Abstract
Reductive acetogenesis via the acetyl coenzyme A (acetyl-CoA) pathway is an alternative hydrogen sink to methanogenesis in the rumen. Functional gene-based analysis is the ideal approach for investigating organisms capable of this metabolism (acetogens). However, existing tools targeting the formyltetrahydrofolate synthetase gene (fhs) are compromised by lack of specificity due to the involvement of formyltetrahydrofolate synthetase (FTHFS) in other pathways. Acetyl-CoA synthase (ACS) is unique to the acetyl-CoA pathway and, in the present study, acetyl-CoA synthase genes (acsB) were recovered from a range of acetogens to facilitate the design of acsB-specific PCR primers. fhs and acsB libraries were used to examine acetogen diversity in the bovine rumen and forestomach of the tammar wallaby (Macropus eugenii), a native Australian marsupial demonstrating foregut fermentation analogous to rumen fermentation but resulting in lower methane emissions. Novel, deduced amino acid sequences of acsB and fhs affiliated with the Lachnospiraceae in both ecosystems and the Ruminococcaeae/Blautia group in the rumen. FTHFS sequences that probably originated from nonacetogens were identified by low “homoacetogen similarity” scores based on analysis of FTHFS residues, and comprised a large proportion of FTHFS sequences from the tammar wallaby forestomach. A diversity of FTHFS and ACS sequences in both ecosystems clustered between the Lachnospiraceae and Clostridiaceae acetogens but without close sequences from cultured isolates. These sequences probably originated from novel acetogens. The community structures of the acsB and fhs libraries from the rumen and the tammar wallaby forestomach were different (LIBSHUFF, P < 0.001), and these differences may have significance for overall hydrogenotrophy in both ecosystems.
Methane is a potent greenhouse gas that is implicated in global warming (35). Of the 600 Tg of methane released into the atmosphere each year, 55 to 70% is anthropogenic (48). Enteric fermentation of ruminant livestock is the largest source of anthropogenic methane, contributing between 20 and 25% (48). During enteric fermentation, archaea in the rumen (methanogens) produce methane mainly through the stepwise reduction of CO2 (4H2 + CO2 → CH4 + 2H2O) (47). As well as contributing to greenhouse gas emissions, methanogenesis is energetically wasteful representing a loss of between 2 and 12% ingested feed energy (23). Reductive acetogenesis is a hydrogenotrophic pathway (4H2 + 2CO2 → CH3COOH + 2H2O) that results in an energy gain for ruminant livestock through the production of acetate (22) and could be an alternative hydrogen sink to methanogenesis if methanogenesis is suppressed (16).
The bacteria capable of reductive acetogenesis via the acetyl coenzyme A (acetyl-CoA) pathway (acetogens) exist in a range of environments, including sediments, wastewater treatment systems, soils, and animal gut systems, and they are likely to be natural microbiota of all ruminants (11, 22). Naturally, however, reductive acetogenesis is not the dominant hydrogenotrophic pathway in the rumen; methanogenesis is (7). Analogous gut fermentation in some native Australian macropod marsupials, such as kangaroos and wallabies, results in lower methane emissions (9, 25, 49), suggesting that alternative hydrogen disposal mechanisms replace methanogenesis in these animals. Understanding hydrogenotrophy in these gut systems may provide insight into mechanisms for redirecting hydrogen away from methanogenesis in ruminants. Acetogenesis may represent a significant hydrogen sink in the foregut of native Australian marsupials (2), and these animals may be a source of novel acetogens.
Attempts at characterizing the acetogen population in complex microbial ecosystems have been hindered by the phylogenetic diversity of this phenotype. A functional gene based molecular approach is ideal; however, existing tools (28) targeting the formyltetrahydrofolate synthetase gene (fhs) are compromised by a lack of specificity due to the presence of formyltetrahydrofolate synthetase (FTHFS) in other biochemical pathways (11, 37). A more appropriate approach may be to use the genes encoding enzymes unique to the acetyl-CoA pathway (a methyltransferase, a corrinoid-iron sulfur protein or the carbon monoxide dehydrogenase/acetyl-CoA synthase complex [CODH/ACS]) as markers (39). However, there is little sequence information available on these proteins and their genes in acetogens. In the present study the gene encoding ACS (acsB, Pierce et al. [37]) was investigated as a potential marker for the acetogens. The primary objective of this investigation was to develop PCR primers targeting acsB in a wide range of acetogens which could be applied in the rumen and other gut ecosystems. The second aim of this study was to use acsB- and fhs-based tools to compare the acetogen diversity in the bovine rumen and the forestomach of a native Australian marsupial, the tammar wallaby (Macropus eugenii).
MATERIALS AND METHODS
Bacterial strains, media, and DNA extraction.
Bacterial strains, media, and growth conditions are listed in Table 1. All confirmed rumen acetogens were included, as well as isolates from other environments representing major acetogen genera (11). Two sulfur-reducing bacteria that contain acetyl-CoA pathway genes were also included: Desulfitobacterium hafniense and Desulfovibrio desulfuricans subsp. desulfuricans. All media were prepared by using standard anaerobic techniques (21), dispensed in an anaerobic chamber with an atmosphere of 95% CO2 and 5% H2 (COY Laboratory Products, Inc., Ann Arbor, MI), and sterilized by autoclaving at 121°C and 100 kPa for 20 min. Modified M8 medium (Table 1) was the M8 medium described by Olsen et al. (36) but also contained 5 g of cellobiose, 10 g of glucose, and 5 ml of Pfennigs metal solution per liter as described by McInerney et al. (33) except modified by the addition of 0.01 g of each of Na2SeO3 and CuCl2·2H2O. Two further modifications of this medium that were sometimes used were the addition of FeSO4·7H2O before autoclaving to a final concentration of 0.5g liter−1, or the addition of sterile prereduced glucose or fructose to final concentration 0.1% (wt/vol) immediately before culture inoculation. Modified cooked meat medium (Table 1) contained per liter: 100 g of CM0081 medium (Oxoid, Basingtoke, Hampshire, United Kingdom), 5 g of yeast extract, 5 g of K2HPO4, 4 g of glucose, 1 g of cellobiose, 1 g of maltose, 1 g of soluble starch, 0.001 g of resazurin indicator, and 0.5 g of cysteine-HCl. Reinforced clostridial medium (Table 1) contained per liter: 38 g of CM0149 (Oxoid), 0.001 g of resazurin indicator, and 0.5 g of cysteine-HCl. Clostridium pfennigii, Syntrophococcus sucromutans, and Sporomusa medium (Table 1) were prepared as outlined online by the Deutsche Sammlung von Mikroorganismen und Zellkulturen (http://www.dsmz.de/microorganisms/medium/pdf/DSMZ_Medium336.pdf, http://www.dsmz.de/microorganisms/medium/pdf/DSMZ_Medium337.pdf, and http://www.dsmz.de/microorganisms/medium/pdf/DSMZ_Medium311.pdf, respectively).
TABLE 1.
Bacterial strains, media, and growth conditions used in this study
| Organism | Growth medium and temp | Original habitat | Reference |
|---|---|---|---|
| Acetitomaculum ruminis (DSM 5522) | Modified M8 medium with glucose at 37°C | Bovine rumen | 19 |
| Acetobacterium woodii (DSM 1030) | Modified M8 medium with fructose at 30°C | Mud | 3 |
| Blautia sp. ser8 | Modified cooked meat medium at 39°C | Rumen of young lamba | 16 |
| Blautia hydrogenotrophica (DSM 10507) | Modified M8 medium at 39°C | Human feces | 4 |
| Blautia producta (DSM 2950) | Modified cooked meat medium at 39°C | Human feces and blood | 14 |
| Blautia schinkii (DSM 10518) | Modified cooked meat medium at 39°C | Rumen of young lamb | 41 |
| Clostridium aceticum (DSM 1496) | Modified M8 medium at 30°C | Mud | 50 |
| Clostridium difficile (DSM 12056) | Reinforced clostridial medium at 37°C | Rumen of young lamb | 40 |
| Eubacterium limosum (ATCC 8486) | Reinforced clostridial medium at 39°C | Human feces | 12 |
| Moorella thermoacetica (DSM 521) | Modified M8 medium with glucose at 55°C | Horse feces | 15 |
| Oxobacter pfennigii (DSM 3222) | Clostridium pfennigii medium at 37°C | Cattle rumen | 26 |
| Sporomusa termitida (DSM 4440) | Sporomusa medium at 30°C | Termite gut | 5 |
| Syntrophococcus sucromutans (DSM 3224) | Syntrophococcus sucromutans medium at 37°C | Cattle rumen | 27 |
| Desulfitobacterium hafniense (DSM 10664) | Modified M8 with FeSO4·7H2O at 37°C | Sewage sludge | 31 |
| Desulfovibrio desulfuricans subsp. desulfuricans (DSM 6949) | Modified M8 with FeSO4·7H2O at 37°C | Sheep rumen | 42 |
This strain was kindly provided by Evelyne Forano, French National Institute for Agricultural Research (INRA), Clermont-Ferrand, France.
In November 2006 rumen contents were collected from five fistulated Brahman (Bos indicus) cross steers that had been feeding a pasture diet of green panic (Panicum maximum var. trichoglume), Buffel grass (Cenchrus ciliaris) and Rhodes grass (Chloris gayana). Forestomach contents were collected from three euthanized female tammar wallabies (Macropus eugenii) in May 2007, as reported by Evans et al. (13). The wallabies had been grazing some pasture grass, predominantly Phalaris spp., and received a diet supplement of a commercial pellet mix (Young Stock Feeds, Young, New South Wales, Australia) comprised of 15% protein, wheat, meals mix (bran and pollard, canola, soy, salt, sodium bicarbonate, bentiote, lime, and vitamin premix), and a coccidiostat-Keymix Keystat Powder (International Animal Health Products, Hungtingwood, New South Wales, Australia) containing 25% amprolium hydrochloride, 1.6% ethopabate, and 73.4% unspecified inert carriers.
DNA was extracted from growing bacterial cultures, stored (frozen at −80°C) rumen contents, or wallaby forestomach digesta by using the cetyltrimethylammonium bromide (CTAB) method of Brookman et al. (8) with minor modifications as follows: samples were centrifuged (13,000 × g for 5 min), and the supernatant was removed before DNA extraction. Cells were homogenized with 200 mg of silica-zirconium beads (1:1 mixture of 0.1- and 1.0-mm beads; Biospec, Bartlesville, OK) and 800 μl of CTAB buffer in a Mini-Beadbeater-8 (Biospec) on maximum speed for 2 min, twice. Samples were incubated at 70°C for 20 min and centrifuged at 10,000 × g for 10 min, and the supernatant was mixed with 500 μl of 25:24:1 phenol-chloroform-isoamyl alcohol (Fluka BioChemika, Buchs, Switzerland).
Recovery of novel acsB sequences.
Putative ACS amino acid sequences that showed BLAST (1) similarity to the ACS of Moorella thermoacetica, accession no. P27988 (34), were downloaded from GenBank and aligned using AlignX in the Vector NTI Advance 10 software (Invitrogen). Degenerate PCR primers were designed around conserved amino acid regions of bacterial ACS sequences and used to recover putative acsB sequences from acetogens listed in Table 1. Forward primer ACSF1 and reverse primer ACSR1 (Table 2) were designed to amplify ∼416 bp of acsB. PCRs (50 μl) contained final concentrations of 1× PCR buffer (10× buffer stock: 200 mM Tris-HCl [pH 8.4] and 500 mM KCl), 3 mM MgCl2, 0.2 mM deoxyribonucleotide triphosphates, 0.14 μM concentrations of each primer, 1 U of Platinum Taq DNA polymerase (Invitrogen), and ∼100 ng of template DNA. The PCR conditions were as follows: initial denaturation at 95°C for 5 min, followed by 30 cycles of denaturation at 95°C for 30 s, followed by annealing at a gradient of temperatures (40 to 55°C) for 30 s and extension at 72°C for 30 s, followed by a final extension at 72°C for 7 min. Aliquots of 5 μl were analyzed by agarose gel electrophoresis to determine the amplicon sizes (44). Amplicons were purified by gel excision (Qiagen, Hilden, Germany), cloned using the pGEM-T Easy vector (Promega Corp., Madison, WI) and TOP10 electrocompetent cells (Invitrogen, Carlsbad, CA) according to the manufacturer's instructions. Clones were sequenced with primers T7 and SP6 (Promega Corp.) using a BigDye Terminator v3.1 cycle sequencing kit (Applied Biosystems, Inc., Foster City, CA) according to the manufacturer's instructions. Sequences were determined by using an ABI3130xl Genetic Analyzer (Applied Biosystems, Inc.), and sequence similarity to ACS was determined by BLASTX analysis (1, 18). The ExPASy online tool (17) was used to translate DNA sequences before alignment in ARB (30). To recover longer acsB sequences from acetogens, less-degenerate PCR primers were designed within the recovered region and used in combination with degenerate PCR primers designed upstream and downstream of the previously recovered region to amplify partial overlapping acsB sequences of ca. 1,340 and 590 bp, respectively. Primer sequences are listed in Table 2. Forward primer ACSF5 was used in combination with ACS_D and reverse primer ACSR4 was used with ACS_U except in the following cases: for Blautia sp. ser8, reverse primer BC_r was used instead of ACSR4; for Blautia schinkii, forward primer BS_f was used instead of ACSF5; and for Acetobacterium woodii, primer AW_f was used as a sequencing primer on genomic DNA instead of PCR using ACSF5 with ACS_D. PCRs were performed as previously described except with increased extension times relative to the expected product length. PCR amplicons were purified, cloned, and sequenced as previously except in the case of A. woodii, where forward primer AW_f (Table 2) was used in a sequencing reaction according to the protocol outlined by the manufacturer for sequencing bacterial artificial chromosome DNA, at an annealing temperature of 45°C. Contigs of overlapping sequences were made using ContigExpress in the Vector NTI Advance 10 software (Invitrogen). Bootstrapped neighbor joining trees of deduced ACS amino acid sequences were constructed in ARB (30) with 100 resamplings. Maximum-likelihood trees of deduced ACS amino acid sequences were constructed by using RAxML version 7.0.3 (46) and the Jones Taylor Thornton (24) model of amino acid substitutions with a gamma rate of substitution and 25 discrete rate categories. Bootstrap analysis was performed for the best-scoring tree topology with 100 resamplings. Putative acsB sequences determined in the present study from pure cultures have been submitted to the GenBank database under accession numbers GU947716 to GU947736.
TABLE 2.
Primers designed in this study
| Primer | Target position in M. thermoacetica acsA/acsBa | Sequenceb (5′-3′) |
|---|---|---|
| ACSF1 | 3836-3855 | CTYTGYCAGTCMTTYGCBCC |
| ACSR1 | 4232-4251 | CCCATAAABCCYGGDGTYTG |
| ACS_U | 2767-2788 | GAYATHCCNGGNGTNGCNGT |
| ACS_D | 4464-4482 | ARNGCNGGRTGNCCYTTYT |
| ACSF5 | 3893-3913 | CTBTGYGGWGCHGTIWSMTGG |
| ACSR4 | 4085-4107 | AARCAWCCRCADGADGTCATBGG |
| BC_r | 3996-4015 | AACGTCCTCGTACTCACCGA |
| BS_f | 3974-3993 | GAAAGACCGATCGACGAGAA |
| AW_f | 4077-4096 | TGGAAGATCCGATGACATCA |
| ACS_f | 3893-3913 | CTBTGYGGDGCIGTIWSMTGG |
| ACS_r | 4085-4107 | AARCAWCCRCADGADGTCATIGG |
acsA and acsB encode CODH and ACS, respectively. The numbering is according to Morton et al. (34).
Degenerate bases follow the IUPAC code.
Final acsB-specific PCR primer design and optimization.
PCR primers, forward ACS_f and reverse ACS_r (Table 2) were designed to amplify a 216-bp fragment of acsB, based on the recovered gene sequences from acetogens. PCRs contained components as before except 0.9 μM each primer, and 100 to 200 ng of template DNA. PCR conditions were optimized on total rumen microbial DNA as follows: initial denaturation at 95°C for 5 min, 30 cycles of denaturation at 95°C for 15 s, annealing at 52°C for 20 s, and extension at 72°C for 13 s.
AcsB and fhs libraries from the bovine rumen and tammar wallaby forestomach.
Partial putative acsB sequences (216 bp) were amplified from total rumen microbial DNA of five pasture-fed steers and the forestomach DNA of three tammar wallabies by using the primers ACS_f and ACS_r. PCR amplicons were pooled according to source on an equal-concentration basis for clone library construction. Phylogenetic trees of deduced ACS amino acid sequences were constructed as outlined previously. ACS amino acid sequences were grouped into operational taxonomic units (OTUs) at a distance of ≤0.035 using MOTHUR (45). Although OTUs for ACS amino acid sequences do not strictly correspond to a species, ≥96.5% sequence identity was chosen in order to group similar sequences while separating the closest ACS amino acid sequences from distinct bacterial species (Blautia schinkii and Ruminococcus obeum, which share 96.3% amino acid identity across the region amplified with ACS_f and ACS_r). Blautia sp. ser8 and Blautia producta ACS amino acid sequences are identical across the region amplified with ACS_f and ACS_r, however, these two bacteria do not separate as individual species at the 16S rRNA gene level (>97% 16S rRNA gene identity). LIBSHUFF in MOTHUR (45) was used to compare the structure of acsB libraries (deduced amino acids). Putative acsB sequences from the rumen (n = 57) and tammar wallaby forestomach (n = 43) have been submitted to the GenBank database under accession numbers HM043971 to HM044070.
Partial fhs sequences were amplified by using the primers (FTHFS_f, 5′-TTYACWGGHGAYTTCCATGC-3′; FTHFS_r, 5′-GTATTGDGTYTTRGCCATACA-3′) and the protocol of Leaphart and Lovell (28) and template DNA as for the acsB libraries. PCR amplicons were pooled according to source and clone libraries constructed as outlined previously. Clones were sequenced by using vector-specific primers for the rumen samples and fhs primers for the tammar wallaby forestomach samples. Maximum-likelihood and neighbor-joining trees of deduced FTHFS amino acid sequences were constructed as outlined previously for ACS amino acid sequences. FTHFS amino acid sequences were grouped into OTUs at a distance of ≤0.025 using MOTHUR (45). Although OTUs for FTHFS amino acid sequences do not strictly correspond to a species, ≥97.5% identity was chosen in order to group similar FTHFS sequences, while separating the closest FTHFS amino acid sequences from distinct species, Blautia hansenii and B. producta (96.65% amino acid identity). LIBSHUFF in MOTHUR (45) was used to compare the structure of fhs libraries (deduced amino acids). Homoacetogen similarity (HS) scores for FTHFS sequences were calculated using the method outlined by Henderson et al. (20) for examining amino acid residues important in FTHFSs from acetogens. Putative fhs sequences from the rumen (n = 61) and from the tammar wallaby forestomach (n = 82) have been submitted to the GenBank database under accession numbers: HM043823 to HM043965.
Comparison of tree topologies for FTHFS amino acids, ACS amino acids, and 16S rRNA genes.
A maximum-likelihood tree of all available ACS amino acid sequences was constructed as outlined previously. FTHFS amino acid sequences and 16S rRNA from the same organisms were downloaded from GenBank. Where FTHFS sequences were unavailable, partial fhs sequences were obtained using the primers of Leaphart and Lovell (28) at an annealing temperature of 52°C or the fhs1 primers (fhs1_f, 5′-GTW TGG GCW AAR GGY GGM GAA GG-3′; reverse, FTHFS_r [see above]) and the conditions described by Xu et al. (52). A maximum-likelihood tree of FTHFS amino acid sequences was constructed as outlined previously. A maximum-likelihood tree of 16S rRNA gene sequences was constructed by using RaXML version 7.0.3 (46) and the general time reversible (43) model of nucleotide substitutions with a gamma model of rate heterogeneity and 25 discrete rate categories. Bootstrap analysis was performed on the best-scoring tree topology with 100 resamplings (46). Bootstrapped neighbor joining trees were constructed in ARB (30) with 100 resamplings.
RESULTS
Recovery of novel putative acsB from acetogens and D. hafniense.
Partial putative acsB sequences recovered from M. thermoacetica (416 bp) and D. hafniense (416 bp) were identical to those determined by genome sequencing for these organisms (GenBank accessions numbers CP000232 and AP008230, respectively). Three distinct partial putative acsB sequences determined for Blautia hydrogenotrophica (1,300, 1,679, and 1,679 bp) showed >99% identity to corresponding sequences in the draft genome for this organism (GenBank accession no. ACBZ00000000). The acsB for Clostridium difficile DSM 12056 (1,676 bp) was unique, although at the amino acid level was identical to ACS from C. difficile strain 630 (GenBank accession no. AM180355). All other putative acsB sequences determined in the present study were novel. There was evidence for at least two distinct copies of acsB (i.e., differing at >10 nucleotide positions across the length of a contig) in 6 of the 13 bacteria evaluated here: Acetitomaculum ruminis, B. hydrogenotrophica, B. schinkii, Oxobacter pfennigii, Sporomusa termitida, and Sy. sucromutans. Differences between multiple copies of acsB within these organisms were also evident at the amino acid level (e.g., see Fig. 1) and were sometimes considerable, as in Sp. termitida, where two copies of putative ACS shared <73% amino acid identity.
FIG. 1.
CLUSTAL W alignment of partial ACS amino acid sequences. Position 1 in this alignment corresponds to amino acid 495 in M. thermoacetica (34) and the start of domain 3 of ACS (10). Primers ACS_f and ACS_r have been marked, and residues important in coordinating the active site of ACS are indicated by an asterisk.
Coverage of acsB-specific PCR primers.
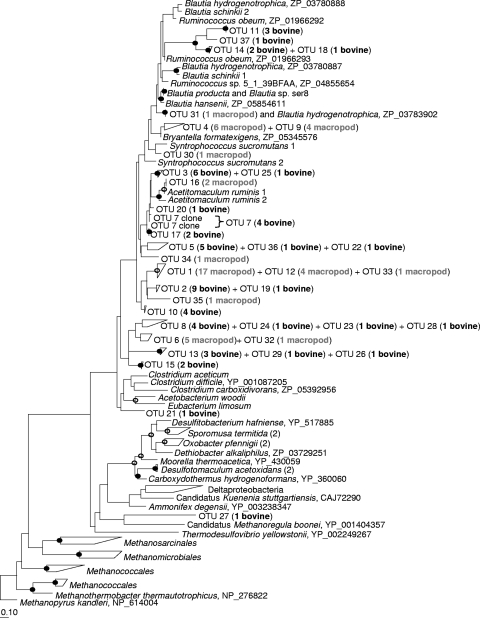
The ACS_f with ACS_r primers amplified an ∼216-bp fragment of acsB in acetogens from at least nine genera. ACS_f targeted a region corresponding to amino acid sequence LCGAVSW, including a cysteine (Cys528 in M. thermoacetica) involved in coordination of the ACS active site and ACS_r targeted the region encoding PMTSCGC including two cysteines and a glycine (Cys595, Gly596, and Cys597 in M. thermoacetica) involved in coordination of the ACS active site (see Fig. 1). The acsB from sulfite-reducing bacterium D. hafniense could not be effectively excluded during primer design and genes of the acetyl-CoA pathway present in this bacterium were similar to those in Clostridiales acetogens. Using ACS_f and ACS_r, a spurious, larger-sized amplicon was generated from the rumen sulfate-reducing bacterium Desulfovibrio desulfuricans subsp. desulfuricans, and acsB was not recovered from this organism with any primer combination used in the present study. Genes showing similarity to acsB were not present in the Desulfovibrio desulfuricans subsp. desulfuricans genome (GenBank accession no. CP001358), although fhs and acsA (encoding CODH) were. acsB sequences from other bacteria belonging to the Deltaproteobacteria (often sulfate-reducing bacteria) and archaea (methanogens) were expected to be excluded with the final PCR primer combination, as all sequences exhibited mismatches across at least one of the primer regions. However, a novel ACS sequence that affiliated distantly with the Deltaproteobacteria was recovered from the rumen using ACS_f and ACS_r (see OTU 27, see Fig. 4). It showed 52.73% amino acid identity to ACS from the Desulfonatronospira thiodismutans. Slight nonspecific amplification from gut samples using ACS_f and ACS_r could be effectively removed by gel excision of amplicons before cloning. Alternatively, spurious products were easily distinguishable when sequenced, showing no BLASTX similarity (1) to ACS amino acid sequences and often with stop codons in the equivalent translation frames that encoded the primer sites.
Comparison of tree topologies: FTHFS amino acid, ACS amino acid, and 16S rRNA gene.
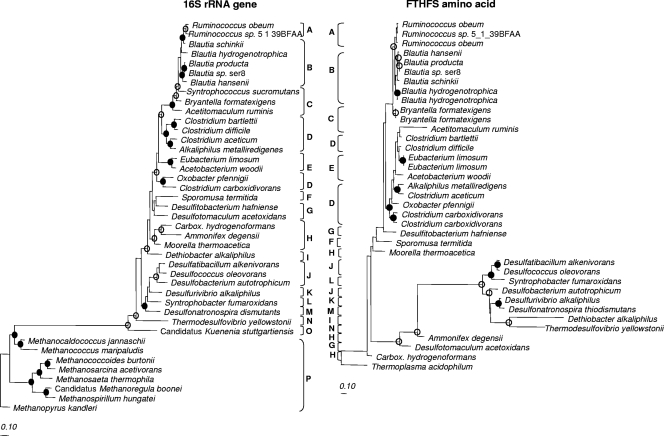
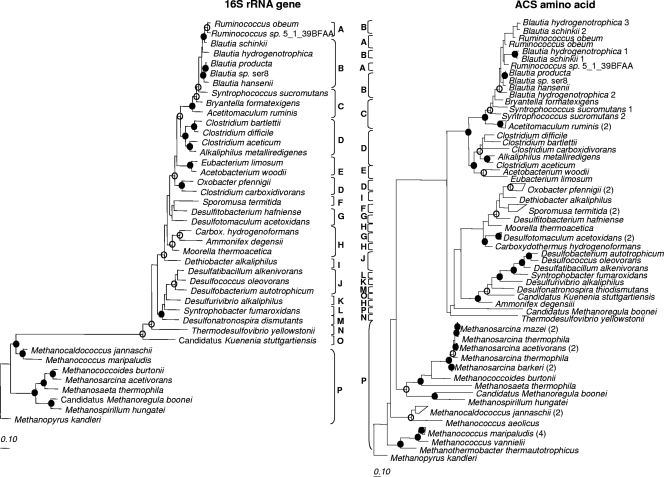
A comparison between ACS amino acid, FTHFS amino acid, and 16S rRNA gene trees (Fig. 2 and 3) showed that many of the family-level relationships observed at the 16S rRNA gene level for acetogens were consistent at the amino acid level for functional genes (e.g., Ruminococcaceae, Blautia group, Eubacteriaceae, and some Clostridiaceae). In some of these groups, intrafamily relationships also held true and were supported by bootstrap analysis. Two acetogen sequences for which phylogeny in the three trees was not similar, were O. pfennigii and Sp. termitida and placement of the latter was not supported by bootstrap analysis in any tree. Amino acid sequences originating from bacteria in the class Deltaproteobacteria were more closely related to some Clostridiales sequences than were 16S rRNA genes. All archaeal ACS sequences except one copy of ACS in “Candidatus Methanoregula boonei” clustered together, away from bacterial sequences in agreement with the 16S rRNA gene tree (Fig. 3). Except for three methanogen ACS sequences (“Candidatus Methanoregula booneii”, YP_001404357; Methanocaldococcus jannaschii, NP_247120; and Methanococcus aeolicus, YP_001325360), all archaeal ACS sequences lacked the N terminus of ACS and were clearly distinguishable from bacterial sequences upon alignment. None of the three methanogen ACS sequences that exhibited an N terminus similar to bacterial sequences affiliated with acetogen ACS sequences. fhs sequences were not found in any of the methanogen genomes investigated (Fig. 2), as expected, because methanogens contain structurally and functionally analogous tetrahydromethanopterin proteins (38, 47).
FIG. 2.
Maximum-likelihood trees of 16S rRNA gene and FTHFS amino acid sequences for comparison of tree topology. Bootstrap values of ≥75% are shown at nodes as closed circles for both tree construction methods or open circles for maximum-likelihood only. In the interest of space, Carboxydothermus has been abbreviated “Carbox.” Marked classifications are indicated by capital letters as follows: A, Ruminococcaceae; B, Blautia group; C, Lachnospiraceae; D, Clostridiaceae; E, Eubacteriaceae; F, Veillonellaceae; G, Peptococcaceae; H, Thermoanaerobacteraceae; I, Syntrophomonadaceae; J, Desulfobacteraceae; K, Desulfobulbaceae; L, Syntrophobacteraceae; M, Desulfohalobiaceae; N, Nitrospiraceae; O, unclassified Planctomycetales; P, methanogenic archaea. The scale bar represents 10% sequence divergence.
FIG. 3.
Maximum-likelihood trees of 16S rRNA gene and ACS amino acid sequences for comparison of tree topology. Bootstrap values of ≥75% are shown at nodes as closed circles for both tree construction methods or open circles for maximum likelihood only. In the interest of space Carboxydothermus has been abbreviated “Carbox.” Marked classifications are indicated by capital letters as follows: A, Ruminococcaceae; B, Blautia group; C, Lachnospiraceae; D, Clostridiaceae; E, Eubacteriaceae; F, Veillonellaceae; G, Peptococcaceae; H, Thermoanaerobacteraceae; I, Syntrophomonadaceae; J, Desulfobacteraceae; K, Desulfobulbaceae; L, Syntrophobacteraceae; M, Desulfohalobiaceae; N, Nitrospiraceae; O, unclassified Planctomycetales; P, methanogenic archaea. The scale bar represents 10% sequence divergence.
ACS and FTHFS diversity in the bovine rumen and tammar wallaby forestomach.
Deduced amino acid sequences of acsB from the rumen grouped into 25 OTUs and from the tammar wallaby forestomach into 12 OTUs. Rarefaction analysis at an approximate species level indicated that there may be more ACS sequences to be recovered from the rumen. Community structure between the two acsB libraries (deduced amino acids) was different based on LIBSHUFF analysis (significance of the ΔCXY score of <0.001); however, sequences from both ecosystems clustered very broadly in the same family groups (Ruminococcaceae/Blautia group and Lachnospiraceae). There were also ACS OTUs from both the tammar wallaby forestomach and the rumen that placed phylogenetically between sequences from the Lachnospiraceae and Clostridiaceae, without a close sequence from a cultured affiliate in the current database. A single ACS sequence from the rumen affiliated distantly with the Eubacteriaceae acetogens (OTU 21, Fig. 4) and another clustered distantly among the Deltaproteobacteria (OTU 27, Fig. 4), whereas no sequences from the tammar wallaby affiliated with either of these groups.
FIG. 4.
Phylogenetic analysis of deduced ACS amino acid sequences from the bovine rumen (bovine) and the tammar wallaby forestomach (macropod). GenBank accession numbers of reference sequences are shown after the species names. Bootstrap values of ≥75% are shown at nodes as closed circles for both tree construction methods or open circles for maximum likelihood only. The scale bar represents 10% sequence divergence.
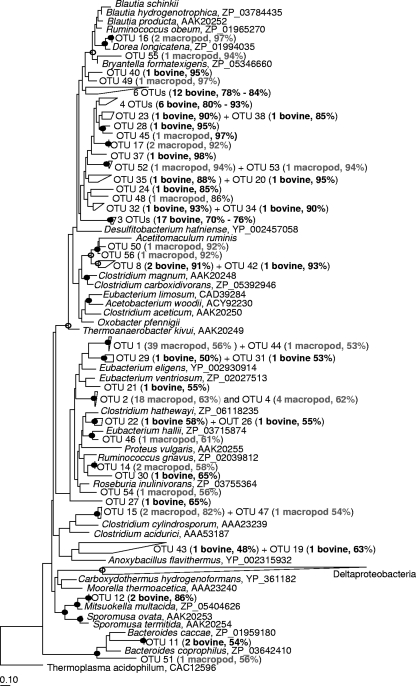
Deduced amino acid sequences of fhs from the rumen grouped into 36 OTUs and from the tammar wallaby forestomach grouped into 20 OTUs. Rarefaction analysis indicated that there may be more FTHFS sequences at an approximate species level to be uncovered from the rumen. The two largest FTHFS OTUs from the tammar wallaby forestomach (OTUs 1 and 2, Fig. 5) placed phylogenetically in the lower half of the FTHFS tree near sequences from nonacetogens. There were also sequences from the rumen scattered in this region of the tree as well as the FTHFSs from acetogens M. thermoacetica, Sp. termitida, and Sporomusa ovata. All recovered FTHFS sequences in this region of the tree showed HS scores of <90%, and some were below the 60% cutoff indicated by Henderson et al. (20) for sequences likely to have originated from nonacetogens, although none affiliated with the Deltaproteobacteria.
FIG. 5.
Phylogenetic analysis of deduced FTHFS amino acid sequences from the bovine rumen (bovine) and the tammar wallaby forestomach (macropod). GenBank accession numbers of reference sequences are shown after the species names. Bootstrap values of ≥75% are shown at nodes as closed circles for both tree construction methods or open circles for maximum likelihood only. HS scores are included in brackets for OTUs recovered in the present study. The scale bar represents 10% sequence divergence.
Of the 35 FTHFS OTUs from the rumen and tammar wallaby forestomach that placed in the top half of the FTHFS tree, six OTUs affiliated with FTHFSs from the Lachnospiraceae (e.g., macropod OTU 55 near Bryantella formatexigens, macropod OTU 16 near Dorea longicatena, macropod OTUs 50 and 56 and bovine OTUs 8 and 42 near Acetitomaculum ruminis, Fig. 5). The remaining diversity from both ecosystems (29 bovine OTUs and 6 macropod OTUs) placed phylogenetically broadly between FTHFSs from the Lachnospiraceae and Clostridiaceaeae, without a close sequence from a cultured affiliate. The community structure of the two fhs libraries (deduced amino acids) was different based on LIBSHUFF analysis (significance of the ΔCXY score of <0.001); however, as with ACS, FTHFSs from both ecosystems affiliated in the same broad groupings (near Lachnospiraceae or between Lachnospiraceae and Clostridiaceae without a close cultured relative).
No FTHFS sequences that affiliated with the Ruminococcaceae/Blautia group were detected in the rumen or tammar wallaby forestomach. Moreover, the fhs primers of Leaphart and Lovell (28) were tested on Blautia sp. ser8, B. hydrogenotrophica, and B. schinkii DNA at the recommended conditions, and no amplicon could be generated. This was also the case for the rumen acetogens Acetitomaculum ruminis, O. pfennigii, and Sy. sucromutans. Partial fhs sequences were recovered from B. hydrogenotrophica and O. pfennigii using the primers of Leaphart and Lovell (28) at an annealing temperature of 52°C and from Acetitomaculum ruminis, Blautia sp. ser8, and B. schinkii using the fhs1 primers of Xu et al. (52). These sequences have been deposited at GenBank under the accession numbers HM043966 to HM043970. The fhs1 primers (52) resulted in nonspecific amplification when tested on rumen microbial DNA and produced multiple-sized amplicons for Sp. termitida and Sy. sucromutans. fhs could not be obtained for Sy. sucromutans.
DISCUSSION
The novel putative acsB sequences recovered in this study significantly expand the public database of functional genes from acetogens. Continued expansion of the database of acsB sequences from acetogens is essential and will improve phylogenetic approximation of novel short sequences from environmental samples and may facilitate refinement of acsB-specific molecular tools. Multiple, distinct copies of acsB were revealed in a range of acetogens in the present study and a similar phenomenon exists for fhs in some acetogens. The origin of multiple copies of acetyl-CoA pathway genes in acetogens is unclear, and whether all are expressed and produce active enzymes is yet to be resolved. It has been suggested for some methanogens and Carboxydothermus hydrogenoformans that multiple copies of CODH perform separate physiological roles (29, 51). Whether this is the case for ACS requires further investigation.
acsB primers ACS_f and ACS_r targeted a wide range of acetogens, recovered novel putative acsB sequences from two environmental samples and excluded currently available Deltaproteobacteria and methanogen ACS sequences. However, it would seem that this combination is not strictly specific for acetogens, since an ACS sequence that affiliated distantly with the Deltaproteobacteria was recovered from the rumen. D. hafniense, a Clostridiales sulfite-reducing bacterium, was also in the target range of the primers, and it is possible that other nonacetogens with acetyl-CoA pathway genes similar to those in acetogens will be targeted. The acetyl-CoA pathway that acetogens use for autotrophic growth and acetate production is also used by some methanogens and sulfur-reducing species for generation of cell carbon (autotrophic growth) and/or acetoclastic growth (39). Although ACS amino acid sequences from methanogens and Deltaproteobacteria cluster away from those found in acetogens phylogenetically (Fig. 2 and 3), there was no obvious method for distinguishing between ACS sequences in acetogens and those in closely related organisms such as Clostridiales sulfur reducing species or Clostridiales bacteria that are not yet known to be acetogens but house acsB in their genome (e.g., Ruminococcus obeum and Clostridium carboxidovorans). Such a distinction is not strictly possible for other enzymes of the acetyl-CoA pathway either, with some CODHs in methanogens showing homology to those in bacteria (29) and some FTHFSs in sulfate-reducing bacteria being very similar to those in acetogens and even sharing conserved residues (20). Also, while acsB is unique to the acetyl-CoA pathway and therefore an appropriate marker for this pathway (39), a limitation with all DNA-based analyses is that the presence of a gene in a genome does not necessarily correlate with expression of that gene and production of an active enzyme. Therefore, it is possible that detected acsB sequences could be remnants from an evolutionary ancestor and not necessarily serving functionally in that microorganism. The use of cultivation studies in combination with molecular techniques will be a start to addressing this limitation in the future and is expected to provide insight into both the identity of potential acetogens and the functioning of the acetyl-CoA pathway in these organisms.
Both FTHFS and ACS amino acid sequences were useful as approximate phylogenetic markers to the family level for a range of acetogens. Until the database of acsB sequences from acetogens expands, novel potential acetogens will ideally be investigated using both acsB and fhs. The presence of ACS is a better indicator for the acetyl-CoA pathway than is FTHFS alone. Phylogenetic analyses involving fhs can be ambiguous, with unclear separation between FTHFS sequences from acetogens and nonacetogens. Statistical models may be useful to confirm which FTHFS sequences in a data set have probably originated from nonacetogens (low HS score to particular FTHFS residues in acetogens [20]). However, the origin of novel FTHFS sequences with intermediate HS scores remains ambiguous. Complementary phylogenetic placement of FTHFS and ACS amino acid sequences supports the likelihood that novel fhs sequences originated from organisms with the acetyl-CoA pathway. Also, since the current fhs primers (28) exclude known rumen acetogens, they may not be the best tools alone for assessing acetogen diversity in that environment. The fhs1 primers of Xu et al. (52) recovered partial FTHFS sequence from a wider range of rumen acetogens than previous fhs primers; however, primer specificity was significantly compromised with multiple spurious amplicons generated from some acetogens and rumen microbial DNA. Therefore, use of the fhs1 primer set on rumen samples for its intended purpose of quantitative, real-time PCR for acetogens, does not seem appropriate.
A large number of FTHFS sequences from the tammar wallaby forestomach probably originated from nonacetogens since they clustered in the lower half of the FTHFS tree and showed low HS scores. This was not unexpected and has been observed previously for rumen samples (20). However, novel FTHFS sequences that affiliated with those from acetogens and demonstrated HS scores of >90% were present in both the tammar wallaby and the rumen and probably originated from novel acetogens affiliating with the Lachnospiraceae. Equivalently, placed ACS sequences were also found to support this likelihood. Novel ACS sequences that affiliated with bacteria in the Ruminococcaceae/Blautia group were found in the rumen but not revealed by FTHFS analysis, and these may have originated from acetogens in that group that were not targeted by the current fhs primers (28). The greatest diversity of FTHFS and ACS sequences in both the rumen and the tammar wallaby forestomach had no close cultured affiliate in the public databases and placed phylogenetically broadly between the Lachnospiraceae and Clostridiaceae. Although not all FTHFS sequences in that region showed HS scores of >90%, the cutoff defined by Henderson et al. (20) to distinguish acetogen FTHFSs, we expect that many of them originated from novel acetogens due to the diversity of ACS sequences with similar phylogenetic placement recovered from both samples. FTHFS sequences that cluster in this region of the FTHFS tree have also been detected by Matsui et al. (32) from the rumen of a Holstein cow fed a mixture of pasture and grain and by Henderson et al. (20) from the rumen of a Fresian-Jersey cross cow fed pasture. Therefore, these novel, diverse gene sequences indicate that uncultivated potential acetogens are present in the rumen regardless of diet or location. This novel population in both the rumen and the tammar wallaby forestomach requires further investigation if reductive acetogenesis is to be exploited as an alternative to methanogenesis in the rumen.
The differences between acsB and fhs libraries from the tammar wallaby and the rumen were significant (LIBSHUFF, P < 0.001); however, these differences are likely to be species or genus level differences as sequences from both ecosystems affiliated broadly with the same family groups. Due to their novelty, these recovered sequences cannot be adequately resolved below family level with the current database of sequences from acetogens. It is possible that the species or genus level differences in potential acetogen populations in the tammar wallaby forestomach and the rumen contribute to differences in hydrogenotrophy in these ecosystems. Potentially, the acetogens in the tammar wallaby are more effective hydrogenotrophs than those in the rumen and possibly even competitive with methanogens, which may account for lower methane emissions (9, 25, 49) and methanogen numbers (13) in foregut-fermenting native Australian marsupials relative to ruminants. For example, acetogens employing mixotrophic growth (harnessing the acetyl-CoA pathway to consume hydrogen simultaneously with growth on organic substrates) may be competitive with methanogens, since mixotrophic growth is energetically more favorable than methanogenesis in some cases (6).
The differences between the marsupial forestomach and rumen are worth continued investigation to clarify the reasons for apparent differences in hydrogenotrophy and methanogenesis. Development of a specific quantitative monitoring tool for the acetogens will be important for this end, as will cultivation studies to characterize and compare novel acetogens in both ecosystems. The acsB primers developed here will also be useful in other studies investigating potential rumen acetogens. For example, ruminants that produce various levels of methane (either naturally or after methane inhibition) may be an area for future research, to identify acetogens that could be useful in strategies to redirect ruminal hydrogen away from methanogenesis.
Acknowledgments
We thank Lyn Hinds, CSIRO Division of Entomology, Black Mountain, for providing tammar wallaby forestomach samples. Emma Gagen was a recipient of scholarships from The University of Queensland and CSIRO Livestock Industries.
This study was partly supported by the Australian Greenhouse Office and Meat and Livestock Australia.
Footnotes
Published ahead of print on 1 October 2010.
REFERENCES
- 1.Altschul, S. F., W. Gish, W. Miller, E. W. Myers, and D. J. Lipman. 1990. Basic local alignment search tool. J. Mol. Biol. 215:403-410. [DOI] [PubMed] [Google Scholar]
- 2.Attwood, G., and C. S. McSweeney. 2008. Methanogen genomics to discover targets for methane mitigation technologies and options for alternative H2 utilisation in the rumen. Aust. J. Exp. Agric. 48:28-37. [Google Scholar]
- 3.Balch, W. E., S. Schoberth, R. Tanner, and R. S. Wolfe. 1977. Acetobacterium, a new genus of hydrogen-oxidizing, carbon dioxide-reducing, anaerobic bacteria. Int. J. Syst. Bacteriol. 27:355-361. [Google Scholar]
- 4.Bernalier, A., A. Willems, M. Leclerc, V. Rochet, and M. D. Collins. 1996. Ruminococcus hydrogenotrophicus sp. nov., a new H2/CO2-utilizing acetogenic bacterium isolated from human feces. Arch. Microbiol. 166:176-183. [DOI] [PubMed] [Google Scholar]
- 5.Breznak, J., J. Switzer, and H. Seitz. 1988. Sporomusa termitida sp. nov., an H2/CO2-utilizing acetogen isolated from termites. Arch. Microbiol. 150:282-288. [Google Scholar]
- 6.Breznak, J. A., and J. S. Blum. 1991. Mixotrophy in the termite gut acetogen, Sporomusa termitida. Arch. Microbiol. 156:105-110. [Google Scholar]
- 7.Breznak, J. A., and J. M. Switzer. 1986. Acetate synthesis from H2 plus CO2 by termite gut microbes. Appl. Environ. Microbiol. 52:623-630. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Brookman, J. L., and M. J. Nicholson. 2005. Anaerobic fungal populations, p. 139-150. In H. P. S. Makkar and C. S. McSweeney (ed.), Methods in gut microbial ecology for ruminants. Springer, Amsterdam, Netherlands.
- 9.Dellow, D. W., I. D. Hume, R. T. J. Clarke, and T. Bauchop. 1988. Microbial activity in the forestomach of free-living macropodid marsupials: comparisons with laboratory studies. Aust. J. Zool. 36:383-395. [Google Scholar]
- 10.Doukov, T. I., T. M. Iverson, J. Seravalli, S. W. Ragsdale, and C. L. Drennan. 2002. A Ni-Fe-Cu center in a bifunctional carbon monoxide dehydrogenase/acetyl-CoA synthase. Science 298:567-572. [DOI] [PubMed] [Google Scholar]
- 11.Drake, H. L., A. S. Gössner, and S. L. Daniel. 2008. Old acetogens, new light. Ann. N. Y. Acad. Sci. 1125:100-128. [DOI] [PubMed] [Google Scholar]
- 12.Eggerth, A. H. 1935. The gram-positive non-spore-bearing anaerobic bacilli of human feces. J. Bacteriol. 30:277-299. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Evans, P. N., L. A. Hinds, L. Sly, C. S. McSweeney, M. Morrison, and A.-D. G. Wright. 2009. Community composition and density of methanogens in the foregut of the tammar wallaby (Macropus eugenii). Appl. Environ. Microbiol. 75:2598-2602. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Ezaki, T., N. Li, Y. Hashimoto, H. Miura, and H. Yamamoto. 1994. 16S ribosomal DNA sequences of anaerobic cocci and proposal of Ruminococcus hansenii comb. nov. and Ruminococcus productus comb. nov. Int. J. Syst. Bacteriol. 44:130-136. [DOI] [PubMed] [Google Scholar]
- 15.Fontaine, F. E., W. H. Peterson, E. McCoy, M. J. Johnson, and G. J. Ritter. 1942. A new type of glucose fermentation by Clostridium thermoaceticum n. sp. J. Bacteriol. 43:701-715. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Fonty, G., K. N. Joblin, M. Chavarot, R. Roux, G. E. Naylor, and F. Michallon. 2007. Methanogen-free lambs: establishment and development of ruminal hydrogenotrophs. Appl. Environ. Microbiol. 73:6391-6403. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Gasteiger, E., A. Gattiker, C. Hoogland, I. Ivanyi, R. D. Appel, and A. Bairoch. 2003. ExPASy: the proteomics server for in-depth protein knowledge and analysis. Nucleic Acids Res. 31:3784-3788. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Gish, W., and D. J. States. 1993. Identification of protein coding regions by database similarity search. Nat. Genet. 3:266-272. [DOI] [PubMed] [Google Scholar]
- 19.Greening, R. C., and J. A. Z. Leedle. 1989. Enrichment and isolation of Acetitomaculum ruminis, gen. nov., sp. nov.: acetogenic bacteria from the bovine rumen. Arch. Microbiol. 151:399-406. [DOI] [PubMed] [Google Scholar]
- 20.Henderson, G., G. E. Naylor, S. C. Leahy, and P. H. Janssen. 2010. Presence of novel, potentially homoacetogenic bacteria in the rumen as determined by analysis of formyltetrahydrofolate synthetase sequences from ruminants. Appl. Environ. Microbiol. 76:2058-2066. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Hungate, R. E. 1969. A roll tube method for cultivation of strict anaerobes, p. 117-132. In J. R. Norris and D. W. Ribbons (ed.), Methods in microbiology, vol. 3B. Academic Press, Inc., New York, NY. [Google Scholar]
- 22.Joblin, K. N. 1999. Ruminal acetogens and their potential to lower ruminant methane emissions. Aust. J. Agric. Res. 50:1307-1313. [Google Scholar]
- 23.Johnson, K. A., and D. E. Johnson. 1995. Methane emissions from cattle. J. Anim. Sci. 73:2483-2492. [DOI] [PubMed] [Google Scholar]
- 24.Jones, D. T., W. R. Taylor, and J. M. Thornton. 1992. A new approach to protein fold recognition. Nature 358:86-89. [DOI] [PubMed] [Google Scholar]
- 25.Kempton, T. J., R. M. Murray, and R. A. Leng. 1976. Methane production and digestibility measurements in the grey kangaroo and sheep. Aust. J. Biol. Sci. 29:209-214. [DOI] [PubMed] [Google Scholar]
- 26.Krumholz, L. R., and M. P. Bryant. 1985. Clostridium pfennigii sp. nov. uses methoxyl groups of monobenenzoids and produces butyrate. Int. J. Syst. Bacteriol. 35:454-456. [Google Scholar]
- 27.Krumholz, L. R., and M. P. Bryant. 1986. Syntrophococcus sucromutans sp. nov. gen. nov. uses carbohydrates as electron donors and formate, methoxymonobenzenoids or Methanobrevibacter as electron acceptor system. Arch. Microbiol. 143:313-318. [Google Scholar]
- 28.Leaphart, A. B., and C. R. Lovell. 2001. Recovery and analysis of formyltetrahydrofolate synthetase gene sequences from natural populations of acetogenic bacteria. Appl. Environ. Microbiol. 67:1392-1395. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Lindahl, P. A. 2002. The Ni-containing carbon monoxide dehydrogenase family: light at the end of the tunnel? Biochemistry 41:2097-2105. [DOI] [PubMed] [Google Scholar]
- 30.Ludwig, W., O. Strunk, R. Westram, L. Richter, H. Meier, Yadhukumar, A. Buchner, T. Lai, S. Steppi, G. Jobb, W. Förster, I. Brettske, S. Gerber, A. W. Ginhart, O. Gross, S. Grumann, S. Hermann, R. Jost, A. König, T. Liss, R. Lüßmann, M. May, B. Nonhoff, B. Reichel, R. Strehlow, A. Stamatakis, N. Stuckmann, A. Vilbig, M. Lenke, T. Ludwig, A. Bode, and K.-H. Schleifer. 2004. ARB: a software environment for sequence data. Nucleic Acids Res. 32:1363-1371. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Madsen, T., and D. Licht. 1992. Isolation and characterization of an anaerobic chlorophenol-transforming bacterium. Appl. Environ. Microbiol. 58:2874-2878. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Matsui, H., N. Kojima, and K. Tajima. 2008. Diversity of the formyltetrahydrofolate synthetase gene (fhs) a key enzyme for reductive acetogenesis, in the bovine rumen. Biosci. Biotechnol. Biochem. 72:3273-3276. [DOI] [PubMed] [Google Scholar]
- 33.McInerney, M. J., M. P. Bryant, and N. Pfennig. 1979. Anaerobic bacterium that degrades fatty acids in syntrophic association with methanogens. Arch. Microbiol. 122:129-135. [DOI] [PubMed] [Google Scholar]
- 34.Morton, T. A., J. A. Runquist, S. W. Ragsdale, T. Shanmugasundaram, H. G. Wood, and L. G. Ljungdahl. 1991. The primary structure of the subunits of carbon monoxide dehydrogenase/acetyl-CoA synthase from Clostridium thermoaceticum. J. Biol. Chem. 266:23824-23828. [PubMed] [Google Scholar]
- 35.Moss, A. R., J.-P. Jouany, and J. Newbold. 2000. Methane production by ruminants: its contribution to global warming. Ann. Zootech. 49:231-253. [Google Scholar]
- 36.Olsen, M. A., T. H. Aagnes, and S. D. Mathiesen. 1994. Digestion of herring by indigenous bacteria in the minke whale forestomach. Appl. Environ. Microbiol. 60:4445-4455. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Pierce, E., G. Xie, R. D. Barabote, E. Saunders, C. S. Han, J. C. Detter, P. Richardson, T. S. Brettin, A. Das, L. G. Ljungdahl, and S. W. Ragsdale. 2008. The complete genome sequence of Moorella thermoacetica (f. Clostridium thermoaceticum). Environ. Microbiol. 10:2550-2573. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Ragsdale, S. W. 1997. The eastern and western branches of the Wood/Ljungdahl pathway: how the east and west were won. Biofactors 6:3-11. [DOI] [PubMed] [Google Scholar]
- 39.Ragsdale, S. W. 1991. Enzymology of the acetyl-CoA pathway of CO2 fixation. Crit. Rev. Biochem. Mol. Biol. 26:261-300. [DOI] [PubMed] [Google Scholar]
- 40.Rieu-Lesme, F., C. Dauga, G. Fonty, and J. Dore. 1998. Isolation from the rumen of a new acetogenic bacterium phylogenetically closely related to Clostridium difficile. Anaerobe 4:89-94. [DOI] [PubMed] [Google Scholar]
- 41.Rieu-Lesme, F., B. Morvan, M. D. Collins, G. Fonty, and A. Willems. 1996. A new H2/CO2-using acetogenic bacterium from the rumen: description of Ruminococcus schinkii sp. nov. FEMS Microbiol. Lett. 140:281-286. [DOI] [PubMed] [Google Scholar]
- 42.Roberts, R. J. 1985. Restriction and modification enzymes and their recognition sequences. Nucleic Acids Res. 13(Suppl.):r165-r200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Rodriguez, F., J. Oliver, A. Marin, and J. Medina. 1990. The general stochastic model of nucleotide substitution. J. Theor. Biol. 142:485-501. [DOI] [PubMed] [Google Scholar]
- 44.Sambrook, J., and D. W. Russell. 2001. Molecular cloning: a laboratory manual, 3rd ed. Cold Spring Harbor Laboratory Press, Cold Spring Harbor, NY.
- 45.Schloss, P. D., S. L. Westcott, T. Ryabin, J. R. Hall, M. Hartmann, E. B. Hollister, R. A. Lesniewski, B. B. Oakley, D. H. Parks, C. J. Robinson, J. W. Sahl, B. Stres, G. G. Thallinger, D. J. Van Horn, and C. F. Weber. 2009. Introducing mothur: open-source, platform-independent, community-supported software for describing and comparing microbial communities. Appl. Environ. Microbiol. 75:7537-7541. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Stamatakis, A. 2006. RAxML-VI-HPC: maximum likelihood-based phylogenetic analyses with thousands of taxa and mixed models. Bioinformatics 22:2688-2690. [DOI] [PubMed] [Google Scholar]
- 47.Thauer, R. K., R. Hedderich, and R. Fischer. 1993. Reactions and enzymes involved in methanogenesis from CO2 and H2, p. 209-252. In J. G. Ferry (ed.), Methanogenesis: ecology, physiology, biochemistry and genetics. Chapman & Hall, London, England.
- 48.Thorpe, A. 2009. Enteric fermentation and ruminant eructation: the role (and control?) of methane in the climate change debate. Clim. Change 93:407-431. [Google Scholar]
- 49.von Engelhardt, W., S. Wolter, H. Lawrenz, and J. A. Hemsley. 1978. Production of methane in two non-ruminant herbivores. Comp. Biochem. Physiol. 60:309-311. [Google Scholar]
- 50.Wieringa, K. T. 1940. The formation of acetic acid from carbon dioxide and hydrogen by anaerobic spore-forming bacteria. Antonie Van Leeuwenhoek 6:251-262. [Google Scholar]
- 51.Wu, M., Q. Ren, A. S. Durking, S. C. Daugherty, L. M. Brinkac, R. J. Dodson, R. Madupu, S. A. Sullivant, J. F. Kolonay, W. C. Nelson, L. J. Tallon, K. M. Jones, L. E. Ulrich, J. M. Gonzalez, I. B. Zhulin, F. T. Robb, and J. A. Eisen. 2005. Life in hot carbon monoxide: the complete genome sequence of Carboxydothermus hydrogenoformans Z-2901. PLoS Genet. 1:e65. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Xu, K., H. Liu, G. Du, and J. Chen. 2009. Real-time PCR assays targeting formyltetrahydrofolate synthetase gene to enumerate acetogens in natural and engineered environments. Anaerobe 15:204-213. [DOI] [PubMed] [Google Scholar]