Abstract
Xylella fastidiosa strain riv11 harbors a 25-kbp plasmid (pXF-RIV11) belonging to the IncP-1 incompatibility group. Replication and stability factors of pXF-RIV11 were identified and used to construct plasmids able to replicate in X. fastidiosa and Escherichia coli. Replication in X. fastidiosa required a 1.4-kbp region from pXF-RIV11 containing a replication initiation gene (trfA) and the adjacent origin of DNA replication (oriV). Constructs containing trfA and oriV from pVEIS01, a related IncP-1 plasmid of the earthworm symbiont Verminephrobacter eiseniae, also were competent for replication in X. fastidiosa. Constructs derived from pXF-RIV11 but not pVEIS01 replicated in Agrobacterium tumefaciens, Xanthomonas campestris, and Pseudomonas syringae. Although plasmids bearing replication elements from pXF-RIV11 or pVEIS01 could be maintained in X. fastidiosa under antibiotic selection, removal of selection resulted in plasmid extinction after 3 weekly passages. Addition of a toxin-antitoxin addiction system (pemI/pemK) from pXF-RIV11 improved plasmid stability such that >80 to 90% of X. fastidiosa cells retained plasmid after 5 weekly passages in the absence of antibiotic selection. Expression of PemK in E. coli was toxic for cell growth, but toxicity was nullified by coexpression of PemI antitoxin. Deletion of N-terminal sequences of PemK containing the conserved motif RGD abolished toxicity. In vitro assays revealed a direct interaction of PemI with PemK, suggesting that antitoxin activity of PemI is mediated by toxin sequestration. IncP-1 plasmid replication and stability factors were added to an E. coli cloning vector to constitute a stable 6.0-kbp shuttle vector (pXF20-PEMIK) suitable for use in X. fastidiosa.
Xylella fastidiosa is a Gram-negative gammaproteobacterium inhabiting the xylem of host plants (42). Infection by X. fastidiosa may lead to blockage of xylem transport, resulting in scorching and dieback symptoms of perennial crops, ornamentals, and landscape plants (23). In California, introduction of the polyphagous glassy-winged sharpshooter vector (6, 24) led to epidemics of Pierce's disease in grapes (21, 41, 33) and emergence of leaf scorch/dieback diseases of oleander (25), olive (16), sweetgum (15), and mulberry (17).
X. fastidiosa strains recovered from mulberry trees exhibiting leaf scorch symptoms harbor closely related 25-kbp plasmids sharing extensive nucleotide sequence identity with pVEIS01 (GenBank accession no. CP000543), a 31-kbp plasmid from the earthworm symbiont Verminephrobacter eiseniae (35). Annotation of the 25,105-bp sequence of pXF-RIV11 (GenBank accession no. GU938457) revealed that the plasmid contained homologues of incompatibility group P-1 (IncP-1) replication elements (trfA and oriV). Replication initiation of IncP-1 plasmids requires one plasmid-encoded protein (TrfA) (4). Plasmid replication is initiated by binding of TrfA protein to iterated sequences of oriV (19, 34).
Stable inheritance of plasmids may be achieved through toxin-antitoxin (TA) addiction systems (1, 3, 9, 14, 28, 29, 36, 40). The toxin disrupts an essential cellular process; antitoxin neutralizes toxin activity by direct binding (5, 7, 31, 39). Because the toxin is stable and persists longer than labile antitoxin, plasmid-free daughter cells are eliminated from the population even in the absence of other means of selection (such as antibiotic resistance or catabolic pathways conferring environmental adaptation). TA modules are grouped into at least eight families based on sequence and structure-function similarity (11, 20). The sequence of pXF-RIV11 includes homologues of the pemI/pemK TA system that has been characterized for pR100 (37, 38, 39, 40). PemK toxin encoded by pR100 inhibits protein synthesis by cleaving mRNAs at specific sites; addition of PemI antitoxin negates toxic properties of PemK (43).
Strains of X. fastidiosa harbor a variety of other plasmids (8, 13, 22, 26, 32). Derivatives of some these plasmids have been used as shuttle vectors able to replicate in both Escherichia coli and X. fastidiosa (13, 26). However, constructs bearing replication elements from these endogenous X. fastidiosa plasmids were unstable in the absence of antibiotic selection, precluding use in complementation experiments conducted with X. fastidiosa in plants. Here, we describe the functional characterization of replication elements from pXF-RIV11 and pVEIS01. We also characterize the pemI/pemK TA module from pXF-RIV11 that conferred stable inheritance of shuttle vector plasmids based on minimal IncP-1 replicons in X. fastidiosa in the absence of antibiotic selection.
MATERIALS AND METHODS
Bacterial strains and growth conditions.
E. coli strain JM109 (Promega) was grown in TB medium at 37°C. X. fastidiosa strain Temecula1 was grown in PD3 medium at 28°C (27). Agrobacterium tumefaciens strain GV3101 was grown in LB medium at 28°C. Pseudomonas syringae pv. tomato strain DC3000 was grown in King's B medium at 28°C. Xanthomonas campestris pv. campestris strain Xcc8004 was grown in NYG medium (5 g of peptone, 3 g of yeast extract, and 20 g of glycerol per liter) at 28°C. When required, antibiotics were added as follows: ampicillin, 100 μg/ml; kanamycin, 50 μg/ml (for E. coli), 30 μg/ml (for A. tumefaciens, P. syringae, and X. campestris), or 5 μg/ml (for X. fastidiosa); chloramphenicol, 30 μg/ml; spectinomycin, 50 μg/ml.
Construction of plasmids.
For the constructs diagrammed in Fig. 1A and B, the indicated sections of pXF-RIV11 or pVEIS01 were obtained by PCR amplification and cloned into pCR2.1 using a TA cloning kit (Invitrogen). Identities of cloned PCR products were confirmed by sequencing. PCR amplification also was used to obtain and add NotI sites to the fragment containing pemI/pemK (pXF-RIV11 nucleotides [nt] 1221 to 1906), with the product cloned into the NotI site in the pCR2.1 portion of plasmids pXF19, pXF20, pVEL4, and pVES3. For pemK and pemI expression in E. coli, various regions of pemI/pemK were PCR amplified and cloned into the pET28a vector (Novagen); pemI was amplified and cloned into pCDF1b (Novagen). For coexpression experiments, inserts were cloned into pETDuet-1 (Novagen). Proteins expressed from pET28a and pCDF1b included N-terminal His6 tags; proteins coexpressed from pETDuet-1 included N-terminal His6 tags (PemK or green fluorescent protein [GFP]) or a C-terminal S tag (PemI).
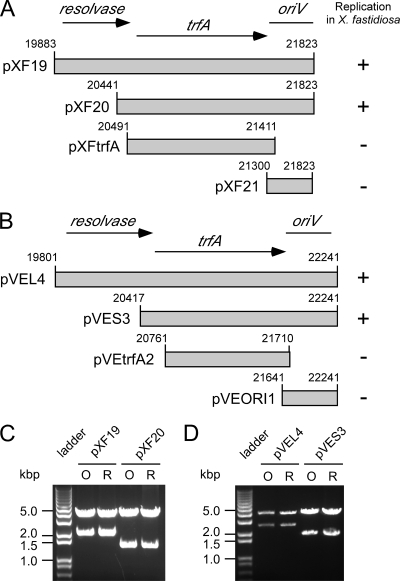
FIG. 1.
IncP-1 replicons from X. fastidiosa and V. eiseniae replicate in X. fastidiosa. DNA fragments derived from pXF-RIV11 (A) or pVEIS01 (B) were PCR amplified and inserted into E. coli cloning vector pCR2.1 bearing the nptII gene for kanamycin resistance (vector sequences not shown). Only constructs bearing both trfA and oriV were competent for replication in X. fastidiosa. Panels C and D show EcoRI digests of plasmids used to transform X. fastidiosa (lanes O) or rescued from X. fastidiosa by transformation of E. coli (lanes R).
Plasmid stability analysis.
Transformation of X. fastidiosa was performed as described previously (18), using a GenePulser Xcell electroporator (Bio-Rad). Electroporation of A. tumefaciens, P. syringae, and X. campestris was performed according to Bio-Rad protocols for P. aeruginosa. X. fastidiosa transformants were picked from PD3-kanamycin plates and grown in PD3 liquid with kanamycin for 7 to 10 days until turbid. Strains were subcultured (1:10 dilution) into PD3 liquid without kanamycin. Every 7 days, culture was dilution plated onto PD3 agar (with and without kanamycin) and subcultured (1:10 dilution) into fresh PD3 liquid. A total of five passages in nonselective PD3 liquid medium were performed. Colonies of A. tumefaciens, P. syringae, and X. campestris transformants were picked directly from kanamycin-supplemented plates into liquid medium without kanamycin and dilution plated and subcultured (1:100 dilution) every other day.
Protein expression, purification, and Western blot analysis.
E. coli strain BL21(DE3)pLysS (Stratagene) was used for all protein expression experiments. For extraction of recombinant proteins, expression was induced at an optical density at 600 nm (OD600) of 0.6 by adding 1 mM isopropyl-β-d-thiogalactopyranoside (IPTG); cultures were harvested 3 h after induction. For experiments where culture growth was monitored over a 5-h postinduction period, protein expression was induced by adding 1 mM IPTG at an OD600 of 0.3. His6-tagged PemK and His6-tagged GFP were purified from soluble cell extracts using Ni-nitrilotriacetic acid (NTA) affinity chromatography as described by the manufacturer (Qiagen). Protein electrophoresis (sodium dodecyl sulfate-polyacrylamide gel electrophoresis [SDS-PAGE]) and Western blot assays were performed using standard methods (30). Primary antibodies, anti-His (Calbiochem) and anti-S (Novagen), were used at a 1:2,000 dilution; horseradish peroxidase-conjugated goat anti-mouse secondary antibody (Santa Cruz Biotechnology) was used at a 1:15,000 dilution and detected using a Fast ECL kit (Thermo Scientific Pierce).
RESULTS
Minimal IncP-1 replicons require trfA and oriV for replication in X. fastidiosa.
Previously (35), a 5.3-kbp fragment of pXF-RIV11 was shown to be sufficient for plasmid replication in X. fastidiosa. As the 5.3-kbp fragment contained homologues (trfA and oriV) known to be required for replication of other IncP-1 plasmids (4), subclones of pXF-RIV11 (Fig. 1A) or the corresponding region of pVEIS01 (Fig. 1B) were generated and evaluated for replication competence in X. fastidiosa. Plasmid replication in X. fastidiosa required both trfA and oriV. Constructs bearing only trfA or oriV from pXF-RIV11 or pVEIS01 were not competent for replication in X. fastidiosa. The open reading frame (ORF) encoding a resolvase homologue and located upstream of trfA for both pXF-RIV11 and pVEIS01 was not required for plasmid replication in X. fastidiosa. Thus, minimal IncP-1 replicons mapped to pXF-RIV11 nt 20441 to 21823 (1,383 bp) or pVEIS01 nt 20417 to 22241 (1,825 bp). Restriction enzyme digests of plasmids used to transform X. fastidiosa and subsequently rescued from X. fastidiosa by transformation of E. coli yielded identical patterns, indicating that five serial passages (under antibiotic selection) of transformed X. fastidiosa bearing IncP-1 replicons derived from pXF-RIV11 (Fig. 1C) or pVEIS01 (Fig. 1D) did not result in gross alteration (large insertions or deletions) of plasmid sequences.
The pemI/pemK plasmid addiction system confers stable inheritance of IncP-1 replicons in X. fastidiosa.
The putative pemI/pemK plasmid addiction system from pXF-RIV11 was added to IncP-1 replicons derived from pXF-RIV11 (Fig. 2A) or pVEIS01 (Fig. 2B). Restriction enzyme digests of plasmid samples before transformation of, and after rescue from, X. fastidiosa yielded identical patterns. Thus, addition of pemI/pemK did not cause gross alteration of IncP-1 replicons derived from pXF-RIV11 (Fig. 2C) or pVEIS01 (Fig. 2D) during passage in X. fastidiosa.
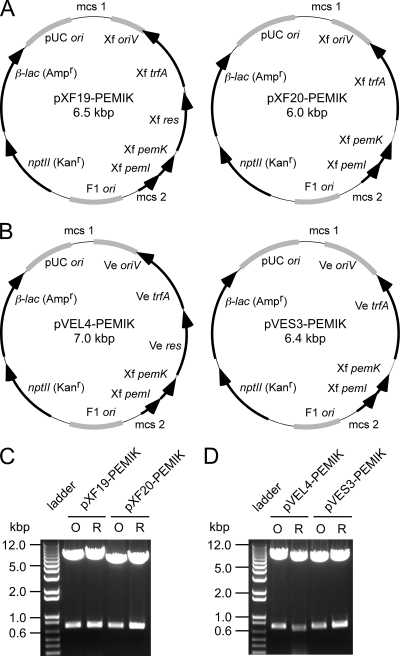
FIG. 2.
Inc-P1 replicon constructs bearing the pemI/pemK plasmid addiction system. Maps of plasmids derived from pXF-RIV11 (A) or pVEIS01 (B) are depicted. ORFs are denoted by black arrows, and various origins of replication are indicated as gray arcs. The larger constructs (pXF19-PEMIK and pVEL4-PEMIK) differ from the corresponding smaller constructs (pXF20-PEMIK and pVES3-PEMIK) by containing the complete resolvase ORF (res) of the respective IncP-1 plasmid donor located upstream of the cognate trfA ORF. Locations of multiple cloning sites (mcs1 and mcs2) are indicated. Panels C and D show NotI digests (excising pemI/pemK) of plasmids used to transform X. fastidiosa (lanes O) or rescued from X. fastidiosa by transformation of E. coli (lanes R).
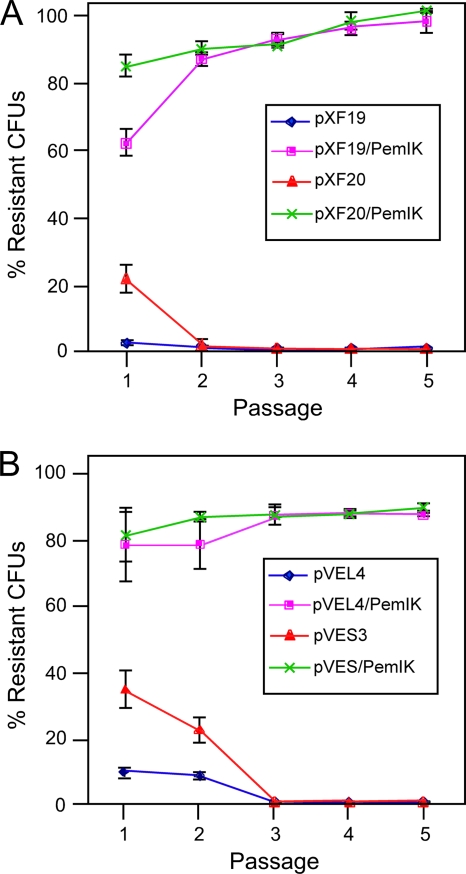
To evaluate plasmid inheritance over an extended period in the absence of antibiotic selection, X. fastidiosa transformed with IncP-1 replicons (containing or lacking pemI/pemK) were grown in liquid medium without kanamycin for five serial passages (Fig. 3). Stability of plasmid inheritance by daughter cells was assessed by plotting ratios of numbers of CFU recovered by plating aliquots of liquid cultures (lacking kanamycin) onto solid medium containing or lacking kanamycin. After three serial passages in liquid culture without antibiotic selection, the number of kanamycin-resistant X. fastidiosa (previously transformed with IncP-1 replicons lacking pemI/pemK) CFU recovered declined to zero. In contrast, the ratio of the number of CFU recovered from X. fastidiosa (transformed with IncP-1 replicons containing pemI/pemK) on plates with or without kanamycin remained stable during passages 3 to 5 at >80% to >90%. These results were considered phenotypic evidence that pemI/pemK constitutes a plasmid addiction system that is functional in X. fastidiosa.
FIG. 3.
The pemI/pemK addiction system (pemI/K) confers stable inheritance in the absence of antibiotic selection. X. fastidiosa transformed with minimal replicons derived from pXFRIV11 (A) or pVEIS01 (B) with or without pemI/K was grown in the absence of antibiotic selection (five passages over a 5-week period). Periodically, samples were plated onto medium with or without kanamycin. Stability of plasmid constructs was estimated by the ratio of the number of CFU on kanamycin-containing medium to the number of CFU on medium lacking kanamycin (% resistant CFU). Error bars denote standard deviations.
IncP-1 replicons derived from pXF-RIV11 have a broader experimental host range than those derived from pVEIS01.
Plant pathogenic bacteria unrelated to X. fastidiosa were evaluated as hosts of IncP-1 replicons derived from pXF-RIV11 or pVEIS01 (Table 1). Electroporation of P. syringae pv. tomato or A. tumefaciens with the minimal IncP-1 replicon (pXF20) derived from pXF-RIV11 resulted in kanamycin-resistant colonies from which the plasmid could be rescued by subsequent transformation of E. coli. In contrast, transformation of (and plasmid rescue from) X. campestris pv. campestris required a larger construct (pXF19) derived from pXF-RIV11 that included the upstream resolvase ORF in addition to trfA and oriV. As expected, the plasmid bearing only oriV from pXF-RIV11 (pXF21) was unable to replicate in all three species. Transformation of X. campestris pv. campestris, P. syringae pv. tomato, and A. tumefaciens with IncP-1 replicons derived from pVEIS01 did not yield kanamycin-resistant colonies from which plasmids could be rescued by subsequent transformation of E. coli. IncP-1 replicons derived from pXF-RIV11 were unstable in X. campestris pv. campestris, P. syringae pv. tomato, and A. tumefaciens in the absence of antibiotic selection (Table 1). Despite addition of the pemI/pemK plasmid addiction system, only a minority of the cells (<5%) retained kanamycin resistance after two passages of growth in liquid medium lacking kanamycin.
TABLE 1.
Experimental host ranges of IncP-1 plasmids derived from pXF-RIV11 or pVEIS01
| Plasmid |
Xanthomonas campestris pv. campestris Xcc8004 |
Pseudomonas syringae pv. tomato PstDC3000 |
Agrobacterium tumefaciens GV3101 |
|||
|---|---|---|---|---|---|---|
| Replicates?a | Stable?b | Replicates? | Stable? | Replicates? | Stable? | |
| pXF19 | Yes | No | Yes | No | Yes | No |
| pXF20 | No | Yes | No | Yes | No | |
| pXF21 | No | No | No | |||
| pXF19/PemIK | Yes | <5% | Yes | <5% | Yes | <5% |
| pXF20/PemIK | No | Yes | <5% | Yes | <5% | |
| pVEL4 | No | No | No | |||
| pVES3 | No | No | No | |||
Yes, numerous colonies after transformation and successful rescue of plasmid by transformation of E. coli. No, few to no colonies after transformation and no rescue of plasmid by transformation of E. coli.
No, no kanamycin-resistant colonies recovered after one passage on solid medium lacking kanamycin; <5%, kanamycin-resistant colonies recovered after one passage on solid medium lacking kanamycin, but liquid culture stability tests indicated that ≤5% of the colonies retained resistance to kanamycin after two passages in liquid medium lacking kanamycin.
Expression of PemI counteracts toxicity of PemK.
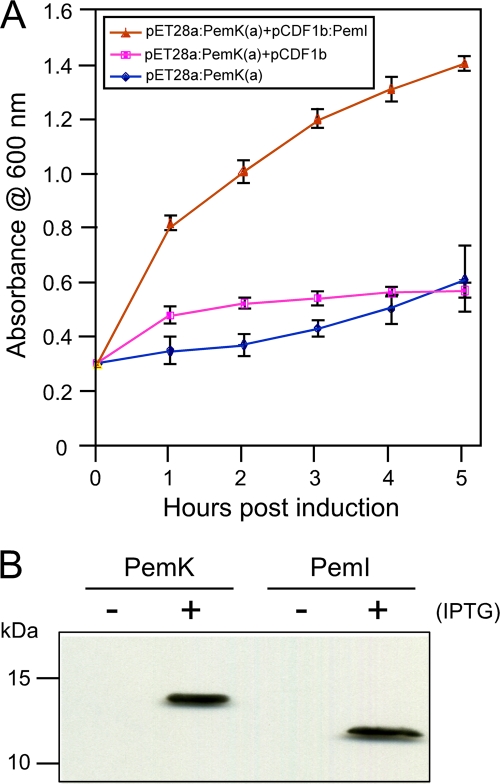
Expression of His6-tagged PemK in E. coli transformed with pET28a:PemK(a) resulted in growth inhibition following IPTG induction (Fig. 4A), a phenotype consistent with that of a toxin expressed in the absence of a cognate antitoxin. In contrast, growth of E. coli cotransformed with pET28:PemK(a) and pCDF1b:PemI expressing His6-tagged PemI was not inhibited following IPTG induction. Restoration of growth following IPTG induction was not observed in E. coli cotransformed with pET28a:PemK(a) and the expression vector pCDF1b lacking pemI. These results indicate that the pemI/pemK plasmid addiction system derived from pXF-RIV11 functions as a TA module, with PemI serving as the cognate antitoxin of PemK toxin. To verify the expression of each protein upon IPTG induction, His6-tagged proteins from E. coli (transformed individually with each expression vector) were purified from total soluble proteins by Ni-NTA affinity chromatography and detected in Western blot assays using anti-His antibodies (Fig. 4B).
FIG. 4.
Effect of PemK and PemI expression on growth of E. coli. Growth of E. coli transformed with expression plasmids was measured by mean culture absorbance at 600 nm (error bars denote standard deviations) following IPTG induction at 0 h (A). Growth inhibition resulting from expression of His6-tagged PemK toxin alone [pET28a:PemK(a)] was reversed by coexpression with His6-tagged PemI antitoxin (pCDF1b:PemI) but not by coexpression with the empty vector pCDF1b. Western blot assay detection (with anti-His antibodies) of His6-tagged PemK toxin and His6-tagged PemI antitoxin (expressed individually in separate cultures) extracted from E. coli with (+) or without (−) IPTG induction and purified by Ni-NTA affinity chromatography (B). Sizes of protein standards are indicated on the left.
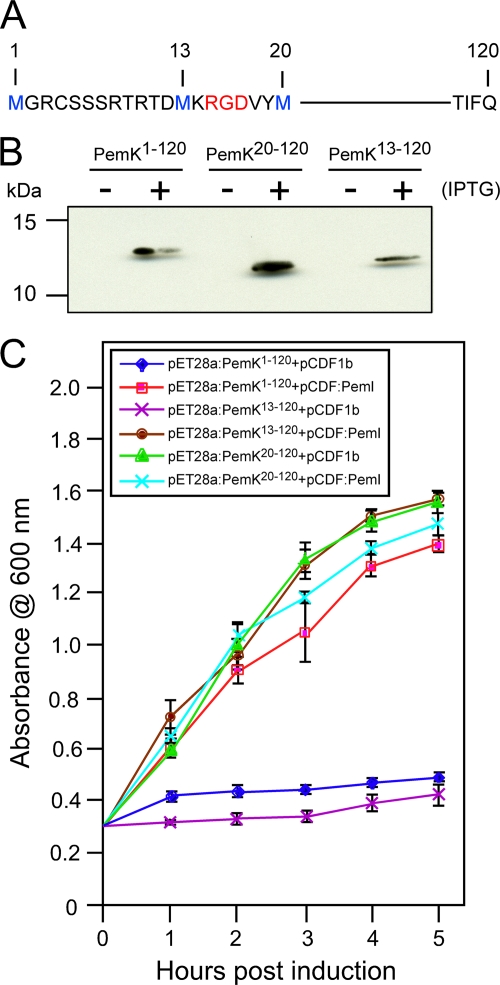
Twelve amino-terminal amino acid residues of PemK are dispensable for toxicity.
The pemK ORF partially overlaps the upstream pemI ORF. There are three in-frame methionine codons that potentially may serve as start codons. Therefore, 5′-terminal nested deletions were constructed in pET28a (Fig. 5A) such that translation of PemK would include methionine at amino acid position 1 (pET28a:PemK1-120), 13 (pET28a:PemK13-120), or 20 (pET28a:PemK20-120) downstream of the His6 tag. Following IPTG induction of E. coli transformed with the PemK constructs, all three His6-tagged proteins were purified by Ni-NTA affinity chromatography and detected by Western blotting with anti-His antibodies (Fig. 5B). Cultures transformed with each PemK construct also were cotransformed with pCDF1b:PemI or pCDF1b empty vector (Fig. 5C). Growth of cotransformed cultures following IPTG induction indicated that deletion of PemK amino acids 1 to 12 did not alter the growth inhibition phenotype, whereas deletion of PemK amino acids 1 to 19 resulted in no inhibition of growth. As expected, coexpression of PemI abolished growth inhibition. The largest deletion lacked RGD at amino acid positions 15 to 17 (Fig. 5A) that constitute a conserved motif (RGD/E) present in a similar position among numerous PemK homologues identified in GenBank through protein BLAST searches (data not shown).
FIG. 5.
Expression and growth inhibition properties of amino-terminal deletion mutant forms of PemK toxin. The sequence of the PemK amino-terminal region is presented as one-letter code (A); methionine residues in blue indicate the beginning of PemK sequences expressed as fusions with amino-terminal His6 tags (not shown); numbers denote amino acid coordinates with the first upstream and in-frame methionine designated 1; red indicates a motif (RGD/E) conserved among PemK homologues. Western blot assay detection (B) with anti-His antibodies of His6-tagged full-length PemK (PemK1-120) or His6-tagged, truncated PemK (PemK13-120, PemK20-120) extracted from E. coli with (+) or without (−) IPTG induction and purified by Ni-NTA affinity chromatography. Sizes of protein standards are indicated on the left. Growth of E. coli transformed with expression plasmids (C) was measured by mean culture absorbance at 600 nm (error bars denote standard deviations) following IPTG induction at 0 h. Growth inhibition resulting from expression of His6-tagged PemK constructs retaining toxin activity (PemK1-120, PemK13-120) was reversed by coexpression with His6-tagged PemI antitoxin (pCDF1b:PemI) but not by coexpression with the empty vector pCDF1b.
Copurification of PemK and PemI indicates that the toxin and antitoxin bind to one another.
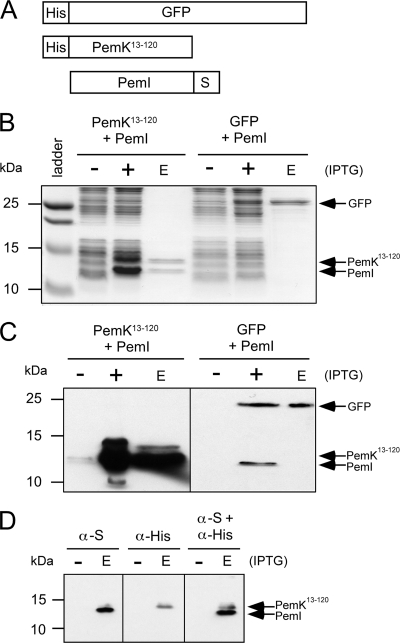
Toxin and antitoxin proteins from other TA systems have been shown to directly interact. Therefore, interaction of pXF-RIV11 PemK and PemI was tested. The genes were cloned into a pETDuet-1 vector, resulting in N-terminally His6-tagged PemK13-120 and C-terminally S-tagged PemI (Fig. 6A). As a negative control, His6-tagged GFP was coexpressed with S-tagged PemI, also in pETDuet-1. E. coli cell extracts were passed over a Ni-NTA affinity column to bind the His6-tagged proteins. As demonstrated by SDS-PAGE (Fig. 6B) and confirmed by Western blot assays (Fig. 6C and D), S-tagged PemI was retained on the Ni-NTA column only in the presence of His6-tagged PemK13-120; coexpression with His6-tagged GFP did not result in retention of S-tagged PemI on the Ni-NTA affinity column. Therefore, antitoxin PemI specifically binds to toxin PemK, a result consistent with toxin sequestration as demonstrated in other TA systems (7, 11, 20).
FIG. 6.
Copurification of PemK13-120 and PemI in vitro. The dual-expression vector pETDuet-1 was used to coexpress His6-tagged PemK13-120 toxin or His6-tagged GFP with S-tagged PemI antitoxin (A). Coomassie-stained SDS-PAGE of cell extracts (B) from uninduced cells (−), IPTG-induced cells (+), and Ni-NTA column eluate (E) indicated that S-tagged PemI is retained on the Ni-NTA column only in the presence of His6-tagged PemK13-120. S-tagged PemI is not retained on the column when coexpressed with His6-tagged GFP. Sizes of protein standards are indicated on the left. Western blot assay detection (with both anti-His and anti-S antibodies) confirms that S-tagged PemI is present in both induced cell extracts (+) but is detected in the Ni-NTA column eluate (E) only when coexpressed with His6-tagged PemK (C). Western blotting (D) with anti-His and anti-S antibodies singly and then both together confirms the specificity of the antibodies and the identities of His6-tagged PemK13-120 and S-tagged PemI.
DISCUSSION
Functional analysis of sequences derived from pXF-RIV11 and pVEIS01 indicated that trfA and oriV are the only plasmid-borne elements necessary for replication in X. fastidiosa, as expected for IncP-1 plasmids. Whereas the natural host (V. eiseniae) of pVEIS01 is a betaproteobacterium, X. fastidiosa is a gammaproteobacterium. As the TrfA proteins encoded by the two plasmids share 87% amino acid sequence identity and since the iterated elements of oriV are similar (35), it is perhaps not unexpected that replicons derived from pVEIS01 were able to replicate in X. fastidiosa. Whether minimal replicons derived from pXF-RIV11 are competent for replication in V. eiseniae was not determined, as a transformation protocol for V. eiseniae has not been developed.
Additional host range experiments indicated that the minimal replicon (pXF20) derived from pXF-RIV11, but not the minimal replicon (pVES3) derived from pVEIS01, was able to replicate in two gammaproteobacteria, P. syringae pv. tomato and A. tumefaciens. However, a larger construct (pXF19) was needed for replication in X. campestris pv. campestris. These two constructs differ in that only pXF19 contained the complete resolvase ORF located upstream of trfA. Stable maintenance of certain low-copy-number plasmids such as pRP4 (12) requires a partitioning (par) system that includes a functional resolvase. Therefore, the simplest interpretation of differential host range data for pXF19 and pXF20 is that X. fastidiosa, P. syringae pv. tomato, and A. tumefaciens (but not X. campestris pv. campestris) encode resolvases able to resolve plasmid dimers such that monomeric progeny molecules may be distributed to daughter cells. As inclusion of the resolvase ORF derived from pVEIS01 did not result in replication of a pVEIS01 derivative (pVEL4) in P. syringae pv. tomato, A. tumefaciens, or X. campestris pv. campestris, host range differences of derivatives from pXF-RIV11 versus pVEIS01 were not simply due to the presence or absence of the cognate resolvase gene.
Phenotypic properties of pemI/pemK homologues of pXF-RIV11 were consistent with that of a TA module: sole expression of PemK toxin inhibited cell growth, addition of PemI antitoxin restored growth, and PemK and PemI directly interacted with one another. Furthermore, addition of the pXF-RIV11 pemI/pemK module conferred stability of plasmid inheritance in X. fastidiosa grown for prolonged periods in the absence of antibiotic selection, precisely as expected for a TA module addiction system. That addition of pemI/pemK did not substantially increase the stability of plasmids in A. tumefaciens, P. syringae, and X. campestris indicates host background affects the efficiency of stability conferred by the pXF-RIV11 plasmid addiction system. Although the specific mode of action of PemK toxin encoded by pXF-RIV11 remains to be determined, it is likely that pXF-RIV11 PemK toxin acts in a fashion similar to that of other PemK toxins: an RNase targeting cellular RNAs (2, 43).
The pXF-RIV11 pemK ORF (nt 1513 to 1872) partially overlaps that of the 5′-proximal pemI ORF (nt 1289 to 1549). Within the 5′-proximal region of the pemK ORF, the first in-frame methionine codon (pXF-RIV11 coordinates 1513 to 1515) completely overlaps the pemI ORF, the second in-frame pemK methionine codon (1549 to 1551) partially overlaps the pemI stop codon (1550 to 1552), and the third in-frame pemK methionine codon (nt 1570 to 1572) is positioned downstream of the overlapping region. As amino-terminal truncation of PemK (deletion of amino acids 1 to 19) resulted in loss of toxin activity, translation of PemK likely initiates at the first or second in-frame methionine codons. That deletion of the first 12 codons of the pemK ORF had no effect on toxin activity suggests that either PemK translation initiates at the second in-frame methionine codon or the first 12 in-frame amino acids are dispensable for PemK toxic properties and binding of PemI antitoxin. The conserved motif RGD (pXF-RIV11 PemK amino acids 15 to 17) was located downstream of sequences dispensable for toxin activity. Interestingly, conserved RGD/E motifs have been found in a variety of cell adhesion proteins (10) but a specific function for this motif in PemK homologues has yet to be identified.
The research reported here was initiated, in part, with a practical objective: development of a plasmid shuttle vector for X. fastidiosa that is stably inherited over extended time periods in the absence of antibiotic selection. Such stability is required to conduct mutant complementation assays in plants, as expression of disease symptoms often requires many months after inoculation (23). Although an exogenous plasmid (pBBRIMCS5 bearing a replication origin from Bordetella) has been used as a shuttle vector to complement a nonpathogenic tolC mutant of X. fastidiosa, only 50% of cells retained the plasmid after 60 days in planta (27). Furthermore, it is unclear whether pBBRIMCS5 would be stable in plants in cases where the mutant retained pathogenicity.
We selected an endogenous plasmid of X. fastidiosa as starting material since pXF-RIV11 could (i) replicate in X. fastidiosa and (ii) was stable despite lacking a selectable marker. The V. eiseniae plasmid (pVEIS01) was selected as alternative starting material given the extensive sequence identity it shares with pXF-RIV11. We anticipated that stable inheritance would require more than the minimal gene complement needed for replication. As bioinformatic analysis of pXF-RIV11 identified the pemI/pemK TA homologue (35), we used this endogenous plasmid addiction system to confer stability of shuttle vectors. Because transformation of X. fastidiosa with derivatives of pXF-RIV11 was consistently more efficient than that with derivatives of pVEIS01 (data not shown), pXF20-PEMIK (Fig. 2) represents the most suitable shuttle vector construct described herein. Ten unique endonuclease restriction sites are clustered in two multiple cloning regions such that foreign DNA may be readily inserted into pXF20-PEMIK.
Acknowledgments
We thank Keira Neumann, Mark Schreiber, Stephanie Underwood, and Kunbo Zhang for technical assistance.
Mention of proprietary or brand names is necessary to report factually on available data; however, the USDA neither guarantees nor warrants the standard of the product, and the use of the name by USDA implies no approval to the exclusion of others that also may be suitable.
Footnotes
Published ahead of print on 8 October 2010.
REFERENCES
- 1.Afif, H., N. Allali, M. Couturier, and L. Van Melderen. 2001. The ratio between CcdA and CccB modulates the transcriptional repression of the ccd poison-antidote system. Mol. Microbiol. 41:73-82. [DOI] [PubMed] [Google Scholar]
- 2.Agarwal, S., N. K. Mishra, S. Bhatnagar, and R. Bhatnagar. 2010. PemK toxin of Bacillus anthracis is a ribonuclease: an insight into its active site, structure, and function. J. Biol. Chem. 285:7254-7270. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Aizenman, E., H. Engelberg-Kulka, and G. Glaser. 1996. An Escherichia coli chromosomal “addiction molecule” regulated by guanosine 3′,5′-bispyrophosphate: a model for programmed bacterial cell death. Proc. Nat. Acad. Sci. U. S. A. 93:6059-6063. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Ayres, E. K., V. J. Thomson, G. Merino, D. Balderes, and D. H. Figurski. 1993. Precise deletions in large bacterial genomes by vector-mediated excision (VEX). The trfA gene of promiscuous plasmid RK2 is essential for replication in several gram-negative hosts. J. Mol. Biol. 230:174-185. [DOI] [PubMed] [Google Scholar]
- 5.Bahassi, E. M., M. H. O'Dea, N. Allai, J. Messens, M. Gellert, et al. 1999. Interactions of CcdB with DNA gyrase. Inactivation of GyrA, poisoning of the gyrase-DNA complex, and the antidote action of CcdA. J. Biol. Chem. 274:10936-10944. [DOI] [PubMed] [Google Scholar]
- 6.Blua, M. J. A., P. A. Phillips, and R. A. Redak. 1999. A new sharpshooter threatens both crops and ornamentals. Calif. Agric. 53:22-25. [Google Scholar]
- 7.Brown, J. M., and K. J. Shaw. 2003. A novel family of Escherichia coli toxin-antitoxin gene pairs. J. Bacteriol. 185:6600-6608. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Chen, J., C. J. Chang, and R. Jarret. 1992. Plasmids from Xylella fastidiosa strains. Can. J. Microbiol. 38:993-995. [Google Scholar]
- 9.Dao-Thi, M. H., D. Charlier, R. Loris, D. Maes, J. Messens, et al. 2002. Intricate interactions within the ccd plasmid addiction system. J. Biol. Chem. 277:3733-3742. [DOI] [PubMed] [Google Scholar]
- 10.D'Souza, S. E., M. H. Ginsberg, and F. F. Plow. 1991. Arginyl-glycyl-aspartic acid (RGD): a cell adhesion motif. Trends Biochem. Sci. 16:246-250. [DOI] [PubMed] [Google Scholar]
- 11.Gerdes, K., S. K. Christensen, and A. Lobner-Olesen. 2005. Prokaryotic toxin-antitoxin stress response loci. Nat. Rev. Microbiol. 3:371-382. [DOI] [PubMed] [Google Scholar]
- 12.Gerlitz, M., O. Hrabak, and H. Schwab. 1990. Partitioning of broad-host-range plasmid RP4 is a complex system involving site-specific recombination. J. Bacteriol. 172:6194-6203. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Guilhabert, M. R., V. J. Stewart, and B. C. Kirkpatrick. 2006. Characterization of putative rolling-circle plasmids from the Gram-negative bacterium Xylella fastidiosa and their use as shuttle vectors. Plasmid 55:70-80. [DOI] [PubMed] [Google Scholar]
- 14.Hargreaves, D., S. Santos-Sierra, R. Giraldo, R. Sabariegos-Jareno, and R. Diaz-Orejas. 2002. Structural and functional analysis of the kid toxin protein from E. coli plasmid R1. Structure 10:1425-1433. [DOI] [PubMed] [Google Scholar]
- 15.Hernandez-Martinez, R., D. A. Cooksey, and F. P. Wong. 2009. Leaf scorch of purple-leafed plum and sweetgum dieback: two new diseases in southern California caused by Xylella fastidiosa. Plant Dis. 93:1131-1138. [DOI] [PubMed] [Google Scholar]
- 16.Hernandez-Martinez, R., K. A. de la Cerda, H. S. Costa, D. A. Cooksey, and F. P. Wong. 2007. Phylogenetic relationships of Xylella fastidiosa strains isolated from landscape ornamentals in southern California. Phytopathology 97:857-864. [DOI] [PubMed] [Google Scholar]
- 17.Hernandez-Martinez, R., T. R. Pinckard, H. S. Costa, D. A. Cooksey, and F. P. Wong. 2006. Discovery and characterization of Xylella fastidiosa strains in southern California causing mulberry leaf scorch. Plant Dis. 90:1143-1149. [DOI] [PubMed] [Google Scholar]
- 18.Matsumoto, A., G. M. Young, and M. M. Igo. 2009. Chromosomal-based genetic complementation system for Xylella fastidiosa. Appl. Environ. Microbiol. 75:1679-1687. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Mei, J., S. Nenashski, and W. Firshein. 1995. Interactions of the origin of replication (oriV) and initiation proteins (TrfA) of plasmid RK2 with submembrane domains of Escherichia coli. J. Bacteriol. 177:6766-6772. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Nordström, K., and S. J. Austin. 1989. Mechanisms that contribute to stable segregation of plasmids. Annu. Rev. Genet. 23:37-69. [DOI] [PubMed] [Google Scholar]
- 21.Perring, T. M., C. A. Farrar, and M. J. Blua. 2001. Proximity to citrus influences Pierce's disease in Temecula Valley vineyards. Calif. Agric. 55:13-18. [Google Scholar]
- 22.Pooler, M. R., J. S. Hartung, and R. G. Fenton. 1997. Sequence analysis of a 1296-nucleotide plasmid from Xylella fastidiosa. FEMS Microbiol. Lett. 155:217-222. [DOI] [PubMed] [Google Scholar]
- 23.Purcell, A. H., and D. L. Hopkins. 1996. Fastidious xylem-limited bacterial plant pathogens. Annu. Rev. Phytopathol. 34:131-151. [DOI] [PubMed] [Google Scholar]
- 24.Purcell, A. H., and S. R. Saunders. 1999. Glassy-winged sharpshooter expected to increase plant disease. Calif. Agric. 53:26-27. [Google Scholar]
- 25.Purcell, A. H., S. R. Saunders, M. Hendson, M. E. Grebus, and M. J. Henry. 1999. Causal role of Xylella fastidiosa in oleander leaf scorch disease. Phytopathology 89:53-58. [DOI] [PubMed] [Google Scholar]
- 26.Qin, X., and J. S. Hartung. 2001. Construction of a shuttle vector and transformation of Xylella fastidiosa with plasmid DNA. Curr. Microbiol. 43:158-162. [DOI] [PubMed] [Google Scholar]
- 27.Reddy, J. D., S. L. Reddy, D. L. Hopkins, and D. W. Gabriel. 2007. TolC is required for pathogenicity of Xylella fastidiosa in Vitis vinifera grapevines. Mol. Plant Microbe Interact. 20:403-410. [DOI] [PubMed] [Google Scholar]
- 28.Ruiz-Echevarría, M. J., A. Berzal-Herranz, K. Gerdes, and R. Diaz-Orejas. 1991. The kis and kid genes of the parD maintenance system of plasmid R1 form an operon that is autoregulated at the level of transcription by the co-ordinated action of the Kis and Kid proteins. Mol. Microbiol. 5:2685-2693. [DOI] [PubMed] [Google Scholar]
- 29.Ruiz-Echevarría, M. J., G. Gimenez-Gallego, R. Sabariegos-Jareno, and R. Diaz-Orejas. 1995. Kid, a small protein of the parD stability system of plasmid R1, is an inhibitor of DNA replication acting at the initiation of DNA synthesis. J. Mol. Biol. 247:568-577. [DOI] [PubMed] [Google Scholar]
- 30.Sambrook, J., and D. W. Russell. 2001. Molecular cloning: a laboratory manual, 3rd ed. Cold Spring Harbor Laboratory Press, Cold Spring Harbor, NY.
- 31.Santos-Sierra, S., M. Lemonnier, B. Nunez, D. Hargreaves, J. Rafferty, et al. 2003. Non-cytotoxic variants of the kid protein that retain their auto-regulatory activity. Plasmid 50:120-130. [DOI] [PubMed] [Google Scholar]
- 32.Simpson, A. J. G., F. C. Reinach, P. Arruda, F. A. Abreu, M. Acencio, R. Alvarenga, et al. 2000. The genome sequence of the plant pathogen Xylella fastidiosa. Nature 406:151-159. [DOI] [PubMed] [Google Scholar]
- 33.Sisterson, M. S., R. Yacoub, G. Montez, E. E. Grafton-Cardwell, and R. L. Groves. 2008. Distribution and management of citrus in California: implications for management of glassy-winged sharpshooter. J. Econ. Entomol. 101:1041-1050. [DOI] [PubMed] [Google Scholar]
- 34.Stalker, D. M., C. M. Thomas, and D. R. Helinski. 1981. Nucleotide sequence of the region of the origin of replication of the broad host range plasmid RK2. Mol. Gen. Genet. 181:8-12. [DOI] [PubMed] [Google Scholar]
- 35.Stenger, D. C., M. W. Lee, E. E. Rogers, and J. Chen. 2010. Plasmids of Xylella fastidiosa mulberry-infecting strains share extensive sequence identity and gene complement with pVEIS01 from the earthworm symbiont Verminephrobacter eiseniae. Physiol. Mol. Plant Pathol. 74:238-245. [Google Scholar]
- 36.Tam, J. E., and B. C. Kline. 1989. Control of the ccd operon in plasmid F. J. Bacteriol. 171:2353-2360. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Tsuchimoto, S., Y. Nishimura, and E. Ohtsubo. 1992. The stable maintenance system pem of plasmid R100: degradation of PemI protein may allow PemK protein to inhibit cell growth. J. Bacteriol. 174:4205-4211. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Tsuchimoto, S., and E. Ohtsubo. 1989. Effect of the pem system on stable maintenance of plasmid R100 in various Escherichia coli hosts. Mol. Gen. Genet. 215:463-468. [DOI] [PubMed] [Google Scholar]
- 39.Tsuchimoto, S., and E. Ohtsubo. 1993. Autoregulation by cooperative binding of the PemI and PemK proteins to the promoter region of the pem operon. Mol. Gen. Genet. 237:81-88. [DOI] [PubMed] [Google Scholar]
- 40.Tsuchimoto, S., H. Ohtsubo, and E. Ohtsubo. 1988. Two genes, pemK and pemI, responsible for stable maintenance of resistance plasmid R100. J. Bacteriol. 170:1461-1466. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Tubajika, K. M., E. L. Civerolo, M. A. Ciomperlik, D. A. Luvisi, and J. M. Hashim. 2004. Analysis of the spatial patterns of Pierce's disease incidence in the lower San Joaquin Valley in California. Phytopathology 94:1136-1144. [DOI] [PubMed] [Google Scholar]
- 42.Wells, J. M., B. C. Raju, H.-Y. Hung, W. G. Weisburg, L. Mandelco-Paul, and D. J. Brenner. 1987. Xylella fastidiosa gen. nov., sp. nov: Gram-negative, xylem-limited, fastidious plant bacteria related to Xanthomonas spp. Int. J. Syst. Bacteriol. 37:136-143. [Google Scholar]
- 43.Zhang, J., Y. Zhang, L. Zhu, M. Suzuki, and M. Inouye. 2004. Interference of mRNA function by sequence-specific endoribonuclease PemK. J. Biol. Chem. 279:20678-20684. [DOI] [PubMed] [Google Scholar]