Abstract
The transforming growth factor-β1 (TGF-β1) is a cytokine involved in many biological events inlcuding immunosuppression, angiogenesis, cell growth, and apoptosis. Expression of TGF-β1 at the transcriptional level is controlled by a series of ubiquitous and specialized factors whose activities can be modulated by a variety of signaling events. Here we demonstrate that activity of the TGF-β1 promoter is increased by C/EBPβ, a DNA-binding transcription factor whose activity can be influenced by several immunomodulators, in astrocytes and microglial cells. Interestingly, expression of Smad3 and Smad4, the downstream regulators of the TGF-β1-signaling pathway, impairs the activity of C/EBPβ on the TGF-β1 promoter. Further, we demonstrate that MH2, a common domain among Smads that has protein-binding activities, interacts with C/EBPβ and decreases its association with a region of the TGF-β1 promoter that is responsive to C/EBPβ activation. Interestingly, the p65 subunit of nuclear factor-κB (NF-κB), which also interacts with C/EBPβ, cooperates with MH2 and decreased DNA-binding and transcriptional activities of C/EBPβ on the TGF-β1 promoter. These observations indicate that an autoregulatory mechanism, involving the MH2 domain of Smads, modulates activation of the TGF-β1 promoter by C/EBPβ. Further, our results show that the interplay between NF-κB and C/EBPβ has an impact on the ability of C/EBPβ to stimulate TGF-β1 transcription, hence, suggesting that the cross-communication of signaling pathways that modulate NF-κB and C/EBPβ may dictate the level of TGF-β1 promoter activity.
Many biological events that are regulated by cell signals can be influenced by a variety of cytokines and immunomodulators through autocrine and paracrine pathways. Among these cytokines, transforming growth factor-β1 (TGF-β1), which is produced and secreted by a majority of cells, regulates many different physiological processes such as development, wound healing, chemo-taxis, proliferation, and homeostasis (Shi and Massague 2003). Evidently, the interaction of TGF-β1 with its receptors triggers a cascade of cytoplasmic reactions that results in the phosphorylation of a family of proteins named Smads and promotes their translocation into the nucleus where they bind to DNA and stimulate transcription of specific genes (Wotton and Massague 2001). For example, in human breast cancer cells, Smad3 binds to the human telomerase reverse transcriptase (hTERT) gene promoter directly and inhibits hTERT gene transcription activity. Further, by interacting with c-myc, Smad3 also represses the c-myc gene (Li and Liu 2007). Expression of the TGF-β1 gene at the transcriptional level is regulated by a variety of cellular proteins, some of which can be influenced by other signaling pathways as well as noncellular proteins including viral regulators (Massague and Wotton 2000). For example, transcription of TGF-β1 has been shown to be regulated by NF-AT whose binding site resides between +268 and 288 in the proximal promoter region (Han and others 1998). The promoter of TGF-β1 also contains a series of binding sites for Sp1, NF1, AD1, and Zf-9 that spans between −453 to +1 (Kim and others 1998). Among the viral proteins, human immunodeficiency virus-1 (HIV-1) Tat has been shown to stimulate the TGF-β1 promoter, partly through interaction with Pur-α, a GC-GA-binding protein that has been shown to interact with nucleotides spanning −453 to −323 of the TGF-β1 promoter (Thatikunta and others 1997). HIV-1 Tat is a potent transcriptional activator that, by positioning itself in close proximity to the transcription initiation site, recruits cyclin T1:Cdk9 where they phosphorylate the C-terminus of RNA polymerase II and potentiate its activity (Quivy and others 2007). In earlier studies we demonstrated that Tat can interact with C/EBPβ, a DNA-binding protein that recognizes CAAT as well as several other cellular proteins involved in the regulation of cell cycle progression, cell proliferation, and other cellular functions (Coyle-Rink and others 2002; Abraham and others 2005; Mameli and others 2007).
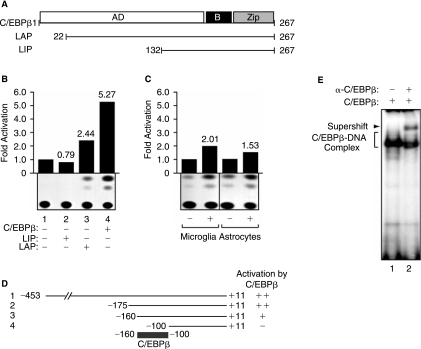
C/EBPβ belongs to a family of highly conserved transcription factors composed of multiple functional domains including a basic-leucine zipper at the C-terminus in juxtaposition of a basic domain that is responsible for DNA binding (Ramji and Foka 2002). The N-terminus of the protein encompasses the activation domain. The most studied members of this family include the 38-kDa C/EBPβ, and its two smaller variants LAP (liver-enriched activator protein) isoform (40 and 35 kDa) and two truncated 14 and 21 kDa LIP (liver-enriched inhibitory protein) (Descombes and Schibler 1991) as schematized in Figure 1A. Because LIP is translated from the third in-frame AUG start codon, it lacks most of transactivation domain and has a higher binding affinity for DNA compared with LAP isoforms. LIP functions as a dominant-negative mutant of LAP by formation of LIP-LAP heterodimer or competing for the binding to the promoter regions of target genes as a LIP homodimer. An increase in the ratio of LIP/LAP negatively regulates C/EBPβ-LAP-mediated gene expression. It has been shown that LIP overexpression, resulting in a LIP/LAP ratio of ∼1, exerted an inhibitory effect on adipogenesis (Hamm and others 2001).
FIG. 1.
Regulation of TGF-β1 by C/EBPβ. (A) A schematic diagram of C/EBPβ and its deletion mutants LIP and LAP. The human astroglioma cell lines, U-87MG (B and D) or primary human microglia and astrocytes (C), were transfected with 5 μg of TGF-β1-CAT reporter plasmid [full length (B and C) or deletion mutants (D)] alone or cotransfected with 5.0 μg of C/EBPβ, LIP, or LAP expression plasmids, as described previously (Amini and others 2002). The amount of DNA used for transfection was normalized with pcDNA3 plasmid. Cell extracts were prepared 48 h after transfection, and CAT assays were performed, as we have previously described (Sawaya and others 1998). The values shown on the top of each bar represent the fold activation over the basal promoter activity arbitrarily set at one (B and C). The values shown on the right represent significant activation (++), modest activation (+), or no effect (−) of C/EBPβ over the basal promoters (D). The data represent the mean value of at least three separate transfection experiments (SE ± 15%). (E) Gel electrophoretic mobility shift assay (EMSA) was performed, as described previously (Amini and others 2002). Ten micrograms of nuclear extract from C/EBPβ-transfected U-87MG cells (lanes 1 and 2) were used in DNA-binding experiments. For supershift assays, antibodies directed against C/EBPβ (lane 2) (Santa Cruz Biotechnology) were mixed with nuclear proteins for 1 h at 4°C prior to the addition of the probe.
Here, we demonstrated that activation of the TGF-β1 promoter by C/EBPβ was abolished by MH2 domain of Smads or p65, which could be beneficial to keep the levels of TGF-β1 under control.
The ability of Tat to physically and functionally interact with C/EBPβ and the capacity of C/EBPβ to communicate with Smads prompted us to investigate the possible regulation of the TGF-β1 promoter by C/EBPβ and assess the impact of Smads and Tat on this event. In the first series of studies we evaluated the impact of C/EBPβ, LIP, and LAP on transcription of the TGF-β1 promoter encompassing nucleotides −453 to +11, in the human astroglioma cell line, U-87MG. As shown in Figure 1B, both C/EBPβ and LAP enhanced the activity of the TGF-β1 promoter in the transfected cells (compare lanes 3 and 4 to lane 1). However, expression of LIP showed no effect on TGF-β1 promoter activity (lane 2). Expression of C/EBPβ also elevated transcription of the TGF-β1 promoter in human primary culture of astrocytes and microglial cells (Fig. 1C). Interestingly, we observed that activation of the TGF-β1 promoter by C/EBPβ is higher in cell lines than in primary cultures. Despite the fact that the numbers presented on the top of each bar represent the average of three separate experiments, however, this difference might be due partially to the efficiency of transfection and to the fact that primary cultures are in the stage of development while cell lines derive from well-developed adult brain. In addition, it is well known that the level of TGF-β1 messenger RNA and protein varies between cells in stage of development and adult brain cells, which could also contribute to the observed difference.
To identify the region within the TGF-β1 promoter that is responsive to C/EBPβ activation, a series of promoter deletion mutants of TGF-β1 containing the various regions of the upstream regulatory sequence of TGF-β1 in fusion with the reporter CAT gene was introduced, either alone or together with a plasmid expressing C/EBPβ, into U-87MG cells (Fig. 1D). Results from CAT assay demonstrated that all the deletion mutants except the mutant containing the DNA fragment spanning nucleotides −100 to +11 were responsive to activation by C/EBPβ (compare lanes 1–3 to lane 4). These results indicate that the region required for activation of the TGF-β1 promoter by C/EBPβ resides between nucleotides −160 to −100 (gray box). Close examination of the sequence spanning this region identified several DNA elements that may serve as binding sites for C/EBPβ.
To examine the ability of these sequences to bind to C/EBPβ, we performed band-shift assay using nuclear extracts from U-87MG and [γ32P]-labeled oligonucleotide derived from the TGF-β1 promoter. To further identify the complex that contains C/EBPβ, we included anti-C/EBPβ antibody in the binding reaction. As seen in Figure 1E, a distinct band, which is typically observed in C/EBPβ band-shift assay (Abraham and others 2005), was detected when the reaction mixture was analyzed by gel electrophoresis, pointing to the binding of C/EBPβ to the TGF-β1 promoter sequence (lane 1). The specificity of C/EBPβ:DNA association was further confirmed using anti-C/EBPβ antibody (lane 2).
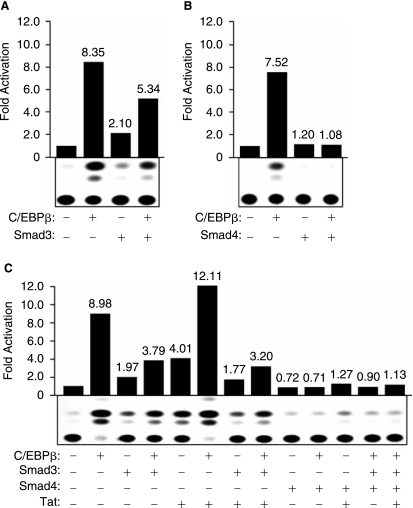
In earlier studies, we demonstrated that the downstream activators of TGF-β1-signaling pathway including members of the Smad family, by interacting with C/EBPβ can modulate its activity C/EBPβ on transcription of MCP-1 (Abraham and others 2005). Thus, in the next set of experiments, we performed a series of transcription assays to assess the impact of Smad3 and Smad4 on transcriptional activation of the TGF-β1 promoter. U87MG cells were transfected with the TGF-β1 promoter alone or in the presence of C/EBPβ, Smad3, or Smad4 expression plasmids. As shown in Figure 2, expression of C/EBPβ (panels A and B, lanes 2), Smad3 (panel 3, lane 3), or Smad 4 (panel B, lane 3) alone activates the TGF-β1 promoter. However, expression of Smad3 and Smad4 in combination with C/EBPβ decreased the level of TGF-β1 activation by C/EBPβ (panels A and B, lanes 4).
FIG. 2.
Functional interplay between C/EBPβ and Smad3/4 in the presence of HIV-1 Tat. U-87MG cells were transfected with 5.0 μg of TGF-β1-CAT reporter plasmid (full length) alone or cotransfected with 5.0 μg of C/EBPβ (A–B), Smad3 (A and C), Smad4 (B and C), or Tat (C) expression plasmids using various combinations. The amount of DNA used for transfection was normalized with pcDNA3 plasmid. Cell extracts were prepared 48 h after transfection, and CAT assays were performed. The values shown on the top of each bar represent the fold activation over the basal promoter activity arbitrarily set at one. The data represent the mean value of at least three separate transfection experiments (SE ± 15%).
The ability of HIV-1 Tat protein to activate the TGF-β1 promoter and to physically and functionally cooperate with Samd3 and C/EBPβ in activating the HIV-1 and MCP-1 gene expression (Thatikunta and others 1997; Coyle-Rink and others 2002; Abraham and others 2005) gave us rationale to evaluate the effect of Smad3 and Smad4 on the ability of Tat, either alone or in cooperation with C/EBPβ, to stimulate TGF-β1 transcription. Results from transfection studies revealed that both Smads, particularly Smad4, suppressed Tat and Tat plus C/EBPβ activation of the TGF-β1 promoter in U-87MG cells (Fig. 2C).
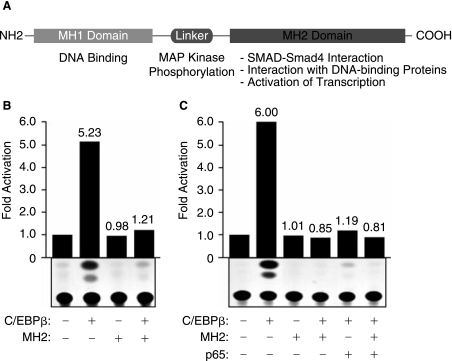
A common feature of the Smad family of transcription factors is the presence of two major domains MH1 and MH2, which are separated by a linker domain (Wotton and Massague 2001). The MH1 domain is known for its DNA-binding activity, whereas the MH2 domain functions as the activator domain that interacts with other DNA binding proteins (Fig. 3A). To gain more information regarding the functional interaction of Smad and C/EBPβ on the TGF-β1 promoter, we used a plasmid expressing the MH2 domain of Smad3 in the transfection assay and demonstrated that expression of MH2 inhibits the ability of C/EBPβ to activate the TGF-β1 promoter (Fig. 3B). Under similar conditions, MH1 had less impact on TGF-β1 activation by C/EBPβ (data not shown).
FIG. 3.
Functional interaction of p65 and MH2 with C/EBPβ. (A) A schematic representation of Smad3 protein and its domains. (B and C) U-87MG cells were transfected with 5 μg of TGF-β1-CAT reporter plasmid alone or cotransfected with 5.0 μg of C/EBPβ, MH2, or p65 expression plasmids using various combinations. The amount of DNA used for transfection was normalized with pcDNA3 plasmid. Cell extracts were prepared 48 h after transfection, and CAT assays were performed. The values shown on the top of each bar represent the fold activation over the basal promoter activity arbitrarily set at one. The data represent the mean value of at least three separate transfection experiments (SE ± 15%).
In addition to MH2, we also sought to suppress C/EBPβ-activation of the TGF-β1 promoter using a cellular protein. In this regard, we previously demonstrated that the physical and functional interaction of C/EBPβ with the NF-κB sub-unit p65 could modulate transcription of several C/EBPβ responsive genes including the BKV promoter (Gorrill and Khalili 2005). Thus, in light of our results on the capacity of C/EBPβ to enhance the TGF-β1 promoter, we evaluated the ability of p65 to affect C/EBPβ-mediated activation of the TGF-β1 promoter in the absence and presence of MH2. U-87MG cells were transfected with the TGF-β1 promoter in the presence of C/EBPβ, p65, or MH2 expression plasmids using various combinations. As shown in Figure 3C, expression of p65 interfered with C/EBPβ activation of the TGF-β1 promoter and had no impact on the inhibitory effect of MH2. This observation suggests that activation of the TGF-β1 promoter by C/EBPβ can be modulated by both Smads and the p65 subunit of NF-κB, both of which are responsive to TGF-β1 and TNF-α-signaling pathways, respectively. Similar to MH2, p65 failed to activate the TGF-β1 promoter (data not shown). Of note, the observed effect may not be attributed to the impact of these regulators on expression of each other's promoter in the transfected cells as all expressor plasmids are driven by a cytomegalovirus promoter and the levels of their expression are not affected by each other.
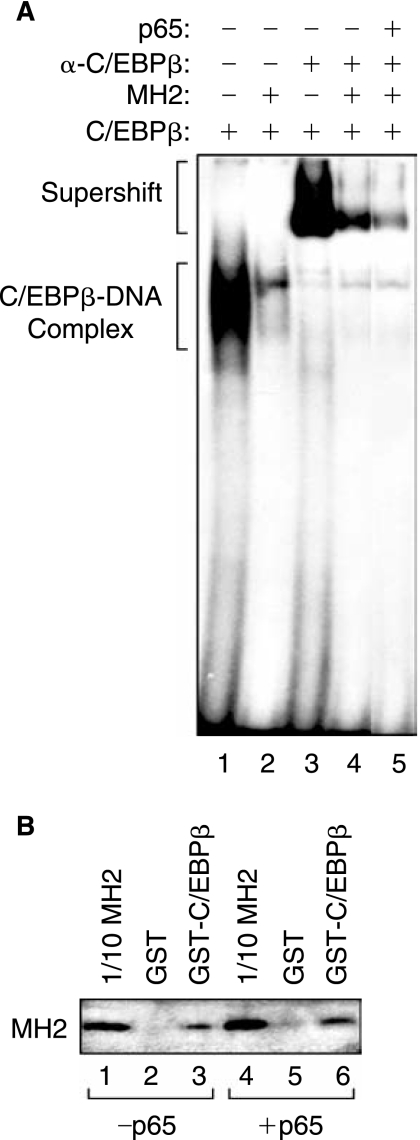
In the next set of experiments, we tested the DNA-binding activity of C/EBPβ in the presence of MH2 and MH2 plus p65 in U-87MG cell extract. As shown in Figure 4A, results from DNA-binding activity showed that expression of MH2 decreases binding of C/EBPβ to the TGF-βl DNA sequence (compare lanes 1 and 2). Association of C/EBPβ with the DNA formed a complex, which was shifted in the presence of anti-C/EBPβ antibody (compare lanes 1 and 3), and was further decreased in nuclear extracts transfected with MH2 (compare lanes 3 and 4). Of interest was the observation that p65 further decreased C/EBPβ interaction with DNA in cells expressing MH2 (lane 5). These observations corroborate the results from transcription assay (shown in Fig. 3B) where p65 expression further decreased TGF-β1 activation by C/EBPβ in cells expressing MH2. Interestingly, association of C/EBPβ with the DNA did not disappear in extracts where p65 and MH2 were coexpressed (lane 5). However, activation of the TGF-β1 promoter by C/EBPβ was abolished in the presence of p65 and/or MH2 (compare Fig. 3C to lanes 2 and 5 of Fig. 4A). These results indicate that despite its association with the DNA, a minimum number of C/EBPβ is required for activation of the TGF-β1 promoter.
FIG. 4.
Physical association between C/EBPβ and MH2 or p65. (A) Gel electrophoretic mobility shift assay (EMSA) was performed using 10 μg of nuclear extracts from U-87MG cells transfected with C/EBPβ (lanes 1–5) alone, with C/EBPβ + MH2 (lanes 2, 4, and 5), or with C/EBPβ + p65 (lane 5). For supershift assays, antibodies directed against C/EBPβ (lanes 3–5) were mixed with nuclear proteins for 1 h at 4°C prior to the addition of the probe. The positions of the C/EBPβ-DNA complexes are indicated with brackets. (B) GST pull-down. Whole cell extract from cells transfected with 2.5 μg CMV-C/EBPβ was incubated with either glutathione-S-transferase (GST) bound to glutathione beads (lanes 2 and 5) or full length GST-C/EBPβ fusion protein bound to glutathione beads (lanes 3 and 6). Lanes 1 and 4 represent direct analysis of the input protein extract by Western blot, used as a positive control for migration of C/EBPβ on the gel.
As the interaction of MH2 with C/EBPβ can interfere with C/EBPβ association with DNA, we then tested C/EBPβ interaction with MH2 in the absence and presence of p65 using GST pull-down assay. As shown in Figure 4B, MH2 interacts with C/EBPβ (lane 3). This interaction was slightly increased in the presence of p65 in the extract (compare lane 6 to lane 3). No interaction was observed in the presence of GST alone (lanes 2 and 4).
Altogether, our results provide evidence for the activation of the TGF-β1 promoter in astrocytes by C/EBPβ and modulation of this activity by Smad and NF-κB proteins. C/EBPβ activates TGF-β1 gene expression, but this activation was abrogated when the p65 subunit of NF-κB or the MH2 domain of Smad3 were coexpressed. Further, these observations pointed to the functional interplay between viral and cellular proteins in modulating viral and cellular transcription.
The ability of C/EBPβ to induce the TGF-β1 promoter is not without a precedent. In this regard, it was shown that activation of the TGF-β1 promoter by C/EBPβ in stellate cells could be inhibited by oltipraz, which provides a molecular target for pharmacological treatment of liver cirrhosis (Kang and others 2002). Further, C/EBPβ was found to be essential for TGF-β1 induction of the cell cycle inhibitor p15INK4b by a FoxO-Smad complex in human epithelial cells. These results led the authors to suggest that C/EBPβ plays a key role in the coordination of TGF-β1 cytostatic gene responses in breast cancer (Gomis and others 2006). On the other hand, the negative interplay between C/EBPβ and Smad3 has also been previously described. It has been shown that Smad3 prevents the cooperative induction of the iNOS promoter by C/EBPβ and NF-κB, pointing to an essential role of Smad3 in mediating TGF-β1 anti-inflammatory responses in vascular smooth muscle cells (Feinberg and others 2004). Smad3 was also shown to abolish transcriptional activation of the haptoglobin promoter by C/EBPβ and STAT3 (Zaubermann and others 2001). In a separate study, we demonstrated the ability of C/EBPβ to associate with HIV-1 Tat protein leading to activation of the HIV-1 gene (Coyle-Rink and others 2002). In another study, we demonstrated that the elevated levels of TGF-β1 in HIV-1 infected cells might be due to the ability of HIV-1 Tat protein to activate the TGF-β1 promoter (Thatikunta and others 1997). Further, we previously demonstrated that MH2 has the ability to suppress Tat-induction of cytokines and chemokines, as well as HIV-1 replication (Eldeen and others 2006). Altogether, these observations identify C/EBPβ as a new partner for Tat in stimulating TGF-β1 transcription in astrocytes and suggest that the delicate balance among the downstream regulatory proteins of several cytokines and immunomodulators can dictate the level of expression of cytokine and chemokines, including MCP-1 and TGF-β1. Hence, inappropriate expression and function of regulatory proteins such as C/EBPβ and Smads by Tat may induce TGF-β1 production in astrocytes and contribute to the neuropathogenesis of acquired immunodeficiency syndrome (AIDS) through stimulation of inflammation in the central nervous system. Therefore, one might hypothesize that the use of MH2 domain of Smads might provide a molecular target against C/EBPβ induction of genes and for pharmacological treatment of several diseases including AIDS.
Acknowledgments
We thank the past and present members of the Department of Neuroscience and Center for Neurovirology for their support, and sharing of reagents and ideas. We especially thank Dr. Martyn White for his contributions during the preparation and critical reading of this manuscript. This work is supported by grants awarded by National Institutes of Health to B.E.S. and S.A.
References
- Abraham S. Sweet T. Sawaya BE. Rappaport J. Khalili K. Amini S. Cooperative interaction of C/EBP beta and Tat modulates MCP-1 gene transcription in astrocytes. J Neuroimmunol. 2005;160:219–227. doi: 10.1016/j.jneuroim.2004.11.009. [DOI] [PubMed] [Google Scholar]
- Amini S. Clavo A. Nadraga Y. Giordano A. Khalili K. Sawaya BE. Interplay between cdk9 and NF-kappaB factors determines the level of HIV-1 gene transcription in astrocytic cells. Oncogene. 2002;21:5797–5803. doi: 10.1038/sj.onc.1205754. [DOI] [PubMed] [Google Scholar]
- Coyle-Rink J. Sweet T. Abraham S. Sawaya B. Batuman O. Khalili K. Amini S. Interaction between TGFbeta signaling proteins and C/EBP controls basal and Tat-mediated transcription of HIV-1 LTR in astrocytes. Virology. 2002;299:240–247. doi: 10.1006/viro.2002.1439. [DOI] [PubMed] [Google Scholar]
- Descombes P. Schibler U. A liver-enriched transcriptional activator protein, LAP, and a transcriptional inhibitory protein, LIP, are translated from the same mRNA. Cell. 1991;67:569–579. doi: 10.1016/0092-8674(91)90531-3. [DOI] [PubMed] [Google Scholar]
- Eldeen MB. Deshmane SL. Simbiri K. Khalili K. Amini S. Sawaya BE. MH2 domain of Smad3 reduces HIV-1 Tat-induction of cytokine secretion. J Neuroimmunol. 2006;176:174–180. doi: 10.1016/j.jneuroim.2006.04.004. [DOI] [PubMed] [Google Scholar]
- Feinberg MW. Watanabe M. Lebedeva MA. Depina AS. Hanai J. Mammoto T. Frederick JP. Wang XF. Sukhatme VP. Jain MK. Transforming growth factor-beta1 inhibition of vascular smooth muscle cell activation is mediated via Smad3. J Biol Chem. 2004;279:16388–16393. doi: 10.1074/jbc.M309664200. [DOI] [PubMed] [Google Scholar]
- Gomis RR. Alarcon C. Nadal C. Van Poznak C. Massague J. C/EBPbeta at the core of the TGFbeta cytostatic response and its evasion in metastatic breast cancer cells. Cancer Cell. 2006;10:203–214. doi: 10.1016/j.ccr.2006.07.019. [DOI] [PubMed] [Google Scholar]
- Gorrill TS. Khalili K. Cooperative interaction of p65 and C/EBPbeta modulates transcription of BKV early promoter. Virology. 2005;335:1–9. doi: 10.1016/j.virol.2005.02.006. [DOI] [PubMed] [Google Scholar]
- Hamm JK. Park BH. Farmer SR. A role for C/EBPβ in regulating peroxisome proliferator-activated receptor gamma activity during adipogenesis in 3T3-L1 preadipocytes. J Biol Chem. 2001;276:18464–18471. doi: 10.1074/jbc.M100797200. [DOI] [PubMed] [Google Scholar]
- Han SH. Yea SS. Jeon YJ. Yang KH. Kaminski NE. Transforming growth factor-beta 1 (TGF-beta1) promotes IL-2 mRNA expression through the up-regulation of NF-kappaB, AP-1 and NF-AT in EL4 cells. J Pharmacol Exp Ther. 1998;287:1105–1112. [PubMed] [Google Scholar]
- Kang KW. Kim YG. Cho MK. Bae SK. Kim CW. Lee MG. Kim SG. Oltipraz regenerates cirrhotic liver through CCAAT/enhancer binding protein-mediated stellate cell inactivation. FASEB J. 2002;16:1988–1990. doi: 10.1096/fj.02-0406fje. [DOI] [PubMed] [Google Scholar]
- Kim Y. Ratziu V. Choi SG. Lalazar A. Theiss G. Dang Q. Kim SJ. Friedman SL. Transcriptional activation of transforming growth factor beta1 and its receptors by the Kruppel-like factor Zf9/core promoter-binding protein and Sp1. Potential mechanisms for autocrine fibrogenesis in response to injury. J Biol Chem. 1998;273:33750–33758. doi: 10.1074/jbc.273.50.33750. [DOI] [PubMed] [Google Scholar]
- Li H. Liu JP. Mechanisms of action of TGF-beta in cancer: evidence for Smad3 as a repressor of the hTERT gene. Ann NY Acad Sci. 2007;1114:56–68. doi: 10.1196/annals.1396.016. [DOI] [PubMed] [Google Scholar]
- Mameli G. Deshmane SL. Ghafouri M. Cui J. Simbiri K. Khalili K. Mukerjee R. Dolei A. Amini S. Sawaya BE. C/EBPbeta regulates human immunodeficiency virus 1 gene expression through its association with cdk9. J Gen Virol. 2007;88:631–640. doi: 10.1099/vir.0.82487-0. [DOI] [PubMed] [Google Scholar]
- Massague J. Wotton D. Transcriptional control by the TGF-beta/Smad signaling system. EMBO J. 2000;19:1745–1754. doi: 10.1093/emboj/19.8.1745. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Quivy V. De Walque S. Van Lint C. Chromatin-associated regulation of HIV-1 transcription: implications for the development of therapeutic strategies. Subcell Biochem. 2007;41:371–396. [PubMed] [Google Scholar]
- Ramji DP. Foka P. CCAAT/enhancer-binding proteins: structure, function and regulation. Biochem J. 2002;365:561–575. doi: 10.1042/BJ20020508. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sawaya BE. Khalili K. Mercer WE. Denisova L. Amini S. Cooperative actions of HIV-1 Vpr and p53 modulate viral gene transcription. J Biol Chem. 1998;273:20052–20057. doi: 10.1074/jbc.273.32.20052. [DOI] [PubMed] [Google Scholar]
- Shi Y. Massague J. Mechanisms of TGF-beta signaling from cell membrane to the nucleus. Cell. 2003;113:685–700. doi: 10.1016/s0092-8674(03)00432-x. [DOI] [PubMed] [Google Scholar]
- Thatikunta P. Sawaya BE. Denisova L. Cole C. Yusibova G. Johnson EM. Khalili K. Amini S. Identification of a cellular protein that binds to Tat-responsive element of TGF beta-1 promoter in glial cells. J Cell Biochem. 1997;67:466–477. [PubMed] [Google Scholar]
- Wotton D. Massague J. Smad transcriptional corepressors in TGF beta family signaling. Curr Top Microbiol Immunol. 2001;254:145–164. [PubMed] [Google Scholar]
- Zauberman A. Lapter S. Zipori D. Smad proteins suppress CCAAT/enhancer-binding protein (C/EBP) beta- and STAT3-mediated transcriptional activation of the haptoglobin promoter. J Biol Chem. 2001;276:24719–24725. doi: 10.1074/jbc.M005813200. [DOI] [PubMed] [Google Scholar]