. O'Leary JJ. Gene discovery in cervical cancer: towards diagnostic and therapeutic biomarkers. Mol Diagn Ther. 2007;11(5):277–290. doi: 10.1007/BF03256249. [DOI] [PubMed] [Google Scholar]
- Mita AC. Mita MM. Nawrocki ST. Giles FJ. Survivin: key regulator of mitosis and apoptosis and novel target for cancer therapeutics. Clin Cancer Res. 2008;14(16):5000–5005. doi: 10.1158/1078-0432.CCR-08-0746. [DOI] [PubMed] [Google Scholar]
- Ochsenbein AF. Sierro S. Odermatt B. Pericin M. Karrer U. Hermans J. Hemmi S. Hengartner H. Zinkernagel RM. Roles of tumour localization, second signals and cross priming in cytotoxic T-cell induction. Nature. 2001;411(6841):1058–1064. doi: 10.1038/35082583. [DOI] [PubMed] [Google Scholar]
- Papa F. Delia M. Trentadue R. Panelli D. Bellomo F. Serpico R. Petruzzi M. De Benedittis M. Scacco S. Differential effects of all-trans retinoic acid on the growth of human keratinocytes and mouth carcinoma epidermoid cultures. Involvement of GRIM-19 and complex I of the respiratory chain. Int J Immunopathol Pharmacol. 2007;20(4):719–729. doi: 10.1177/039463200702000407. [DOI] [PubMed] [Google Scholar]
- Reeves MB. Davies AA. McSharry BP. Wilkinson GW. Sinclair JH. Complex I binding by a virally encoded RNA regulates mitochondria-induced cell death. Science. 2007;316(5829):1345–1348. doi: 10.1126/science.1142984. [DOI] [PubMed] [Google Scholar]
- Reuter S. Delius H. Kahn T. Hofmann B. zur Hausen H. Schwarz E. Characterization of a novel human papillomavirus DNA in the cervical carcinoma cell line ME180. J Virol. 1991;65(10):5564–5568. doi: 10.1128/jvi.65.10.5564-5568.1991. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Santin AD. Zhan F. Bignotti E. Siegel ER. Cane S. Bellone S. Palmieri M. Anfossi S. Thomas M. Burnett A. Kay HH. Roman JJ. O'Brien TJ. Tian E. Cannon MJ. Shaughnessy J., Jr Pecorelli S. Gene expression profiles of primary HPV16- and HPV18-infected early stage cervical cancers and normal cervical epithelium: identification of novel candidate molecular markers for cervical cancer diagnosis and therapy. Virology. 2005;331(2):269–291. doi: 10.1016/j.virol.2004.09.045. [DOI] [PubMed] [Google Scholar]
- Schaefer LK. Ren Z. Fuller GN. Schaefer TS. Constitutive activation of Stat3alpha in brain tumors: localization to tumor endothelial cells and activation by the endothelial tyrosine kinase receptor (VEGFR-2) Oncogene. 2002;21(13):2058–2065. doi: 10.1038/sj.onc.1205263. [DOI] [PubMed] [Google Scholar]
- Scheffner M. Munger K. Huibregtse JM. Howley PM. Targeted degradation of the retinoblastoma protein by human papilloma-virus E7-E6 fusion proteins. EMBO J. 1992;11(7):2425–2431. doi: 10.1002/j.1460-2075.1992.tb05307.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Scheffner M. Werness BA. Huibregtse JM. Levine AJ. Howley PM. The E6 oncoprotein encoded by human papillomavirus types 16 and 18 promotes the degradation of p53. Cell. 1990;63(6):1129–1136. doi: 10.1016/0092-8674(90)90409-8. [DOI] [PubMed] [Google Scholar]
- Schiffman M. Castle PE. Jeronimo J. Rodriguez AC. Wacholder S. Human papillomavirus and cervical cancer. Lancet. 2007;370(9590):890–907. doi: 10.1016/S0140-6736(07)61416-0. [DOI] [PubMed] [Google Scholar]
- Seo T. Lee D. Shim YS. Angell JE. Chidambaram NV. Kalvakolanu DV. Choe J. Viral interferon regulatory factor 1 of Kaposi's sarcoma-associated herpesvirus interacts with a cell death regulator, GRIM19, and inhibits interferon/retinoic acid-induced cell death. J Virol. 2002;76(17):8797–8807. doi: 10.1128/JVI.76.17.8797-8807.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sidky YA. Borden EC. Inhibition of angiogenesis by interferons: effects on tumor- and lymphocyte-induced vascular responses. Cancer Res. 1987;47(19):5155–5161. [PubMed] [Google Scholar]
- Speliotes EK. Uren A. Vaux D. Horvitz HR. The survivin-like C. elegans BIR-1 protein acts with the Aurora-like kinase AIR-2 to affect chromosomes and the spindle midzone. Mol Cell. 2000;6(2):211–223. doi: 10.1016/s1097-2765(00)00023-x. [DOI] [PubMed] [Google Scholar]
- Terenzi F. Saikia P. Sen GC. Interferon-inducible protein, P56, inhibits HPV DNA replication by binding to the viral protein E1. EMBO J. 2008;27(24):3311–3321. doi: 10.1038/emboj.2008.241. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang L. Luo J. He S. Induction of MMP-9 release from human dermal fibroblasts by thrombin: involvement of JAK/STAT3 signaling pathway in MMP-9 release. BMC Cell Biol. 2007;8:14. doi: 10.1186/1471-2121-8-14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wei D. Le X. Zheng L. Wang L. Frey JA. Gao AC. Peng Z. Huang S. Xiong HQ. Abbruzzese JL. Xie K. Stat3 activation regulates the expression of vascular endothelial growth factor and human pancreatic cancer angiogenesis and metastasis. Oncogene. 2003;22(3):319–329. doi: 10.1038/sj.onc.1206122. [DOI] [PubMed] [Google Scholar]
- Wheatley SP. Carvalho A. Vagnarelli P. Earnshaw WC. INCENP is required for proper targeting of Survivin to the centromeres and the anaphase spindle during mitosis. Curr Biol. 2001;11(11):886–890. doi: 10.1016/s0960-9822(01)00238-x. [DOI] [PubMed] [Google Scholar]
- Xie TX. Huang FJ. Aldape KD. Kang SH. Liu M. Gershenwald JE. Xie K. Sawaya R. Huang S. Activation of stat3 in human melanoma promotes brain metastasis. Cancer Res. 2006;66(6):3188–3196. doi: 10.1158/0008-5472.CAN-05-2674. [DOI] [PubMed] [Google Scholar]
- Xie TX. Wei D. Liu M. Gao AC. Ali-Osman F. Sawaya R. Huang S. Stat3 activation regulates the expression of matrix metal-loproteinase-2 and tumor invasion and metastasis. Oncogene. 2004;23(20):3550–3560. doi: 10.1038/sj.onc.1207383. [DOI] [PubMed] [Google Scholar]
- Xu Q. Briggs J. Park S. Niu G. Kortylewski M. Zhang S. Gritsko T. Turkson J. Kay H. Semenza GL. Cheng JQ. Jove R. Yu H. Targeting Stat3 blocks both HIF-1 and VEGF expression induced by multiple oncogenic growth signaling pathways. Oncogene. 2005;24(36):5552–5560. doi: 10.1038/sj.onc.1208719. [DOI] [PubMed] [Google Scholar]
- Zhang J. Yang J. Roy SK. Tininini S. Hu J. Bromberg JF. Poli V. Stark GR. Kalvakolanu DV. The cell death regulator GRIM-19 is an inhibitor of signal transducer and activator of transcription 3. Proc Natl Acad Sci USA. 2003;100(16):9342–9347. doi: 10.1073/pnas.1633516100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang L. Gao L. Li Y. Lin G. Shao Y. Ji K. Yu H. Hu J. Kalvakolanu DV. Kopecko DJ. Zhao X. Xu DQ. Effects of plasmid-based Stat3-specific short hairpin RNA and GRIM-19 on PC-3M tumor cell growth. Clin Cancer Res. 2008;14(2):559–568. doi: 10.1158/1078-0432.CCR-07-1176. [DOI] [PubMed] [Google Scholar]
- Zhang X. Huang Q. Yang Z. Li Y. Li CY. GW112, a novel antiapoptotic protein that promotes tumor growth. Cancer Res. 2004;64(7):2474–2481. doi: 10.1158/0008-5472.can-03-3443. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.