Abstract
Although many species have similar total distributional ranges, they might be restricted to very different habitats and might have different phylogeographic histories. In the European Alps, our excellent knowledge of the evolutionary history of silicate-dwelling (silicicole) plants is contrasted by a virtual lack of data from limestone-dwelling (calcicole) plants. These two categories exhibit fundamentally different distribution patterns within the Alps and are expected to differ strongly with respect to their glacial history. The calcicole Ranunculus alpestris group comprises three diploid species of alpine habitats. Ranunculus alpestris s. str is distributed over the southern European mountain system, while R. bilobus and R. traunfellneri are Southern Alpine narrow endemics. To explore their phylogenetic relationships and phylogeographic history, we investigated the correlation between information given by nuclear and chloroplast DNA data. Analyses of AFLP fingerprints and matK sequences gave incongruent results, indicative for reticulate evolution. Our data highlight historical episodes of range fragmentation and expansion, occasional long distance dispersal and on-going gene flow as important processes shaping the genetic structure of the group. Genetic divergence, expressed as a rarity index (“frequency-down-weighted marker values”) seems a better indicator of historical processes than patterns of genetic diversity, which rather mirror contemporary processes as connectivity of populations and population sizes. Three phylogeographical subgroups have been found within the R. alpestris group, neither following taxonomy nor geography. Genetic heterogeneity in the Southern Alps contrasts with Northern Alpine uniformity. The Carpathians have been stepwise colonised from the Eastern Alpine lineage resulting in a marked diversity loss in the Southern Carpathians. The main divergence within the group, separating the ancestor of the two endemic species from R. alpestris s. str., predates the Quaternary. Therefore, range shifts produced by paleoclimatic oscillations seem to have acted on the genetic structure of R. alpestris group on a more regional level, e.g. triggering an allopatric separation of R. traunfellneri from R. bilobus.
Keywords: AFLP, endemism, hybridisation, phylogeography, Ranunculus alpestris group, refugia
Introduction
Climatic fluctuations during the past three million years have not only had a significant impact on the distribution of biota by enforcing massive range shifts and changes in abundance (Hewitt 2000, 2004), but also enforced speciation (Bennett 2004). For instance, for biota lacking adaptations for surviving in the tundra steppes prevailing in the lowlands of Central Europe during the Last Glacial Maximum (Frenzel et al. 1992), cold periods are thought to have fostered allopatric diversification recurrently as well as speciation through fragmentation of distribution ranges combined with local adaptation. Subsequent warmer periods led to hybridisation via secondary contact (Comes & Kadereit 1998; Kadereit et al. 2004). Signatures of these historical processes have thereby been imprinted and are often still reflected in the genetic structure of present-day biota (Hewitt 1996, 2000, 2004; Comes & Kadereit 1998, 2003). Recent molecular clues in combination with paleo-environmental reconstruction have aided locating glacial refugia and tracing the routes of postglacial spread. Comparative studies across ecologically similar, but taxonomically unrelated taxa have partly revealed congruent patterns of glacial history, but unexpected incongruences have also been found (e.g., Soltis et al. 1997, 2006; Taberlet et al. 1998; Schönswetter et al. 2005). Such differences might have arisen due to chance events, but also due to intrinsic characteristics of species, such as breeding system, dispersal ability or adaptation to ecologically divergent habitats (e.g., Taberlet et al. 1998; Schönswetter et al. 2005).
The impact of the Pleistocene climatic oscillations on phylogeography and evolution of mountain plants has been extensively studied in the European Alps (for a review see Schönswetter et al. 2005), an area which functions as ideal study system for research in the causes and consequences of patterns of genetic variation, phylogeography, and ecology. Most species of the Alps are, although often co-distributed, either strictly calcicole or silicicole (i.e., restricted to calcareous or siliceous bedrock; Ozenda 1988), and the availability of a high number of studies of silicicole plants contrasts with the near absence of studies of calcicole plants with a sampling design comparable to that of silicicole species. In the taxa studied so far (Anthyllis montana, Kropf et al. 2002; Rumex nivalis Stehlik, 2002; Erinus alpinus, Stehlik et al. 2002; Pritzelago alpina, Kropf et al. 2003; Dryas octopetala, Skrede et al. 2006), the sampling either covered only a part of the Alps or did not include enough populations to allow detailed phylogeographic inferences. Additionally, only Kropf et al. (2003) and Skrede et al. (2006) included samples from the Carpathians, from where detailed information about glacial refugia is still scarce (but see Mráz et al. 2007). Due to the uneven distribution of calcareous and siliceous bedrock especially in the Eastern Alps – a siliceous core being flanked by peripheral limestone ranges (Egger et al. 2000) – glacial histories are expected to be fundamentally different for both groups of species (Schönswetter et al. 2005).
The Ranunculus alpestris group (Ranunculaceae) is almost exclusively restricted to calcareous bedrock, making it a good model to test hypotheses on the location of Pleistocene refugia for calcicole plants of the European Alps. In its recent-most circumscription based on DNA sequence data (Paun et al. 2005) which corroborated previous morphological studies (e.g., Baltisberger & Müller 1981; Müller & Baltisberger 1984; Huber 1988; Baltisberger 1994), the group comprises three white-flowered species characterised by petaloid honey-leaves without nectary scale. They are distributed in subalpine to alpine limestone areas of the southern European mountain system (Fig. 1). Ranunculus alpestris L. is widespread from the Cordillera Cantabrica over Pyrenees, Alps, and Central Apennines to the Carpathians, whereas R. bilobus Bertol. and R. traunfellneri Hoppe are narrowly distributed endemics of the southeastern Alps. The R. alpestris group forms a strongly supported, early-diverged lineage within Ranunculus that evolved already in the early Miocene (Paun et al. 2005). Irrespective of the rather ancient origin of the group, diversification among R. alpestris s.str., R. bilobus and R. traunfellneri might have started as recently as 1.1 mya (Paun et al. 2005).
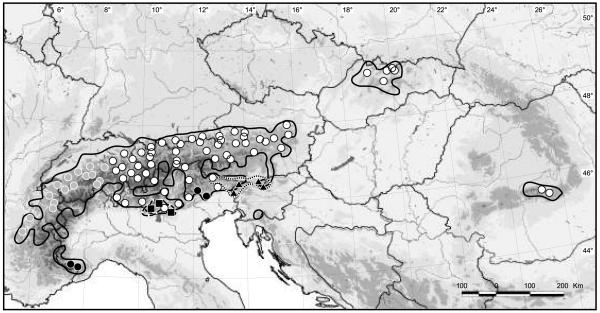
Figure 1.
Distribution of the Ranunculus alpestris group in the Alps and Carpathians, geographic location of the 89 sampled populations and their phylogeographical grouping according to the Bayesian clustering analyses of AFLP phenotypes conducted with the programs BAPS and STRUCTURE. White symbols, group E; grey symbols, group W; black symbols, group S and SW. Sampled locations/distribution of species: circles/solid line, R. alpestris s.str.; triangles/dotted line, R. traunfellneri; squares/dashed line, R. bilobus. More details to the investigated populations are given in Appendices 1-2.
Here, we explore the phylogenetic and phylogeographic history of the R. alpestris group in the European Alps and the Carpathians, using amplified fragment length polymorphism (AFLP) and sequence variation of chloroplast DNA (cpDNA) towards evaluating the impact of Pleistocene climatic fluctuations on evolutionary divergence of a calcicole plant. In particular, we aim to answer the following questions: Are the main genetic lineages within the R. alpestris group corresponding to taxonomical entities? Are the phylogeographical structure and the pattern of genetic diversity in the Alps and the Carpathians explained by hypothesised Pleistocene refugia for calcicole plants (Schönswetter et al. 2005) and thus markedly different from those of silicicole species? Is there evidence for lineage sorting or hybridisation and how significant are consequences of gene flow in the evolution of this group?
Material and Methods
The study species
The three species constituting the Ranunculus alpestris group are diploid (2n = 16, e.g., Goepfert 1974) and their karyotype exhibits no significant difference (Müller & Baltisberger 1984). Crossing experiments between all pair combinations of the three species have produced fertile hybrids (Müller & Baltisberger 1984). The three species are mainly distinguished by the shape of the basal leaves (i.e., depth of incisions and shape of the leaf basis). They are perennial herbs restricted to similar habitats on calcareous bedrock or (rarely) basic silicates above the treeline. Ranunculus alpestris is insect pollinated (Schroeter 1926; Wertlen 2006) and self-incompatible (Baltisberger & Müller 1981; Müller & Baltisberger 1984). Gravity, snowmelt water and epizoochory are probably the main vectors of seed dispersal (Müller-Schneider 1986; Gerber et al. 2004). Adult plants produce tightly clustered ramets; however, no extensive vegetative proliferation occurs (Gerber et al. 2004).
Sampling
The three species were sampled across the Alps and Carpathians. Following the strategy adopted for the IntraBioDiv project (Gugerli et al. in press), leaf samples were collected from at least one locality in every second 12′ latitude × 20′ longitude regular grid cell based on Niklfeld (1971; Fig. 1, Appendices 1-2). Three individuals per sampling locality (named hereafter population) were collected randomly at a distance of at least 10 m, and dried in silica gel. This sampling scheme resulted in 89 sampled populations (R. alpestris, 82 locations; R. bilobus, three and R. traunfellneri, four; Fig. 1, Appendix 1). Analysing data from only three individuals per sampling locality was counter-balanced by using a large number of genetic markers (Nei 1987) and a large number of localities. Four individuals consistently failed to produce reliable AFLP patterns and were excluded from all analyses (see Appendix 1). Twenty-three plants (from 20 populations) were sampled and extracted twice to test the reproducibility of AFLP fingerprinting (Bonin et al. 2004). Two samples were used as replicates between PCR plates, and were replicated more than twice (in total 31 replicates have been performed). Voucher specimens are deposited at the herbarium of the University of Vienna (WU).
DNA extraction and AFLP fingerprinting
Total genomic DNA was extracted from similar amounts of dried tissue (ca. 10 μg) with the DNeasy 96 plant mini kit (Qiagen, Hilden, Germany) following the manufacturer’s protocol. AFLP profiles were generated following established procedures (Vos et al. 1995) with minor modifications (Gugerli et al. in press). Genomic DNA (c. 200 ng) was digested with 1 U MseI (New England BioLabs, Ipswich, USA) and 5 U EcoRI (Promega, Madison, USA) and ligated (with 1.2 U of T4 DNA-Ligase; Promega) to double-stranded adapters in a thermal cycler (GeneAmp PCR System 9700; Applied Biosystems, Foster City, CA, USA) for 2.5 h at 37 °C. Preselective amplification was performed using primer pairs with a single or two selective nucleotides. Three selective primer combinations were chosen after a primer trial (fluorescent dye in brackets): EcoRI AAG (VIC)-MseI CATA; EcoRI ACA (6-FAM)-MseI CTGA; EcoRI ACC (NED)-MseI CAT. For each individual, 1.2 μl 6-FAM, 2 μl VIC and 3 μl NED labelled, Sephadex purified, selective PCR products were combined with 0.2 μl GeneScan ROX 500 (PE Applied Biosystems) internal size standard and 9.8 μl formamide and run on a capillary sequencer ABI 3170 (Applied Biosystems). Blind samples were included to test for contamination (Bonin et al. 2004). Fragments in the range 65-500 bp were aligned with ABI Prism GeneScan 2.1 Analysis Software (PE Applied Biosystems) and visualized, scored and exported as binary presence/absence matrix using Genographer 1.6 (available from http://hordeum.msu.montana.edu/genographer/).
cpDNA sequencing
Chloroplast DNA haplotype variation was explored to complement the information given by the mainly nuclear (Bussell et al. 2005) AFLPs. A part of the matK region, plus a part of the 3′ flanking trnK intron that were previously found to be variable in Ranunculus (Paun et al. 2005) were sequenced for 37 accessions representing the three species and the groups indicated by the AFLP analysis. After an initial screening that revealed the virtual lack of variation over large parts of the range of R. alpestris s.l., we examined in detail the pattern of cpDNA variation within the two endemic species R. bilobus and R. traunfellneri, and in Southern and Southwestern Alpine populations of R. alpestris. The plastid region was amplified and sequenced using the primers trnK345f and trnK3r according to Paun et al. (2005). GenBank accession numbers of the obtained sequences are indicated in Appendix 1.
Analysis of AFLP data
All monomorphic fragments and the ones present/absent in all but one individual were removed from the data set to avoid biased parameter estimates (Bonin et al. 2004). Average gene diversity over loci (πn, in the following termed “genetic diversity”) corrected for small sample size and its standard deviation for both sampling and stochastic processes was computed for populations and phylogroups with ARLEQUIN 3.0 (Excoffier et al. 2005), available at http://cmpg.unibe.ch/software/arlequin3. The same program was used to conduct analyses of molecular variance (AMOVAs). We estimated the number of private fragments, defined as markers confined to a single population or a single phylogroup. We calculated a rarity index, i.e. a measure of population genetic divergence, by computing “frequency-down-weighted marker values” per population or phylogroup (DW, Schönswetter & Tribsch 2005; in the following termed “rarity”) equivalent to range-down-weighted species values in historical biogeographical research (Crisp et al. 2001). For each population or phylogroup, the frequency of each AFLP marker in that population was divided by the number of occurrences of that particular marker in the total dataset. Finally these values were summed up, and corrected for the total number of markers and individuals. DW is expected to be higher in those populations or groups that harbour a high number of rare markers and to be independent from sample size. R scripts to calculate DW and additional documentation are available from http://www.intrabiodiv.eu/spip.php?article77. In order to test whether molecular indices are related to historical patterns of potential refugia, based on the map of presumed glacial refugia published in Schönswetter et al. (2005) and given in Fig. 2a,d, each individual was denoted as sampled (i) in or very close to refugia including peripheral nunatak refugia below the Last Glacial Maximum (LGM) snow line (i.e. a thermal boundary connecting points above which snow does not melt in a climatically average year, Körner 2003), or (ii) in once heavily glaciated areas within the LGM snow line. In order to test whether genetic diversity and rarity (πn & DW) were correlated with regional abundance of populations, frequency data of occurrences of the Ranunculus species per grid cell were used. These were gathered across the entire grid, including additional information on the number of occurrences (frequency) within 16 sub-squares per cell (IntraBioDiv Consortium-Floristic Group, unpublished: The IntraBioDiv Floristic Database, ver. 4.0 [June 2007]. Mapping the Distribution of High-Mountain taxa of the Alps and Carpathians. Maintained at Univ. Vienna, Dept. of Biogeography). Correlation coefficients between πn, DW, and frequency were calculated with SPSS 15.0 (SPSS, Chicago).
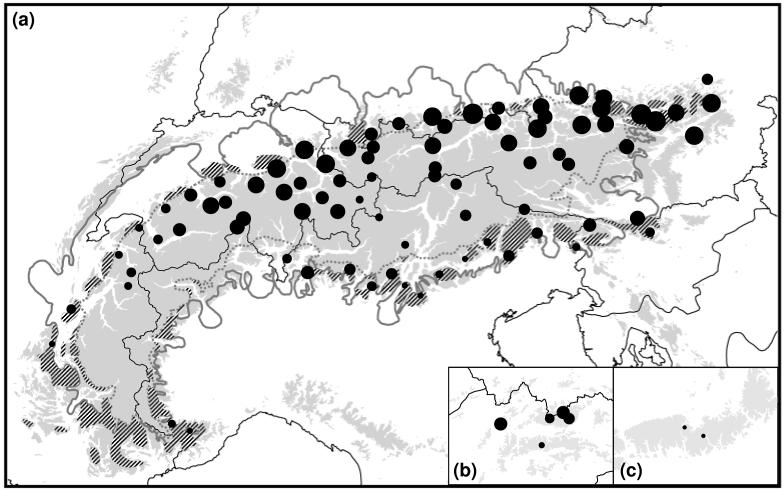
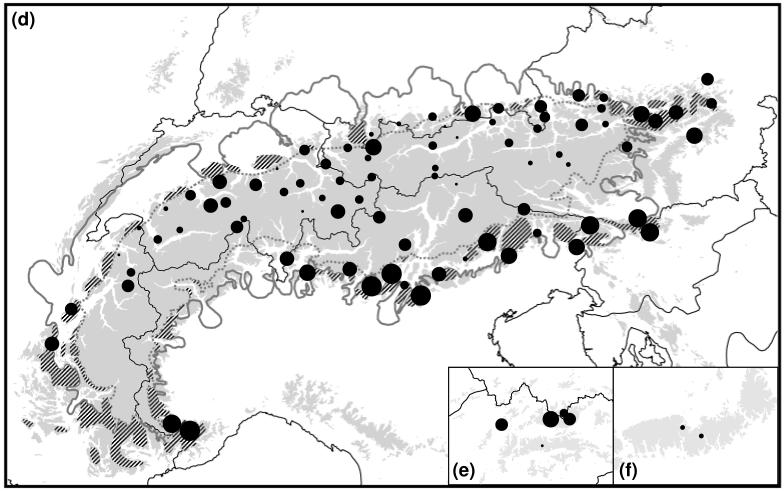
Figure 2.
Patterns of within-population genetic diversity (πn, a-c) and rarity (DW, d-f) in the AFLP data in 89 sampled populations of the Ranunculus alpestris group. The size of the dots is directly proportional to the degree of genetic diversity, or rarity, respectively. (a, d) Alps; (b, e) Western Carpathians; (c, f) Southern Carpathians (see also Fig. 1 and Appendix 2). Within the Alps, the hatched areas indicate presumed glacial refugia (Schönswetter et al. 2005); the continuous grey line shows the maximum extent of the ice sheet; and the broken grey line indicates represents the Last Glacial Maximum snow line.
A midpoint-rooted neighbour joining dendrogram (NJ) based on the genetic distance of Nei & Li (1979) was generated and bootstrapped (Felsenstein 1985) using 1000 replicates with TREECON 1.3b (Van de Peer & De Wachter 1997). To visualize the pattern of population differentiation based on pair-wise FST values computed with ARLEQUIN 3.0, we constructed a Principal Coordinate Analysis (PCoA) using the modules ‘Dcenter’ and ‘Eigen’ from NTSYS-Pc (Rohlf 1997). We estimated the goodness of fit of the analysis by generating a model distance matrix from the eigenvector matrix (with ‘Simint’) and comparing it to the original FST matrix (using ‘Mxcomp’ and 1000 permutations).
Both spatial and non-spatial genetic mixture analysis implemented in the program Bayesian Analysis of Population Structure (BAPS 4.13, Corander et al. 2003, 2004, 2008; Corander & Marttinen 2006; available at http://web.abo.fi/fak/mnf//mate/jc/software/baps.html) was used to cluster individuals into “panmictic” groups. BAPS was run with the maximal number of groups (K) set to 1-10 and each run was replicated three times. The partition with the highest log marginal likelihood was plotted onto the distribution map (Fig. 1). Another model-based clustering approach, implemented in STRUCTURE 2.0 (Pritchard et al. 2000), was used to test the results obtained from BAPS. The algorithm implemented in STRUCTURE, though developed for codominant markers, can be also used for dominant markers under a “no admixture” model. The number of clusters was estimated using 106 iterations, with a burn-in period of 105 iterations. The number of clusters (K) was used as a prior value; two replicates for each K were analysed from K = 1 to K = 10. All analyses using STRUCTURE were carried out at the Bioportal of the University of Oslo (www.bioportal.uio.no).
Analysis of cpDNA sequence data
cpDNA sequences were edited using AutoAssembler 1.4.0 (Applied Biosystems, Foster City, USA) and manually aligned, together with accessions of R. bilobus and R. traunfellneri available in GenBank (part of accessions AY954220 and AY954222). One informative 6 bp insertion/deletion (indel) from the trnK intron region was coded as present-absent and the region itself was excluded from further analysis (following the recommendations of Simmons & Ochoterena 2000). A minimum spanning tree (MST) of the haplotypes was drawn manually. Additionally, phylogenetic analyses were conducted using PAUP* 4.0b10 for Macintosh (Swofford 2003). Maximum parsimony analyses were performed under a heuristic search strategy, using stepwise addition, TBR branch swapping, the MULTREES option on, and simple addition of sequences. The same options were selected for a non-parametric bootstrap analysis (Felsenstein 1985) with 1000 replicates. Sequences from R. aconitifolius, R. crenatus and R. parnassifolius ssp. parnassifolius (part of GenBank Accession Nos. AY954228, AY954217 and AY954224) were used for outgroup rooting in accordance with Hörandl et al. (2005) and Paun et al. (2005). The number of substitutions between the two major ingroup lineages in the amplified coding region was used to estimate the age of the respective split, based on a matK substitution rate calculated for Ranunculus of 2.2 × 10−9 per site per year, with a standard deviation of 0.68 × 10−9 (Paun et al. 2005).
Results
AFLP data
The three AFLP primer combinations yielded 438 clear polymorphic fragments. In the AFLP profiles from replicated samples, 46 differences were observed out of 13.578 phenotypic comparisons, resulting in an error rate of 0.34%. All 263 individuals had different AFLP phenotypes. Average gene diversity over loci (πn; Appendix 1) varied from 0.003 in population 82 (S Carpathians) to 0.104 in population 52 (N Alps). Rarity (DW; Appendix 1) varied from 0.47 in population 25 (C Alps) to 3.62 in population 83 (R. bilobus). Values of πn, DW and private fragments calculated for the phylogroups defined below are given in Table 1. Genetic diversity followed a clear N-S distribution with highest diversity in the Northern Alps and lowest diversity in the S Carpathians (Fig. 2a-c). In the Alps, rarity (DW) followed the opposite trend, with higher rarity in the S (but also NE) Alps and lower in the Central Alps and the Southern Carpathians (Fig 2d-f). πn and DW were not correlated (Pearson’s r = −0.184; P = 0.084). For the Alpine populations, DW was significantly higher in potential refugia than in once heavily glaciated areas inside the LGM snow line (Kruskal-Wallis test, χ2 = 13.147; P < 0.001), whereas the pattern of πn did not correlate with the location of potential refugia (χ2 = 1.801; P = 0.180). Frequency of populations in a grid cell was significantly correlated with πn (Spearman’s r = 0.311; P = 0.003), but not with DW (Spearman’s r = −0.110; P = 0.310).
Table 1.
Overview of genetic diversity and rarity estimates of the regional groups/gene pools/taxonomic entities investigated. N, number of population samples; πn, average gene diversity over loci; NPRIV, number of private AFLP markers; DW, within-population rarity of markers. Information on the groups is given in Fig. 1
| N | π n | NPRIV | DW | |
|---|---|---|---|---|
| W Carpathians only |
5 | 0.076 ± 0.039 | 4 | 0.789 |
| S Carpathians only | 2 | 0.012 ± 0.008 | 0 | 0.563 |
| All Carpathian populations |
7 | 0.073 ± 0.037 | 4 | 0.721 |
| E Alps only | 55 | 0.102 ± 0.049 | 65 | 0.771 |
| E phylogroup | 62 | 0.102 ± 0.049 | 93 | 0.766 |
| W phylogroup | 16 | 0.080 ± 0.040 | 12 | 0.747 |
| S phylogroup | 11 | 0.134 ± 0.066 | 76 | 2.442 |
| R. alpestris S only | 4 | 0.117 ± 0.061 | 24 | 2.377 |
| R. traunfellneri | 4 | 0.081 ± 0.043 | 16 | 1.836 |
| R. bilobus | 3 | 0.118 ± 0.064 | 22 | 3.336 |
| R. alpestris s.str. | 82 | 0.108 ± 0.052 | 41 | 0.842 |
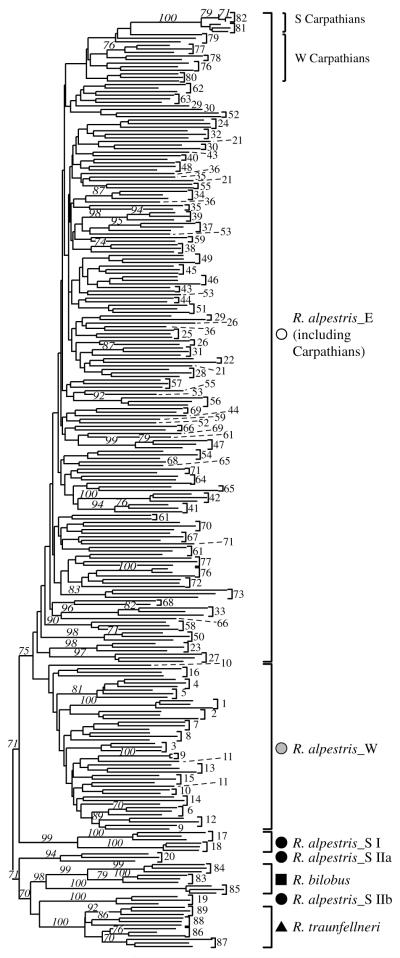
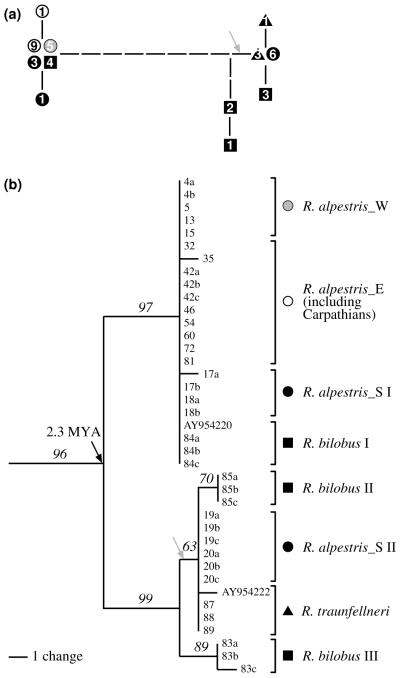
In the NJ analysis (Fig. 3), three major phylogroups were separated with moderate or no bootstrap support. One group, termed phylogroup E in the following, is represented by Eastern Alpine R. alpestris (including the Carpathian populations), and another by the Western Alpine R. alpestris (phylogroup W). The relationships within these two groups were unresolved. A third phylogroup (S; bootstrap support – BS, 75%) comprised the clearly separated R. bilobus (BS, 98%), and R. traunfellneri (BS, 100%), plus four populations of R. alpestris from the S and SW Alps. Consequently, Ranunculus alpestris was not resolved as monophyletic in this analysis. The three dimensional PCoA (Fig. 4) showed a congruent pattern with the NJ tree. The goodness of fit analysis for the scatter-plot indicated a matrix correlation value of r = 0.91 at P = 0.002 (one-tailed probability). A PCoA derived under the same conditions, but on a reduced data set including a comparable number of populations from each of the three groups, indicated a similar pattern (not shown).
Figure 3.
Midpoint rooted Neighbour Joining dendrogram based on Nei & Li (1979) distances among AFLP phenotypes of 263 individuals of the Ranunculus alpestris group. Numbers above branches are NJ bootstrap values (1000 replicates) higher than 70%, numbers at the tips of branches are population identifiers explained in Appendix 1.
Figure 4.
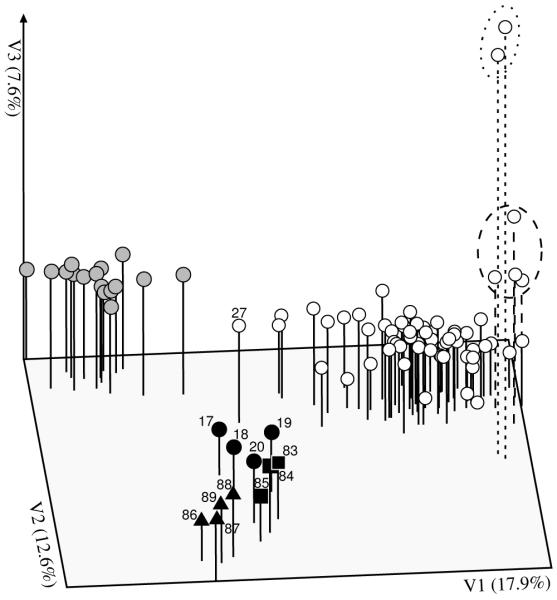
Principal Coordinate Analysis (goodness of fit 0.91 at P = 0.002) of the pair-wise FST matrix of 89 populations of the Ranunculus alpestris group, based on AFLP phenotypes. Symbols as in Fig. 1; the dotted line encircles populations 81 and 82 from the S Carpathians; the dashed line populations 76 to 80 from the Tatras. Sampling localities mentioned in the discussion are labelled with population identifiers explained in Appendix 1. The three ordination factors together explain 38.1% of the variation in the AFLP data matrix.
The partition with the highest log marginal likelihood (spatial −26227.1, non-spatial −25852.3) produced by BAPS consisted of three clusters that corresponded to the three groups in the NJ analysis (Fig. 1). An identical grouping was indicated also by STRUCTURE. The likelihood values for K reached a clear plateau at K =3 (Appendix 3); only “empty” clusters (i.e. with < 0.1% membership) were added when K ≥ 4.
AMOVAs (Table 2) attributed 44.4% of the overall genetic variation to the among-population component. In nested AMOVA analyses, the variation between the three species accounted for 29% and the variation among the three groups indicated by the NJ, PCoA, BAPS and STRUCTURE analyses for 20.1% of the overall variation. Lower values were found for the differentiation between phylogroups E and W (18.9%), and between the Alpine and the Carpathian samples of phylogroup E (12.6%). Two-level AMOVAs calculated for each of the three phylogroups (E, W and S) indicated higher differentiation within the S group.
Table 2.
Analyses of molecular variance (AMOVA) for AFLP phenotypes in Ranunculus alpestris s.l. The letters E, W and S indicate phylogroups (see text)
| Source of variation | d.f. | % of variation | F ST * |
|---|---|---|---|
| Among all populations | 88 | 44.37 | 0.44 |
| Within populations | 174 | 55.63 | |
| Among R. alpestris, R. bilobus and R. traunfellneri | 2 | 29.03 | 0.58 |
| Among populations within groups | 86 | 29.04 | |
| Within populations | 174 | 41.93 | |
|
Among R. alpestris E, R. alpestris W and (R. alpestris S, R.
bilobus and R. traunfellneri) |
2 | 20.14 | 0.50 |
| Among populations within groups | 86 | 30.13 | |
| Within populations | 174 | 49.73 | |
| Among Alpine R. alpestris E and W | 1 | 18.73 | 0.43 |
| Among populations within groups | 69 | 24.56 | |
| Within populations | 139 | 56.71 | |
| Among R. alpestris and (R. bilobus and R. traunfellneri) | 1 | 23.16 | 0.55 |
| Among populations within groups | 87 | 32.18 | |
| Within populations | 174 | 44.66 | |
| Among R. bilobus and R. traunfellneri | 1 | 28.63 | 0.64 |
| Among populations within groups | 5 | 35.21 | |
| Within populations | 14 | 36.16 | |
| Among R. alpestris E and W | 1 | 18.86 | 0.45 |
| Among populations within groups | 76 | 26.72 | |
| Within populations | 152 | 54.41 | |
| Among Alpine and Carpathian populations of R. alpestris E | 1 | 12.57 | 0.40 |
| Among populations within groups | 60 | 27.85 | |
| Within populations | 121 | 59.58 | |
| Among populations of R. alpestris E | 61 | 33.74 | 0.34 |
| Within populations | 121 | 66.26 | |
| Among populations of R. alpestris W | 15 | 28.57 | 0.29 |
| Within populations | 31 | 71.43 | |
| Among populations of R. alpestris S | 10 | 62.60 | 0.63 |
| Within populations | 22 | 37.40 |
All P values were < 0.001.
cpDNA sequence data
Eighteen substitutions out of 1422 aligned positions, plus one coded indel were parsimony informative. Nine haplotypes have been identified within the 39 individual sequences. The method by Dixon (2006, on-line interface available at http://www.botanik.univie.ac.at/plantchorology/haplo.htm), which calculates a posterior probability distribution of the total number of haplotypes, including those which may have not been sampled, indicates a “very likely” (P = 84%) complete haplotype sampling, with a 95% confidence interval of [9, 10] haplotypes. A single most parsimonious tree (50 steps) was found (Fig. 5) with consistency index (CI) of 0.96 and retention index (RI) of 0.99. The R. alpestris group was resolved as monophyletic (BS, 96%). Within the ingroup, two main lineages were found, but none of the three species was monophyletic. Ranunculus alpestris and R. bilobus were partitioned to both clades. The two cpDNA lineages differed by six point mutations (substitutions) out of 1179 bp sequenced from matK. The resulting sequence divergence for the main lineages pair of 0.509% would imply an average divergence time of 2.3 mya, i.e., within the interval 3.3–1.7 mya.
Figure 5.
The minimum-spanning tree (MST, a) and the single most parsimonious tree (MPT, b) of 50 steps (CI = 0.96 and RI = 0.99) found in the analysis of matK data (1423 characters of which 19 were parsimony informative) from selected accessions of the Ranunculus alpestris group (see Appendix 1). Numbers in MST indicate how many individuals were found in the respective group. Numbers above branches in MPT are maximum-parsimony bootstrap (1000 replicates) percentages. The heuristic tree was rooted with R. crenatus, R. parnassifolius and R. aconitifolius as outgroup. The grey arrow indicates a synapomorphic 6 bp deletion in the trnK intron. The black arrow indicates the split between two major haplotype families within the Ranunculus alpestris group estimated to have happened c. 2.3 MYA.
Discussion
Diversification of the Ranunculus alpestris group
Neither the two well-supported clades displayed by cpDNA sequences (Fig. 5) nor the three clusters of individuals evidenced by AFLPs (Figs. 1, 3-4) parallel the recognised taxa of the R. alpestris group. Incongruence between the two markers is caused by the populations at the southern border of the Alpine distribution, an area characterised by (1) the occurrence of all three species, (2) high cpDNA variation compared to the rest of the distribution area (seven haplotypes from both main clades vs. two haplotypes from one clade), and (3) the occurrence of strongly differentiated AFLP lineages (Fig. 3).
One of the main cpDNA lineages was restricted to R. alpestris and one population of R. bilobus (population 84), whereas the second main cpDNA lineage mostly comprised accessions of R. bilobus and R. traunfellneri, but also two populations of R. alpestris from the southeastern Alps (Fig. 5). In the AFLP data, in contrast, R. bilobus and R. traunfellneri formed monophyletic entities, and only the placement of population 19 from the southwestern Alps prevented R. alpestris from being resolved as monophyletic (Fig. 3).
Assuming a proper inference of the mutation rate of matK for Ranunculus (Paun et al. 2005), cpDNA suggests a main divergence event within the R. alpestris group during the Pliocene or early Pleistocene. This correlates with evidence for some European alpine plants that had already started diversifying in the Late Tertiary or Early Quaternary (e.g., Globularia, Soldanella, Gentiana sect. Ciminalis, Kadereit et al. 2004). The split between R. traunfellneri and R. bilobus as well as the formation of phylogroups E and W (see also below) that show no cpDNA haplotype divergence can be safely dated to the Pleistocene, as these events must have happened after the formation of the two cpDNA clades. With a probably complete haplotype sampling (see results), the presence of a synapomorphic 6 bp deletion in the trnK intron in all individuals analysed of R. traunfellneri and its absence in population 83 of R. bilobus (Fig. 5) suggests an allopatric origin of the former from the latter. Recurrent isolation in refugia might have caused this allopatry and was suggested to have strongly fostered allopatric diversification and speciation also in other plant groups that radiated in the Alps (Comes & Kadereit 1998).
There are several potential explanations for the inconsistency between the information from cytoplasmic and nuclear signals, such as differential lineage sorting of ancestral polymorphisms in chloroplast and nuclear genes (Comes & Abbott 2001) or evolutionary convergence (i.e., homoplasy, Davis et al. 1998). The conflict between the present data derived from chloroplast and nuclear genomes, however, is most likely the result of recent chloroplast capture via hybridisation and introgression, as previously reported in numerous taxa such as Eucalyptus, Gossypium, Helianthus, Pinus, and Quercus (Rieseberg & Soltis 1991; Tsitrone et al. 2003). Species of the R. alpestris group are able to hybridise as crossing experiments in all combinations of the three taxa were successful (Müller & Baltisberger 1984). Introgressive hybridisation after allopatric differentiation might have led to transfer of cpDNA from R. alpestris to R. bilobus in the case of population 84. The same but in opposite direction seems to apply to populations 19 and 20 of R. alpestris. Frequent local hybridisation is corroborated by the Bayesian cluster analysis of AFLP data (Fig. 1), which combined individuals from southern Alpine R. alpestris with R. bilobus and R. traunfellneri into one (“panmictic”) gene pool. Mixing of previously isolated populations has long been related to Quaternary climatic changes (e.g., Stebbins’ “secondary contact” model, Stebbins 1984). Independent evidence for hybridisation comes from population 85, which is morphologically intermediate between R. bilobus and R. alpestris (F. Prosser, Rovereto, pers. comm.), but genetically pertains to R. bilobus, as revealed by AFLPs (Fig. 3). In conclusion, our genetic data along with the reciprocal interfertility (Müller & Baltisberger 1984) suggest that the three investigated taxa should be best treated as subspecies, R. alpestris L. subsp. alpestris, R. alpestris L. subsp. traunfellneri (Hoppe) Nyman, and R. alpestris L. subsp. bilobus (Bertol.) Paun comb. nov. (≡ R. bilobus Bertol. in Misc. Bot. 19: 5, 1858, basionym). A thorough taxonomic study in the Southern and South-western Alps is necessary, because more subspecies might be still found there.
Phylogeography of the R. alpestris group in the Alps: southern heterogeneity in refugia vs. northern uniformity
Genetic variation and rarity, the latter measured as frequency-down-weighted marker values which highlight populations with rare markers, show a contrasting pattern in the R. alpestris group in the Alps. High rarity is found in potential refugia (Fig. 2d) whereas high genetic diversity is present in populations from the northern part of the Alps most of which has been fully glaciated, but where R. alpestris is very abundant, populations are large and well connected (Fig. 2a). Patterns of genetic variation seem strongly influenced by present factors such as actual population size which results in a correlation of diversity and regional abundance. Rare markers (e.g., “endemic” fragments) representative for genetic divergence, appear to be a much better indicator of historical processes and are correlated with refugia (Comps et al. 2001; Widmer & Lexer 2001).
The heterogeneous southern/southwestern Alpine phylogroup (S, pops. 17-20, and 83-89) is disjunctly but narrowly following the southern border of the Alps. This phylogroup includes all three taxa of the R. alpestris group. It corresponds well to known refugia, and nowadays, areas of endemism for vascular plants in the Alps (Pawłowski 1970; Tribsch 2004). The southern margin of the Alps has been previously suggested to be one of the main refugia for alpine plants depending on calcareous bedrock (e.g., Tribsch & Schönswetter 2003). Besides exhibiting stronger internal genetic substructure (Table 2; Fig. 3), this group is characterized by a high number of private and rare markers (Fig. 2d, Appendix 1) and high differentiation as a whole (76 of the 260 present fragments in this group are private, Table 1), but rather low levels of within-population genetic diversity (Fig. 2a). Therefore, we favour the view that the genetic pattern in the Southern Alps is mainly a result of diversification due to vicariance in multiple refugia. Although hybridisation of lineages occurred (see above), historical episodic restriction to few high alpine areas produced an isolation effect resulting in high levels of rarity (Fig. 2d). After survival in multiple refugia this group hardly contributed to postglacial colonisation of the Alps.
The Western phylogroup (pops. 1-16) has average levels of diversity and local differentiation (Table 2, Fig. 2a,d). As a group, it has the lowest differentiation (12 private fragments of 190 present, Table 1). The lack of supported substructure (Fig. 3) and the relatively evenly distributed patterns of rarity (Fig. 2d) do not give exact clues for elucidating its glacial history. The group might well have survived the last glaciation in a northern-Alpine peripheral refugium in central Switzerland (refugium V from Schönswetter et al. 2005), where we find at present increased levels of genetic variation (Fig. 2a, Appendix 1). From there genetic diversity decreases in parallel to the presumed migration route towards southwest. A similar pattern has previously been found in Saponaria pumila in the eastern Central Alps (Tribsch et al. 2002). The Northern Swiss Alps had long been postulated to represent a glacial refugium (Briquet 1906) and were suggested to have provided suitable habitats during the last glaciation for two other species (Rumex nivalis, Stehlik 2002; and Erinus alpinus, Stehlik et al. 2002) growing also on calcareous bedrock.
The Eastern phylogroup (E, pops. 21-82) is the geographically most widely distributed of the three AFLP groups, and is generally characterized by the presence of rather high levels of within-population genetic variation (Table 2, Fig. 2a, Appendix 1), and, as a phylogroup, by the highest number of private fragments (93 out of 345 present, Table 1). Both, rarity and genetic diversity show higher levels in the north and east of the Alps and become lower towards the southwest. Moreover, a high level of gene flow among populations and thus rather weak differentiation compared to group S is indicated by the AMOVA results (Table 2). This supports the classical view of an important north-eastern Alpine refugium for calcicolous alpine plants that has been suggested based on paleo-environmental data and patterns of endemism (see e.g., Merxmüller 1954; Tribsch & Schönswetter 2003; Tribsch 2004). Additionally, two localities at the northern border of the Alps (pops. 38 and 52) show increased levels of rarity (Fig. 2d) and are located in formerly less glaciated areas. Based on species’ distribution patterns, it was hypothesised earlier that small areas between glacier tongues have acted as refugia (Merxmüller 1954). Recently, the existence of glacial refugia for calcicolous plants at the northern border of the Alps has been indicated in two studies (Stehlik 2002; Stehlik et al. 2002; see also Schönswetter et al. 2005), one of which postulates a refugium in the Bavarian Alps for Rumex nivalis, close to pop. 38 from this study. It seems, however, that populations in refugia remained less isolated than in the southern part of the Alps and that rapid post-glacial colonisation of Central and Eastern Alps from a main north-eastern refugium has lead to a fairly uniform phylogeographic pattern without internal support of populations/subgroups (Fig. 3).
The Carpathians were step-wise colonised from the Eastern Alps
Phylogroup E comprises also two disjunct areas in the Carpathians with a single cpDNA haplotype (the most common in R. alpestris, Fig. 5). AFLP data suggest that in an early colonisation event R. alpestris reached the Tatra mountains in the Western Carpathians. The populations from the Tatras are not genetically bottlenecked, exhibit a level of rare markers similar to many Alpine populations (Fig 2, Appendix 1) and are somewhat divergent from the Alpine populations of phylogroup E as illustrated by NJ (Fig. 3), PCoA (Fig. 4) as well as AMOVA (Table 2). A similar biogeographic history of Carpathian and Alpine populations has been suggested for another largely calcicole plant species, Pritzelago alpina (Kropf et al. 2003). Range expansion in the opposite direction, i.e. from the Tatras to the Eastern Alps, was previously suggested for the arctic-alpine Comastoma tenellum (Schönswetter et al. 2004), the rare arctic-alpine R. pygmaeus (Schönswetter et al. 2006) and the widespread Arabis alpina (Ehrich & al., 2007). Colonization of the Alps from the Tatras appears, however, unlikely for R. alpestris given the (though unsupported) topology of the NJ dendrogram and the high number of private fragments in the Alpine part of the phylogroup E (65 vs. four in the Tatras, Table 1). The Western Carpathians were only partly glaciated during the Pleistocene, and are loosely connected with the Eastern Alps via a 300 km long ridge with elevations of up to c. 1000 m. Thus, a rather continuous distribution of alpine habitats during glacial periods appears possible. Moreover, a close biogeographic relationship is also documented by a large number of eastern Alpine-Carpathian endemics (e.g., Pawłowski 1970).
The isolated populations in the Southern Carpathians are nested within the samples from the Tatras as a distinct, genetically extremely depauperate group (Appendix 1) with almost absent genetic differentiation (Fig. 2, Table 1). Such a pattern indicates a long-distance dispersal from the Tatras to the Southern Carpathians in fairly recent times, probably during or even after the last glaciation, which was accompanied by a strong founder effect.
Altogether, our data suggest that historical processes mainly shaped allelic richness in the R. alpestris group, while genetic variation is rather reflecting present-day processes such as gene flow. Thus, in genetic fingerprinting studies rarity should be used to unravel refugia rather than genetic diversity. Nevertheless, to fully understand plant evolution and speciation, both aspects have to been seen in a framework of a changing environment caused by climatic fluctuations. To discriminate between hypotheses of ongoing speciation in historical refugia and of gene flow as driver for diversity, other approaches complementing a phylogeographic or phylogenetic approach are needed. Such approaches might involve comparison of the ecology of sympatric and parapatric populations or sister species in order to test, whether these populations are already adapted to their habitat or whether just drift in allopatric refugia has caused the neutral phylogeographic pattern.
Acknowledgements
We thank Thorsten Englisch for help with producing Fig. 1 and for providing frequency data from the unpublished IntraBioDiv floristic database. Harald Niklfeld and Filippo Prosser are thanked for comments on earlier drafts of the manuscript especially on taxonomy, distribution of the taxa and corrections of toponymes. We are grateful to Christian Lexer, Elvira Hörandl and three anonymous referees for commenting on a previous version of the manuscript. Financial support from the European Commission for Science and Research, under the 6th Framework Programme (project STREP, SUSTDEV-2002-3.III.1.4: IntraBioDiv) and from the Austrian Science Fund (Erwin-Schrödinger fellowship to O.P., J26406-B03) is acknowledged.
Appendix 1
Details of the 89 populations of Ranunculus alpestris s.l. investigated in the present study.
Appendix 2
Sampling localities codes indicated with numbers on a geographical map.
Appendix 3
Log-likelihood values of each number of groups (K) as estimated by STRUCTURE 2.0 plotted against the K values. Standard deviation of L(K) between runs at a given K varied between 0 (K = 2) and 33.9 (K = 9).
References
- Baltisberger M. Ranunculus cacuminis and R. crenatus, representatives of the R. alpestris-group (Ranunculaceae) on the Balkan Peninsula. Plant Systematics and Evolution. 1994;190:231–244. [Google Scholar]
- Baltisberger M, Müller M. Comparative cytotaxonomical studies in Ranunculus seguieri and the R. alpestris group. Plant Systematics and Evolution. 1981;138:47–60. [Google Scholar]
- Bennett KD. Continuing the debate on the role of Quaternary environmental change for macroevolution. Philosophical Transactions of the Royal Society of London Series B. 2004;359:295–303. doi: 10.1098/rstb.2003.1395. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bonin A, Bellemain E, Bronken Eidesen P, et al. How to track and assess genotyping errors in population genetic studies. Molecular Ecology. 2004;13:3261–3273. doi: 10.1111/j.1365-294X.2004.02346.x. [DOI] [PubMed] [Google Scholar]
- Briquet J. Le développement des flores dans les Alpe occidentales, avec aperçu sur les Alpes en général. In: Wettstein R, Wiesner J, Zahlbruckner A, editors. Résultats Scientifiques du Congrés International de Botanique Vienne 1905. Fischer; Jena: 1906. pp. 130–172. [Google Scholar]
- Bussell JD, Waycott M, Chappill JA. Arbitrarily amplified DNA markers as characters for phylogenetic inference. Perspectives in Plant Ecology Evolution and Systematics. 2005;7:3–26. [Google Scholar]
- Comes HP, Abbott RJ. Molecular phylogeny, reticulation, and lineage sorting in Mediterranean Senecio sect. Senecio (Asteraceae) Evolution. 2001;55:1943–1962. [PubMed] [Google Scholar]
- Comes HP, Kadereit JW. The effect of Quaternary climatic changes on plant distribution and evolution. Trends in Plant Science. 1998;3:432–438. [Google Scholar]
- Comes HP, Kadereit JW. Spatial and temporal patterns in the evolution of the flora of the European Alpine System. Taxon. 2003;52:451–462. [Google Scholar]
- Comps B, Gömöry D, Letouzey J, Thiébaut B, Petit RJ. Diverging trends between heterozygosity and allelic richness during postglacial colonization in the European beech. Genetics. 2001;157:389–397. doi: 10.1093/genetics/157.1.389. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Corander J, Marttinen P. Bayesian identification of admixture events using multi-locus molecular markers. Molecular Ecology. 2006;15:2833–2843. doi: 10.1111/j.1365-294X.2006.02994.x. [DOI] [PubMed] [Google Scholar]
- Corander J, Sirén J, Arjas E. Bayesian spatial modeling of genetic population structure. Computational Statistics. 2008;23:111–129. [Google Scholar]
- Corander J, Waldmann P, Marttinen P, Sillanpää MJ. BAPS 2: enhanced possibilities for the analysis of genetic population structure. Bioinformatics. 2004;20:2363–2369. doi: 10.1093/bioinformatics/bth250. [DOI] [PubMed] [Google Scholar]
- Corander J, Waldmann P, Sillanpää MJ. Bayesian analysis of genetic differentiation between populations. Genetics. 2003;163:367–374. doi: 10.1093/genetics/163.1.367. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Crisp MD, Laffan S, Linder HP, Monro A. Endemism in the Australian flora. Journal of Biogeography. 2001;28:183–198. [Google Scholar]
- Davis JI, Simmons MP, Stevenson DW, Wendel JF. Data decisiveness, data quality, and incongruence in phylogenetic analyses: an example from the monocotyledons using mitochondrial atpA sequences. Systematic Botany. 1998;47:282–310. doi: 10.1080/106351598260923. [DOI] [PubMed] [Google Scholar]
- Dixon CJ. A means of estimating the completeness of haplotype sampling using the Stirling probability distribution. Molecular Ecology Notes. 2006;6:650–652. [Google Scholar]
- Egger H, Krenmayr HG, Mandl GW, Matura A, Nowotny A, Pascher G, Pestal G, Pistotnik J, Rockenschaub M, Schnabel W. Geological map of Austria (1:1500000) Geological Survey of Austria; Vienna: 2000. [Google Scholar]
- Ehrich D, Gaudeul M, Assefa A, Koch MA, Mummenhoff K, Nemomissa S, IntraBioDiv Consortium. Brochmann C. Genetic consequences of Pleistocene range shifts: contrast between the Arctic, the Alps and the East African mountains. Molecular Ecology. 2007;16:2542–2559. doi: 10.1111/j.1365-294X.2007.03299.x. [DOI] [PubMed] [Google Scholar]
- Excoffier L, Laval G, Schneider S. Arlequin ver. 3.0: An integrated software package for population genetics data analysis. Evolutionary Bioinformatics Online. 2005;1:47–50. [PMC free article] [PubMed] [Google Scholar]
- Felsenstein J. Confidence limits on phylogenetics: an approach using the bootstrap. Evolution. 1985;39:783–791. doi: 10.1111/j.1558-5646.1985.tb00420.x. [DOI] [PubMed] [Google Scholar]
- Frenzel B, Pécsi M, Velichko AA. Atlas of Paleoclimates and Paleoenvironments of the Northern Hemisphere, Late Pleistocene – Holocene. G. Fischer, Stuttgart; 1992. [Google Scholar]
- Gerber J-D, Baltisberger M, Leuchtmann A. Effects of a snowmelt gradient on the population structure of Ranunculus alpestris (Ranunculaceae) Botanica Helvetica. 2004;114:67–78. [Google Scholar]
- Goepfert D. Karyotypes and DNA content in species of Ranunculus L. and related genera. Botaniska Notiser. 1974;127:464–489. [Google Scholar]
- Gugerli F, Tribsch A, Niklfeld H, et al. Relationships among levels of biodiversity and the relevance of intraspecific diversity in conservation a project synopsis. Perspectives in Plant Ecology, Evolution and Systematics. in press. [Google Scholar]
- Hewitt GM. Some genetic consequences of ice ages, and their role in divergence and speciation. Biological Journal of the Linnean Society. 1996;58:247–276. [Google Scholar]
- Hewitt GM. The genetic legacy of the Quaternary ice ages. Nature. 2000;405:907–913. doi: 10.1038/35016000. [DOI] [PubMed] [Google Scholar]
- Hewitt GM. Genetic consequences of climatic oscillations in the Quaternary. Philosophical Transactions of the Royal Society of London, Series B. 2004;359:183–195. doi: 10.1098/rstb.2003.1388. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hörandl E, Paun O, Johansson JT, et al. Phylogenetic relationships and evolutionary traits in Ranunculus s.l. (Ranunculaceae) inferred from ITS sequence analysis. Molecular Phylogenetics and Evolution. 2005;36:305–327. doi: 10.1016/j.ympev.2005.02.009. [DOI] [PubMed] [Google Scholar]
- Huber W. Natural hybridization between white-flowered species of Ranunculus in the Alps. Veröffentlichungen des Geobotanischen Institutes der ETH, Stiftung Rübel. 1988;100:1–160. [Google Scholar]
- Kadereit JW, Griebleler EM, Comes HP. Quaternary diversification in European alpine plants: pattern and process. Philosophical Transactions of the Royal Society of London, Series B. 2004;359:265–274. doi: 10.1098/rstb.2003.1389. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Körner C. Alpine Plant Life. Springer; Berlin: 2003. [Google Scholar]
- Kropf M, Kadereit JW, Comes HP. Late Quaternary distributional stasis in the submediterranean mountain plant Anthyllis montana L. (Fabaceae) inferred from ITS sequences and amplified fragment length polymorphism markers. Molecular Ecology. 2002;11:447–463. doi: 10.1046/j.1365-294x.2002.01446.x. [DOI] [PubMed] [Google Scholar]
- Kropf M, Kadereit JW, Comes HP. Differential cycles of range contraction and expansion in European high mountain plants during the Late Quaternary: insights from Pritzelago alpina (L.) O. Kuntze (Brassicaceae) Molecular Ecology. 2003;12:931–949. doi: 10.1046/j.1365-294x.2003.01781.x. [DOI] [PubMed] [Google Scholar]
- Merxmüller H. Untersuchungen zur Sippengliederung und Arealbildung in den Alpen. III. Jahrbuch des Vereins zum Schutze der Alpenpflanzen und Tiere. 1954;19:97–139. [Google Scholar]
- Mráz P, Gaudeul M, Rioux D, Gielly L, Choler P, Taberlet P, IntraBioDiv Consortium Genetic structure of Hypochaeris uniflora (Asteraceae) suggests vicariance in the Carpathians and rapid post-glacial colonization of the Alps from an eastern Alpine refugium. Journal of Biogeography. 2007;34:2100–2114. [Google Scholar]
- Müller M, Baltisberger M. Cytotaxonomical studies in the group of Ranunculus alpestris (Ranunculaceae) Plant Systematics and Evolution. 1984;145:269–289. [Google Scholar]
- Müller-Schneider P. Diasporology of the Spermatophytes of the Grisons (Switzerland) Veröffentlichungen des Geobotanischen Institutes der ETH, Stiftung Rübel. 1986;85:1–263. [Google Scholar]
- Nei M. Molecular Evolutionary Genetics. Columbia University Press; New York: 1987. [Google Scholar]
- Nei M, Li WH. Mathematical model for studying genetic variation in terms of restriction endonucleases. Proceedings of the National Academy of Sciences, USA. 1979;76:5269–5273. doi: 10.1073/pnas.76.10.5269. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Niklfeld H. Bericht über die Kartierung der Flora Mitteleuropas. Taxon. 1971;20:545–571. [Google Scholar]
- Ozenda P. Die Vegetation der Alpen im europäischen Gebirgsraum. Gustav Fischer, Stuttgart; 1988. [Google Scholar]
- Paun O, Lehnebach C, Johansson JT, Lockhart PJ, Hoerandl E. Phylogenetic relationships and biogeography of Ranunculus and allied genera (Ranunculaceae) in the Mediterranean region and in the European Alpine System. Taxon. 2005;54:911–930. [Google Scholar]
- Pawłowski B. Remarques sur l’endémisme dans la flore des Alpes et des Carpates. Vegetatio. 1970;21:181–243. [Google Scholar]
- Pritchard JK, Stephens M, Donnelly P. Inference of population structure using multilocus genotype data. Genetics. 2000;155:945–959. doi: 10.1093/genetics/155.2.945. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rieseberg LH, Soltis DE. Phylogenetic consequences of cytoplasmic gene flow in plants. Evolutionary Trends in Plants. 1991;5:65–84. [Google Scholar]
- Rohlf FJ. NTSYS-Pc: Numerical Taxonomy and Multivariate Analysis System, Version 2.0. 2. Exeter Software; New York, USA: 1997. [Google Scholar]
- Schönswetter P, Popp M, Brochmann C. Rare arctic-alpine plants of the European Alps have different immigration histories: the snow bed species Minuartia biflora and Ranunculus pygmaeus. Molecular Ecology. 2006;15:709–720. doi: 10.1111/j.1365-294X.2006.02821.x. [DOI] [PubMed] [Google Scholar]
- Schönswetter P, Stehlik I, Holderegger R, Tribsch A. Molecular evidence for glacial refugia of mountain plants in the European Alps. Molecular Ecology. 2005;14:3547–3555. doi: 10.1111/j.1365-294X.2005.02683.x. [DOI] [PubMed] [Google Scholar]
- Schönswetter P, Tribsch A. Vicariance and dispersal in the alpine perennial Bupleurum stellatum L. (Apiaceae) Taxon. 2005;54:725–732. [Google Scholar]
- Schönswetter P, Tribsch A, Niklfeld H. Amplified fragment length polymorphism (AFLP) suggests old and recent immigration into the Alps by the arctic-alpine annual Comastoma tenellum (Gentianaceae) Journal of Biogeography. 2004;31:1673–1681. [Google Scholar]
- Schroeter C. Das Pflanzenleben der Alpen. Verlag von Albert Raustein; Zürich, Switzerland: 1926. [Google Scholar]
- Simmons MP, Ochoterena H. Gaps as characters in sequence-based phylogenetic analyses. Systematic Biology. 2000;49:369–381. [PubMed] [Google Scholar]
- Skrede I, Bronken Eidesen P, Piñeiro Portela R, Brochmann C. Refugia, differentiation and postglacial migration in arcticalpine Eurasia, exemplified by the mountain avens (Dryas octopetala L.) Molecular Ecology. 2006;15:1827–1840. doi: 10.1111/j.1365-294X.2006.02908.x. [DOI] [PubMed] [Google Scholar]
- Soltis DE, Gitzendanner MA, Strenge DD, Soltis PS. Chloroplast DNA intraspecific phylogeography of plants from the Pacific Northwest of North America. Plant Systematics and Evolution. 1997;206:353–373. [Google Scholar]
- Soltis DE, Morris AB, McLachlan JS, Manos PS, Soltis PS. Comparative phylogeography of unglaciated eastern North America. Molecular Ecology. 2006;15:4261–4293. doi: 10.1111/j.1365-294X.2006.03061.x. [DOI] [PubMed] [Google Scholar]
- Stebbins GL. Polyploidy and the distribution of the arctic-alpine flora: new evidence and a new approach. Botanica Helvetica. 1984;94:1–13. [Google Scholar]
- Stehlik I. Glacial history of the alpine herb Rumex nivalis (Polygonaceae): a comparison of common phylogeographic methods with nested clade analysis. American Journal of Botany. 2002;89:2007–2016. doi: 10.3732/ajb.89.12.2007. [DOI] [PubMed] [Google Scholar]
- Stehlik I, Schneller JJ, Bachmann K. Immigration and in situ glacial survival of the low-alpine Erinus alpinus (Scrophulariaceae) Biological Journal of the Linnean Society. 2002;77:87–103. [Google Scholar]
- Swofford DL. PAUP*:Phylogenetic Analysis Using Parsimony (*and other methods), Version 4.0b10. Sinauer Associates; Sunderland, MA: 2003. [Google Scholar]
- Taberlet P, Fumagalli L, Wust-Saucy A-G, Cosson J-F. Comparative phylogeography and postglacial colonization routes in Europe. Molecular Ecology. 1998;7:453–464. doi: 10.1046/j.1365-294x.1998.00289.x. [DOI] [PubMed] [Google Scholar]
- Tribsch A. Areas of endemism of vascular plants in the Eastern Alps in relation to Pleistocene glaciation. Journal of Biogeography. 2004;31:747–760. [Google Scholar]
- Tribsch A, Schönswetter P. Patterns of endemism and comparative phylogeography confirm palaeo-environmental evidence for Pleistocene refugia in the Eastern Alps. Taxon. 2003;52:477–497. [Google Scholar]
- Tribsch A, Schönswetter P, Stuessy TF. Saponaria pumila (Caryophyllaceae) and the ice age in the European Alps. American Journal of Botany. 2002;89:2024–2033. doi: 10.3732/ajb.89.12.2024. [DOI] [PubMed] [Google Scholar]
- Tsitrone A, Kirkpatrick M, Levin DA. A model for chloroplast capture. Evolution. 2003;57:1776–1782. doi: 10.1111/j.0014-3820.2003.tb00585.x. [DOI] [PubMed] [Google Scholar]
- Van de Peer Y, De Wachter R. Construction of evolutionary distance trees with TREECON for Windows: accounting for variation in nucleotide substitution rate among sites. Computer Applications in Biosciences. 1997;13:227–230. doi: 10.1093/bioinformatics/13.3.227. [DOI] [PubMed] [Google Scholar]
- Vos P, Hogers R, Bleeker M, et al. AFLP – a new technique for DNA–fingerprinting. Nucleic Acids Research. 1995;23:4407–4414. doi: 10.1093/nar/23.21.4407. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Widmer A, Lexer C. Glacial refugia: sanctuaries for allelic richness, but not for gene diversity. Trends in Ecology and Evolution. 2001;16:267–269. doi: 10.1016/s0169-5347(01)02163-2. [DOI] [PubMed] [Google Scholar]
- Wertlen AM. Evolution of Flower Colours: Choice Strategies of Pollinating Hymenoptera as Selection Factors. Free University of Berlin; 2006. PhD Dissertation. [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.