Abstract
mRNA lacking stop codons (“nonstop mRNA”) can arise from errors in gene expression, and encode aberrant proteins whose accumulation could be deleterious to cellular function 1,2. In bacteria, such “nonstop proteins” become co-translationally tagged with a peptide encoded by tmRNA/ssrA, which signals their degradation by energy-dependent proteases 1,3. How eukaryotic cells eliminate nonstop proteins has remained unknown. Here we show that the S. cerevisiae Ltn1 RING domain-type E3 ubiquitin ligase acts in the quality control of nonstop proteins, in a process that is mechanistically distinct but conceptually analogous to the one carried out by ssrA: Ltn1 is predominantly associated with ribosomes, and marks nascent nonstop proteins with ubiquitin to signal their proteasomal degradation. Ltn1-mediated ubiquitylation of nonstop proteins appears to be triggered by their stalling in ribosomes upon translation through the poly(A) tail. The biological relevance of this process is underscored by the finding that loss of Ltn1 function conferred sensitivity to stress caused by increased nonstop protein production. We speculate that defective protein quality control may underlie the neurodegenerative phenotype that results from mutation of the mouse Ltn1 homolog, Listerin.
The accidental generation of nonstop mRNA appears to be prevalent across all organisms 1–3. In addition to specifying termination of protein coding sequence, stop codons also signal recruitment of nascent peptide release factors. Thus, translation of nonstop mRNA could give rise to stalled ribosome•peptide complexes, sequestration of ribosomes due to failure to recycle, and decreased translation efficiency. Furthermore, nonstop mRNAs encode aberrant polypeptides (“nonstop proteins”) that cannot be corrected by the chaperones responsible for protein quality control (PQC). As a result, systems in place from bacteria to humans enable degradation of both nonstop transcripts and proteins, suggesting an important biological function for these processes 1–5. However, the mechanisms responsible for nonstop protein degradation in eukaryotes remain unknown. Evidence for a direct role of the ubiquitin-proteasome system is lacking 6,7—in particular, it has yet to be determined which, if any, of the ~650 human and ~80 budding yeast E3 ubiquitin ligases 8,9 confer specificity to the process.
Here we demonstrate that Ltn1 is an E3 that mediates quality control of nonstop proteins (Fig.S1a). Ltn1 is the yeast homolog of Listerin, whose mutation we and our collaborators had reported to cause neurodegeneration in mice 10. Listerin orthologs are 150–190 kDa proteins conserved across eukaryotes (Fig.S2). The exact role of Listerin/Ltn1 has remained to be elucidated, although genetic analyses with yeast have implicated LTN1/RKR1/YMR247c in some yet unknown aspect of chromatin regulation 11. LTN1 was also identified in a screen for nonstop mRNA decay (NSD) pathway components, as a gene whose deletion led to increased expression of an NSD reporter protein 6.
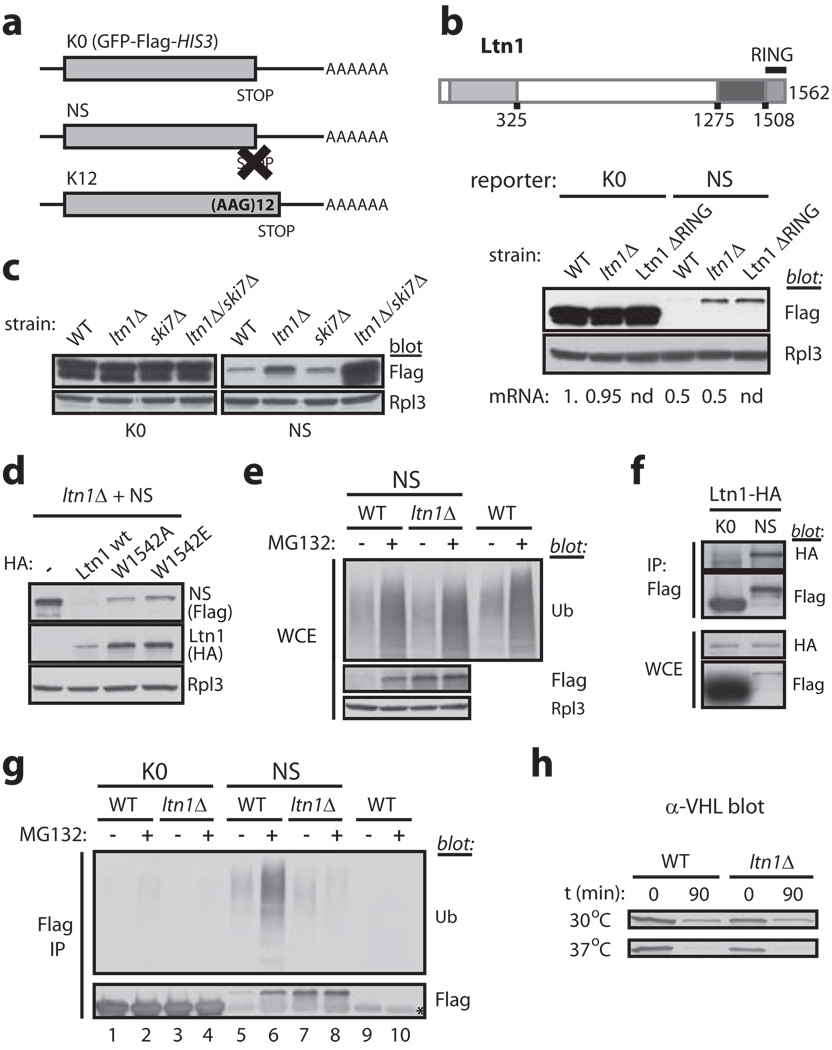
We set out to elucidate Ltn1’s role in NSD. Using reporter constructs based on GFP-Flag-HIS3 (referred to as K0; Fig.1a and 7) and Protein A, we first confirmed that the proteins encoded by the corresponding nonstop mutants (NS and Protein A-NS, respectively) were expressed at lower levels in a wild type (WT) strain, that deletion of LTN1 increased steady-state levels of NS and Protein A-NS, but not of K0 or Protein A, and that it did so without increasing the abundance of NS or Protein A-NS mRNA (Figs.1b and S3a-c). Notably, NS expression in ltn1Δ strains was not completely restored relative to K0 (Fig.1b). To test whether this resulted from translation inhibition and/or degradation of the nonstop mRNA mediated by the NSD machinery 2,4,12, we examined strains also lacking Ski7, an essential factor for NSD. As predicted, the double ltn1/ski7 deletion fully restored NS expression (Figs.1c and S3d; see also Fig.3a).
Figure 1. The yeast Listerin/Ltn1 E3 ligase functions in quality control of nonstop proteins.
a, mRNA encoding GFP-Flag-HIS3 (K0), a nonstop (NS) protein and a protein fused to 12 lysines (K12). b, Regulation of NS protein levels is Ltn1 RING domain-dependent. Top, Ltn1 structure. Conserved regions are shaded. Bottom, K0 and NS protein expression in a wild type (WT) strain, an LTN1 deletion strain (ltn1Δ) and a strain whose endogenous Ltn1 lacks the RING domain. Rpl3 immunoblot controls for loading. Below, relative levels of the corresponding mRNAs (from Fig.S3b; nd, not determined). c, Ski7 and Ltn1 independently control NS protein expression. Immunoblot of K0 and NS in various strains. d, RING domain point mutations impaired Ltn1’s ability to downregulate NS expression. NS levels in a ltn1Δ strain expressing plasmidborne HA-Ltn1 wildtype or Trp1542 mutants. e, Ubiquitin (Ub) blot, LTN1 deletion does not exert a general effect on ubiquitylation; Flag blot, proteasomal degradation of NS proteins is Ltn1-dependent. Immunoblots of whole cell extracts (WCE). Cells were treated (+) or not (−) with the proteasome inhibitor MG132. f, Ltn1 and NS specifically co-IP. Strains expressing endogenous HALtn1 and K0 or NS were used for anti-Flag IP, followed by anti-HA or Flag blot. g, NS proteins are ubiquitylated, and this depends on Ltn1. SDS-boiled lysates of WT or or ltn1Δ strains expressing K0 or NS were used for Flag IP and immunoblots. The WT strain expressing no Flag-tagged proteins was a negative control. The corresponding WCEs are shown in “e”. Asterisk, cross-reacting faint band. h, Ltn1 is not required for VHL degradation. VHL immunoblot in WT or ltn1Δ strains at steady state (t=0; 22°C) or 90 min following cycloheximide addition and switch to 30°C or 37°C.
Figure 3. Nascent poly(Lys) peptides stall in ribosomes, cause translational arrest and trigger Ltn1-mediated ubiquitylation.
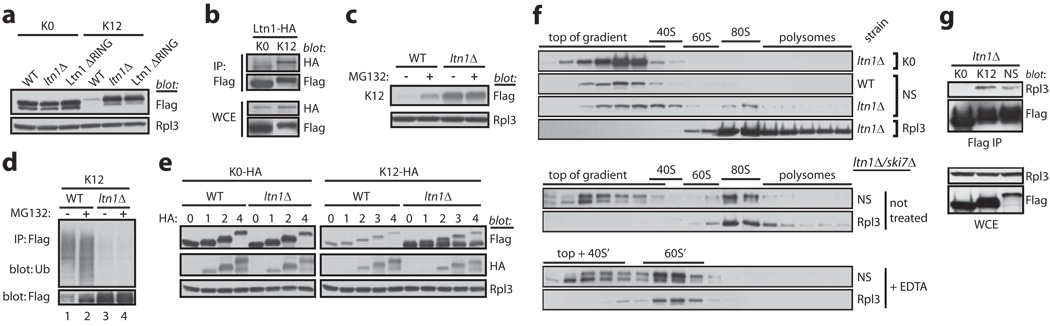
a, K12 levels are regulated in a Ltn1 RING-dependent manner. As in Fig.1b. b, Ltn1 and K12 specifically co-IP. These results are part of the set in Fig.1f. c, Proteasomal degradation of K12 is Ltn1-dependent. Immunoblot of WCEs. d, K12 ubiquitylation and degradation is Ltn1-dependent. K12 expressed in WT or ltn1Δ strains was Flag IP’ed and used for immunoblots, as in Fig.1g. e, A nascent Lys12 tract located at various distances from the C-terminus mediates translational arrest and Ltn1-dependent degradation. Expression of K0 and K12 constructs with 0–4 C-terminal HA tags, in WT and ltn1Δ strains. f, Nascent NS protein stalls in 80S ribosomes and is cleared by Ltn1. Sucrose gradient fractions of lysates expressing K0 or NS proteins were analyzed by anti-Flag immunoblot. Exposures were adjusted to facilitate comparison of the proteins’ distribution. The sedimentation of ribosomal particles was inferred from the A254 profile (e.g., Figs.4c and S8c) and confirmed by reprobing blots for the 60S component, Rpl3. Lower 4 panels, NS-expressing ltn1Δ/ski7Δ cell lysate was treated or not with EDTA prior to centrifugation. EDTA dissociates 80S ribosomes and promotes loss of certain ribosomal components, slowing down sedimentation of 40S and 60S subunits (indicated by S’). Rpl3 co-fractionates mostly with 80S ribosomes in the absence, and 60S’ subunits in the presence, of EDTA (see also Fig.S8c). g, NS and K12, but not K0, co-IP with the 60S protein, Rpl3. Lysates of ltn1Δ strains expressing the reporters were Flag IP’ed, followed by anti-Rpl3 blot.
RING domains underlie E3 activity by binding to ubiquitin-charged E2 conjugases 9. If Ltn1 regulates NS expression via ubiquitylation, its RING domain should be required. Accordingly, deletion of the RING domain encoded by endogenous LTN1 (Ltn1 ΔRING) was as efficient as deletion of the entire gene in restoring NS levels (Fig.1b; Ltn1 ΔRING was expressed at higher levels than Ltn1; e.g., Fig.4a). We next introduced a point mutation in the RING domain of HA-tagged Ltn1 (Trp1542 mutated to Ala or Glu) that is predicted to selectively decrease E2 binding affinity, without perturbing structure (e.g., 9). Like RING-deleted Ltn1, W1542A and W1542E mutants were expressed at higher levels than Ltn1 (Fig.1d). Despite this fact, both were defective in negatively regulating NS expression (Fig.1d). Thus, Ltn1 controls NS levels in a RING domain- and E3 activity-dependent manner.
Figure 4. Ltn1 is predominantly associated with ribosomes.
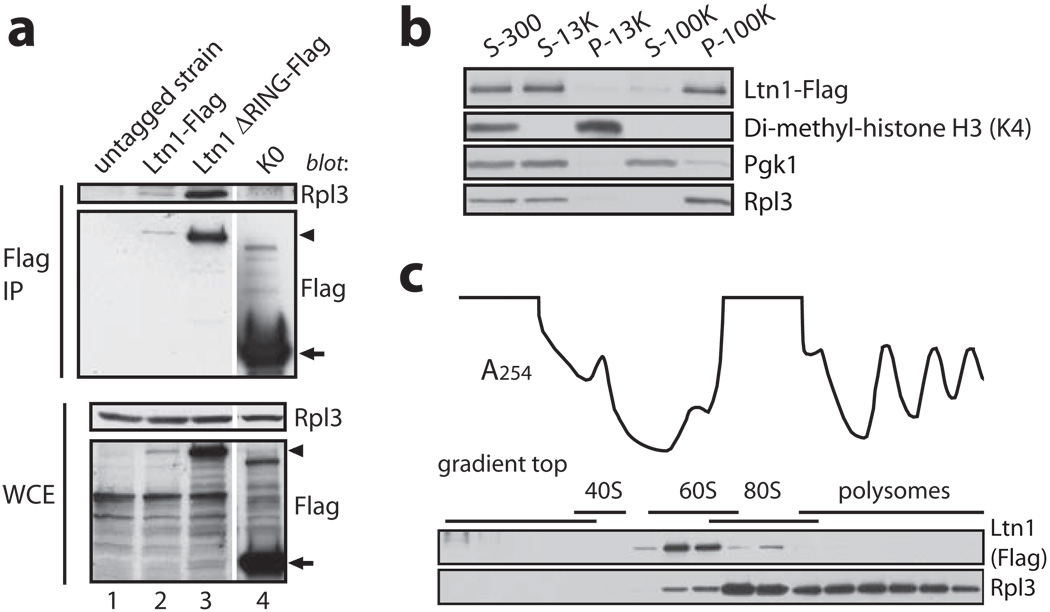
Strains expressing C-terminally Flag-tagged endogenous Ltn1 (a-c) or Ltn1 ΔRING (a) were used in this figure. a, Ltn1 specifically co-IP’s with Rpl3. Indicated lysates were Flag IP’ed, followed by anti-Rpl3 blot. K0 and the untagged WT strain were negative controls. Arrowhead, Ltn1 and Ltn1 ΔRING; arrow, K0. b, Ltn1 is predominantly cytoplasmic. Pellet (P) and supernatant (S) samples were taken following centrifugation of lysate at 300, 13K and 100K × g. Blots were probed for Flag, Lys4-di-methylated histone H3, Pgk1, and Rpl3. c, Ltn1 is predominantly 60S-bound in steady-state. Ltn1’s distribution in sucrose gradient fractions analyzed by immunoblot. Line tracing, A254 profile.
Previous reports had shown that Ltn1 co-immunoprecipitated (co-IP’ed) with the 19S proteasome 13, and that deletion of proteasome assembly factors led to increased nonstop protein levels 6. This raised the possibility that Ltn1’s effect on NS levels might be indirect, mediated by a general decrease in proteasomal function. That this was not the case is suggested by the findings that both 20S proteolytic activity (Fig.S4a) and ubiquitylated protein levels in cells treated or not with the proteasome inhibitor MG132 (Fig.1e) were comparable in WT and ltn1Δ whole cell extracts (WCE). Furthermore, expression of an unrelated unstable protein was not affected by LTN1 deletion (see VHL below).
These results led to the hypothesis that Ltn1 might ubiquitylate nonstop proteins to signal their proteolysis. As predicted, Ltn1 and NS could be co-IP’ed, indicating their interaction (Fig.1f). Moreover, NS —but not the K0 control— was ubiquitylated in WT cells (Fig.1g), and this was Ltn1-dependent since less ubiquitin co-IP’ed with NS from ltn1Δ cells (compare ubiquitin-to-flag ratios in lanes 5 and 7). Moreover, MG132 induced accumulation of ubiquitylated NS in WT but not ltn1Δ cells (lanes 6,8), and failed to further increase NS protein levels in ltn1Δ cells (lanes 7, 8 and Fig.1e).
To evaluate Ltn1’s specificity, we examined its requirement for degradation of an unrelated PQC substrate, the human Von Hippel Lindau (VHL) protein. VHL expressed in yeast is misfolded, ubiquitylated and degraded 14. LTN1 deletion had no effect on VHL levels either at steady state or after 90 min of cycloheximide treatment (Fig.1h). Moreover, heat stress (37°C) accelerated VHL’s degradation to a similar extent in WT and ltn1Δ strains (Fig.1h). Thus, our results suggest an essential and direct role for Ltn1 in nonstop protein quality control, but not a general essential function in PQC.
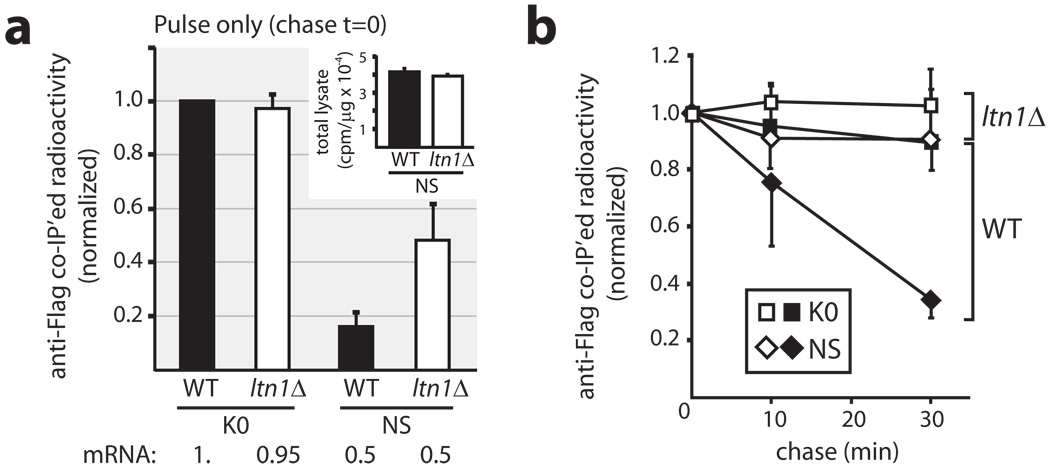
We next sought further evidence that Ltn1 regulates NS stability. WT and ltn1Δ strains expressing K0 or NS proteins were labelled with 35S-Met for 1 min (which should be just long enough for those proteins’ synthesis to be completed) and chased with cold Met and cycloheximide for up to 30 min. The results show an effect of Ltn1 as early as after 1 min of labelling (i.e., chase t=0; Fig.2a). At that time point, NS protein expression was >5-fold lower than K0 in WT yeast, while this difference was reduced to ~2-fold upon deletion of LTN1. (NS mRNA levels were ~2-fold lower than K0’s in both strains; Fig.S3b). Thus, Ltn1 may mediate rapid ubiquitylation and degradation of newly-synthesized NS proteins.
Figure 2. Ltn1 targets newly-synthesized nonstop proteins.
a, NS expression during a 1-min labeling with 35S-Met was increased in response to LTN1 deletion. K0 and NS expression in WT and ltn1Δ strains, normalized to K0 expression in WT cells. The inset shows similar labeling efficiency of total cellular proteins (cpm/µg WCE) in WT and ltn1Δ strains expressing NS protein. Below, corresponding mRNA levels (from Fig.S3b). b, Degradation of newly-synthesized NS protein is Ltn1-dependent. Cells labelled with a 1-min pulse (panel “a”) were chased with cold Met and cycloheximide before lysis and Flag IP. Labels: K0, squares; NS, diamonds; WT strain, black; ltn1Δ, white. Error bars shown below the labels for the WT and above for the ltn1Δ train. Each curve was normalized to chase time=0. a–b, The data is an average of two experiments (n=2) performed in duplicates each, and are representative of four independent experiments. Error bars show s.d.
While a putative additional effect of LTN1 deletion on nonstop mRNA translation rate could have contributed to the above result, a clear role for Ltn1 in regulating NS stability became evident upon examination of the chase (Fig.2b). Both K0 (Fig.2b) and the bulk of newly synthesized cellular proteins (Fig.S4b) were stable throughout, whether in WT or ltn1Δ trains. On the other hand, NS continued to be degraded in WT but not ltn1Δ trains (Fig.2b). Thus, newly-synthesized NS protein degradation depended on Ltn1.
The above findings raised the question of what triggers NS ubiquitylation by Ltn1. In the absence of stop codons, translation can proceed through the poly(A) tail, resulting in appendage of a poly(Lys) tract to the protein’s C-terminus (see Figs.1a and S1). A role for the poly(Lys) tract in triggering nonstop protein degradation was first suggested by the observation that hard coding 12 Lys residues in a stop codon-containing reporter protein sufficed to destabilize the latter 7 (12 Arg had a similar effect, suggesting the involvement of electrostatic interactions with the poly-basic sequences 15). Consistent with this model, the degradation of a GFP-Flag-HIS3-K12 reporter (K12; Fig.1a and 7) was also mediated by Ltn1: K12 expression was reduced compared to K0 and was completely restored upon deletion of either LTN1 or Ltn1’s RING domain (Fig.3a), but not of the E3 NOT4 (Fig.S5); LTN1 deletion did not affect K12 mRNA levels (Fig.S3b); K12 co-IP’ed with Ltn1 (Fig.3b); and K12 was ubiquitylated and degraded by the proteasome in an Ltn1-dependent manner (Figs.3c,d).
As poly(Lys) is appended to the C-terminus of nonstop proteins, we next examined whether Ltn1 targeting depended on the tract’s C-terminal location. Fusion of 1–4 HA tags to K12 immediately following the 12 lysines did not interfere with K12’s effect, such that expression of all K12-HA proteins in WT cells was reduced compared to K0-HA controls (Fig.3e). Moreover, in ltn1Δ cells K12-HAn protein expression was restored to near K0-HAn levels. However, the former were truncated, and of comparable size to the parental K12 (full-length proteins appeared not to be normally targeted for degradation by Ltn1). These results are consistent with previous reports implicating nascent poly-basic tracts in translational pausing and arrest 15,16, and suggest that translationally-arrested nonstop polypeptides may be targeted by Ltn1.
Nascent poly-basic tract-mediated translation arrest has been attributed to electrostatic interactions with the negatively-charged ribosomal polypeptide exit tunnel16. We reasoned that if such interactions were strong enough to arrest translation, nascent nonstop proteins might also remain stably associated with ribosomes. To test this, subcellular components were fractionated by sucrose gradient centrifugation. While in WT cell lysates both K0 and NS proteins sedimented at the top of the gradient, in ltn1Δ strain lysates, accumulated NS was also found in 80S fractions (Figs.3f and S6a). This effect was poly(Lys)-mediated, since K12 also co-fractionated with ribosomes in the absence of Ltn1 (Fig.S6d). To confirm that NS sedimentation in 80S fractions was mediated by its association with ribosomes, we first used EDTA to dissociate translating ribosomes into their 60S and 40S subunits. As expected, upon EDTA treatment, NS no longer sedimented in the 80S fraction (Fig.3f; fast-sedimenting K12 complexes were also dissociated by EDTA, Fig.S6e). Notably, in the presence of EDTA much of NS and K12 now co-fractionated with 60S ribosomes rather than being driven to the top of the gradient. This is consistent with the interaction of the nascent nonstop polypeptide with the 60S subunit being stable enough to resist EDTA-induced ribosomal dissociation. Lastly, we used an independent approach—co-IP—to obtain further evidence for association of poly(Lys)-containing proteins with ribosomes, and found that both NS and K12, but not K0, specifically co-IP’ed with Rpl3 (Fig.3g). Together, the results in Figs.3e–g suggest that nascent nonstop proteins stably stall in ribosomes upon synthesis of a poly(Lys) tract, and are subsequently cleared by Ltn1.
The above findings suggested that nonstop protein ubiquitylation begins in ribosomes, and predicted that Ltn1 might bind to the latter. We found several lines of evidence to support this possibility: first, Flag-tagged endogenous Ltn1 and Ltn1 ΔRING both specifically co-IP’ed with Rpl3 (Fig.4a); no Rpl3 signal was detected in the IP of an untagged strain or co-IP’ed with a Flag-tagged negative control expressed at higher levels than Ltn1. Second, Ltn1 was predominantly cytoplasmic and co-fractionated with the bulk of Rpl3 upon differential centrifugation of subcellular fractions (Figs.S7 and 4b). Lastly, Ltn1 mostly co-fractionated with 60S and possibly 80S ribosomes in sucrose gradient (Figs.4c and S8; indeed, endogenous Ltn1 had been previously reported in an inventory of proteins co-sedimenting in a sucrose gradient 60S fraction 17). Consistently, nearly all 60S ribosomal proteins and half of the 40S ribosomal proteins co-IP’ed with Ltn1 ΔRING but not with a control sample (Supplementary Table I). However, given that NS accumulated in translating (80S) ribosomes in the absence of Ltn1, it was surprising to find that most of the E3 was 60S-bound in steady-state. Among the possibilities we are currently examining is that Ltn1 promotes dissociation of 80S ribosomes following binding (see also Fig.S1a and S9 legends).
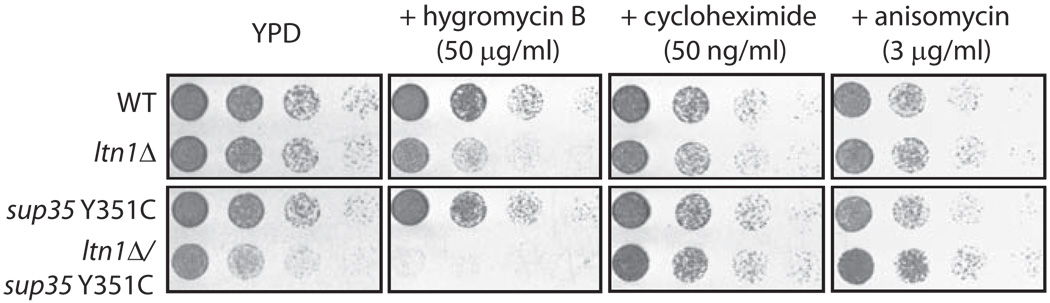
We next investigated Ltn1’s biological relevance in nonstop protein degradation. ltn1Δ cells had no obvious growth defect in rich media (YPD; Fig.5). Our discoveries raised the possibility that a phenotype for LTN1 deletion might be revealed under conditions of increased nonstop protein production. To test this, we first analyzed the sensitivity of ltn1Δ yeast to hygromycin B, an aminoglycoside antibiotic that affects translational fidelity and increases readthrough of stop codons (e.g., 18). Cells lacking Ltn1 exhibited greater sensitivity to the drug, and this effect appeared specific, as ltn1Δ cells were insensitive to other inhibitors of translation elongation that do not cause readthrough, namely cycloheximide and anisomycin (e.g.,19; Fig.5). More importantly, we also analyzed a strain harboring a hypomorphic mutation (Y351C) of the translation termination factor eRF3/Sup35 which selectively weakens stop codon recognition and nonsense suppression (e.g., 20). (Thus, in this strain, nonstop proteins are produced from normal, stop codon-containing mRNA.) LTN1 deletion led to a strong synthetic phenotype in this background (viability was reduced by 5–25-fold and growth was slower; Fig.5, YPD, lower row), underscoring the E3’s biological relevance in the QC of endogenous nonstop proteins. Moreover, the combination of ltn1Δ with the sup35 Y351C mutation and hygromycin B treatment led to complete synthetic lethality. Thus, Ltn1 confers resistance to stress caused by elevated nonstop protein production.
Figure 5. Ltn1 confers resistance to stress caused by nonstop protein production.
Cultures of the indicated strains normalized to equal cell density were spotted in 5-fold serial dilutions onto plates with rich media (YPD), containing or not the indicated drugs.
We propose a model for the elimination of nonstop proteins in eukaryotes that has analogous features to the ssrA-mediated process in bacteria 1—both involve tagging of stalled nascent polypeptides followed by degradation by ATP-dependent proteases (Fig.S1). The specific mechanisms utilized by bacteria and eukaryotes are, nonetheless, distinct. Translation of the eukaryote-specific mRNA poly(A) tail in mRNA lacking stop codons results in proteins C-terminally-extended with poly(Lys). This serves as an indirect proteolytic signal that induces Ltn1-dependent tagging of nonstop proteins with ubiquitin. How might this signaling occur? It is plausible, e.g., that interactions of the nascent poly(Lys) sequence in the ribosomal exit tunnel elicit conformational changes that are transduced to the ribosomal surface 21, revealing a binding site for Ltn1. Such a regulated mechanism allowing identification of ribosomes engaged in synthesis of nonstop proteins would explain Ltn1’s efficacy, in light of estimates that it is present at ~200 molecules per haploid yeast cell in steady-state 22—far substoichiometric to the ~170,000 translationally-active ribosomes 23.
Methods summary
Reagents and DNA constructs, yeast genetic methods, conditions for cell growth, cell extract preparation, immunoprecipitation, immunoblot, and pulse-labeling are described in detail in Methods (Supplemental Information). All strains were isogenic to BY4741, except for those in the experiments presented in Figs.5 and S3c, which are isogenic to 74-D694 (Methods). Sucrose gradient sedimentation, differential centrifugation, real-time PCR, immunofluorescence, and proteasome activity assay were performed as referenced in Methods. MG132 treatment of yeast was for 2h before lysis.
Supplementary Material
Acknowledgements
We thank M.C. Sogayar (U. of Sao Paulo, Brazil) for continued mentorship, I. Kwan for technical help, A. Bacconi for help with microscopy, T. Inada (Nagoya U., Japan), A. van Hoof (UT-Houston), S. Liebman (U. of Illinois) and J. Frydman (Stanford U.) for reagents, the Williamson and Saez laboratories (TSRI) for help with sucrose gradient fractionation and real-time PCR, respectively, E. Peters (GNF) for mass spectrometry analyses, M. Smolka (Cornell U.) for help with yeast methods, and R. Deshaies (Caltech), E.P. Geiduschek (UCSD) and T. Hunter (Salk Institute) for discussions. In loving memory of Zenaide P. Joazeiro.
Footnotes
Full Methods and any associated references are available in the Supplementary Information.
Supplementary Information accompanies the paper on www.nature.com/nature.
Author Contributions. C.A.P.J. and M.H.B. designed the studies, interpreted the data and wrote the manuscript. M.H.B. conducted experiments.
Author Information. Reprints and permissions information is available at www.nature.com/reprints.
References
- 1.Moore SD, Sauer RT. The tmRNA system for translational surveillance and ribosome rescue. Annu. Rev. Biochem. 2007;76:101–124. doi: 10.1146/annurev.biochem.75.103004.142733. [DOI] [PubMed] [Google Scholar]
- 2.Frischmeyer PA, van Hoof A, O'Donnell K, et al. An mRNA surveillance mechanism that eliminates transcripts lacking termination codons. Science. 2002;295:2258–2261. doi: 10.1126/science.1067338. [DOI] [PubMed] [Google Scholar]
- 3.Dulebohn D, Choy J, Sundermeier T, et al. Trans-translation: the tmRNA-mediated surveillance mechanism for ribosome rescue, directed protein degradation, and nonstop mRNA decay. Biochemistry. 2007;46:4681–4693. doi: 10.1021/bi6026055. [DOI] [PubMed] [Google Scholar]
- 4.van Hoof A, Frischmeyer PA, Dietz HC, et al. Exosome-mediated recognition and degradation of mRNAs lacking a termination codon. Science. 2002;295:2262–2264. doi: 10.1126/science.1067272. [DOI] [PubMed] [Google Scholar]
- 5.Doma MK, Parker R. RNA quality control in eukaryotes. Cell. 2007;131:660–668. doi: 10.1016/j.cell.2007.10.041. [DOI] [PubMed] [Google Scholar]
- 6.Wilson MA, Meaux S, van Hoof A. A genomic screen in yeast reveals novel aspects of nonstop mRNA metabolism. Genetics. 2007;177:773–784. doi: 10.1534/genetics.107.073205. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Ito-Harashima S, Kuroha K, Tatematsu T, et al. Translation of the poly(A) tail plays crucial roles in nonstop mRNA surveillance via translation repression and protein destabilization by proteasome in yeast. Genes Dev. 2007;21:519–524. doi: 10.1101/gad.1490207. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Li W, Bengtson MH, Ulbrich A, et al. Genome-wide and functional annotation of human E3 ubiquitin ligases identifies MULAN, a mitochondrial E3 that regulates the organelle's dynamics and signaling. PLoS One. 2008;3:e1487. doi: 10.1371/journal.pone.0001487. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Deshaies RJ, Joazeiro CA. RING domain E3 ubiquitin ligases. Annu. Rev. Biochem. 2009;78:399–434. doi: 10.1146/annurev.biochem.78.101807.093809. [DOI] [PubMed] [Google Scholar]
- 10.Chu J, Hong NA, Masuda CA, et al. A mouse forward genetics screen identifies LISTERIN as an E3 ubiquitin ligase involved in neurodegeneration. Proc. Natl. Acad. Sci. U S A. 2009;106:2097–2103. doi: 10.1073/pnas.0812819106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Braun MA, Costa PJ, Crisucci EM, et al. Identification of Rkr1, a nuclear RING domain protein with functional connections to chromatin modification in Saccharomyces cerevisiae. Mol. Cell. Biol. 2007;27:2800–2811. doi: 10.1128/MCB.01947-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Inada T, Aiba H. Translation of aberrant mRNAs lacking a termination codon or with a shortened 3'-UTR is repressed after initiation in yeast. EMBO J. 2005;24:1584–1595. doi: 10.1038/sj.emboj.7600636. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Verma R, Chen S, Feldman R, et al. Proteasomal proteomics: identification of nucleotide-sensitive proteasome-interacting proteins by mass spectrometric analysis of affinity-purified proteasomes. Mol. Biol. Cell. 2000;11:3425–3439. doi: 10.1091/mbc.11.10.3425. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.McClellan AJ, Scott MD, Frydman J. Folding and quality control of the VHL tumor suppressor proceed through distinct chaperone pathways. Cell. 2005;121:739–748. doi: 10.1016/j.cell.2005.03.024. [DOI] [PubMed] [Google Scholar]
- 15.Dimitrova LN, Kuroha K, Tatematsu T, et al. Nascent peptide-dependent translation arrest leads to Not4p-mediated protein degradation by the proteasome. J. Biol. Chem. 2009;284:10343–10352. doi: 10.1074/jbc.M808840200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Lu J, Deutsch C. Electrostatics in the ribosomal tunnel modulate chain elongation rates. J. Mol. Biol. 2008;384:73–86. doi: 10.1016/j.jmb.2008.08.089. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Fleischer TC, Weaver CM, McAfee KJ, et al. Systematic identification and functional screens of uncharacterized proteins associated with eukaryotic ribosomal complexes. Genes Dev. 2006;20:1294–1307. doi: 10.1101/gad.1422006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Brodersen DE, Clemons WMJ, Carter AP, et al. The structural basis for the action of the antibiotics tetracycline, pactamycin, and hygromycin B on the 30S ribosomal subunit. Cell. 2000;103:1143–1154. doi: 10.1016/s0092-8674(00)00216-6. [DOI] [PubMed] [Google Scholar]
- 19.Kirillov S, Porse BT, Vester B, et al. Movement of the 3'-end of tRNA through the peptidyl transferase centre and its inhibition by antibiotics. FEBS Lett. 1997;406:223–233. doi: 10.1016/s0014-5793(97)00261-5. [DOI] [PubMed] [Google Scholar]
- 20.Bradley ME, Bagriantsev S, Vishveshwara N, et al. Guanidine reduces stop codon read-through caused by missense mutations in SUP35 or SUP45. Yeast. 2003;20:625–632. doi: 10.1002/yea.985. [DOI] [PubMed] [Google Scholar]
- 21.Rospert S. Ribosome function: governing the fate of a nascent polypeptide. Curr. Biol. 2004;14:R386–R388. doi: 10.1016/j.cub.2004.05.013. [DOI] [PubMed] [Google Scholar]
- 22.Ghaemmaghami S, Huh WK, Bower K, et al. Global analysis of protein expression in yeast. Nature. 2003;425:737–741. doi: 10.1038/nature02046. [DOI] [PubMed] [Google Scholar]
- 23.Zenklusen D, Larson DR, Singer RH. Single-RNA counting reveals alternative modes of gene expression in yeast. Nature Struct. Mol. Biol. 2008;15:1263–1271. doi: 10.1038/nsmb.1514. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.