Abstract
Purpose
The JBR.10 trial demonstrated benefit from adjuvant cisplatin/vinorelbine (ACT) in early-stage non–small-cell lung cancer (NSCLC). We hypothesized that expression profiling may identify stage-independent subgroups who might benefit from ACT.
Patients and Methods
Gene expression profiling was conducted on mRNA from 133 frozen JBR.10 tumor samples (62 observation [OBS], 71 ACT). The minimum gene set that was selected for the greatest separation of good and poor prognosis patient subgroups in OBS patients was identified. The prognostic value of this gene signature was tested in four independent published microarray data sets and by quantitative reverse-transcriptase polymerase chain reaction (RT-qPCR).
Results
A 15-gene signature separated OBS patients into high-risk and low-risk subgroups with significantly different survival (hazard ratio [HR], 15.02; 95% CI, 5.12 to 44.04; P < .001; stage I HR, 13.31; P < .001; stage II HR, 13.47; P < .001). The prognostic effect was verified in the same 62 OBS patients where gene expression was assessed by qPCR. Furthermore, it was validated consistently in four separate microarray data sets (total 356 stage IB to II patients without adjuvant treatment) and additional JBR.10 OBS patients by qPCR (n = 19). The signature was also predictive of improved survival after ACT in JBR.10 high-risk patients (HR, 0.33; 95% CI, 0.17 to 0.63; P = .0005), but not in low-risk patients (HR, 3.67; 95% CI, 1.22 to 11.06; P = .0133; interaction P < .001). Significant interaction between risk groups and ACT was verified by qPCR.
Conclusion
This 15-gene expression signature is an independent prognostic marker in early-stage, completely resected NSCLC, and to our knowledge, is the first signature that has demonstrated the potential to select patients with stage IB to II NSCLC most likely to benefit from adjuvant chemotherapy with cisplatin/vinorelbine.
INTRODUCTION
Recent clinical trials have led to the adoption of adjuvant cisplatin-based chemotherapy (ACT) for patients with resected stages IB to IIIA non–small-cell lung cancer (NSCLC). The 5-year survival advantage conferred by ACT in these studies ranged from 4% in the International Adjuvant Lung Trial to 15% in National Cancer Institute of Canada Clinical Trials Group JBR.10.1,2 No trial showed a significant survival benefit in stage IB.1,3 The Lung Adjuvant Cisplatin Evaluation meta-analysis pooled individual patient data from five trials of cisplatin-based chemotherapy1,2 and found a 5.4% 5-year survival advantage (hazard ratio [HR], 0.89; 95% CI, 0.82 to 0.96; P = .005). Subgroup analysis confirmed the lack of significant benefit in stage IB (HR, 0.93; 95% CI, 0.78 to 1.10). Moreover, a potential detrimental effect was observed in stage IA (HR, 1.40; 95% CI, 0.95 to 2.06). Therefore, the current standard of treatment for patients with stage I NSCLC remains surgery alone. However, 30% to 40% of stage I patients will relapse,4 indicating that some of these patients have a poorer prognosis, and that potentially, they might benefit from ACT.
The lack of consistent prognostic molecular markers for early-stage NSCLC led to attempts to identify novel gene expression signatures using genome-wide microarray platforms. It was hypothesized that multigene signatures might be stronger than individual genes to predict prognosis, and that poor prognosis patients potentially could benefit from ACT. Numerous studies have identified prognostic signatures in NSCLC with minimal overlap in their gene sets.5–13 Only recently published signatures have been subjected to independent validation.6,7,9,11 Importantly, none could be evaluated for the ability to predict benefit from adjuvant chemotherapy as none of the testing or validation sets included randomly assigned treated and untreated control groups. In National Cancer Institute of Canada Clinical Trials Group JBR.10, snap-frozen tumor tissues were collected prospectively from Canadian patients. We report here our microarray study of these samples, and present a gene signature that is both prognostic for survival and predictive of benefit from ACT.
PATIENTS AND METHODS
Patients and Samples
JBR.10, a randomized controlled trial of adjuvant vinorelbine/cisplatin versus observation alone, included prospective collection of snap-frozen or formalin-fixed paraffin-embedded tumor samples for RAS mutation analysis (stratification variable) and banking for future laboratory studies.1 Although 445 of 482 randomly assigned patients consented to banking, frozen tissue was collected from only 169 (Data Supplement), of which only 166 contained more than 20% tumor cellularity; gene expression profiling was completed in 133 of these samples, using the U133A oligonucleotide microarrays (Affymetrix, Santa Clara, CA). Of patients with microarray profiles, 62 were in the observation (OBS) group, while 71 received ACT. A quantitative reverse-transcriptase polymerase chain reaction (RT-qPCR) verification and validation was carried out in these 133 cases and an additional 30 (19 OBS, 21 ACT) cases not microarray-profiled initially.
RNA Isolation and Microarray Profiling
The University Health Network research ethics board approved this study. Total RNA was isolated from frozen tumors by homogenization in guanidium isothiocyanate solution and acid phenol-chloroform extraction, purified by RNeasy mini kit (Qiagen, Mississauga, Ontario, Canada) and checked by Agilent Bioanalyzer (Agilent, Palo Alto, CA) for quality. Ten-μg total RNA was processed, labeled, and hybridized to Affymetrix HG-U133A at the Center for Cancer Genome Discovery, Dana-Farber Cancer Institute.14,15 Microarray data are available at National Center for Biotechnology Information Gene Expression Omnibus (GSE14814) and from the Director's Challenge Project (Data Supplement).15
Microarray Data Analysis and Gene Annotation
Raw microarray data were preprocessed using RMAexpress version 0.3.16 Probe sets were annotated using the NetAffx version 4.2 annotation tool, and only probe sets with grade A annotation17 (NA22) were included for further analysis. Microarray profiling was batched at two different times, and unsupervised heuristic K-means clustering using Genesis version 1.7.5 identified a systematic difference between the batches (Data Supplement). Therefore, distance-weighted discrimination (https://genome.unc.edu/pubsup/dwd/index.html) was used to adjust for the difference. The data were then transformed to a Z-score by centering to the mean and scaling to the standard deviation.
Selection of the Prognostic Gene Expression Signature
We employed the Maximizing R Square Algorithm approach to identify a minimum set of genes that had the highest independent ability to classify patients into high- and low-risk subgroups (Data Supplement). The probe sets were preselected by univariate survival analysis at P < .005, and transformed to a risk score (expression level weighted by the coefficient from the Cox regression model), then selected by using exclusion and inclusion selection procedures. The exclusion procedure removed one probe set at a time based on the resultant R square (R2, goodness-of-fit) of the Cox model.18,19 The procedure was repeated until there was only one probe set left. Starting with this probe set, the inclusion procedure was performed by including one probe set at a time based on the resultant R2 of the Cox model. After plotting the R2 against the probe set, the minimum number of probe sets having the largest R2 was chosen as the candidate signature for further testing and validation. The gene signature was established after internal validation by leave-one-out-cross-validation.
In addition, the expression levels of the 15 genes were verified by RT-qPCR using the HT7900 Fast Real-time PCR System (Applied Biosystems) on microarray profiled cases (Data Supplement).
Statistical Analysis
Principle component analysis was used to synthesize information from the 15 selected gene expressions. The first four principal components (PCs) with Eigen value ≥ 1 were introduced to the Cox regression model using disease-specific survival (from date of random assignment to death from disease or treatment complication). The risk score was derived using the four coefficients from the Cox regression and the four PCs of the 15-gene expressions (Data Supplement). Using the median of the risk score as the cutoff point, patients were divided into high- and low-risk prognostic groups. Since cause of death was not available in the validation data sets, overall survival was used as the outcome. Kaplan-Meier product-limit methods and log-rank tests were used to estimate and test differences in survival distributions between risk groups and treatment arms. Multivariate Cox regression models were used to validate the prognostic and predictive effects of risk groups on survival while adjusting for predefined baseline factors. All statistical analyses were performed using SAS version 9.1 (SAS Institute, Cary, NC).
Validation in Independent Microarray Data Sets
Our signature was tested in four independent microarray data sets, including a subset of the National Cancer Institute Director's Challenge Consortium (DCC) for the Molecular Classification of Lung Adenocarcinoma.15 As 43 JBR.10 tumors were included in the DCC study as part of the 82 Canada/Dana-Farber samples (Data Supplement), only data from the other three institutions (University of Michigan, H.L. Moffitt Cancer Center, and Memorial Sloan-Kettering Cancer Center) were used for validation. To reflect the JBR.10 population, validation was restricted to stage IB to II patients who received neither ACT nor radiotherapy (Data Supplement). Therefore, the DCC validation data set included 96 patients (27 University of Michigan, 38 H.L. Moffitt Cancer Center, 31 Memorial Sloan-Kettering Cancer Center). Other validation datasets of stage IB to II patients who received no adjuvant therapy included 48 from Duke University,8 79 patients with squamous carcinoma from University of Michigan,9 and 133 NSCLC patients from the Netherlands Cancer Institute (NLCI)20 (Data Supplement). Probe set matching from Affymetrix U133A to the Agilent 44K platform (NLCI study) was based on Unigene ID mapping from NetAffx annotation (NA22), and annotation provided by Roepman et al20 (http://research.agendia.com/), respectively (Data Supplement). Expression level was averaged if multiple matching probe sets were found in the NLCI data. The RT-qPCR validation was performed in 30 additional JBR.10 cases (Data Supplement).
RESULTS
Table 1 compares the demographic features of 133 JBR.10 patients with microarray profiling to 349 without profiling. All factors are similarly distributed, except for stage, where more stage IB patients are present in the microarray-profiled cohort (55% v 42%; P = .01). There was no significant difference in the overall survival of patients with or without microarray profiling (Data Supplement). Furthermore, a similar beneficial effect of ACT was observed in the 133 cases (HR, 0.80; 95% CI, 0.48 to 1.32), although the difference was not significant due to the reduced sample size (P = .38; Data Supplement).
Table 1.
Baseline Demographics of JBR.10 Patients With and Without Microarray Profiles
| Factor | All Patients (N = 482) | Microarray Profiled (n = 133) |
No Microarray (n = 349) |
P | ||
|---|---|---|---|---|---|---|
| No. | % | No. | % | |||
| Treatment received | .14 | |||||
| Adjuvant chemotherapy | 231 | 71 | 53 | 160 | 46 | |
| Observation alone | 251 | 62 | 47 | 189 | 54 | |
| Age, years | .6 | |||||
| < 65 | 324 | 87 | 65 | 237 | 68 | |
| ≥ 65 | 158 | 46 | 35 | 112 | 32 | |
| Sex | .35 | |||||
| Male | 314 | 91 | 68 | 223 | 64 | |
| Female | 168 | 42 | 32 | 126 | 36 | |
| Performance status | .72 | |||||
| 0 | 236 | 67 | 50 | 169 | 49 | |
| 1 | 245 | 66 | 50 | 179 | 51 | |
| Stage of disease | .01 | |||||
| IB | 219 | 73 | 55 | 146 | 42 | |
| II | 263 | 60 | 45 | 203 | 58 | |
| Surgery | .66 | |||||
| Pneumonectomy | 113 | 33 | 25 | 80 | 23 | |
| Other resection | 369 | 100 | 75 | 269 | 77 | |
| Pathologic type | .56 | |||||
| Adenocarcinoma | 256 | 71 | 53 | 185 | 53 | |
| Squamous | 179 | 52 | 39 | 127 | 36 | |
| Other | 47 | 10 | 8 | 37 | 11 | |
| RAS mutation status | .12* | |||||
| Present | 117 | 28 | 21 | 89 | 26 | |
| Absent | 333 | 105 | 79 | 228 | 65 | |
| Unknown | 32 | 0 | 0 | 32 | 9 | |
P value: without including those with missing or unknown values.
Derivation of the Gene Expression Signature
Using P < .005 as the cutoff, 172 of 19,619 U133A probe sets were significantly associated with survival in 62 OBS patients (Data Supplement). Maximizing R Square Algorithm identified a 15-gene/probe-set prognostic signature (Table 2) having the largest R2 of 0.78 (Data Supplement). The first four PCs of these signature genes were used to derive the high-risk and low-risk groups (HR, 15.02; 95% CI, 5.12 to 44.04; P < .001; Fig 1A), with 31 of 62 OBS patients classified into each group. The signature was not associated with stage, patient age or sex, RAS or p53 mutation, or p53 protein level (Data Supplement). The prognostic value of this 15-gene signature was stage-independent, and separated risk groups in stage I (HR, 13.32; 95% CI, 2.86 to 62.11; P < .001; Fig 1B) and stage II (HR, 13.47; 95% CI, 3.00 to 60.43; P < .001; Fig 1C). Multivariate analysis adjusting for preselected significant prognostic factors showed that the signature was an independent prognostic marker (HR, 18.00; 95% CI, 5.78 to 56.05; P < .001, Table 3).
Table 2.
Genes and Probe Sets That Constitute the 15-Gene Signature
| Gene Symbol | Probe Set | Gene Title |
|---|---|---|
| ATP1B1 | 201243_s_at | ATPase, Na+/K+ transporting, beta 1 polypeptide |
| TRIM14 | 203147_s_at | Tripartite motif-containing 14 |
| FAM64A | 221591_s_at | Family with sequence similarity 64, member A |
| FOSL2 | 218881_s_at | FOS-like antigen 2 |
| HEXIM1 | 202814_s_at | Hexamethylene bis-acetamide inducible 1 |
| MB | 204179_at | Myoglobin |
| L1CAM | 204584_at | L1 cell adhesion molecule |
| UMPS | 202707_at | Uridine monophosphate synthetase |
| EDN3 | 208399_s_at | Endothelin 3 |
| STMN2 | 203001_s_at | Stathmin-like 2 |
| MYT1L | 210016_at | Myelin transcription factor 1-like |
| IKBKAP | 202490_at | Inhibitor of kappa light polypeptide gene enhancer in B-cells, kinase complex-associated protein |
| MLANA | 206426_at | Melan-A |
| MDM2 | 205386_s_at | Mdm2, transformed 3T3 cell double minute 2 |
| ZNF236 | 219171_s_at | Zinc finger protein 236 |
Fig 1.

Disease-specific survival outcome based on the 15-gene signature in the JBR.10 training set. (A) Observation all; (B) observation stage IB; (C) observation stage II. HR, hazard ratio; ACT, adjuvant chemotherapy arm.
Table 3.
Validation of the Independent Prognostic Value of the 15-Gene Signature in Four Other Separate Stage IB-II Patient Cohorts Who Received No Adjuvant Treatment
| Cohort | Tumor Type | Platform | No. | Hazard Ratio* | 95% CI | Adjusted P |
|---|---|---|---|---|---|---|
| Training set | ||||||
| JBR.10 | NSCLC | U133A | 62 | 18.00 | 5.78 to 56.05 | < .001 |
| JBR.10 | NSCLC | RT-qPCR | 62 | 2.29 | 1.06 to 4.94 | .034 |
| Validation sets | ||||||
| DCC | ADC | U133A | 96 | 2.26 | 1.02 to 4.97 | .044 |
| NLCI | NSCLC | 44K | 133 | 2.27 | 1.18 to 4.35 | .014 |
| Duke | NSCLC | U133 + 2 | 48 | 1.96 | 0.87 to 4.42 | .11 |
| UM-SQ | SQC | U133A | 79 | 3.57 | 1.48 to 8.58 | .005 |
| JBR.10† | NSCLC | RT-qPCR | 19 | 7.65 | 0.85 to 69.04 | .037 |
Abbreviations: NSCLC, non–small-cell lung cancer; U133A, Affymetrix U133A chip; RT-qPCR, quantitative reverse-transcriptase polymerase chain reaction; DCC, Director's Challenge Consortium adenocarcinoma data set; ADC, adenocarcinoma; NLCI, Netherlands Cancer Institute; 44K, Agilent 44K gene expression array; Duke, Duke University; U133 + 2, Affymetrix U133 plus2 chip; UM-SQ, University of Michigan, squamous cell carcinoma data set; SQC, squamous cell carcinoma.
HR compares the overall survival of the high-risk (poor prognosis) patient group to that of the low-risk (good prognosis) group, after adjustment for tumor histologic subtype, stage, age, and sex.
Values were not adjusted for clinical factors due to small sample size. The model was not adjusted for histology for UM-SQ. Since the NLCI data set did not contain information on sex this covariate was not included in the model.
Validation of Prognostic Signature
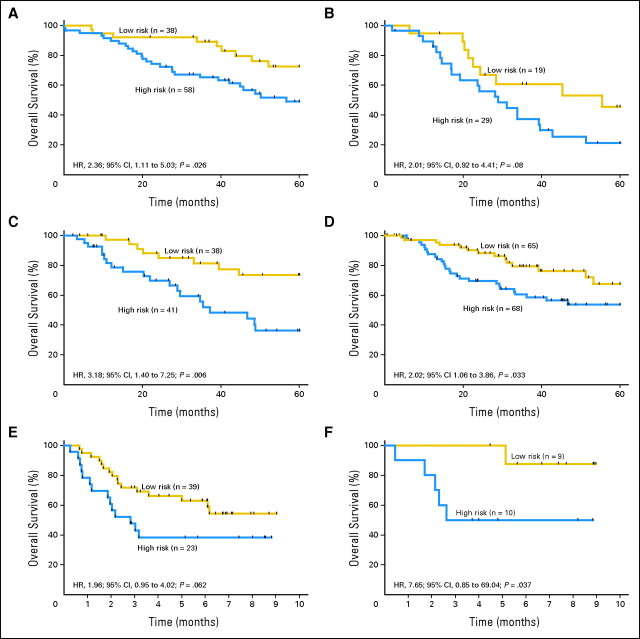
The 15-gene signature was tested for its prognostic significance in four independent published microarray data sets containing patients with completely resected stage IB to II NSCLC who had not received any adjuvant therapy (total n = 356). The risk score was the four PCs weighted by the coefficients of the four PCs derived from the training set, where the four PCs of validation set were derived using the same rotation matrix from the training set (Data Supplement). When the risk score was dichotomized at −0.1, the median of the risk score from the training set (Data Supplement), our 15-gene signature classified samples into low- and high-risk groups, respectively in 38 and 58 of 96 ADC patients from DCC (P = .026, Fig 2A); 19 and 29 of 48 NSCLC Duke University patients (P = .08, Fig 2B); and 38 and 41of 79 University of Michigan patients with squamous cancer (P = .006, Fig 2C). In addition, cross-platform validation of the signature was achieved in 133 NLCI patients profiled using the Agilent 44K platform, classifying 65 and 68 patients into low- and high-risk groups, respectively with significantly different survival (P = .033, Fig 2D). Moreover, multivariate analysis demonstrated that the signature was an independent prognostic factor in these four validation data sets after adjusting for other prognostic factors (DCC: HR, 2.26; 95% CI, 1.02 to 4.97; P = .044; NLCI: HR, 2.27; 95% CI, 1.18 to 4.35; P = .014; Duke: HR, 1.96; 95% CI, 0.9 to 4.4, P = .11; University of Michigan patients with squamous cancer: HR, 3.57; 95% CI, 1.48 to 8.58; P = .005; Table 3).
Fig 2.
In silico and quantitative reverse-transcriptase polymerase chain reaction (RT-qPCR) validation of the signature in stage IB to II patients who received no adjuvant therapy. (A) Director's Challenge Consortium adenocarcinoma data set; (B) Duke University data set; (C) University of Michigan squamous cancer data set; (D) Netherlands Cancer Institute data set; (E) observation with RT-qPCR; (F) observation with RT-qPCR with additional samples. HR, unadjusted hazard ratio.
We employed RT-qPCR to verify the 15-gene signature risk group classifying algorithm in our 133 cases that had been microarray profiled, and to validate further in another 30 JBR.10 cases that were not profiled by microarray (Data Supplement). For 62 OBS of 133 microarray-profiled patients, qPCR result demonstrated significant difference in survival of high- and low-risks patients (HR, 1.96; 95% CI, 0.95 to 4.02; P = .062; Fig 2E). Multivariate analysis showed that the signature remained an independent prognostic factor (HR, 2.29; 95% CI, 1.06 to 4.94; P = .034; Table 3). In the 30 additional JBR.10 cases that were not profiled by microarray, 19 were OBS patients. The signature classified nine as low-risk and 10 as high-risk patients with significantly different survival (HR, 7.65; 95% CI, 0.85 to 69.04; P = .037 by log-rank test; P = .07 by Wald's test; Fig 2F and Table 3).
Predictive Value of the Signature for Adjuvant Chemotherapy Benefit
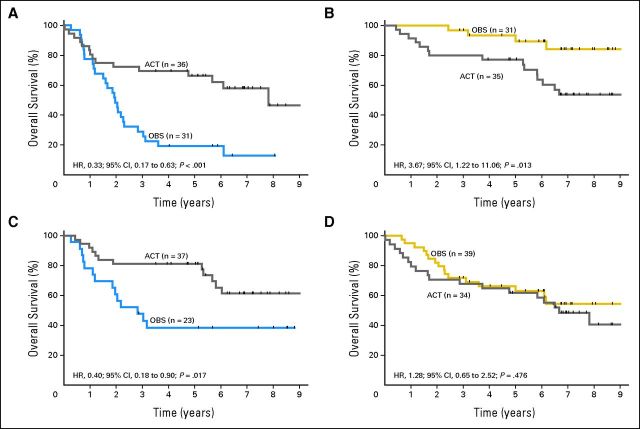
The signature classified 35 and 36 of 71 patients in the ACT arm into low- and high-risk groups, respectively. Comparison of survival between these two subgroups showed that there was no significant difference (HR, 1.15; 95% CI, 0.56 to 2.37; P = .694, Data Supplement). However, ACT significantly prolonged survival of high-risk patients (HR, 0.33; 95% CI, 0.17 to 0.63; P = .0008; Fig 3A), but was not beneficial and potentially even detrimental to low-risk patients (HR, 3.67; 95% CI, 1.22 to 11.06; P = .021; Fig 3B). The interaction between risk group and ACT was highly significant (P = .001). Similar differential treatment effects between high-risk and low-risk groups were observed in stage IB and II (Data Supplement). Subsequent qPCR verification confirmed that only high-risk patients benefited from ACT (HR, 0.40; 95% CI, 0.18 to 0.87; P = .017; Fig 3C) with no benefit from ACT in low-risk (HR, 1.28; 95% CI, 0.65 to 2.52; P = .476; Fig 3D). Multivariate analysis confirmed the signature by qPCR as predictive (Interaction P = .025).
Fig 3.
Predictive effect of the signature to adjuvant chemotherapy. Only high-risk group benefits from adjuvant chemotherapy. (A) High risk (microarray); (B) low risk (microarray); (C) high risk quantitative reverse-transcriptase polymerase chain reaction (RT-qPCR); (D) low risk RT-qPCR.
For the additional 30 JBR.10 samples, 11 patients received chemotherapy, with four patients classified in low-risk group and seven in high-risk group. Due to the small number, no conclusion could be reached. However, for 17 of 30 patients classified in high-risk group (10 OBS, seven ACT), the risk of death was reduced by 46% for those who received ACT (HR, 0.54; 95% CI, 0.10 to 2.80; P = .452).
DISCUSSION
To our knowledge, we present the first NSCLC prognostic gene expression signature generated from microarray studies using samples collected prospectively in a randomized phase III ACT trial. The samples from the untreated control group led to the identification of a stage independent 15-gene signature that separated the cohort into good and poor prognosis patients (adjusted HR, 18.00; P < .001). Our signature was validated for its prognostic significance in independent microarray data sets of 96 patients with stage IB to II adenocarcinoma15 and 79 patients with squamous cancer.9 Further, cross-platform validation of the signature was demonstrated in 133 NLCI patients profiled by the Agilent array,20 and showed a trend of similar magnitude in a smaller Duke data set (n = 48).8 Importantly, the 15-gene signature and its prognostic value also were verified by RT-qPCR both in the original 133 samples profiled by microarray and in 30 additional JBR.10 samples not profiled by microarray.
ACT for resected NSCLC was not considered standard until recently, when the results of several trials, including JBR.10 became available.1,2 To date, there is no laboratory or clinical marker other than stage that can identify patients who are likely to benefit from ACT. Therefore, this is the first study, to our knowledge, that has shown that a prognostic microarray signature can also be predictive of benefit from ACT. Patients identified as high-risk benefited significantly (HR, 0.33; P = .0008). In contrast, low-risk patients did not benefit from ACT (HR, 3.67; P = .021), and the interaction P value was highly significant (P < .001). As no other microarray studies performed using tumor samples from randomized trials with treated and untreated patients are available we could not validate the predictive value of our signature, either in the data sets used to validate its prognostic strength or in other data sets. We realize, therefore, that while promising, these results cannot be considered conclusive, and that they must be validated prospectively in future trials.
The benefit of ACT in stage IB remains controversial.1,3 We showed that stage IB patients also could be separated into low-risk and high-risk groups using our signature (HR, 13.22; 95% CI, 2.86 to 62.11; P < .001). Furthermore, the survival of high-risk IB patients was greatly improved with ACT (HR, 0.44; 95% CI, 0.18 to 1.09; P = .07), whereas low-risk patients had no survival benefit when treated with chemotherapy. Although JBR.10 did not demonstrate benefit from ACT in stage IB overall,1 this study suggests that a subset of patients within stage IB may have the potential to benefit from adjuvant therapy. The Lung Adjuvant Cisplatin Evaluation meta-analysis3 reported a small but clinically and statistically nonsignificant benefit from chemotherapy in stage IB (HR, 0.93; 95% CI, 0.78 to 1.10). It is possible that this modest benefit arose almost entirely from a small subset of patients with a poorer prognosis, who potentially could have been identified by our gene signature.
The current practice is to treat all stage II patients with ACT. Our signature identified stage II patients who had a good prognosis and who did not appear to benefit from chemotherapy or potentially could be affected adversely by ACT (HR, 2.93; 95% CI, 0.63 to 13.57; P = .15).
As there are no other frozen tumor banks associated with a clinical trial containing patients randomly assigned to treatment and no treatment, we attempted to evaluate the predictive value of previously published prognostic signatures when applied to treated and untreated patients in the JBR.10 data set. We were able to replicate eight of these signatures (reviewed in Zhu21) in JBR.10 cases.6,12,13,15,22–25 Only the six-gene signature identified by Boutros22 was significant in JBR.10 OBS patients. This signature also showed that only high-risk patients benefited from chemotherapy in JBR.10 (HR, 0.21; 95% CI, 0.05 to 0.84; P = .027; interaction P = .0323; Data Supplement). A significant beneficial effect of ACT in the high-risk group was also achieved by the three-gene signature of Lau13 (HR, 0.48; 95% CI, 0.23 to 1.00; P = .05; Data Supplement), but the interaction with low-risk group did not reach significance. However, the signatures reported by Chen,6 Gordon,23 Hsu,24 Shedden (method E in DCC),15 Skrzypski,25 and Sun12 were neither significantly prognostic when applied to the observation arm of JBR.10, nor predictive of benefit from adjuvant chemotherapy. The results demonstrate the significant challenges faced in validating prognostic gene expression signatures. More importantly, the JBR.10 microarray data set now available with this report may be used for testing future predictive markers.
Functional annotation for the 15 genes reveals properties that may elucidate their role in lung cancer biology. Using annotation from Gene Ontology26 and KEGG pathways,27 cellular localization and functions that predominate among these genes (six of 15) are nuclear proteins or transcription regulators MDM2, ZNF236, FOSL2, HEXIM1, MYT1L and IKBKAP. MDM2 is an E3 ubiquitin ligase that targets p53 for proteasomal degradation,28,29 and may represses transcriptional activity of p53.30–32 MDM2 is amplified in 6.2% of lung adenocarcinoma33,34 and overexpression was associated with poor prognosis in NSCLC.35 MDM2 amplification also appears mutually exclusive with p53 mutation33,34 further demonstrating the importance of MDM2 in the p53 pathway. This supports the notion that a minimal signature would include genes that regulate additional key genes and pathways, rather than belonging to a single dominant process or pathway. The second subset of genes includes MLANA, ATP1B1, L1CAM, andSTMN2, which encode for transmembrane- or membrane-associated proteins, potentially involved in signaling pathways.36 Finally, ATP1B1 and UMPS are involved in purine and pyramidine metabolism, respectively, suggesting dependency of NSCLC on these pathways.
Direct comparison of the 15 genes in our signature with those in the published signatures we evaluated (Data Supplement) shows no overlap but they are connected at the protein level (Data Supplement; eg, ATP1B1 and HEXIM1 with epidermal growth factor receptor25 and MDM2 with hypoxia-inducible factor 1α),13 suggesting they may share common signaling networks.
To our knowledge, this is the first report of a gene signature that is both independently prognostic in patients with untreated NSCLC, and predictive of survival benefit from ACT. With this signature, we have provided the algorithm to classify individual patients. The predictive role of our signature should be tested in prospectively planned adjuvant chemotherapy trials.
Supplementary Material
Footnotes
See accompanying editorial on page 4404
Supported by grants from the Canadian Cancer Society (clinical trial, microarray data and correlative analysis), the US National Cancer Institute (microarray studies), Grants No. 12301 and 203383 from the Canada Foundation for Innovation (lab infrastructure), and the Canada Research Chair Program (I.J.). This research was funded in part by the Ontario Ministry of Health and Long Term Care. We thank GlaxoSmithKline for supporting the establishment of the JBR.10 frozen tumor bank.
The views expressed do not necessarily reflect those of the Ontario Ministry of Health and Long-Term Care.
Presented in part at the 44th Annual Meeting of the American Society of Clinical Oncology, Chicago, IL, May 30-June 3, 2008.
Authors' disclosures of potential conflicts of interest and author contributions are found at the end of this article.
AUTHORS' DISCLOSURES OF POTENTIAL CONFLICTS OF INTEREST
Although all authors completed the disclosure declaration, the following author(s) indicated a financial or other interest that is relevant to the subject matter under consideration in this article. Certain relationships marked with a “U” are those for which no compensation was received; those relationships marked with a “C” were compensated. For a detailed description of the disclosure categories, or for more information about ASCO's conflict of interest policy, please refer to the Author Disclosure Declaration and the Disclosures of Potential Conflicts of Interest section in Information for Contributors.
Employment or Leadership Position: None Consultant or Advisory Role: Roman K. Thomas, Sequenom (C), Roche (C) Stock Ownership: None Honoraria: Roman K. Thomas, Infinity, Boehringer Ingelheim Research Funding: Roman K. Thomas, AstraZeneca; Sandy Der, Med BioGene; Lesley Seymour, GlaxoSmithKline (funding to National Cancer Institute of Canada Clinical Trials Group for JBR10); Frances A. Shepherd, Med BioGene; Ming-Sound Tsao, Med BioGene Expert Testimony: None Other Remuneration: None
AUTHOR CONTRIBUTIONS
Conception and design: Chang-Qi Zhu, Keyue Ding, Matthew Meyerson, Melania Pintilie, Lesley Seymour, Frances A. Shepherd, Ming-Sound Tsao
Financial support: Matthew Meyerson, Sandy Der, Frances A. Shepherd, Ming-Sound Tsao
Administrative support: Lesley Seymour, Frances A. Shepherd, Ming-Sound Tsao
Provision of study materials or patients: Matthew Meyerson, Lesley Seymour, Frances A. Shepherd, Ming-Sound Tsao
Collection and assembly of data: Chang-Qi Zhu, Keyue Ding, Dan Strumpf, Barbara A. Weir, Matthew Meyerson, Nathan Pennell, Roman K. Thomas, Katsuhiko Naoki, Christine Ladd-Acosta, Ni Liu, Lesley Seymour, Frances A. Shepherd, Ming-Sound Tsao
Data analysis and interpretation: Chang-Qi Zhu, Keyue Ding, Dan Strumpf, Melania Pintilie, Sandy Der, Igor Jurisica, Frances A. Shepherd, Ming-Sound Tsao
Manuscript writing: Chang-Qi Zhu, Keyue Ding, Dan Strumpf, Melania Pintilie, Sandy Der, Igor Jurisica, Frances A. Shepherd, Ming-Sound Tsao
Final approval of manuscript: Chang-Qi Zhu, Keyue Ding, Dan Strumpf, Barbara A. Weir, Matthew Meyerson, Nathan Pennell, Roman K. Thomas, Katsuhiko Naoki, Christine Ladd-Acosta, Ni Liu, Melania Pintilie, Sandy Der, Lesley Seymour, Igor Jurisica, Frances A. Shepherd, Ming-Sound Tsao
REFERENCES
- 1.Winton T, Livingston R, Johnson D, et al. Vinorelbine plus cisplatin vs. observation in resected non-small-cell lung cancer. N Engl J Med. 2005;352:2589–2597. doi: 10.1056/NEJMoa043623. [DOI] [PubMed] [Google Scholar]
- 2.Pisters KM, Evans WK, Azzoli CG, et al. Cancer Care Ontario and American Society of Clinical Oncology adjuvant chemotherapy and adjuvant radiation therapy for stages I-IIIA resectable non small-cell lung cancer guideline. J Clin Oncol. 2007;25:5506–5518. doi: 10.1200/JCO.2007.14.1226. [DOI] [PubMed] [Google Scholar]
- 3.Pignon JP, Tribodet H, Scagliotti GV, et al. Lung adjuvant cisplatin evaluation: A pooled analysis by the LACE Collaborative Group. J Clin Oncol. 2008;26:3552–3559. doi: 10.1200/JCO.2007.13.9030. [DOI] [PubMed] [Google Scholar]
- 4.Nesbitt JC, Putnam JB, Jr, Walsh GL, et al. Survival in early-stage non-small cell lung cancer. Ann Thorac Surg. 1995;60:466–472. doi: 10.1016/0003-4975(95)00169-l. [DOI] [PubMed] [Google Scholar]
- 5.Beer DG, Kardia SL, Huang CC, et al. Gene-expression profiles predict survival of patients with lung adenocarcinoma. Nat Med. 2002;8:816–824. doi: 10.1038/nm733. [DOI] [PubMed] [Google Scholar]
- 6.Chen HY, Yu SL, Chen CH, et al. A five-gene signature and clinical outcome in non-small-cell lung cancer. N Engl J Med. 2007;356:11–20. doi: 10.1056/NEJMoa060096. [DOI] [PubMed] [Google Scholar]
- 7.Lu Y, Lemon W, Liu PY, et al. A gene expression signature predicts survival of patients with stage I non-small cell lung cancer. PLoS Med. 2006;3:e467. doi: 10.1371/journal.pmed.0030467. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Potti A, Mukherjee S, Petersen R, et al. A genomic strategy to refine prognosis in early-stage non-small-cell lung cancer. N Engl J Med. 2006;355:570–580. doi: 10.1056/NEJMoa060467. [DOI] [PubMed] [Google Scholar]
- 9.Raponi M, Zhang Y, Yu J, et al. Gene expression signatures for predicting prognosis of squamous cell and adenocarcinomas of the lung. Cancer Res. 2006;66:7466–7472. doi: 10.1158/0008-5472.CAN-06-1191. [DOI] [PubMed] [Google Scholar]
- 10.Wigle DA, Jurisica I, Radulovich N, et al. Molecular profiling of non-small cell lung cancer and correlation with disease-free survival. Cancer Res. 2002;62:3005–3008. [PubMed] [Google Scholar]
- 11.Bianchi F, Nuciforo P, Vecchi M, et al. Survival prediction of stage I lung adenocarcinomas by expression of 10 genes. J Clin Invest. 2007;117:3436–3444. doi: 10.1172/JCI32007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Sun Z, Wigle DA, Yang P. Non-overlapping and non-cell-type-specific gene expression signatures predict lung cancer survival. J Clin Oncol. 2008;26:877–883. doi: 10.1200/JCO.2007.13.1516. [DOI] [PubMed] [Google Scholar]
- 13.Lau SK, Boutros PC, Pintilie M, et al. Three-gene prognostic classifier for early-stage non small-cell lung cancer. J Clin Oncol. 2007;25:5562–5569. doi: 10.1200/JCO.2007.12.0352. [DOI] [PubMed] [Google Scholar]
- 14.Bhattacharjee A, Richards WG, Staunton J, et al. Classification of human lung carcinomas by mRNA expression profiling reveals distinct adenocarcinoma subclasses. Proc Natl Acad Sci U S A. 2001;98:13790–13795. doi: 10.1073/pnas.191502998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Shedden K, Taylor JM, Enkemann SA, et al. Gene expression-based survival prediction in lung adenocarcinoma: A multi-site, blinded validation study. Nat Med. 2008 doi: 10.1038/nm.1790. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Bolstad BM, Irizarry RA, Astrand M, et al. A comparison of normalization methods for high density oligonucleotide array data based on variance and bias. Bioinformatics. 2003;19:185–193. doi: 10.1093/bioinformatics/19.2.185. [DOI] [PubMed] [Google Scholar]
- 17.Affymetrix. Santa Clara, CA: Affymetrix Inc; 2006. Transcript assignment for NetAffxTM annotation, Affymetrix GeneChip IVT array whitepaper collection 2006. http://media.affymetrix.com/support/technical/whitepapers/netaffxannot_whitepaper.pdf. [Google Scholar]
- 18.Kent J, O'Quigley J. Measures of dependence for censored survival data. Biometrika. 1988;75:525–534. [Google Scholar]
- 19.Heinzl H. Using SAS to calculate the Kent and O'Quigley measure of dependence for Cox proportional hazards regression model. Comput Methods Programs Biomed. 2000;63:71–76. doi: 10.1016/s0169-2607(00)00073-0. [DOI] [PubMed] [Google Scholar]
- 20.Roepman P, Jassem J, Smit EF, et al. An immune response enriched 72-gene prognostic profile for early-stage non-small-cell lung cancer. Clin Cancer Res. 2009;15:284–290. doi: 10.1158/1078-0432.CCR-08-1258. [DOI] [PubMed] [Google Scholar]
- 21.Zhu CQ, Pintilie M, John T, et al. Understanding prognostic gene expression signatures in lung cancer. Clin Lung Cancer. 2009;10:331–340. doi: 10.3816/CLC.2009.n.045. [DOI] [PubMed] [Google Scholar]
- 22.Boutros PC, Lau SK, Pintilie M, et al. Prognostic gene signatures for non-small-cell lung cancer. Proc Natl Acad Sci U S A. 2009;106:2824–2828. doi: 10.1073/pnas.0809444106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Gordon GJ, Richards WG, Sugarbaker DJ, et al. A prognostic test for adenocarcinoma of the lung from gene expression profiling data. Cancer Epidemiol Biomarkers Prev. 2003;12:905–910. [PubMed] [Google Scholar]
- 24.Hsu YC, Yuan S, Chen HY, et al. A four-gene signature from NCI-60 cell line for survival prediction in non-small cell lung cancer. Clin Cancer Res. 2009;15:7309–7315. doi: 10.1158/1078-0432.CCR-09-1572. [DOI] [PubMed] [Google Scholar]
- 25.Skrzypski M, Jassem E, Taron M, et al. Three-gene expression signature predicts survival in early-stage squamous cell carcinoma of the lung. Clin Cancer Res. 2008;14:4794–4799. doi: 10.1158/1078-0432.CCR-08-0576. [DOI] [PubMed] [Google Scholar]
- 26.Ashburner M, Ball CA, Blake JA, et al. Gene ontology: Tool for the unification of biology—The Gene Ontology Consortium. Nat Genet. 2000;25:25–29. doi: 10.1038/75556. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Kanehisa M, Araki M, Goto S, et al. KEGG for linking genomes to life and the environment. Nucleic Acids Res. 2008;36:D480–484. doi: 10.1093/nar/gkm882. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Haupt Y, Maya R, Kazaz A, et al. Mdm2 promotes the rapid degradation of p53. Nature. 1997;387:296–299. doi: 10.1038/387296a0. [DOI] [PubMed] [Google Scholar]
- 29.Kubbutat MH, Jones SN, Vousden KH. Regulation of p53 stability by Mdm2. Nature. 1997;387:299–303. doi: 10.1038/387299a0. [DOI] [PubMed] [Google Scholar]
- 30.Oliner JD, Pietenpol JA, Thiagalingam S, et al. Oncoprotein MDM2 conceals the activation domain of tumour suppressor p53. Nature. 1993;362:857–860. doi: 10.1038/362857a0. [DOI] [PubMed] [Google Scholar]
- 31.Momand J, Zambetti GP, Olson DC, et al. The mdm-2 oncogene product forms a complex with the p53 protein and inhibits p53-mediated transactivation. Cell. 1992;69:1237–1245. doi: 10.1016/0092-8674(92)90644-r. [DOI] [PubMed] [Google Scholar]
- 32.Xirodimas DP, Saville MK, Bourdon JC, et al. Mdm2-mediated NEDD8 conjugation of p53 inhibits its transcriptional activity. Cell. 2004;118:83–97. doi: 10.1016/j.cell.2004.06.016. [DOI] [PubMed] [Google Scholar]
- 33.Ding L, Getz G, Wheeler DA, et al. Somatic mutations affect key pathways in lung adenocarcinoma. Nature. 2008;455:1069–1075. doi: 10.1038/nature07423. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Weir BA, Woo MS, Getz G, et al. Characterizing the cancer genome in lung adenocarcinoma. Nature. 2007;450:893–898. doi: 10.1038/nature06358. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Higashiyama M, Doi O, Kodama K, et al. MDM2 gene amplification and expression in non-small-cell lung cancer: Immunohistochemical expression of its protein is a favourable prognostic marker in patients without p53 protein accumulation. Br J Cancer. 1997;75:1302–1308. doi: 10.1038/bjc.1997.221. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Lee HS, Lee DC, Park MH, et al. STMN2 is a novel target of beta-catenin/TCF-mediated transcription in human hepatoma cells. Biochem Biophys Res Commun. 2006;345:1059–1067. doi: 10.1016/j.bbrc.2006.05.017. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.