Abstract
Epstein-Barr virus (EBV) efficiently immortalizes human B cells and is associated with several human malignancies. The EBV transcriptional activating protein EBNA2 and the EBNA2 coactivator EBNA-leader protein (EBNA-LP) are important for B cell immortalization. Recent observations from our laboratory indicate that EBNA-LP coactivation function is mediated through interactions with the interferon-inducible gene (ISG) Sp100, resulting in displacement from its normal location in promyelocytic leukemia nuclear bodies (PML NBs) into the nucleoplasm. The EBNA-LP- and interferon-mediated mechanisms that regulate Sp100 subnuclear localization and transcriptional function remain undefined. To clarify these issues, we generated a panel of Sp100 mutant proteins to ascertain whether EBNA-LP induces Sp100 displacement from PML NBs by interfering with Sp100 dimerization or through other domains. In addition, we tested EBNA-LP function in interferon-treated cells. Our results indicate that Sp100 dimerization, PML NB localization, and EBNA-LP interaction domains overlap significantly. We also show that IFN-β does not inhibit EBNA-LP coactivation function. The results suggest that EBNA-LP might play a role in EBV-evasion of IFN-mediated antiviral responses.
Introduction
Epstein-Barr virus (EBV), a gammaherpesvirus, is a primary cause of infectious mononucleosis and is linked with several human malignancies including, endemic Burkitt's lymphoma (eBL), nasopharyngeal carcinoma (NPC), and lymphomas in the immunosuppressed (Blaho and Aaronson 1999; Bornkamm and Hammerschmidt 2001; Thorley-Lawson 2001; Young and Rickinson 2004). EBV efficiently immortalizes human B cells (Rowe 2001; Thorley-Lawson 2001). The gene-expression program associated with B cell immortalization includes the EBV nuclear antigens (EBNAs) and latent membrane proteins (LMPs) (Thorley-Lawson 2001). Genetic and biochemical evidence has shown that EBNA2 and EBNA-leader protein (EBNA-LP) are important for initiating and maintaining EBV-mediated B cell immortalization (Hammerschmidt and Sugden 1989; Mannick and others 1991; Kempkes and others 1995; Gordadze and others 2004). EBNA2 is a transcription-activating protein, which regulates viral latency genes through mimicry of Notch signaling pathways (Fahraeus and others 1990; Tsang and others 1991; Laux and others 1994; Hsieh and others 1996; Gordadze and others 2001; Hofelmayr and others 2001) and EBNA-LP coactivates EBNA2, preferentially enhancing the expression of the major viral oncoprotein LMP1 (Allday and others 1989; Rooney and others 1989; Harada and Kieff 1997; Nitsche and others 1997; Peng and others 2005).
EBNA-LP is an unusual protein comprising a variable number of 66 amino acid repeats and a unique 45-residue carboxyl-terminus (Sample and others 1986; Speck and others 1986) (Fig. 1). EBNA-LP plays an important role in establishment of B cell immortalization as mutant viruses lacking the Y1Y2 exons, which code for the unique carboxy-terminal domain, are much less efficient at immortalizing B cells relative to wild-type viruses (Hammerschmidt and Sugden 1989; Mannick and others 1991). Interestingly, EBNA2 coactivation function does not require the carboxy-terminal unique domain (Harada and Kieff 1997; Nitsche and others 1997). The EBNA2 coactivation function of EBNA-LP requires a minimum of two repeats, and deletion of evolutionarily conserved region 3 (CR3) in EBNA-LP greatly diminishes its coactivation function (Nitsche and others 1997; Peng and others 2000). Recent evidence has suggested that the promyelocytic leukemia nuclear body (PML NB)-associated protein Sp100 is an important mediator of EBNA-LP coactivation function (Ling and others 2005). EBNA-LP specifically relocalizes Sp100 from PML NBs, and this activity correlates positively with EBNA2 coactivation.
FIG. 1.
Schematic of the Epstein-Barr Virus (EBV) genome and the sequence of EBNA-LP. The EBV genome is shown in linear form with relevant structural motifs and location of exons (black boxes) encoding latency proteins, which are labeled above. The internal repeat region (IR1) is also shown by the gray box. The viral transcript encoding EBNA-LP is shown below and is derived from the latency W promoter (Wp). The first exon transcribed from Wp is W0 and is noncoding. Alternative splicing from W0 to W1 can result in formation of an initiation codon (W1′) or no initiation codon (W1). Splicing that generates W1′ gives rise to transcripts encoding EBNA-LP. EBNA-LP is thus composed of repeated W1 and W2 exons as well as two unique exons known as Y1 and Y2. The transcript is bi-cistronic as it also contains the EBNA2 ORF at the 3′ end. The dashed line from the EBNA2 protein indicates promoters that are activated by EBNA2. The viral bidirectional promoter, which controls expression of LMP-1 and LMP2B, is indicated by asterisks. Transcripts encoding LMP-1 are shown below. A sequence comparison between EBV EBNA-LP and other nonhuman primate lymphocryptoviruses is shown below indicating presence of conserved regions (CR). Location of nuclear localization signals (NLS) and regions required for EBNA2 coactivation are indicated. Inverted triangles above the sequence indicate conserved serine residues.
Promyelocytic leukemia nuclear bodies have been shown to regulate a broad range of cellular processes including apoptosis, genomic stability, cell cycle regulation, transcriptional regulation, and antiviral responses (Everett and others 1998; Muller and Dejean 1999; Seeler and Dejean 1999; Adamson and Kenney 2001; Borden 2002; Bernardi and Pandolfi 2003; Ling and others 2005; Bernardi and Pandolfi 2007; Everett and Chelbi-Alix 2007). Proteins endogenously expressed in PML NBs include Sp100, PML, Daxx, and small ubiquitin-related modifiers such as SUMO (Negorev and Maul 2001). The PML protein is a definitive marker for PML NBs and is required for their proper assembly (Ishov and others 1999). Other proteins conditionally expressed in or in close proximity to PML NBs, include heterochromatin protein 1α (HP-1α), CREB-binding protein (CBP), and the tumor suppressors p53 and pRb (Negorev and Maul 2001).
Several herpesviruses have been shown to interact with or degrade PML and PML NB-associated proteins, suggesting that these nuclear organelles are important modulators of virus infection. For example, expression of the immediate early gene product ICP0 by herpes simplex virus-1 (HSV-1) has been shown to induce the proteosome-dependent degradation of PML (Everett and others 1998; Chelbi-Alix and de The 1999). This results in the transient association of PML NBs that are first found associated with incoming genomes (Everett and others 2003). HSV-1 viruses expressing ICP0 mutants unable to induce PML degradation have a much lower probability of progressing into productive lytic infection at low MOIs (Everett and others 2006). In addition, extensive depletion of PML in human fibroblast cells significantly increases gene expression and plaque formation of ICP0 null viruses (Everett and others 2006). Moreover, viruses with mutations that inactivate ICP0 do not enter productive replication after low multiplicity infection of human fibroblasts, but instead are retained in a repressed quiescent state that in some respects resembles latency. The CMV immediate early protein IE1 appears to induce the loss of sumoylated forms of PML, resulting in the destruction of PML NBs (Korioth and others 1996; Wilkinson and others 1998; Muller and Dejean 1999; Ahn and Hayward 2000; Lee and others 2004). Like HSV-1, CMV mutants impaired for inducing PML NB destruction are less efficient at establishing productive infection at low MOIs and these mutants can be complemented in human fibroblast cells depleted for PML protein (Mocarski and others 1996; Greaves and Mocarski 1998; Ahn and Hayward 2000; Lee and others 2004; Tavalai and others 2006). CMV also encodes a tegument protein known as pp71 that induces the degradation of the PML NB-associated protein hDaxx (Hwang and Kalejta 2007). Depletion of hDaxx enhances hCMV gene expression, especially by pp71-deficient viruses (Ishov and others 2002; Cantrell and Bresnahan 2005; Cantrell and Bresnahan 2006; Saffert and Kalejta 2007). Collectively, the data suggest that PML NB-associated proteins have a role in inhibiting virus gene expression during productive lytic infection. In addition, some of these studies suggest that PML NBs may play a role in establishment or maintenance of latency.
Both Sp100 and PML promoters contain interferon stimulated response elements and are up-regulated at the transcriptional level by interferon (Guldner and others 1992; ChelbiAlix and others 1995; Grotzinger and others 1996; Negorev and others 2006). HSV-1 replicates more efficiently in interferon-treated PML-negative fibroblasts relative to interferon-treated wild-type fibroblasts (Chee and others 2003). Interferon was also able to inhibit immediate early viral gene expression and PML degradation in human cells (Taylor and others 2000). Together, these data suggest that PML carries out interferon-mediated antiviral activities. Whether Interferon modifies the function of Sp100, however, remains undefined.
Sp100A contains several functional domains including a carboxyl-terminal nuclear localization signal (NLS), a heterochromatin-1α interaction site, an amino-terminal dimerization domain, and a PML NB-targeting domain (Seeler and others 1998; Sternsdorf and others 1999; Negorev and others 2001; Ling and others 2005). Three additional alternatively spliced isoforms of Sp100A are made in lower abundance relative to Sp100A and localize predominantly outside of PML NBs in the nucleoplasm (Sp100-B, -C, -HMG) (Negorev and others 2006). EBNA-LP coactivation function has been linked predominantly with the PML NB-associated Sp100A isoform (Ling and others 2005). The Sp100 dimerization, PML-targeting, and EBNA-LP interacting domains reside within the amino-terminal 150 amino acid residues, but their relationship to each other remains unclear (Sternsdorf and others 1999; Negorev and others 2001; Ling and others 2005). Previous attempts to separate the PML NB localization and dimerization domains were unsuccessful. These studies utilized truncated Sp100 fusion proteins, which might not have adopted physiological conformations required for these domains to function correctly (Sternsdorf and others 1999). However, another study suggested that the Sp100 dimerization and PML NB-localization domains are distinct because fusion of Sp100 residues 29–152 to the p53-binding domain of hMDM2 resulted in loss of self-aggregation, but retention of PML-NB localization (Negorev and others 2001). To clarify this issue, we generated a series of small deletions within residues 3–152 in the context of the full length Sp100 protein. The mutant Sp100 proteins were also used to determine the EBNA-LP binding domain relative to the dimerization and PML NB localization domains. In addition, we tested whether Interferon could modulate EBNA-LP-mediated relocalization of Sp100 from PML NBs and whether this might have an effect on EBNA-LP coactivation function.
Materials and Methods
Plasmids
A flag epitope-tagged Sp100A protein expressing the amino terminal 182 residues of Sp100 fused with the SV40 NLS was generated by PCR (1-182-Flag). Fifteen amino acid deletions were introduced into full length, hemagglutinintagged Sp100A by PCR as described previously (Gordadze and others 2004; Ling and others 2005). EBNA2, wild-type EBNA-LP, and ΔCR3 EBNA-LP expression plasmids, and the multimerized Cp promoter luciferase reporter plamid (8xCp-LUC) have been described previously (Harada and Kieff 1997; Fuentes-Panana and Ling 1998). The ICP0 expression plasmid (pGH92) was a gift from Dr. Gary Hayward (Johns Hopkins University School of Medicine, Baltimore, MD, USA). The ICP0-luc plasmid was kindly provided by Dr. Gerd Maul (The Wistar Institute, Philadelphia, PA, USA).
Cell culture, interferon treatment, and transfections
EBV-positive Daudi and EBV-negative DG75 cells were grown in RPMI 1640 media supplemented with 10% fetal bovine serum (Hyclone) and maintained at 37°C in 5% CO2. The BL41-LP cell line (a gift from Dr. Elliott Kieff, Harvard Medical School, Boston, MA, USA) was maintained in tetracycline-free media. EBNA-LP expression was induced in BL41-LP cells by adding 2 ng/mL doxycycline (Clontech). Recombinant human interferon beta (Chemicon, IF014) was added directly to the cells where indicated, at a final concentration of 100 U/mL. All transfections were carried out by electroporation (BioRad Gene Pulser) as described previously (Gordadze and others 2004). In brief, 1 × 107 cells plus 30 μg total plasmid DNA were resuspended in 0.5 mL serum-free RPMI. Cells were pulsed at 200V and 975F, placed in 2.5 mL RPMI media plus 10% FBS, and maintained for 48 h at 37°C in 5% CO2.
Luciferase reporter assays
Two days posttransfection, cells were solubilized and luciferase activity measured using the dual-luciferase reporter assay system (Promega) according to the manufacturer's recommendations. The remainder of the cell lysates were used to determine protein expression levels by immunoblot assays.
Coimmunoprecipitation and immunoblot assays
For coimmunoprecipitation assays, cells were harvested in 1% NP40 (Calbiochem) containing protease inhibitor cocktail tablets (Roche). Soluble lysates were immunoprecipitated with anti-Flag antibodies at 4°C overnight. Immune complexes were pulled down with Immobilized Protein G beads (Pierce) for 1 h at room temperature. Protein complexes were recovered by boiling for 5 min in Laemmli buffer (4% SDS, 20% glycerol, 0.125 M Tris-Cl, pH 6.8, 0.004% bromphenol blue, 3% 2-mercaptoethanol). For immunoblot analyses, lysates were resolved on a 7.5% SDS-PAGE gel, immobilized onto nitrocellulose membranes, and blocked in 5% milk in phosphate-buffered saline (PBS). EBNA2 and EBNA-LP protein expression were detected using rat monoclonal anti-EBNA2 antibodies or mouse monoclonal anti-EBNA-LP antibodies respectively (Finke and others 1987; Kremmer and others 1995). Transfected Sp100A proteins were detected using anti-HA (Covance) or Flag antibodies (M2, Sigma). Endogenous Sp100 protein expression was determined by probing the blots with rabbit polyclonal anti-Sp100 antibodies, which were generated by immunizing rabbits with a His-tagged Sp100 protein comprised of the amino-terminal 252 residues purified from Escherichia coli. PML protein expression was monitored using rabbit polyclonal anti-PML (H-238, Santa Cruz), while α-tubulin expression was determined using mouse monoclonal anti α-tubulin antibodies (Sigma). All primary antibody incubations were carried out in 0.5% milk in PBS, while secondary antibody reactions were carried out using antirat (EBNA2), antimouse (HA, Flag, EBNA-LP, α-tubulin) or antirabbit (Sp100) HRP-conjugated antibodies (Jackson Immunolaboratories) in a solution of 0.01% Tween-20/PBS. Blots were developed using a chemiluminescence detection kit (Pierce).
Immunofluorescence and co-localization
Transfected or IFN-treated cells were spun onto coverslips at 1,000 rpm for 1 min. The cells were fixed in 4% formaldehyde, and permeabilized in 0.5% Triton X-100 (Fisher) (Ling and others 2005). Permeabilized cells were incubated at 4°C overnight in 5% milk/TBST/0.02% Sodium azide (blocking buffer). Mouse monoclonal anti-HA antibodies (Covance), followed by Alexa Fluor 594 goat antimouse IgG (Molecular probes) secondary antibodies were used to detect localization of HA-tagged Sp100A proteins. Endogenous Sp100 expression and localization was monitored using rabbit -polyclonal anti-Sp100 antibodies (Calbiochem) followed by Alexa Fluor 488 goat antirabbit IgG secondary antibodies (Molecular Probes). PML localization was detected using primary anti-PML monoclonal or polyclonal antibodies (Santa Cruz) followed by Alexa Fluor 594 goat antimouse or antirabbit secondary antibodies. ICP0 expression and localization was detected using mouse monoclonal antibodies (Eastcoast Bio). The cells were visualized and counted using a Zeiss AxioPlan fluorescence microscope (Integrated Microscopy Core, BCM). To determine the numbers of cells displaying punctate or diffuse Sp100 expression, we counted 100 cells each, in triplicate experiments. The numbers of Sp100-associated dots were also determined as a function of time and IFN-β dose in BL41-LP cells.
Results
Sp100 dimerization and EBNA-LP-interaction domains reside between amino acid residues 46–150
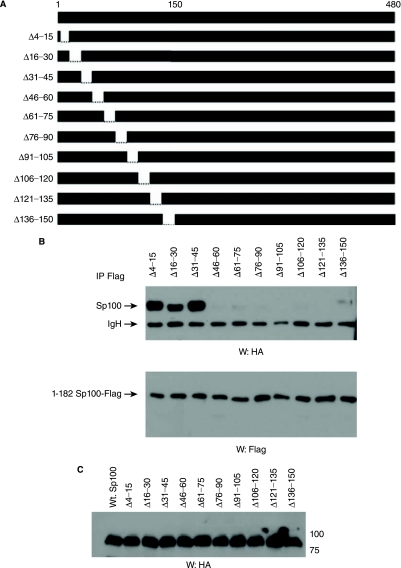
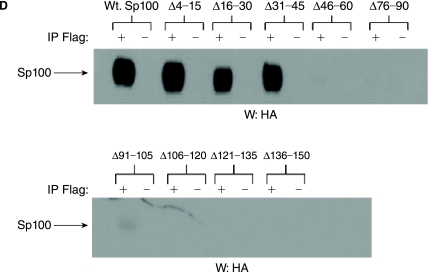
Biochemical characterization of Sp100A has revealed that the amino-terminal 152 residues mediate its dimerization, interaction with EBNA-LP, and localization to PML NBs (Sternsdorf and others 1999; Negorev and others 2001; Ling and others 2005). To further define the Sp100A residues most important for these functions, we generated a panel of Sp100A proteins with 15 amino acid deletions within the amino-terminal 150 amino acid residues (Fig. 2A). Dimerization was assessed by analyzing the ability of a coexpressed truncated Sp100 protein (1-182-Flag) to coimmunoprecipitate wild-type or mutant Sp100A proteins as described previously (Sternsdorf and others 1999). Deletions spanning amino acid residues 46–150 were deficient for Sp100A self-association, while all other deletions tested did not affect Sp100 self-association (Fig. 2B). Control blots indicated that all of the proteins were expressed at similar levels (Figs. 2B and C). To determine the specific EBNA-LP binding domain, EBNA-LP-Flag was coexpressed with the Sp100 mutant proteins and their association with each other was analyzed by coimmunoprecipitation. EBNA-LP was able to coimmunoprecipitate Sp100 with deletions in residues 4–45, but failed to associate with Sp100A mutants with deletions spanning residues 46–150 (Fig. 2D). Collectively, these data suggest that Sp100A dimerization and EBNA-LP-binding domains localize between amino acid residues 46–150.
FIG. 2.
Sp100A dimerization and EBNA-LP interaction domains are between amino acid residues 46–150. (A) Schematic representation of wild-type and mutant Sp100A. Boundaries of each deletion are indicated. (B) EBV-negative DG75 cells were cotransfected with 1–182 Sp100-Flag and wild-type or mutant Sp100A plasmids represented in (A). Following immunoprecipitation with anti-Flag antibodies, coprecipitated Sp100A was detected by Western blot using anti-HA antibodies. Anti-Flag antibodies were used to confirm 1–182-Flag precipitation. (C) Western blot detection of wild-type and mutant Sp100A proteins expressed in transfected DG75 cells. (D) DG75 cells were cotransfected with wild-type or mutant Sp100A and flag-tagged EBNA-LP expression plasmids. Immunoprecipitations and Western blots were carried out as outlined above, using anti-Flag and anti-HA antibodies.
The Sp100A PML NB targeting domain maps to residues 46–150
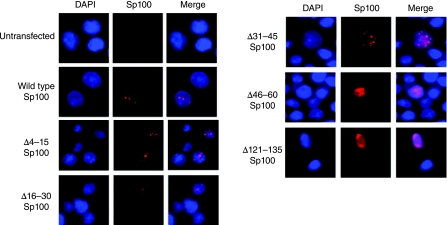
Immunofluorescence assays were carried out to determine the cellular localization of the Sp100A deletion mutants described in Figure 2A. Wild-type Sp100A was localized in punctate nuclear dots characteristic of PML NBs (Fig. 3). Amino acid deletions within Sp100A residues 4–45 showed similar staining patterns as wild-type Sp100A while Sp100A mutants with deletions spanning residues 46–150 showed diffuse nuclear localization (Fig. 3, selected mutants shown to illustrate general pattern). The percentage of cells expressing wild-type or mutant Sp100 proteins as punctuate nuclear dots or diffuse nuclear staining, is shown in Table 1. Co-staining with a PML antibody confirmed that those proteins with a punctate staining pattern were within PML NBs (data not shown). These data suggest that the amino terminal 45 residues of Sp100A are not important for Sp100A localization to PML nuclear bodies, and PML NB targeting function is mediated by a domain(s) contained between amino acid residues 46–150.
FIG. 3.
The Sp100A PML NB targeting domain is between amino acid residues 46–150. Immunofluorescence detection of transiently expressed Sp100A proteins in DG75 cells. Sp100A (red) was monitored using mouse monoclonal anti-HA antibodies. Cells were co-stained with DAPI to detect the nucleus. Merged panels are shown in the far right hand column as indicated.
Table 1.
Wild-Type and Mutant Sp100 Expression Patterns in EBV-Negative DG 75 Cells
| |
Expression patterns (n = 300) |
|
|---|---|---|
| Sp100 | Punctate (%) | Diffuse (%) |
| Wild-type | 96 | 4 |
| Δ4–15 | 98 | 2 |
| Δ16–30 | 96 | 4 |
| Δ31–45 | 97 | 3 |
| Δ46–60 | 0 | 100 |
| Δ121–135 | 0 | 100 |
EBNA-LP function is not inhibited by interferon
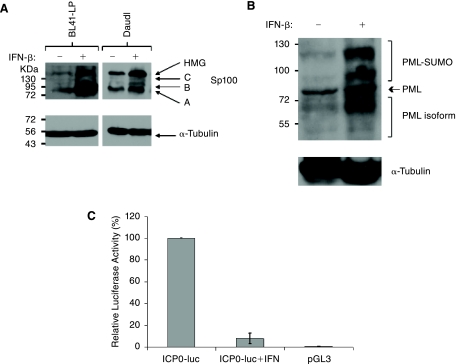
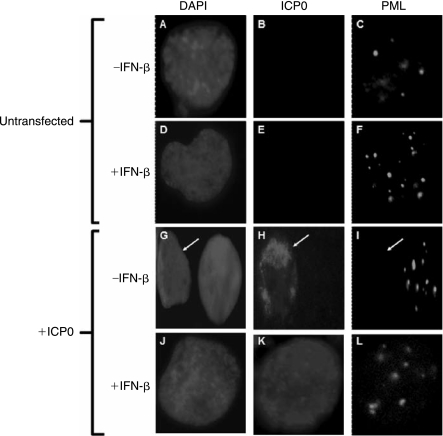
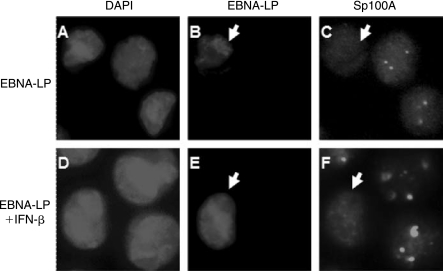
Type I and II IFNs induce Sp100 and PML expression, resulting in an increase in the number and size of PML NBs (Guldner and others 1992; ChelbiAlix and others 1995; Grotzinger and others 1996). Previous studies have shown that PML contributes to IFN-induced effects that inhibit HSV-1 replication (Chee and others 2003; Everett and others 2006). HSV-1 ICP0 and EBV EBNA-LP functions have been linked to their association with, and modulation of PML NBs (Ling and others 2005; Everett and others 2006). We therefore questioned whether IFN-induced Sp100A accumulation in PML NBs might inhibit EBNA-LP function. The IFN-inducibility of Sp100 was first confirmed in EBV-positive Daudi and BL41-LP cell lines (Fig. 4A). We also showed that the numbers of Sp100-associated nuclear dots increased as a function of IFN-β dose (Table 2) and treatment time (Table 3). As expected, IFN-β also induced PML (Figs. 4B and 5F). In addition, ICP0 induced PML degradation in untreated cells but not in those treated with IFN-β (Figs. 5I and L). Finally, IFN-β also inhibited the stimulation of the ICP0 promoter in transient transfection assays in agreement with previous reports (Negorev and others 2006) (Fig. 4C). To determine the effects of IFN-β on EBNA-LP-induced Sp100A relocalization from PML NBs, we analyzed Sp100A by immunofluorescence in Daudi cells following IFN-β treatment and transient expression of EBNA-LP. Transient expression of EBNA-LP in Daudi cells showed characteristic diffuse nuclear staining for EBNA-LP in both untreated and IFN-treated cells (Figs. 6B and E). Surprisingly, EBNA-LP displaced Sp100 from PML NBs in IFN-treated cells (Fig. 6F).
FIG. 4.
IFN-β regulates Sp100 and PML expression and the ICP0 promoter. (A) Western blot of endogenous Sp100A expression in BL41-LP and Daudi cells treated (+) or untreated (−) with 100 U/mL IFN-β for 24 h. Molecular weight markers are shown to the left side of the blot and Sp100 isoforms are indicated on the right side. α-Tubulin controls are shown in the panel below. (B) Western blot of endogenous PML expression in Daudi cells treated (+) or untreated (−) with 100 U/mL IFN-β for 24 h. Molecular weight markers are shown to the left side of the blot and PML isoforms indicated on the right side. α-Tubulin controls are shown in the panel below. (C) ICP0 promoter activity is inhibited by IFN-β. ICP0 promoter-driven luciferase plasmids were transfected into Daudi cells treated (+) and untreated (−) with IFN-β. A control sample was transfected with empty vector (pGL3). Relative luciferase activity for each of the transfections is indicated on the y-axis. Standard errors are indicated.
Table 2.
Effect of IFN-β Dose on Sp100 Accumulation in BL41-LP Cells
| |
Percentage of Sp100-associated nuclear dots per cell (n = 300) |
||
|---|---|---|---|
| IFN-β dose (U/mL) | ≤2 | ≥3≤4 | ≥5 |
| Untreated | 88 | 9 | 3 |
| 10 | 84 | 14 | 2 |
| 50 | 69 | 19 | 12 |
| 100 | 17 | 22 | 61 |
| 500 | 8 | 19 | 73 |
| 1000 | 7 | 27 | 66 |
Table 3.
Time-Dependent Response of Sp100 to IFN-β Treatment in BL41-LP Cells
| |
Percentage of Sp100-associated nuclear dots per cell (n = 300) |
||
|---|---|---|---|
| Time after IFN-β treatment (h) | ≤2 | ≥3≤4 | ≥5 |
| 0 | 85 | 9 | 6 |
| 4 | 76 | 15 | 9 |
| 8 | 62 | 22 | 16 |
| 16 | 49 | 19 | 32 |
| 24 | 20 | 18 | 62 |
FIG. 5.
HSV-1 ICP0-mediated PML degradation is inhibited by IFN-β. Immunofluorescence detection of ICP0 and Sp100A in Daudi cells. Untransfected Daudi cells without IFN (−) or treated with IFN (+) for 24 h (+) are indicated on the left side of the panels. Daudi cells transfected with an ICP0 expression plasmid without IFN (−) or treated with IFN (+) for 24 h (+) are indicated similarly.
FIG. 6.
EBNA-LP displaces Sp100 from PML NBs in IFN-treated cells. Immunofluorescence of Daudi cells transfected with an EBNA-LP expression plasmid with or without IFN treatment. EBNA-LP and Sp100A were detected using monoclonal and polyclonal antibodies respectively as described in the Materials and Methods. DAPI staining of each field is shown in the panels on the left.
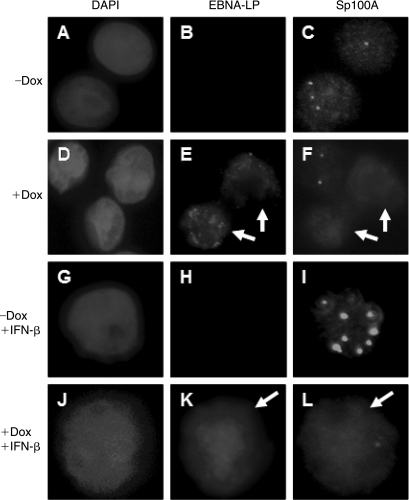
To alleviate concerns that this observation was due to EBNA-LP overexpression, we analyzed Sp100A localization in an EBNA-LP inducible cell line BL41-LP. EBNA-LP induction resulted in the displacement of Sp100A from PML NBs in both untreated and IFN-β-treated cells (Figs. 7F and L). These results suggest that EBNA-LP's ability to relocalize Sp100 may be refractory to the effects of IFN-β on Sp100A accumulation in nuclear bodies.
FIG. 7.
Induction of EBNA-LP expression in BL41-LP cells displaces Sp100A. BL41-LP cells were treated with 2 ng/mL doxycycline (Dox), resulting in the induction of EBNA-LP expression in about 50–80% of the cells. EBNA-LP and Sp100A were detected with monoclonal anti-EBNA-LP and polyclonal Sp100 antibodies respectively. Addition of Dox and/or IFN-β is shown on the left of each series of panels. White arrows indicate cells expressing EBNA-LP.
To determine the effects of IFN-β on EBNA-LP coactivation function, we transfected untreated or IFN-β-treated Daudi cells with an EBNA2-responsive luciferase reporter. In untreated and IFN-β-treated cells, EBNA-LP coactivated EBNA2 by about 16-fold (Figs. 8A and B). No coactivation was observed when EBNA2 was coexpressed with a previously characterized coactivation deficient EBNA-LP mutant (ΔCR3LP).
FIG. 8.
EBNA-LP coactivation function is not inhibited by IFN-β. Cp promoter luciferase reporter plasmids were transfected with EBNA2, EBNA-LP (LP), EBNA2 + LP, or EBNA2 + ΔCR3LP expression plasmids. Transfected cells were either untreated (A) or pulsed and maintained in 100 U/mL IFN-β for 24 h posttransfection (B). Relative fold-induction is shown on the left of each graph. Standard errors of the mean are indicated for each bar.
Discussion
In this study, we found that the Sp100A EBNA-LP binding domain lies between residues 46–150, which overlap with its dimerization and PML NB localization domains. Furthermore, under conditions in which IFN-β inhibited HSV-1 ICP0-induced PML degradation and ICP0 promoter acitivty, EBNA-LP coactivation function was unimpaired. The results implicate a previously uncharacterized role for EBNA-LP in resistance against host innate immune responses.
The results we obtained from characterizing Sp100A dimerization, EBNA-LP binding, and PML NB localization domains are consistent with previous findings (Sternsdorf and others 1999; Negorev and others 2001; Ling and others 2005). We were able to extend these studies by using more refined mutations in the context of the full length Sp100 protein. Although we were unable to separate domains for each of these functions, our data do not preclude the possibility that they are distinct. One explanation for our findings is that the Sp100A amino acid residues 46–150 may form interdependent tertiary structures, such that mutations in one region affect the function of adjacent regions. Therefore, more targeted subtle mutations may need to be introduced within residues 46–150 in order to functionally distinguish these domains. Alternatively, the Sp100A dimerization domain could mediate heterodimerization with EBNA-LP and/or cellular factors that modulate Sp100A localization within PML NBs (i.e., there is in fact only a single domain that mediates all three functions).
The ability of EBNA-LP to overcome the cellular response induced by IFN-β may play an important role during the establishment of latent infection in B cells. Several previous studies have shown that preincubation or simultaneous treatment with IFN relative to EBV exposure inhibits EBV-induced immortalization (Adams and others 1975b; Aman and Vongabain 1990; Kanda and others 1992). However, other studies have found that addition of IFN 24–48 h following exposure to EBV had little to no effect on EBV-induced immortalization (Adams and others 1975a; Menezes and others 1976; Lotz and others 1985). Furthermore, EBV-transformed lymphoblasts are refractory to IFN-induced growth inhibition.
Several EBV proteins or gene products have been implicated in counteracting IFN-induced cellular responses, including LMP-1, EBNA2, and the small noncoding EBER RNAs (Aman and Vongabain 1990; Zhang and Pagano 2000; Nanbo and others 2002). Our work suggests that EBNA-LP function is also refractory to IFN, which may contribute to the IFN-insensitive phenotype of EBV immortalized B cells. Although interferons are able to induce antiproliferative effects on target cells, including some malignant cell types, the potential widespread application of IFNs in controlling aberrant growth of malignant cells is hindered by IFN resistance (Adams and others 1975b; Strander 1986). Identifying viral or cellular proteins that contribute to IFN resistance will provide more insight into the mechanisms that counteract the antiproliferative actions of IFNs. Determining more definitive mechanisms of EBNA-LP action will reveal specific factors that help mediate EBNA-LP's coactivation function as well as its involvement in IFN resistance. In addition, since the gene expression pattern seen in lymphoblastoid cell lines (LCLs) mimics in vivo cases of EBV-associated post-transplant lymphoproliferative diseases (Straus and others 1993), our results may have some therapeutic applications. Identification of factors which mediate IFN resistance may be important in designing therapeutic agents against EBV-associated malignancies.
Acknowledgments
We would like to thank Drs. Andy Rice and Richard Sutton for insightful discussions and critical reading of this manuscript. This work was supported by NIH RO1 grants CA124309 (P.D.L.) and F31 CA126523-02 (C.W.E.)
References
- Adams A. Lidin B. Strander H. Cantell K. Spontaneous interferon-production and Epstein-Barr virus antigen expression in human lymphoid-cell lines. J Gen Virol. 1975a;28:219–223. doi: 10.1099/0022-1317-28-2-219. [DOI] [PubMed] [Google Scholar]
- Adams A. Strander H. Cantell K. Sensitivity of Epstein-Barr Virus transformed human lymphoid-cell lines to interferon. J Gen Virol. 1975b;28:207–217. doi: 10.1099/0022-1317-28-2-207. [DOI] [PubMed] [Google Scholar]
- Adamson AL. Kenney S. Epstein-Barr virus immediate-early protein BZLF1 is SUMO-1 modified and disrupts promyelocytic leukemia bodies. J Virol. 2001;75:2388–2399. doi: 10.1128/JVI.75.5.2388-2399.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ahn JH. Hayward GS. Disruption of PML-associated nuclear bodies by IE1 correlates with efficient early stages of viral gene expression and DNA replication in human cytomegalovirus infection. Virology. 2000;274:39–55. doi: 10.1006/viro.2000.0448. [DOI] [PubMed] [Google Scholar]
- Allday MJ. Crawford DH. Griffin BE. Epstein-Barr virus latent gene-expression during the initiation of B-cell immortalization. J Gen Virol. 1989;70:1755–1764. doi: 10.1099/0022-1317-70-7-1755. [DOI] [PubMed] [Google Scholar]
- Aman P. Vongabain A. An Epstein-Barr virus immortalization associated gene segment interferes specifically with the IFN-induced anti-proliferative response in human B-lymphoid cell-lines. EMBO J. 1990;9:147–152. doi: 10.1002/j.1460-2075.1990.tb08090.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bernardi R. Pandolfi PP. Role of PML and the PML-nuclear body in the control of programmed cell death. Oncogene. 2003;22:9048–9057. doi: 10.1038/sj.onc.1207106. [DOI] [PubMed] [Google Scholar]
- Bernardi R. Pandolfi PP. Structure, dynamics and functions of promyelocytic leukaemia nuclear bodies. Nat Rev Mol Cell Biol. 2007;8:1006–1016. doi: 10.1038/nrm2277. [DOI] [PubMed] [Google Scholar]
- Blaho JA. Aaronson SA. Convicting a human tumor virus: Guilt by association? Proc Natl Acad Sci USA. 1999;96:7619–7621. doi: 10.1073/pnas.96.14.7619. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Borden KLB. Pondering the promyelocytic leukemia protein (PML) puzzle: Possible functions for PML nuclear bodies. Mol Cell Biol. 2002;22:5259–5269. doi: 10.1128/MCB.22.15.5259-5269.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bornkamm GW. Hammerschmidt W. Molecular virology of Epstein-Barr virus. Philos Trans R Soc London Ser B. 2001;356:437–459. doi: 10.1098/rstb.2000.0781. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cantrell SR. Bresnahan WA. Interaction between the human cytomegalovirus UL82 gene product (pp71) and hDaxx regulates immediate-early gene expression and viral replication. J Virol. 2005;79:7792–7802. doi: 10.1128/JVI.79.12.7792-7802.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cantrell SR. Bresnahan WA. Human cytomegalovirus (HCMV) UL82 gene product (pp71) relieves hDaxx-mediated repression of HCMV replication. J Virol. 2006;80:6188–6191. doi: 10.1128/JVI.02676-05. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chee AV. Lopez P. Pandolfi PP. Roizman B. Promyelocytic leukemia protein mediates interferon-based anti-herpes simplex virus 1 effects. J Virol. 2003;77:7101–7105. doi: 10.1128/JVI.77.12.7101-7105.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chelbi-Alix MK. de The H. Herpes virus induced proteasome-dependent degradation of the nuclear bodies-associated PML and Sp100 proteins. Oncogene. 1999;18:935–941. doi: 10.1038/sj.onc.1202366. [DOI] [PubMed] [Google Scholar]
- ChelbiAlix MK. Pelicano L. Quignon F. Koken MHM. Venturini L. Stadler M. Pavlovic J. Degos L. Dethe H. Induction of the PML protein by interferons in normal and APL cells. Leukemia. 1995;9:2027–2033. [PubMed] [Google Scholar]
- Everett RD. Chelbi-Alix MK. PML and PML nuclear bodies: Implications in antiviral defence. Biochimie. 2007;89:819–830. doi: 10.1016/j.biochi.2007.01.004. [DOI] [PubMed] [Google Scholar]
- Everett RD. Freemont P. Saitoh H. Dasso M. Orr A. Kathoria M. Parkinson J. The disruption of ND10 during herpes simplex virus infection correlates with the Vmw11O- and proteasome-dependent loss of several PML isoforms. J Virol. 1998;72:6581–6591. doi: 10.1128/jvi.72.8.6581-6591.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Everett RD. Rechter S. Papior P. Tavalai N. Stamminger T. Orr A. PML contributes to a cellular mechanism of repression of herpes simplex virus type 1 infection that is inactivated by ICP0. J Virol. 2006;80:7995–8005. doi: 10.1128/JVI.00734-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Everett RD. Sourvinos G. Orr A. Recruitment of herpes simplex virus type 1 transcriptional regulatory protein ICP4 into foci juxtaposed to ND10 in live, infected cells. J Virol. 2003;77:3680–3689. doi: 10.1128/JVI.77.6.3680-3689.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fahraeus R. Jansson A. Ricksten A. Sjoblom A. Rymo L. Epstein-Barr virus-encoded nuclear antigen-2 activates the viral latent membrane-protein promoter by modulating the activity of a negative regulatory element. Proc Natl Acad Sci USA. 1990;87:7390–7394. doi: 10.1073/pnas.87.19.7390. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Finke J. Rowe M. Kallin B. Ernberg I. Rosen A. Dillner J. Klein G. Monoclonal and polyclonal antibodies against Epstein-Barr-virus nuclear antigen-5 (EBNA-5) detect multiple protein species in Burkitts-lymphoma and lymphoblastoid cell-lines. J Virol. 1987;61:3870–3878. doi: 10.1128/jvi.61.12.3870-3878.1987. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fuentes-Panana EM. Ling PD. Characterization of the CBF2 binding site within the Epstein-Barr virus latency C promoter and its role in modulating EBNA2-mediated transactivation. J Virol. 1998;72:693–700. doi: 10.1128/jvi.72.1.693-700.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gordadze AV. Onunwor CW. Peng RS. Poston D. Kremmer E. Ling PD. EBNA2 amino acids 3 to 30 are required for induction of LMP-1 and immortalization maintenance. J Virol. 2004;78:3919–3929. doi: 10.1128/JVI.78.8.3919-3929.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gordadze AV. Peng RS. Tan J. Liu GZ. Sutton R. Kempkes B. Bornkamm GW. Ling PD. Notch1IC partially replaces EBNA2 function in B cells immortalized by Epstein-Barr virus. J Virol. 2001;75:5899–5912. doi: 10.1128/JVI.75.13.5899-5912.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Greaves RF. Mocarski ES. Defective growth correlates with reduced accumulation of a viral DNA replication protein after low-multiplicity infection by a human cytomegalovirus ie1 mutant. J Virol. 1998;72:366–379. doi: 10.1128/jvi.72.1.366-379.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Grotzinger T. Jensen K. Will H. The interferon (IFN)-stimulated gene Sp100 promoter contains an IFN-gamma activation site and an imperfect IFN-stimulated response element which mediate type I IFN inducibility. J Biol Chem. 1996;271:25253–25260. doi: 10.1074/jbc.271.41.25253. [DOI] [PubMed] [Google Scholar]
- Guldner HH. Szostecki C. Grotzinger T. Will H. IFN enhance expression of Sp100, an autoantigen in primary biliary-cirrhosis. J Immunol. 1992;149:4067–4073. [PubMed] [Google Scholar]
- Hammerschmidt W. Sugden B. Genetic-analysis of immortalizing functions of Epstein-Barr virus in human lymphocytes-B. Nature. 1989;340:393–397. doi: 10.1038/340393a0. [DOI] [PubMed] [Google Scholar]
- Harada S. Kieff E. Epstein-Barr virus nuclear protein LP stimulates EBNA-2 acidic domain-mediated transcriptional activation. J Virol. 1997;71:6611–6618. doi: 10.1128/jvi.71.9.6611-6618.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hofelmayr H. Strobl LJ. Marschall G. Bornkamm GW. Zimber-Strobl U. Activated Notch1 can transiently substitute for EBNA2 in the maintenance of proliferation of LMP1-expressing immortalized B cells. J Virol. 2001;75:2033–2040. doi: 10.1128/JVI.75.5.2033-2040.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hsieh JJD. Henkel T. Salmon P. Robey E. Peterson MG. Hayward SD. Truncated mammalian Notch1 activates CBF1/RBPJk-repressed genes by a mechanism resembling that of Epstein-Barr virus EBNA2. Mol Cell Biol. 1996;16:952–959. doi: 10.1128/mcb.16.3.952. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hwang JW. Kalejta RF. Proteasome-dependent, ubiquitin-independent degradation of Daxx by the viral pp71 protein in human cytomegalovirus-infected cells. Virology. 2007;367:334–338. doi: 10.1016/j.virol.2007.05.037. [DOI] [PubMed] [Google Scholar]
- Ishov AM. Sotnikov AG. Negorev D. Vladimirova OV. Neff N. Kamitani T. Yeh ETH. Strauss JF. Maul GG. PML is critical for ND10 formation and recruits the PML-interacting protein Daxx to this nuclear structure when modified by SUMO-1. J Cell Biol. 1999;147:221–233. doi: 10.1083/jcb.147.2.221. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ishov AM. Vladimirova OV. Maul GG. Daxx-mediated accumulation of human cytomegalovirus tegument protein pp71 at ND10 facilitates initiation of viral infection at these nuclear domains. J Virol. 2002;76:7705–7712. doi: 10.1128/JVI.76.15.7705-7712.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kanda K. Decker T. Aman P. Wahlstrom M. Vongabain A. Kallin B. The EBNA2-related resistance towards alpha interferon (IFN-alpha) in Burkitt-lymphoma cells effects induction of IFN-induced genes but not the activation of transcription factor ISGF-3. Mol Cell Biol. 1992;12:4930–4936. doi: 10.1128/mcb.12.11.4930. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kempkes B. Spitkovsky D. Jansendurr P. Ellwart JW. Kremmer E. Delecluse HJ. Rottenberger C. Bornkamm GW. Hammerschmidt W. B-Cell proliferation and induction of early G(1)-regulating proteins by Epstein-Barr-virus mutants conditional for EBNA2. EMBO J. 1995;14:88–96. doi: 10.1002/j.1460-2075.1995.tb06978.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Korioth F. Maul GG. Plachter B. Stamminger T. Frey J. The nuclear domain 10 (ND10) is disrupted by the human cytomegalovirus gene product IE1. Exp Cell Res. 1996;229:155–158. doi: 10.1006/excr.1996.0353. [DOI] [PubMed] [Google Scholar]
- Kremmer E. Kranz BR. Hille A. Klein K. Eulitz M. Hoffmannfezer G. Feiden W. Herrmann K. Delecluse HJ. Delsol G. Bornkamm GW. Muellerlantzsch N. Grasser FA. Rat monoclonal-antibodies differentiating between the Epstein-Barr-virus nuclear antigens 2A (Ebna2A) and 2B (Ebna2B) Virology. 1995;208:336–342. doi: 10.1006/viro.1995.1157. [DOI] [PubMed] [Google Scholar]
- Laux G. Dugrillon F. Eckert C. Adam B. Zimberstrobl U. Bornkamm GW. Identification and characterization of an Epstein-Barr-virus nuclear antigen 2-responsive cis-element in the bidirectional promoter region of latent membrane-protein and terminal protein-2 genes. J Virol. 1994;68:6947–6958. doi: 10.1128/jvi.68.11.6947-6958.1994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lee HR. Kim DJ. Lee JM. Choi CY. Ahn BY. Hayward GS. Ahn JH. Ability of the human cytomegalovirus IE1 protein to modulate sumoylation of PML correlates with its functional activities in transcriptional regulation and infectivity in cultured fibroblast cells. J Virol. 2004;78:6527–6542. doi: 10.1128/JVI.78.12.6527-6542.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ling PD. Peng RS. Nakajima A. Yu JH. Tan J. Moses SM. Yang WH. Zhao B. Kieff E. Bloch KD. Bloch DB. Mediation of Epstein-Barr virus EBNA-LP transcriptional coactivation by Sp100. EMBO J. 2005;24:3565–3575. doi: 10.1038/sj.emboj.7600820. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lotz M. Tsoukas CD. Fong S. Carson DA. Vaughan JH. Regulation of Epstein-Barr virus-infection by recombinant interferons—selected sensitivity to interferon-gamma. Eur J Immunol. 1985;15:520–525. doi: 10.1002/eji.1830150518. [DOI] [PubMed] [Google Scholar]
- Mannick JB. Cohen JI. Birkenbach M. Marchini A. Kieff E. The Epstein-Barr-virus nuclear-protein encoded by the leader of the EBNA RNAs is important in lymphocyte-B transformation. J Virol. 1991;65:6826–6837. doi: 10.1128/jvi.65.12.6826-6837.1991. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Menezes J. Patel P. Dussault H. Joncas J. Leibold W. Effect of interferon on lymphocyte-transformation and nuclear antigen production by Epstein-Barr virus. Nature. 1976;260:430–432. doi: 10.1038/260430a0. [DOI] [PubMed] [Google Scholar]
- Mocarski ES. Kemble GW. Lyle JM. Greaves RF. A deletion mutant in the human cytomegalovirus gene encoding IE1(491aa) is replication defective due to a failure in autoregulation. Proc Natl Acad Sci USA. 1996;93:11321–11326. doi: 10.1073/pnas.93.21.11321. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Muller S. Dejean A. Viral immediate-early proteins abrogate the modification by SUMO-1 of PML and Sp100 proteins, correlating with nuclear body disruption. J Virol. 1999;73:5137–5143. doi: 10.1128/jvi.73.6.5137-5143.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nanbo A. Inoue K. Adachi-Takasawa K. Takada K. Epstein-Barr virus RNA confers resistance to interferon-alpha-induced apoptosis in Burkitt's lymphoma. EMBO J. 2002;21:954–965. doi: 10.1093/emboj/21.5.954. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Negorev D. Ishov AM. Maul GG. Evidence for separate ND10-binding and homo-oligomerization domains of Sp100. J Cell Sci. 2001;114:59–68. doi: 10.1242/jcs.114.1.59. [DOI] [PubMed] [Google Scholar]
- Negorev D. Maul GG. Cellular proteins localized at and interacting within ND10/PML nuclear bodies/PODs suggest functions of a nuclear depot. Oncogene. 2001;20:7234–7242. doi: 10.1038/sj.onc.1204764. [DOI] [PubMed] [Google Scholar]
- Negorev DG. Vladimirova OV. Ivanov A. Rauscher F. Maul GG. Differential role of Sp100 isoforms in interferon-mediated repression of herpes simplex virus type 1 immediate-early protein expression. J Virol. 2006;80:8019–8029. doi: 10.1128/JVI.02164-05. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nitsche F. Bell A. Rickinson A. Epstein-Barr virus leader protein enhances EBNA-2-mediated transactivation of latent membrane protein 1 expression: a role for the W1W2 repeat domain. J Virol. 1997;71:6619–6628. doi: 10.1128/jvi.71.9.6619-6628.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Peng RS. Moses SC. Tan J. Kremmer E. Ling PD. The Epstein-Barr virus EBNA-LP protein preferentially coactivates EBNA2-mediated stimulation of latent membrane proteins expressed from the viral divergent promoter. J Virol. 2005;79:4492–4505. doi: 10.1128/JVI.79.7.4492-4505.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Peng RS. Tan J. Ling PD. Conserved regions in the Epstein-Barr virus leader protein define distinct domains required for nuclear localization and transcriptional cooperation with EBNA2. J Virol. 2000;74:9953–9963. doi: 10.1128/jvi.74.21.9953-9963.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rooney C. Howe JG. Speck SH. Miller G. Influences of Burkitts-lymphoma and primary B-cells on latent gene-expression by the nonimmortalizing P3J-Hr-1 strain of Epstein-Barr virus. J Virol. 1989;63:1531–1539. doi: 10.1128/jvi.63.4.1531-1539.1989. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rowe M. Cell transformation induced by Epstein-Barr virus—living dangerously. Semin Cancer Biol. 2001;11:403–405. doi: 10.1006/scbi.2000.0406. [DOI] [PubMed] [Google Scholar]
- Saffert RT. Kalejta RF. Human cytomegalovirus gene expression is silenced by Daxx-mediated intrinsic immune defense in model latent infections established in vitro. J Virol. 2007;81:9109–9120. doi: 10.1128/JVI.00827-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sample J. Hummel M. Braun D. Birkenbach M. Kieff E. Nucleotide-sequences of messenger-RNAs encoding Epstein-Barr-virus nuclear proteins—a probable transcriptional initiation site. Proc Natl Acad Sci USA. 1986;83:5096–5100. doi: 10.1073/pnas.83.14.5096. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Seeler JS. Dejean A. The PML nuclear bodies: actors or extras? Curr Opin Genet Dev. 1999;9:362–367. doi: 10.1016/s0959-437x(99)80054-9. [DOI] [PubMed] [Google Scholar]
- Seeler JS. Marchio A. Sitterlin D. Transy C. Dejean A. Interaction of SP100 with HP1 proteins: A link between the promyelocytic leukemia-associated nuclear bodies and the chromatin compartment. Proc Natl Acad Sci USA. 1998;95:7316–7321. doi: 10.1073/pnas.95.13.7316. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Speck SH. Pfitzner A. Strominger JL. An Epstein-Barr-virus transcript from a latently infected, growth-transformed B-cell line encodes a highly repetitive polypeptide. Proc Natl Acad Sci USA. 1986;83:9298–9302. doi: 10.1073/pnas.83.24.9298. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sternsdorf T. Jensen K. Reich B. Will H. The nuclear dot protein Sp100, characterization of domains necessary for dimerization, subcellular localization, and modification by small ubiquitin-like modifiers. J Biol Chem. 1999;274:12555–12566. doi: 10.1074/jbc.274.18.12555. [DOI] [PubMed] [Google Scholar]
- Strander H. Interferon treatment of human neoplasia. Adv Cancer Res. 1986;46:1–265. [PubMed] [Google Scholar]
- Straus SE. Cohen JI. Tosato G. Meier J. Epstein-Barr-virus infections—biology, pathogenesis, and management. Ann Intern Med. 1993;118:45–58. doi: 10.7326/0003-4819-118-1-199301010-00009. [DOI] [PubMed] [Google Scholar]
- Tavalai N. Papior P. Rechter S. Leis M. Stamminger T. Evidence for a role of the cellular ND10 protein PML in mediating intrinsic immunity against human cytomegalovirus infections. J Virol. 2006;80:8006–8018. doi: 10.1128/JVI.00743-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Taylor JL. Unverrich D. O'Brien WJ. Wilcox KW. Interferon coordinately inhibits the disruption of PML-positive ND10 and immediate-early gene expression by herpes simplex virus. J Interferon Cytokine Res. 2000;20:805–815. doi: 10.1089/10799900050151076. [DOI] [PubMed] [Google Scholar]
- Thorley-Lawson DA. Epstein-Barr virus: exploiting the immune system. Nat Rev Immunol. 2001;1:75–82. doi: 10.1038/35095584. [DOI] [PubMed] [Google Scholar]
- Tsang SF. Wang F. Izumi KM. Kieff E. Delineation of the cis-acting element mediating EBNA-2 transactivation of latent infection membrane-protein expression. J Virol. 1991;65:6765–6771. doi: 10.1128/jvi.65.12.6765-6771.1991. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wilkinson GWG. Kelly C. Sinclair JH. Rickards C. Disruption of PML-associated nuclear bodies mediated by the human cytomegalovirus major immediate early gene product. J Gen Virol. 1998;79:1233–1245. doi: 10.1099/0022-1317-79-5-1233. [DOI] [PubMed] [Google Scholar]
- Young LS. Rickinson AB. Epstein-Barr virus: 40 years on. Nat Rev Cancer. 2004;4:757–768. doi: 10.1038/nrc1452. [DOI] [PubMed] [Google Scholar]
- Zhang LW. Pagano JS. Interferon regulatory factor 7 is induced by Epstein-Barr virus latent membrane protein 1. J Virol. 2000;74:1061–1068. doi: 10.1128/jvi.74.3.1061-1068.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]