Abstract
Among the lactic acid bacteria (LAB) present in the oenological microbial ecosystem, Oenococcus oeni, an acidophilic lactic acid bacterium, is essential during winemaking. It outclasses all other bacterial species during malolactic fermentation (MLF). Oenological performances, such as malic acid degradation rate and sensorial impact, vary significantly according to the strain. The genetic diversity of the O. oeni species was evaluated using a multilocus sequence typing (MLST) scheme. Seven housekeeping genes were sequenced for a collection of 258 strains that had been isolated all over the world (particularly Burgundy, Champagne, and Aquitaine, France, Chile, South Africa, and Italy) and in several wine types (red wines, white wines, and champagne) and cider. The allelic diversity was high, with an average of 20.7 alleles per locus, many of them being rare alleles. The collection comprised 127 sequence types, suggesting an important genotypic diversity. The neighbor-joining phylogenetic tree constructed from the concatenated sequence of the seven housekeeping genes showed two major phylogenetic groups, named A and B. One unique strain isolated from cider composed a third group, rooting the phylogenetic tree. However, all other strains isolated from cider were in group B. Eight phylogenetic subgroups were statistically differentiated and could be delineated by the analysis of only 32 mutations instead of the 600 mutations observed in the concatenated sequence of the seven housekeeping genes. Interestingly, in group A, several phylogenetic subgroups were composed mostly of strains coming from a precise geographic origin. Three subgroups were identified, composed of strains from Chile, South Africa, and eastern France.
Oenococcus oeni is a Gram-positive bacterium belonging to the lactic acid bacteria (LAB) group and is mainly encountered on grapes and in wine. During the winemaking process, from grape must to wine, it gradually becomes the dominant bacterial species. It is also the main species involved in malolactic fermentation (MLF). MLF is an essential winemaking step occurring after alcoholic fermentation (AF). It contributes to deacidification and flavor changes in wine (16). After AF, carried out by Saccharomyces cerevisiae, the high-stress conditions in wine induce a decrease in microbial diversity, allowing only the most resistant species of yeast and LAB to survive. Among LAB, O. oeni is the most adapted to these stress conditions and consequently is most commonly associated with MLF. The oenological properties of O. oeni vary highly with respect to the considered strain. Indeed, O. oeni strains do not have the same ability to grow and survive in wine. These properties are mainly related to low pH and high ethanol tolerance but also to resistance to other toxic compounds, such as phenolic compounds. This phenotypic variability is linked to genotypic differences. It may be characterized by the presence or absence of some genes (22) which could be implicated in the response to environmental stress or by the variation in gene expression (3).
Among the LAB group, the O. oeni species, formerly Leuconostoc oenos, has been shown to have evolved rapidly from the other lactic acid bacteria (18), forming a new genus, Oenococcus, as described by Dicks et al. (7). Today, this genus is composed of two species, Oenococcus oeni and Oenococcus kitaharae (9). The high level of phylogenetic divergence of the genus Oenococcus compared to that of other lactic acid bacteria has been correlated to the absence of the mismatch mutation repair system. This contributes to a high mutation rate, an excess of recombination, and a rapid genetic evolution and is characterized by a lack of certain genes and highly divergent sequences (19). However, several studies based on the sequencing of the 16S, 23S, and 16S-to-23S intergenic spacer region (ISR) sequences suggested that the species O. oeni is genetically homogenous (14, 20, 21, 26). This was reinforced by the levels of DNA-DNA homology and by the similarities of genetic maps observed for various strains (8, 27).
Originally, evaluation of O. oeni diversity was performed by analysis of restriction fragments of entire genomes (restriction endonuclease analysis-pulsed-field gel electrophoresis [REA-PFGE]) (13). However, multilocus sequence typing (MLST) has been shown to be the most suitable and reliable approach (17) for studying genetic diversity and population structure. MLST targeting housekeeping genes presents the advantage to generate data that can be compared in different laboratories. Moreover, sequencing of housekeeping genes in a population can show the intergenic recombination within the population in addition to giving information on the genetic population structure.
To date, two papers have reported the analysis of O. oeni genetic diversity using MLST but with different results. In the first study, 18 strains were analyzed and 5 loci were sequenced, with a high level of allelic diversity and frequent recombination events within the species (5). In the second paper, 43 strains and 8 loci were used. Two distinct phylogenetic groups appeared, which was in agreement with the finding of two groups by REA-PFGE (4). Several other methods have also delineated two distinct genetic groups within the O. oeni species. These two groups were observed by REA-PFGE (4, 13) as well as random amplified polymorphism DNA (RAPD) (25) and ribotyping analyses (5, 25). However, to date, the existence of these two distinct phylogenetic groups cannot be ascertained, mainly because of the restricted number and low variety of the studied strains.
The aim of this study was to analyze, by the two most discriminating methods used for genetic diversity analysis (REA-PFGE and MLST), a large collection of O. oeni strains taken from broad and various environmental (red and white wines, champagne, and ciders) and geographical origins.
MATERIALS AND METHODS
Strain cultures.
A total of 513 strains were obtained from several collections around the world. They mainly originated from distant wine-producing areas, with strains originating from France (313 isolates) (including 87 from Champagne, 55 from Burgundy, and 41 from Aquitaine), Chile (46 isolates), South Africa (33 isolates), Italy (30 isolates), Germany (27 isolates), and other countries (Spain, United States, Argentina, Brazil, Switzerland, Australia, Greece, Austria, and Portugal) (64 isolates). They were isolated from several wine types: red wine (150 isolates) or white wines (43 isolates), champagne and fortified wine, and several grape varieties. Strains isolated from stone fruit mashes (12 isolates) and ciders (32 isolates) were also included. Overall, the strains were isolated over the 1967-to-2008 period. They were cultivated at 25°C in de Man, Rogosa, and Sharpe broth (MRS) (6) at pH 4.8 supplemented with 1% d-l malic acid (Sigma-Aldrich) and then stored at −80°C.
REA-PFGE analysis.
Strain typing was performed by enzymatic restriction of the total genome, using the endonuclease NotI, followed by pulsed-field gel electrophoresis (i.e., restriction endonuclease analysis-pulsed-field gel electrophoresis [REA-PFGE]) (10). Bacterial cells were collected during mid-exponential phase by centrifugation (10 min at 10,000 × g). They were then washed with Tris-EDTA (TE) buffer (10 mM Tris, 1 mM EDTA, pH 7.5), resuspended in T100E buffer (10 mM Tris, 100 mM EDTA, pH 7.5), and embedded in plugs of Megabase agarose (Bio-Rad). The agarose plugs were incubated at 37°C in T100E containing 10 mg ml−1 lysozyme for 3 h and then in T100E supplemented with 2 mg ml−1 pronase and 15 mg ml−1 N-laurylsarcosine for 16 h. After successive washings in TE buffer and water, the agarose-embedded DNA was digested by incubating the plugs for 16 h at 25°C in 100-μl reaction mixtures containing 10 units of NotI endonuclease and the appropriate buffer (New England Biolabs). The digested DNA fragments were separated by electrophoresis in a 1% PFGE agarose gel (Bio-Rad) in 0.5× Tris-boric acid-EDTA (TBE) buffer (45 mM Tris-HCl, pH 8.0, 45 mM boric acid, 1 mM EDTA) using a CHEF-DRIII apparatus (Bio-Rad) with pulse times of 1 to 25 s, at 6 V cm−1, for 22 h and at 15°C. After electrophoresis, the gel was stained for 30 min with 0.5 μg ml−1 ethidium bromide and observed under UV light. The DNA fingerprint patterns were analyzed using Bionumerics 5.1 software (Applied Maths, Kortrijk, Belgium). The comparison of profiles allowed for a dendrogram to be created using the unweighted-pair group method using arithmetic means (UPGMA), the Dice coefficient of similarity, and a tolerance limit of 2.3%.
DNA extraction and MLST.
For DNA extraction, the Wizard genomic DNA purification (Promega) or the Nucleospin tissue (Macherey-Nagel) kit was used according to the manufacturer's instructions. PCR were performed in a 50-μl reaction volume containing a DNAzyme PCR master mix (Finnzymes), 10 ng of template DNA, and 10 pmol of each primer associated with one of the seven target genes. The seven housekeeping targeted genes were gyrB, g6pd, pgm, dnaE, purK, rpoB, and recP (primers used are listed in Table 1). The PCR program was as follows: 95°C for 5 min, 25 cycles of 95°C for 30 s, 55°C for 30 s, and 72°C for 30 s, with a final extension at 72°C for 5 min. For some loci, for a better amplification, the annealing temperature was lowered to 45°C and the number of cycles increased to 35. For the amplification of the transposon sequence in the purK locus, the elongation time was increased to 1 min. PCR fragment amplification was verified by electrophoresis of the obtained PCR products in 1% agarose gels containing 10 μl GelRed (Biotium) for 100 ml agarose. The PCR products were then sequenced by either the Genotyping and Sequencing Laboratory of Bordeaux 2 University or the GATC Company.
TABLE 1.
Gene name, function, primers, and amplicon size
| Gene | Gene product function | Amplicon size (bp) | Primer set | Sequence (5′-3′) |
|---|---|---|---|---|
| gyrB | Gyrase, β subunit | 674 | gyrB-1 | CTTCGGTTGTTAATGCTTTGTC |
| gyrB-2 | CAACTTGGTTTTTGTCTGCC | |||
| g6pd | Glucose-6-phosphate dehydrogenase | 669 | g6pd-1 | TTATATGTCTGTTGCTCCTCGT |
| g6pd-2 | CCGGTTCTGATGTAAAAAGG | |||
| Pgm | Phosphoglucomutase | 654 | pgm-1 | ATATCTGCCGAAGTGCTAAGAG |
| pgm-2 | AGCAGCAATTTGATTTCCAG | |||
| dnaE | DNA polymerase III, α subunit | 714 | dnaE-1 | CGTATATAGAGCGCTTTGCC |
| dnaE-2 | CGTTCTTATCGCGAGTTGTAC | |||
| purK | Phosphorybosylaminoimidazole carboxylase | 597 | purK-1 | TGGTTATCATGTTGGTATTTTGG |
| purK-2 | GAAGCAGGAGCATAGGAAAGA | |||
| rpoB | RNA polymerase, β subunit | 665 | rpob-1 | CGATATTCTCCTTTCTCCAATG |
| rpoB-2 | CTTTAGCGATCTGTTCCAATG | |||
| recP | Transketolase | 676 | recP-1 | AGCGACAAACCATCCTTTATC |
| recP-2 | CGACAGCTAAGGAATCATGAG |
Phylogenetic analysis.
Storage and analysis of the nucleic acid sequences generated by the MLST analysis were done using the Bionumerics 5.1 software (Applied Maths, Kortrijk, Belgium). A dendrogram was constructed from REA-PFGE data. From the MLST sequencing results, the number of different sequence types (ST) was determined and a minimum spanning tree was built. The software also performed a principal component analysis (PCA) and a multivariate analysis of variance (MANOVA).
Analysis of allelic diversity was done using the DnaSP software (15). This software calculates the following indices: number of alleles for each locus, mean of G+C, number of synonymous and nonsynonymous mutations, ratio between these two values (dN/dS ratio), and allelic diversity.
Phylogenetic and molecular evolutionary analyses were performed using MEGA software, version 4.0 (23). The phylogenetic tree for MLST was constructed by the neighbor-joining method with a Kimura two-parameter distance model. Bootstrap values were obtained after 1,000 iterations.
The software Splitstree (11) was used to analyze the intragenic recombination in the loci, displaying the linear apparition of mutations in alleles of each locus. For the intergenic recombination, the START software was used (12) to calculate the standardized index of association (IsA), a measure of the linkage disequilibrium between alleles.
Allelic diversity and richness of populations were calculated using the GENETIX 4.05 software (2). Allelic richness was expressed as the number of alleles per locus, for populations reduced to 16 samples for each population (16 strains being the lowest number of strains as observed in the German population). Rarefaction of the populations was done by resampling from the initial population. An estimation of the allelic diversity (Hs) was calculated using the formula n(1 − Σpi2)/(n − 1), where n is the size of population and pi the frequency of each allele in the population.
Genotypic richness was calculated with the RAREFACTION 1.3 software (available at http://www.uga.edu/strata), restricted to a population of 16 samples. The Simpson index (or index of genotypic diversity) is the equivalent index of allelic diversity for genotypes. It was calculated using the GENCLONE software (1).
Finally, the differentiation index (θ or Fst) (24), measuring the genetic distance between the different populations, was calculated with the GENETIX software.
MLST data set.
Nucleotide sequences included for MLST analysis from a previous study (4) are available in GenBank under accession numbers FJ392690 to FJ392698 (dnaE), FJ403333 to FJ403343 (g6pd), FJ403344 to FJ403350 (pgm), FJ403351 to FJ403361 (purK), FJ403362 to FJ403370 (recP), FJ403371 to FJ403373 (rpoB), and FJ413033 to FJ413040 (gyrB).
Nucleotide sequence accession numbers.
New nucleotide sequences of MLST loci were deposited in GenBank under accession numbers HQ238155 to HQ238164 (dnaE), HQ238140 to HQ238146 (g6pd), HQ238147 to HQ238154 (pgm), HQ238165 to HQ238180 (purK), HQ238188 to HQ238215 (recP), HQ238181 to HQ238187 (rpoB), and HQ238129 to HQ238139 (gyrB).
RESULTS
REA-PFGE typing.
In this first part of the study, 513 strains of O. oeni were analyzed by REA-PFGE using the NotI restriction enzyme. A total of 363 unique REA-PFGE patterns were obtained and covered the entire collection (data not shown). They corresponded to a combination of 29 differently sized restriction fragments.
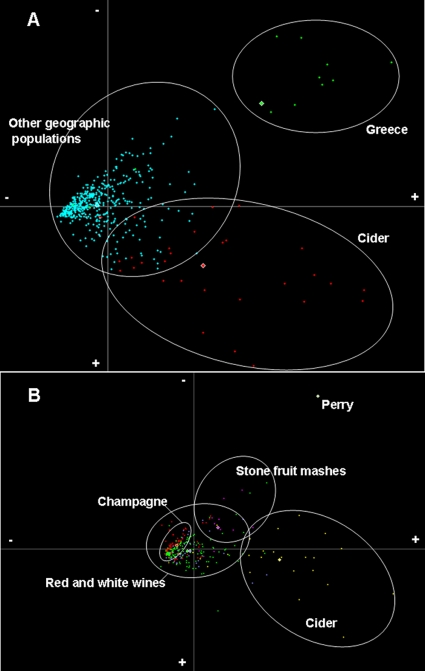
A MANOVA analysis was performed on the 513 REA-PFGE profiles, therefore comparing populations from 16 different geographic origins. Each population was characterized by three to four discriminating restriction fragments. The Greek population was significantly different from the others, with a notable distance from the origin (Fig. 1A). No other populations could be significantly differentiated according to their geographic origins. However, several strains isolated in France and Spain were grouped far from the other profiles. Interestingly, all of them corresponded to cider isolates.
FIG. 1.
MANOVA analysis of PFGE profiles of 513 region-related isolates (A) and 314 product-related isolates (B). Stars represent each individual profile. (A) Red stars, cider strains; green stars, Greek strains; blue stars, all other strains. (B) Yellow stars, cider strains; red stars, champagne strains; green stars, red wine strains; blue stars, white wine strains; purple stars, stone fruit mash; white stars, perry.
In addition, a MANOVA analysis taking into account the product type led to more distinct populations (Fig. 1B). In this context, due to the lack of information concerning the origin of several strains, the analysis was performed on 314 samples corresponding to six populations. The obtained results indicated that 77% of the variance was supported by the x axis, 11% by the y axis, and 7% by the z axis, all three with significant values (P < 0.001). The average for the cider population was positioned far from the origin, in the positive part of the x axis. The observations were the same for the stone fruit mash population and the only strain isolated from perry (a product of pear juice fermentation). This suggested a clear discrimination of these populations in comparison to those isolated from white, red, and champagne wines. The later populations formed a major group situated close to the center that was constituted of strains which were not well differentiated. However, although the champagne population was very close to the other wine-related isolate populations, it was concentrated in a small part of the graph and could be discriminated from the red and white wine populations.
MLST typing.
Based on the REA-PFGE results, 235 strains were selected for MLST typing. The criteria of choice were, first, their different REA-PFGE profiles to get maximal diversity and, second, for similar selected REA-PFGE patterns, their different geographic origins, year of isolation, and type of product. The strains were also chosen to be the most diverse for each criterion, geographic origin, type of product, and year of isolation. In addition, 43 of these strains that had already been analyzed by MLST (4) in a previous study were included in the phylogenetic analysis, finally leading to 278 strains typed by MLST.
Seven housekeeping genes were targeted in this MLST analysis (gyrB, g6pd, pgm, dnaE, purK, rpoB, recP). These genes were spread along the chromosome of Oenococcus oeni. The gene functions and the amplicon sizes are presented in Table 1. Finally, for each strain, a concatenated sequence of 4,085 bp was constructed. Only 258 strains out of the 278 previously selected for MLST analysis allowed us to obtain the complete data. Twenty sequences failed to amplify by PCR despite using several primer couples and varying the PCR conditions. The loci recP and purK were especially difficult to amplify in 19 wine strains and 1 cider strain. These 20 strains were removed from the analysis.
The average allelic diversity was 20.7 alleles per locus, from 10 alleles for rpoB to 37 for recP, with a nucleotidic diversity varying between 0.002 for pgm and 0.036 for rpoB (Table 2). Almost all the observed mutations corresponded to substitutions, the only exceptions being an 860-bp insertion sequence in the purK sequence and a 1-bp deletion in recP. Both mutations disrupted the open reading frame (ORF). Comparing the sequences of each locus, the number of polymorphic sites was variable, ranging from 14 for pgm to 148 for recP, which is a very high value, for sequence lengths ranging from 531 bp (pgm) to 665 bp (dnaE). However, this high level of mutation was due to only one strain, genetically very distant from the other strains. For this strain, DIV 5.7, isolated from cider, the respective sequences of the gyrB, g6pd, purK, rpoB, and recP genes were very distinct. Therefore, when the whole collection is considered, the number of mutations in the concatenated sequences varied from 337 bp, without the DIV 5.7 strain, to 600 bp, when this strain was considered. The absence of intermediate sequences between alleles of strain DIV 5.7 and those of other strains suggested a horizontal transfer of specific alleles from unknown species. Moreover, the nucleotidic diversity, showing the average of nucleotide difference per site from two randomly selected sequences, remained very low for five loci (gyrB, g6pd, pgm, dnaE, and purK). This suggested that the insertion of distinct sequences does not interfere in the global genetic diversity for all loci. Even for rpoB and recP, which presented a high level of nucleotidic diversity, the absence of distinct sequences did not modify the diversity values. Interestingly, the majority of mutations were silent mutations. Indeed, the dN/dS ratio remained low (average, 0.204), suggesting an accumulation of synonymous mutations and a high selection pressure.
TABLE 2.
Average allelic and nucleotidic diversity
| Locus | Fragment size (bp) | No. of alleles | Mean G+C content (%) | No. of polymorphic sitesb | Nucleotide diversityc | dN/dSd |
|---|---|---|---|---|---|---|
| gyrb | 611 | 19 | 40.7 | 88 (11) | 0.006 | 0.077 |
| g6pd | 551 | 18 | 41.2 | 95 (19) | 0.008 | 0.275 |
| pgm | 531 | 15 | 41.4 | 14 (6) | 0.002 | 0.431 |
| dnaE | 665 | 19 | 38.9 | 30 (11) | 0.009 | 0.243 |
| purK | 537 (1,397)a | 27 | 42.2 | 122 (37) | 0.010 | 0.196 |
| rpoB | 598 | 10 | 43.4 | 103 (14) | 0.036 | 0.062 |
| recP | 592 | 37 | 45.8 | 148 (43) | 0.029 | 0.148 |
Length of purK locus with the insertion sequence.
The number of informative sites is in parentheses.
Represented in number of nucleotide substitutions per nucleotide site.
Representing the ratio of nonsynonymous to synonymous mutations.
Construction of neighbor-joining phylogenetic and minimum spanning trees.
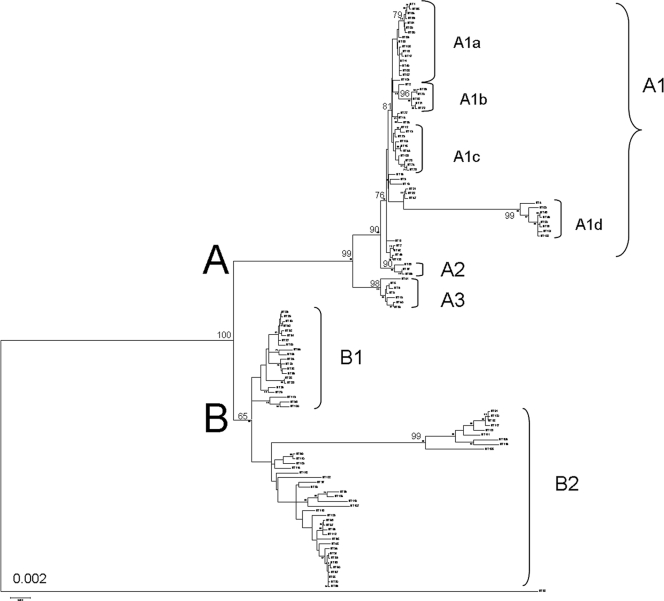
The concatenation of the obtained sequences for each of the seven housekeeping genes formed a sequence of 4,085 bp, and 127 different sequence types (ST) were determined. A phylogenetic tree was then constructed, using MEGA software (Fig. 2). The tree showed the existence of two major groups, A and B. The same two groups were delimited in the REA-PFGE analysis. Both approaches, although based on completely different principles of analysis, led to the same grouping of strains. Concerning the DIV 5.7 strain isolated from cider, its sequence was so different from the others that it formed a third branch. Six main subgroups formed the A group, and two subgroups appeared in group B. However, when the most diverse locus, recP, was removed from the analysis, group B proved to be very homogenous and group A lost some subgroups, reducing it to three subgroups (data not shown).
FIG. 2.
Neighbor-joining phylogenetic tree constructed from the concatenated sequence (seven loci) of 127 sequence types. Different subgroups have been highlighted and named. Some ST in group A1 could not be integrated in the subgroups because of too-low bootstrap values.
The subgroup A1a contained the highest number of strains, with 72 strains coming from various countries. The subgroup A1b was special in the fact that it was mostly composed of Chilean strains, and the subgroup A1c mainly comprised strains from South Africa. The group A1d was created by the presence of an insertion sequence in the locus purK. Champagne and Burgundy were almost the only regions for which this insertion was found in the genome of O. oeni strains. The insertion was also found in a strain isolated in Jura (close to Burgundy) as well as in an Italian strain. A recP deletion was also found exclusively in strains from eastern France (Burgundy, Champagne, and Jura). The A2 and A3 groups were formed due to divergent recP alleles. Interestingly, all the strains from cider were located in the B group and mainly in subgroup B2.
The observed diversity seems to be associated with the lack of some genotypes in a few diversified geographic regions rather than with the apparition of new and specific genotypes in specific geographic areas (except for the Chilean population).
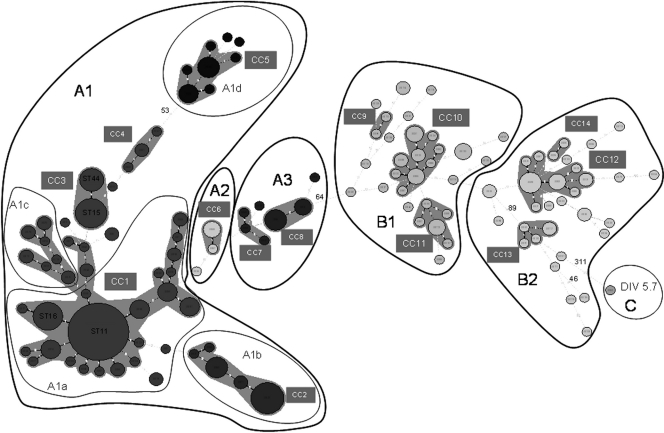
Finally, a minimum spanning tree constructed using the Bionumerics software showed the same results as the neighbor-joining tree (Fig. 3). Concatenated sequences differing by less than four mutations were grouped into 14 distinct clonal complexes. The two groups, A and B, were again well separated. The group A1 included three major clonal complexes corresponding to the large group comprising the strains of the subgroups A1a, A1c, and some others, to the group A1d (strains carrying the insertion sequence in purK), and to the group A1b (formed by the strains from Chile). The dominant group A1a included 37 strains presenting ST 11, the most represented ST. Only a few strains were not grouped in a clonal complex (9 out of 175). The B group was subdivided in two major clonal complexes, plus four small clonal groups and a lot of isolated strains (31 out of 80), indicating that the genetic diversity is larger in this group.
FIG. 3.
Minimum spanning tree analysis of 258 Oenococcus oeni strains. Each circle represents a different ST, and the circle diameter is correlated with the number of strains presenting the corresponding ST. Shaded ensembles represent the 14 clonal complexes (annotated CC) detected by strains differentiated by at least three mutations in the concatenated sequence. ST differentiated by more than 26 mutations are indicated. ST associations are separated and annotated according to their location in the neighbor-joining phylogenetic tree.
Intragenic and intergenic recombination in the population.
All loci, except pgm and recP, analyzed in this study were organized in the same way. Their alleles were subdivided into three groups, one group with alleles found exclusively in strains of group A, another group with alleles found only in strains of group B, and, interestingly, a third group with alleles found in cider strains (data not shown). As for pgm, no significative discrimination of O. oeni could be performed due to a low sequence diversity of the locus. Concerning the recP locus, which was the most variable, the separation between the alleles of group A and B was less visible (data not shown).
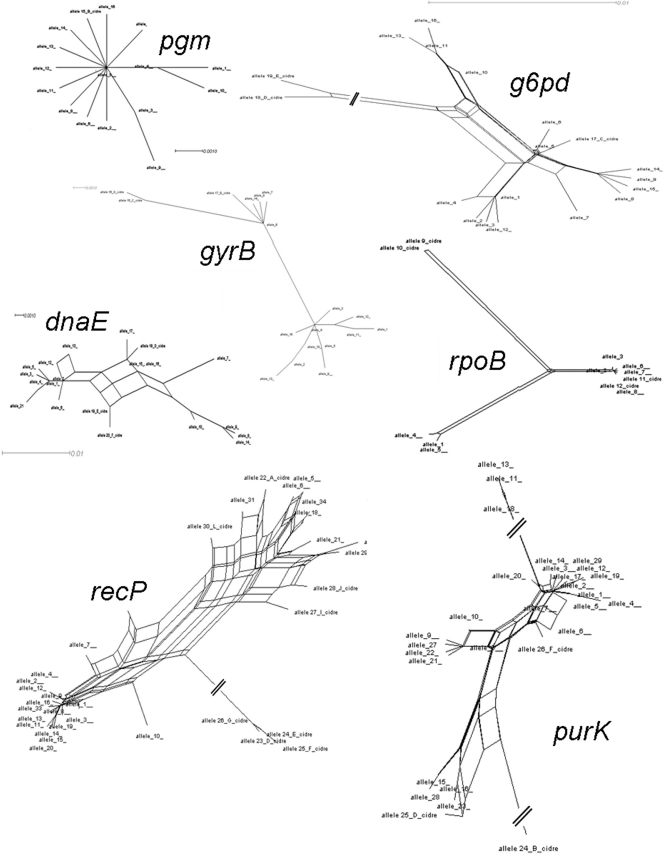
Sequences of the seven loci were analyzed with Splitstree, in order to determine the role of intragenic recombination for allele creation (Fig. 4). The association of allele sequences was transcribed into a tree. A representative network configuration showed the implication of recombination between alleles, for four out of seven loci, namely, g6pd, dnaE, purK, and recP. On the other hand, gyrB, pgm, and rpoB showed a relative linear evolution. The g6pd tree was composed of two major groups representative of phylogenetic groups A and B and alleles from cider in a third part of the tree. The whole formed a network configuration that disappeared when strains from cider were removed from the analysis (data not shown). A network configuration was also obtained for the dnaE tree. This suggests that the appearance of the cider alleles could originate from recombination events between alleles of groups A and B in several genes.
FIG. 4.
Split graph constructed from sequences of analyzed alleles.
Finally, only recP and purK showed network conformation trees, suggesting the role of recombination in the formation of these alleles. Interestingly, when the purK loci of strains from groups A and B were analyzed separately, the network configuration disappeared for the alleles of group B (data not shown). Therefore, only alleles from group A seemed to recombine to form new alleles. For recP, the network configuration corresponded to intragenic recombination but differed from those of other alleles, suggesting a different evolution pathway.
The significance of intergenic recombination was analyzed by measuring the standardized index of association (ISA) in the entire collection and also separately in strains of groups A and B. A low but significant linkage disequilibrium was observed in the collection, with an index of association of 0.199 (P < 0.01), meaning that even if intergenic recombination played an important role in the distribution of alleles, an allelic linkage was preserved, possibly due to the specificity of some populations. As for the index of association of the two groups, both values for A and B were significantly different from 0, with respective ISA values of 0.125 (P < 0.01) and 0.093 (P < 0.01) showing that the linkage disequilibrium for the whole population could not be entirely due to the A/B group separation. However, for group B, when the population without cider strains was analyzed, the observed linkage disequilibrium disappeared, with an ISA of 0.015 (P = 0.25), suggesting a specific allelic content in the two subpopulations of group B, cider strains, on one hand, and wine and stone fruit mashes isolates, on the other hand.
Comparison of populations according to their geographic origin.
Seven geographic populations (each containing >15 strains) were compared. They corresponded to populations from Burgundy (31 strains), Champagne (21 strains), Aquitaine (28 strains), Italy (25 strains), Germany (16 strains), South Africa (23 strains), and Chile (18 strains). The Italian, German, South African, and Aquitaine populations had similar allelic richness, with approximately six alleles per locus. The populations of Burgundy and Champagne were less diverse, but in comparison the Chilean population appeared the least diverse, with only two alleles per locus. The analysis of an allelic diversity measure (Hs), varying from 0 for null diversity to 1 for maximal diverse populations, confirmed these results. Indeed, two groups were again observed, corresponding to, on one hand, the populations from Burgundy, Champagne, and Chile, with Hs varying around 0.5, and, on the other hand, the other populations, with Hs of 0.7 (Table 3).
TABLE 3.
Genetic analysis of geographic and wine populations
| Population | Genotypic richnessa | Simpson index | Allelic richnessa | Allelic diversityb |
|---|---|---|---|---|
| South Africa | 14.1 | 0.98 | 5.91 | 0.71 |
| Chile | 3 | 0.57 | 2.43 | 0.49 |
| Italy | 14.1 | 0.98 | 5.99 | 0.68 |
| Germany | 13 | 0.98 | 4.57 | 0.61 |
| Burgundy, France | 8.8 | 0.88 | 4.36 | 0.53 |
| Champagne, France | 10.2 | 0.91 | 3.4 | 0.48 |
| Aquitaine, France | 12.7 | 0.97 | 7.06 | 0.72 |
| Red wine | 10.9 | 0.95 | 6.81 | 0.76 |
| White wine | 9.5 | 0.91 | 5.92 | 0.64 |
| Champagne | 9 | 0.92 | 3.14 | 0.52 |
| Cider | 13.9 | 0.98 | 5.96 | 0.64 |
Calculated for artificial populations of 16 strains for geographic populations and 14 strains for product populations; expressed as average number of alleles per locus.
Expressed as Hs.
The same tendency was observed for the genotypic diversity, as similar diversity indexes were found in all populations, with around 10 to 14 ST per population (no significant difference) except for the Chilean population (3 ST). This was confirmed by comparing the Simpson index (or index of genotypic diversity), which was around 0.98 for the populations from Germany, Italy, South Africa, and Aquitaine but lower (0.9) for populations from Burgundy and Champagne and even lower for the Chilean population (0.57) (Table 3).
In order to evaluate the relationship between the populations and their respective genetic distance, an unbiased estimation of pairwise genetic differentiation, called theta, was calculated according to geographic origin (data not shown). Almost all populations could be delimited, although with values lower than 0.15. However, Aquitaine, Italian, and South African populations could not be differentiated from each other. The Chilean population was separated from the others, with theta values higher than 0.3. In addition, Champagne and Burgundy populations were differentiated from the German population, with significant values of 0.37 and 0.34 for Champagne and Burgundy strains, respectively.
Comparing the composition of populations and their position in the phylogenetic tree, South African and Chilean populations and populations from Burgundy and Champagne were highlighted (Fig. 5). The South African population was composed of strains dispersed all over the tree, equally in groups A and B. However, nine of them were grouped together in group A1c, composed almost exclusively of South African strains, suggesting an evolution of this population that makes it specific and different from other populations. Their genotype was found in only two strains isolated in Charente, France, and one strain from Greece, infrequently represented in Europe. It seems to be a predominant genotype in the South African strains.
FIG. 5.
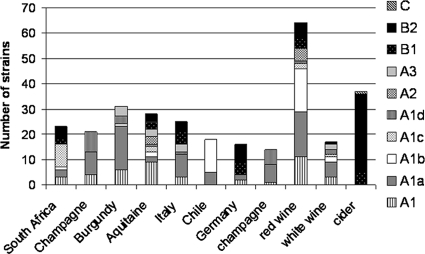
Repartition of strains from analyzed populations according to their localization in the neighbor-joining phylogenetic tree. The figure is divided in two parts: the first seven bars correspond to geographic populations, whereas the final four bars correspond to populations according to their product type.
The same was observed in the Chilean population, with 13 of 18 strains grouped in A1b, a group well represented in this collection but also found in Australia, Burgundy, and Aquitaine.
The Champagne and Burgundy populations, despite being the largest sample, had a great majority of strains belonging to groups A1a and A1d (44 out of 50). Moreover, the insertion and deletion observed, respectively, in purK and recP were exclusively present in these populations. Strains from group B were not found in these areas. This confirms the low diversity of the Champagne population.
Comparison of population according to product type.
Because of some missing data concerning the exact origin of some strains, a reduced panel of 134 strains was analyzed. The strains originated from red wines (64 strains), white wines (17 strains), champagne (15 strains), and other products, namely, cider and stone fruit mashes (38 strains). The red wine population was the most diverse, with an allelic richness of seven alleles per locus, while an allelic richness of about six alleles per locus was observed for strains isolated from white wines and ciders. The champagne population was less diverse, with only three alleles per locus. This tendency was supported by Hs calculation, which gave similar results. However, the genotypic diversity was similar for all these populations, with values around 0.95 for the Simpson index (Table 3). This could be explained, for the champagne population, by a high level of recombination that led to an increase of genotypes independently from the new alleles.
Comparing the composition of the populations in the phylogenetic tree, two major observations have been highlighted. All strains isolated from cider and stone fruit mashes were in group B, equally distributed in groups B1 and B2 (Fig. 5). Comparably, all the strains isolated from champagne were located in group A1 (in particular in groups A1a and A1d). Strains isolated from red and white wines were more diverse and could not be differentiated.
Mutation multivariate analysis of variance and discriminant analysis.
MANOVA was done on all mutations of the concatenated sequence. The significance of phylogenetic-delineated groups (A1a-A1bI-A1c-A1d, A2, A3, B1, B2, and the DIV 5.7 strain) and of discriminating mutations was analyzed.
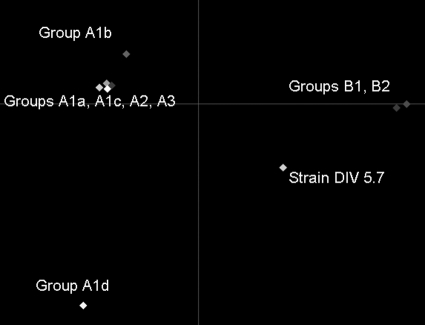
More than 600 mutations were found all over the concatenated sequence, and 251 of them contributed to the discrimination of the phylogenetic groups. The most discriminating values were obtained for 32 mutations.
The first discriminant axis (x axis) could explain 78% of the variance and the second (y axis) 10% (Fig. 6). The groups A1a-A1bI-A1c, A2, and A3 (group A) were very close with each other and clearly distant from B1 and B2 (group B), A1d (group C), and the DIV 5.7 strain. Group B was in the negative half of the x axis, whereas group A was the most positive group on this axis. Mutation positions rpoB 528 and rpoB 87 (pronounced negative position) and mutation positions rpoB 525 and rpoB 384 (pronounced positive position) discriminated group B from group A (data not shown). Group C was positive on the y axis, whereas group A was negative on this axis. Mutation positions purK 462, purK 481, and purK 490 (pronounced negative position) discriminated group C from the two other groups.
FIG. 6.
MANOVA analysis, comparing the mutations in the concatenated sequence of seven genes and phylogenetic groups. Each point represents the medium value of variance for each phylogenetic group.
MANOVA and discriminant analysis were also done for strains isolated from group A (wine) and group B (cider, perry, and stone fruit mashes) (data not shown). The first discriminant axis explained 97% of the variance (data not shown). Eighty percent of wine isolates were in the positive half of the x axis, whereas all group B strains were in the negative part of this axis. The mutations with the most positive positions corresponded to rpoB 384, rpoB 525, and purK 394, and those in the negative part of this discriminant axis were rpoB 528 and recP 239.
DISCUSSION
Relevance of both PFGE and MLST for studying the collection's diversity.
The two methods that were used for bacterial typing provided distinct data and showed distinct genetic evolutionary changes. In accordance with the method (10), PFGE analysis showed great genome rearrangements, representative of major evolutionary changes. On the other hand, MLST gave another view and showed neutral diversity and evolution, since minor mutations were able to better differentiate strains. Nevertheless, these two independent and distinct methods achieved, in the same way, the delineation of the collection into two subpopulations. However, although these two approaches separated the phylogenetic groups A and B, the subgroups that they formed were different. The subgroups set up by MLST that discriminated Chilean, South Africa, and Champagne strains were not obtained in the REA-PFGE analysis.
Existence of two subpopulations that are evolving separately.
In previous studies that used several different methods, two subpopulations were also detected, by REA-PFGE (4, 13), MLST (4), RAPD analysis (25), and ribotyping (5, 25). However, the number of strains that was considered in any of these studies was not sufficient to be representative of the O. oeni genetic structure. In this study, the collection of 278 strains that had been isolated in very distant areas and products is without a doubt more representative of the O. oeni species. As the main result, the hypothesis of the presence of two subpopulations, A and B, was confirmed. In addition, for five loci, two groups of alleles were identified and separated in a neighbor-joining phylogenetic tree. For the pgm locus, the sequence was very well conserved and thus did not produce subgroups in a phylogenetic tree. On the other hand, the recP sequence was very variable, with multiple events of intragenic recombination, which prevented the separation of alleles in two distinct subgroups. As shown by Splitstree analysis, for each locus, alleles of strains found in groups A and B seemed to evolve by the addition of mutations, separately, with no important intragenic recombination between alleles. Each group of alleles had its own mutations and linear evolution. No recombination between alleles from group A and B could be noticed, except for the dnaE locus, where the recombination of alleles from group A and B led to some alleles found in cider strains. The other exception was the recP locus, which seemed to have a different evolution path than those of the other loci.
In fact, groups A and B are very well differentiated, which suggests that they evolved separately, with almost no exchange of genetic material. On the contrary, within each group, genetic exchanges seemed to occur. Intergenic recombination plays a significant role in diversity and the appearance of new genotypes. In addition, these two groups are well differentiated, with populations exclusively represented in each of them. Strains from cider are located exclusively in group B, whereas strains from Champagne and Burgundy are only in group A. The fact that genetic exchanges between these two groups could be very limited is more evidence in favor of the dissociation between the two observed groups. In this context, the possibility of defining these two subpopulations into two corresponding subspecies can be raised.
Possible existence of a third phylogenetic group.
A particular strain, DIV 5.7, isolated in cider, forms a third phylogenetic group. Its concatenated sequence is very different from the others. More than 300 bases of difference were observed when its sequence was compared to the concatenated sequences of other cider strains, and up to 600 bases of distance were found compared to sequences of group A strains. These mutations were distributed in all loci except for pgm and dnaE, and very specific and divergent alleles could be found. It was the only strain to present some specific alleles in the gyrB, g6pd, purK, and rpoB loci. Divergent alleles of recP were also detected in other strains from cider (seven strains) and wine (two strains from Italy and one from Germany, isolated in stone fruit mash). They were only 85% identical to O. oeni alleles. It was verified that this strain could be classified in this species, since the sequence of the 16S DNA was 99% identical to the sequence of the O. oeni 16S sequence. Therefore, it could be possible that some cider strains mutated in such a way that they formed a third phylogenetic group. However, because of the absence of intermediary sequences between those divergent alleles and those from wine, it is hypothesized that the presence of these alleles results from horizontal transfer with some other bacterial species. This hypothesis is well supported by the lack of a mutation mismatch repair system in O. oeni (19). Indeed, this system normally prevents the incorporation of heteroduplex DNA, leading to a reduction of interspecies recombination. At this time, it is not possible to determine from which species the transfer of genomic material was done. However, the rpoB allele of this strain that shows 89% homology with other alleles of O. oeni was found to have a 77% homology with Lactobacillus johnsonii.
Description of subpopulations in each group.
Within the two phylogenetic groups, A and B, some particular subgroups were observed, with interesting particularities. Several strains from Chile formed a specific phylogenetic group (A1b), distinct from the others in the neighbor-joining phylogenetic tree and the minimum spanning tree as well as in the other genetic analyses. This subgroup also included two strains isolated in Australia, one in Greece, and two in France. In reference to the low level of mutations, which was about 10 for the concatenated sequence, these strains were genetically close. However, these 10 mutations were disseminated in all concatenated sequences, leading to specific alleles for each locus. This could be a mark of significant genetic distance, despite the low number of mutations. The same was observed for nine South African strains that formed the A1c phylogenetic subgroup, with one strain from Greece and two from Aquitaine. However, in this case, intergenic recombination should explain the occurrence of these genotypes.
Strains from Champagne and Burgundy were also specific. Several strains were in subgroup A1d, essentially created by the insertion of a transposable element in purK. With their own allelic content, they formed a particular structure. Their genetic diversity was also noticeable. It was lower than for other populations, with respect to allelic and genotypic diversity. All these strains were located in the phylogenetic tree in group A and in only two subgroups (A1a and A1d). Moreover, this similarity in Champagne and Burgundy is interesting and might be linked to their usual environment, which is mainly characterized by lower-pH wines. Acidity may have selected the most resistant strains, leading to a lower diversity in comparison to other populations adapted to more permissive conditions.
Overall, among the seven populations that were more intensively studied, strains from South Africa, Aquitaine, and Italy were the most diverse and also the less differentiated. They exhibited almost all types of genotypes. The only differences between these populations were in the proportion of these genotypes. This suggests that the diversity level in these populations could be representative of the global genetic diversity of O. oeni. Additionally, the specificity of some populations could be the result of adaptation and proliferation of particular genotypes in a corresponding region. This was the case in South Africa, Chile, Champagne, and Burgundy, where most of the strains were in a distinct genetic subgroup. However, for the Chilean population, the strains were isolated from only two areas. Similarly, for the German population, most strains were isolated from stone fruit mashes. This result cannot correctly represent the real diversity of the O. oeni species in this country, but it underlines the specificity of the stone fruit mash population.
The cider populations were particularly original. They were exclusively located in subpopulation B. Moreover, although they were equally shared in the neighbor-joining phylogenetic tree and the minimum spanning tree, they formed a specific population, with specific alleles. They were characterized by 23 different ST, of which 6 were found only in cider strains. They were well differentiated from other populations, with theta values higher than 0.3. This result is in accordance with PFGE profile grouping. Interestingly, strains isolated from other fruit products, such as stone fruit mashes and pear, were also located in group B. Finally, it could be possible that strains of group A could be adapted only to wine conditions. This hypothesis is also consistent with the fact that eight of nine malolactic starters are located in subgroup A. Until now, all strains isolated from fruits other than grapes or wines were located in subgroup B.
Conclusion.
In conclusion, the analysis of a large collection of Oenococcus oeni evidenced in this work the existence of two major phylogenetic groups. The two completely different approaches, MLST and REA-PFGE, led to the same conclusion. The analysis of allelic diversity in the two groups suggested that they may evolve separately, an observation which could ultimately lead to the delimitation of two subspecies. In addition the position of a cider strain forming a specific and distant phylogenetic group could be a clue to the existence of another subpopulation. Several phylogenetic subgroups appeared, and specific populations, like those from Chile, Champagne, and cider, could be highlighted. According to the MLST results, several mutations were typical of some populations or phylogenetic subgroups. From there, it is possible to envisage the development of a method by high-resolution melting temperature (HRM)-PCR or single nucleotide polymorphism (SNP) sequencing, for the rapid assignment of strains to specific groups of O. oeni. This work is going on, with a study of physiological and biochemical characters of strains, seeking possible relationships between these phylogenetic data and phenotypic data. Ultimately the results could help in the understanding of the possible impact of environmental conditions on the genetic content and evolution of the species.
Acknowledgments
This work was funded by the French National Research Agency (ANR) and the Fondation pour la recherche sur la biodiversité (FRB; project Divoeni [ANR-07-BDIV-011]).
We are grateful to Lallemand SAS (Toulouse, France), ADRIA-Normandie (Villers-Bocage, France), IFVV (Beaune, France), SARCO SAS (Bordeaux, France), all partners of Divoeni, and to A. Ganga (Universidad de Santiago de Chile, Chile), P. Pramateftaki (Wine Institute of Athens, Greece), G. Zapparoli (Università degli Studi di Verona, Italy), and M. du Toit (Institute for Wine Biotechnology, University of Stellenbosch, South Africa) for providing the O. oeni strains from their respective collections.
Footnotes
Published ahead of print on 8 October 2010.
REFERENCES
- 1.Arnaud-Haond, S., and K. Belkhir. 2007. Genclone: a computer program to analyse genotypic data, test for clonality and describe spatial clonal organization. Mol. Ecol. Notes 7:15-17. [Google Scholar]
- 2.Belkhir, K., P. Borsa, L. Chikhi, N. Raufaste, and F. Bonhomme. 1996. GENETIX 4.05, logiciel sous Windows TM pour la génétique des populations. http://www.genetix.univ-montp2.fr/genetix/intro.htm.
- 3.Beltramo, C., N. Desroche, R. Tourdot-Maréchal, C. Grandvalet, and J. Guzzo. 2006. Real-time PCR for characterizing the stress response of Oenococcus oeni in a wine-like medium. Res. Microbiol. 157:267-274. [DOI] [PubMed] [Google Scholar]
- 4.Bilhere, E., P. M. Lucas, O. Claisse, and A. Lonvaud-Funel. 2009. Multilocus sequence typing of Oenococcus oeni: detection of two subpopulations shaped by intergenic recombination. Appl. Environ. Microbiol. 75:1291-1300. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.De Las Rivas, B., A. Marcobal, and R. Munoz. 2004. Allelic diversity and population structure in Oenococcus oeni as determined from sequence analysis of housekeeping genes. Appl. Environ. Microbiol. 70:7210-7219. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.de Man, J. C., M. Rogosa, and M. E. Sharpe. 1960. A medium for the cultivation of lactobacilli. J. Appl. Microbiol. 23:130-135. [Google Scholar]
- 7.Dicks, L. M., F. Dellaglio, and M. D. Collins. 1995. Proposal to reclassify Leuconostoc oenos as Oenococcus oeni [corrig.] gen. nov., comb. nov. Int. J. Syst. Bacteriol. 45:395-397. [DOI] [PubMed] [Google Scholar]
- 8.Dicks, L. M. T., H. J. J. Van Vuuren, and F. Dellaglio. 1990. Taxonomy of Leuconostoc species, particularly Leuconostoc oenos, as revealed by numerical analysis of total soluble cell protein patterns, DNA base compositions, and DNA-DNA hybridizations. Int. J. Syst. Bacteriol. 40:83-91. [Google Scholar]
- 9.Endo, A., and S. Okada. 2006. Oenococcus kitaharae sp. nov., a non-acidophilic and non-malolactic-fermenting oenococcus isolated from a composting distilled shochu residue. Int. J. Syst. Evol. Microbiol. 56:2345-2348. [DOI] [PubMed] [Google Scholar]
- 10.Gindreau, E., A. Joyeux, G. De Revel, O. Claisse, and A. Lonvaud-Funel. 1997. Evaluation of the settling of malolactic starters within the indigenous microflora of wines. J. Int. Sci. Vigne Vin 31:197-202. [Google Scholar]
- 11.Huson, D. H., and D. Bryant. 2006. Application of phylogenetic networks in evolutionary studies. Mol. Biol. Evol. 23:254-267. [DOI] [PubMed] [Google Scholar]
- 12.Jolley, K. A., E. J. Feil, M. S. Chan, and M. C. Maiden. 2001. Sequence type analysis and recombinational tests (START). Bioinformatics 17:1230-1231. [DOI] [PubMed] [Google Scholar]
- 13.Larisika, M., H. Claus, and H. König. 2008. Pulsed-field gel electrophoresis for the discrimination of Oenococcus oeni isolates from different wine-growing regions in Germany. Int. J. Food Microbiol. 123:171-176. [DOI] [PubMed] [Google Scholar]
- 14.Le Jeune, C., and A. Lonvaud-Funel. 1997. Sequence of DNA 16S/23S spacer region of Leuconostoc oenos (Oenococcus oeni): application to strain differentiation. Res. Microbiol. 148:79-86. [DOI] [PubMed] [Google Scholar]
- 15.Librado, P., and J. Rozas. 2009. DnaSP v5: a software for comprehensive analysis of DNA polymorphism data. Bioinformatics 25:1451-1452. [DOI] [PubMed] [Google Scholar]
- 16.Lonvaud-Funel, A. 1999. Lactic acid bacteria in the quality improvement and depreciation of wine. Antonie Van Leeuwenhoek 76:317-331. [PubMed] [Google Scholar]
- 17.Maiden, M. C. J. 2006. Multilocus sequence typing of bacteria. Annu. Rev. Microbiol. 60:561-588. [DOI] [PubMed] [Google Scholar]
- 18.Makarova, K., A. Slesarev, Y. Wolf, A. Sorokin, B. Mirkin, E. Koonin, A. Pavlov, N. Pavlova, V. Karamychev, N. Polouchine, V. Shakhova, I. Grigoriev, Y. Lou, D. Rohksar, S. Lucas, K. Huang, D. M. Goodstein, T. Hawkins, V. Plengvidhya, D. Welker, J. Hughes, Y. Goh, A. Benson, K. Baldwin, J. Lee, I. Díaz-Muñiz, B. Dosti, V. Smeianov, W. Wechter, R. Barabote, G. Lorca, E. Altermann, R. Barrangou, B. Ganesan, Y. Xie, H. Rawsthorne, D. Tamir, C. Parker, F. Breidt, J. Broadbent, R. Hutkins, D. O'Sullivan, J. Steele, G. Unlu, M. Saier, T. Klaenhammer, P. Richardson, S. Kozyavkin, B. Weimer, and D. Mills. 2006. Comparative genomics of the lactic acid bacteria. Proc. Natl. Acad. Sci. U. S. A. 103:15611-15616. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Marcobal, A. M., D. A. Sela, Y. I. Wolf, K. S. Makarova, and D. A. Mills. 2008. Role of hypermutability in the evolution of the genus Oenococcus. J. Bacteriol. 190:564-570. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Martinez-Murcia, A. J., and M. D. Collins. 1990. A phylogenetic analysis of the genus Leuconostoc based on reverse transcriptase sequencing of 16S rRNA. FEMS Microbiol. Lett. 58:73-83. [DOI] [PubMed] [Google Scholar]
- 21.Martinez-Murcia, A. J., N. M. Harland, and M. D. Collins. 1993. Phylogenetic analysis of some Leuconostocs and related organisms as determined from large-subunit rRNA gene sequences: assessment of congruence of small- and large-subunit rRNA derived trees. J. Appl. Bacteriol. 74:532-541. [PubMed] [Google Scholar]
- 22.Renouf, V., A. Delaherche, O. Claisse, and A. Lonvaud-Funel. 2008. Correlation between indigenous Oenococcus oeni strain resistance and the presence of genetic markers. J. Ind. Microbiol. Biotechnol. 35:27-33. [DOI] [PubMed] [Google Scholar]
- 23.Tamura, K., J. Dudley, M. Nei, and S. Kumar. 2007. MEGA4: Molecular Evolutionary Genetics Analysis (MEGA) software version 4.0. Mol. Biol. Evol. 24:1596-1599. [DOI] [PubMed] [Google Scholar]
- 24.Weir, B. S., and C. C. Cockerham. 1984. Estimating F-statistics for the analysis of population structure. Evolution 38:1358-1370. [DOI] [PubMed] [Google Scholar]
- 25.Zavaleta, A. I., A. J. Martínez-Murcia, and F. Rodríguez-Valera. 1997. Intraspecific genetic diversity of Oenococcus oeni as derived from DNA fingerprinting and sequence analyses. Appl. Environ. Microbiol. 63:1261-1267. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Zavaleta, A. I., A. J. Martinez-Murcia, and F. Rodriguez-Valera. 1996. 16S-23S rDNA intergenic sequences indicate that Leuconostoc oenos is phylogenetically homogeneous. Microbiology 142:2105-2114. [DOI] [PubMed] [Google Scholar]
- 27.Zé-Zé, L., R. Tenreiro, and H. Paveia. 2000. The Oenococcus oeni genome: physical and genetic mapping of strain GM and comparison with the genome of a “divergent” strain, PSU-1. Microbiology 146:3195-3204. [DOI] [PubMed] [Google Scholar]