Abstract
Interferons (IFNs) are critical to the host innate immune response by inducing the expression of a family of early response genes, denoted as IFN-stimulated genes (ISGs). The role of tyrosine phosphorylation of STAT proteins in the transcription activation of ISGs is well-documented. Recent studies have indicated that other transcription factors (TFs) are likely to play a role in regulating ISG expression. Here, we describe a novel integrative approach that combines gene expression profiling, promoter sequence analysis, and literature mining to screen candidate regulatory factors in the IFN signal transduction pathway. Application of this method identified the nuclear factor κB (NFκB) protein, cRel, as a candidate regulatory factor for a subset of ISGs in mouse embryo fibroblasts. Chromatin immunoprecipitation (ChIP) and real-time PCR assays confirmed that cRel directly binds to the promoters of several ISGs, including Cxcl10, Isg15, Gbp2, Ifit3, and Ifi203, and regulates their expression. Thus, our studies identify cRel as an important TF for ISGs, and validate the approach of using Latent Semantic Indexing (LSI)-based methods to identify regulatory factors from microarray data.
Introduction
Interferons (IFNs) were discovered by virtue of their antiviral activity; however, IFNs are multifunctional proteins that also affect cell proliferation, cell differentiation, apoptosis (programmed cell death), and the immune response. Type I IFNs, consisting of IFN-α, IFN-β, IFN-ω, IFN-ɛ, IFN-κ, IFN-δ, and IFN-τ, regulate their diverse cellular functions by modulating the expression of IFN-stimulated genes (ISGs) through the activation of a signal transduction pathway involving the JAK tyrosine kinases and STAT proteins (Friedman and Stark 1985; Larner and others 1986; Schindler and others 1992; Darnell and others 1994). Although the JAK/STAT signal transduction pathway is critical in mediating IFNs' antiviral and antiproliferative activities, IFNs also activate the nuclear factor κB (NFκB) signaling pathway, which also plays an important role in the biological actions of IFN (Yang and others 2000; Yang and others 2001; Pfeffer and others 2004).
The NFκB transcription factor (TF) family regulates the expression of genes involved in cell survival and immune responses (Beg and others 1995; Beg and Baltimore 1996; Van Antwerp and others 1996; Wang and others 1996). In mammals, the NFκB family of related proteins includes NFκB1 (p105 processed to p50), NFκB2 (p100 processed to p52), RelA (p65), RelB, and cRel. Both p50 and p52 lack a transcription activation domain, and as homodimers function as repressors. In contrast, p65, cRel, and RelB have a transcription activation domain, and thus when complexed with p50 or p52 are capable of activating transcription. Although p50:p65 and p52:RelB heterodimers are the NFκB complexes most often observed in cells, other Rel heterodimers also form. Recent studies identified that IFN induces NFκB activation through both a classical pathway that results in the formation of p50:NFκB dimers through IκB degradation, and an alternative pathway that results in the formation of p52:NFκB dimers through a NFκB-inducing kinase (NIK)/tumor necrosis factor (TNF) receptor-associated factors (TRAF)-dependent pathway (Yang and others 2001; Pfeffer and others 2004; Yang and others 2005a; Wei and others 2006). Moreover, a subset of ISGs are regulated by NFκB, and appear to play important roles in IFNs' biological actions.
The coordinated regulation of gene expression by extracellular signals requires the interplay between multiple TFs that selectively bind to gene promoters with spatial and temporal precision (Bluthgen and others 2005). Comprehensive analysis of motifs in gene promoters by phylogenetic footprinting or Bayesian clustering has been useful in constructing regulatory networks in mammalian cells (Qin and others 2003; Xie and others 2005). Combining gene expression profiling with promoter analysis may significantly improve identification of novel gene regulatory networks based on the assumption that co-regulated genes share common TF-binding sites and regulatory factors. However, due to the complexity of mammalian promoters, genes often contain potential binding sites for several TFs, making identification of regulatory programs very difficult. More sophisticated probabilistic graphical models have been explored to identify critical regulators, which are themselves transcriptionally regulated by differing stimuli (Segal and others 2003; Li and others 2005). Also, combination of promoter analysis and gene function information in human curated Gene Ontology database has had some success in identifying meaningful gene regulatory mechanisms (Bluthgen and others 2005). However, as with any human curated index, Gene Ontology may have limited usefulness because it is incomplete and contains broad functional index terms. Therefore, integration of functional information extracted from the primary literature would substantially improve these approaches.
Classical information retrieval and information extraction methods have recently been employed to mine the biomedical literature to elucidate gene/protein function and regulatory networks (Krallinger and others 2005; Rebholz-Schuhmann and others 2005). Information retrieval involves term matching and may include Boolean method or lexical matching methods common to natural language processing (Jenssen and others 2001; Yandell and Majoros 2002). However, these methods are limited in that they rely solely on known relationships. On the other hand, information extraction involves mathematical modeling of text, for example, by vector-space or statistical (Bayesian) approaches (Shatkay and Feldman 2003), which can deduce relationships from the literature even in the absence of a direct link. We previously developed a method using Latent Semantic Indexing (LSI) to identify gene relationships with high precision from titles and abstracts in MEDLINE citations (Homayouni and others 2005). In addition, we showed that this method identified both known (explicit) as well as unknown (implicit) gene relationships from the literature.
In the present report, we have applied a novel algorithm that combines expression profiling, promoter analysis and LSI to infer relationships between co-regulated genes and the TFs that are shared in gene promoters (outlined in Fig. 1). To test this method, we interrogated a dataset of ISGs from mouse embryonic fibroblasts (MEFs) (Pfeffer and others 2004). Our algorithm identified the cRel as a potential TF for a subset of ISGs. Chromatin immunoprecipitation (ChIP) assays demonstrated that cRel directly binds to the promoters of many of the ISGs identified, including Cxcl10, G1p2/Isg15, Gbp2, Ifit3, and Ifi203. Moreover, real-time PCR analysis demonstrated that induction of these ISGs by IFN was diminished in cRel-deficient MEFs as compared to MEFs from their wild-type (WT) littermates.
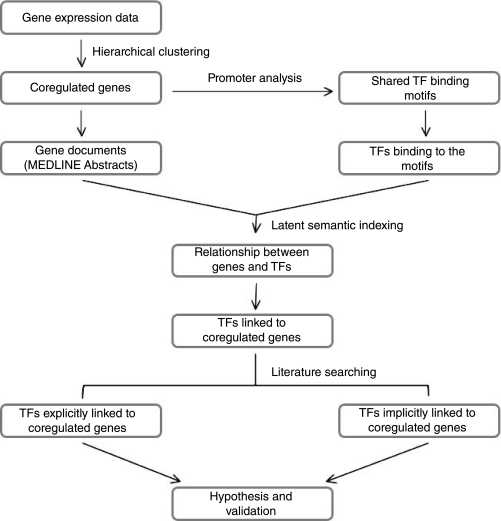
FIG. 1.
Overview of the method used to identify candidate regulatory transcription factors (TFs) from microarray data. Small groups of genes that are similarly affected by experimental treatments are selected for promoter sequence analysis to identify shared TF-binding motifs. A matrix consisting of pairwise literature relationships between each TF and co-regulated gene is constructed by Latent Semantic Indexing (LSI). The relationship dvalues for genes and TFs are then subjected to hierarchical clustering and the TFs that have high literature relationship to the co-regulated genes are selected for manual analysis of the literature to determine an explicit or implicit relationship.
Materials and Methods
Biological reagents and cell culture
Highly purified recombinant rat IFN-β was obtained from Biogen-Idec, Inc. (2 × 108 IU/mg protein) (Arduini and others 2004). Polyclonal anti-cRel was obtained from Santa Cruz Biotechnology (Santa Cruz, CA, USA). 3T3 immortalized MEFs generated from E12.5 to 14.5 embryos from C57BL6/J mice and cRel-deficient littermates were generously provided by Dr. Alexander Hoffmann (University of Caliornia, San Diego) (Hoffmann and others 2003). MEFs were plated at 3 × 105 cells/60-mm dish every 3 days in Dulbecco's modified Eagle's medium supplemented with 10% fetal calf serum (FCS) (Hyclone Laboratories, Logan, UT, USA), and 100 μg/mL of penicillin G and streptomycin.
Microarray analysis
Microarray experiments and data analysis were performed as described previously (Pfeffer and others 2004). In brief, total cellular RNA from untreated and IFN-β-treated (2500 units/mL for 5 h) MEFs was extracted with TRIzol reagent (Invitrogen) and submitted to Genome Explorations Inc. (Memphis, TN, USA) for labeling and hybridization to the murine U74Av2 GeneChip (Affymetrix Inc.). The data were deposited in Gene Expression Omnibus (GEO) at NCBI (Accession GSM76653-76658). Expression values were determined using Affymetrix Microarray Suite Version 5.0 software and analyzed using GeneSpring 7.2 (Silicon Genetics, Inc.). A subset of genes were selected whose expression was altered greater than 2-fold (p < 0.05, Welch's t-test, n = 3 for each group) and then subjected to hierarchical clustering using standard correlation coefficients.
Promoter sequence analysis
Genomic sequences corresponding to 3-kb upstream and 50-bp downstream of transcription start site (TSS) of each gene was obtained from the UCSC Genome Browser (http://genome.ucsc.edu/). DNA-binding motifs and TF-binding sites were identified using MOTIF (http://motif.genome.jp) and the information in TRANSFAC 6.0 database (Matys and others 2003).
LSI-based literature analysis
The text representation was performed as described previously (Homayouni and others 2005). In brief, the information for each gene was generated by concatenation of titles and abstracts in the MEDLINE citations cross-referenced in the mouse, rat and human Entrez Gene entries. To lower the false-positive rate, all references to sequencing or other high-throughput projects were removed from the database. Relationships between ISGs and TFs were determined by calculating the cosine of vector angles between gene document vectors derived by a rank-300 approximation [single value decomposition (SVD)-based] to the original term-by-gene document matrix. The relationship of each gene to potential TFs was normalized by calculating the Z-score distribution of the similarity scores. The normalized similarity scores were imported into GeneSpring 7.2 and subjected to hierarchical clustering using standard correlation coefficients.
ChIP assays
ChIP experiments were performed using the ChIP-IT™ Chromatin Immunoprecipitation Kit (Active Motif, Carlsbad, CA, USA) according to the manufacturer's instructions. In brief, cells were fixed with 1% formaldehyde at 22°C for 15 min to generate protein-DNA cross-links, and chromatin DNA was fragmented to an average size of 600 bp. Immunoprecipitation with anti-cRel or anti-immunoglobulin (Ig) of sheared chromatin was performed on precleared cell lysates for 1 h at 4°C. PCR was performed for 38 cycles using 2 μL out of a 100 μL DNA extraction and the following forward and reverse primers corresponding to the upstream regions of ISGs flanking cRel-binding sites that are close to TSSs were used:
Cxcl10: 5′-CCTGTAAACCGAGGGCATTG-3′, 5′-CACGCTT TGGAAAGTGAAAC-3′;
Isg15: 5′-CCTTCTCTCCTTCCACTTTG-3′, 5′-AGGTGAGATGGGAGGTAGAG-3′;
Gbp2: 5′-GTCTCAGTTTTGACAGTGGC-3′, 5′-GTGGAGTT TCCAGTCATTTG-3′;
Ifit3: 5′-CTGTCAGGCTGGAGGAAATG-3′, 5′-TCAACCAGAAGAGGAAAGTG-3′;
Ifit1: 5′-TGATGCAGAGAACACAGCCA-3′, 5′-CTTCTTTCCTTTTGGTCTTC-3′;
Ifi203: 5′-CTTGGAAACCCATGAAATTG-3′, 5′-TTTTGGAATGAAAGTAACCA-3′
Quantitative real-time PCR
Total RNA was isolated from untreated and IFN-β-treated (1000 IU/mL, 5 h) MEFs using TRIzol reagent (Invitrogen, Carlsbad, CA, USA). Quantitative RT-PCR was performed on an iCycler (BioRad) with 60 ng of total RNA in 15 μL of reaction mixture using the AccessQuick™ RT-PCR system (Promega, Madison, WI, USA), and SYBR green I (Molecular Probes, Eugene, OR, USA) according to the manufacturer's instructions. The one-step RT-PCR cycling was as follows: reverse transcription at 48°C for 45 min, denaturation at 95°C for 2 min, amplification for 35 cycles at 94°C for 30 s, and 62°C for 30 s. The product size was initially monitored by agarose gel electrophoresis and melting curves were analyzed to control for the specificity of PCR reactions. Gene expression data was normalized to the expression of the housekeeping gene β-actin. The relative units were calculated from a standard curve, plotting three different concentrations against the PCR cycle number at the cycle threshold (with a 10-fold increment equivalent to ~3.1 cycles).
The following forward and reverse primers were used for each gene, respectively:
Cxcl10: 5′-CGTGTTGAGATCATTGCCAC-3′, 5′-TTAAGGA GCCCTTTTAGACC-3′
Gbp2: 5′-CCTCTTCCTTCAAATGAGAC-3′, 5′-GTGTTTCA ACAACATCTGCC-3′
Ifi203: 5′-AGTGGTGGTTTATGGACGAC-3′, 5′-CTGTGCCTTACAGACCTCAG-3′
Ifit1: 5′-AAGAGAAGTCCTTTGCTTGG-3′, 5′-TGCCCTTTCAGTTTGTAGAC-3′
Ifit3: 5′-GACGATTAACGATGGAGTTC-3′, 5′-GGGCTCTCCTTACTGATGAC-3′
Isg15: 5′-ATGAGGTCTTTCTGACGCAG-3′, 5′-AGCAGCTC CTTGTCCTCCAT-3′
β-actin: 5′-AAGGAGATTACTGCTCTGGC-3′, 5′-ACATCTGCTGGAAGGTGGAC-3′.
Results
Identification of promoter elements in co-regulated genes
In a previous study, we identified 124 genes whose expression levels were significantly changed in MEFs after IFN-β treatment (Pfeffer and others 2004). Hierarchical clustering using standard correlation coefficients revealed several clusters of ISGs with highly similar expression profiles (Fig. 2). The basic hypothesis for our approach is that genes displaying closely related expression profiles are regulated by a common set of TFs. However, neighboring gene clusters are distinguished from one another by their utilization of different TFs. To test this approach, we selected two closely related clusters that were identified from this ISG dataset for promoter sequence analysis. Cluster A included the ISGs: Cxcl10, Isg15/G1p2, Gbp2, Ifit3, Ifi203, and Ifit1. Cluster B included the ISGs: Igtp, Stat1, Irf9, and Lgals3bp. The expression of the ISGs in both clusters were highly induced by IFN, ranging from 8-fold for Irf9 to 669-fold for Ifit1 (Pfeffer and others 2004). The expression values for these ISGs where then normalized across all of the arrays. The ISGs were clustered based on the expression pattern rather than the amplitude of the expression changes. As illustrated in the schematic in Figure 1, we next examined the 3-kb upstream region of each gene in Clusters A and B using the genomic sequence information at UCSC Genome Browser and the web-based search tool MOTIF. The gene promoters contained an average of 381 (ranging from 334 to 420) motifs, corresponding to 67 (ranging from 62 to 74) different TF-binding sites. Forty-eight TFs were common to at least 5 out of the 6 genes in cluster A, and 46 TFs were common to 3 out of 4 genes in the Cluster B. Interestingly, only 11 TFs were unique to Cluster A, and 9 TFs were unique to Cluster B.
FIG. 2.
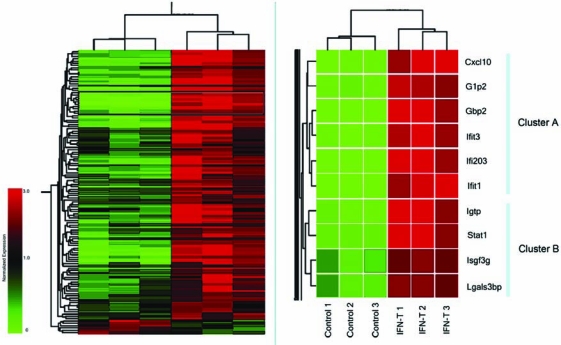
Gene expression clustering of 124 ISGs. ISGs (rows) were subjected to hierarchical clustering based on expression values obtained from three IFN-treated and three IFN-untreated samples (columns). Two related gene clusters (boxed in left panel) were identified as Cluster A and Cluster B gene clusters (right panel). Color bar indicates the normalized expression values below (green) and above (red) the median expression (set to 1.0). The brackets at the top of each panel reveal the similarities among samples, showing that control and IFN-treated triplicates clustered together, respectively.
Literature-based prediction of candidate TFs
As outlined in Figure 1, the next task in our approach was to identify which TFs are already known to regulate the genes in Clusters A and B, and which TFs are likely to regulate them based on implied information in the literature. Previous studies have established a role for STAT and IFN regulatory factor (IRF) proteins in ISG regulation (Barnes and others 2002; Pfeffer and others 2004). However, the differences in the expression patterns among the ISGs suggest that other TFs are also involved in their regulation. To predict which TFs may play a role in ISG expression, we implemented LSI to identify both explicit and implicit (probable) relationships from MEDLINE abstracts for subsets of ISGs and the TFs that are shared in their promoters. First, an abstract document was constructed for each ISG and TF by concatenating titles and abstracts of the citations cross-referenced in their Entrez Gene entries. The number of abstracts used in our corpus for the ISGs ranged from 1 (Ifi203) to 162 (Stat1) and for the TFs ranged from 4 (Tcfap4) to 1025 (Trp53). The gene documents were parsed into a dictionary of terms (tokens) and weighted frequencies (mathematical values used to describe the correlation between terms and the corresponding MEDLINE documents), which were used to construct a term-by-gene document (sparse) matrix.
In the LSI model, term and document vectors are generated by truncating the singular value decomposition of the term-by-gene document matrix to a preselected number of factors. Fewer factors may be used for broad (more conceptual) comparisons, whereas a larger number of factors may be used for specific (more literal) comparisons (Berry and others 1999). Thus, LSI produces a rank-reduced space in which two gene documents can be compared at different conceptual levels. Landauer and colleagues have previously demonstrated that maximal performance of LSI is achieved between 250 and 400 dimensions (factor space) (Landauer and others 2004). Therefore, in the present study, we calculated the relationships between ISGs and TFs using 300 factors. The resulting relationship values, which describe rank distribution, were then normalized and subjected to hierarchical clustering. We found that cRel, Stat2, Irf4, Icsbp1, Irf1, Irf2, and Irf9/Isgf3g were highly associated in the literature with the genes in Cluster A (Fig. 3A). On the other hand, STAT1, STAT4, IRF1, Myb, Ets1, Yy1, Cutl1, and Zfpn1a1 appeared to be highly associated with the genes in Cluster B (Fig. 3B). These associations appeared to be specific because several well-studied TFs such as Trp53, myc, and NFκB1, which have large abstract representations, were not found to be highly associated with either ISG cluster. The classical type I IFN signaling pathway leads to the formation of the ISGF3 complex that consists of STAT1, STAT2, and IRF9. ISGF3 binds to the highly conserved ISRE promoter element present in ISGs, which leads to their transcriptional activation. Therefore, the identification of STAT and IRF proteins with co-regulated genes was expected. Remarkably, there is no known regulatory relationship between the other candidate TFs and co-regulated genes in Clusters A and B, suggesting that there is only an implicit relationship between them as determined by LSI.
FIG. 3.
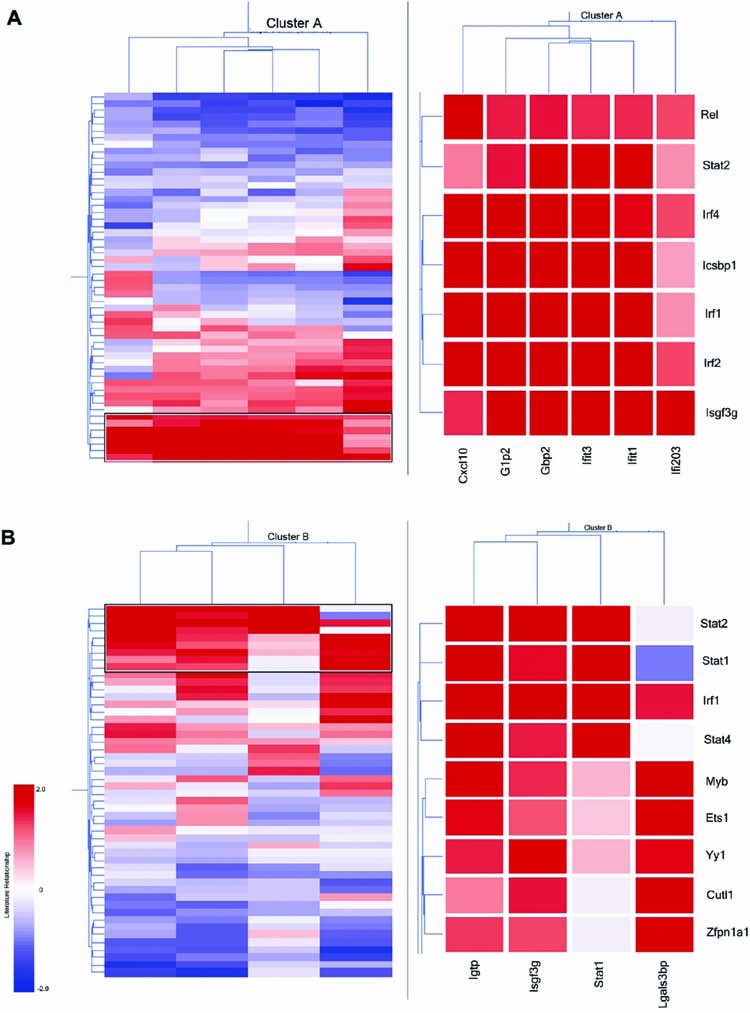
Literature-based clustering of ISGs and TFs. (A) Hierarchical clustering of 6 Gbp2 co-regulated genes (columns) and 48 TFs common to their promoters (rows) using the normalized LSI literature similarity values. (B) Hierarchical clustering of 4 Stat1 co-regulated genes (columns) and 46 TFs common to their promoters (rows) using the normalized LSI literature similarity values. Right panel shows a magnified view of the boxed area in the left panel.
The role of cRel in regulation of an ISG subset
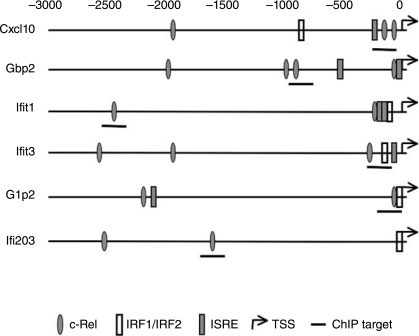
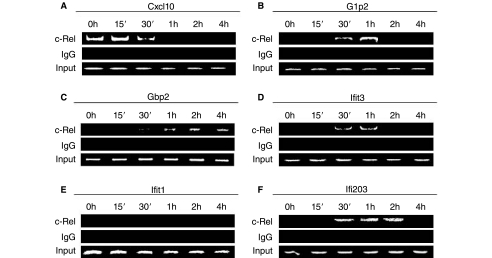
We showed high literature association of six ISGs in Cluster A (Cxcl10, Isg15, Gbp2, Ifit1, Ifit3, and Ifi203) with seven TFs (cRel, Stat2, Irf1, Irf2, Irf4, Irf8/Icsbp1, and Irf9/Isgf3g). These TFs bind to three different DNA-binding motifs (Fig. 4). Since STAT and IRF proteins are well described in the regulation of ISGs in MEFs, we examined the potential role of a less well-described TF in ISG expression. Previous studies have determined that the family of NFκB proteins regulates ISG expression (Pfeffer and others 2004; Wei and others 2006; Yang and others 2007). However, the role of cRel in ISG expression is relatively unknown. We determined whether cRel interacted directly with the promoters of the six ISGs in Cluster A by ChIP assays. As shown in Figure 5, IFN induced the recruitment of cRel to promoters of Isg15, Gbp2, Ifit3, and Ifi203 within 30 min of addition. cRel remained bound to the promoters of these ISGs for different lengths of time, ranging from 1 h for Ifit3 and Isg15 to 4 h for Gbp2. In contrast, cRel was basally bound to the promoter of Cxcl10 and IFN induced cRel detachment from the Cxcl10 promoter by 1 h after addition. However, we were unable to detect cRel binding to the Ifit1 promoter either basally or upon IFN addition.
FIG. 4.
Putative TF-binding motifs in Cluster A ISGs. Genomic sequences corresponding to 3-kb upstream and 50-bp downstream of transcription start site (TSS) of each gene was obtained from the UCSC Genome Browser. TF-binding sites were identified using MOTIF program and the information in TRANSFAC 6.0 database.
FIG. 5.
The binding of cRel to ISG promoters. ChIP assays were performed on extracts from MEFs treated with IFN for different durations. Specific antibodies were used to selectively precipitate cRel, or IgG (negative control). DNA was sheared to an average size of 600 bp fragments, and 38 cycles of amplification were used for PCR using primers that targeted upstream of Cxcl10 (A), G1p2/Isg15 (B), Gbp2 (C), Ifit3 (D), Ifit1 (E), and Ifi203 (F) transcription start sites. Bottom panel shows the amount of input DNA prior to ChIP analysis.
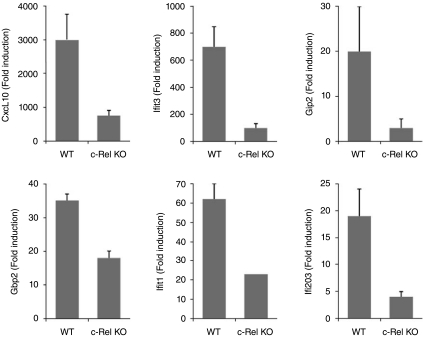
Since these data indicated a role for cRel in regulating ISG expression, quantitative real-time PCR assays were performed for Cxcl10, Isg15, Gbp2, Ifit1, Ifit3, and Ifi203 using RNA from WT and cRel-deficient MEFs treated with IFN-β. As shown in Figure 6, the expression of all six ISGs in response to IFN addition was reduced in cRel-deficient MEFs. For example, while Cxcl10 was induced over 3000-fold by IFN in WT MEFs, Cxcl10 was induced by IFN only 800-fold in cRel-deficient MEFs. Although we did not detect cRel binding to Ifit1 promoter as shown in Figure 5E, we found reduced induction of Ifit1 in cRel-deficient MEFs. Taken together these results suggest that cRel plays distinct roles in the regulation of the IFN-induced expression of this cluster of ISGs. Moreover, since cRel was found to regulate the expression of all six ISGs in this cluster, our text-mining approach identified previously unknown regulatory relationships.
FIG. 6.
Regulation by cRel of ISG induction. Real-time PCR was performed for Cxcl10, G1p2/Isg15, Gbp2, Ifit3, Ifit1, and Ifi203 using cDNAs prepared from WT and cRel-deficient MEFs treated with IFN-β at 1000 U/mL for 5 h. Gene expression was normalized to actin expression in each sample. Data are shown as fold-induction relative to untreated fibroblasts, and are mean values ± the SEM (n = 3).
Discussion
The cRel TF is a member of the NFκB family, which also includes NFκB1 (p105 processed to p50), NFκB2 (p100 processed to p52), RelA (p65), RelB. The family of NFκB TFs regulates gene expression by binding to cis-exacting κB sites in its promoters (Beg and others 1995; Beg and Baltimore 1996; Van Antwerp and others 1996; Wang and others 1996). NFκB-regulated genes play important roles in immunity, inflammation, cell growth, and cell survival, which are all processes affected by IFN and this led us to examine a role for NFκB in IFN signal transduction. Both p50 and p52 lack a transcription activation domain, and as homodimers function as repressors. In contrast, p65, cRel, and RelB have a transcription activation domain, and thus when complexed with p50 or p52 are capable of activating transcription. Under most circumstances, NFκB is bound to IκB inhibitory proteins in the cytoplasm of unstimulated cells. Many cytokines including IFN promote the dissociation of the cytosolic inactive NFκB/IκB complexes via the serine phosphorylation and degradation of IκB, leading to NFκB translocation to the nucleus and DNA binding (Yang and others 2000), which is denoted as the classical NFκB pathway. Recent studies have identified an alternative NFκB signaling pathway, which does not involve IκB degradation (Senftleben and others 2001; Xiao and others 2001; Claudio and others 2002; Coope and others 2002; Dejardin and others 2002; Pomerantz and Baltimore 2002; Muller and Siebenlist 2003; Luftig and others 2004; Xiao and others 2004). This pathway involves the linkage of TRAFs to the activation of the MAP3K-related kinase, NIK, which results in the ubiquitinylation and proteolytic processing of p100/NFκB2 protein and nuclear translocation of p52:RelB dimers to regulate specific NFκB target genes (Bonizzi and others 2004). We previously established that IFN induces NFκB activity in a variety of cells through both classical and alternative pathways, which promote cell survival (Yang and others 2000; Yang and others 2001; Pfeffer and others 2004; Yang and others 2005a; Yang and others 2005b; Wei and others 2006). In addition, we have identified a number of ISGs that are regulated through a NFκB-dependent pathway, but this is the first instance of identification of an ISG regulated by cRel (Pfeffer and others 2004; Wei and others 2006).
Constitutive cRel expression has been previously found in cells of the mature monocytic and lymphocytic lineages (Liou and others 1994). cRel is critical for the normal function of the host immune system as evidenced by the finding that cRel knockout mice develop normally but have severe defects in lymphocyte proliferation and humoral immunity (Kontgen and others 1995). cRel exists as homodimers or as heterodimers with p50 in a variety of cell types (Hoffmann and others 2003). cRel binds to a set of related 9–10 bp DNA sequences (κB sites) and regulates the expression of genes involved in cell development, proliferation, and survival (Gilmore and others 2004). In this study, we implemented a novel integrative approach to identify cRel as a candidate regulatory factor for a subset of ISGs including Isg15/G1p2, Gbp2, Ifit3, and Ifi203. These genes were previously identified to be potential NFκB-regulated ISG targets, but the specific NFκB proteins responsible for the regulation of their expression was unknown (Ohmori and others 1994; Kumar and others 1997; Baker and others 2003; Pfeffer and others 2004; Sizemore and others 2004; Wei and others 2006). Using ChIP assays we determined that IFN rapidly induced the recruitment of cRel to the promoters of Isg15, Gbp2, Ifit3, and Ifi203, and that cRel remained bound to the promoters of these ISGs for different lengths of time. Our preliminary studies suggest that the binding of cRel to the promoters of these genes correlates well with their transcriptional activation. In contrast, we determined that while cRel was basally bound to the Cxcl10 promoter, IFN induced cRel detachment from the promoter so that by 1 h after IFN addition no cRel binding was observed. Since Cxcl10 expression is induced within this time frame, our results are consistent with the hypothesis that cRel homodimers or heterodimers inhibit Cxcl10 gene expression.
Our analysis revealed several novel TFs to be highly associated with ISG expression. It is important to note that not all of the relationships predicted by this method were confirmed, nor were all experimentally documented relationships in the literature predicted by our method. A possible source of false-positive prediction by LSI may be due to technical limitations of the follow-up ChIP assays, for instance in selection of appropriate primer pairs for the target gene promoters. On the other hand, false-negative predictions by LSI may be caused by the information biases in the scientific literature. For instance, some genes are studied in a very specific context whereas others may be studied in much broader context. Although the LSI model generally performs well in these circumstances, we have observed that broadly studied genes generally show low association scores with a large set of genes and lack high association scores with any specific set of genes.13 Nonetheless, we have provided evidence that LSI-based algorithms can serve as a valuable exploratory tool for analysis of gene datasets.
Recent advances in genomic technologies allow investigators to identify novel associations between genes in a high-throughput manner. However, interpretation of genomic data is relatively low-throughput because of the requirement for expert knowledge and the ability of humans to identify implicit (hypothetical) connections between genes. Automated information extraction methods such as LSI will undoubtedly be useful in interpretation of high-throughput genomic data and may play a significant role in taking discovery-based approaches to hypothesis-driven science.
Acknowledgments
We thank Drs. E. Chesler and K. Manly for their helpful comments. We thank Dr. A. Hoffmann (University of California, San Diego) for providing MEFs, and Dr. D. Baker (Biogen-Idec) for providing rat IFN. This research was supported in part by UT Center for Genomics and Bioinformatics (R.H.), UT Center for Neurobiology of Brain Diseases (R.H.), NIH subcontract LM007292-03 (R.H.), CA73753 (L.M.P), Muirhead Chair Endowment (L.M.P) and UT Center for Information Technology Research (K.H. and M.W.B.).
References
- Arduini RM. Li Z. Rapoza A. Gronke R. Hess DM. Wen D. Miatkowski K. Coots C. Kaffashan A. Viseux N. Delaney J. Domon B. Young CN. Boynton R. Chen LL. Betzenhauser M. Miller S. Gill A. Pepinsky RB. Hochman PS. Baker DP. Expression, purification, and characterization of rat interferon-beta, and preparation of an N-terminally PEGylated form with improved pharmacokinetic parameters. Protein Expr Purif. 34(2):229–242. doi: 10.1016/j.pep.2003.11.004. [DOI] [PubMed] [Google Scholar]
- Baker MS. Chen X. Rotramel AR. Nelson JJ. Kaufman DB. Interferon regulatory factor-1 down-regulates cytokine-induced IP-10 expression in pancreatic islets. Surgery. 2003;134(2):134–41. doi: 10.1067/msy.2003.236. [DOI] [PubMed] [Google Scholar]
- Barnes B. Lubyova B. Pitha PM. On the role of IRF in host defense. J Interferon Cytokine Res. 2002;22(1):59–71. doi: 10.1089/107999002753452665. [DOI] [PubMed] [Google Scholar]
- Beg AA. Baltimore D. An essential role for NF-κB in preventing TNF-a-induced cell death. Science. 1996;274:782–784. doi: 10.1126/science.274.5288.782. [DOI] [PubMed] [Google Scholar]
- Beg AA. Sha WC. Bronson RT. Ghosh S. Baltimore D. Embryonic lethality and liver degeneration in mice lacking the RelA component of NF-κB. Nature. 1995;376:167–170. doi: 10.1038/376167a0. [DOI] [PubMed] [Google Scholar]
- Berry MW. Drmac Z. Jessup E. Matrices, vector spaces, and information retrieval. SIAM Rev. 1999;41:335–362. [Google Scholar]
- Bluthgen N. Kielbasa SM. Herzel H. Inferring combinatorial regulation of transcription in silico. Nucleic Acids Res. 2005;33(1):272–279. doi: 10.1093/nar/gki167. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bonizzi G. Bebien M. Otero DC. Johnson-Vroom KE. Cao Y. Vu D. Jegga AG. Aronow BJ. Ghosh G. Rickert RC and others. Activation of IKKalpha target genes depends on recognition of specific kappaB binding sites by RelB:p52 dimers. EMBO J. 2004;23(21):4202–4210. doi: 10.1038/sj.emboj.7600391. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Claudio E. Brown K. Park S. Wang H. Siebenlist U. BAFF-induced NEMO-independent processing of NF-kappa B2 in maturing B cells. Nat Immunol. 2002;3(10):958–965. doi: 10.1038/ni842. [DOI] [PubMed] [Google Scholar]
- Coope HJ. Atkinson PG. Huhse B. Belich M. Janzen J. Holman MJ. Klaus GG. Johnston LH. Ley SC. CD40 regulates the processing of NF-kappaB2 p100 to p52. Embo J. 2002;21(20):5375–5385. doi: 10.1093/emboj/cdf542. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Darnell JEJ. Kerr IM. Stark GR. Jak-STAT pathways and transcriptional activation in response to IFNs and other extracellular signaling proteins. Science. 1994;264:1415–1421. doi: 10.1126/science.8197455. [DOI] [PubMed] [Google Scholar]
- Dejardin E. Droin NM. Delhase M. Haas E. Cao Y. Makris C. Li ZW. Karin M. Ware CF. Green DR. The lymphotoxin-beta receptor induces different patterns of gene expression via two NF-kappaB pathways. Immunity. 2002;17(4):525–535. doi: 10.1016/s1074-7613(02)00423-5. [DOI] [PubMed] [Google Scholar]
- Friedman RL. Stark GR. a-Interferon-induced transcription of HLA and metallothionein genes containing homologous upstream sequences. Nature. 1985;314:637–639. doi: 10.1038/314637a0. [DOI] [PubMed] [Google Scholar]
- Gilmore TD. Kalaitzidis D. Liang MC. Starczynowski DT. The c-Rel transcription factor and B-cell proliferation: a deal with the devil. Oncogene. 2004;23(13):2275–2286. doi: 10.1038/sj.onc.1207410. [DOI] [PubMed] [Google Scholar]
- Hoffmann A. Leung TH. Baltimore D. Genetic analysis of NF-kappaB/Rel transcription factors defines functional specificities. EMBO J. 2003;22(20):5530–5539. doi: 10.1093/emboj/cdg534. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Homayouni R. Heinrich K. Wei L. Berry MW. Gene clustering by latent semantic indexing of MEDLINE abstracts. Bioinformatics. 2005;21(1):104–115. doi: 10.1093/bioinformatics/bth464. [DOI] [PubMed] [Google Scholar]
- Jenssen TK. Laegreid A. Komorowski J. Hovig E. A literature network of human genes for high-throughput analysis of gene expression. Nat Genet. 2001;28(1):21–28. doi: 10.1038/ng0501-21. [DOI] [PubMed] [Google Scholar]
- Kontgen F. Grumont RJ. Strasser A. Metcalf D. Li R. Tarlinton D. Gerondakis S. Mice lacking the c-rel proto-oncogene exhibit defects in lymphocyte proliferation, humoral immunity, and interleukin-2 expression. Genes Dev. 1995;9(16):1965–1977. doi: 10.1101/gad.9.16.1965. [DOI] [PubMed] [Google Scholar]
- Krallinger M. Erhardt RA. Valencia A. Text-mining approaches in molecular biology and biomedicine. Drug Discov Today. 2005;10(6):439–445. doi: 10.1016/S1359-6446(05)03376-3. [DOI] [PubMed] [Google Scholar]
- Kumar A. Yang YL. Flati V. Der S. Kadereit S. Deb A. Haque J. Reis L. Weissmann C. Williams BR. Deficient cytokine signaling in mouse embryo fibroblasts with a targeted deletion in the PKR gene: role of IRF-1 and NF-kappaB. EMBO J. 1997;16(2):406–416. doi: 10.1093/emboj/16.2.406. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Landauer TK. Laham D. Derr M. From paragraph to graph: latent semantic analysis for information visualization. Proc Natl Acad Sci USA. 2004;101(Suppl 1):5214–5219. doi: 10.1073/pnas.0400341101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Larner AC. Chaudhuri A. Darnell JEJ. Transcriptional induction by interferon: new protein(s) determine the length and extent of induction. J Biol Chem. 1986;261:453–459. [PubMed] [Google Scholar]
- Li H. Lu L. Manly KF. Chesler EJ. Bao L. Wang J. Zhou M. Williams RW. Cui Y. Inferring gene transcriptional modulatory relations: a genetical genomics approach. Hum Mol Genet. 2005;14(9):1119–1125. doi: 10.1093/hmg/ddi124. [DOI] [PubMed] [Google Scholar]
- Liou HC. Sha WC. Scott ML. Baltimore D. Sequential induction of NF-kappa B/Rel family proteins during B-cell terminal differentiation. Mol Cell Biol. 1994;14(8):5349–5359. doi: 10.1128/mcb.14.8.5349. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Luftig M. Yasui T. Soni V. Kang MS. Jacobson N. Cahir-McFarland E. Seed B. Kieff E. Epstein-Barr virus latent infection membrane protein 1 TRAF-binding site induces NIK/IKK alpha-dependent noncanonical NF-kappaB activation. Proc Natl Acad Sci USA. 2004;101(1):141–146. doi: 10.1073/pnas.2237183100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Matys V. Fricke E. Geffers R. Gossling E. Haubrock M. Hehl R. Hornischer K. Karas D. Kel AE. Kel-Margoulis OV and others. TRANSFAC: transcriptional regulation, from patterns to profiles. Nucleic Acids Res. 2003;31(1):374–378. doi: 10.1093/nar/gkg108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Muller JR. Siebenlist U. Lymphotoxin beta receptor induces sequential activation of distinct NF-kappa B factors via separate signaling pathways. J Biol Chem. 2003;278(14):12006–12012. doi: 10.1074/jbc.M210768200. [DOI] [PubMed] [Google Scholar]
- Ohmori Y. Tebo J. Nedospasov S. Hamilton TA. Kappa B binding activity in a murine macrophage-like cell line. Sequence-specific differences in kappa B binding and transcriptional activation functions. J Biol Chem. 1994;269(26):17684–17690. [PubMed] [Google Scholar]
- Pfeffer LM. Kim JG. Pfeffer SR. Carrigan DJ. Baker DP. Wei L. Homayouni R. The role of NF-κB in the antiviral action of interferon and interferon-regulated gene expression. J Biol Chem. 2004;279(30):31304–31311. doi: 10.1074/jbc.M308975200. [DOI] [PubMed] [Google Scholar]
- Pomerantz JL. Baltimore D. Two pathways to NF-kappaB. Mol Cell. 2002;10(4):693–695. doi: 10.1016/s1097-2765(02)00697-4. [DOI] [PubMed] [Google Scholar]
- Qin ZS. McCue LA. Thompson W. Mayerhofer L. Lawrence CE. Liu JS. Identification of co-regulated genes through Bayesian clustering of predicted regulatory binding sites. Nat Biotechnol. 2003;21(4):435–439. doi: 10.1038/nbt802. [DOI] [PubMed] [Google Scholar]
- Rebholz-Schuhmann D. Kirsch H. Couto F. Facts from text—is text mining ready to deliver? PLoS Biol. 2005;3(2):e65. doi: 10.1371/journal.pbio.0030065. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schindler C. Fu X-Y. Improta T. Aebersold R. Darnell JEJ. Proteins of transcription factor ISGF-3: one gene encoded the 91- and 84-kDa ISGF-3 that are activated by interferon a. Proc Natl Acad Sci USA. 1992;89:7836–7839. doi: 10.1073/pnas.89.16.7836. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Segal E. Shapira M. Regev A. Pe'er D. Botstein D. Koller D. Friedman N. Module networks: identifying regulatory modules and their condition-specific regulators from gene expression data. Nat Genet. 2003;34(2):166–176. doi: 10.1038/ng1165. [DOI] [PubMed] [Google Scholar]
- Senftleben U. Cao Y. Xiao G. Greten FR. Krahn G. Bonizzi G. Chen Y. Hu Y. Fong A. Sun SC and others. Activation by IKKalpha of a second, evolutionary conserved, NF-kappa B signaling pathway. Science. 2001;293(5534):1495–1499. doi: 10.1126/science.1062677. [DOI] [PubMed] [Google Scholar]
- Shatkay H. Feldman R. Mining the biomedical literature in the genomic era: an overview. J Comput Biol. 2003;10(6):821–855. doi: 10.1089/106652703322756104. [DOI] [PubMed] [Google Scholar]
- Sizemore N. Agarwal A. Das K. Lerner N. Sulak M. Rani S. Ransohoff R. Shultz D. Stark GR. Inhibitor of kappaB kinase is required to activate a subset of interferon gamma-stimulated genes. Proc Natl Acad Sci USA. 2004;101(21):7994–7998. doi: 10.1073/pnas.0401593101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Van Antwerp DJ. Martin SJ. Kafri T. Green DR. Verma IM. Suppression of TNF-a-induced apotosis by NF-κB. Science. 1996;274:787–789. doi: 10.1126/science.274.5288.787. [DOI] [PubMed] [Google Scholar]
- Wang C-Y. Mayo MW. Baldwin AS., Jr TNF- and cancer therapy-induced apoptosis: potentiation by inhibition of NF-κB. Science. 1996;274:784–787. doi: 10.1126/science.274.5288.784. [DOI] [PubMed] [Google Scholar]
- Wei L. Sandbulte MR. Thomas PG. Webby RJ. Homayouni R. Pfeffer LM. NFκB negatively regulates interferon-induced gene expression and anti-influenza activity. J Biol Chem. 2006;281(17):11678–11684. doi: 10.1074/jbc.M513286200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xiao G. Fong A. Sun SC. Induction of p100 processing by NF-kappaB-inducing kinase involves docking IkappaB kinase alpha (IKKalpha) to p100 and IKKalpha-mediated phosphorylation. J Biol Chem. 2004;279(29):30099–30105. doi: 10.1074/jbc.M401428200. [DOI] [PubMed] [Google Scholar]
- Xiao G. Harhaj EW. Sun SC. NF-kappaB-inducing kinase regulates the processing of NF-kappaB2 p100. Mol Cell. 2001;7(2):401–409. doi: 10.1016/s1097-2765(01)00187-3. [DOI] [PubMed] [Google Scholar]
- Xie X. Lu J. Kulbokas EJ. Golub TR. Mootha V. Lindblad-Toh K. Lander ES. Kellis M. Systematic discovery of regulatory motifs in human promoters and 3′ UTRs by comparison of several mammals. Nature. 2005;434(7031):338–345. doi: 10.1038/nature03441. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yandell MD. Majoros WH. Genomics and natural language processing. Nat Rev Genet. 2002;3(8):601–610. doi: 10.1038/nrg861. [DOI] [PubMed] [Google Scholar]
- Yang CH. Murti A. Basu L. Kim JG. Pfeffer LM. IFNα/β promotes cell survival by activating NF-κB. Proc Natl Acad Sci USA. 2000;97:13631–13636. doi: 10.1073/pnas.250477397. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yang CH. Murti A. Pfeffer SR. Kim JG. Donner DB. Pfeffer LM. Interferon α/β promotes cell survival by activating NF-κB through phosphatidylinositol-3 kinase and Akt. J Biol Chem. 2001;276:13756–13761. doi: 10.1074/jbc.M011006200. [DOI] [PubMed] [Google Scholar]
- Yang CH. Murti A. Pfeffer LM. IFN induces NIK/TRAF dependent NF-κB activation to promote cell survival. J Biol Chem. 2005a;280(36):31530–31536. doi: 10.1074/jbc.M503120200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yang CH. Murti A. Valentine WJ. Du Z. Pfeffer LM. Interferon alpha activates NF-κB in JAK1-deficient cells through a TYK2-dependent pathway. J Biol Chem. 2005b;280(27):25849–25853. doi: 10.1074/jbc.M413721200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yang CH. Wei L. Pfeffer SR. Du Z. Murti A. Valentine WJ. Zheng Y. Pfeffer LM. Identification of CXCL11 as a STAT3-dependent gene induced by IFN. J Immunol. 2007;178(2):986–992. doi: 10.4049/jimmunol.178.2.986. [DOI] [PubMed] [Google Scholar]