Diffuse large B-cell lymphoma (DLBCL) is a disease marked by heterogeneity in clinical presentation, morphology, and underlying biology. Importantly, this translates into variability of patient response to chemotherapy. While a majority of patients can be cured effectively with modern chemotherapy regimens, many present with or acquire resistance to treatment. Characterizing the mechanism(s) responsible for poor patient outcome is critical for identifying future targets to improve treatment.
Major histocompatibility complex class II (MHCII) molecules are cell-surface glycoproteins that present peptides for antigen recognition, and are important for the adaptive immune response. Studies have shown that loss of MHCII expression correlates with poor outcome in DLBCL following CHOP or R-CHOP therapy [1–3], as well as in primary mediastinal B-cell lymphoma (PMBCL) [4]. This loss has been associated with deficiencies in tumor-infiltrating T-lymphocytes [2], suggesting a loss of immune surveillance.
The expression of classical MHCII, non-classical MHCII, and invariant chain molecules are coordinately regulated by the class II transactivator (CIITA), the master regulator of MHCII transcription [5–7]. CIITA is essential for MHCII expression; thus, control of MHCII expression depends on control of and by CIITA expression [5,8]. MHCII cell-specific and temporal expression is controlled by the differential activation of four different promoters within the CIITA gene [8]. The CIITA promoters pIII and pIV are important for MHCII expression in B-cells and hematopoietic tumors [6]. We have shown that expression of each of the classical, non-classical, and invariant chain MHCII molecules and CIITA correlate with one another in DLBCL patient samples, that intervening genes are not affected, and that large deletions within MHCII genes do not explain lost expression [9]. Our demonstration that MHCII is consistently downregulated in concert with CIITA suggests that altered transcription of MHCII via the master transactivator CIITA is a mechanism for MHCII down-regulation in DLBCL, a finding confirmed in DLBCL cell culture studies [7]. We have further shown that mutations of CIITA do not explain the coordinate down-regulation [10].
DNA methylation of CIITA pIII and pIV is one mechanism of MHCII suppression in hematopoietic and other tumors [6,11,12]. Treatment with demethylating agents has been shown to re-express CIITA and MHCII [11,12], suggesting a potential for such agents in therapy. We therefore asked whether CpG methylation of CIITA is associated with loss of MHCII expression in DLBCL. As we have shown that CIITA expression correlates with MHCII expression in DLBCL [9], and because loss of MHCII is a meaningful endpoint in DLBCL, we chose to correlate methylation of CIITA with MHCII expression. We hypothesized that methylation of CIITA pIII and pIV would be more extensive in MHCII(−) samples. To determine whether CpG methylation (Me-CpG) of CIITA pIII and pIV correlated with loss of MHCII expression in DLBCL, we performed bisulfite sequencing [13].
Approval for this study was obtained from the University of Arizona Institutional Review Board in accordance with the Declaration of Helsinki. Forty-six patient samples were obtained from the Leukemia/Lymphoma Molecular Profiling Project (LLMPP), of which we are principle participants. These cases had been previously reviewed by a panel of hematopathologists and each contained a minimum of 70% tumor. MHCII expression had been determined by gene expression profiling [1]. As described previously, the patients in the lowest tenth percentile of MHCII expression by gene expression profiling were considered MHCII(−) [9]. Twenty-three LLMPP samples comprised 20 de novo DLBCL and three PMBCL in the lowest 10% of MHCII expressers and were considered MHCII(−). Two transformed DLBCL, one relapsed DLBCL, and one relapsed PMBCL with expression of MHCII in the range of the low 10% expressers were considered MHCII(−). Four de novo DLBCL in the range of 10–25% of MHCII expressers were considered MHCII(+). DLBCL cases represented both ABC and GCB subtypes [Supplemental Figure 1]. The remaining 15 LLMPP samples were PMBCL samples with levels of MHCII(+) expression in the 25–100% range [4].
Fourteen tissue samples were obtained from the Arizona Lymphoid Tissue and Blood Repository. Four were lymphoma cases: one MHCII(+) DLBCL; one MHCII(−) DLBCL; and two MHCII(−) peripheral T-cell lymphoma (PTCL), unspecified. MHCII expression was determined by immunohistochemistry (data not shown); we have previously shown that cases in the tenth percentile of MHCII by gene expression profiling, which correlate with poor outcome, are also negative by immunohistochemistry [2]. Ten additional samples were reactive tissues: seven lymph nodes and three tonsils.
MHCII expression in twelve cell lines was determined by flow cytometry [7] (and data not shown); cell lines with peaks that did not differ substantially from isotype controls were considered MHCII(−). Jurkat, JAR, Raji, and DB lines were from the American Type Culture Collection (Manassas, Virginia). The RJ2.2.5 cell line was a gift from Dr. Jeremy Boss (Emory University, Atlanta, Georgia) with the permission of Dr. Robert Accolla (University of Insubria, Varese, Italy) [14]. Other lines were gifted from Dr. Louis Staudt (coauthor).
Bisulfite modification of purified DNA (50 ng) was performed using the Epitect Bisulfite Kit (Qiagen) with 40 μL yield. Bisulfite-modified DNA was amplified by nested polymerase chain reaction (PCR) using the Multiplex PCR Kit with Q-solution (Qiagen). CIITA pIII and pIV were amplified individually using primers specific for bisulfite-modified DNA (i.e., assuming C converted to T). Initial amplification was performed using 3 μl bisulfite-modified DNA and 20 nM primers per 25 μl reaction. The second, nested amplification was performed using 3 μl DNA from the initial PCR reaction, with 20 nM primers per 50 μl reaction. Primer sequences for nested PCR: P3forward: 5′-TTAAGGGAGTGTG GTAAAATTAGAGGGTG-3′; P3reversenested: 5′-AAACACAAACTCCTATTCCCATCCTCAC-3′; P3reverseouter: 5′-AAACAA CTCTTTCACATCTTCCAATAACCTAC-3′; P4forwardouter: 5′-GGTTG GATTGAGTTGGAGAGAA ATAGAGAT-3′; P4forwardnested: 5′-TGGGGATAAGTTTTTTGTAA TTTAGGA-3′; P4reverse: 5′-CTACTAATAACCTCTCCCTCCAGCCAA-3′. CIITA pIV primers, as well as PCR conditions, were as described by Morris, et al., 2002 [15]. Amplicons were purified with the QiaQuick PCR Purification Kit (Qiagen), ligated into the pGEM-T Easy plasmid (Promega, Madison, WI), and transformed into DH5α competent E. coli cells (Invitrogen). Single colonies were expanded and plasmids purified with the Wizard Plus SV Miniprep DNA Purification system (Promega). Sequencing was performed at Arizona’s Genomic Analysis and Technology Core using primer PCM13R = 5′-TCACACAGGAAACAGCTATGAC, which binds within the plasmid lacz gene, to sequence the locus of amplicon inserts. Ten pIII and pIV amplicons were reported per sample.
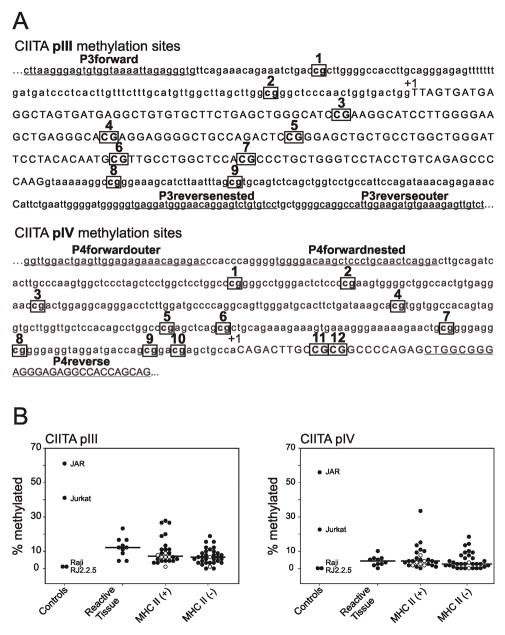
The methylation status of CIITA pIII and pIV was determined at nine and twelve CpG sites, respectively [Figure 1A]. Tallies of methylation marks within each of the ten amplicons per sample, per promoter, were divided by the number of possible methylation marks in ten samples (i.e., 90 for pIII, and 120 for pIV), to give a percentage methylation for each sample. Raji and RJ2.2.5 were used as negative controls. Raji, a Burkitt line, expresses CIITA and MHCII constitutively, and does not demonstrate methylation at the CIITA pIV promoter [11,12]. RJ2.2.5, a derivative of Raji [14], is MHCII(−) due to a deletion and an inactivating mutation of the CIITA alleles [5]. Both Raji and RJ2.2.5 revealed minimal methylation of pIII (1.1%) and pIV (0.0%) [Figure 1B]. Jurkat and JAR were used as positive controls. Jurkat, a T-cell leukemia line, does not express CIITA or MHCII, and has been shown to be methylated at CIITA pIV [12]. JAR, a human placental choriocarcinoma line, does not express CIITA or MHCII, and has been shown to be extensively methylated at CIITA pIV [11]. We confirmed extensive methylation, with Jurkat having 41.1% and 22.5%, and JAR having 61.1% and 55.8% methylation at pIII and pIV, respectively [Figure 1B].
Figure 1.
(A) CIITA promoter sites tested for CpG methylation and primers used for nested PCR. Possible CpG sites for methylation of CIITA promoters pIII and pIV are boxed and numbered. Sites for primer binding are underlined. Exons are in capitals and indicated by +1 [8]. (B) Percentage methylation of CIITA promoters pIII and pIV. Tallies of methylation marks within each of the ten amplicons per sample, per promoter, were divided by the number of possible methylation marks in ten samples (i.e., 90 for pIII, and 120 for pIV), to give a percentage methylation for each sample. Raji and RJ2.2.5 do not demonstrate CIITA methylation, as described in the text, and were used as negative controls. Jurkat and JAR demonstrate CIITA methylation, as described in the text, and were used as positive controls. Kruskal-Wallis One Way Analysis of Variance on Ranks and Dunn’s method were used to determine significant differences in methylation of CIITA pIII and pIV between reactive tissues, MHCII(+), and MHCII(−) samples using SigmaPlot 11.0 (San Jose, CA). Cell lines within the MHCII(+) and MHCII(−) groups are indicated with open circles.
The reactive tissues contain a variety of cell types, including B-cells, T-cells, dendritic cells, macrophages, and stromal cells, which likely have varying levels of MHCII expression, with varying underlying methylation of the CIITA promoter. Median methylation of pIII in benign tissues was 12.2% (range: 4.4% to 23.3%). Median methylation of the pIV was 4.2% (range: 0.0% to 10.0%) [Figure 1B].
The 20 MHCII(+) cases comprised five de novo DLBCL and 15 PMBCL. Six MHCII(+) DLBCL lines were included: SUDHL4; SUDHL6; SUDHL10; OCI-Ly3; OCI-Ly7; and OCI-Ly19. Median methylation of pIII in MHCII(+) cases and lines was 7.2% (range: 1.1% to 27.8%). Median methylation of pIV was 4.2% (range: 0.0% to 33.3%) [Figure 1B].
The 30 MHCII(−) cases comprised 21 de novo DLBCL, three relapsed or transformed DLBCL, two PTCL, three PMBCL, and one relapsed PMBCL. Two MHCII(−) lines were included: DB and OCI-Ly10. Median methylation of pIII in MHCII(−) cases and lines was 6.7% (range: 0.0% to 18.8%). The median percentage of pIII promoter methylation in MHCII(−) cases was significantly lower than benign samples (p<0.05), but not significantly different from MHCII(+) cases. Median methylation of pIV in MHCII(−) cases and lines was 2.50% (range: 0.0% to 18.3%). The median percentage of pIV promoter methylation in MHCII(−) cases was not significantly different from benign samples or MHCII(+) cases [Figure 1B]. Additionally, CIITA pIII and pIV methylation did not correlate with CIITA gene expression data in the MHCII(+) and (−) cases (data not shown).
We conclude that differences in CIITA promoter methylation do not correlate with differences in MHCII expression. Therefore, epigenetic silencing of CIITA by DNA methylation is not a likely mechanism for loss of MHCII expression in DLBCL. Ongoing research will investigate other mechanisms, including CIITA histone acetylation changes and the expression of genes in regulatory pathways upstream of CIITA.
Supplementary Material
Supplemental Figure 1. Distribution of methylation within CIITA promoters pIII and pIV in (A) cell lines, (B) reactive tissues, and (C) lymphoma cases. The methylation status of CIITA pIII and pIV was determined at nine and twelve CpG sites, respectively. Data from ten successfully bisulfite-modified clones was compressed using a color scale for methylation marks at each potential Me-C site (Legend). Abbreviations: LN, lymph node; GCB, germinal center B-cell; Uncl, unclassified; ABC, activated B-cell; PMBL, peripheral mediastinal B-cell lymphoma; DLBCL, diffuse large B-cell lymphoma; PTCL, peripheral T-cell lymphoma; Tf, transformed; Re, relapsed.
Acknowledgments
Funding was from American Cancer Society RSG0605501L1B (L.M.R.), NCI-T32 CA09213 (S.T.W.), and NCI/NIH 1-U10-CA-84967-02 (W.C.C.). We thank Jessica Rimsza, Betty Glinsmann-Gibson, and Robin Roberts for technical work and data analysis.
Footnotes
The authors report no conflicts of interest. The authors alone are responsible for the content and writing of the paper
References
- 1.Rosenwald A, Wright G, Chan WC, Connors JM, Campo E, Fisher RI, Gascoyne RD, Muller-Hermelink HK, Smeland EB, Giltnane JM, Hurt EM, Zhao H, Averett L, Yang L, Wilson WH, Jaffe ES, Simon R, Klausner RD, Powell J, Duffey PL, Longo DL, Greiner TC, Weisenburger DD, Sanger WG, Dave BJ, Lynch JC, Vose J, Armitage JO, Montserrat E, Lopez-Guillermo A, Grogan TM, Miller TP, LeBlanc M, Ott G, Kvaloy S, Delabie J, Holte H, Krajci P, Stokke T, Staudt LM. The use of molecular profiling to predict survival after chemotherapy for diffuse large-B-cell lymphoma. N Engl J Med. 2002;346:1937–47. doi: 10.1056/NEJMoa012914. [DOI] [PubMed] [Google Scholar]
- 2.Rimsza LM, Roberts RA, Miller TP, Unger JM, Leblanc M, Braziel RM, Weisenberger DD, Chan WC, Muller-Hermelink HK, Jaffe ES, Gascoyne RD, Campo E, Fuchs DA, Spier CM, Fisher RI, Delabie J, Rosenwald A, Staudt LM, Grogan TM. Loss of MHC class II gene and protein expression in diffuse large B-cell lymphoma is related to decreased tumor immunosurveillance and poor patient survival regardless of other prognostic factors: a follow-up study from the Leukemia and Lymphoma Molecular Profiling Project. Blood. 2004;103:4251–8. doi: 10.1182/blood-2003-07-2365. [DOI] [PubMed] [Google Scholar]
- 3.Rimsza LM, Leblanc ML, Unger JM, Miller TP, Grogan TM, Persky DO, Martel RR, Sabalos CM, Seligmann B, Braziel RM, Campo E, Rosenwald A, Connors JM, Sehn LH, Johnson N, Gascoyne RD. Gene expression predicts overall survival in paraffin-embedded tissues of diffuse large B-cell lymphoma treated with R-CHOP. Blood. 2008;112:3425–33. doi: 10.1182/blood-2008-02-137372. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Roberts RA, Wright G, Rosenwald AR, Jaramillo MA, Grogan TM, Miller TP, Frutiger Y, Chan WC, Gascoyne RD, Ott G, Muller-Hermelink HK, Staudt LM, Rimsza LM. Loss of major histocompatibility class II gene and protein expression in primary mediastinal large B-cell lymphoma is highly coordinated and related to poor patient survival. Blood. 2006;108:311–8. doi: 10.1182/blood-2005-11-4742. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Steimle V, Otten LA, Zufferey M, Mach B. Complementation cloning of an MHC class II transactivator mutated in hereditary MHC class II deficiency (or bare lymphocyte syndrome) Cell. 1993;75:135–46. [PubMed] [Google Scholar]
- 6.Wright KL, Ting JP. Epigenetic regulation of MHC-II and CIITA genes. Trends Immunol. 2006;27:405–12. doi: 10.1016/j.it.2006.07.007. [DOI] [PubMed] [Google Scholar]
- 7.Cycon KA, Rimsza LM, Murphy SP. Alterations in CIITA constitute a common mechanism accounting for downregulation of MHC class II expression in diffuse large B-cell lymphoma (DLBCL) Exp Hematol. 2009;37:184–94. doi: 10.1016/j.exphem.2008.10.001. [DOI] [PubMed] [Google Scholar]
- 8.Muhlethaler-Mottet A, Otten LA, Steimle V, Mach B. Expression of MHC class II molecules in different cellular and functional compartments is controlled by differential usage of multiple promoters of the transactivator CIITA. EMBO J. 1997;16:2851–60. doi: 10.1093/emboj/16.10.2851. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Rimsza LM, Roberts RA, Campo E, Grogan TM, Bea S, Salaverria I, Zettl A, Rosenwald A, Ott G, Muller-Hermelink HK, Delabie J, Fisher RI, Unger JM, Leblanc M, Staudt LM, Jaffe ES, Gascoyne RD, Chan WC, Weisenburger DD, Greiner T, Braziel RM, Miller TP. Loss of major histocompatibility class II expression in non-immune-privileged site diffuse large B-cell lymphoma is highly coordinated and not due to chromosomal deletions. Blood. 2006;107:1101–7. doi: 10.1182/blood-2005-04-1510. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Rimsza LM, Chan WC, Gascoyne RD, Campo E, Jaffe ES, Staudt LM, Delabie J, Rosenwald A, Murphy SP. CIITA or RFX coding region loss of function mutations occur rarely in diffuse large B-cell lymphoma cases and cell lines with low levels of major histocompatibility complex class II expression. Haematologica. 2009;94:596–8. doi: 10.3324/haematol.2008.000752. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Morris AC, Spangler WE, Boss JM. Methylation of class II trans-activator promoter IV: a novel mechanism of MHC class II gene control. J Immunol. 2000;164:4143–9. doi: 10.4049/jimmunol.164.8.4143. [DOI] [PubMed] [Google Scholar]
- 12.Morimoto Y, Toyota M, Satoh A, Murai M, Mita H, Suzuki H, Takamura Y, Ikeda H, Ishida T, Sato N, Tokino T, Imai K. Inactivation of class II transactivator by DNA methylation and histone deacetylation associated with absence of HLA-DR induction by interferon-gamma in haematopoietic tumour cells. Br J Cancer. 2004;90:844–52. doi: 10.1038/sj.bjc.6601602. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Frommer M, McDonald LE, Millar DS, Collis CM, Watt F, Grigg GW, Molloy PL, Paul CL. A genomic sequencing protocol that yields a positive display of 5-methylcytosine residues in individual DNA strands. Proc Natl Acad Sci U S A. 1992;89:1827–31. doi: 10.1073/pnas.89.5.1827. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Accolla RS. Human B cell variants immunoselected against a single Ia antigen subset have lost expression of several Ia antigen subsets. J Exp Med. 1983;157:1053–8. doi: 10.1084/jem.157.3.1053. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Morris AC, Beresford GW, Mooney MR, Boss JM. Kinetics of a gamma interferon response: expression and assembly of CIITA promoter IV and inhibition by methylation. Mol Cell Biol. 2002;22:4781–91. doi: 10.1128/MCB.22.13.4781-4791.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Supplemental Figure 1. Distribution of methylation within CIITA promoters pIII and pIV in (A) cell lines, (B) reactive tissues, and (C) lymphoma cases. The methylation status of CIITA pIII and pIV was determined at nine and twelve CpG sites, respectively. Data from ten successfully bisulfite-modified clones was compressed using a color scale for methylation marks at each potential Me-C site (Legend). Abbreviations: LN, lymph node; GCB, germinal center B-cell; Uncl, unclassified; ABC, activated B-cell; PMBL, peripheral mediastinal B-cell lymphoma; DLBCL, diffuse large B-cell lymphoma; PTCL, peripheral T-cell lymphoma; Tf, transformed; Re, relapsed.