Abstract
Sesquiterpene synthases are responsible for the cyclization of farnesyl pyrophosphate into a myriad of structurally diverse compounds with various biological activities. We examine here the role of the conserved active site H-α1 loop in catalysis in three previously characterized fungal sesquiterpene synthases. The H-α1 loops of Cop3, Cop4, and Cop6 from Coprinus cinereus were altered by site-directed mutagenesis and the resultant product profiles were analyzed by gas chromatography-mass spectrometry and compared to the wild-type enzymes. In addition, we examine the effect of swapping the H-α1 loop from the promiscuous enzyme Cop4 with the more selective Cop6 and the effect of acidic or basic conditions on loop mutations in Cop4. Directed mutations of the H-α1 loop had a marked effect on the product profile of Cop3 and Cop4, while little to no change was shown in Cop6. Swapping of the Cop4 and Cop6 loops with one another was again shown to influence the product profile of Cop4, while the product profile of Cop6 remained identical to the wild-type enzyme. The loop mutations in Cop4 also implicate specific residues responsible for the pH sensitivity of the enzyme. These results affirm the role of the H-α1 loop in catalysis and provide a potential target to increase the product diversity of terpene synthases.
Sesquiterpene synthases catalyze the cyclization of farnesyl pyrophosphate (FPP) to structurally diverse C15-hydrocarbons. These enzymes belong to the large group of terpene synthases that convert isoprene pyrophosphate substrates into hundreds of described terpenoid compounds by employing some of the most complex carbon-carbon forming reactions known (8). Many terpenoids are biologically active and are produced by plants, bacteria, and fungi such as, for example, antibiotics, toxins, and pheromones (5, 9).
Catalysis in this class of enzymes is dependent on the presence of three Mg2+ ions coordinated by two conserved motifs, an aspartate-rich DDXXD/E and an NSE/DTE motif, flanking the entrance of the active site. This Mg2+ cluster binds the pyrophosphate (PPi) group of FPP and positions the isoprenyl chain in the hydrophobic substrate binding pocket of the enzyme (10). Substrate binding triggers a conformational change that results in the closure of the active site and concurrent PPi cleavage to generate an initial transoid, allylic carbocation (6, 7, 35). This carbocation is then transferred along the isoprenyl chain and eventually quenched either by a water molecule or through proton abstraction. The binding pocket determines folding of the isoprenyl chain and chaperones the reactive carbocation intermediates until the final quenching step (20), thereby defining the product profile of a particular sesquiterpene synthase.
Crystal structures have been solved for several microbial and plant sesquiterpene synthases (1, 7, 14, 30). All enzymes share the same α-helical fold characteristic for ionization-dependent terpene synthases. Plant enzymes possess an additional, catalytically inactive N-terminal domain that has the β-barrel fold of protonation-dependent terpene synthases (7). Structural information has guided several mutagenesis studies aimed at understanding and/or manipulating the cyclization mechanism of sesquiterpene synthases. Residues in the two metal-binding motifs and the active site of these enzymes have been targeted by site-directed mutagenesis (13, 16, 21, 24, 28, 29, 34-36, 40, 41). In most cases, substitution of residues that are in direct contact with the bound substrate had the largest effect on FPP cyclization products, while modification of the metal-binding residues only moderately affected product profiles (17, 18, 40, 41). Significantly altered product profiles of several plant sesquiterpene synthases were obtained by using structure-guided combinatorial mutagenesis of multiple residues located in and surrounding the binding pocket (16, 24, 40, 41).
Until very recently, aristolochene synthase from Aspergillus terreus (AtACS) and trichodiene synthase from Fusarium sporotrichioides (FsTHS) were the only two fungal enzymes for which crystal structures were available in both the open (ligand-free) and closed (FPP-complexed) conformations (27, 28, 30, 31). Structural studies of AtACS showed that the formation of a Mg2+3-PPi complex triggers a large conformational change with a root mean square deviation (rmsd) of 1.9 Å between ligand-free and ligand-bound enzymes (30, 31). A similar change (rmsd 1.4 Å) is seen with FsTHS (28), while the conformational change in plant terpene synthases is much smaller (39). Several helices and loops participate in the transition from open to closed conformation. Of particular importance is the role of the H-α1 loop in capping the active site to protect reactive carbocations from possible premature solvent quenching. This loop is disordered in the unliganded enzymes but moves inward and adopts a well defined conformation that shields the active site upon formation of a Mg2+3-PPi complex (28, 31). Very recently, the first bacterial sesquiterpene synthase, epo-isozizoene synthase (EIZ) from Streptomyces coelicolor A3 (ScEIZ) (2), was crystallized in the open (mutant D99N) and closed conformation (1). As with the fungal terpene synthases, the H-α1 loop of ScEIZ moves to shield the active site upon formation of the Mg2+3-PPi complex. However, despite the structural importance of the H-α1 loop in this group of enzymes, very little is known about its influence on the cyclization mechanism of terpene synthases. In one recent study, homology modeling and site-directed mutagenesis of α-farnesene synthase from apple (Malus x domestica) identified residues in the H-α1 loop as important for monovalent metal binding (15).
In previous work we have characterized several new fungal sesquiterpene synthases (Cop1 to Cop4 and Cop6; Cop5 was not functional) cloned from Coprinus cinereus (2, 3). The four closely related enzymes (Cop1 to Cop4) each synthesized multiple cyclization products, while trichodiene synthase homolog Cop6 was highly specific. In a subsequent study we characterized the cyclization pathways of Cop4 and Cop6 and compared homology models of the two enzymes, indicating that different binding pocket sizes may be responsible for their different catalytic fidelities by allowing FPP to bind in one or several conformations (22). Cop4 rearranges a secondary cisoid, allylic cation derived from FPP after isomerization (cis-trans pathway of catalysis) into multiple products, with δ-cadinene and β-copaene as the major products when expressed in E. coli and germacrene D and cubebol as the major products in vitro. The fidelity of Cop4 was drastically influenced by the pH of the in vitro reaction. Cop6 also generates a secondary allylic cation which it rearranges highly selective into α-cuprenene under all conditions tested. Comparison of homology models of Cop4 and Cop6 also suggested that the H-α1 loop may play a role in determining the product selectivity of Cop4.
Here we investigate for the first time the influence of the H-α1 loop on the cyclization mechanism of sesquiterpene synthases. Structural models of Cop4 and Cop6 in the open and closed conformation predicted interactions between H-α1 loop residues and other residues that line the active site entrance. Site-directed mutagenesis of the H-α1 loop then identified residues in the permissive Cop4 that modulate and drastically increase product selectivity. Analogous mutations introduced into the α-muurolene synthase Cop3, which, like Cop4, makes multiple products but mostly from a primary, transoid farnesyl cation, were found to have a similar effect on product selectivity. Mutations in the H-α1 loop of Cop6, however, did not change the product outcome of this high-fidelity enzyme.
MATERIALS AND METHODS
Chemicals.
(E,E)-FPP was purchased from Sigma-Aldrich (St. Louis, MO). DNA-modifying enzymes were obtained from New England Biolabs (Ipswich, MA). Other chemicals were from suppliers as indicated below or from Sigma-Aldrich.
Strains, plasmids, and growth conditions.
All cloning and investigations of sesquiterpene biosynthesis were carried out in Escherichia coli strain JM109 or E. coli BL21. Plasmids pUC-Cop3, pET21-Cop4, and pHis8-Cop6 (Cop6 and Cop4 contain a His8 tag and a His6 tag, respectively, added to their N termini for affinity purification) for overexpression of sesquiterpene synthases from Coprinus cinereus in E. coli have been described previously (2, 3, 22) (see Table S1 in the supplemental material). E. coli cultures were grown at 30°C and 250 rpm in Luria-Bertani (LB) medium supplemented with 100 μg ml−1 of the appropriate antibiotics (ampicillin for pUC-Cop3 and pET21-Cop4, kanamycin for pHis8-Cop6). Expression of genes controlled by the T7-promoter (pET21-Cop4 and pHis8-Cop6) was induced by adding 1 mM IPTG (isopropyl-β-d-thiogalactopyranoside) to cultures grown to an optical density (OD) of 0.6.
Creation of H-α1 loop mutations.
Single amino acid substitutions in the H-α1 loops of Cop3, Cop4, and Cop6 were created by site-directed mutagenesis according to the QuikChange protocol (Stratagene, La Jolla, CA) using plasmids pHis8-Cop6, pET21-Cop4, and pUC-Cop3 as templates.
H-α1 loop regions of Cop4 and Cop6 were swapped by using overlap extension PCR. Briefly, for Cop4 and Cop6, gene-flanking PCR primers were designed corresponding to the 5′ and 3′ ends of the Cop gene and including appropriate restriction sites for cloning. Internal primers were designed that bind to the 5′ and 3′ regions next to the loop region and contain complementary 5′ overhangs that encode the new loop region to be inserted. For each Cop gene, two PCRs with external and internal primer pairs yielded two overlapping PCR products that were joined in a second PCR. The final PCR products encoding the grafted Cop4 and Cop6 loop mutants (Cop4L6 and Cop6L4) were digested and ligated back into the same expression plasmids that contained their parent genes to yield pHis8-Cop6L4 and pET21-Cop4L6.
In vivo analysis of sesquiterpene product profiles of H-α1 loop mutants.
Cultures (50 ml) of E. coli JM109 transformed with Cop3 wild-type or loop mutants on plasmid pUC were inoculated with 1 ml of overnight cultures and grown for 20 h at 30°C prior to sampling of culture headspace by solid-phase microextraction (SPME) using a 100-μm-pore-size polydimethlysiloxane fiber (Supelco, Bellefonte, PA). The fiber was inserted through the tin foil sealing the top of the flasks into the flask headspace (gas phase). After 10 min of absorption, the fiber was inserted into the injection port of a gas chromatography-mass spectrometry (GC-MS) for thermal desorption. For Cop4 and Cop6 wild-type and loop mutants, 50-ml cultures of E. coli BL21 transformants were grown at 30°C until cultures reached an OD of 0.6 before induction of gene expression with 1 mM IPTG. Induced cultures of Cop6 wild-type and mutant transformants were grown for 14 h at 20°C, while Cop4 cultures were grown for the same time at 30°C before analysis of sesquiterpenes in the culture headspace by SPME and GC-MS.
Protein purification and enzyme kinetics.
For kinetic measurements, Cop4 and Cop6 wild-type and loop mutants were overexpressed as described above, and the histidine-tagged recombinant proteins were purified by affinity chromatography as described previously (2, 3, 22). Protein concentrations were determined by using Bradford reagent (Bio-Rad, Hercules, CA).
Steady-state kinetics of sesquiterpene synthases were determined by varying the concentration (1-100 μM) of (E,E)-FPP and measuring the release of pyrophosphate (PPi) in a fluorometric assay. The PiPer pyrophosphate assay kit supplied by Invitrogen (Carlsbad, CA) was used to detect PPi in a coupled enzyme assay generating resorufin. The fluorescence of resorufin was measured by using an excitation wavelength of 540 nm and an emission wavelength of 590 nm. Known concentrations of PPi were used to obtain a calibration curve. Assays were carried out in 96-well microplates, and fluorescence was measured with a plate reader (Spectramax multi-mode microplate reader; Molecular Devices, Sunnyvale, CA). Reaction mixtures contained 50 μl of PiPer solution, 40 μl of assay buffer (50 mM Tris-HCl, 10 mM MgCl2 [pH 8.0]), and 10 μl of FPP. Blank reactions without substrate and without enzyme were run in parallel. Assay mixtures were allowed to equilibrate for 5 min at 30°C prior to the addition of 5 μl of enzyme (0.2 mg ml−1) to start the reaction. The activity was determined as the difference between the increase of fluorescent per minute of the sample and of the blanks. One unit of activity was defined as the amount of enzyme needed to release 1 μmol of PPi. The Km and Vmax values were determined by using a nonlinear fit of the V versus [S] plot. The analysis was carried out running a macro in Xcel 2007 (11).
In vitro analysis of sesquiterpene product profiles.
Sesquiterpene product profiles of wild-type and mutant Cop enzymes were analyzed in vitro with isolated proteins and (E,E)-FPP as the substrate. Cop enzymes were overexpressed as described above, and harvested cells were resuspended in terpene synthase buffer (10 mM Tris-HCl, 10 mM MgCl2, and 1 mM β-mercaptoethanol at pH 8) prior to lysis by sonication. The cell lysates were cleared by centrifugation and the soluble fractions (100 μg of total protein) incubated in sealed high-pressure liquid chromatography vials with 100 μM FPP in terpene synthase buffer (final reaction volume 200 μl) for 10 h at 25°C. The headspace of the reactions was sampled by SPME (insertion of fiber through the vial septum) for 10 min, followed by GC-MS analysis of absorbed sesquiterpenes.
To measure the influence of the pH of the reaction on the product profiles of Cop4 and Cop4 variants, the terpene synthase buffer was modified by substituting 10 mM Tris-HCl with 10 mM sodium carbonate (pH 10.0) or 10 mM sodium acetate (pH 5.0). Reactions were carried out for 18 h at 25°C prior to the analysis of sesquiterpene hydrocarbon products as described above.
GC-MS analysis.
GC-MS analysis was carried out on an HP GC 7890A coupled to an anion-trap mass spectrometer HP MSD triple axis detector (Agilent Technologies, Santa Clara, CA). Separation was carried out using a HP-5MS capillary column (30 m by 0.25 mm [inner diameter] by 1.0 μm) with an injection port temperature of 250°C and helium as a carrier gas. Mass spectra were recorded in electron impact ionization mode. Volatile compounds adsorbed on a fiber from the enzyme reaction headspace were desorbed for 10 min in the injection port. The temperature program started at 60°C and ramped up 8°C min−1 to a final oven temperature of 250°C. Mass spectra were scanned in the range of 5 to 300 atomic mass units at 1-s intervals.
For product identification, the retention index of each compound peak was determined by calibrating the GC-MS first with a C8-C40 alkane mix. Retention indices and mass spectra of compound peaks were compared to reference data in Mass Finder's (software version 3) terpene library (19). In addition, essential oils with known terpene compositions were used as authentic standards for the major products as previously described (2, 3, 22).
Structural modeling.
Structural models in the open, unliganded conformation were built using the structure of trichodiene synthase from F. sporotrichoides (27) (PDB 1JFA, chain A) for Cop6 (44% amino acid sequence similarity) and of aristolochene synthase from A. terreus (PDB 2E40, chain D) (30) for Cop4 and Cop 3 (39 and 37% amino acid sequence similarity, respectively). Crystal structures of trichodiene (PDB 2Q9z, chain B) (37) and of aristolochene synthase (PDB 2OA6, chain D) (30) in the closed conformation, liganded with Mg2+ and PPi, were used to built the corresponding models for Cop6, Cop4, and Cop3. Models were built using the Swiss Model homology-modeling server and alignment mode (4). Protein models were visualized and aligned with their template structure using PyMol 0.99 developed by DeLano Scientific LLC (San Francisco, CA). Active-site volumes were calculated with CASTp (12) (using CASTpyMol version 2.0).
RESULTS
Modeling of structural changes in the H-α1 loop upon active-site closure.
Homology models of Cop4 and Cop6, and for comparison of Cop3, were built in the open and closed conformation to gain insight into the mechanism of active-site capping (Fig. 1A to C). Models of Cop4 and of Cop3 were built based on the open and closed X-ray structures of aristolochene synthase AtACS (30). Ligand-free and complexed structures of trichodiene synthase (FsTHS) (27) served as a template for Cop6 model generation.
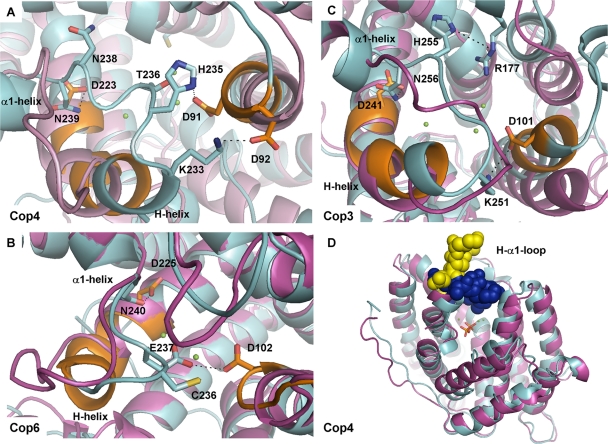
FIG. 1.
Homology models of Cop enzymes. Superimpositions of homology models (see Materials and Methods) in the open (magenta, ligand-free) and closed (cyan, ligand-bound) conformations of Cop4 (A), Cop6 (B), and Cop3 (C) from C. cinereus are presented. Shown are the H-α1 loop and residues of the metal binding DDXXDD and NSE/DTE motifs (orange) with their associated Mg2+ atoms (green spheres). Presumed interactions between side chains of loop residues and residues lining the active site entrance are shown. (D) Superimposition of the open (magenta, H-α1 loop yellow) and closed (cyan, H-α1 loop blue) homology models of Cop4 showing capping of a wide active site entrance by the H-α1 loop upon substrate binding.
Superimposition of the Cop homology models (Fig. 1A to C) show that the H-α1 loop of each enzyme moves closer to the trinuclear Mg2+ cluster in the liganded, closed conformation of the enzyme. The binding pockets of the three Cop models have approximately the same volume (ca. 400 to 500 Å3) in the ligand-bound conformation. However, in the open conformation, the active site cavities of the Cop3 and Cop4 models are about twice larger in volume (∼3,300 Å3) than the cavity of the Cop6 model (∼1,700 Å3) (see Table S2 in the supplemental material). Transition from the open to the closed conformation induces a much greater change in Cop4 and Cop3 than in Cop6. In Cop4, the H-α1 loop appears to undergo a particular large movement upon ligand binding in order to seal the wide open active site cavity of the enzyme (Fig. 1D).
A closer look at the superimposed closed and open model structures (Fig. 1) shows that several polar side chains in the H-α1 loops of Cop4 and Cop3 move closer to side chains in the metal-binding DDXXD motif. In the closed conformation of the Cop4 model, the imidazole side chain of H235 is now positioned close to the carboxyl group of D91 (the first Asp residue in the DDXXD motif) and may form an ionic interaction with this residue. Similarly, the ɛ-amino group of K233 may form a salt bridge with the carboxyl group of D92. Residue N238 also undergoes a conformational change and projects into the active site cavity, but interactions with surrounding residues are not obvious in the model (Fig. 1A). In the closed Cop3 model structure, H255 may hydrogen bond with R177 and K251 (corresponds to K233 in Cop4) may interact with D101 in the DDXXD motif (Fig. 1C).
The H-α1 loop in the Cop6 model undergoes a much smaller conformational change upon substrate binding compared to the other two Cop enzymes. Furthermore, the overall net charge of the Cop6 loop is negative, whereas the loops of Cop3 and Cop4 contain basic residues that potentially interact with acidic residues in the two metal binding motifs. In the closed model of Cop6, only the side chains of two loop residues, E237 and C236, face the active site entrance. Of these two residues, only E237 may form a hydrogen bond with D102 in the aspartate-rich DDXXD-motif (Fig. 1B).
In all Cop models and their respective fungal template structures a conserved Asn residue (N240 in Cop6, N239 in Cop4, and N256 in Cop3) can be identified at the C terminus of the H-α1 loop and may have a role in stabilizing the closed conformation. As shown in Fig. 1, upon ligand binding the side chain of this conserved Asn moves close to the side chain of an aspartic acidic residue in the second metal-coordinating NSE/DTE motif; the NSE/DTE motif is located at the active site entrance opposite of the DDXXD motif. Ionic interactions between N239 and D224 in Cop4, N256 and D241 in Cop3 and N240 and D225 in Cop6 may be important for locking the H-α1 loop in place.
Site-directed mutations in the H-α1 loop alter the product profiles of Cop4 and Cop3.
Residues in the H-α1 loops of Cop4 (K233, H235, T236, N238, and N239) and Cop6 (C236, E237, and N240) identified above were mutated to investigate their influence on catalysis. Cop4 residues T236 and N238, as well as Cop6 residue C236, which do not appear to interact with surrounding residues in the ligand-bound enzymes, were included to investigate the importance of interactions between residues of the loop with those of the two metal-binding motifs. These polar, acidic, or basic residues were substituted for nonpolar residues with similar side chain lengths to disrupt potential polar or ionic interactions while imposing minimal conformational strain.
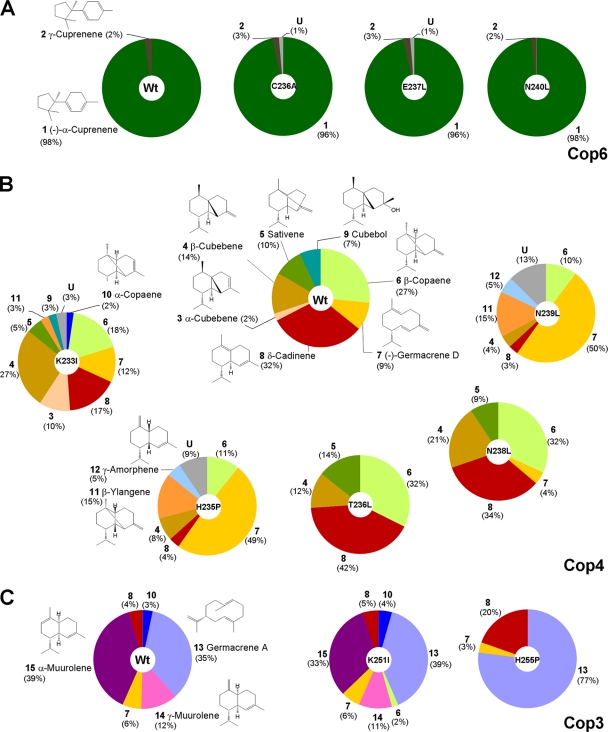
The effect of the H-α1 loop mutations on the product profiles of Cop4 and Cop6 was investigated by analyzing the sesquiterpenes accumulated in the headspace of recombinant E. coli cultures overexpressing the enzymes (Fig. 2) (see Fig. S1 for GC analysis of product profiles, Fig. S2 for mass spectra of identified sesquiterpenoids, and Table S3 for in vitro product profiles with isolated enzymes in the supplemental material). As reported previously for the wild-type enzymes (2, 3, 22), Cop4 makes 10 different sesquiterpene products with δ-cadinene 8 (compounds are numbered; see Fig. 2 and 4 for structures) and β-copaene 6 as the major products in vivo, whereas Cop6 makes almost exclusively (−)-α-cuprenene 1. Comparison of product profiles obtained with wild-type enzymes and loop mutants shows that none of the mutations introduced into the highly selective Cop6 significantly change the fidelity of the enzyme (Fig. 2A). In contrast, several Cop4 loop mutants have drastically altered product profiles (Fig. 2B).
FIG. 2.
Influence of site-specific H-α1 loop mutations on the product profiles of Cop enzymes. The relative ratios (%) of sesquiterpene compounds detected in the headspace of E. coli cultures (see Materials and Methods) expressing wild-type or loop mutants of Cop6 (A), Cop 4 (B), and Cop3 (C) were determined. Sesquiterpene compounds are numbered and structures are shown for each compound (see also Fig. 4 and Fig. S2 in the supplemental material for compound numbers and structures). U, pool of unidentified sesquiterpene products.
Mutations H235P and N239L, which presumably interact with a conserved Asp in the two metal-binding motifs, each convert Cop4 into a much more selective enzyme that makes (−)-germacrene D 7 as the major cyclization product (∼50% of total sesquiterpenes products) (Fig. 2B). Both mutants make β-ylangene 11, which is a diastereomer of β-copaene 6 and not synthesized by wild-type Cop4. Mutation of K233 presumed to interact with the second Asp (D92) in the DDXXD motif of Cop4 does not significantly change the overall product promiscuity of Cop4, though β-cubebene 4 (27% of total sesquiterpene products) does become the major product. Mutagenesis of Cop4 loop residues T236 and N238, which do not seem to interact with any residues of the conserved motifs, cause only subtle changes in the product profile of Cop4 (Fig. 2B).
These results indicate that the H-α1 loop plays an important role in determining the cyclization outcome of the permissive Cop4 but not of the stringent Cop6. Cop4 has a large active-site cavity that undergoes substantial conformational change in the model upon ligand binding. The model structure of the permissive Cop3 also contains a large active-site cavity and ligand-binding triggers a H-α1 loop movement similar to Cop4. The H-α1 loop movement in Cop3 may lead to new polar and ionic interactions between side chains of basic loop residues (K251, H255, and N256) and residues of the metal-binding motifs. Site-directed mutagenesis of these residues was done to compare their effect on FPP cyclization to effects on cyclization seen with Cop4 loop mutations.
Cop3 synthesizes seven different sesquiterpenes with α- muurolene 15 and germacrene A 13 as the major products, each constituting ca. 35 to 40% of the total sesquiterpenes produced (Fig. 2C). As with Cop4, mutation of the only His residue (H255) in the loop has a dramatic effect on product selectivity, increasing synthesis of the prematurely quenched, monocyclic germacrene A 13 (∼80% of total sesquiterpenes products), while abolishing cyclization of FPP to the bicyclic α-muurolene 15. Mutations of the conserved Lys residue (K251I) in Cop3 have only a moderate effect on product selectivity, which is comparable to the effect seen with the corresponding K233I mutation in Cop4. Substitution in Cop3 of N256, which is conserved in all three Cop enzymes, renders Cop3 inactive, whereas mutation of this conserved residue (N239L) had a large impact on the product profile of Cop4.
H-α1 loop swap between Cop4 and Cop6 shifts Cop4 to a germacrene D synthase.
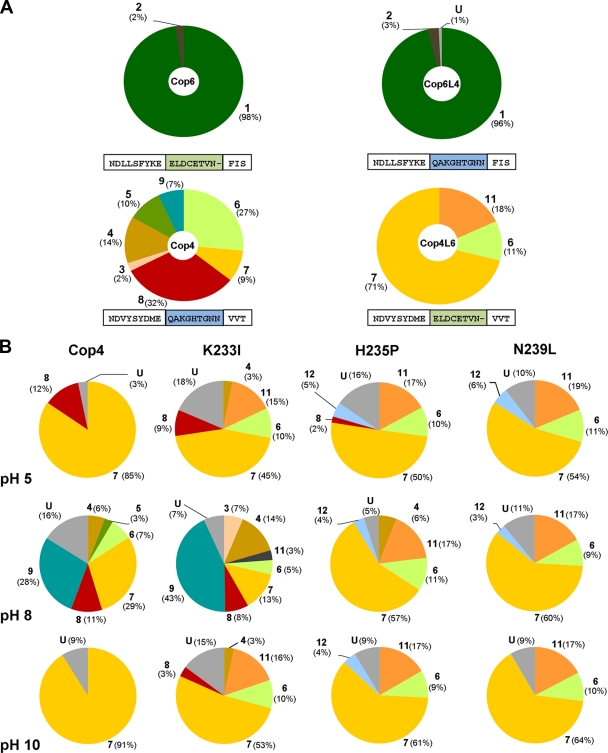
Site-directed mutagenesis of several H-α1 loop residues changed the product profiles of the permissive Cop4 and Cop3, but not of the highly selective Cop6. Maintenance of product fidelity by the Cop6 loop mutants may either be the result of the loop structure, or the fact that the H-α1 loop is less important in Cop6 for the cyclization mechanism. The loops of Cop4 and Cop6 starting at the conserved Glu from the NSE/DTE-motif were swapped to study the effects of the longer and basic Cop4 loop on the activity of Cop6 (Cop6L4 mutant) and vice versa, the effect of the shorter, negatively charged Cop6 loop on the activity of Cop4 (Cop4L6 mutant). Sesquiterpene product profiles (Fig. 3A) of the two loop graft mutants were analyzed as described for the site-directed H-α1 loop mutations of Cop4 and Cop6.
FIG. 3.
Product profiles of Cop4 and Cop6 loop grafted mutants and the influence of H-α1 loop residues on the pH-dependent product profile of Cop4. (A) Comparison of relative amounts of sesquiterpene compounds produced by recombinant cultures (see Materials and Methods) expressing wild type (Cop4 and Cop6) and loop-grafted mutants (Cop6L4 and Cop4L6). Amino sequences of loop regions for wild-type and loop-grafted Cop enzymes are shown, and swapped loop regions are highlighted. (B) Relative amounts of sesquiterpene compounds produced by Cop4 wild-type and loop mutants in in vitro reactions at pH 5, 8, and 10. Numbers and shading of the pie areas of sesquiterpene products correspond to the structures and compound names shown in Fig. 2 and 4. U, pool of unidentified sesquiterpene products.
Replacement of the H-α1 loop in Cop4 with the loop from Cop6 increases the product selectivity of the resulting loop graft mutant Cop4L6 even more than what is observed for the site-specific loop mutants H235P and N239L (Fig. 2 and 3A). Cop4L6 becomes very selective for the synthesis of (−)-germacrene D 7 (70% of total sesquiterpene products), while at the same time loosing its ability to make the bicyclic δ-cadinene 8, which is the major product of the wild type (Fig. 3A). The relative amount of β-copaene 6, the second major product of the wild type, made by Cop4L6 is also drastically reduced. In addition, β-ylangene 11 appears as a new cyclization product of Cop4L6, as observed for the site-specific loop mutants H235P and N239L. In the case of the Cop6 loop graft mutant Cop6L4, loop replacement has no influence on the selectivity of the enzyme for α-cuprenene, confirming that the H-α1 loop does not influence the product spectrum of Cop6.
H-α1 loop mutations affect catalytic efficiency more strongly in Cop4 than in Cop6.
Cop4 and Cop6 wild-type and mutant proteins (site-directed loop and loop grafted mutants) were purified to determine the influence of the various loop mutations on FPP binding (Km) and catalytic turnover (kcat) (Table 1). All site-directed H-α1 loop mutations decrease the catalytic efficiency (kcat/Km) of Cop4 and Cop6, but the decrease is much larger in Cop4 than in Cop6, reducing the catalytic efficiency of Cop4 by two to three orders of magnitude compared to a 3 to 17-fold decrease seen with Cop6. In both enzymes, mutation of the conserved Asn residue located at the C terminus of the H-α1 loop (N239 and N240 for Cop4 and Cop6, respectively) (Fig. 1) causes the largest loss of activity.
TABLE 1.
Kinetic constants of Cop4 and Cop6 wild type and loop mutants with E,E-FPP
| Enzyme | Wild type or mutant | Mean relative kcat (%) ± SD | Mean Km (μM) ± SD | kcat/Km (M−1 s−1) |
|---|---|---|---|---|
| Cop4 | Wild type | 100 | 11 ± 3 | 1,000 |
| K233I | 21 ± 1 | 70.7 ± 6.3 | 35 | |
| H235P | 2.7 ± 0.2 | 11 ± 2.6 | 29 | |
| N239L | 0.47 ± 0.03 | 11 ± 2.5 | 5 | |
| Cop4L6 | 0.53 ± 0.13 | 48 ± 10 | 1 | |
| Cop6 | Wild type | 100 | 7.6 ± 2.4 | 88,000 |
| C236A | 41.2 ± 3.1 | 12.5 ± 2.1 | 21,300 | |
| E237L | 59.4 ± 3.1 | 13.6 ± 1.4 | 28,300 | |
| N240L | 31 ± 4 | 40 ± 14 | 5,246 | |
| Cop6L4 | 15 ± 2 | 7.4 ± 3.5 | 13,200 |
The negative effect of the H-α1 loop mutations on the catalytic efficiency (kcat/Km) of Cop4 and Cop6 is largely caused by a decrease in their catalytic turnover rates, which is three orders of magnitude reduced in the Cop4 N239L mutant compared to a 3-fold reduction for the corresponding Cop6 N240L mutant. Most site-directed loop mutations have little or no effect on FPP binding (Km); only mutation of K233 in Cop4 and of N240 in Cop6 decreases binding affinity 5- to 6-fold.
Loop swapping in Cop4 severely decreases catalytic turnover (kcat) and substrate affinity (Km), resulting in a mutant protein Cop4L6 with a 1,000-fold-decreased catalytic efficiency compared to the wild-type enzyme. In contrast, the catalytic efficiency of the Cop6 loop graft mutant Cop6L4 is only ∼6-fold reduced compared to wild-type Cop6. Loop replacement in Cop6 has no affect on FPP binding but decreases catalytic turnover (kcat).
Influence of H-α1 loop residues on pH dependence of Cop4 product profile.
In previous work we showed that the product profile of Cop4, but not of Cop6, is strongly dependent on pH (22). Acidic or basic reaction conditions render Cop4 much more selective for (−)-germacrene D 7 synthesis by promoting carbocation deprotonation at an earlier step in the cyclization pathway. Cop4 loop mutants H235P and N239L also strongly favor (−)-germacrene D 7 synthesis, suggesting that basic or acidic pH conditions may induce similar changes in the H-α1 loop than the two loop mutations.
Cyclization of FPP by purified Cop4 wild-type and loop mutants (K233I, H235P, and N239L) was analyzed at pH 5, 8, and 10 to investigate whether residues of the H-α1 loop are responsible for the observed pH dependence of Cop4's cyclization mechanism (Fig. 3B). As expected from previous results (22), wild-type Cop4 becomes a (−)-germacrene D 7 synthase at pH 5 or pH 10. Cop4 loop mutant K2331 also becomes more selective for (−)-germacrene D 7 under acidic or basic reaction conditions, although less so than the wild-type enzyme. The product profiles of loop mutants Cop4 H235P and N239L, however, are not affected by the pH of the reaction, suggesting that H235 and N239 are responsible for the pH induced changes in the product profile of Cop4.
DISCUSSION
Structural studies of sesquiterpene synthases have provided insights into the rearrangements that occur upon binding of metal cofactor (Mg2+) and substrate (FPP) to the active site (1, 27, 28, 30, 32). Despite the identification of several loops and helices involved in this conformational rearrangement, only one study has specifically targeted residues of the H-α1 loop that caps the active site upon ligand binding (15). Here we used structural modeling and site-directed mutagenesis to investigate the role of the H-α1 loop in the cyclization mechanism of three fungal sesquiterpene synthases (Cop3, Cop4, and Cop6) that have different product profiles and fidelities.
Superimposition of the three Cop homology models in the open and closed conformations shows that the N-terminal region of the H-α1 loop becomes part of the H-helix upon substrate binding, causing the loop and the conserved metal-binding NSE/DTE motif to close over the binding cavity as has been observed for the AtACS and FsTHS structures (27, 30) (Fig. 1A to C). The H-α1 loop of Cop4 undergoes the largest conformational change upon substrate binding (Fig. 1D). The binding pocket volumes of the un-liganded Cop4 and Cop3 model structures are twice that of Cop6 (see Table S2 in the supplemental material), suggesting that the two enzymes may be able to bind FPP in different conformations that lead to multiple cyclization products. Substrate binding triggers a significant contraction of the Cop4 and Cop3 binding pockets so that their volumes become comparable to that of the ligand-bound Cop6. However, comparison of the active site contours of the ligand-bound Cop4 and Cop6 homology models (see Fig. S3 in the supplemental material) shows a narrow binding pocket for Cop6, whereas that of Cop4 is much wider, allowing the initial cyclic Z,E-germacradienyl carbocation to rearrange along multiple cyclization paths. In contrast, the narrow binding pocket of Cop6 restrains rearrangement of the initial cyclic 6-(R)-bisabolyl carbocation along only one cyclization route to yield (−)-α-cuprenene 1.
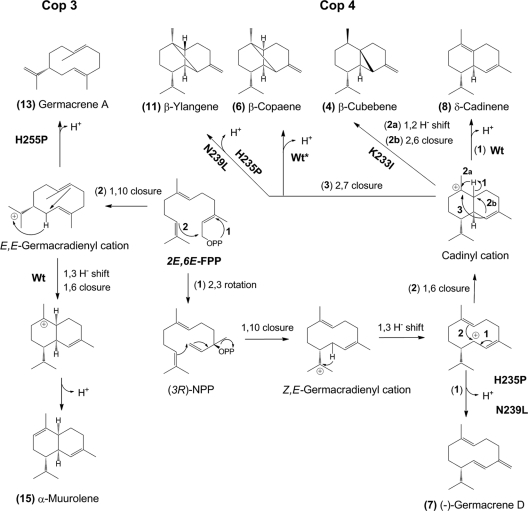
In ligand-bound Cop4 and Cop3, ionic and polar interactions between side chains of H-α1 loop residues and of residues lining the active site may facilitate a tight closure of the active-site cavity (Fig. 1). Disruption of the presumed interaction between H235-D91 in Cop4 and H255-R177 in Cop3 leads to premature deprotonation in the normal cyclization pathways of the two enzymes. The mutant enzymes now each produce one major monocyclic product which are (−)-germacrene D 7 and germacrene A 13 for Cop4 and Cop3, respectively (Fig. 4). The mutated H-α1 loops in these enzymes likely no longer efficiently close and shield the active site from the environment, leading to premature deprotonation. Incomplete closure may also affect contraction of the binding pocket, resulting in a binding cavity that is less able to chaperone reaction intermediates to subsequent reaction pathways by lowering energy barriers via conformational restraints. This is consistent with the relative increased synthesis by Cop3 mutant H255P of the cis pathway product δ-cadinene 8, which requires accommodation of bulkier reaction intermediates resulting from the isomerization of the C2-C3 bond in all-trans FPP (Fig. 4). In Cop4 mutant H235P, a larger binding cavity allows the generation of a cadinyl cation that can have both configurations at C6, which after 2,7-cyclization yields the diastereomers β-copaene 6 and β-ylangene 11; the latter is not made by the wild-type enzyme (Fig. 4). Aberrant cyclization products and premature quenching has previously been observed in other sesquiterpene synthases when residues of the conserved metal-binding motifs were mutated (28, 35).
FIG. 4.
Proposed cyclization pathways of wild-type and H-α1-loop mutants of Cop3 (left side) and Cop4 (right side). Cyclization of E,E-FPP by Cop4 involves an ionization/isomerization step to yield (3R)-nerolidyl diphosphate [(3R)-NPP] (22) which, after ionization and 1,10-ring closure, gives a Z,E-germacradienyl cation that is further rearranged to the different cyclization products of Cop4 wild type and loop mutants. The major cyclization products of Cop3 and its loop mutants are derived from a E,E-germacradienyl cation formed after ionization and 1,10 cyclization of E,E-FPP (2, 3). Shown are cyclization pathways to major products of Cop3 and Cop4 wild type and loop mutants that significantly impact product outcomes of the two enzymes. Compound numbers correspond to sesquiterpenes in the product profiles shown in Fig. 2 to 4. Numbered reaction arrows indicate different branch points in the cyclization reaction.
In the ligand-bound Cop6, a polar interaction may be formed between loop residue E237 and the first conserved D102 in the DDXXD motif (Fig. 1), similar to the presumed interaction between H235 and D91 in Cop4. However, hydrogen bond formation between the two acidic residues in Cop6 may not be as efficient as salt bridge formation between the corresponding D91 and H235 in Cop4. Mutation of E237 in Cop6 has no influence on product profile and reduces only moderately the catalytic efficiency of Cop6 (Fig. 2, Table 1).
Mutation of a K233 in Cop4 and K251 in Cop3 only moderately affects FPP cyclization (Fig. 2). Cop4 mutant K233I now prefers a reaction pathway leading from a cadinyl cation to β-cubebene 4 instead of to δ-cadinene 8 as in the wild-type enzyme (Fig. 4). The side chain of K233 presumably forms a salt bridge with the second Asp residue in the DDXXD motif that has been shown to not be involved in Mg2+ binding in AtACS or FsTHS (27, 30) (Fig. 1A). Mutation of K233 in Cop4 causes the least decrease in activity compared to the other Cop4 loop mutations in the present study (Table 1), suggesting that K233 in Cop4 and K251 in Cop3 do not play a major role in the network of side chain and ligand interactions formed during active-site closure.
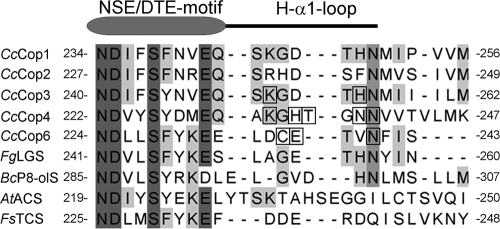
Alignment of functionally characterized fungal sesquiterpene synthase sequences shows that C. cinereus sesquiterpene synthases (2, 3, 22), longifolene synthase from Fusarium graminearum (23), and presilphiperfolan-8-β-ol synthase from Botrytis cinerea (26, 38), but not aristolochene synthase AtACS (30) and trichodiene synthase FsTHS (27), contain a conserved Asn in the H-α1 loop (Fig. 5). Green et al. (15) recently reported that this residue is also conserved as Asn, Lys, or Ser in the H-1α-loops of a large subgroup of plant terpene synthases. A conserved Ser was found to participate in K+ binding in α-farnesene synthase from apple and pinene synthase from spruce; replacement of Ser with Lys abolished the dependence of these enzymes on K+ for activity (15). In the closed Cop models, the amide group of the conserved Asn hydrogen bonds with the carboxyl group of a conserved Asp residue (D225 in Cop6, D223 in Cop4, and D241 in Cop3) in the NSE/DTE metal-binding motif (NDxxS/TxxxE/D) and may therefore function as a lock during active-site closure. In plant terpene synthases, side chains of the conserved Asn and Lys residues or Ser, perhaps through coordination with K+, may in a similar manner stabilize capping of the active site. In fact, in the ligand-bound model of apple α-farnesene synthase (15), the side chains of S487 and E479 are positioned for interaction as in the Cop models.
FIG. 5.
Alignment of H-α1 loop regions and preceding conserved NSE/DTE-motifs of fungal sesquiterpene synthases. The following protein sequences of experimentally characterized fungal sesquiterpene synthases were aligned by using the CLUSTAL W algorithm (33): CcCop1, Cop1 from Coprinus cinereus (XP_001832573); CcCop2, Cop2 from Coprinus cinereus (XP_001836556); CcCop3, Cop3 from Coprinus cinereus (XP_001832925);. CcCop4, Cop4 from Coprinus cinereus (XP_001836356); CcCop6, Cop6 from Coprinus cinereus (XP001832549); FgLGS, longiborneol synthase from Fusarium graminearum (ACY69978); BcP8-olS, presilphiperfolan-8-β-ol synthase from Botrytis cinerea (AAQ16575); and AtACS, aristolochene synthase from Aspergillus terreus (AAF13263). FsTHS, trichodiene synthase from Fusarium sporotrichioides (P13513).
Disruption of the presumed hydrogen bonding between the Asn and Asp side chains (N239-D223 in Cop4 and N240-D225 in Cop6) severely compromises catalytic efficiency of the resulting Cop4 and Cop6 mutants (Cop4 N239L and Cop6 N240L) (Table 1), indicating that this interaction may be critical in positioning side chains for optimal PPi/Mg2+ coordination and subsequent diphosphate cleavage for catalysis. This mutation also strongly affects the product profile of Cop4 (but not of Cop6); causing premature deprotonation to yield (−)-germacrene D as a major cyclization product as in Cop4 mutant H235P.
Mutation of Cop6 loop residues reduce catalytic activities to different extents, but none of these substitutions change the product fidelity of Cop6, suggesting that the H-1α loop in Cop6 plays a role in optimal PPi/Mg2+ coordination but not in preventing premature quenching and in the substrate induced conformational changes that narrow the active site of this enzyme. These Cop6 loop mutants likely maintain the same cavity contour as the wild-type enzyme. The active site cavity in Cop6 restricts substrate and reaction intermediate binding almost exclusively to one productive conformation that yields (−)-α-cuprenene 1. As in trichodiene synthase, the pyrophosphate group of the substrate likely shields in Cop6 its narrow binding cleft from solvent and prevents premature quenching of reaction intermediates (28). Complete substitution of the H-α1 loop of Cop6 with the one residue longer and positively charged loop from Cop4, reduces activity but again does not change the product profile of Cop6 (Fig. 2, Table 1), supporting the above conclusions that the H-α1 loop is not important for the substrate induced conformational change and shielding of the active site. In contrast, the H-α1 loop exchange in Cop4 not only severely decreases catalytic activity (Table 1) but also prevents active-site shielding and adoption of a productive cavity conformation. The Cop4 loop exchange mutant has an even more drastically altered product profile than loop mutants H235P and N239L in that it only makes three products: (−)-germacrene D 7 as the main product, along with smaller amounts of the diastereomers β-copaene 6 and β-ylangene 11 (Fig. 2 and 4).
The product fidelity of Cop4 was previously found to be unusually sensitive to the pH of its environment (22), which is in contrast to Cop6 (22) and to amorphadiene-4,11-diene synthase, another terpene synthase for which product formation was studied under various pH conditions (25). Similar to Cop4 loop mutants H235P, N239L, and Cop4L6 in the present study, the wild-type enzyme becomes, under basic or acidic conditions, prone to premature quenching of cyclization intermediates and produces mostly (−)-germacrene D 7 (Fig. 3B). Comparison of product profiles of loop mutants obtained at pH 5, 8, and 10 shows that only the product profile of the Cop4 mutant K233I is as pH dependent as the wild-type enzyme. Mutation of loop residues H235 or N239 abolishes the pH sensitivity, indicating that in the wild-type enzyme changes in pH disrupt the ionic interactions of these two residues, causing the same conformational perturbations as in the loop mutants.
In summary, we have shown that the H-α1 loop plays a major role in determining the product profile of low-fidelity fungal sesquiterpene synthases with large active-site cavities. Structural modeling of Cop enzymes pointed out potential interactions of H-α1 loop residues with conserved residues of the two metal-binding motifs; one of the potential interactions between conserved Asp/Glu and Arg, Asn, and Lys (or Ser in monovalent cation-dependent terpene synthase) is conserved in several fungal and plant terpene synthases and may play an important role in stabilizing the closed enzyme conformation by locking the H-α1 loop in place. Mutagenesis of the H-α1 loop may provide a new approach to alter the product profiles of terpene synthases and study the reaction mechanism of this class of enzymes.
Supplementary Material
Acknowledgments
This research was supported by National Institute of Health grant GM080299 (C.S.-D.).
Footnotes
Published ahead of print on 1 October 2010.
Supplemental material for this article may be found at http://aem.asm.org/.
REFERENCES
- 1.Aaron, J. A., X. Lin, D. E. Cane, and D. W. Christianson. 2010. Structure of epi-isozizaene synthase from Streptomyces coelicolor A3(2), a platform for new terpenoid cyclization templates. Biochemistry 49:1787-1797. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Agger, S., F. Lopez-Gallego, and C. Schmidt-Dannert. 2009. Diversity of sesquiterpene synthases in the basidiomycete Coprinus cinereus. Mol. Microbiol. 72:1307-1308. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Agger, S., F. Lopez-Gallego, and C. Schmidt-Dannert. 2009. Diversity of sesquiterpene synthases in the basidiomycete Coprinus cinereus. Mol. Microbiol. 72:1181-1195. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Bordoli, L., F. Kiefer, K. Arnold, P. Benkert, J. Battey, and T. Schwede. 2009. Protein structure homology modeling using SWISS-MODEL workspace. Nat. Protoc. 4:1-13. [DOI] [PubMed] [Google Scholar]
- 5.Buckingham, J. 2009. Dictionary of natural products (online), version 17.1. CHEMnetBASE, Chapman & Hall, New York, NY.
- 6.Cane, D. E., and I. Kang. 2000. Aristolochene synthase: purification, molecular cloning, high-level expression in Escherichia coli, and characterization of the Aspergillus terreus cyclase. Arch. Biochem. Biophys. 376:354-364. [DOI] [PubMed] [Google Scholar]
- 7.Christianson, D. W. 2006. Structural biology and chemistry of the terpenoid cyclases. Chem. Rev. 106:3412-3442. [DOI] [PubMed] [Google Scholar]
- 8.Christianson, D. W. 2008. Unearthing the roots of the terpenome. Curr. Opin. Chem. Biol. 12:141-150. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Croteau, R., and D. E. Cane. 1985. Steroids and isoprenoids (part A). Methods Enzymol. 110:383-405. [PubMed] [Google Scholar]
- 10.Davis, E. M., and R. Croteau. 2000. Cyclization enzymes in the biosynthesis of monoterpenes, sesquiterpenes, and diterpenes. Top. Curr. Chem. 209:54-92. [Google Scholar]
- 11.de Levie, R. 2001. How to use Excel in analytical chemistry and general scientific data analysis. Cambridge University Press, Cambridge, United Kingdom.
- 12.Dundas, J., Z. Ouyang, J. Tseng, A. Binkowski, Y. Turpaz, and J. Liang. 2006. CASTp: computed atlas of surface topography of proteins with structural and topographical mapping of functionally annotated residues. Nucleic Acids Res. 34:W116-W118. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Felicetti, B., and D. E. Cane. 2004. Aristolochene synthase: mechanistic analysis of active site residues by site-directed mutagenesis. J. Am. Chem. Soc. 126:7212-7221. [DOI] [PubMed] [Google Scholar]
- 14.Gennadios, H. A., V. Gonzalez, L. Di Costanzo, A. Li, F. Yu, D. J. Miller, R. K. Allemann, and D. W. Christianson. 2009. Crystal structure of (+)-delta-cadinene synthase from Gossypium arboreum and evolutionary divergence of metal binding motifs for catalysis. Biochemistry 48:6175-6183. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Green, S., C. J. Squire, N. J. Nieuwenhuizen, E. N. Baker, and W. Laing. 2009. Defining the potassium binding region in an apple terpene synthase. J. Biol. Chem. 284:8661-8669. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Greenhagen, B. T., P. E. O'Maille, J. P. Noel, and J. Chappell. 2006. Identifying and manipulating structural determinates linking catalytic specificities in terpene synthases. Proc. Natl. Acad. Sci. U. S. A. 103:9826-9831. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Koellner, T. G., J. Gershenzon, and J. Degenhardt. 2009. Molecular and biochemical evolution of maize terpene synthase 10, an enzyme of indirect defense. Phytochemistry 70:1139-1145. [DOI] [PubMed] [Google Scholar]
- 18.Koellner, T. G., P. E. O'Maille, N. Gatto, W. Boland, J. Gershenzon, and J. Degenhardt. 2006. Two pockets in the active site of maize sesquiterpene synthase TPS4 carry out sequential parts of the reaction scheme resulting in multiple products. Arch. Biochem. Biophys. 448:83-92. [DOI] [PubMed] [Google Scholar]
- 19.Koenig, W. A., N. Bulow, and Y. Saritas. 1999. Identification of sesquiterpene hydrocarbons by gas phase analytical methods. Flavour Fragrance J. 14:367-378. [Google Scholar]
- 20.Lesburg, C. A., J. M. Caruthers, C. M. Paschall, and D. W. Christianson. 1998. Managing and manipulating carbocations in biology: terpenoid cyclase structure and mechanism. Curr. Opin. Struct. Biol. 8:695-703. [DOI] [PubMed] [Google Scholar]
- 21.Little, D. B., and R. B. Croteau. 2002. Alteration of product formation by directed mutagenesis and truncation of the multiple-product sesquiterpene synthases delta-selinene synthase and gamma-humulene synthase. Arch. Biochem. Biophys. 402:120-135. [DOI] [PubMed] [Google Scholar]
- 22.Lopez-Gallego, F., S. A. Agger, D. Abate-Pella, M. D. Distefano, and C. Schmidt-Dannert. 2010. Sesquiterpene synthases Cop4 and Cop6 from Coprinus cinereus: catalytic promiscuity and cyclization of farnesyl pyrophosphate geometric isomers. Chembiochem 11:1093-1106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.McCormick, S. P., N. J. Alexander, and L. J. Harris. 2010. CLM1 of Fusarium graminearum encodes a longiborneol synthase required for culmorin production. Appl. Environ. Microbiol. 76:136-141. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.O'Maille, P. E., A. Malone, N. Dellas, B. Andes Hess, Jr., L. Smentek, I. Sheehan, B. T. Greenhagen, J. Chappell, G. Manning, and J. P. Noel. 2008. Quantitative exploration of the catalytic landscape separating divergent plant sesquiterpene synthases. Nat. Chem. Biol. 4:617-623. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Picaud, S., M. Brodelius, and P. E. Brodelius. 2005. Expression, purification and characterization of recombinant (E)-β-farnesene synthase from Artemisia annua. Phytochemistry 66:961-967. [DOI] [PubMed] [Google Scholar]
- 26.Pinedo, C., C. M. Wang, J. M. Pradier, B. Dalmais, M. Choquer, P. Le Pecheur, G. Morgant, I. G. Collado, D. E. Cane, and M. Viaud. 2008. Sesquiterpene synthase from the botrydial biosynthetic gene cluster of the phytopathogen Botrytis cinerea. ACS Chem. Biol. 3:791-801. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Rynkiewicz, M. J., D. E. Cane, and D. W. Christianson. 2001. Structure of trichodiene synthase from Fusarium sporotrichioides provides mechanistic inferences on the terpene cyclization cascade. Proc. Natl. Acad. Sci. U. S. A. 98:13543-13548. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Rynkiewicz, M. J., D. E. Cane, and D. W. Christianson. 2002. X-ray crystal structures of D100E trichodiene synthase and its pyrophosphate complex reveal the basis for terpene product diversity. Biochemistry 41:1732-1741. [DOI] [PubMed] [Google Scholar]
- 29.Seemann, M., G. Zhai, J. W. de Kraker, C. M. Paschall, D. W. Christianson, and D. E. Cane. 2002. Pentalenene synthase: analysis of active site residues by site-directed mutagenesis. J. Am. Chem. Soc. 124:7681-7689. [DOI] [PubMed] [Google Scholar]
- 30.Shishova, E. Y., L. Di Costanzo, D. E. Cane, and D. W. Christianson. 2007. X-ray crystal structure of aristolochene synthase from Aspergillus terreus and evolution of templates for the cyclization of farnesyl diphosphate. Biochemistry 46:1941-1951. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Shishova, E. Y., F. Yu, D. J. Miller, J. A. Faraldos, Y. Zhao, R. M. Coates, R. K. Allemann, D. E. Cane, and D. W. Christianson. 2008. X-ray crystallographic studies of substrate binding to aristolochene synthase suggest a metal ion binding sequence for catalysis. J. Biol. Chem. 283:15431-15439. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Starks, C. M., K. W. Back, J. Chappell, and J. P. Noel. 1997. Structural basis for cyclic terpene biosynthesis by tobacco 5-epi-aristolochene synthase. Science 277:1815-1820. [DOI] [PubMed] [Google Scholar]
- 33.Thompson, J. D., T. J. Gibson, and D. G. Higgins. 2003. Multiple sequence alignment using ClustalW and ClustalX. Curr. Protoc. Bioinformatics 2:2.3.1-2.3.22. [DOI] [PubMed] [Google Scholar]
- 34.Vedula, L. S., D. E. Cane, and D. W. Christianson. 2005. Role of arginine-304 in the diphosphate-triggered active site closure mechanism of trichodiene synthase. Biochemistry 44:12719-12727. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Vedula, L. S., J. Jiang, T. Zakharian, D. E. Cane, and D. W. Christianson. 2008. Structural and mechanistic analysis of trichodiene synthase using site-directed mutagenesis: probing the catalytic function of tyrosine-295 and the asparagine-225/serine-229/glutamate-233-Mg2+ B motif. Arch. Biochem. Biophys. 469:184-194. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Vedula, L. S., M. J. Rynkiewicz, H. J. Pyun, R. M. Coates, D. E. Cane, and D. W. Christianson. 2005. Molecular recognition of the substrate diphosphate group governs product diversity in trichodiene synthase mutants. Biochemistry 44:6153-6163. [DOI] [PubMed] [Google Scholar]
- 37.Vedula, L. S., Y. X. Zhao, R. M. Coates, T. Koyama, D. E. Cane, and D. W. Christianson. 2007. Exploring biosynthetic diversity with trichodiene synthase. Arch. Biochem. Biophys. 466:260-266. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Wang, C. M., R. Hopson, X. Lin, and D. E. Cane. 2009. Biosynthesis of the sesquiterpene botrydial in Botrytis cinerea: mechanism and stereochemistry of the enzymatic formation of presilphiperfolan-8β-ol. J. Am. Chem. Soc. 131:8360-8361. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Whittington, D. A., M. L. Wise, M. Urbansky, R. M. Coates, R. B. Croteau, and D. W. Christianson. 2002. Bornyl diphosphate synthase: structure and strategy for carbocation manipulation by a terpenoid cyclase. Proc. Natl. Acad. Sci. U. S. A. 99:15375-15380. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Yoshikuni, Y., T. E. Ferrin, and J. D. Keasling. 2006. Designed divergent evolution of enzyme function. Nature 440:1078-1082. [DOI] [PubMed] [Google Scholar]
- 41.Yoshikuni, Y., V. J. Martin, T. E. Ferrin, and J. D. Keasling. 2006. Engineering cotton (+)-delta-cadinene synthase to an altered function: germacrene D-4-ol synthase. Chem. Biol. 13:91-98. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.