Abstract
Two commonly used culture systems in hepatic tissue engineering are the collagen sandwich (CS) and monolayers of cells. In this study, genome-wide gene expression profiles of primary hepatocytes were measured over an 8-day period for each cell culture system using Affymetrix GeneChips and compared via gene set enrichment analysis to elicit biologically meaningful information at the level of gene sets. Our results demonstrate that gene expression in hepatocytes in CS cultures steadily and comprehensively diverges from that in monolayer cultures. Gene sets up-regulated in CS cultures include several associated with liver metabolic and synthesis functions, such as metabolism of lipids, amino acids, carbohydrates, and alcohol, and synthesis of bile acids. Monooxygenases such as Cytochrome-P450 enzymes do not show any change between the culture systems after 1 day, but exhibit significant up-regulation in CS cultures after 3 days in comparison to hepatocyte monolayers. These data provide insights into the up- and down-regulation of several liver-critical gene sets and their subsequent effects on liver-specific functions. These results provide a baseline for further explorations into the systems biology of engineered liver mimics.
Introduction
As one of the important organs in our bodies, the liver performs many essential functions such as metabolism, synthesis, secretion, and detoxification.1 Hepatocytes are the principal cells in the liver, comprising over 80% of its mass. Hepatocytes perform several characteristic functions of the liver, such as lipid metabolism, glucose homeostasis, regulation of urea, production of plasma proteins, alcohol clearance, and biotransformation of xenobiotics.1 In hepatic tissue engineering, two widely used culture systems are hepatocyte monolayers (HMs) and the collagen sandwich (CS).2,3 In HMs, hepatocytes are cultured on a single-collagen gel. Such cells progressively lose their phenotypic characteristics over time. In CS cultures, hepatocytes are maintained between two collagen gels and remain stable over extended periods.4,5 Studies have indicated that CS cultures exhibit the preservation of differentiated functions, including secretion of urea, expression of plasma proteins such as albumin and fibrinogen, polygonal morphology, the presence of bile canaliculi, as well as the synthesis of gap junction and tight junction proteins.4,5 Although morphological and physiological characteristics of hepatocytes in CS cultures have been studied extensively, comprehensive evaluations of temporal genome-wide gene expression programs in these culture systems have not been reported. Global gene expression of human hepatocellular carcinoma cells (HepG2) in monolayer and spheroidal cultures revealed up-regulated metabolic functions in spheroids but not in monolayer cultures.6 Since these data were taken at a single time point, they did not reveal temporal variations. Another study that monitored temporal gene expression in HMs cultured over a 3-day period revealed the down-regulation of cytochrome-P450 expression.7 However, neither did this study investigate longer time points nor did it compare monolayers to other, more stable culture conditions. DNA microarray measurements have also been used to study specific pathways through which toxicity was conferred in human hepatoblastoma cells8 and to understand the effects of nonparenchymal cells in 2D cocultures of hepatocytes with fibroblasts or sinusoidal endothelial cells.9,10
We hypothesized that the enhanced in vivo liver-like phenotypes in CS cultures were a result of the underlying differences in the transcriptional program between hepatocytes cultured in CS and HMs. Accordingly, genome-wide gene expression profiles of primary hepatocytes were measured at four different time points over an 8-day period for each cell culture system using Affymetrix GeneChips. Among the wide range of techniques that are available to analyze DNA microarray data, a method was desired that would summarize, at the level of predefined biological pathways, the differences between the culture conditions at each time point. Gene set enrichment analysis (GSEA)11 was selected since it satisfies this criterion. GSEA is one among a family of techniques that can summarize differential expression at the level of gene sets.12 GSEA is widely used, generates detailed information on the results, and has shown very good performance in a comparison of methods that compute enrichment at the level of gene sets.13 Further, GSEA has been used to identify pathways involved in liver toxicity in human hepatoblastoma cells.8 GSEA is designed to identify predefined gene sets that are differentially expressed in a treatment and a control. All the genes expressed on each gene chip are ranked based upon their differential expression in CS and HM cultures. Therefore, a gene set could be important if its members are clustered within the ranked gene list. GSEA measures the statistical significance of the distribution of ranks within the gene set against the background of the ranks of all the genes.
Over the 8-day culture period, the gene expression program of hepatocytes in CS cultures monotonically diverged from cells cultured as a monolayer. Gene sets that were up-regulated to a statistically significant extent in CS cultures included those associated with liver-specific functions such as bile acid synthesis and lipid, amino acid, carbohydrate, and alcohol metabolism. Nuclear receptors, which play a key role in controlling the transcriptional activation of target proteins, were up-regulated in CS cultures on day 1 in culture. Sets containing genes whose expression is mediated by nuclear receptors were up-regulated in CS systems after 1 day. Gene sets related to xenobiotic metabolism and monoxygenase activity were not differentially expressed after 1 or 2 days, but showed highly significant up-regulation after 3 days, suggesting a recovery in expression of the genes in these sets. Numerous gene sets related to the cell cycle were down-regulated, suggesting that the cell cycle was arrested in hepatocytes maintained in CS culture systems in comparison to HMs. These findings recapitulated well-known aspects of liver function, thereby suggesting that DNA microarrays are a powerful tool for shedding light on the transcriptional signatures that underlie differences between these two culture systems. The DNA microarray data generated in this study are available at NCBI's Gene Expression Omnibus under accession number GSE20659 at www.ncbi.nlm.nih.gov/geo/query/acc.cgi?acc=GSE20659. All our results are available at the following supplementary Web site: http://bioinformatics.cs.vt.edu/∼murali/supplements/2010-kim-tissue-engineering.
Materials and Methods
Materials
Dulbecco's modified Eagle's medium (DMEM) containing 4.5 g/L glucose, phosphate-buffered saline, penicillin, streptomycin, and trypsin–ethylenediaminetetraacetic acid were obtained from Invitrogen Life Technologies. Type IV collagenase, HEPES [4-(2-hydroxyethyl) piperazine-1-ethanesulfonic acid], glucagon, and hydrocortisone were obtained from Sigma-Aldrich. Unless otherwise noted, all chemicals were used as received from Fisher Scientific.
Hepatocyte isolation and culture
Primary rat hepatocytes were harvested from female Lewis rats (Harlan) that weighed between 170 and 200 g. Animal care and surgical procedures were conducted as per procedures approved by Virginia Polytechnic Institute and State University's Institutional Animal Care and Use Committee. A two-step in situ collagenase perfusion method was utilized to excise the liver.4,5 Briefly, animals were anesthetized with 3 L/min of a gas mixture of 3% (v/v) isofluorane/97% oxygen (Veterinary Anesthesia Systems Co.). The liver was perfused through the portal vein with Krebs Ringer Buffer (7.13 g/L sodium chloride, 2.1 g/L sodium bicarbonate, 1 g/L glucose, 4.76 g/L HEPES, and 0.42 g/L potassium chloride) that contained 1 mM ethylenediaminetetraacetic acid, followed by serial perfusion with a 0.075% w/v and a 0.1% w/v collagenase (Type IV; Sigma-Aldrich) in Krebs Ringer Buffer containing 5 mM calcium chloride. Cell suspensions were filtered through nylon meshes with porosity ranging from 250 to 62 μm (Small Parts, Inc.). Hepatocytes were separated using a Percoll (Sigma-Aldrich) density centrifugation technique. Cell viability was determined by trypan blue exclusion. Hepatocytes were cultured on collagen-coated 6-well sterile tissue culture plates (Becton Dickinson Labware) and were maintained in a culture medium that consisted of DMEM supplemented with 10% heat-inactivated fetal bovine serum (Hyclone), 200 U/mL penicillin, 200 μg/mL streptomycin, 20 ng/mL epidermal growth factor (BD Biosciences), 0.5 U/mL insulin (USP), 14 ng/mL glucagon, and 7.5 μg/mL hydrocortisone. A collagen gelling solution was prepared by mixing nine parts of type I collagen (BD Biosciences) solution and one part of 10× DMEM. Sterile 6-well tissue culture plates were coated with 0.5 mL of the gelling solution and incubated at 37°C for 1 h to promote gel formation. Isolated hepatocytes were suspended in the hepatocyte culture medium at a concentration of 1 × 106 cells/mL and seeded on the collagen-coated wells at a density of 1 million cells/well. CS cultures were formed by the deposition of a second layer of collagen 1 day after the hepatocytes were seeded.4,5 Hepatocytes maintained in stable CS and in unstable confluent HM cultures served as positive and negative controls, respectively. Hepatocyte cultures were maintained at 37°C in a humidified gas mixture of 90% air/10% CO2. The culture medium was replaced every 24 h.
RNA extraction and gene chip hybridization
Primary rat hepatocytes cultured in CS and HM cultures were maintained for an 8-day culture period. The samples were analyzed at four time points: days 1, 2, 3, and 8 after deposition of the second layer of collagen gel on hepatocytes. Total RNA was extracted and purified from cells for each culture system using an RNeasy mini kit (Qiagen) following the manufacturer's protocol. Isolated RNA samples in triplicate at each time point were labeled according to the Affymetrix Standard Target labeling process, hybridized to the GeneChip Rat Genome 230 2.0 array (Affymetrix), and scanned as described by the manufacturer. Complementary RNA (cRNA) synthesis, hybridization, and scanning were performed at the Virginia Bioinformatics Institute Core Laboratory facility as follows. Briefly, total RNA was converted into double-stranded complementary DNA using a T7-oligo (dT) primer (5′-GGCCAGTGAATTGTAATACGACTCACTATAGGGAGGCGG–(dT)24–3′) and reverse transcription. Synthesized cDNA was converted into biotinylated cRNA by transcription using T7 RNA polymerase. Randomly fragmented cRNA was hybridized to GeneChip and the arrays were washed and stained according to Affymetrix's protocols. The arrays were scanned using an Affymetrix 7G scanner.
Microarray data analysis
The Bioconductor package14 was used to perform initial statistical analysis of the DNA microarray data. The data from 24 chips (2 culture conditions × 4 time points × 3 replicates) were normalized using the Robust Multichip Average method for further analysis. The affylmGUI interface to linear models for microarray data (LIMMA)15 was used to perform differential gene expression analysis for the contrasts shown in Table 1. Specifically, for each contrast, LIMMA was used to compute a p-value for each probe set that indicated the statistical significance of the difference of the expression levels of that probe set between the two conditions in the contrast.
Table 1.
Contrasts Analyzed Using Gene Set Enrichment Analysis
| Contrast name | Treatment | Control |
|---|---|---|
| CS vs. monolayer cultures | ||
| CS vs. HM 1 day | CS 1 day | HM 1 day |
| CS vs. HM 2 days | CS 2 days | HM 2 days |
| CS vs. HM 3 days | CS 3 days | HM 3 days |
| CS vs. HM 8 days | CS 8 days | HM 8 days |
| Within CS | ||
| CS 8 days vs. 1 day | CS 8 days | CS 1 day |
| CS 8 days vs. 2 days | CS 8 days | CS 2 days |
| CS 8 days vs. 3 days | CS 8 days | CS 3 days |
CS, collagen sandwich; HM, hepatocyte monolayer.
Gene set enrichment analysis
The normalized gene expression data were analyzed using GSEA.11 Given replicate gene expression measurements for a control phenotype (e.g., HM at day 1) and for a treatment phenotype (e.g., CS at day 1), GSEA starts by ranking all genes by the extent of their differential expression in the two phenotypes. Thus, the lower the rank of a gene, the more up-regulated it is in the treatment than the control. Next, given a gene set of interest (e.g., the genes involved in metabolism of xenobiotics), GSEA uses a modified Kolmogorov–Smirnov test16 to determine if the genes in this set have surprisingly high or low ranks in the list of differentially expressed genes. GSEA computes an enrichment score that summarizes the ranks of genes in the gene set. This score has the following interpretation: the more positive the score, the more up-regulated the genes in the gene set are in the treatment (compared to the control), and the more negative the score, the more down-regulated the genes in that gene set are. Since the size of a gene set may influence its enrichment score, GSEA controls this bias by performing a permutation test and calculating a p-value that represents the statistical significance of the enrichment score. Finally, GSEA converts the p-value into a q-value that measures the false discovery rate, after adjusting for multiple hypothesis testing. Note that the q-value is unsigned but the enrichment score is signed (positive for overall up-regulation and negative for overall down-regulation). We applied GSEA using the following criteria:
Sort genes in decreasing order of the signal-to-noise measure.
Compute p-values using 10,000 permutations of the sample-to-phenotype associations.
Report all gene sets with q-value (false discovery rate) at most 0.2. Note that with this cutoff, we expect one out of five gene sets to be a false discovery.
Results
LIMMA and GSEA were applied to compare the two culture conditions, as shown in Table 1. The first set of four contrasts compared the hepatocyte transcriptional program in CS cultures to that in HMs, at each of the four time-points analyzed. These contrasts were expected to reveal time-dependent differences between these two culture conditions. The second set of three contrasts compared CS samples to each other: 8 days to 1 day, 8 days to 2 days, and 8 days to 3 days. Such contrasts were expected to provide information on how transcriptional programs may vary within CS cultures condition over time.
The transcriptional program in CS cultures steadily and comprehensively diverges from that in HMs
For each of the first four contrasts in Table 1, the number of differentially expressed probe sets was counted after applying different cutoffs on the p-values computed by LIMMA. The first column in Table 2 indicates the p-value cutoff, while each of the other four columns show the number of probe sets whose p-value meets the cutoff specified in each row. An important feature revealed by these data is the monotonic divergence between the transcriptional programs of CS and HM samples over the 8-day culture period. For each cutoff, the number of differentially expressed probe sets increased steadily from day 1 to 8. Further, this trend was maintained even over a variation of four orders of magnitude in the p-value cutoff. On day 8, as many as 6185 probe sets had a p-value of at most 0.01 (2242 had a p-value of at most 10−5). Since, the Affymetrix Rat230_2 GeneChip has 31,099 probe sets, these results suggest widespread transcriptional perturbation in CS cultures compared to HM cultures.
Table 2.
The Number of Differentially Expressed Genes in the Each of the Four Collagen Sandwich Versus Hepatocyte Monolayer Contrasts at Different p-Value Cutoffs
| |
CS vs. HM |
|||
|---|---|---|---|---|
| p-Value cutoff | 1 day | 2 days | 3 days | 8 days |
| 10−5 | 31 | 224 | 1046 | 2242 |
| 0.0001 | 61 | 362 | 1535 | 3092 |
| 0.001 | 118 | 569 | 2277 | 4287 |
| 0.01 | 276 | 1095 | 3497 | 6185 |
| 0.05 | 552 | 1812 | 5134 | 8551 |
Upon the identification of the global trends, GSEA was employed to study patterns of differential expression in specific gene sets. GSEA was applied to the gene expression data obtained through our experiments and to the following gene sets in the Molecular Signature DataBase (MSigDB): 1892 curated gene sets from various sources such as online pathway databases, publications in PubMed, and knowledge of domain experts; 837 motif gene sets containing genes that share a cis-regulatory motif that is conserved across the human, mouse, rat, and dog genomes; and 1454 gene sets corresponding to genes annotated by different Gene Ontology (GO) terms. For each contrast in Table 1 and for each of these gene sets, GSEA was used to compute a q-value.
Gene sets were filtered to those that exhibited a monotonic up-regulation in the CS–HM comparison. Specifically, gene sets were restricted to those whose q-values decreased monotonically from day 1 to 8 and whose enrichment scores were positive in all four CS–HM contrasts (the first four contrasts in Table 1). Gene sets that were monotonically down-regulated in CSs over the 8-day period were also identified (q-values decreasing monotonically and negative enrichment scores in all four CS–HM contrasts). Since MSigDB collates data from several sources, many gene sets in it have high degrees of overlap. When an overlapping group of gene sets is up-regulated or down-regulated in our data, only one gene set per group is discussed below. The complete sets of results are available on our supplementary Web page.
Liver-specific gene sets are up-regulated starting on day 1 or 2 in CS cultures
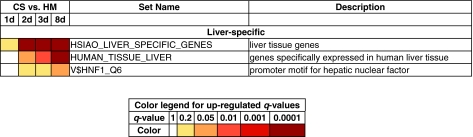
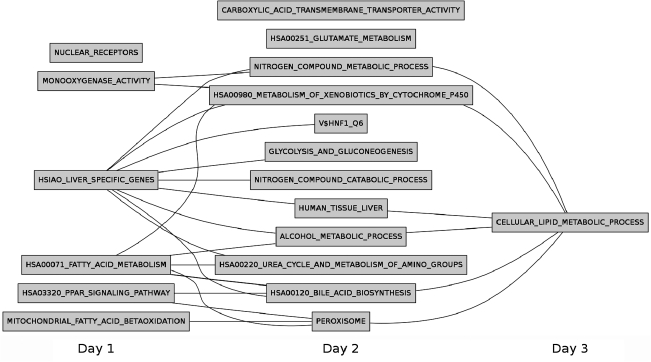
Hsiao et al.17 created a compendium of gene expression in normal human tissues with the goal of defining a reference for basic organ systems biology. They identified 251 genes expressed selectively in the liver, which are included in MSigDB in the HSIAO_LIVER_SPECIFIC_GENES gene set. In a similar study, Su et al.18 profiled gene expression from 91 human and mouse samples across a diverse array of tissues, organs, and cell lines. They identified 37 genes that were expressed specifically in human liver tissue samples; these genes belong to the HUMAN_TISSUE_LIVER gene set. The gene sets HSIAO_LIVER_SPECIFIC_GENES and LIVER_SPECIFIC_GENES were up-regulated significantly at day 1 and 2, respectively. They were monotonically up-regulated on subsequent days. Both these gene sets had insignificant q-values in the CS–CS contrasts, suggesting that liver-specific genes are up-regulated on day 1 in CS cultures, and that they continue to be monotonically up-regulated on subsequent days (Fig. 1). The presence and concentration of albumin is often used as a marker of phenotypic function of in vitro hepatic models.4,5 The albumin gene was expressed in several gene sets such as HSIAO_LIVER_SPECIFIC_GENES and V$HNF1_Q6 that were up-regulated over the 8-day culture period (Fig. 1). The promoter regions of genes in the set V$HNF1_Q6 contain binding sites for hepatic nuclear factor (HNF1), a transcription factor that activates gene expression of albumin.19,20 This gene set has an overlap of 25 genes with the gene set HSIAO_ LIVER_ SPECIFIC_GENES, indicating that HNF1 monotonically up-regulates expression of albumin and other liver-specific genes in CS cultures but not in HMs. These observations support the conclusion that transcriptional programs that have been identified in other datasets to be liver-specific are active through the 8-day period in CS cultures but are not active in HMs.
FIG. 1.
Liver-specific up-regulated gene sets. The bottom table shows the q-value ranges for each color. The color scheme used in this figure is RdYlGn from Color Brewer (http://colorbrewer2.org). CS, collagen sandwich; HM, hepatocyte monolayer. Color images available online at www.liebertonline.com/ten.
Gene sets involved in cholesterol, fatty-acid, alcohol, and carbohydrate metabolism are significantly up-regulated starting on day 1 or 2 in CS cultures
Cholesterol metabolism
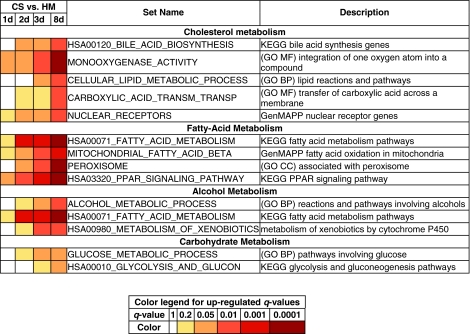
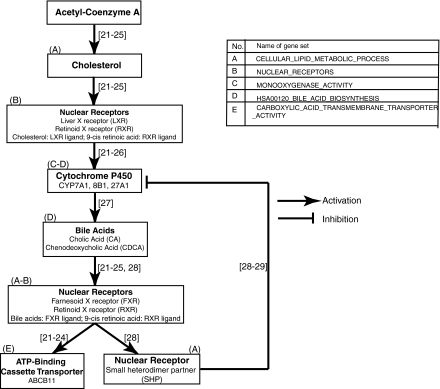
Cholesterol metabolism is an important component of hepatic phenotypic function.1 The trends exhibited by gene sets linked to cholesterol metabolism in our data were investigated. Multiple gene sets involved in cholesterol metabolism were up-regulated in CS cultures compared to HMs. These gene sets include HSA00120_BILE_ACID_BIOSYNTHESIS, MONOOXYGENASE_ACTIVITY, CELLULAR_LIPID_METABOLIC_PROCESS, NUCLEAR_RECEPTORS, and CARBOXYLIC_ACID_TRANSMEMBRANE_TRANSPORTER_ACTIVITY (Fig. 2). Bile acids mediate cholesterol metabolism, and the synthesis of bile acids is initiated through the activity of CYP7A1, CYP8B1, and CYP27A1 enzymes21–23 (Fig. 3). In CS samples, the three CYP enzymes mentioned above are present either in the gene set HSA00120_BILE_ACID_BIOSYNTHESIS or in the gene set MONOOXYGENASE_ACTIVITY, both of which are up-regulated in CS cultures. Gene expression of CYP enzymes is activated by nuclear receptors—specifically, the retinoid X receptor (RXR) and the liver X receptor (LXR).24–27 The gene set NUCLEAR_RECEPTORS, which contains nuclear receptors involved in the activation of hepatic functions, behaved similarly to the liver-specific gene sets discussed earlier: it had insignificant q-values in the CS–CS contrasts, suggesting that nuclear receptors were up-regulated on day 1 in CS cultures and remained up-regulated on subsequent days. The nuclear receptor Farnesoid X receptor (FXR) plays a critical role in liver functioning. FXR is responsible for regulating the concentration of bile acids.21–25,28,29 Bile-acid-mediated activation of FXR leads to the transcriptional activation of the ATP-binding cassette transporter B11 (ABCB11, also known as bile salt export pump), a process that is crucial for cholesterol secretion into the bile canaliculi.27,28 In CS samples, the transcription of ABCB11, present in gene set CARBOXYLIC_ACID_TRANSMEMBRANE_TRANSPORTER_ACTIVITY, was shown to be up-regulated over the culture period in comparison to HMs. This gene set contains genes annotated with the Gene Ontology molecular function that involves the catalysis of the transfer of carboxylic acids from one side of the membrane to the other. These trends and data indicate that genes responsible for the formation, transformation, and transport of bile acids are up-regulated in CS cultures, thereby promoting cholesterol metabolism.
FIG. 2.
Up-regulated gene sets involved in cholesterol, fatty-acid, alcohol, and carbohydrate metabolism. The bottom table shows the q-value ranges for each color. Abbreviations: CARBOXYLIC_ACID_TRANSM_TRANSP, CARBOXYLIC_ACID_TRANSMEMBRANE_TRANSPORTER_ACTIVITY; MITOCHONDRIAL_FATTY_ACID_BETA, MITOCHONDRIAL_FATTY_ACID_BETAOXIDATION; HSA00980_METABOLISM_OF_XENOBIOTICS, HSA00980_METABOLISM_OF_XENOBIOTICS_BY_CYTOCHROME_P450; HSA00010_GLYCOLYSIS_AND_GLUCON, HSA00010_GLYCOLYSIS_AND_GLUCONEOGENESIS. Color images available online at www.liebertonline.com/ten.
FIG. 3.
Pathway for cholesterol metabolism that shows gene sets involved in this process.
Fatty acid metabolism (peroxisome-proliferator-activated receptor α–mediated metabolism)
Peroxisome-proliferator-activated receptor α (PPARα) is a nuclear receptor that activates gene expression of enzymes linked to fatty acid metabolism.30–33 PPARα-mediated fatty acid metabolism initiates transcriptional activation of liver fatty-acid-binding protein (L-FABP or FABP1), which deliver fatty acids to its cognitive nuclear receptor, PPARα, and promote expression of two transporters, ABCD2 and ABCD3, which are necessary to transport fatty acids into peroxisomes, where target enzymes catalyze the clearance of fatty acids.21,30,32,33 PPARα, being dependent on intracellular FABP concentrations, regulates expression of Acyl-CoA oxidases (ACOXs), short/branched-, long-, and very long-chain Acyl-CoA dehydrogenase (ACADs), and mitochondrial enzymes involved in β-oxidation.34–38 In our data, the gene sets involved in PPARα-mediated fatty acid metabolism are HSA00071_FATTY_ACID_METABOLISM, PEROXISOME, HSA03320_PPAR_ SIGNALING_PATHWAY, and MITOCHONDRIAL_FATTY_ACID_BETAOXIDATION (Fig. 2). All these gene sets were monotonically up-regulated in CS samples over the 8-day period in contrast to HMs. The gene FABP1, which belongs to the gene set HSA03320_PPAR_SIGNALING_PATHWAY, was up-regulated in CS cultures and its expression increased over time. In response to expression of FABP1, the gene PPARα is expressed.30 The PPARα signaling pathway promotes the transcriptional activation of fatty acid metabolic enzymes [ACOX (acyl-CoA oxidase), ACAD (acyl-CoA dehydrogenase), CPT (carnitine palmitoyltransferase), LPL (lipoprotein lipase), and ACAT (acetyl-CoA acetyl transferase)].30,34–36 These genes are members of the HSA00071_FATTY_ACID_METABOLISM, PEROXISOME, and MITOCHONDRIAL_FATTY_ACID_BETAOXIDATION gene sets. The combination of expression of key enzymes responsible for fatty acid metabolism as well as expression of two members of the ABC transporter family (ABCD2 and ABCD3) indicate that PPARα-mediated metabolism was up-regulated in CS samples.
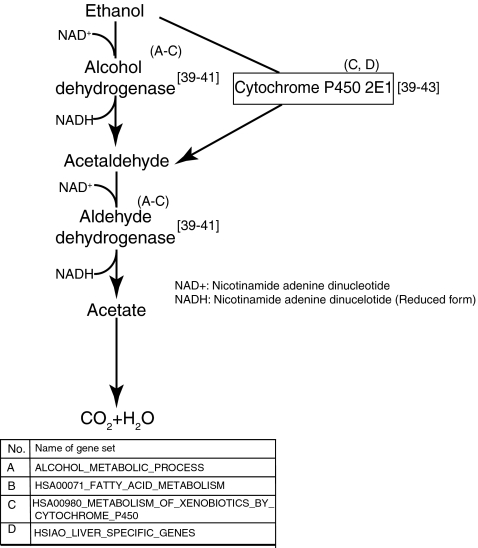
Alcohol metabolism
Alcohol, specifically ethanol, is metabolized in the liver by several enzymes present in the subcellular compartments of hepatocytes. Alcohol dehydrogenase (ADH), a key cytoplasmic enzyme, plays an important role in converting ethanol to acetaldehyde.39–41 Acetaldehyde, a toxic molecule, is subsequently converted to nontoxic acetates by mitochondrial acetaldehyde dehydrogenase.39,41 Additionally, CYP2E1 enables the clearance of ethanol through an oxidative reaction41–43 (Fig. 4). The gene sets ALCOHOL_METABOLIC_PROCESS, HSA00980_METABOLISM_OF_XENOBIOTICS_BY_CYTOCHROME_P450, and HSA00071_FATTY_ACID_METABOLISM were up-regulated over time in CS cultures in comparison to HMs (Fig. 2). These gene sets include three alcohol dehydrogenase genes, ADH, ADH1A, and ADH7, that mediate the transformation of alcohol.
FIG. 4.
Pathway for alcohol metabolism that shows gene sets involved in this process.
Carbohydrate metabolism
Gluconeogenesis and glycolysis are essential to maintain glucose homeostasis.44–47 The maintenance of a healthy glucose level is dependent upon the presence and concentration of insulin and glucagon.44–46 In our gene expression data, genes for key enzymes involved in the formation and metabolism of glucose are up-regulated in CS cultures in contrast to HMs. The relevant gene sets include HSA00010_GLYCOLYSIS_AND_GLUCONEOGENESIS and GLUCOSE_METABOLIC_PROCESS (Fig. 2). Specifically, genes corresponding to enzymes implicated in glycolysis are hexokinase (HK), glucose phosphate isomerase (or phosphoglucose isomerase, GPI, or PGI), phosphofructokinase (PFK), aldolase (ALDOB and ALDOC), triosephosphate isomerase (TPI1), phosphoglycerate kinase 1 (PGK1), phosphoglycerate mutase (PGAM), pyruvate kinase (PKLR), and lactate dehydrogenase C (LDH) (terminal).47 PKLR catalyzes the transphosphorylation of phosphoenolpyruvate into pyruvate and adenosine triphosphate (ATP), which is the rate-limiting step of glycolysis. LDH catalyzes the terminal step in glycolysis. Genes that code for enzymes involved in gluconeogenesis are phosphoenolpyruvate carboxykinase 1 (PCK or PEPCK), glucose-6-phosphatase (G6PC), pyruvate carboxylase (PC), and fructose-1, 6-bisphosphatase 1 (FBP1).46–48 In the gene sets related to carbohydrate metabolism, genes such as ALDOB, ALDOC, PKLR, PFK, PGK1, and GPI, which are involved in glycolysis, and genes such as G6PC, FBP1, PCK1, and PC, which are involved in gluconeogenesis, were up-regulated in CS samples.
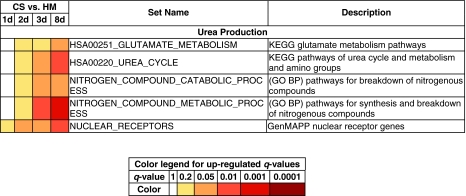
Urea production
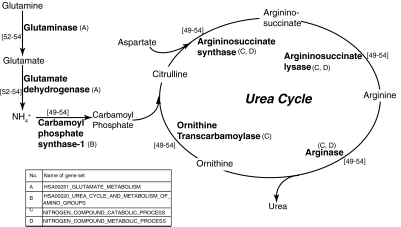
In the liver, the formation of urea is a critical step in ammonia clearance. The metabolism of amino acids results in the formation of urea through the conversion of glutamate, an intermediate metabolite.1 Gene sets involved in glutamate metabolism such as HSA00251_GLUTAMATE_METABOLISM are gradually up-regulated over time in CS cultures (Fig. 5). Urea is formed as a result of the action of five enzymes: carbamoyl phosphate synthetase-1 (CPS-1), ornithine transcarbamoylase (OTC), argininosuccinate synthase (ASS), argininosuccinate lysase (ASL), and arginase (ARG)49–54 (Fig. 6). These five genes are present in the gene set HSA00220_UREA_CYCLE_AND_METABOLISM_OF_AMINO_GROUPS. This gene set is up-regulated in CS cultures over the 8-day period. In addition, the gene sets NITROGEN_COMPOUND_CATABOLIC_PROCESS and NITROGEN_COMPOUND_METABOLIC_PROCESS include genes such as ASL, ARG, and ASS. Both gene sets are also monotonically up-regulated in CS cultures. The nuclear receptor HNF-4α (a member of the up-regulated gene set NUCLEAR_RECEPTORS) plays an important role in triggering the transcription of key enzymes for urea production.49 Together, these data provide information on why urea production is stable in CS cultures but not in HMs.
FIG. 5.
Up-regulated gene sets involved in urea production. The bottom table shows the q-value ranges for each color. Abbreviations: GO, gene ontology; BP, biological process; HSA00220_UREA_CYCLE, HSA00220_UREA_CYCLE_AND_METABOLISM_OF_AMINO_GROUPS; KEGG, Kyoto Encyclopedia of Genes and Genomes; GenMAPP, Gene Map Annotator and Pathway Profiler. Color images available online at www.liebertonline.com/ten.
FIG. 6.
Pathway for urea production that shows gene sets involved in this process.
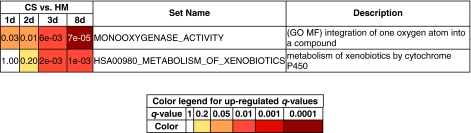
Monooxygenases are initially not differentially expressed but recover after day 3 in CS cultures
Xenobiotic metabolism in the liver is mediated through cytochrome P450 enzymes.55–58 Expression of these enzymes has been shown to decrease upon the isolation of hepatocytes from the liver.57,58 The gene set MONOOXYGENASE_ACTIVITY contains several cytochrome P450 genes and flavin containing monooxygenase. This gene set had q-values of 0.03, 0.01, 6 × 10−3, and 7 × 10−5 at days 1, 2, 3, and 8, respectively (Fig. 7). The q-values at days 1 and 2 were nearly identical but decreased by an order of magnitude at day 3 and by another order of magnitude at day 8. These trends were examined further by computing the q-values for this gene set in the CS–CS contrasts. This gene set was up-regulated with q-value 0.13 for the day 8–day 1 contrast, 0.11 for the day 8–day 2 contrast, and 1.00 for the day 8–day 3 contrast. The statistical significance values for corresponding contrasts among the HM samples were also computed. This gene set was down-regulated in all three contrasts. The q-values were 0.13, 0.06, and 0.13, respectively. Thus, the variation in expression of this gene set arises from a combination of up-regulation in CS cultures and down-regulation in HM cultures. Taken together, these trends indicate that the genes in this set recovered or became up-regulated at day 3 and later within CS cultures, in comparison to HM cultures. This trend is of significance since genes in this set include those that encode for CYP3A, CYP4A, CYP1A, and CYP2C enzymes. CYP3A and 4A enzymes metabolize a wide range of pharmaceuticals and drugs, and the CYP2C and CYP1A enzymes break down toxins and xenobiotics. The gene set HSA00980_ METABOLISM_OF_XENOBIOTICS_BY_ CYTOCHROME_P450, which contains cytochrome P450s, phase II metabolizing enzymes such as UDP-glucuronosyltransferase (UDP-GT) isoforms, and glutathione S-transferase (GST), also exhibited a similar trend. It showed no significant regulation at day 1, had a q-value of 0.20 at day 2, but had q-values an order of magnitude less at days 3 and 8 (2 × 10−3 and 10−3, respectively). In the three CS–CS contrasts, the q-values for this gene set were 0.12, 0.05, and 0.13 (all for up-regulation), while they were 0.1, 0.12, and 1 in the HM–HM contrasts (all for down-regulation). In a previous study, expression of a single-CYP enzyme, specifically, CYP1A1, was monitored and was shown to recover on day 3.59 However, additional time points or CYP enzymes were not investigated. The results in this work point to a more widespread recovery phenomenon among the CYP gene family.
FIG. 7.
Gene sets that show recovery after day 3. We show the q-values to underscore the recovery. MF, molecular function. Color images available online at www.liebertonline.com/ten.
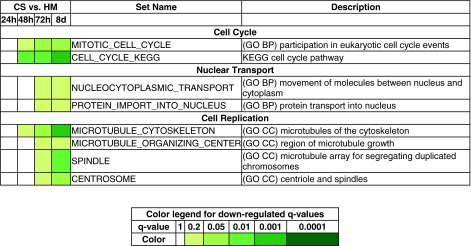
Cell-cycle activity decreases significantly in CS cultures
Analysis of the down-regulated gene sets presented interesting insights into cellular response within CS and HM systems. Our results suggested a significant difference in cell cycle activity between the HM and CS samples over the 8-day culture period. The monotonic down-regulation of the gene sets MITOTIC_CELL_CYCLE and CELL_CYCLE_KEGG, coupled with insignificant q-values in the CS-versus-CS comparisons (data not shown), suggested decreasing cell cycle activity within the CS cultures (Fig. 8). Nuclear transport and import functions show decreased activity within CS samples as indicated by the monotonically down-regulated gene sets NUCLEOCYTOPLASMIC_TRANSPORT and PROTEIN_IMPORT_INTO_NUCLEUS. In addition, the down-regulation of processes involved in microtubule organization (gene sets such as MICROTUBULE_CYTOSKELETON, MICROTUBULE_ORGANIZING_CENTER, SPINDLE, and CENTROSOME) were also observed. Our results are similar to a previous study in which the cell cycle and cell proliferation were found to be up-regulated in HMs in comparison to hepatoma cells cultured in a 3D spheroid culture.6 Although hepatocytes cultured in spheroids experience an environment that differs from CS configurations, they have been shown to exhibit metabolic and synthesis profiles similar to CS cultures.6
FIG. 8.
Down-regulated gene sets. The bottom table shows the q-value ranges for each color. CC, cellular component. Color images available online at www.liebertonline.com/ten.
Discussion
In vitro hepatic cultures play an increasingly important role in the design and development of pharmaceutics, toxicity testing, and the design of bio-artificial liver devices. Therefore, obtaining a comprehensive understanding of the global trends in transcription over time is critical to the advancement of each of these applications. A common problem associated with in vitro cultures of primary hepatocytes is the onset of de-differentiation soon after the cells are removed from the tissue. The temporal genome-wide transcriptional studies reported in this study provide insights into the up- and down-regulation of various liver-critical gene sets and their subsequent effects on various metabolic activities, synthesis of proteins, and detoxification. A gene set level network was constructed to obtain a systems level perspective of the current results (Fig. 9). Each node in this network is an up-regulated gene set and two gene sets are connected by an edge if they share a statistically significant number of genes. Only those connections between gene sets are included that are significant after correcting for testing multiple hypotheses using the method of Benjamini and Hochberg,60 at an alpha of 0.05. This correction resulted in the omission of several insignificant connections. A gene set was placed at the first time point at which it was significantly up-regulated (i.e., had a q-value reported by GSEA of at most 0.2). Note that day 8 does not appear in Figure 9 since all the genes sets are significantly up-regulated by day 3. Although this network does not reveal the multiple signaling pathways involved in liver-related processes in their entirety, it presents a preliminary high-level view of the temporal cascade of transcriptional activation in CS cultures. For instance, nuclear receptors, liver-specific genes, and the PPAR-α pathway are up-regulated significantly on day 1. Processes related to alcohol metabolism, bile acid biosynthesis, and urea production are up-regulated significantly on day 2. Lipid metabolism is activated at later time points. Although monooxygenase activity and xenobiotic metabolism appear on days 1 and 2, respectively, in this network, these gene sets show much more statistical significance at days 3 and 8, as noted earlier. Nuclear receptors are known to trigger gene expression of CYP 450 enzymes (which are monooxygenases and involved in metabolism of xenobiotics). It is intriguing that nuclear receptors are up-regulated at day 1 but with a q-value of 0.1. At later time points, this gene set becomes much more statistically significant (q-values of 0.01 at days 2 and 3 and a q-value of 3 × 10−3 at day 8), mirroring the trend observed for monooxygenase activity and xenobiotic metabolism. These observations will direct future experimental studies. It is also notable that the gene set NUCLEAR_RECEPTORS is not connected to any gene sets in day 2, suggesting that there is little overlap between the genes that are members of NUCLEAR_RECEPTORS and the gene sets that nuclear receptors may regulate. In future work, we will develop computational techniques that can utilize protein and gene interaction networks to automatically suggest regulatory connections between gene sets.
FIG. 9.
Gene set level network of up-regulated processes in CS cultures.
The time-dependent overview of the transcriptional changes that occur in HM and CS cultures illustrated in Figure 9 enables the investigation of liver phenotypic functions at appropriate time points. For example, in the absence of inducers, measuring the enzymatic activity and kinetics of CYP enzymes would be appropriate after hepatocytes are maintained in CS cultures for 3 days or more. In contrast, investigating metabolic activity could reveal important information when conducted on hepatocytes maintained in CS cultures for a minimum of 2 days. In vitro hepatic cultures can never replace or truly replicate animal models. However, their significance lies in their ability to serve as models to explore a wide range of physiological phenomena. Therefore, obtaining a comprehensive view of the numerous transcriptional changes that occur over a period and characterizing these changes in terms of succinct gene-set-level signatures will be valuable in guiding further investigations of cell signaling, metabolism, and protein synthesis functions in engineered livers.
In summary, the genome-wide gene expression profiles in two major hepatocyte culture systems have been analyzed, the first such dataset to our knowledge. The comparative analysis of transcriptional data obtained from the two culture systems reveals numerous liver-specific transcriptional programs that are up-regulated in CS cultures but not in monolayers. These processes span a wide range of biological processes important to liver function. Taken together, these trends and results provide a baseline for further systems-level modeling of engineered liver tissues.
Acknowledgments
We gratefully acknowledge financial support from the National Science Foundation (CBET-0933225, P.R. and T.M.M.), the National Institutes of Health (NIDDK-1R21DK077802, P.R.), and the Institute for Critical Technology and Applied Sciences (P.R. and T.M.M.) at Virginia Polytechnic Institute and State University. We thank the staff at the Virginia Bioinformatics Institute Core Laboratory facility for their assistance with this project.
Disclosure Statement
No competing financial interests exist.
References
- 1.Arias I.M. Boyer J.L. Chisari F.V. Fausto M. Schachter D. Shafritz D.A. The Liver: Biology and Pathobiology. Philadelphia, PA: Lippincott Williams and Wilkins; 2001. [Google Scholar]
- 2.Dunn J.C. Yarmush M.L. Koebe H.G. Tompkins R.G. Hepatocyte function and extracellular matrix geometry: long-term culture in a sandwich configuration. FASEB J. 1989;3:174. doi: 10.1096/fasebj.3.2.2914628. [DOI] [PubMed] [Google Scholar]
- 3.Dunn J.C. Tompkins R.G. Yarmush M.L. Long-term in vitro function of adult hepatocytes in a collagen sandwich configuration. Biotechnol Prog. 1991;7:237. doi: 10.1021/bp00009a007. [DOI] [PubMed] [Google Scholar]
- 4.Dunn J.C. Tompkins R.G. Yarmush M.L. Hepatocytes in collagen sandwich: evidence for transcriptional and translational regulation. J Cell Biol. 1992;116:1043. doi: 10.1083/jcb.116.4.1043. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Moghe P.V. Berthiaume F. Ezzell R.M. Toner M. Tompkins R.G. Yarmush M.L. Culture matrix configuration and composition in the maintenance of hepatocyte polarity and function. Biomaterials. 1996;17:373. doi: 10.1016/0142-9612(96)85576-1. [DOI] [PubMed] [Google Scholar]
- 6.Chang T.T. Hughes-Fulford M. Monolayer and spheroid culture of human liver hepatocellular carcinoma cell line cells demonstrate distinct global gene expression patterns and functional phenotypes. Tissue Eng Part A. 2008;15:559. doi: 10.1089/ten.tea.2007.0434. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Baker T.K. Carfagna M.A. Gao H. Dow E.R. Li Q. Searfoss G.H. Ryan T.P. Temporal gene expression analysis of monolayer cultured rat hepatocytes. Chem Res Toxicol. 2001;14:1218. doi: 10.1021/tx015518a. [DOI] [PubMed] [Google Scholar]
- 8.Li Z. Srivastava S. Yang X. Mittal S. Norton P. Resau J. Haab B. Chan C. A hierarchical approach employing metabolic and gene expression profiles to identify the pathways that confer cytotoxicity in HepG2 cells. BMC Syst Biol. 2007;1:21. doi: 10.1186/1752-0509-1-21. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Khetani S.R. Szulgit G. Del Rio J.A. Barlow C. Bhatia S.N. Exploring interactions between rat hepatocytes and nonparenchymal cells using gene expression profiling. Hepatology. 2004;40:545. doi: 10.1002/hep.20351. [DOI] [PubMed] [Google Scholar]
- 10.Khetani S.R. Bhatia S.N. Microscale culture of human liver cells for drug development. Nat Biotechnol. 2008;26:120. doi: 10.1038/nbt1361. [DOI] [PubMed] [Google Scholar]
- 11.Subramanian A. Tamayo P. Mootha V.K. Mukherjee S. Ebert B.L. Gillette M.A. Paulovich A. Pomeroy S.L. Golub T.R. Lander E.S. Mesirov J.P. Gene set enrichment analysis: a knowledge-based approach for interpreting genome-wide expression profiles. Proc Natl Acad Sci USA. 2005;102:15545. doi: 10.1073/pnas.0506580102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Dinu I. Potter J.D. Mueller T. Liu Q. Adewale A.J. Jhangri G.S. Einecke G. Famulski K.S. Halloran P. Yasui Y. Gene-set analysis and reduction. Brief Bioinform. 2009;10:24. doi: 10.1093/bib/bbn042. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Abatangelo L. Maglietta R. Distaso A. D'Addabbo A. Creanza T.M. Mukherjee S. Ancona N. Comparative study of gene set enrichment methods. BMC Bioinformatics. 2009;10:275. doi: 10.1186/1471-2105-10-275. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Gentleman R.C. Carey V.J. Bates D.M. Bolstad B. Dettling M. Dudoit S. Ellis B. Gautier L. Ge Y. Gentry J. Hornik K. Hothorn T. Huber W. Iacus S. Irizarry R. Leisch F. Li C. Maechler M. Rossini A.J. Sawitzki G. Smith C. Smyth G. Tierney L. Yang J.Y. Zhang J. Bioconductor: open software development for computational biology and bioinformatics. Genome Biol. 2004;5:R80. doi: 10.1186/gb-2004-5-10-r80. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Smyth G.K. LIMMA: linear models for microarray data. In: Gentlemen R., editor; Carey V., editor; Dudoit S., editor; Irizarry R., editor; Huber W., editor. Bioinformatics and Computational Biology Solutions Using R and Bioconductor. New York: Springer; 2005. pp. 397–420. [Google Scholar]
- 16.Knuth D.E. The Art of Computer Programming, Volume 2: Seminumerical Algorithms. 3rd. Reading, MA: Addison Wesley; 1998. [Google Scholar]
- 17.Hsiao L.L. Dangond F. Yoshida T. Hong R. Jensen R.V. Misra J. Dillon W. Lee K.F. Clark K.E. Haverty P. Weng Z. Mutter G.L. Frosch M.P. Macdonald M.E. Milford E.L. Crum C.P. Bueno R. Pratt R.E. Mahadevappa M. Warrington J.A. Stephanopoulos G. Gullans S.R. A compendium of gene expression in normal human tissues. Physiol Genomics. 2001;7:97. doi: 10.1152/physiolgenomics.00040.2001. [DOI] [PubMed] [Google Scholar]
- 18.Su A.I. Cooke M.P. Ching K.A. Hakak Y. Walker J.R. Wiltshire T. Orth A.P. Vega R.G. Sapinoso L.M. Moqrich A. Patapoutian A. Hampton G.M. Schultz P.G. Hogenesch J.B. Large-scale analysis of the human and mouse transcriptomes. Proc Natl Acad Sci USA. 2002;99:4465. doi: 10.1073/pnas.012025199. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Angrand P.O. Rousset J.P. Weiss M.C. Cell phenotype, binding affinity and promoter structure modulate transactivation by HNF1 and LAP. J Cell Sci. 1992;103(Pt 4):1083. doi: 10.1242/jcs.103.4.1083. [DOI] [PubMed] [Google Scholar]
- 20.Wu K.J. Wilson D.R. Shih C. Darlington G.J. The transcription factor HNF1 acts with C/EBP alpha to synergistically activate the human albumin promoter through a novel domain. J Biol Chem. 1994;269:1177. [PubMed] [Google Scholar]
- 21.Chawla A. Repa J.J. Evans R.M. Mangelsdorf D.J. Nuclear receptors and lipid physiology: opening the X-files. Science. 2001;294:1866. doi: 10.1126/science.294.5548.1866. [DOI] [PubMed] [Google Scholar]
- 22.Chiang J.Y. Bile acid regulation of hepatic physiology: III. Bile acids and nuclear receptors. Am J Physiol Gastrointest Liver Physiol. 2003;284:G349. doi: 10.1152/ajpgi.00417.2002. [DOI] [PubMed] [Google Scholar]
- 23.Ory D.S. Nuclear receptor signaling in the control of cholesterol homeostasis: have the orphans found a home? Circ Res. 2004;95:660. doi: 10.1161/01.RES.0000143422.83209.be. [DOI] [PubMed] [Google Scholar]
- 24.Kalaany N.Y. Mangelsdorf D.J. LXRS and FXR: the yin and yang of cholesterol and fat metabolism. Annu Rev Physiol. 2006;68:159. doi: 10.1146/annurev.physiol.68.033104.152158. [DOI] [PubMed] [Google Scholar]
- 25.Russell D.W. Nuclear orphan receptors control cholesterol catabolism. Cell. 1999;97:539. doi: 10.1016/s0092-8674(00)80763-1. [DOI] [PubMed] [Google Scholar]
- 26.Zelcer N. Tontonoz P. Liver X receptors as integrators of metabolic and inflammatory signaling. J Clin Invest. 2006;116:607. doi: 10.1172/JCI27883. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Lefebvre P. Cariou B. Lien F. Kuipers F. Staels B. Role of bile acids and bile acid receptors in metabolic regulation. Physiol Rev. 2009;89:147. doi: 10.1152/physrev.00010.2008. [DOI] [PubMed] [Google Scholar]
- 28.Lu T.T. Makishima M. Repa J.J. Schoonjans K. Kerr T.A. Auwerx J. Mangelsdorf D.J. Molecular basis for feedback regulation of bile acid synthesis by nuclear receptors. Mol Cell. 2000;6:507. doi: 10.1016/s1097-2765(00)00050-2. [DOI] [PubMed] [Google Scholar]
- 29.Goodwin B. Jones S.A. Price R.R. Watson M.A. McKee D.D. Moore L.B. Galardi C. Wilson J.G. Lewis M.C. Roth M.E. Maloney P.R. Willson T.M. Kliewer S.A. A regulatory cascade of the nuclear receptors FXR, SHP-1, and LRH-1 represses bile acid biosynthesis. Mol Cell. 2000;6:517. doi: 10.1016/s1097-2765(00)00051-4. [DOI] [PubMed] [Google Scholar]
- 30.Wolfrum C. Borrmann C.M. Borchers T. Spener F. Fatty acids and hypolipidemic drugs regulate peroxisome proliferator-activated receptors alpha- and gamma-mediated gene expression via liver fatty acid binding protein: a signaling path to the nucleus. Proc Natl Acad Sci USA. 2001;98:2323. doi: 10.1073/pnas.051619898. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Gervois P. Torra I.P. Fruchart J.C. Staels B. Regulation of lipid and lipoprotein metabolism by PPAR activators. Clin Chem Lab Med. 2000;38:3. doi: 10.1515/CCLM.2000.002. [DOI] [PubMed] [Google Scholar]
- 32.Willson T.M. Brown P.J. Sternbach D.D. Henke B.R. The PPARs: from orphan receptors to drug discovery. J Med Chem. 2000;43:527. doi: 10.1021/jm990554g. [DOI] [PubMed] [Google Scholar]
- 33.Francis G.A. Fayard E. Picard F. Auwerx J. Nuclear receptors and the control of metabolism. Annu Rev Physiol. 2003;65:261. doi: 10.1146/annurev.physiol.65.092101.142528. [DOI] [PubMed] [Google Scholar]
- 34.Tugwood J.D. Issemann I. Anderson R.G. Bundell K.R. McPheat W.L. Green S. The mouse peroxisome proliferator activated receptor recognizes a response element in the 5′ flanking sequence of the rat acyl CoA oxidase gene. EMBO J. 1992;11:433. doi: 10.1002/j.1460-2075.1992.tb05072.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Bardot O. Aldridge T.C. Latruffe N. Green S. PPAR-RXR heterodimer activates a peroxisome proliferator response element upstream of the bifunctional enzyme gene. Biochem Biophys Res Commun. 1993;192:37. doi: 10.1006/bbrc.1993.1378. [DOI] [PubMed] [Google Scholar]
- 36.Hashimoto T. Fujita T. Usuda N. Cook W. Qi C. Peters J.M. Gonzalez F.J. Yeldandi A.V. Rao M.S. Reddy J.K. Peroxisomal and mitochondrial fatty acid beta-oxidation in mice nullizygous for both peroxisome proliferator-activated receptor alpha and peroxisomal fatty acyl-CoA oxidase. Genotype correlation with fatty liver phenotype. J Biol Chem. 1999;274:19228. doi: 10.1074/jbc.274.27.19228. [DOI] [PubMed] [Google Scholar]
- 37.Kersten S. Seydoux J. Peters J.M. Gonzalez F.J. Desvergne B. Wahli W. Peroxisome proliferator-activated receptor alpha mediates the adaptive response to fasting. J Clin Invest. 1999;103:1489. doi: 10.1172/JCI6223. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Brandt J.M. Djouadi F. Kelly D.P. Fatty acids activate transcription of the muscle carnitine palmitoyltransferase I gene in cardiac myocytes via the peroxisome proliferator-activated receptor alpha. J Biol Chem. 1998;273:23786. doi: 10.1074/jbc.273.37.23786. [DOI] [PubMed] [Google Scholar]
- 39.Gyamfi M.A. Kocsis M.G. He L. Dai G. Mendy A.J. Wan Y.J. The role of retinoid X receptor alpha in regulating alcohol metabolism. J Pharmacol Exp Ther. 2006;319:360. doi: 10.1124/jpet.106.108175. [DOI] [PubMed] [Google Scholar]
- 40.Lieber C.S. ALCOHOL: its metabolism and interaction with nutrients. Annu Rev Nutr. 2000;20:395. doi: 10.1146/annurev.nutr.20.1.395. [DOI] [PubMed] [Google Scholar]
- 41.Nagy L.E. Molecular aspects of alcohol metabolism: transcription factors involved in early ethanol-induced liver injury. Annu Rev Nutr. 2004;24:55. doi: 10.1146/annurev.nutr.24.012003.132258. [DOI] [PubMed] [Google Scholar]
- 42.Liu C. Russell R.M. Seitz H.K. Wang X.D. Ethanol enhances retinoic acid metabolism into polar metabolites in rat liver via induction of cytochrome P4502E1. Gastroenterology. 2001;120:179. doi: 10.1053/gast.2001.20877. [DOI] [PubMed] [Google Scholar]
- 43.Lieber C.S. Cytochrome P-4502E1: its physiological and pathological role. Physiol Rev. 1997;77:517. doi: 10.1152/physrev.1997.77.2.517. [DOI] [PubMed] [Google Scholar]
- 44.Cariou B. Duran-Sandoval D. Kuipers F. Staels B. Farnesoid X receptor: a new player in glucose metabolism? Endocrinology. 2005;146:981. doi: 10.1210/en.2004-1595. [DOI] [PubMed] [Google Scholar]
- 45.Stayrook K.R. Bramlett K.S. Savkur R.S. Ficorilli J. Cook T. Christe M.E. Michael L.F. Burris T.P. Regulation of carbohydrate metabolism by the farnesoid X receptor. Endocrinology. 2005;146:984. doi: 10.1210/en.2004-0965. [DOI] [PubMed] [Google Scholar]
- 46.Konno Y. Negishi M. Kodama S. The roles of nuclear receptors CAR and PXR in hepatic energy metabolism. Drug Metab Pharmacokinet. 2008;23:8. doi: 10.2133/dmpk.23.8. [DOI] [PubMed] [Google Scholar]
- 47.Claudel T. Staels B. Kuipers F. The Farnesoid X receptor: a molecular link between bile acid and lipid and glucose metabolism. Arterioscler Thromb Vasc Biol. 2005;25:2020. doi: 10.1161/01.ATV.0000178994.21828.a7. [DOI] [PubMed] [Google Scholar]
- 48.Yamagata K. Daitoku H. Shimamoto Y. Matsuzaki H. Hirota K. Ishida J. Fukamizu A. Bile acids regulate gluconeogenic gene expression via small heterodimer partner-mediated repression of hepatocyte nuclear factor 4 and Foxo1. J Biol Chem. 2004;279:23158. doi: 10.1074/jbc.M314322200. [DOI] [PubMed] [Google Scholar]
- 49.Desvergne B. Michalik L. Wahli W. Transcriptional regulation of metabolism. Physiol Rev. 2006;86:465. doi: 10.1152/physrev.00025.2005. [DOI] [PubMed] [Google Scholar]
- 50.Morris S.M., Jr. Regulation of enzymes of the urea cycle and arginine metabolism. Annu Rev Nutr. 2002;22:87. doi: 10.1146/annurev.nutr.22.110801.140547. [DOI] [PubMed] [Google Scholar]
- 51.Schimke R.T. Adaptive characteristics of urea cycle enzymes in the rat. J Biol Chem. 1962;237:459. [PubMed] [Google Scholar]
- 52.Krebs H.A. Metabolism of amino-acids: The synthesis of glutamine from glutamic acid and ammonia, and the enzymic hydrolysis of glutamine in animal tissues. Biochem J. 1935;29:1951. doi: 10.1042/bj0291951. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Haussinger D. Nitrogen metabolism in liver: structural and functional organization and physiological relevance. Biochem J. 1990;267:281. doi: 10.1042/bj2670281. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Saheki T. Katunuma N. Analysis of regulatory factors for urea synthesis by isolated perfused rat liver. I. Urea synthesis with ammonia and glutamine as nitrogen sources. J Biochem. 1975;77:659. doi: 10.1093/oxfordjournals.jbchem.a130768. [DOI] [PubMed] [Google Scholar]
- 55.Omiecinski C.J. Remmel R.P. Hosagrahara V.P. Concise review of the cytochrome P450s and their roles in toxicology. Toxicol Sci. 1999;48:151. doi: 10.1093/toxsci/48.2.151. [DOI] [PubMed] [Google Scholar]
- 56.Thummel K.E. Wilkinson G.R. In vitro and in vivo drug interactions involving human CYP3A. Annu Rev Pharmacol Toxicol. 1998;38:389. doi: 10.1146/annurev.pharmtox.38.1.389. [DOI] [PubMed] [Google Scholar]
- 57.Rendic S. Di Carlo F.J. Human cytochrome P450 enzymes: a status report summarizing their reactions, substrates, inducers, and inhibitors. Drug Metab Rev. 1997;29:413. doi: 10.3109/03602539709037591. [DOI] [PubMed] [Google Scholar]
- 58.Wang X.J. Hodgkinson C.P. Wright M.C. Paine A.J. Temperature-sensitive mRNA degradation is an early event in hepatocyte de-differentiation. Biochem J. 1997;328(Pt 3):937. doi: 10.1042/bj3280937. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Tuschl G. Mueller S.O. Effects of cell culture conditions on primary rat hepatocytes-cell morphology and differential gene expression. Toxicology. 2006;218:205. doi: 10.1016/j.tox.2005.10.017. [DOI] [PubMed] [Google Scholar]
- 60.Benjamini Y. Hochberg Y. Controlling the false discovery rate: a practical and powerful approach to multiple testing. J Royal Stat Soc Ser B (Methodol) 1995;57:289. [Google Scholar]