Summary
Current research suggests that a number of newly identified helper T-cell subsets retain a degree of context-dependent plasticity in their signature cytokine expression patterns. To understand this process, a major challenge is to determine the molecular mechanisms by which lineage-defining transcription factors regulate gene expression profiles in helper T cells. This mechanistic information will aid in our interpretation of whether a T-helper cell state that expresses or retains the capacity to re-express a combination of lineage-defining transcription factors will have a stable or more flexible gene expression profile. Studies examining the developmental T-box transcription factor T-bet demonstrate the powerful information that is gained from combining in vivo analysis with basic biochemical and molecular mechanism approaches. Significantly, T-bet's ability to physically recruit epigenetic modifying complexes, in particular a Jmjd3 H3K27-demethylase and a Set7/9 H3K4-methyltransferase complex, to its target genes allows T-bet to effectively reverse and establish new epigenetic states. This observation suggests that until T-bet is permanently extinguished, helper T cells will retain some plasticity towards a T-helper 1-like program. Therefore, insight into the complexity of T-helper cell commitment decisions will be aided by determining the molecular mechanisms for lineage-defining transcription factors.
Keywords: T-bet, T-helper cell, lineage-defining transcription factors, epigenetic, chromatin
Introduction
Each cell in the body starts out with the same DNA or genetic material, yet how that material is expressed in specific cell types forms the basis for the functional capacity of an individual cell. This concept is apparent when thinking about the diverse cell types that make up a fully functioning human being. Without the proper expression of the genome in a cell type and activation state appropriate manner, diverse organ systems cannot develop and perform their precise effector functions. Therefore, defining the mechanisms that are responsible for the appropriate expression profile of the genome in a temporal and cell type-dependent manner is a critical area of research.
Cellular differentiation involves a complex series of events that progressively commits a cell to a defined lineage. The precise mechanisms by which this occurs are still being uncovered, but we have made great progress over the last several years. As discussed in more detail below, the mechanisms that help to define developmentally appropriate gene expression patterns appear to be conserved in a number of different organ systems. That is, mechanisms that are being uncovered in the immune system are also utilized in early embryonic development and vice versa (1, 2). This is an extremely important point, because much knowledge has already been gained examining a variety of developmental systems and each one provides valuable information about unique aspects of the mechanistic events that are required for cellular differentiation. Indeed, insight into the complex issue of helper T-cell lineage plasticity can be gained from combining information known about early embryonic development with our knowledge of T-helper cells.
What defines a lineage?
In recent discussions in immunology, the question of what characteristics define a lineage have become somewhat blurred for helper T-cell subtypes (3-5). Previously, the paradigm in the field was that naive CD4+ T cells differentiate into either the T-helper 1 (Th1) or Th2 lineage in response to environmental cues (6). Th1 cells secrete interferon-γ (IFNγ) to communicate with other cells of the immune system to aid in the clearance of intracellular pathogens, while Th2 cells secrete interleukin-4 (IL-4), IL-5, and IL-13 to help alleviate extracellular infections. This paradigm correlated with the data garnered from in vivo pathogen clearance studies and disease models mimicking dysregulated immune responses. However, several lines of evidence have now suggested that the Th1/Th2 paradigm is too simplistic and cannot fully account for the complex series of events that occur to both clear a pathogen and also dampen the immune response to prevent autoimmune states. Namely, the T-helper cell cytokine expression profile that is responsible for regulating an appropriate immune response involves many more players than those originally recognized as the prototypic Th1 and Th2 cytokines (7). Emerging evidence suggests that numerous other cytokines including IL-17, transforming growth factor β (TGFβ), and IL-21 to name just a few, are all required to mount a proper immune response, and the expression pattern of these cytokines does not fall neatly within the Th1/Th2 paradigm (5, 8).
The discovery that categorizing helper T cells is not as simple as two lineages composed of an IFNγ-producing Th1 cell and an IL-4-producing Th2 cell has led to a reexamination of our views on helper T-cell differentiation in general (4). One thing that has become clear is that there are a number of overlapping cytokine expression patterns that can be produced depending on the acute and sustained environmental signals that each cell receives. For instance, CD4+ or CD8+ T cells can express IFNγ and IL-17 in either a mutually exclusive or simultaneous manner dependent upon the circumstances (3, 9, 10). Significantly, this phenomenon has also been observed for IFNγ and IL-4 as well as IL-17 and IL-9 (11-13). It has been tempting to define each of these helper T-cell populations as a newly defined lineage, but with the ever-expanding number of overlapping patterns of cytokines, it may be time to reassess defining a stable T-helper cell lineage based solely upon the expression of a few signature genes. Another way to start to address this complex issue is through a more comprehensive examination of the molecular series of events that establish the gene expression programs found in each subset (14). This will allow us to assess whether the cell is merely altering the expression of a small number of genes to respond to a new stimulus in its immediate environment or rather it is stably changing its gene expression profile in a heritable way to become a new lineage. Importantly, the transcription factors that regulate these processes have been identified for the major subsets of helper T cells. Below, we discuss in detail this topic from the perspective of the lineage-defining transcription factor T-bet, which is critical for Th1 cells.
Molecular mechanisms that define lineage commitment
There are a number of molecular events that need to occur at cellular transitions to progressively commit a cell to a terminally differentiated lineage with distinct effector cell functions. Significantly, this series of events must result in alterations to the packaging of the genome so that the expression profile of the cell will be heritable and will not revert back to a previous more pluripotent state (15). Examples of this can be drawn from early embryogenesis, where the expression of the combination of key transcription factors that are required for the pluripotent nature of a stem cell are permanently repressed as cells exit this uncommitted state and begin to progressively commit to the lineages that define each organ system (16, 17). This is a very important point, because the reversion of gene expression profiles to a less differentiated state can result in uncontrolled cellular proliferation and cancer (18, 19). However, the commitment of a cell into a terminally differentiated lineage does not necessitate a completely inflexible gene expression program. Indeed, terminally committed effector cells still respond to environmental cues and these responses require minor changes in the epigenetic environment and the genes that are expressed in the cell. Therefore, a certain degree of flexibility in gene expression patterns occur in all cell types, even those that are considered a fully committed lineage.
Epigenetic regulation
Advances in epigenetic research have aided in our understanding of the events that are required to establish cell lineages. Epigenetic control, including DNA methylation, nucleosome positioning, and histone modifications, regulates gene expression independent from the underlying DNA sequence per se. That is, epigenetic regulation relies primarily on creating states of chromatin that differentially affect accessibility to DNA-binding sites. The genome is packaged into the nucleus by wrapping the DNA around a histone octamer composed of two subunits each of histone H2A, H2B, H3, and H4. This basic unit of chromatin creates a dynamic template to read the genome in a precise and regulated manner. The N-terminal tails of the histone proteins extend beyond the globular core that the DNA is wound around, and these tails can be covalently modified. Several modifications have been described on both the H3-H4 tetramer and the H2A-H2B dimers that comprise the histone octamer (20). Covalent modifications that have been characterized at this point include acetylation, methylation, phosphorylation, ubiquitination, and sumoylation (21).
Thhe majority of research on histone modifications has focused on the function of acetylation and methylation as well as the enzymes that catalyze the addition and removal of these marks. Acetyl modifications are added to lysine residues by histone acetyltransferases (HATs) and are removed by deacetylases (HDACs) (20). These proteins are frequently found in larger protein complexes that change the chromatin environment in multiple ways. HATs are often associated with the transcription machinery itself, and HDACs are often found in polycomb complexes (22-24). While the presence of acetylation is most commonly associated with accessible or active regions of the genome, histone methylation is far more complicated (25-27). Lysine residues in the histone tails can be mono-, di-, or tri-methylated, and the number of methyl moieties present as well as the specific residue modified have been differentially linked with either permissive or repressive chromatin. For example, trimethylation of lysine 4 of histone 3 (H3K4me3) is associated with active transcription (28-30). Conversely, H3K27 di- and trimethylation is linked to repressed genes and heterochromatin (31-33). As briefly highlighted, there are a number of mechanisms by which the chromatin environment can be regulated to achieve specific gene expression patterns, but the focus here is on the role that two specific histone tail modifications, H3K4-methylation and H3K27-methylation, play in this process. Understanding how these modifications contribute to regulating both environmentally dynamic and developmentally appropriate gene expression patterns requires determining the functional significance for these modifications as well as the mechanisms by which they are established. Both of these topics are challenging to address and are areas of ongoing studies.
Research profiling the epigenetic modifications that are found in embryonic stem cells and the changes that occur as a cell commits to a more differentiated state have provided much insight into the nature of histone tail modifications during cellular commitment (16, 34). The correlation between H3K4-methylation and active transcription as well as H3K27-methylation and inactive genes is well established. The current view based upon available data is that lineage-specific genes are inactive in multi-potential progenitor cells, because they are marked by repressive epigenetic states such as H3K27-trimethylation (H3K27me3) (35). During developmental transitions, the repressive H3K27me3 modification is removed, and the active H3K4me3 modification is added. Thus, changes in the epigenetic state coincide with the developmentally appropriate expression of the gene. However, unbiased genome wide epigenetic profiling of embryonic stem cells has revealed that these correlations are not so simple. Interestingly, in stem cells, the genes for some lineage-defining transcription factors required for later developmental transitions are contained within regions displaying both the H3K27me3 and H3K4me3 modifications (16, 34). This composition of modifications has been termed a bivalent domain and is thought to represent a poised state that can be resolved into either permissive or repressive chromatin, depending upon the environmental stimuli or developmental cue. Another finding from unbiased studies that combine histone modifications with gene expression profiling is that there are several genes that are depleted of repressive modifications and not expressed (18, 36). The opposite also occurs where promoters have permissive marks but gene expression remains silent (18, 36). Taken together, unbiased genome-wide studies have provided much insight into epigenetic states during development but have also made it clear that the regulation of the chromatin environment is a very complex process.
Histone methyltransferases and demethylases
To better understand histone methylation modifications and their contribution to gene regulation, a great deal of research has been focused on the enzymes that catalyze the addition or removal of the methyl group. Histone methyltransferases have long been recognized as the enzymes responsible for the addition of methyl modifications to specific residues within the histone tails (28, 37-39). Not only are these proteins specific for individual lysines, but they can also be restricted in the number of methyl moieties they can add. Methyltransferases are defined by a SET protein domain that catalyzes the methylation reaction, but they frequently contain additional domains (21, 28). Many of these regions mediate interactions with other proteins that are important in gene regulation. Indeed, methyltransferases are found in a variety of large multi-protein complexes, usually with similar types of functional activities contained within the same complex. For example, EZH2, the methyltransferase that catalyzes the repressive H3K27me3 modification, is a member of the polycomb group (PcG) complex which is best known for silencing Hox genes during embryogenesis (24, 32, 40). In addition, SET proteins that contribute to the activation of genes have been detected in protein complexes with other histone-modifying enzymes including those that remove repressive methyl marks and that add the permissive acetyl modification (41, 42). Importantly, the characterized methyltransferases are not known to have specific DNA recognition sequences in mammalian cells. Therefore, uncovering the mechanisms by which they interact with DNA binding transcription factors becomes integral in understanding how these catalytic activities are targeted to the proper genomic locations at developmental transitions.
The recent discovery of the demethylases, starting with LSD1 and then expanding to the Jumonji family, revealed the dynamic nature of histone methylation (21, 43-51). Like the methyltransferases, demethylases are also highly specific for the individual lysine residues and the number of methyl groups they remove (42, 43, 46, 48, 52, 53). Although defined by the catalytic Jmjc domain, these proteins tend to be very large and often contain several different protein domains (46). Some of these domains have been shown to recognize general aspects of the DNA or chromatin, such as AT-rich repeats or specific histone modifications (46). However, like other histone-modifying proteins, none of these domains are known to bind to specific DNA sequences. This makes it extremely unlikely that demethylases individually target themselves to specific subsets of genes in a developmentally appropriate manner. Like the methyltransferases, demethylase proteins have been detected in multimolecular complexes. UTX, a demethylase that removes the repressive H3K27-methyl modification, has been found in complex with MLL proteins that catalyze the addition of permissive methyl modifications (41). These types of dual complexes have the potential to be very powerful, because they have the ability to both resolve a repressive modification and transition to a more permissive chromatin environment. It is even more critical for the cell to target such complexes appropriately or risk widespread dysregulation of gene expression patterns. Therefore, an important area of ongoing research is identifying the specific transcription factors that target these complexes to regulatory regions. As will be discussed below, studies in helper T-cell differentiation examining the lineage-defining T-box transcription factor family member T-bet has shed light on this topic.
Lineage-defining transcription factors
Lineage-defining transcription factors control the differentiation of individual cell types and are responsible for regulating a very precise gene expression program. These proteins are fundamentally different from general transcription factors, in that they must be able to both activate lineage-specific genes as well as repress genes for opposing lineages. In many circumstances, lineage-defining transcription factors create chromatin environments de novo. That is, other proteins that contribute to gene expression typically achieve their functions in the context of promoter or enhancer regions that have already been made accessible by other means. However, lineage-defining transcription factors must by definition be able to bind to repressed targets and induce changes to the chromatin environment of its target promoters in order to make the binding sites of other factors available. In addition to this critical role at transition points, many of these transcription factors function in the developed lineage as well. The functional requirements for gene activation and repression are likely different at the developmental transition versus within the committed cell. This suggests that lineage-defining proteins might have multiple functions that are differentially utilized when they are establishing versus maintaining gene expression profiles.
Identifying the transcription factors required to establish specific cell types has been a major focus for developmental biology research (54-57). Mutations in these factors are also commonly found in developmental diseases. Currently, there is a great deal of experimental data identifying the key transcription factor(s) for cells at every level of development ranging from embryonic stem cells to cardiac cells, neurons, and various immune lineages (55, 57-63). A surprisingly small number of transcription factor families control the majority of cell fate decisions, and members of these families tend to be commonly utilized to regulate similar aspects of diverse developmental transitions. One such example of this is the T-box transcription factor family, which has family members that regulate aspects of almost every developmental system (see below). However, although key lineage-defining transcription factors for a variety of cell types have been identified, the molecular mechanisms by which they regulate developmental transitions in many cases have not yet been well defined. There has been some insight into the specific cofactors utilized by some of the known lineage-defining transcription factors, but creating a comprehensive profile of the functional activities coordinated by an individual factor or a family of factors has been challenging.
Cellular differentiation in the immune system
Research in the immune system has clear relevance to human health for our understanding of infectious diseases and autoimmunity, but the immune system also provides a unique model for studying cellular differentiation. There are multiple well-defined lineages that develop from a single hematopoietic stem cell, including CD4+ and CD8+ T cells, B cells, NK, and NKT cells and dendritic cells (64-67). Not only do these lineages differentiate in response to pathogens within the context of the whole animal, but cells can also be isolated, skewed toward an effecter type, and expanded in tissue culture systems for study. In addition, there are well-defined cell lines that are very amenable to experimental manipulation. This combination of experimental tools provides a means to uncover molecular aspects of developmental processes, and the opportunity to determine how these mechanisms are utilized in a physiological setting.
Many of the cell lineages in the immune system can develop into multiple, distinct subsets in response to specific environmental stimuli. For example, CD4+ T cells can be subdivided into several effecter cell types (3, 5). The development of each of these sub-lineages depends on the integrated action of key transcription factors. Similar to other developmental systems, no one transcription factor acting alone can ensure the proper patterning of gene expression for a defined cell type (14, 68-71). This makes it imperative to understand the molecular capabilities of each factor and how they might work in combination to determine what encompasses a true lineage. This will provide a way to mechanistically determine how a lineage is established, and in turn, understand which cell subsets encompass distinct lineages versus effector subsets.
Key lineage-defining transcription factors in CD4+ T cells
Several transcription factor families with known functions in development also play a role in CD4+ T-cell differentiation. In part, the definition for distinct CD4+ T-cell subsets has come from the identity of the lineage-defining transcription factors that are required for that subset to be produced. Namely, the T-box transcription factor T-bet is required for Th1 cells, GATA3 is important for Th2 cells, RORγt is needed for Th17 cells, Foxp3 is required for regulatory T cells, and Bcl6 is important for follicular helper T-cell development (5, 62, 72-75). The predominance of these factors in specific CD4+ T-cell populations has helped to solidify the hypothesis that there are distinct terminally committed effector cell types. However, recent studies have shown that the expression of these lineage-defining transcription factors are not entirely mutually exclusive, but instead their expression profiles overlap in a number of circumstances (3). As discussed in more detail below, if we understand the mechanisms by which these lineage-defining transcription factors establish gene expression profiles, we can then predict the functional outcome for their expression in an unexpected subset. Importantly, this will shed light into the potential plasticity for the cells in which they are expressed. To illustrate the utility of using molecular mechanisms to define helper T-cell differentiation, we will outline our current understanding of how T-bet regulates gene expression patterns and the implications this has on helper T-cell plasticity.
T-bet's role as a lineage-defining transcription factor
T-bet, or T-box expressed in T cells, was first characterized in a screen for transcription factors that were differentially expressed in Th1 versus Th2 cells (62). Further analysis revealed that T-bet is absolutely required for the differentiation of Th1 cells as defined by the expression of signature Th1 genes including the cytokine Ifng and the cell surface receptor Cxcr3 (68, 76-81). The importance of T-bet's role in immune responses has been demonstrated in several different autoimmune and infectious disease models in inbred mouse strains (78, 80-85). Dysregulation of T-bet has also been associated with the development of human autoimmune diseases including asthma, colitis, and diabetes (86-90). However, these disease states may not be entirely caused by malfunctioning Th1 cells.
T-bet is also expressed in a variety of immune cells and plays critical roles in every cell type where it has been detected. T-bet has been shown to be important for class switching in B cells, NK cell development, and dendritic cell function (62, 68, 78, 80, 82, 91-94). It was hypothesized that T-bet would have different target genes in each of these cell types in order to achieve these disparate functions. Surprisingly, T-bet was found to bind to the same target genes in all cell types examined (77). However, binding to the promoter did not completely correlate with changes in gene expression (76, 77). Therefore, T-bet must be involved in aspects of gene regulation that occur downstream of the DNA-binding event that are required to induce the expression of its target genes in a cell type-specific manner. A more comprehensive understanding of potential interactions between T-bet and cell-type specific cofactors, including but not limited to chromatin remodeling complexes (see below), may provide insight into this topic.
T-box transcription factors
T-bet is a member of the T-box transcription factor family. The T-box family of developmental transcription factors is one group of proteins that has conserved roles in body patterning and organogenesis from fish to humans (95, 96). These proteins are critically involved in the development of numerous tissues as has been demonstrated by the association of mutations in T-box factors with human genetic diseases resulting in birth defects (97-104). The tissue where each T-box protein is expressed predicts the type of disease associated with that factor's mutation. For example, there are many T-box factors involved in the development of cardiac cells and the patterning of the heart. Tbx1, Tbx3, Tbx5, and Tbx20 are all expressed in the developing heart and mutations in each of these genes have been associated either with congenital heart defects in isolation or as part of larger syndromes (54, 95, 97, 98, 100-102, 104-106). In addition, genetic knockouts of T-box factors in mice frequently result in an embryonic lethal phenotype (95). Here again, the embryo arrests at the stage where the tissue type should have formed. Taken together, this high level of association with developmental diseases makes the T-box family important for study and a good model for determining the mechanisms of lineage-defining transcription factors in general.
The T-box family is defined by the central DNA-binding domain that is also termed the T-box (95). This region is large, comprising approximately 180 amino acids of the protein (95). The DNA contact residues are highly conserved, and outside of these amino acids there are both regions of complete homology as well as sequence divergence (2). It is also important to note that the N- and C-termini of T-box proteins are more variable between factors both in sequence and overall length than the conserved T-box domain itself (76). This combination of conserved and divergent regions strongly suggests that T-box factors will have both common and factor-specific functions in gene regulation. Indeed, this must be the case, because several T-box factors are expressed in overlapping patterns in the development of the limb buds where they have non-redundant roles (95, 107, 108).
The high level of conservation in the DNA contact residues accounts for the fact that all T-box factors recognize the same consensus sequence termed the T-site (95). Although there have been some reports that different T-box factors might have slight preferences for different sequences, several divergent T-box factors have been shown to bind the same promoter regions (2, 76, 77). However, not every T-box factor can activate gene expression at all of the promoters where it binds (76, 77). This finding suggests that there are events independent of DNA binding that are required for gene regulation. In the immune system, two related T-box factors, T-bet and Eomesodermin (Eomes), have some common functions but cannot fully compensate for each other (61, 93). T-bet is exclusively found in immune cells, while Eomes has roles in the immune system as well as other organ systems (61, 62, 68, 109, 110). In immune cells, both T-bet and Eomes can bind to the Ifng promoter and induce its transcription to a degree (61, 68, 76, 77, 93). However, Ifng production and Th1 cell development is severely impaired in T-bet-deficient mice, even though there is still some expression of Eomes in CD4+ T cells (62, 68). In contrast, CD8+ T cells produce almost normal levels of Ifng in T-bet-deficient mice but are completely deficient in the absence of both T-bet and Eomes (111). These data suggest a context-dependent quality for the compensation between T-bet and Eomes in T cells and that there are both common and differential mechanisms by which T-bet and Eomes regulate gene expression.
Mechanisms by which T-bet establishes appropriate gene expression states
To understand how interactions between transcription factors and their cofactors might inhibit or enhance their activities, it is important to characterize the mechanisms that have the potential to be altered. Since the binding of T-bet and other T-box factors to DNA is not alone sufficient to induce gene expression changes (76, 77), epigenetic alterations represent the next level of regulatory control for this family. There have been studies showing that alterations in the chromatin environment at T-bet target promoters occur in a T-bet-dependent manner (2, 76, 112-114). Both permissive acetylation and methylation modifications are altered in a manner that is consistent with the T-bet-dependent activation of prototypic targets. Specifically, H3K9-acetylation increases and the permissive H3K4me2 modification, but not H3K4me3, is enhanced when T-bet is present (76, 77, 114-116). It is equally possible that epigenetic changes are either directly modulated by a required transcription factor or that they are merely coincident with active transcription. Therefore, one of the open questions has been whether T-bet physically interacts with proteins that catalyze these changes or if T-bet is only functionally required for an upstream event. Also, it is unclear whether changes in histone modifications are necessary for T-bet's ability to activate target genes. It is possible that these changes typically occur at activated target genes but are dispensable for transcription. There is precedence for this possibility based upon the data from genome-wide studies where histone modification changes have been observed in the absence of any detectable functional consequence (18, 26, 27, 34, 36, 117). Another consideration is that histone modifications are only one aspect of gene regulation, so that these changes are needed but not sufficient for promoter activation. Until more is understood about the functions of histone modifications and the enzymes that catalyze them, this will remain unclear. The studies with T-bet and the T-box protein family have been informative in beginning to address these topics (2, 76, 77).
T-bet's role in recruiting epigenetic modifying complexes during Th1 differentiation
A structure-function analysis revealed that in addition to family member-specific transactivation potential, T-bet and other T-box factors are able to functionally recruit H3K4me2-methyltransferase activity to target promoters (76). Interestingly, this is due to a conserved physical interaction between the T-box family and an H3K4me2-methyltransferase complex. This interaction localizes to the T-box DNA binding domain (2, 76). The conservation within this region likely accounts for the shared capability of T-box factors to induce this permissive modification. It also presents an evolutionary rationale for maintaining such a large DNA-binding domain. Namely, the T-box domain is 180 amino acids, and the majority of these residues are not directly involved in DNA contacts. Coupling an interaction with histone-modifying activity to the DNA-binding domain is a powerful way to ensure that a developmental transcription factor will be able to specifically target the activity to regulatory regions. Therefore, this exciting finding demonstrating that T-bet plays a role in functionally regulating epigenetic events in the immune system has provided insight into a common mechanism that regulates developmental transitions. Indeed, current data highly suggest that this is a conserved activity for the T-box family in development (2).
The regions within the T-box domain that are required for the interaction with H3K4me2-methyltransferase activity are separable from the DNA contact residues. To define these regions, the location of mutations associated with T-box-mediated genetic diseases as well as areas of high conservation were examined (2). The rationale for this approach was that mutations in the T-box domain that result in developmental diseases but do not impede DNA binding are likely to disrupt the required functional activity to modify the epigenetic environment. This analysis revealed two distinct regions in the T-box, which were termed T-box domain 1 and 2, that are located on the opposite face from the DNA contact residues (2). Both domains are in fact required for the functional addition of the H3K4me2 modification, but surprisingly, only T-box domain 1 is necessary for the physical interaction with the H3K4-methyltransferase complex (2). These data suggested that another required functional activity that precedes the establishment of the permissive H3K4me2 state localizes to T-box domain 2. This means that two functional activities that effectively alter the epigenetic state of regulatory regions are spatially contained within the T-box DNA-binding domain.
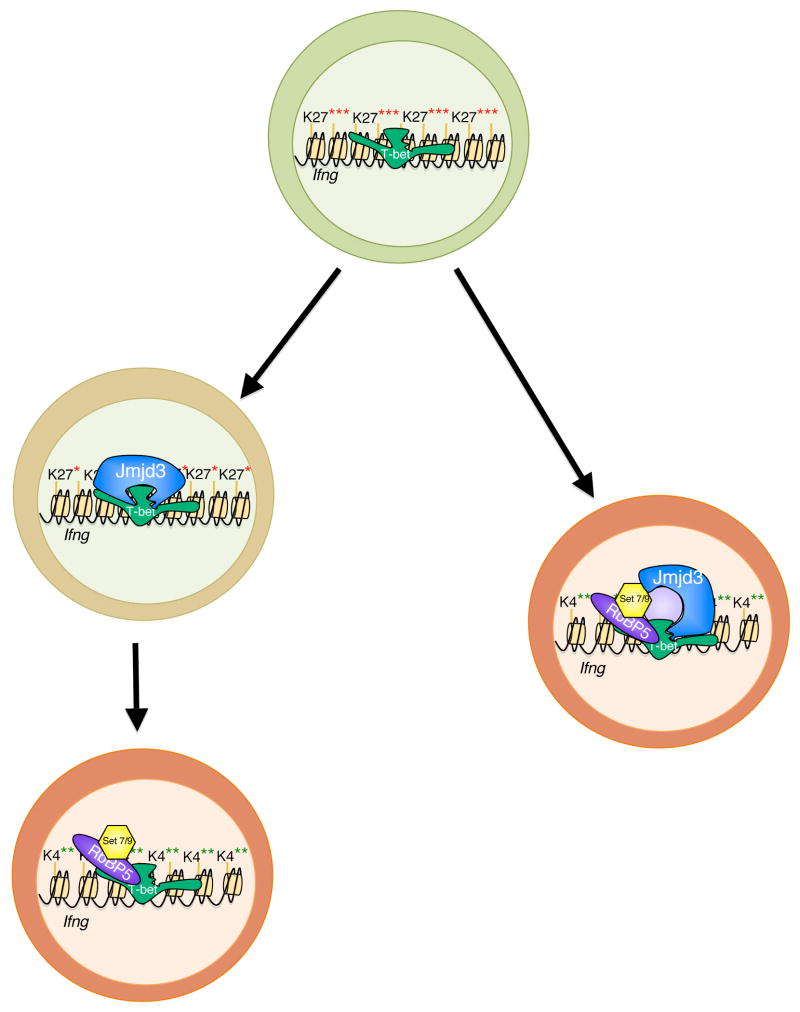
These data led to the hypothesis that T-box domain 2 is required to mediate the functional removal of the repressive H3K27me3 modification as a prerequisite to the addition of the permissive H3K4me2 modification. To establish a permissive chromatin state, especially at developmental transitions, repressive chromatin modifications must first be removed. The repressive H3K27me3 modification is preferentially found at the Ifng promoter in Th2 versus Th1 cells (118). Significantly, T-bet is able to mediate the removal of the repressive H3K27me3 mark at the Ifng and Cxcr3 promoters through a physical interaction with an H3K27-demethylase complex (2). One of the H3K27-demethylases that is important for T-bet-dependent transcriptional regulation is Jmjd3 (Fig. 1). The physical interaction with the H3K27-demethylase complex requires both T-box domain 1 and 2, and this interaction is also a common function for the T-box family (2). Collectively, these data suggest that the functional resolution of the repressive H3K27-methyl mark and the addition of the permissive H3K4me2 modification are utilized by T-box factors to mediate cellular transitions in several tissue systems. These studies presented the first evidence that a single developmental transcription factor can target the activity of an H3K27-demethylase and H3K4-methyltransferase to promoters to regulate gene expression profiles during cellular differentiation (2) (Fig. 1).
Fig. 1. T-bet interacts with H3K27-demethylase and H3K4-methyltransferase complexes in helper T-cell differentiation.
In naive helper T cells (green), loci that are expressed upon differentiation are kept in a repressed epigenetic state (i.e. H3K27me3). T-bet is able to physically recruit an H3K27-demethylase complex (such as Jmjd3) to its target loci during T-helper cell differentiation to effectively reverse the repressive epigenetic state. Subsequently or alternatively in a simultaneous manner, T-bet has the ability to physically interact with and recruit a permissive H3K4-methyltransferase complex (such as Set7/9) to target genes to establish a permissive epigenetic environment (i.e. H3K4me2) compatible with transcription. T-bet's ability to influence the epigenetic environment of the cell gives it the potential to alter the expression of its target genes in any of the T-helper cells in which it is expressed (see Fig. 2).
This functional capability of the T-box family is very powerful, especially at developmental transitions where lineage-defining transcription factors must bind to repressed gene loci and alter the chromatin environment to create unique gene expression programs appropriate for the newly committed cell type. It also suggests that altering one histone modification is not sufficient to induce the collective changes that need to occur at cellular transitions and that at least some developmental transcription factors must interact with several different complexes in order to execute their functions. However, a number of developmental transcription factors, including T-box proteins, are needed both at cell fate transitions as well as in the differentiated lineage. Once the histone modifications are established during the transition, it is not clear whether T-bet and other T-box factors are needed to maintain them or if there are other functions for which these proteins are required in the differentiated cell.
What do these mechanisms mean in the context of T-helper cell lineages?
One line of evidence that has been used to suggest that various helper T-cell subsets represent stable, terminally committed lineages is the marking of signature cytokine genes with the H3K27me3 epigenetic modification (118, 119). The H3K27-methylated state, which is mediated by the polycomb family, is most often associated with developmentally repressed genes, except when it is found in a poised, bivalent state with H3K4me3 in pluripotent cells (16). The functional capacity for T-bet to regulate epigenetic events, and in particular mediating the removal of the H3K27me3 modification, provides the opportunity for T-bet to change the fate of its target genes in any T-helper cell subset in which it is expressed (2). T-bet's direct targets include Th1 signature genes, including Ifng and Cxcr3 (77). Therefore, T-bet's mechanistic potential to mediate developmentally appropriate epigenetic states allows for plasticity in the signature gene expression program of a T-helper cell until T-bet expression is permanently extinguished. In this scenario, many of the recently defined helper T-cell subsets may not represent terminally committed cells but rather subsets that retain flexibility due to their capacity to re-express T-bet in response to environmental cues.
With this new understanding of the mechanisms by which T-bet establishes chromatin states, a plausible hypothesis is that the terminal commitment of a T-helper cell lies in the events that are upstream of T-bet expression. That is, after the polycomb-mediated H3K27me3 modification is established in a T-helper cell type, if T-bet is permanently repressed, the T-helper cell lineage will likely lose the capacity to express the Th1 signature gene profile. In contrast, if T-bet can be re-expressed, the H3K27me3 state can be functionally reversed by T-bet, providing a mechanism for the cell to again possess the potential to upregulate Th1 genes. This makes understanding the functional regulation of Tbx21 (the gene encoding T-bet) an important area of research in defining T-helper cell plasticity as it relates to the Th1 phenotype.
Recent studies have shown that many T-helper cell types have the ability to re-express T-bet both in vitro and in vivo (13, 14, 120). Some progress has been made addressing the mechanisms that account for the flexibility of Tbx21 expression (14). Importantly, the Tbx21 locus is kept in a poised, bivalent epigenetic state in many T-helper cell subsets (14). Mechanistically, this means that if the cell receives the proper stimuli, the developmentally poised bivalent modifications at the Tbx21 locus can be resolved into a permissive chromatin state (Fig. 2). The Tbx21 locus is also thought to be a direct target of T-bet. Therefore, even low levels of T-bet can then potentially act in a positive feedback mechanism to directly reinforce its own expression, possibly by aiding in the removal of the repressive H3K27 modification. This means that even small environmental alterations that induce minor changes in Tbx21 transcription may produce greater downstream effects in T-bet expression and ultimately the T-helper cell program. As noted above, the flexibility to re-express T-bet then provides the T-helper cell with a certain degree of plasticity, because T-bet has the capacity to direct histone-modifying complexes to its target genes to remove repressive H3K27-methylation modifications and establish the permissive H3K4-methylation state (2). Therefore, T-bet-dependent epigenetic patterns consistent with Th1 cells may be found in a number of different T-helper cell subsets that have now been shown to have the capacity to re-express T-bet. In particular, if environmental cues trigger T-bet to be re-expressed in a Th2 cell, this would allow for a time when T-bet reverses the repressive epigenetic state at the Ifng locus prior to IL-4 being extinguished. In support of this mechanism, in a cell subset that has been termed a Th2-1 cell, the co-expression of Ifnγ and IL-4 has been observed, and the dual expression of Ifnγ and IL-17 has been found in T-helper cells that were previously thought to strictly be Th17 cells (10, 13).
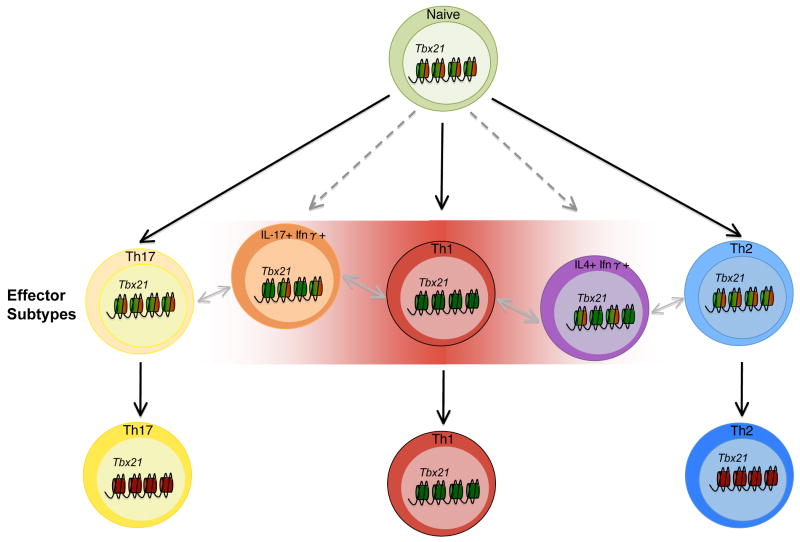
Fig. 2. Model for how T-bet expression patterns regulate T-helper cell commitment.
T-bet has the ability to recruit H3K27-demethylase and H3K4-methyltransferase complexes to its target genes to establish their epigenetic profile. Therefore, if T-bet is expressed, or re-expressed in response to an environmental stimuli, a Th1-like epigenetic signature will be established in the cell. Illustrated here is a gradient model for helper T cell commitment based upon the levels of T-bet (shaded red box). The epigenetic modification at the Tbx21 locus (the gene that encodes T-bet) can be contained within either a permissive (green), repressive (red), or a poised, bivalent (red and green) state in various helper T-cell populations. Importantly, until the Tbx21 locus is permanently extinguished in repressive chromatin, T-bet has the potential to be re-expressed and alter the epigenetic profile of the cell to contain a Th1-like signature. This model is focused on T-bet, but as more studies assess the functional capabilities of the other important transcription factors in T-helper cells, similar and overlapping gradients may exist for their mechanistic role in commitment decisions as well.
Conserved mechanisms within transcription factor families
The current mechanisms that have been defined for T-bet's role in regulating epigenetic events are conserved within the larger T-box transcription factor family (2). These findings imply that there are a common series of changes that occur at cellular transitions in the development of the organism (1, 121). It also suggests that other T-box family members will likely compensate for the absence of T-bet, at least to a degree, if they have overlapping expression patterns. Significantly, in addition to T-bet, another T-box factor, Eomes, has been shown to play an important role in the immune system (61, 93). Eomes expression overlaps with T-bet in many circumstances, including in CD8+ T cells, NK cells, and B cells (61, 77, 91, 93). The loss of T-bet does not have severe phenotypic consequences in the cell types that also express high levels of Eomes (61, 91). The most likely explanation for these results is that T-bet and Eomes can perform similar functional activities, including the ability to establish the appropriate epigenetic state. In fact, experiments have confirmed that Eomes also has the ability to physically and functionally interact with H3K27-demethylase and H3K4-methyltransferase activities (2). Taken together, when assessing the ability of a cell to alter a T-bet-dependent epigenetic profile, it is also important to determine what other T-box factors are present that can perform in a functionally redundant manner.
Mechanistically, deciphering the specific versus conserved functional activities for transcription factor family members is a critical area of research, especially as it pertains to immune cell development and function. It is clear that T-bet and Eomes are functionally redundant in many ways; both are able to directly regulate a number of common target genes including Ifng and Cxcr3 (2). However, in vivo studies examining T-bet−/−, a conditional Eomes−/−, or T-bet and Eomes double deficient mice have made it clear that these factors do indeed play both overlapping and nonredundant roles in immune cells (111). In particular, effector versus memory cell formation appears to be related to the balance between T-bet and Eomes (93). Molecular studies examining T-bet and Eomes have not been able to determine their mechanistic differences that can account for these unique phenotypic roles. This presents one of the outstanding challenges in the field—pinpointing the molecular mechanisms that account for the specialized functions of closely related transcription factor family members.
Molecular mechanisms for key lineage-defining transcription factors in the immune system
Here, we have highlighted our current understanding of the molecular mechanisms that mediate the functional activities of the T-box transcription factor family. In particular, the mechanisms by which T-bet regulates the epigenetic states of its target genes provides a great deal of insight into the topic of helper T-cell plasticity. We now have plausible hypotheses to explore to understand the nature of cellular commitment decisions in relationship to helper T-cell differentiation. However the notion of a single transcription factor acting in isolation as the sole determinant of a cell's phenotype is much too simplistic. Clearly, it is the combination of transcription factors that are present in the cell along with their functional capabilities and the presence or absence of required cofactors that will ultimately determine the overall phenotypic outcome. Therefore, a significant challenge in the field is to uncover the mechanistic details by which all of the key lineage-defining transcription factors work in each cellular context. This will aid in our ability to predict the functional consequence for the expression of combinations of transcription factors in immune cell types.
Conclusions: solving molecular mechanisms for T-helper cell differentiation
A major challenge in immunology research in the coming years will be to increase our understanding of the molecular mechanisms by which the known lineage-defining transcription factors regulate gene expression profiles in immune cells. Over the past decade, we have discovered many of the critical factors that are required for the differentiation and effector functions of a number of cell types in the immune system. In CD4+ helper T cells, we have identified a role for T-bet in Th1 cells, GATA3 in Th2 cells, RORγt in Th17 cells, and Foxp3 in regulatory T cells (5). By analyzing mice engineered to be deficient in each of these factors, we better understand the role each plays in establishing phenotypes during an immune response and the types of pathology that occur if their activities are dysregulated. Currently, however, there is a paucity of studies defining the biochemical and molecular characteristics of these key transcription factors in immune cells. To correct this deficiency, we need to learn from other disciplines to address what mechanistic capabilities each factor possesses so that we can then examine the functional activities in the context of a natural immune response. For instance, traditional biochemistry and molecular biology studies in cell culture and in vitro systems provide powerful tools to determine enzymatic potentials, partner proteins, complex formation, whether multiple activities are contained within a single protein, and the minimal requirements for functional activity. Using the approaches that have successfully defined regulatory activities in other systems, we can then apply this knowledge to better understand the series of events that occur in the course of a natural immune response. The collective studies that have been performed examining the molecular mechanisms for T-bet's role in transcriptional regulation as well as the characterization of its requirement in immune responses in vivo provide a model for what can be achieved with this integrated approach.
Acknowledgments
We thank the members of the Weinmann lab and our colleagues for insightful discussions and comments. Research in the authors' lab is supported by grants from the NIAID (AI061061) and (AI07272-061A) and the American Cancer Society (RSG-09-045-01-DDC) to A.S.W. and S.A.M was supported by a predoctoral training grant from the NIGMS (PHS NRSA 2T32 GM007270).
References
- 1.Miller SA, Weinmann AS. Common themes emerge in the transcriptional control of T helper and developmental cell fate decisions regulated by the T-box, GATA and ROR families. Immunology. 2009;126:306–315. doi: 10.1111/j.1365-2567.2008.03040.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Miller SA, Huang AC, Miazgowicz MM, Brassil MM, Weinmann AS. Coordinated but physically separable interaction with H3K27-demethylase and H3K4-methyltransferase activities are required for T-box protein-mediated activation of developmental gene expression. Genes Dev. 2008;22:2980–2993. doi: 10.1101/gad.1689708. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.O'Shea JJ, Paul WE. Mechanisms Underlying Lineage Commitment and Plasticity of Helper CD4+ T cells. Science. 2010;327:1098–1102. doi: 10.1126/science.1178334. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Bluestone JA, Mackay CR, O'Shea JJ, Stockinger B. The functional plasticity of T cell subsets. Nat Rev Immunol. 2009;9:811–816. doi: 10.1038/nri2654. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Zhou L, Chong MM, Littman DR. Plasticity of CD4+ T cell lineage differentiation. Immunity. 2009;30:646–655. doi: 10.1016/j.immuni.2009.05.001. [DOI] [PubMed] [Google Scholar]
- 6.Mosmann TR, Cherwinski H, Bond MW, Giedlin MA, Coffman RL. Two types of murine helper T cell clone. I. Definition according to profiles of lymphokine activities and secreted proteins. J Immunol. 1986;136:2348–2357. [PubMed] [Google Scholar]
- 7.Steinman L. A brief history of T(H)17, the first major revision in the T(H)1/T(H)2 hypothesis of T cell-mediated tissue damage. Nat Med. 2007;13:139–145. doi: 10.1038/nm1551. [DOI] [PubMed] [Google Scholar]
- 8.Littman DR, Rudensky AY. Th17 and regulatory T cells in mediating and restraining inflammation. Cell. 2010;140:845–858. doi: 10.1016/j.cell.2010.02.021. [DOI] [PubMed] [Google Scholar]
- 9.Curtis MM, Way SS, Wilson CB. IL-23 promotes the production of IL-17 by antigen-specific CD8 T cells in the absence of IL-12 and type-I interferons. J Immunol. 2009;183:381–387. doi: 10.4049/jimmunol.0900939. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Wilson NJ, Boniface K, Chan JR, McKenzie BS, Blumenschein WM, Mattson JD, et al. Development, cytokine profile and function of human interleukin 17-producing helper T cells. Nat Immunol. 2007;8:950–957. doi: 10.1038/ni1497. [DOI] [PubMed] [Google Scholar]
- 11.Veldhoen M, et al. Transforming growth factor-beta ‘reprograms’ the differentiation of T helper 2 cells and promotes an interleukin 9-producing subset. Nat Immunol. 2008;9:1341–1346. doi: 10.1038/ni.1659. [DOI] [PubMed] [Google Scholar]
- 12.Beriou G, et al. TGF-{beta} induces IL-9 production from human Th17 cells. J Immunol. 2010 doi: 10.4049/jimmunol.1000356. Epub. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Hegazy AN, et al. Interferons direct Th2 cell reprogramming to generate a stable GATA3(+) T-bet(+) cell subset with combined Th1 and Th2 cell functions. Immunity. 2010;32:116–128. doi: 10.1016/j.immuni.2009.12.004. [DOI] [PubMed] [Google Scholar]
- 14.Wei G, Wei L, Zhu J, Zang C, Hu-Li J, Yao Z, et al. Global mapping of H3K4me3 and H3K27me3 reveals specificity and plasticity in lineage fate determination of differentiating CD4+ T cells. Immunity. 2009;30:155–167. doi: 10.1016/j.immuni.2008.12.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Cui K, et al. Chromatin signatures in multipotent human hematopoietic stem cells indicate the fate of bivalent genes during differentiation. Cell Stem Cell. 2009;4:80–93. doi: 10.1016/j.stem.2008.11.011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Bernstein BE, et al. A bivalent chromatin structure marks key developmental genes in embryonic stem cells. Cell. 2006;125:315–326. doi: 10.1016/j.cell.2006.02.041. [DOI] [PubMed] [Google Scholar]
- 17.Hemberger M, Dean W, Reik W. Epigenetic dynamics of stem cells and cell lineage commitment: digging Waddington's canal. Nat Rev Mol Cell Biol. 2009;10:526–537. doi: 10.1038/nrm2727. [DOI] [PubMed] [Google Scholar]
- 18.Ting AH, McGarvey KM, Baylin SB. The cancer epigenome - components and functional correlates. Genes Dev. 2006;20:3215–3231. doi: 10.1101/gad.1464906. [DOI] [PubMed] [Google Scholar]
- 19.Ben-Porath I, et al. An embryonic stem cell-like gene expression signature in poorly differentiated aggressive human tumors. Nat Genet. 2008;40:499–4507. doi: 10.1038/ng.127. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Strahl BD, Allis CD. The language of covalent histone modification. Nature. 2000;403:41–45. doi: 10.1038/47412. [DOI] [PubMed] [Google Scholar]
- 21.Shi Y. Histone lysine demethylases: emerging roles in development, physiology and disease. Nat Rev Genet. 2007;8:829–833. doi: 10.1038/nrg2218. [DOI] [PubMed] [Google Scholar]
- 22.Hebbes T, Clayton A, Thorne A, Crane-Robinson C. Core histone hyperacetylation co-maps with gerneralized DNaseI sensitivity in the chicken b-globin chromosomal domain. J Embryol. 1994;13:1823–1830. doi: 10.1002/j.1460-2075.1994.tb06451.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Braunstein M, Rose AB, Holmes SG, Allis CD, Broach JR. Transcriptional silencing in yeast is associated with reduced nucleosome acetylation. Genes Dev. 1993;4:592–604. doi: 10.1101/gad.7.4.592. [DOI] [PubMed] [Google Scholar]
- 24.Bantignies F, Cavalli G. Cellular memory and dynamic regulation of polycomb group proteins. Curr Opin Cell Biol. 2006;18:275–283. doi: 10.1016/j.ceb.2006.04.003. [DOI] [PubMed] [Google Scholar]
- 25.Shilatifard A. Chromatin modifications by methylation and ubiquitination: implications in the regulation of gene expression. Annu Rev Biochem. 2006;75:243–269. doi: 10.1146/annurev.biochem.75.103004.142422. [DOI] [PubMed] [Google Scholar]
- 26.Wang Z, et al. Genome-wide mapping of HATs and HDACs reveals distinct functions in active and inactive genes. Cell. 2009;138:1019–1031. doi: 10.1016/j.cell.2009.06.049. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Wang Z, et al. Combinatorial patterns of histone acetylations and methylations in the human genome. Nat Genet. 2008;40:897–903. doi: 10.1038/ng.154. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Dou Y, et al. Regulation of MLL1 H3K4 methyltransferase activity by its core components. Nature Structural and Molecular Biology. 2006;13:713–719. doi: 10.1038/nsmb1128. [DOI] [PubMed] [Google Scholar]
- 29.Ruthenburg AJ, Allis CD, Wysocka J. Methylation of Lysine 4 on histone H3: intracacy of writing and reading a single epigenetic mark. Mol Cell. 2007;25:15–30. doi: 10.1016/j.molcel.2006.12.014. [DOI] [PubMed] [Google Scholar]
- 30.Wysocka J, et al. WDR5 associates with histone H3 methylated at K4 and is essential for H3 K4 methylation and vertebrate development. Cell. 2005;121:859–872. doi: 10.1016/j.cell.2005.03.036. [DOI] [PubMed] [Google Scholar]
- 31.Cao R, et al. Role of Histone H3 Lysine 27 methylation in Polycomb-Group silencing. Science. 2002;298:1039–1043. doi: 10.1126/science.1076997. [DOI] [PubMed] [Google Scholar]
- 32.Czermin B, Melfi R, McCabe D, Seitz V, Imhof A, Pirrotta V. Drosophila Enhancer of Zeste/ESC complexes have a histone H3 methyltransferase activity that marks chromosomal polycomb sites. Cell. 2002;111:185–196. doi: 10.1016/s0092-8674(02)00975-3. [DOI] [PubMed] [Google Scholar]
- 33.Koyanagi M, Baguet A, Martens J, Margueron R, Jenuwein T, Bix M. EZH2 and histone 3 trimethyl lysine 27 associated with Il4 and Il13 gene silencing in Th1 cells. J Biol Chem. 2005;280:31470–31477. doi: 10.1074/jbc.M504766200. [DOI] [PubMed] [Google Scholar]
- 34.Mikkelsen TS, Ku M, Jaffe DB, Issac B, Lieberman E, Giannoukos G, et al. Genome-wide maps of chromatin state in pluripotent and lineage-committed cells. Nature. 2007;448:553–560. doi: 10.1038/nature06008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Boyer LA, Plath K, Zeitlinger J, Brambrink T, Medeiros LA, Lee TI, et al. Polycomb complexes repress developmental regulators in murine embryonic stem cells. Nature. 2006;441:349–353. doi: 10.1038/nature04733. [DOI] [PubMed] [Google Scholar]
- 36.Araki Y, Wang Z, Zang C, Wood WHr, Schones D, Cui K, et al. Genome-wide analysis of histone methylation reveals chromatin state-based regulation of gene transcription and function of memory CD8+ T cells. Immunity. 2009;30:912–925. doi: 10.1016/j.immuni.2009.05.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Kuzmichev A, Nishioka K, Erdjument-Bromage H, Tempst P, Reinberg D. Histone methyltransferase activity associated with human multiprotein complex containing the Enhancer of Zeste protein. Genes Dev. 2002;16:2893–2905. doi: 10.1101/gad.1035902. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Cho YW, et al. PTIP associates with MLL3- and MLL4-containing histone H3 lysine 4 methyltransferase complex. J Biol Chem. 2007;282:20395–20406. doi: 10.1074/jbc.M701574200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Lee JH, Tate CM, You JS, Skalnik DG. Identification and characterization of the human Set1B histone H3-Lys4 methyltransferase complex. J Biol Chem. 2007;282:13419–13428. doi: 10.1074/jbc.M609809200. [DOI] [PubMed] [Google Scholar]
- 40.Muller J, et al. Histone Methyltransferase activity of a Drosophila polycomb group repressor complex. Cell. 2002;111:197–208. doi: 10.1016/s0092-8674(02)00976-5. [DOI] [PubMed] [Google Scholar]
- 41.Lee MG, et al. Demethylation of H3K27 regulates polycomb recruitment and H2A ubiquitination. Science. 2007;318:447–450. doi: 10.1126/science.1149042. [DOI] [PubMed] [Google Scholar]
- 42.Agger K, Cloos PA, Christensen J, Pasini D, Rose S, Rappsilber J, et al. UTX and JMJD3 are histone H3K27 demethylases involved in HOX gene regulation and development. Nature. 2007;449:731–734. doi: 10.1038/nature06145. [DOI] [PubMed] [Google Scholar]
- 43.Lan F, et al. A histone H3 lysine 27 demethylase regulates animal posterior development. Nature. 2007;449:689–695. doi: 10.1038/nature06192. [DOI] [PubMed] [Google Scholar]
- 44.Smith ER, et al. Drosophila UTX is a Histone H3 Lys27 demethylase that colocalizes with the elongating form of RNA polymerase II. Mol Cell Biol. 2008;28:1041–1046. doi: 10.1128/MCB.01504-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Agger K, Christensen J, Cloos PAC, Helin K. The emerging functions of histone demethylases. Curr Opin Genet Dev. 2008;18:1–10. doi: 10.1016/j.gde.2007.12.003. [DOI] [PubMed] [Google Scholar]
- 46.Lan F, Nottke AC, Shi Y. Mechanisms involved in the regulation of histone lysine demethylases. Curr Opin Cell Biol. 2008;20:1–10. doi: 10.1016/j.ceb.2008.03.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Chang B, Chen Y, Zhao Y, Bruick RK. JMJD6 is a histone arginine demethylase. Science. 2007;318:444–447. doi: 10.1126/science.1145801. [DOI] [PubMed] [Google Scholar]
- 48.Loh YH, Zhang W, Chen X, George J, Ng HH. Jmjd1a and Jmjd2c histone H3 Lys 9 demethylases regulate self-renewal in embryonic stem cells. Genes Dev. 2007;21:2545–257. doi: 10.1101/gad.1588207. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Hong S, Cho YW, Yu LR, Yu H, Veenstra TD, Ge K. Identification of JmjC domain-containing UTX and JMJD3 as histone H3 lysine 27 demethylases. Proc Natl Acad Sci USA. 2007;104:18439–18444. doi: 10.1073/pnas.0707292104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Shi Y, et al. Histone demethylation mediated by the nuclear amine oxidase homolog LSD1. Cell. 2004;119:941–953. doi: 10.1016/j.cell.2004.12.012. [DOI] [PubMed] [Google Scholar]
- 51.Culhane JC, Cole PA. LSD1 and the chemistry of histone demethylation. Curr Opin Chem Biol. 2007;11:561–568. doi: 10.1016/j.cbpa.2007.07.014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.De Santa F, Totaro MG, Prosperini E, Notarbartolo S, Testa G, Natoli G. The histone H3 lysine-27 demethylase Jmjd3 links inflammation to inhibition of polycomb-mediated gene silencing. Cell. 2007;130:1083–1094. doi: 10.1016/j.cell.2007.08.019. [DOI] [PubMed] [Google Scholar]
- 53.Wilson JR. Targeting the JMJD2A histone lysine demethylase. Nat Struct Mol Biol. 2007;14:682–684. doi: 10.1038/nsmb0807-682. [DOI] [PubMed] [Google Scholar]
- 54.Bimber B, Dettman RW, Simon HG. Differential regulation of Tbx5 protein expression and sub-cellular localization during heart development. Dev Biol. 2007;301:230–242. doi: 10.1016/j.ydbio.2006.09.023. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Zagami CJ, Zusso M, Stifani S. Runx transcription factors: lineage-specific regulators of neuronal precursor cell proliferation and post-mitotic neuron subtype development. J Cell Biochem. 2009;107:1063–1072. doi: 10.1002/jcb.22221. [DOI] [PubMed] [Google Scholar]
- 56.Yu J, et al. Induced pluripotent stem cell lines derived from human somatic cells. Science. 2007;318:1917–1920. doi: 10.1126/science.1151526. [DOI] [PubMed] [Google Scholar]
- 57.Boyer LA, et al. Core transcriptional regulatory circuitry in human embryonic stem cells. Cell. 2005;122:947–956. doi: 10.1016/j.cell.2005.08.020. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Chang JT, Reiner SL. Specifying helper T cell fates during immunity. J Ped Gastroenterol Nutr. 2008;46(Suppl):E19–20. doi: 10.1097/01.mpg.0000313832.47207.0e. [DOI] [PubMed] [Google Scholar]
- 59.Hevner RF, et al. Tbr1 regulates differentiation of the preplate and layer 6. Neuron. 2001;29:353–366. doi: 10.1016/s0896-6273(01)00211-2. [DOI] [PubMed] [Google Scholar]
- 60.Ho IC, Pai SY. GATA-3 - not just for Th2 cells anymore. Cell Mol Immunol. 2007;4:15–29. [PubMed] [Google Scholar]
- 61.Pearce EL, et al. Control of effector CD8+ T cell function by the transcription factor eomesodermin. Science. 2003;302:1041–1043. doi: 10.1126/science.1090148. [DOI] [PubMed] [Google Scholar]
- 62.Szabo SJ, Kim ST, Costa GL, Zhang X, Fathman CG, Glimcher LH. A novel transcription factor, T-bet directs Th1 lineage commitment. Cell. 2000;100:655–669. doi: 10.1016/s0092-8674(00)80702-3. [DOI] [PubMed] [Google Scholar]
- 63.Wernig M, et al. In vitro reprogrammin of fibroblasts into a pluripotent ES-cell-like state. Nature. 2007;448:318–324. doi: 10.1038/nature05944. [DOI] [PubMed] [Google Scholar]
- 64.Rolink AG, Schaniel C, Busslinger M, Nutt SL, Melchers F. Fidelity and infidelity in commitment to B-lymphocyte lineage development. Immunol Rev. 2000;175:104–111. [PubMed] [Google Scholar]
- 65.Liu K, et al. In vivo analysis of dendritic cell development and homeostasis. Science. 2009;324:392–397. doi: 10.1126/science.1170540. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Sitnicka E. From the bone marrow to the thymus: the road map of early stages of T-cell development. Crit Rev Immunol. 2009;29:487–530. doi: 10.1615/critrevimmunol.v29.i6.30. [DOI] [PubMed] [Google Scholar]
- 67.Papathanasiou P, et al. Self-renewal of the long-term reconstituting subset of hematopoietic stem cells is regulated by Ikaros. Stem Cells. 2009;27:3082–3092. doi: 10.1002/stem.232. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Szabo SJ, Sullivan BM, Stenmann C, Satoskar AR, Sleckman BP, Glimcher LH. Distinct effects of T-bet in Th1 lineage commitment and IFN gamma production in CD4 and CD8 T cells. Science. 2002;295:338–342. doi: 10.1126/science.1065543. [DOI] [PubMed] [Google Scholar]
- 69.Zheng W, Flavell RA. The transcription factor GATA-3 is necessary and sufficient for Th2 cytokine gene expression in CD4 T cells. Science. 1997;89:587–596. doi: 10.1016/s0092-8674(00)80240-8. [DOI] [PubMed] [Google Scholar]
- 70.Schulz SM, Köhler G, Holscher C, Iwakura Y, Alber G. IL-17A is produced by Th17, gammadelta T cells and other CD4- lymphocytes during infection with Salmonella enterica serovar Enteritidis and has a mild effect in bacterial clearance. Int Immunol. 2008;20:1129–1138. doi: 10.1093/intimm/dxn069. [DOI] [PubMed] [Google Scholar]
- 71.Harrington LE, Hatton RD, Mangan PR, Turner H, Murphy TL, Murphy KM, et al. Interleukin 17-producing CD4+ effector T cells develop via a lineage distinct from the T helper type 1 and 2 lineages. Nat Immunol. 2005;6:1123–1132. doi: 10.1038/ni1254. [DOI] [PubMed] [Google Scholar]
- 72.Zhou L, et al. TGF-beta-induced Foxp3 inhibits Th17 cell differentiation by antagonizing ROR gamma T function. Nature. 2008;453:236–240. doi: 10.1038/nature06878. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73.Yu D, et al. The transcriptional repressor Bcl-6 directs T follicular helper cell lineage commitment. Immunity. 2009;31:457–468. doi: 10.1016/j.immuni.2009.07.002. [DOI] [PubMed] [Google Scholar]
- 74.Nurieva RI, et al. Bcl6 mediates the development of T follicular helper cells. Science. 2009;325:1001–1005. doi: 10.1126/science.1176676. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75.Fontenot JD, Gavin MA, Rudensky AY. Foxp3 programs the development and function of CD4+CD25+ regulatory T cells. Nat Immunol. 2003;4:330–336. doi: 10.1038/ni904. [DOI] [PubMed] [Google Scholar]
- 76.Lewis MD, Miller SA, Miazgowicz MM, Beima KM, Weinmann AS. T-bet's ability to regulate individual target genes requires the conserved T-box domain to recruit histone methyltransferase activity and a separate family member-specific transactivation domain. Mol Cell Biol. 2007;27:8510–8521. doi: 10.1128/MCB.01615-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 77.Beima KM, Miazgowicz MM, Lewis MD, Yan PS, Huang TH, Weinmann AS. T-bet binding to newly identified target gene promoters is cell type-independent but results in variable context-dependent functional effects. J Biol Chem. 2006;281:11992–12000. doi: 10.1074/jbc.M513613200. [DOI] [PubMed] [Google Scholar]
- 78.Lord GM, et al. T-bet is required for optimal proinflammatory CD4+ T-cell trafficking. Blood. 2005;106:3432–3439. doi: 10.1182/blood-2005-04-1393. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 79.Cho JY, Grigura V, Murphy TL, Murphy KM. Identification of cooperative monomeric Brachyury sites conferring T-bet responsiveness to the proximal IFN-g promoter. Int Immunol. 2003;15:1149–1160. doi: 10.1093/intimm/dxg113. [DOI] [PubMed] [Google Scholar]
- 80.Lugo-Villarino G, Maldonado-Lopez R, Possemato R, Penaranda C, Glimcher LH. T-bet is required for optimal production of IFNg and antigen-specific T cell activation by dendritic cells. Proc Natl Acad Sci USA. 2003;100:7749–7754. doi: 10.1073/pnas.1332767100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 81.Peng SL. The T-box transcription factor T-bet in immunity and autoimmunity. Cell Mol Immunol. 2006;3:87–95. [PubMed] [Google Scholar]
- 82.Wang J, et al. Transcription factor T-bet regulates inflammatory arthritis through its function in dendritic cells. J Clin Invest. 2006;116:414–421. doi: 10.1172/JCI26631. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 83.Dorfman DM, Hwang ES, Shahsafaei A, Glimcher LH. T-bet, a T cell-associated transcription factor, is expressed in Hodgkin's lymphoma. Hum Pathol. 2005;36:10–15. doi: 10.1016/j.humpath.2004.10.006. [DOI] [PubMed] [Google Scholar]
- 84.Dorfman DM, Hwang ES, Shahsafaei A, Glimcher LH. T-bet, a T-cell-associated transcription factor, is expressed in a subset of B-cell lymphoproliferative disorders. Am J Clin Pathol. 2004;122:292–297. doi: 10.1309/AQQ2-DVM7-5DVY-0PWP. [DOI] [PubMed] [Google Scholar]
- 85.Hwang ES, Hong JH, Glimcher LH. IL-2 production in developing Th1 cells is regulated by heterodimerization of RelA and T-bet and requires T-bet serine residue 508. J Exp Med. 2005;202:1289–1300. doi: 10.1084/jem.20051044. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 86.Sasaki Y, Ihara K, Matsuura N, Kohno H, Nagafuchi S, Kuromaru R, et al. Identification of a novel type 1 diabetes susceptibility gene, T-bet. Hum Genet. 2004;115:177–184. doi: 10.1007/s00439-004-1146-2. [DOI] [PubMed] [Google Scholar]
- 87.Garrett WS, et al. Colitis-associated colorectal cancer driven by T-bet deficiency in dendritic cells. Cancer Cell. 2009;16:208–219. doi: 10.1016/j.ccr.2009.07.015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 88.Park JW, Min HJ, Sohn JH, Kim JY, Hong JH, Sigrist KS, et al. Restoration of T-box-containing protein expressed in T cells protects against allergen-induced asthma. J Allergy Clin Immunol. 2009;123:479–485. doi: 10.1016/j.jaci.2008.10.035. [DOI] [PubMed] [Google Scholar]
- 89.Raby BA, et al. T-bet polymorphisms are associated with asthma and airway hyperresponsiveness. Am J Respir Crit Care Med. 2006;173:64–70. doi: 10.1164/rccm.200503-505OC. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 90.You J, et al. Association of TBX21 gene haplotypes in a Chinese population with systemic lupus erythematosus. Scand J Rheumatol. 2010;39:254–258. doi: 10.3109/03009740903347983. [DOI] [PubMed] [Google Scholar]
- 91.Townsend MJ, et al. T-bet regulates the terminal maturation and homeostasis of NK and Valpha14i NKT cells. Immunity. 2004;20:477–494. doi: 10.1016/s1074-7613(04)00076-7. [DOI] [PubMed] [Google Scholar]
- 92.Sullivan BM, Juedes A, Szabo SJ, von Herrath M, Glimcher LH. Anitgen driven effector CD8 T cell function regulated by T-bet. Proc Natl Acad Sci USA. 2003;100:15818–15823. doi: 10.1073/pnas.2636938100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 93.Intlekofer AM, et al. Effector and memory CD8+ T cell fate coupled by T-bet and eomesodermin. Nat Immunol. 2005;12:1236–1244. doi: 10.1038/ni1268. [DOI] [PubMed] [Google Scholar]
- 94.Peng SL, Szabo SJ, Glimcher LH. T-bet regulates IgG class switching and pathogenic autoantibody production. Proc Natl Acad Sci USA. 2002;99:5545–5550. doi: 10.1073/pnas.082114899. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 95.Naiche LA, Harrelson Z, Kelly RG, Papaioannou VE. T-box genes in vertebrate development. Annu Rev Genet. 2005;17:219–239. doi: 10.1146/annurev.genet.39.073003.105925. [DOI] [PubMed] [Google Scholar]
- 96.Goering LM, Hoshijima K, Hug B, Bisgrove B, Kispert A, Grunwald DJ. An interacting network of T-box genes directs gene expression and fate in zebrafish mesoderm. Proc Natl Acad Sci USA. 2003;100:9410–9415. doi: 10.1073/pnas.1633548100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 97.Fan C, Liu M, Wang Q. Functional analysis of Tbx5 missense mutations associated with Holt-Oram syndrome. J Biol CHem. 2003;278:8780–8785. doi: 10.1074/jbc.M208120200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 98.Mori AD, Bruneau BG. TBX5 mutations and congenital heart disease: Holt-Oram syndrome revealed. Curr Opin Cardiol. 2004;19:211–215. doi: 10.1097/00001573-200405000-00004. [DOI] [PubMed] [Google Scholar]
- 99.Andreou AM, et al. TBX22 missense mutations found in patients with X-linked cleft palate affect DNA binding, sumoylation, and transcriptional repression. Am J Hum Genet. 2007;81:700–712. doi: 10.1086/521033. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 100.Bamshad M, et al. The spectrum of mutations in TBX3: Genotype/Phenotype relationship in ulnar-mammary syndrome. Am J Hum Genet. 1999;64:1550–1562. doi: 10.1086/302417. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 101.Reamon-Buettner SM, Borlak J. TBX5 mutations in non-Holt-Oram syndrome (HOS) malformed hearts. Hum Mutation. 2004;24:1–7. doi: 10.1002/humu.9255. [DOI] [PubMed] [Google Scholar]
- 102.Kirk EP, et al. Mutations in cardiac T-box factor gene TBX20 are associated with diverse cardiac pathologies, including defects of septation and valvulogenesis and cardiomyopathy. Am J Hum Genet. 2007;81:280–291. doi: 10.1086/519530. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 103.Bongers EM, et al. Mutations in the human TBX4 gene cause small patella syndrome. Am J Hum Genet. 2004;74:1239–1248. doi: 10.1086/421331. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 104.Bamshad M, et al. Mutations in human TBX3 alter limb, apocrine and genital development in ulnar-mammary syndrome. Nat Genetics. 1997;16:311–315. doi: 10.1038/ng0797-311. [DOI] [PubMed] [Google Scholar]
- 105.Bruneau BG, et al. A murine model of Holt-Oram syndrome defines roles of the T-box transcription factor Tbx5 in cardiogenesis. Cell. 2001;106:709–721. doi: 10.1016/s0092-8674(01)00493-7. [DOI] [PubMed] [Google Scholar]
- 106.Plageman TF, Yutzey KE. Microarray analysis of Tbx5-induced genes expressed in the developing heart. Dev Dynamics. 2006;235:2868–2880. doi: 10.1002/dvdy.20923. [DOI] [PubMed] [Google Scholar]
- 107.Packham EA, Brook JD. T-box genes in human disorders. Hum Mol Genet. 2003;12:R37–44. doi: 10.1093/hmg/ddg077. [DOI] [PubMed] [Google Scholar]
- 108.Showell C, Binder O, Conlon FL. T-box genes in early embryogenesis. Dev Dynamics. 2004;229:201–218. doi: 10.1002/dvdy.10480. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 109.Baala L, et al. Homozygous silencing of T-box transcription factor EOMES leads to microcephaly with polymicrogyria and corpus callosum agenesis. Nat Genet. 2007;39:454–456. doi: 10.1038/ng1993. [DOI] [PubMed] [Google Scholar]
- 110.Russ AP, et al. Eomesodermin is required for mouse trophoblast development and mesoderm formation. Nature. 2000;404:95–99. doi: 10.1038/35003601. [DOI] [PubMed] [Google Scholar]
- 111.Intlekofer AM, et al. Anomalous type 17 response to viral infection by CD8+ T-bet and eomesodermin. Science. 2008;321:408–411. doi: 10.1126/science.1159806. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 112.Agarwal S, Rao A. Modulation of chromatin structure regulates cytokine gene expression during T cell differentiation. Immunity. 1998;9:765–775. doi: 10.1016/s1074-7613(00)80642-1. [DOI] [PubMed] [Google Scholar]
- 113.Fields PE, Kim ST, Flavell RA. Cutting edge: changes in histone acetylation at the IL-4 and IFNg loci accompany Th1/Th2 differentaition. J Immunol. 2002;169:647–650. doi: 10.4049/jimmunol.169.2.647. [DOI] [PubMed] [Google Scholar]
- 114.Avni O, Lee D, Macian F, Szabo SJ, Glimcher LH, Rao A. T(H) cell differentiation is accompanied by dynamic changes in histone acetylation of cytokine genes. Nat Immunol. 2002;3:643–651. doi: 10.1038/ni808. [DOI] [PubMed] [Google Scholar]
- 115.Ansel KM, Lee DU, Rao A. An epigenetic view of helper T cell differentiation. Nat Immunol. 2003;4:616–623. doi: 10.1038/ni0703-616. [DOI] [PubMed] [Google Scholar]
- 116.Aune TM, Collins PL, Chang S. Epigenetics and T helper 1 differentiation. Immunology. 2009;126:299–305. doi: 10.1111/j.1365-2567.2008.03026.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 117.De Santa F, et al. Jmjd3 contributes to the control of gene expression in LPS-activated macrophages. EMBO J. 2009;28:3341–3352. doi: 10.1038/emboj.2009.271. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 118.Schoenborn JR, et al. Comprehensive epigenetic profiling identifies multiple distal regulatory elements directing transcription of the gene encoding interferon-gamma. Nat Immunol. 2007;8:732–742. doi: 10.1038/ni1474. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 119.Ansel KM, Djuretic I, Tanasa B, Rao A. Regulation of Th2 differentitation and IL4 locus accessibility. Annu Rev Immunol. 2006;24:607–656. doi: 10.1146/annurev.immunol.23.021704.115821. [DOI] [PubMed] [Google Scholar]
- 120.Koch MA, Tucker-Heard G, Perdue NR, Killebrew JR, Urdahl KB, Campbell DJ. The transcription factor T-bet controls regulatory T cell homeostasis and function during type 1 inflammation. Nat Immunol. 2009;10:595–602. doi: 10.1038/ni.1731. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 121.Miller SA, Weinmann AS. An essential interaction between T-box proteins and histone-modifying enzymes. Epigenetics. 2009;4:85–88. doi: 10.4161/epi.4.2.8111. [DOI] [PubMed] [Google Scholar]