Abstract
The biological effects of all-trans-retinoic acid (RA), a major active metabolite of retinol, are mainly mediated through its interactions with retinoic acid receptor (RARs α, β, γ) and retinoid X receptor (RXRs α, β, γ) heterodimers. RAR/RXR heterodimers activate transcription by binding to RA-response elements (RAREs or RXREs) in the promoters of primary target genes. Murine F9 teratocarcinoma stem cells have been widely used as a model for cellular differentiation and RA signaling during embryonic development. We identified and characterized genes that are differentially expressed in F9 wild type (Wt) and F9 RAR γ−/− cells, with and without RA treatment, through the use of oligonucleotide based microarrays. Our data indicate that RARγ, in the absence of exogenous RA, modulates gene expression. Genes such as Sfrp2, Tie1, Fbp2, Emp1, and Emp3 exhibited higher transcript levels in RA treated Wt, RARα−/− and RARβ2−/− lines than in RA-treated RARγ−/− cells, and represent specific RARγ targets. Other genes, such as Runx1, were expressed at lower levels in both F9 RARβ2−/− and RARγ−/− cell lines then in F9 Wt and RARα−/−. Genes specifically induced by RA at 6h with the protein synthesis inhibitor cycloheximide in F9 Wt, but not in RARγ−/− cells, included Hoxa3, Hoxa5, Gas1, Cyp26a1, Sfrp2, Fbp2, and Emp1. These genes represent specific primary RARγ targets in F9 cells. Several genes in the Wnt signaling pathway were regulated by RARγ. Delineation of the receptor specific actions of RA with respect to cell proliferation and differentiation should result in more effective therapies with this drug.
Keywords: retinoic acid receptor, gene expression profiles, differentiation, retinol, sfrp5, Tie1
1. INTRODUCTION
Retinoids are a group of natural or synthetic derivatives of vitamin A (retinol) which include all-trans-, 9-cis- and 13-cis-retinoic acids (RA, 9cRA and 13cRA, respectively). Synthesis of RA, the main biologically active metabolite of vitamin A, involves the irreversible oxidation of retinol in target cells [1, 2]. Retinoids are involved in multiple physiological processes including reproduction, embryonic development, epithelial differentiation, immune function, and vision [3]. In addition, retinoids modulate both normal and neoplastic cell growth through the regulation of cell differentiation and/or induction of apoptosis [4, 5].
The biological effects of RA are thought to be primarily mediated through interactions with retinoic acid receptors (RARs) or retinoic X receptors (RXRs), which function as transcription factors that activate transcription by binding to RA-response elements (RAREs or RXREs) in the promoter regions of target genes [6]. Both RARs and RXRs exhibit three isotypes, α, β, and γ, which are encoded by different genes. Each isotype has several isoforms produced from differential splicing and alternative use of promoters [7, 8]. All-trans retinoic acid (RA) binds and activates RARs, while 9-cis-retinoic acid binds and activates both RARs and RXRs. The multiple RARs and RXRs are conserved in vertebrate evolution and display distinct spatial-temporal expression patterns in developing embryos and adult tissues [9, 10]. Although gene disruption studies in mice have revealed some functional redundancy among the different RAR isotypes [11, 12], each RAR isotype performs unique functions in development and differentiation which can not be replaced by the actions of the other isotypes [11, 12]. RARγ plays a major role in axial rotation and vertebra specification [13], hindbrain patterning (specification of rhombomere 5/6 territory) [14], epidermal hyperplasia [15, 16], formation of otocysts, pharyngeal arches and the forelimb bud, closure of the primitive gut, cardiac looping morphogenesis [17], alveoli regeneration [18], hematopoietic stem cell self-renewal [19], and inflammatory cytokine production [20]. In addition, activation of RARγ is essential for induction of apoptosis in melanoma cells [21], neuroblastoma cells [22], neoplastic epidermal cells [23], and human pancreatic cancer cells [24]. In melanoma cells, RARγ selective compounds were able to induce apoptosis and differentiation [25]. However, the molecular mechanisms by which RARγ induces apoptosis and growth inhibition in these tumor cells are not well defined.
To delineate the specific roles of RARγ in retinoid signaling, the identification of specific RARγ target genes is crucial. The F9 teratocarcinoma stem cell line is a good model for investigating the mechanism by which RA induces cell differentiation and controls cell proliferation [26]. Upon exposure to RA, F9 Wt cells differentiate into primitive endoderm-like cells, with differentiation markers that resemble those in early embryos [27–29]. In our laboratory F9 RARα−/−, RARβ2−/− and RARγ−/− cell lines have been generated by homologous recombination [30–33]. The F9 RARγ null cells are unable to differentiate fully [30, 34]. Based on our previous findings and those of other researchers that the expression of some RA-inducible genes, such as Hoxa1, Hoxa3, laminin B1, Cdx1, collagen IV (α1), GATA-4, BMP-2, and Cyp26a1, is reduced in the RARγ−/− F9 cells [11, 30, 34, 35], we wanted to explore the effect of the loss of RARγ on gene transcription on a genome-wide scale. Thus, we compared the global gene expression profiles of F9 Wt versus RARγ−/− cells using Affymetrix oligonucleotide arrays (MG-U74Av2 and MG-430.2). These experiments uncovered new molecular actions of RARγ in F9 cell differentiation and provided insights into the molecular mechanisms by which RA causes cell differentiation.
2. MATERIALS AND METHODS
2.1. Chemicals
RA and cycloheximide were purchased from Sigma, St. Louis, MO.
2.2. Cell Culture, RA, cycloheximide treatment and RNA preparation
F9 Wt, RARγ−/−[30], RARα−/−[32] and RARβ2−/−[31] cells were grown in Dulbecco’s modified Eagle’s (DMEM) supplemented with 5% bovine calf serum, 5% fetal calf serum and 2 mM glutamine. Tissue culture flasks and plates were gelatinized with 0.3% gelatin solution for at least 5 min prior to plating. 2.0 ×106 cells were plated in 100 mm tissue culture plates for 16 hours before RA or vehicle (ethanol) treatment. For cyclohexmide treatment, cells were pretreated with cycloheximide at 1 μg/ml for 20 minutes before RA or vehicle (ethanol) was added for a further incubation for 5.5 h. RA was dissolved in 100% ethanol, filter (0.2 μm) sterilized, and diluted in cell culture medium to a final concentration of 1 × 10−6 M. Control samples were cultured in the same volume of ethanol (vehicle) used for the RA-treated samples. The final concentration of ethanol in all experiments did not exceed 0.1% volume. All experiments involving RA were performed in dim light. Total cellular RNA was extracted using Trizol (Invitrogen, P/N 15596026) by following the manufacturer’s protocol.
2.3. DNA Microarray Analyses
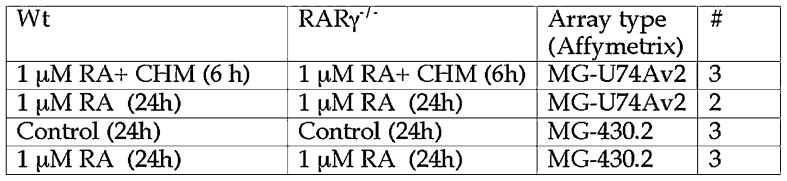
Microarray analysis was carried out according to the Affymetrix Genechip expression analysis technical manual. The extracts of total RNA were cleaned up with RNeasy Mini Kit (QIAGEN) according to the manufactors’s protocol. Total RNA (30 μg) from RA-treated (1 μM RA for 24 h), or RA plus cycloheximide –treated (1 μg/mL for 6 h) F9 Wt and F9 RARγ−/− cells was reverse transcribed into cDNA. After second-strand synthesis, cDNAs were then in vitro transcribed into cRNAs with biotinylated ribonucleotides (Enzo Diagnostics, Farmingdale, NY). cRNA (20 μg) was fragmented by heating at 94°C for 35 minutes. A cocktail containing fragmented cRNA, control oligonucleotide B2, control cRNA (Biotin B, Biotin C, Biotin D, and Cre, Hybridization Control Kit, Affymetrix, P/N 900454),), and herring sperm DNA was hybridized to microarray chip MG-U74Av2 (Affymetrix, P/N 900344), which covers 36,000 mouse transcripts and variants representing 12,488 genes, for 16 hours at 45°C. In a second study, 15 μg of total RNA from vehicle (ethanol) treated or RA (1 μM for 24 h) treated F9 Wt and F9 RARγ−/− cells was reverse transcribed into cDNA and then in vitro transcribed into cRNAs. Fragmented cRNAs (15 μg) were hybridized to microarray chip MG-430 2.0 Array (Affymetrix, P/N 900496) which covers the transcribed mouse genome with over 45,000 transcripts representing 34,000 well-substantiated mouse genes. The hybridized microarray chips were washed and stained in a Fluidics station, and were scanned by the GeneArray™ Scanner in the Microarray Core Facility at Weill Cornell Medical College. The resultant images were processed with MAS (Microarray Suite) 5.0 software (Affymetrix, Santa Clara, CA). The microarray experiments were repeated two or three times with different RNA preparations, and the overall data from the three independent experiments were further analyzed using the GeneSpring v7.0 (Silicon Genetics) software package. These data have been deposited in the GEO database (www.ncbi.nlm.nih.gov/geo, Accession # GSE8431).
A three-step normalization algorithm was used in the GeneSpring software analysis. In the first step, data transformation of all data less than 0.01 was reset to 0.01. In the second normalization step, each chip was normalized to the 50th percentile intensity of total intensity. In the third step, per gene normalization was done by dividing the median intensity of that gene in several control samples. A Welch’s T-test with a cutoff of a P-value ≤ 0.05 in a cross-gene error model was applied to select those statistically significant genes between two groups.
2.4. Semi-Quantitative and Quantitative Real-time RT-PCR
Total RNA (5 μg) isolated from F9 cells was reverse transcribed in a total 20 μl volume reaction with 200 units of Superscript™ Reverse transcritase II (Invitrogen Life Technologies) and oligo dT (12–18) primer (Invitrogen Life Technologies). The cDNA thus produced was diluted to 100 μl with diethyl pyrocarbonate/water, of which 2 μl was used in a PCR as follows. The PCR was performed using the following conditions: 94°C for 30 s, 58°C for 30 s, and 72°C for 80 s, with a final extension at 72°C for 10 min. Taq polymerase was from Invitrogen (catalog number 18038-042). The PCR products were subjected to 1% agarose gel electrophoresis. The gel images were stained with ethidium bromide and were recorded with a FluorChem 8800 system (Alpha Innotech, San Leandro, CA). Real-time PCR was performed using Bio-Rad iQ™ SYBR Green Supermix following the manufacturer’s instructions, on a Bio-Rad MyiQ™ Single Color Real-time PCR Detection System. Primers used in this study are listed in Appendix 3.
Appendix 3.
Primers used for RT-PCR.
| Common Name | Sequence | Product Size (bp) | GeneBank Accession No. |
|---|---|---|---|
| Hoxa1 (sense) | 5′ TGG AGG AAG TGA GAA AGT TGG C 3′ | 485 | NM_010449 |
| (antisense) | 5′ATG GGA GTC GAG AGG TTT CC 3′ | ||
| FBP-2 (sense) | 5′ TCC TGT ATG AAT GCA ATC CTG T 3′ | 232 | D42083 |
| (antisense) | 5′CAA TTG ACA AAG ACA AAG GGA AG 3′ | ||
| Tie1 (sense) | 5′ CCT TTG CTC AGA TCG CAC TA 3′ | 381 | X80764 |
| (antisense) | 5′ ATG CTG CTT TAG GTG GAG GA 3′ | ||
| EMP1 (sense) | 5′ CCT CTC CAT CAT CTT CTC CAT C 3′ | 620 | X98471 |
| (antisense) | 5′ GAC GTC AAG GAA GCA TCA GCA T 3′ | ||
| EMP3 (sense) | 5′ CCT CTT CAT GTT CCA ACT CTA 3′ | 238 | NM_010129 |
| (antisense) | 5′ TTT CCG CAG GTG GAT GTA GAC 3′ | ||
| Sfrp2 (sense) | 5′ CAA CCT GCT GGG CCA CGA GAC C 3′ | 695 | NM_009144 |
| (antisense) | 5′ GCT TGC GGA TGC TGC GGG AGA T 3′ | ||
| Sfrp5 (sense) | 5′ CTA TCC CTG TTC CCT CTA CTA C 3′ | 240 | NM_018780 |
| (antisense) | 5′ AGA ACC CTT CAG TCA AAG AGG G 3′ | ||
| Coch5B2 (sense) | 5′ GGG CAG TCC TAT GAT GAT GT 3′ | 243 | AF006741 |
| (antisense) | 5′ CCT TGCACG TAT TCC TTG AG 3′ | ||
| mOTT3 (sense) | 5′ ATC TCA TCA TGA CCA CTC AGG G 3′ | 267 | NM_011022 |
| (antisense) | 5′ TTC TTC GAT GTT CCT GTA CCC A 3′ | ||
| Slc27a2 (sense) | 5′ AGT TCT ACG CAT CCA CTG AAG 3′ | 655 | AF072757 |
| (antisense) | 5′ TGA CTG TGG GAT TGA AGC CCT CTT 3′ | ||
| Xlr3b (sense) | 5′ AGG CTG CCT TGT GGA GAG 3′ | 231 | NM_011727 |
| (antisense) | 5′ CTG TTG CCT CTC TGT TCC TG 3′ | ||
| Runx-1 (sense) | 5′ CCA GCA AGC TGA GGA GCG GCG 3′ | 348 | NM_009821 |
| (antisense) | 5′ CGG ATT TGT AAA GAC GGT GA 3′ | ||
| Crygc (sense) | 5′ CTG ACT ACC AGC AGT GGA TGG G 3′ | 171 | NM_007775 |
| (antisense) | 5′ CCT CAC TGA GGT GGA AGC GAT C 3′ | ||
| Zfp503 (sense) | 5′ GCC TTT TGT GCA CGC TGT 3′ | 243 | AK032903 |
| (antisense) | 5′ ACCGAG AGT TTG GAA GA3′ | ||
| Raet1a (sense) | 5′GCC ACT CTA CTT CTA AGA AAG GAT T 3′ | 307 | NM_009016 |
| (antisense) | 5′GTG AAG CTT ACT GTG GGA CTT 3′ | ||
| Peg1/Mest(sense) | 5′ ATT CGC AAC AAT GAC GGC 3′ | 441 | BC006639 |
| (antisense) | 5′ TGA GGT GGA CTA TTG TGT CAC C 3′ | ||
| RARα (sense) | 5′ ATC GAG ACC CAG AGC AGC AG 3′ | 409 | NM_009024 |
| (antisense) | 5′ CCT GGT GCG CTT TGC GAA CC 3′ | ||
| RARβ (sense) | 5′ GCA GAG TTT GAT GGA GTT CGT 3′ | 507 | NM_011243 |
| (antisense) | 5′ CCC ACT TCA AAG CAC TTC TGC A3′ | ||
| RARγ (sense) | 5′ CAA TAA GGA GAG ACT CTT TGC G 3′ | 324 | NM_011244 |
| (antisense) | 5′ TAC CAC TAT GGG GTC AGC TCC TGT G 3′ | ||
| Neo-RARγ (sense) | 5′-ATT CGC AGC GCA TCG CCT TCT AT3′ | 612 | NM_011244 |
| (antisense) | 5′-TTG CTG ACC TTG GTG ATG AGT 3′ | ||
| Actin (sense) | 5′ AAG TGT GAC GTT GAC ATC CG 3′ | 222 | NM_007393 |
| (antisense) | 5′ GAT CCA CAT CTG CTG GAA GG 3′ | ||
3. RESULTS
3.1. Comparison of the gene expression profiles in F9 Wt versus F9 RARγ−/− cell lines after treatment with RA plus a protein synthesis inhibitor, cycloheximide, for 6 h and after treatment with RA for 24 h
To identify the immediate, primary RARγ-regulated genes, F9 Wt and F9 RARγ−/− cells were treated with RA and the protein synthesis inhibitor, cycloheximide, for 6 h, and the gene expression profiles in the two cell lines were then compared using the U74Av2 affymetrix arrays. No untreated F9 Wt or F9 RARγ−/− samples were analyzed (Fig. 1). The rationale for this is that since RARγ is constitutively expressed in F9 cells, the primary target genes of RARγ should be transcriptionally activated by RA even in the absence of new protein synthesis. Thus, these transcripts can be detected in RA plus cycloheximide treated F9 Wt cells, but not in the F9 cells which lack RARγ. A total number of 5885 probe sets which were flagged with Presence in at least 3 chips were subjected to statistical analysis using GeneSpring V7 software. The resulting 94 genes exhibiting a 2-fold or greater net intensity ratio difference (p ≤0.05, Welch’s t-test) are shown (Table 1). These genes are putative RARγ primary target genes.
Figure 1. Design of microarray experiment.
Four conditions were used to examine gene expression in F9 Wt and F9 RARγ−/− cells. A“#” indicates the number of times the microarray analysis was performed under each condition, starting with fresh cells. RA, all-trans retinoic acid; CHM, cycloheximide.
Table 1. RARγ primary target genes identified using microarrays U74Av2.
Wt and RARγ−/− F9 cells were treated for 6 h with 1 μg/ml cycloheximide and 1 μM RA. Only those genes with at least a 2.0-fold change in F9 Wt cells relative to F9 RARγ−/− cells in both sets of microarrays with p values ≤0.05 are shown. Fold change=F9 Wt/F9 RARγ−/−.
| Affymetrix ID | Fold Changeab | Gene Symbol | Gene Title |
|---|---|---|---|
| 103086_at | 10.94 | Hoxa5 | homeo box A5 |
| 102087_at | 10.6 | Hoxa3 | homeo box A3 |
| 97426_at | 9.359 | Emp1 | epithelial membrane protein 1 |
| 94813_at | 7.189 | Gas1 | growth arrest specific 1 |
| 97379_at | 5.973 | Fbp2 | fructose bisphosphatase 2 |
| 104378_at | 5.521 | Pon2 | paraoxonase 2 |
| 104464_s_at | 4.512 | Kdelr3 | KDEL (Lys-Asp-Glu-Leu) endoplasmic reticulum protein retention receptor 3 |
| 101861_at | 4.397 | Sgce | sarcoglycan, epsilon |
| 98320_at | 4.226 | Cyp26a1 | cytochrome P450, family 26, subfamily a, polypeptide 1 |
| 104406_at | 4.208 | Ptges | Prostaglandin E synthase |
| 100914_at | 4.204 | Transcribed sequences | |
| 104580_at | 4.168 | Plcd | phospholipase C, delta |
| 93503_at | 4.129 | Sfrp2 | Secreted frizzled-related sequence protein 2 |
| 95016_at | 4.056 | Nrp | Neuropilin |
| 102649_s_at | 4.051 | Raet1a | retinoic acid early transcript 1, alpha |
| 103653_at | 3.88 | Mras | muscle and microspikes RAS |
| 99665_at | 3.666 | Satb1 | special AT-rich sequence binding protein 1 |
| 92502_at | 3.588 | Plagl1 | pleiomorphic adenoma gene-like 1 |
| 161013_f_at | 3.435 | 4930422J18Rik | RIKEN cDNA 4930422J18 gene |
| 161817_f_at | 3.405 | 4930422J18Rik | RIKEN cDNA 4930422J18 gene |
| 103761_at | 3.403 | Tcfcp2l1 | transcription factor CP2-like 1 |
| 104480_at | 3.328 | Dsg2 | desmoglein 2 |
| 104701_at | 3.322 | Bhlhb2 | basic helix-loop-helix domain containing, class B2 |
| 96662_at | 3.195 | Ppap2b | phosphatidic acid phosphatase type 2B |
| 99045_at | 3.152 | Eno2 | enolase 2, gamma neuronal |
| 95474_at | 3.087 | F2r | coagulation factor II (thrombin) receptor |
| 104375_at | 3.062 | Spock2 | sparc/osteonectin, cwcv and kazal-like domains proteoglycan 2 |
| 92501_s_at | 2.963 | Plagl1 | pleiomorphic adenoma gene-like 1 |
| 93994_at | 2.941 | Chpt1 | choline phosphotransferase 1 |
| 92582_at | 2.776 | Slc1a5 | solute carrier family 1 (neutral amino acid transporter), member 5 |
| 94027_at | 2.737 | CDNA clone IMAGE:30033444, partial cds | |
| 97353_at | 2.706 | Dab2ip | Disabled homolog 2 (Drosophila) interacting protein |
| 102712_at | 2.706 | Saa3 | serum amyloid A 3 |
| 100011_at | 2.575 | Klf3 | Kruppel-like factor 3 (basic) |
| 160961_at | 2.57 | Sipa1l2 | signal-induced proliferation-associated 1 like 2 |
| 102886_at | 2.553 | Gpc4 | glypican 4 |
| 92241_at | 2.507 | 1110019L22Rik | RIKEN cDNA 1110019L22 gene |
| 100410_at | 2.47 | C330027G06Rik | RIKEN cDNA C330027G06 gene |
| 94418_at | 2.467 | Elovl6 | ELOVL family member 6, elongation of long chain fatty acids (yeast) |
| 98016_at | 2.46 | D3Wsu161e | DNA segment, Chr 3, Wayne State University 161, expressed |
| 100528_at | 2.45 | Ube2h | ubiquitin-conjugating enzyme E2H |
| 95161_at | 2.409 | Ctdsp2 | CTD (carboxy-terminal domain, RNA polymerase II, polypeptide A) small phosphatase 2 |
| 98535_at | 2.402 | Comt | catechol-O-methyltransferase |
| 93083_at | 2.392 | Anxa5 | annexin A5 |
| 160255_at | 2.376 | 1110004P15Rik | RIKEN cDNA 1110004P15 gene |
| 104576_at | 2.371 | Ski | Sloan-Kettering viral oncogene homolog |
| 97509_f_at | 2.37 | Fgfr1 | Fibroblast growth factor receptor 1 |
| 97138_at | 2.369 | 9430077C05Rik | RIKEN cDNA 9430077C05 gene |
| 92833_at | 2.365 | Hal | histidine ammonia lyase |
| 99994_at | 2.335 | Cidea | Cell death-inducing DNA fragmentation factor, alpha subunit-like effector A |
| 97711_at | 2.322 | B430320C24Rik | RIKEN cDNA B430320C24 gene |
| 160188_at | 2.29 | Nudt4 | nudix (nucleoside diphosphate linked moiety X)-type motif 4 |
| 104210_at | 2.285 | Itga3 | integrin alpha 3 |
| 103288_at | 2.276 | Nrip1 | nuclear receptor interacting protein 1 |
| 96207_at | 2.264 | Rbms1 | RNA binding motif, single stranded interacting protein 1 |
| 160393_at | 2.167 | Etnk1 | Ethanolamine kinase 1 |
| 92796_at | 2.162 | Akp2 | alkaline phosphatase 2, liver |
| 103614_at | 2.128 | Nfkb2 | nuclear factor of kappa light polypeptide gene enhancer in B-cells 2, p49/p100 |
| 92368_at | 2.047 | Ramp3 | receptor (calcitonin) activity modifying protein 3 |
| 100475_at | 2.044 | Trim25 | B6-derived CD11 +ve dendritic cells cDNA, RIKEN full-length enriched library, clone:F730014H10 product:unclassifiable, full insert sequence |
| 100962_at | 2.039 | Nab2 | Ngfi-A binding protein 2 |
| 97452_at | 2.038 | H2afy | H2A histone family, member Y |
| 160887_at | 2.036 | Hes1 | hairy and enhancer of split 1 (Drosophila) |
| 94408_at | 2.025 | Nab1 | Ngfi-A binding protein 1 |
| 103551_at | 2 | 2810431N21Rik | RIKEN cDNA 2810431N21 gene |
| 160546_at | 0.487 | Aldo3 | aldolase 3, C isoform |
| 94829_at | 0.466 | 1110020A09Rik | RIKEN cDNA 1110020A09 gene |
| 92635_at | 0.464 | Tuba4 | tubulin, alpha 4 |
| 98109_at | 0.462 | Mrpl55 | Mitochondrial ribosomal protein L55 |
| 103549_at | 0.459 | Nes | Nestin |
| 93483_at | 0.458 | Hck | hemopoietic cell kinase |
| 98544_at | 0.455 | Guk1 | Guanylate kinase 1 |
| 93543_f_at | 0.452 | Gstm1 | glutathione S-transferase, mu 1 |
| 93836_at | 0.439 | Bnip3 | BCL2/adenovirus E1B 19kDa-interacting protein 1, NIP3 |
| 97995_at | 0.431 | Tcf7 | transcription factor 7, T-cell specific |
| 160495_at | 0.423 | Ahr | Aryl-hydrocarbon receptor |
| 103581_at | 0.407 | Cte1 | cytosolic acyl-CoA thioesterase 1 |
| 94334_f_at | 0.392 | Ina | internexin neuronal intermediate filament protein, alpha |
| 103405_at | 0.388 | 2610019A05Rik | RIKEN cDNA 2610019A05 gene |
| 160085_at | 0.384 | Tst | thiosulfate sulfurtransferase, mitochondrial |
| 92786_at | 0.359 | Efhd1 | EF hand domain containing 1 |
| 104652_at | 0.349 | Kcnk2 | potassium channel, subfamily K, member 2 |
| 94335_r_at | 0.346 | Ina | internexin neuronal intermediate filament protein, alpha |
| 95669_g_at | 0.345 | Stmn2 | stathmin-like 2 |
| 93680_at | 0.344 | Stk10 | serine/threonine kinase 10 |
| 99812_at | 0.338 | Capn3 | calpain 3 |
| 95670_at | 0.296 | Stmn2 | stathmin-like 2 |
| 95603_at | 0.268 | Gldc | glycine decarboxylase |
| 95706_at | 0.267 | Lgals3 | lectin, galactose binding, soluble 3 |
| 96244_at | 0.239 | Uchl1 | ubiquitin carboxy-terminal hydrolase L1 |
| 104286_at | 0.183 | Slc38a4 | solute carrier family 38, member 4 |
| 161068_at | 0.12 | 3830422N12Rik | RIKEN cDNA 3830422N12 gene |
| 100967_at | 0.0994 | Slc27a2 | solute carrier family 27 (fatty acid transporter), member 2 |
| 104544_at | 0.0981 | 4930517K11Rik | RIKEN cDNA 4930517K11 gene |
Average from three experiments.
Some genes have multiple distinct probes on the GeneChip® resulting in different fold-changes from different probes.
The global gene expression patterns in F9 Wt and F9 RARγ−/− cells after a 24-hour exposure to 1 μM RA were also compared on U74 Av2 arrays. The overall presence call was 6171/12422=49.68%. After the Welch T-test, 241 genes with P-values ≤0.05 were selected for further fold change analysis. The results from 66 genes exhibiting a 2.0-fold or greater net intensity ratio difference are shown (Table 2). These genes encode cell membrane proteins, cell-cell adhesion molecules, signal transducers, and surface tyrosine kinases involved in cell development. By comparing the 94 genes (6 h treatment with RA plus cycloheximide, Table 2) with those altered at 24 hour after RA exposure (Table 2), we found that only 8 genes overlapped. These genes are the epithelial memebrane protein (emp)-1, secreted frizzled-related sequence protein 2 (Sfrp2), alkaline phosphatase 2 (AKP2), Stmn2, solute carrier family 38 member 4 (Slc38a4), Slc27a2, and lectin-galactose binding soluble 3 (Lgals3). One obvious reason why the changes in most of the early response genes were not seen at the 24 h RA treatment is that the kinetics of the transcriptional activation of the early vs. late genes induced by RARγ are different.
Table 2. Genes differentially expressed between F9 Wt and F9 RARγ−/− cells after RA treatment for 24 h, identified using U74AV2 microarrays.
Only those genes with at least a 2.0-fold change in F9 Wt cells relative to F9 RARγ−/− cells with p values ≤0.05 are shown. Fold change=F9 Wt/F9 RARγ−/−.
| Affymetrix ID | Fold Changeab | Gene Symbol | Gene Ttile |
|---|---|---|---|
| 99936_at | 25.08 | Tie1 | tyrosine kinase receptor 1 |
| 160943_at | 15.3 | Stag3 | stromal antigen 3 |
| 97426_at | 13.86 | Emp1 | epithelial membrane protein 1 |
| 93503_at | 4.634 | Sfrp2 | secreted frizzled-related sequence protein 2 |
| 101027_s_at | 4.142 | Pttg1 | pituitary tumor-transforming 1 |
| 103506_f_at | 3.773 | Dsc2 | Desmocollin 2 |
| 104146_at | 3.567 | Rasip1 | Ras interacting protein 1 |
| 97379_at | 3.534 | Fbp2 | fructose bisphosphatase 2 |
| 99669_at | 3.532 | Lgals1 | Lectin, galactose binding, soluble 1 |
| 161482_f_at | 3.427 | Prph1 | Peripherin 1 |
| 104580_at | 3.385 | Plcd | phospholipase C, delta |
| 100610_at | 2.998 | Capns1 | calpain, small subunit 1 |
| 95426_at | 2.946 | Echs1 | enoyl Coenzyme A hydratase, short chain, 1, mitochondrial |
| 101358_at | 2.885 | Plcb3 | phospholipase C, beta 3 |
| 104746_at | 2.821 | Fkbp7 | FK506 binding protein 7 |
| 160978_at | 2.808 | D630035O19Rik | RIKEN cDNA D630035O19 gene |
| 96865_at | 2.677 | Marcks | myristoylated alanine rich protein kinase C substrate |
| 98435_at | 2.631 | Adss | adenylosuccinate synthetase, muscle |
| 100828_at | 2.325 | --- | |
| 160388_at | 2.305 | Sc4mol | sterol-C4-methyl oxidase-like |
| 160215_at | 2.299 | Aes | amino-terminal enhancer of split |
| 104232_at | 2.289 | Gjb3 | gap junction membrane channel protein beta 3 |
| 96016_at | 2.27 | 2700094K13Rik | RIKEN cDNA 2700094K13 gene |
| 102993_at | 2.265 | Ggta1 | glycoprotein galactosyltransferase alpha 1, 3 |
| 98535_at | 2.247 | Comt | catechol-O-methyltransferase |
| 92586_at | 2.242 | Glud | Glutamate dehydrogenase |
| 99184_at | 2.232 | Csad | cysteine sulfinic acid decarboxylase |
| 101107_at | 2.222 | Calu | Calumenin |
| 102926_at | 2.193 | Gfra3 | Glial cell line derived neurotrophic factor family receptor alpha 3 |
| 93750_at | 2.16 | Gsn | Gelsolin |
| 104673_at | 2.137 | Epha4 | Eph receptor A4 |
| 103085_at | 2.133 | Hebp1 | Heme binding protein 1 |
| 92607_at | 2.103 | Peg1 | Mus musculus Peg1/MEST protein |
| 93522_at | 2.094 | Rad9 | RAD9 homolog (S. pombe) |
| 95733_at | 2.048 | Slc29a1 | solute carrier family 29 (nucleoside transporters), member 1 |
| 97717_at | 2.041 | Tcf15 | transcription factor 15 |
| 92796_at | 2.04 | Akp2 | alkaline phosphatase 2, liver |
| 96038_at | 2.038 | Rnase4 | ribonuclease, RNase A family 4 |
| 93373_at | 2.023 | Naglu | alpha-N-acetylglucosaminidase (Sanfilippo disease IIIB) |
| 103056_at | 0.496 | 6230425C22Rik | RIKEN cDNA 6230425C22 gene |
| 103423_at | 0.495 | Cyb561 | cytochrome b-561 |
| 93038_f_at | 0.486 | Anxa1 | annexin A1 |
| 95756_at | 0.486 | Ftsj3 | FtsJ homolog 3 (E. coli) |
| 100600_at | 0.467 | Cd24a | CD24a antigen |
| 92306_at | 0.465 | Ott | Ovary testis transcribed |
| 101030_at | 0.426 | Rhob | ras homolog gene family, member B |
| 102841_at | 0.414 | Similar to ribosomal protein L40 (LOC216818), mRNA | |
| 96151_at | 0.407 | Mocos | molybdenum cofactor sulfurase |
| 160236_at | 0.383 | 9630044O09Rik | RIKEN cDNA 9630044O09 gene |
| 95954_at | 0.379 | D7Ertd143e | DNA segment, Chr 7, ERATO Doi 143, expressed |
| 93864_s_at | 0.36 | Enah | enabled homolog (Drosophila) |
| 95883_at | 0.349 | Phf17 | PHD finger protein 17 |
| 104486_at | 0.349 | A2m | alpha-2-macroglobulin |
| 102344_s_at | 0.345 | Tcea3 | transcription elongation factor A (SII), 3 |
| 93680_at | 0.343 | Stk10 | serine/threonine kinase 10 |
| 102418_at | 0.339 | Tex19 | testis expressed gene 19 |
| 96704_at | 0.339 | Sfn | Stratifin |
| 99577_at | 0.33 | Kitl | kit ligand |
| 104063_at | 0.327 | Srcasm | Src activating and signaling molecule |
| 95670_at | 0.308 | Stmn2 | stathmin-like 2 |
| 99034_at | 0.266 | Irx3 | Iroquois related homeobox 3 (Drosophila) |
| 104338_r_at | 0.25 | 1200008D14Rik | RIKEN cDNA 1200008D14 gene |
| 93028_at | 0.246 | H19 | H19 fetal liver mRNA |
| 101883_at | 0.212 | Xlr3b | XLR related protein, Mouse A12 mRNA |
| 93294_at | 0.211 | Ctgf | connective tissue growth factor |
| 103317_at | 0.189 | Coch5B2 | Coagulation factor C homolog (Limus polyphemus) cds=(68,1726) |
| 100967_at | 0.189 | Slc27a2 | solute carrier family 27 (fatty acid transporter), member 2 |
| 104126_at | 0.188 | Cstf2t | cleavage stimulation factor, 3′ pre-RNA subunit 2, tau |
| 161081_at | 0.158 | Cpeb2 | cytoplasmic polyadenylation element binding protein 2 |
| 104286_at | 0.106 | Slc38a4 | solute carrier family 38, member 4 |
| 93013_at | 0.0853 | Idb2 | inhibitor of DNA binding 2 |
| 95706_at | 0.0819 | Lgals3 | lectin, galactose binding, soluble 3 |
Average from two experiments.
Some genes have multiple distinct probes on the GeneChip® resulting in different fold-changes from different probes.
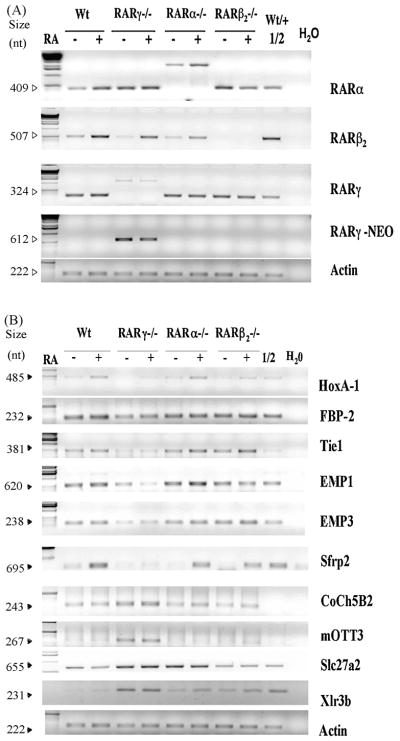
To validate the microarray data, new F9 cell samples were treated with RA for 24 h to confirm the results. Several differentially expressed genes were selected for examination by semi-quantitative RT-PCR. In addition to the F9 Wt and RARγ−/− lines, F9 RARα−/− and RARβ2−/− cell lines were also tested during the validation studies in order to determine if the genes were specifically regulated by RARγ. The mRNA expression of the different RAR isotypesα, β, and γ in each cell line was examined. The RARα−/−, RARβ2−/−, and RARγ−/− cell lines were each established by homologous recombination with a targeting vector to their B domains [30–32]. Both alleles of the mutant RAR in each cell line contain the neomycin resistance gene, which disrupts the encoded RARα, RARβ2, and RARγ transcripts and protein. Using primers specific for each isotype which span the neomycin resistance gene, the transcripts from the wild type allele and mutant allele were amplified. Since each of the mutant alleles contains the neomycin resistance gene, the mutant allele gives rise to a larger transcript than that from the wild type allele. RARα mRNA was detected in Wt, RARγ−/− and RARβ2−/− cells, but not in F9 RARα−/− cells (Fig. 2A). The expression of RARβ2 transcripts was strongly induced F9 Wt cells, F9 RARα−/−, and RARγ−/− cells, but not in RA treated RARβ2−/− cells (Fig. 2A). RARγ mRNA could not be detected in F9 RARγ−/− cells, though RARγ mRNA was detected in F9 Wt, RARα−/− and RARβ2−/− cells (Fig. 2A). Transcripts from the mutant RARγ (RARγ-NEO) were only detected in F9 RARγ−/−, but not in F9 Wt, RARα−/− and RARβ2−/− cells (Fig. 2A). Thus, the identities of the cell lines were confirmed (Fig. 2A).
Figure 2. Semi-quantitative RT-PCR analyses of selected genes, FBP-2, Tie1, EMP1, EMP3, Sfrp2, Coch5B2, mOTT3, mFATP2, and Xlr3b, in F9 Wt, RARγ−/−, RARα−/− and RARβ2−/− cell lines in response to RA.
A and B, total RNA was extracted from the F9 Wt, RAR γ−/−, RARα−/−, RARβ2−/− cells cultured in the presence or absence of 1 μM RA for 24 h. An equivalent amount of RNA (5 μg) was subjected to RT-PCR with oligonucleotide primers specific for the indicated genes. The PCR-reactions were stopped between the 22ndand 32nd cycles depending on the gene being tested to ensure linear amplification ranges. PCR-amplified products were electrophoresed in 1% agarose gels and the products were stained with ethidium bromide. The 1:2 dilution of cDNA from RA-treated F9 Wt or RA-treated F9 RARγ−/− samples was used to indicate the linear amplification ranges for genes which showed increased mRNA levels in RARγ−/− cells or decreased levels in RARγ−/− cells, respectively. The sizes of the amplified bands are indicated on the left. These assays were repeated using three different RNA preparations, and similar results were obtained.
Hoxa1, a known RARγ-regulated gene, was examined as a positive control. Hoxa1 mRNA levels increased after exposure to RA in F9 Wt, RARα−/− and RARβ2−/− cells, but the RA associated increase in Hoxa1 mRNA was greatly reduced in F9 RARγ−/− cells as compared to F9 Wt cells (Fig. 2B).
The nine genes chosen for validation by semi-quantitative RT-PCR analyses were FBP-2, Tie1, EMP1, EMP3, Sfrp2, Coch5B2, mOTT3, mFATP2 (Slc27a2), and Xlr3b (Fig. 2B). These genes were selected for validation because they exhibited relatively high fold changes in mRNA levels and/or interesting biological functions.
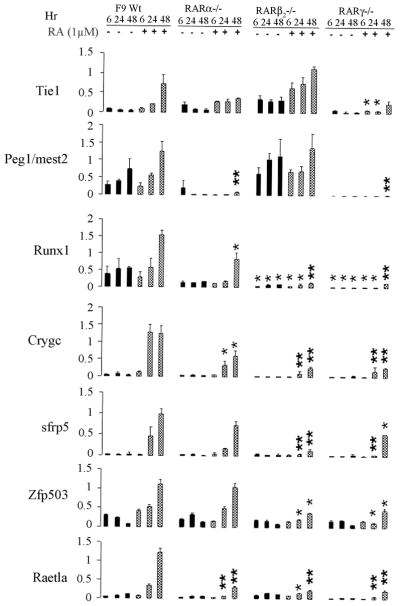
FBP-2 (fructose 1,6-bisphosphatase), via altering levels of F26P2 (Fructose-2,6-bisphosphate), influences glucose and lipid metabolism and provides cooperative regulation of fuel metabolism [36]. F26P2 can in turn regulate transcription factors and certain key proteins (enzymes) of signaling and/or energy sensing [36]. For example, F26P2 can regulate the amount and/or phosphorylation state of transcription factors such as hepatic nuclear factor 1-alpha (HNF1α), peroxisome proliferator-activated receptor alpha (PPARα), and PPARγ co-activator 1beta (PGC1β), and the kinases Akt and AMP-activated protein kinase (AMPK) [36]. Fbp2 mRNA levels were 5.9 ± 2.0-fold and 3.5 ± 1.1-fold higher in F9 Wt cells than in F9 RARγ−/− cells cultured in the presence of RA plus cycloheximide for 6 h, and in the presence of RA for 24 h, respectively. Semi-quantitative RT-PCR confirmed that Fbp2 mRNA was present at higher levels in F9 Wt than in F9 RARγ−/− cells (Fig. 2B).
Tie1 is a member of the receptor tyrosine kinase family [37, 38]. It is essential for the development and maintenance of vascular vascular system and hematopoiesis [39]. Tie1 mRNA levels were 25.1 ± 8.2-fold higher in F9 Wt than in F9 RARγ−/− cells cultured in the presence of RA for 24 h. Semi-quantitative RT-PCR confirmed that Tie1 mRNA levels were higher in F9 Wt cells than in F9 RARγ−/− cells (Fig. 2B).
Emp1 (epithelial membrane protein 1) and Emp3 (epithelial membrane protein 3) are members of the peripheral myelin protein 22 (PMP22) family [40]. Emp1 mRNA levels were 9.4 ± 2.4-fold and 13.7 ± 3.5-fold higher in F9 Wt cells than in F9 RARγ−/− cells cultured in the presence of RA plus cycloheximide for 6 h, and cultured in the presence of RA for 24 h, respectively. Semi-quantitative RT-PCR confirmed that Emp1 and Emp3 transcripts are present at higher levels in F9 Wt than in F9 RARγ−/− cells (Fig. 2B).
Sfrp2 (secreted frizzled related protein 2) is a secreted Wnt antagonist that directly interacts with the Wnt ligand to inhibit Wnt signaling [41]. Sfrp2 is required for anteroposterior (AP) axis elongation and somitogenesis in the thoracic region during mouse embryogenesis. Sfrp2 mRNA levels were 4.1 ± 1.7-fold higher in F9 Wt than in F9 RARγ−/− cells cultured in the presence of RA plus cycloheximide for 6 h, and 4.6 ± 2.0-fold cultured in the presence of RA for 24h, respectively. Semi-quantitative RT-PCR confirmed that Sfrp2 mRNA levels are higher in F9 Wt cells than in F9 RARγ−/− cells (Fig. 2B).
Coch5B2 encodes a protein with regions highly homologous to the collagen-binding type A domains of von Willebrand factor [42]. Coch5B2 is highly expressed in fetal inner ear structures, the cochlea, and the vestibule [43]. Mutations in Coch5B2 cause DFNA9, a human nonsyndromic deafness with vestibular dysfunction [44, 45]. Coch 5B2 mRNA levels were 5.3 ± 1.4-fold higher in the F9 RARγ−/− cells than in F9 Wt cells cultured in the presence of RA for 24 h, and this was confirmed by semi-quantitative RT-PCR (Fig. 2B).
OTT3 is an X-linked gene expressed in testis and spermatogonia, but not in somatic tissues [46]. A role for OTT3 in splicing regulation has been demonstrated [47]. Ott3 mRNA levels were 2.2 ± 0.7-fold higher in F9 RARγ−/− cells than in F9 Wt cells cultured in the presence of RA for 24 h, and this was validated by semi-quantitative RT-PCR (Fig. 2B).
Slc27a2 (solute carrier family 27 (fatty acid transporter), member 2) belongs to the long chain fatty acid transporter (FATP) family. These proteins facilitate fatty acid uptake from the medium [48]. Slc27a2 mRNA levels were 2.2 ± 0.3-fold higher in F9 RARγ−/− cells than in F9 Wt cells cultured in the presence of RA for 24 h, and this was confirmed by semi-quantitative RT-PCR (Fig. 2B).
Xlr3b (X-linked lymphocyte regulated, 3b) is an X-linked, maternally expressed, imprinted gene [49] which may mediate X-linked imprinting effects on cognitive processes [50, 51]. Xlr3b mRNA levels were 4.7±1.3-fold higher in F9 RARγ−/− cells than in F9 Wt cells cultured in the presence of RA for 24 h and semi-quantitative RT-PCR confirmed that Xlr3b mRNA levels are higher in F9 RARγ−/− cells than in F9 Wt cells (Fig. 2B).
The mRNA levels of FBP-2, Tie1, EMP1, EMP3 and Sfrp2 were lower in F9 RARγ−/− cells than in F9 Wt cells, but these transcripts were not expressed at lower levels in F9 RARα−/− and RARβ2−/− cells, either in the presence or absence of RA. Thus, these genes represent specific RARγ target genes. In contrast, Coch5B2, mOTT3, Slc27a2, and Xlr3b transcripts were expressed at higher levels in F9 RARγ−/− cells than in Wt, RARα−/− (with the exception of Slc27a2), and RARβ2−/− cells. Coch5B2, Xlr3b and mOTT3 transcripts, therefore, are specifically increased in F9 RARγ−/− cells (Fig. 2B). A list of these genes is in the GEO database (www.ncbi.nlm.nih.gov/geo, Accession # GSE8431).
3.2. Comparison of the gene expression profiles associated with F9 Wt and F9 RARγ−/− cells cultured in the absence vs. presence of RA
The initial microarray analyses we performed identified the genes that are differentially expressed between RA-treated F9 Wt and RA-treated F9 RARγ−/− cells at 6 h and 24 h, but no control (untreated) F9 Wt or F9 RARγ−/− samples were analyzed (Fig. 1). Thus, the genes identified in these initial studies also included a subset of genes that were not responsive to RA. In order to identify additional RA responsive genes and to incorporate a larger fraction of the mouse genome into our survey of RARγ-regulated genes, we performed another microarray analysis utilizing Affymetrix MG-430.2 gene chips, which contain 45,037 transcripts representing 34,000 well-substantiated mouse genes.
F9 Wt and F9 RARγ−/− cells were treated with either 1 μM RA or vehicle (ethanol) for 24 hours. The gene expression profiles of RA-treated F9 Wt cells, vehicle–treated (control) F9 Wt cells, RA-treated F9 RARγ−/− cells, and vehicle-treated (control) F9 RARγ−/− cells were then compared using MG-430.2 gene chips. The genes that were differentially expressed between F9 Wt cells and F9 RARγ−/− cells in the presence or absence of RA were identified. Since a protein synthesis inhibitor was not used in this set of experiments, both primary and secondary RARγ dependent RA response genes were identified.
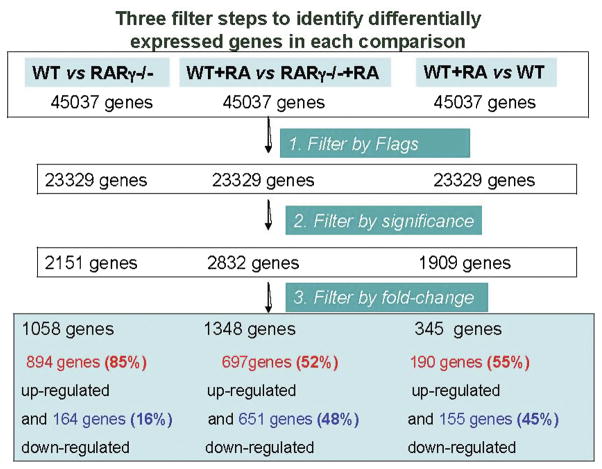
In order to identify the subset of RARγ regulated, RA-responsive genes, a strategy involving multiple two-condition comparisons was employed (Fig. 3). Genes were only considered differentially expressed between the two conditions if they exhibited at least a twofold difference in expression with a p value of less than 0.05 (Welch’s t-test). Genes were defined as RA responsive genes if they exhibited at least a two fold difference from the control (no RA) condition (P≤0.05). For the analysis with MG-430.2 chips, all of the genes with a “Present” call in at least three chips were selected and were subjected to statistical analysis for two condition comparisons using GeneSpring v7.0. The overall presence call (# of genes with a Present call/total # of probe sets on the array) for those samples was 23329/45037 = 51.78 %. For filtering the data for significant changes, the parameteric Welch’s t-test was performed individually for each of the following pair comparisons: Vehicle-treated (control) F9 Wt cells vs vehicle-treated (control) F9 RARγ−/− cells (Wt vs RARγ−/−); RA-treated Wt cells vs RA-treated RARγ−/− cells (Wt +RA vs RARγ−/−+RA); RA-treated Wt cells vs vehicle-treated control Wt cells (Wt +RA vs Wt); resulting in 2151 genes, 2832 genes, and 1909 genes that had statistically significant p values ≤ 0.05, respectively. Genes with statistically significant differences in expression (p< 0.05) were then further subjected to a fold change filter application, resulting in 1058 genes (Wt vs RARγ−/−), 1348 genes (Wt +RA vs RARγ−/−+RA), and 345 genes (Wt +RA vs Wt), respectively, with a differential expression of at least two fold for each pair comparison (Fig. 3).
Figure 3. An overview of the strategy to select differentially expressed genes.
Beginning with all Affymetrix IDs on the MG-430 2.0 GeneChip (45037 genes), the first filter selected the genes with a “Present” call in at least 3 microarrays. The second step filtered out statistically insignificant means based on the Welch’s t- test. The third step filtered out genes with less than a 2-fold difference in expression between the two conditions.
3.3. Distinctive expression profiles associated with the F9 Wt and F9 RARγ−/− cell lines cultured in the absence of RA
There were a total of 1058 probe sets differentially expressed by at least 2-fold between the vehicle-treated (control) F9 Wt and vehicle-treated control F9 RARγ−/− cells at 24 hours (Fig. 3). This indicates a fundamental difference in the gene expression profiles between the two cell types in the absence of RA. The differentially expressed genes with the highest fold changes are shown (Table 3A, B). After a 24 h-RA treatment, a total of 1348 probe sets exhibited at least a 2–fold difference in expression between the RA-treated Wt and RA-treated RARγ−/− F9 cells (Fig. 3), and the differentially expressed genes with the highest fold changes are listed (Table 4A, B).
Table 3. A.
Top 53 genes with at least 2-fold higher expression in F9 Wt cells relative to F9 RARγ−/− cells at 24 h in the absence of RA. Fold change =WT/RARγ−/−.
| Affymetrix ID | Fold Changeab | Gene Symbol | Gene Title |
|---|---|---|---|
| 1431094_at | 43.31 | Rtl1 | Retrotransposon-like 1 |
| 1456250_x_at | 42.39 | Tgfbi | BB533460 RIKEN full-length enriched, 0 day neonate lung Mus musculus cDNA clone E030030E06 3′ similar to L19932 Mouse (beta ig-h3) mRNA, mRNA sequence. |
| 1417426_at | 38.65 | Prg1 | Proteoglycan 1, secretory granule |
| 1452183_a_at | 38.62 | Gtl2 | GTL2, imprinted maternally expressed untranslated mRNA |
| 1448123_s_at | 34.09 | H2-Ab1 | Histocompatibility 2, class II antigen A, beta 1 |
| 1422865_at | 31.5 | Spp1 | Secreted phosphoprotein 1 |
| 1439380_x_at | 29.61 | Gtl2 | BB093563 RIKEN full-length enriched, 12 days embryo, embryonic body between diaphragm region and neck Mus musculus cDNA clone 9430042P15 3′ similar to Y13832 Mus musculus mRNA for GT12 protein, mRNA sequence. |
| 1415871_at | 27.87 | H2-Ab1 | Histocompatibility 2, class II antigen A, beta 1 |
| 1448926_at | 27.49 | Hoxa5 | Homeo box A5 |
| 1441075_at | 25.77 | LOC329416 | nitric oxide synthase trafficker |
| 1418215_at | 25.05 | Mep1b | Meprin 1 beta |
| 1432159_a_at | 24.87 | Il2 | Interleukin 2 |
| 1452905_at | 24.52 | Gtl2 | GTL2, imprinted maternally expressed untranslated mRNA |
| 1455626_at | 23.25 | Hoxa9 | homeo box A9 |
| 1425109_at | 20.16 | Slc44a3 | Solute carrier family 44, member 3 |
| 1422864_at | 19.68 | Spp1 | Secreted phosphoprotein 1 |
| 1423294_at | 18.68 | Mest | Transcribed sequence with moderate similarity to protein sp:Q9UBF2 (H. sapiens) CPG2_HUMAN Coatomer gamma-2 subunit |
| 1435989_x_at | 18.51 | Krt2-8 | keratin complex 2, basic, gene 8 |
| 1427580_a_at | 18.48 | Rian | 15 days embryo head cDNA, RIKEN full-length enriched library, clone:D930050K13 product:unclassifiable, full insert sequence |
| 1426990_at | 18.39 | Cubn | cubilin (intrinsic factor-cobalamin receptor) |
| 1429273_at | 18.02 | Bmper | BMP-binding endothelial regulator |
| 1452400_a_at | 17.57 | Hoxa11s | homeo box A11, opposite strand transcript |
| 1419537_at | 17.39 | Ctsd | Cathepsin D |
| 1460550_at | 16.3 | BC051083 | cDNA sequence BC051083 |
| 1417787_at | 16.24 | Prnd | Prion protein dublet |
| 1451332_at | 16.03 | Zfp521 | Zinc finger protein 521; synonyms: Evi3, B930086A16Rik; isoform 2 is encoded by transcript variant 2; ecotropic viral integration site 3; Mus musculus zinc finger protein 521 (Zfp521), transcript variant 2, mRNA. |
| 1448804_at | 15.79 | Cyp11a1 | Cytochrome P450, family 11, subfamily a, polypeptide 1 |
| 1430637_at | 15.68 | 2210016H18Rik | Adult male stomach cDNA, RIKEN full-length enriched library, clone:2210016H18 product:hypothetical protein, full insert sequence |
| 1418713_at | 15.56 | Pcbd1 | Pterin 4 alpha carbinolamine dehydratase/dimerization cofactor of hepatocyte nuclear factor 1 alpha (TCF1) 1 |
| 1431475_a_at | 15.11 | Hoxa10 | Homeo box A10 |
| 1458232_at | 15.1 | Dkk1 | dickkopf homolog 1 (Xenopus laevis) |
| 1434647_at | 14.92 | AU040377 | expressed sequence AU040377 |
| 1427433_s_at | 14.79 | 5730446D14Rik | RIKEN cDNA 5730446D14 gene |
| 1428781_at | 13.99 | 1110014F24Rik | RIKEN cDNA 1110014F24 gene |
| 1451687_a_at | 12.91 | Tcf2 | transcription factor 2 |
| 1451191_at | 12.51 | Crabp1 | Cellular retinoic acid binding protein I |
| 1418672_at | 12.46 | Lrrc1 | Leucine rich repeat containing 1 |
| 1418187_at | 12.46 | Ramp2 | Receptor (calcitonin) activity modifying protein 2 |
| 1429693_at | 12.41 | Dab2 | Disabled homolog 2 (Drosophila) |
| 1424393_s_at | 12.14 | Syt11 | Synaptotagmin XI |
| 1453782_at | 11.99 | 3021401C12Rik | RIKEN cDNA 0610012A05 gene |
| 1443696_s_at | 11.95 | Habp2 | hyaluronic acid binding protein 2 |
| 1433670_at | 11.65 | Emp2 | epithelial membrane protein 2 |
| 1450555_at | 11.56 | Il2 | Interleukin 2 |
| 1460722_at | 11.52 | Soat2 | Sterol O-acyltransferase 2 |
| 1449568_at | 11.4 | Klb | Klotho beta |
| 1440878_at | 11.38 | Runx1 | Runt related transcription factor 1 |
| 1420360_at | 11.05 | Dkk1 | Dickkopf homolog 1 (Xenopus laevis) |
| 1420565_at | 10.88 | Hoxa1 | Homeo box A1 |
| 1417828_at | 10.62 | Aqp8 | Aquaporin 8 |
| 1449088_at | 10.47 | Eef2 | Eukaryotic translation elongation factor 2 |
| 1456733_x_at | 10.36 | Serpinh1 | BB329489 RIKEN full-length enriched, 4 days neonate thymus Mus musculus cDNA clone B630008L19 3′, mRNA sequence. |
| 1417920_at | 10.22 | Creb1 | CAMP responsive element binding protein 1 |
| 1438558_x_at | 9.989 | Foxq1 | forkhead box Q1 |
Average from three experiments.
Some genes have multiple distinct probes on the GeneChip® resulting in different fold-changes from different probes.
Table 3. B.
Top 50 genes with at least 2-fold lower expression in F9 Wt cells relative to F9 RARγ−/− cells after 24 h in the absence of RA. Fold change =WT/RARγ−/−.
| Affymetrix ID | Fold Changeab | Gene Symbol | Gene Title |
|---|---|---|---|
| 1436796_at | 0.307 | 1110061A14Rik | matrin 3 |
| 1422557_s_at | 0.305 | Mt1 | Metallothionein 1 |
| 1450292_a_at | 0.305 | Ap3b1 | Adaptor-related protein complex 3, beta 1 subunit |
| 1448991_a_at | 0.302 | Ina | Internexin neuronal intermediate filament protein, alpha |
| 1450417_a_at | 0.301 | Rps20 | ribosomal protein S20 |
| 1457195_at | 0.299 | Plekhm1 | cDNA sequence BC038943 |
| 1424490_at | 0.292 | Atp5o | ATP synthase, H+ transporting, mitochondrial F1 complex, O subunit |
| 1454681_at | 0.288 | 2210008M09Rik | cDNA sequence BC031468 |
| 1429490_at | 0.286 | Rif1 | Rap1 interacting factor 1 homolog (yeast) |
| 1417751_at | 0.285 | Mapk7 | Mitogen activated protein kinase 7 |
| 1418994_at | 0.283 | 2410116G06Rik | RIKEN cDNA 2410116G06 gene |
| 1437311_at | 0.279 | A930034L06Rik | RIKEN cDNA A930034L06 gene |
| 1425415_a_at | 0.255 | Slc1a1 | Solute carrier family 1 (neuronal/epithelial high affinity glutamate transporter, system Xag), member 1 |
| 1416149_at | 0.252 | Akt2 | Thymoma viral proto-oncogene 2 |
| 1436619_at | 0.251 | H3077A05-3 NIA Mouse 15K cDNA Clone Set Mus musculus cDNA clone H3077A05 3′, mRNA sequence. | |
| 1460204_at | 0.251 | Tsc2 | Tuberous sclerosis 2 |
| 1427046_at | 0.246 | 1810073N04Rik | RIKEN cDNA 1810073N04 gene |
| 1423281_at | 0.246 | Stmn2 | stathmin-like 2 |
| 1440997_at | 0.246 | 9930033H14Rik | RIKEN cDNA 9930033H14 gene |
| 1455688_at | 0.245 | AI838577 | Adult male urinary bladder cDNA, RIKEN full-length enriched library, clone:9530036D08 product:unknown EST, full insert sequence |
| 1439880_at | 0.238 | D630023F18Rik | Transcribed sequences |
| 1453219_a_at | 0.231 | L1td1 | LINE-1 type transposase domain containing 1 |
| 1425035_s_at | 0.23 | Dnmt3l | DNA (cytosine-5-)-methyltransferase 3-like |
| 1421299_a_at | 0.229 | Lef1 | Lymphoid enhancer binding factor 1 |
| 1423280_at | 0.226 | Stmn2 | stathmin-like 2 |
| 1458599_at | 0.223 | Transcribed sequences | |
| 1443858_at | 0.218 | Trim34 | RIKEN cDNA 9230105E10 gene |
| 1448299_at | 0.217 | Slc1a1 | Solute carrier family 1 (neuronal/epithelial high affinity glutamate transporter, system Xag), member 1 |
| 1436388_a_at | 0.213 | 3830406C13Rik | RIKEN cDNA 3830406C13 gene |
| 1425926_a_at | 0.199 | Otx2 | Orthodenticle homolog 2 (Drosophila) |
| 1460386_a_at | 0.197 | Slc1a1 | solute carrier family 1 (neuronal/epithelial high affinity glutamate transporter, system Xag), member 1 |
| 1418825_at | 0.195 | Psmd11 | Proteasome (prosome, macropain) 26S subunit, non-ATPase, 11 |
| 1433789_at | 0.158 | Rnu17d | 18 days pregnant adult female placenta and extra embryonic tissue cDNA, RIKEN full-length enriched library, clone:3830421G02 product:unknown EST, full insert sequence |
| 1445271_at | 0.151 | 9230105E10Rik | RIKEN cDNA 9230105E10 gene |
| 1445617_at | 0.15 | Transcribed sequences | |
| 1439207_at | 0.149 | Similar to paraneoplastic antigen like 5; paraneoplastic antigen family 5 (LOC386569), mRNA | |
| 1423327_at | 0.133 | D730048I06Rik | RIKEN cDNA D730048I06 gene |
| 1429203_at | 0.131 | 2410076I21Rik | RIKEN cDNA 2410076I21 gene |
| 1433792_at | 0.131 | AW491344 | Transcribed sequence with weak similarity to protein ref:NP_113662.1 (H. sapiens) hypothetical protein DKFZp761G1913 [Homo sapiens] |
| 1448558_a_at | 0.118 | Pla2g4a | Phospholipase A2, group IVA (cytosolic, calcium-dependent) |
| 1422617_at | 0.102 | Top1 | Topoisomerase (DNA) I |
| 1429701_at | 0.101 | 2410003J06Rik | RIKEN cDNA 2410003J06 gene |
| 1434739_at | 0.0949 | 3830422N12Rik | RIKEN cDNA 3830422N12 gene |
| 1453544_at | 0.0876 | 4933424F23Rik | RIKEN cDNA 4933424F23 gene |
| 1449625_at | 0.0707 | Transcribed sequence with weak similarity to protein ref:NP_570953.1 (M. musculus) CAMP [Mus musculus] | |
| 1419241_a_at | 0.0671 | Aire | Autoimmune regulator (autoimmune polyendocrinopathy candidiasis ectodermal dystrophy) |
| 1420357_s_at | 0.0663 | Fndc6 | Fibronectin type III domain containing 6 |
| 1447021_at | 0.0447 | CDNA clone MGC:61032 IMAGE:30024827, complete cds | |
| 1448889_at | 0.0344 | Slc38a4 | Solute carrier family 38, member 4 |
| 1428111_at | 0.028 | Slc38a4 | Solute carrier family 38, member 4 |
| 1423523_at | 0.0225 | Aass | aminoadipate-semialdehyde synthase |
Average from three experiments.
Some genes have multiple distinct probes on the GeneChip® resulting in different fold-changes from different probes.
Table 4, A.
Top 50 genes with at least 2-fold higher expression in RA-treated Wt F9 cells relative to RA-treated RARγ−/− cells at 24 h. Fold change=F9 Wt/F9 RARγ−/−.
| Affymetrix ID | Fold Changeab | Gene Symbol | Gene Title |
|---|---|---|---|
| 1455498_at | 122.6 | Transcribed sequence with weak similarity to protein pir:A43932 (H. sapiens) A43932 mucin 2 precursor, intestinal – human | |
| 1451332_at | 71.76 | Zfp521 | Zinc finger protein 521; synonyms: Evi3, B930086A16Rik; isoform 2 is encoded by transcript variant 2; ecotropic viral integration site 3; Mus musculus zinc finger protein 521 (Zfp521), transcript variant 2, mRNA. |
| 1441075_at | 51 | LOC329416 | nitric oxide synthase trafficker |
| 1452183_a_at | 48.69 | Gtl2 | GTL2, imprinted maternally expressed untranslated mRNA |
| 1428055_at | 47.37 | 15 days embryo head cDNA, RIKEN full-length enriched library, clone:D930050K13 product:unclassifiable, full insert sequence | |
| 1417426_at | 35.01 | Prg1 | Proteoglycan 1, secretory granule |
| 1452905_at | 34.73 | Gtl2 | GTL2, imprinted maternally expressed untranslated mRNA |
| 1432159_a_at | 31.56 | Il2 | Interleukin 2 |
| 1450555_at | 25.66 | Il2 | Interleukin 2 |
| 1423294_at | 25.26 | Mest | Transcribed sequence with moderate similarity to protein sp:Q9UBF2 (H. sapiens) CPG2_HUMAN Coatomer gamma-2 subunit |
| 1423023_at | 24.61 | Sfrp5 | Secreted frizzled-related sequence protein 5 |
| 1439380_x_at | 24.04 | Gtl2 | BB093563 RIKEN full-length enriched, 12 days embryo, embryonic body between diaphragm region and neck Mus musculus cDNA clone 9430042P15 3′ similar to Y13832 Mus musculus mRNA for GT12 protein, mRNA sequence. |
| 1415871_at | 24 | H2-Ab1 | Histocompatibility 2, class II antigen A, beta 1 |
| 1460550_at | 23.48 | BC051083 | cDNA sequence BC051083 |
| 1429273_at | 23.48 | Bmper | BMP-binding endothelial regulator |
| 1448926_at | 22.68 | Hoxa5 | Homeo box A5 |
| 1437347_at | 21.85 | Ednrb | endothelin receptor type B |
| 1430637_at | 20.7 | 2210016H18Rik | Adult male stomach cDNA, RIKEN full-length enriched library, clone:2210016H18 product:hypothetical protein, full insert sequence |
| 1421952_at | 19.86 | Capn6 | calpain 6 |
| 1421224_a_at | 19.8 | Tcf2 | Transcription factor 2 |
| 1456250_x_at | 19.4 | Tgfbi | BB533460 RIKEN full-length enriched, 0 day neonate lung Mus musculus cDNA clone E030030E06 3′ similar to L19932 Mouse (beta ig-h3) mRNA, mRNA sequence. |
| 1436172_at | 19.3 | Adult male testis cDNA, RIKEN full-length enriched library, clone:4921508F21 product:similar to HISTOCOMPATIBILITY 2, CLASS II ANTIGEN E BETA [Mus musculus], full insert sequence | |
| 1455626_at | 17.4 | Hoxa9 | homeo box A9 |
| 1426758_s_at | 17.38 | Gtl2 | GTL2, imprinted maternally expressed untranslated mRNA |
| 1422866_at | 17.33 | Col13a1 | Procollagen, type XIII, alpha 1 |
| 1436713_s_at | 16.96 | L0922F05-3 NIA Mouse Newborn Kidney cDNA Library (Long) Mus musculus cDNA clone NIA:L0922F05 IMAGE:30002176 3′, mRNA sequence. | |
| 1425273_s_at | 15.6 | Emp2 | Epithelial membrane protein 2 |
| 1440878_at | 15.18 | Runx1 | Runt related transcription factor 1 |
| 1459742_at | 14.46 | BB800744 RIKEN full-length enriched, 15 days embryo brain Mus musculus cDNA clone G630032A08 3′, mRNA sequence. | |
| 1420603_s_at | 14.36 | Sgk3 | Serum/glucocorticoid regulated kinase 3 |
| 1422865_at | 12.97 | Spp1 | Secreted phosphoprotein 1 |
| 1450759_at | 12.89 | BC019943 | CDNA sequence BC019943 |
| 1433428_x_at | 12.67 | Tgm2 | transglutaminase 2, C polypeptide |
| 1416855_at | 12.61 | Gas1 | BB550400 RIKEN full-length enriched, 2 days pregnant adult female oviduct Mus musculus cDNA clone E230022A16 3′ similar to X65128 M. musculus gas1 mRNA, mRNA sequence. |
| 1448123_s_at | 12.58 | H2-Ab1 | Histocompatibility 2, class II antigen A, beta 1 |
| 1421917_at | 12.28 | Pdgfra | platelet derived growth factor receptor, alpha polypeptide |
| 1452400_a_at | 12.15 | Hoxa11s | homeo box A11, opposite strand transcript |
| 1438558_x_at | 11.59 | Foxq1 | forkhead box Q1 |
| 1432673_at | 11.11 | 2300010F08Rik | 12 days embryo embryonic body between diaphragm region and neck cDNA, RIKEN full-length enriched library, clone:9430087B15 product:unclassifiable, full insert sequence |
| 1450624_at | 10.81 | Surf4 | Surfeit gene 4 |
| 1429177_x_at | 10.6 | Mafg | V-maf musculoaponeurotic fibrosarcoma oncogene family, protein G (avian) |
| 1423805_at | 10.3 | Dab2 | Disabled homolog 2 (Drosophila) |
| 1424649_a_at | 10.22 | Tspan8 | Tetraspanin 8 |
| 1449088_at | 10.12 | Eef2 | Eukaryotic translation elongation factor 2 |
| 1429693_at | 9.983 | Dab2 | Disabled homolog 2 (Drosophila) |
| 1451191_at | 9.949 | Crabp1 | Cellular retinoic acid binding protein I |
| 1433615_at | 9.937 | B930062P21Rik | RIKEN cDNA B930062P21 gene |
| 1417787_at | 9.648 | Prnd | Prion protein dublet |
| 1429310_at | 9.583 | Flrt3 | fibronectin leucine rich transmembrane protein 3 |
| 1426341_at | 9.02 | Slc1a3 | solute carrier family 1 (glial high affinity glutamate transporter), member 3 |
Average from three experiments.
Some genes have multiple distinct probes on the GeneChip® resulting in different fold-changes from different probes.
Table 4, B.
Top 50 genes with at least 2-fold lower expression in RA-treated Wt F9 cells relative to RA-treated RARγ−/− cells at 24 h. Fold change=F9 Wt/F9 RARγ−/−.
| Affymetrix ID | Fold Changeab | Gene Symbol | Gene Title |
|---|---|---|---|
| 1419227_at | 0.195 | Cct6b | Chaperonin subunit 6b (zeta) |
| 1424531_a_at | 0.191 | Tcea3 | Transcription elongation factor A (SII), 3 |
| 1449005_at | 0.188 | Itgav | Integrin alpha V |
| 1436657_at | 0.187 | LOC380738 (LOC380738), mRNA | |
| 1455015_at | 0.187 | 4933431N12Rik | RIKEN cDNA 4933431N12 gene |
| 1452384_at | 0.187 | Enpp3 | ectonucleotide pyrophosphatase/phosphodiesterase 3 |
| 1455881_at | 0.186 | 2610524G09Rik | RIKEN cDNA 2610524G09 gene |
| 1416626_at | 0.185 | Pla2g1b | Phospholipase A2, group IB, pancreas |
| 1441971_at | 0.184 | Transcribed sequences | |
| 1436677_at | 0.184 | 1810032O08Rik | sialyltransferase 7 ((alpha-N-acetylneuraminyl 2,3-beta-galactosyl-1,3)-N-acetyl galactosaminde alpha-2,6-sialyltransferase) B |
| 1416953_at | 0.183 | Ctgf | Connective tissue growth factor |
| 1426808_at | 0.181 | Lgals3 | Lectin, galactose binding, soluble 3 |
| 1450989_at | 0.18 | Tdgf1 | teratocarcinoma-derived growth factor |
| 1428942_at | 0.178 | Mt2 | metallothionein 2 |
| 1434719_at | 0.175 | A2m | Alpha-2-macroglobulin |
| 1434094_at | 0.162 | 6330530A05Rik | RIKEN cDNA 6330530A05 gene |
| 1423281_at | 0.161 | Stmn2 | stathmin-like 2 |
| 1453219_a_at | 0.149 | L1td1 | LINE-1 type transposase domain containing 1 |
| 1425035_s_at | 0.144 | Dnmt3l | DNA (cytosine-5-)-methyltransferase 3-like |
| 1431633_x_at | 0.142 | 4930526L06Rik | Adult male testis cDNA, RIKEN full-length enriched library, clone:4930526L06 product:unknown EST, full insert sequence |
| 1423280_at | 0.137 | Stmn2 | stathmin-like 2 |
| 1439207_at | 0.135 | Similar to paraneoplastic antigen like 5; paraneoplastic antigen family 5 (LOC386569), mRNA | |
| 1433789_at | 0.13 | Rnu17d | 18 days pregnant adult female placenta and extra embryonic tissue cDNA, RIKEN full-length enriched library, clone:3830421G02 product:unknown EST, full insert sequence |
| 1450943_at | 0.128 | Ndufa11 | NADH dehydrogenase (ubiquinone) 1 alpha subcomplex 11 |
| 1439746_at | 0.126 | C130085G02Rik | RIKEN cDNA C130085G02 gene |
| 1423327_at | 0.122 | D730048I06Rik | RIKEN cDNA D730048I06 gene |
| 1423378_at | 0.115 | Adam23 | a disintegrin and metalloprotease domain 23 |
| 1436388_a_at | 0.114 | 3830406C13Rik | RIKEN cDNA 3830406C13 gene |
| 1417156_at | 0.104 | Krt19 | Keratin 19 |
| 1437786_at | 0.095 | C80008 | Transcribed sequences |
| 1453874_at | 0.0878 | 4933401B06Rik | RIKEN cDNA 4933401B06 gene |
| 1456035_at | 0.0827 | Transcribed sequences | |
| 1425926_a_at | 0.0799 | Otx2 | Orthodenticle homolog 2 (Drosophila) |
| 1450177_at | 0.0773 | Ngfr | Nerve growth factor receptor (TNFR superfamily, member 16) |
| 1417122_at | 0.0767 | Vav3 | Vav 3 oncogene; synonyms: AA986410, MGC27838, A530094I06Rik; isoform 2 is encoded by transcript variant 2; Mus musculus vav 3 oncogene (Vav3), transcript variant 2, mRNA. |
| 1454254_s_at | 0.072 | Itgb1 | Integrin beta 1 (fibronectin receptor beta) |
| 1431711_a_at | 0.07 | Apbb2 | Amyloid beta (A4) precursor protein-binding, family B, member 2 |
| 1429701_at | 0.0689 | 2410003J06Rik | RIKEN cDNA 2410003J06 gene |
| 1434739_at | 0.0629 | 3830422N12Rik | RIKEN cDNA 3830422N12 gene |
| 1429203_at | 0.06 | 2410076I21Rik | RIKEN cDNA 2410076I21 gene |
| 1423523_at | 0.0587 | Aass | aminoadipate-semialdehyde synthase |
| 1436799_at | 0.0559 | D230005D02Rik | RIKEN cDNA D230005D02 gene |
| 1447021_at | 0.0509 | CDNA clone MGC:61032 IMAGE:30024827, complete cds | |
| 1419540_at | 0.0496 | Clec5a | C-type lectin domain family 5, member a |
| 1420357_s_at | 0.0474 | Fndc6 | Fibronectin type III domain containing 6 |
| 1448889_at | 0.0334 | Slc38a4 | Solute carrier family 38, member 4 |
| 1419241_a_at | 0.0192 | Aire | Autoimmune regulator (autoimmune polyendocrinopathy candidiasis ectodermal dystrophy) |
| 1453544_at | 0.0184 | 4933424F23Rik | RIKEN cDNA 4933424F23 gene |
| 1428111_at | 0.0165 | Slc38a4 | Solute carrier family 38, member 4 |
Average from three experiments.
Some genes have multiple distinct probes on the GeneChip® resulting in different fold-changes from different probes.
The mRNA levels of these genes were greater than 10-fold higher in F9 Wt cells than in F9 RARγ−/− cells cultured in the absence of RA: retrotransposon-like 1, meprin 1 beta, nitric oxide synthase trafficker, runt related transcription factor 1(Runx-1), transcription factor 2, Zfp521 (Evi3), GTL2 (the imprinted maternally expressed untranslated mRNA), prg (proteoglycan, secretory granule), tex13 (testis expressed gene 13), Mest, Tgfbi, Bmper (BMP-binding endothelial regulator), Dab2, Col13a1, BMP6, Foxq1, aquaporin 8, EMP2 and Amot (angiomotin). Since some of these genes, such as Dab2, BMP6, Foxq1, Hoxa1, and Col13a1, play critical roles in F9 endoderm differentiation, the lower expression of these genes in the F9 RARγ−/− cells relative to F9 Wt cells could reflect a reduced differentiation potential of the F9 RARγ−/− cells. The F9 RARγ−/− cells have been shown to exhibit reduced RA associated endoderm differentiation [30].
Genes exhibiting greater than 5-fold higher mRNA levels in F9 RARγ−/− cells than in F9 Wt cells in the absence of RA included Aass (aminoadipate-semialdehyde synthase), Slc38a4, Xlr3a, Aire, RIKEN cDNA 3830422N12 gene, RIKEN cDNA 4933424F23 gene, topoisomerase (DNA) I, Pla2g4a (Phospholipase A2, group IVA (cytosolic, calcium-dependent)), Ifi1 (Proteasome (prosome, macropain) 26S subunit, non-ATPase, 11), Slc1a1, and Otx2.
The number of genes expressed at higher levels in the F9 Wt cells relative to the F9 RARγ−/− cells was 5-fold greater than the number of genes expressed at reduced levels (894 versus 164) in the control condition (vehicle treated, 24 h) (Fig. 3). This could indicate that RARγ can mediate a substantial level of transcriptional activation in the absence of RA. A complete list of these genes is in the GEO database (www.ncbi.nlm.nih.gov/geo, Accession # GSE8431).
3.4. Distinctive Expression Profiles in the F9 Wt cell line following RA addition: RA-regulated genes
RA-responsive genes in this F9 stem cell differentiation system were identified by comparing F9 Wt cells treated with 1 μM RA for 24 h versus F9 Wt cells treated with vehicle only (ethanol). Of the 45,037 gene transcripts, 345 gene transcripts (0.76 %) exhibited at least a two-fold difference in expression after RA treatment (p ≤ 0.05, Welch’s t-test). Among these, 190 (0.42%) of the gene transcripts are up-regulated and 155 (0.34%) of the gene transcripts are down-regulated by at least two-fold after RA addition. The complete list of these 345 genes, including ESTs, can be found in the GEO database (www.ncbi.nlm.nih.gov/geo, Accession # GSE8431).
Many known RA responsive genes with RAREs were identified in this analysis. They include Cdx1, a number of Hox genes (Hox-a1, b1, a4), Cyp26A1, CRABPII, and RARβ (appendix 1). The list also includes genes known to be RA-responsive, but which lack a well characterized RARE; they include AKP2, Stra6, CRABP1, Ptge1, CRBPII, (RBP2), KcNK6, Kitl, Lgals3, Lgals1, PDGFα, and Pik3r1(P85α) [52]. We also found that some genes whose expression was known to be regulated by RA in other cell lines are also regulated by RA in F9 Wt cells. These genes include Gas1 [53], IL12b, Zfp503 [54], PMP22 [53, 55], Nrip1 (Rip140) [56], C3 complement [57], keratin8, Gfra3, and Myosin light regulatory polypeptide 7.
Appendix 1. RA responsive genes in the F9 Wt cells identified using MG-430.2.
F9 Wt cells were treated for 24 h with 1 μM RA or vehicle control. Only those genes with at least a 2.0-fold change in RA treated F9 Wt cells relative to control treated cells with p values ≤0.05 are shown. Fold change=F9 Wt+RA/F9 Wt control.
| Affymetrix ID | Fold Changeab | Gene Symbol | Gene Title |
|---|---|---|---|
| 1449582_at | 43.69 | Cdx1 | Caudal type homeo box 1 |
| 1427354_at | 40.16 | Hoxa4 | homeo box A4 |
| 1449397_at | 35.68 | Hoxb2 | Homeo box B2 |
| 1418415_at | 26.59 | Il12b | Interleukin 12b |
| 1455498_at | 23.18 | Transcribed sequence with weak similarity to protein pir:A43932 (H. sapiens) A43932 mucin 2 precursor, intestinal-human | |
| 1423836_at | 21.98 | Zfp503 | zinc finger protein 503 |
| 1422723_at | 16.94 | Stra6 | Stimulated by retinoic acid gene 6 |
| 1422674_s_at | 15.55 | Crygb | Crystallin, gamma B |
| 1453501_at | 13.08 | Il12b | Interleukin 12b |
| 1422617_at | 11.5 | Top1 | Topoisomerase (DNA) I |
| 1435693_at | 10.2 | BC012256 | cDNA sequence BC012256 |
| 1451191_at | 9.91 | Crabp1 | Cellular retinoic acid binding protein I |
| 1454906_at | 8.227 | Rarb | retinoic acid receptor, beta |
| 1419430_at | 7.714 | Cyp26a1 | Cytochrome P450, family 26, subfamily a, polypeptide 1 |
| 1456229_at | 7.222 | H3122A02-3 NIA Mouse 15K cDNA Clone Set Mus musculus cDNA clone H3122A02 3′, mRNA sequence. | |
| 1416855_at | 7.03 | Gas1 | BB550400 RIKEN full-length enriched, 2 days pregnant adult female oviduct Mus musculus cDNA clone E230022A16 3′ similar to X65128 M. musculus gas1 mRNA, mRNA sequence. |
| 1420603_s_at | 6.49 | Sgk3 | Serum/glucocorticoid regulated kinase 3 |
| 1418880_at | 6.354 | Gfra3 | Glial cell line derived neurotrophic factor family receptor alpha 3 |
| 1448926_at | 6.102 | Hoxa5 | Homeo box A5 |
| 1460379_at | 6.055 | Hoxb4 | homeo box B4 |
| 1420568_at | 5.652 | Msc | Musculin |
| 1455037_at | 5.247 | Plxna2 | plexin A2 |
| 1422007_at | 5.069 | Krt8 | Keratin 8 |
| 1420565_at | 5.057 | Hoxa1 | Homeo box A1 |
| 1451481_s_at | 5.049 | Cul7 | Cullin 7 |
| 1433428_x_at | 4.912 | Tgm2 | transglutaminase 2, C polypeptide |
| 1436075_at | 4.842 | Sfrp5 | secreted frizzled-related sequence protein 5 |
| 1416149_at | 4.695 | Akt2 | Thymoma viral proto-oncogene 2 |
| 1427233_at | 4.662 | Sdccag33 | serologically defined colon cancer antigen 33 |
| 1446358_at | 4.603 | 0 day neonate cerebellum cDNA, RIKEN full-length enriched library, clone:C230033K16 product:unknown EST, full insert sequence | |
| 1418084_at | 4.478 | Nrp1 | Neuropilin 1 |
| 1438512_at | 4.414 | LOC210321 | epididymal protein Av381126 |
| 1418445_at | 4.405 | Slc16a2 | solute carrier family 16 (monocarboxylic acid transporters), member 2 |
| 1423023_at | 4.404 | Sfrp5 | Secreted frizzled-related sequence protein 5 |
| 1421474_a_at | 4.372 | Capzb | Capping protein (actin filament) muscle Z-line, beta |
| 1448494_at | 4.321 | Gas1 | BB550400 RIKEN full-length enriched, 2 days pregnant adult female oviduct Mus musculus cDNA clone E230022A16 3′ similar to X65128 M. musculus gas1 mRNA, mRNA sequence. |
| 1449089_at | 4.31 | Nrip1 | synonyms: RIP140, 9630050P12, 6030458L20Rik; go_component: nucleus [goid 0005634] [evidence IDA] [pmid 9774688]; go_function: transcription co-repressor activity [goid 0003714] [evidence IDA] [pmid 9774688]; go_function: protein binding [goid 0005515] [evidence IPI] [pmid 9774688]; go_process: negative regulation of transcription from Pol II promoter [goid 0000122] [evidence IDA] [pmid 9774688]; Mus musculus nuclear receptor interacting protein 1 (Nrip1), mRNA. |
| 1437277_x_at | 4.262 | Tgm2 | transglutaminase 2, C polypeptide |
| 1434384_at | 4.237 | 8430438I05Rik | nuclear receptor interacting protein 1 |
| 1423835_at | 4.207 | Zfp503 | zinc finger protein 503 |
| 1418379_s_at | 4.133 | 1700034M03Rik | RIKEN cDNA 1700034M03 gene |
| 1418496_at | 4.116 | Foxa2 | Forkhead box A2 |
| 1435261_at | 4.087 | 4732416N19Rik | Adult male epididymis cDNA, RIKEN full-length enriched library, clone:9230114I18 product:unknown EST, full insert sequence |
| 1427433_s_at | 4.024 | 5730446D14Rik | RIKEN cDNA 5730446D14 gene |
| 1460605_at | 4.02 | AA606869 | Similar to Homeobox protein goosecoid-like (GSC-2) (LOC381848), mRNA |
| 1435184_at | 4.013 | B430320C24Rik | 0 day neonate lung cDNA, RIKEN full-length enriched library, clone:E030020K21 product:weakly similar to HYPOTHETICAL 13.8 KDA PROTEIN [Homo sapiens], full insert sequence |
| 1422099_a_at | 3.99 | Oprl1 | Opioid receptor-like 1 |
| 1448327_at | 3.961 | AI747699 | Expressed sequence AI747699 |
| 1417937_at | 3.932 | Ctsb | Cathepsin B |
| 1439341_at | 3.903 | Transcribed sequences | |
| 1422008_a_at | 3.886 | Krt8 | Keratin 8 |
| 1417133_at | 3.777 | Pmp22 | Peripheral myelin protein |
| 1439561_at | 3.759 | 2010012O05Rik | BB322051 RIKEN full-length enriched, adult male adrenal gland Mus musculus cDNA clone B330007D01 3′, mRNA sequence. |
| 1420753_at | 3.719 | Kitl | Kit ligand |
| 1419735_at | 3.71 | C3 | Complement component 3 |
| 1449370_at | 3.619 | Sox4 | SRY-box containing gene 4 |
| 1447211_at | 3.611 | Nrip1 | nuclear receptor interacting protein 1 |
| 1449071_at | 3.572 | Myl7 | Myosin, light polypeptide 7, regulatory |
| 1422597_at | 3.572 | Mmp15 | Matrix metallopeptidase 15 |
| 1452303_at | 3.485 | Arhgef10 | ceroid-lipofuscinosis, neuronal 8 |
| 1426927_at | 3.473 | Ap3b2 | adaptor-related protein complex 3, beta 2 subunit |
| 1418446_at | 3.453 | Slc16a2 | solute carrier family 16 (monocarboxylic acid transporters), member 2 |
| 1453286_at | 3.445 | Plxna2 | BB085537 RIKEN full-length enriched, adult male diencephalon Mus musculus cDNA clone 9330200E06 3′, mRNA sequence. |
| 1440143_at | 3.434 | F630022B06Rik | RIKEN cDNA F630022B06 gene |
| 1447845_s_at | 3.404 | Vnn1 | vanin 1 |
| 1454677_at | 3.373 | Timp2 | tissue inhibitor of metalloproteinase 2 |
| 1417500_a_at | 3.367 | Tgm2 | Transglutaminase 2, C polypeptide |
| 1418469_at | 3.359 | Nrip1 | synonyms: RIP140, 9630050P12, 6030458L20Rik; go_component: nucleus [goid 0005634] [evidence IDA] [pmid 9774688]; go_function: transcription co-repressor activity [goid 0003714] [evidence IDA] [pmid 9774688]; go_function: protein binding [goid 0005515] [evidence IPI] [pmid 9774688]; go_process: negative regulation of transcription from Pol II promoter [goid 0000122] [evidence IDA] [pmid 9774688]; Mus musculus nuclear receptor interacting protein 1 (Nrip1), mRNA. |
| 1433662_s_at | 3.353 | Timp2 | tissue inhibitor of metalloproteinase 2 |
| 1429134_at | 3.347 | 2900056N03Rik | RIKEN cDNA 2900056N03 gene |
| 1430425_at | 3.328 | 5330435L01Rik | RIKEN cDNA 5330435L01 gene |
| 1417956_at | 3.325 | Cidea | Cell death-inducing DNA fragmentation factor, alpha subunit-like effector A |
| 1420150_at | 3.316 | 4930422J18Rik | RIKEN cDNA 4930422J18 gene |
| 1450460_at | 3.233 | Krt8 | Keratin 8 |
| 1437347_at | 3.217 | Ednrb | endothelin receptor type B |
| 1448754_at | 3.201 | Rbp1 | Retinol binding protein 1, cellular |
| 1420831_at | 3.19 | Klhl22 | Kelch-like 22 (Drosophila) |
| 1456329_at | 3.171 | A230098A12Rik | RIKEN cDNA A230098A12 gene |
| 1456481_at | 3.169 | D9Ertd280e | hypothetical protein D930024E11 |
| 1418486_at | 3.073 | Edg2 | Endothelial differentiation, lysophosphatidic acid G-protein-coupled receptor, 2 |
| 1449364_at | 3.066 | Aurkc | Aurora kinase C |
| 1438682_at | 3.054 | Pik3r1 | Hypothetical protein C530050K14 (C530050K14), mRNA |
| 1432269_a_at | 2.995 | Sh3kbp1 | SH3-domain kinase binding protein 1 |
| 1439747_at | 2.987 | Ptges | prostaglandin E synthase |
| 1460204_at | 2.974 | Tsc2 | Tuberous sclerosis 2 |
| 1450624_at | 2.955 | Surf4 | Surfeit gene 4 |
| 1419959_s_at | 2.882 | C330003B14Rik | RIKEN cDNA C330003B14 gene |
| 1435743_at | 2.882 | C130068N17Rik | RIKEN cDNA C130068N17 gene |
| 1449925_at | 2.88 | Cxcr3 | Chemokine (C-X-C motif) receptor 3 |
| 1416674_at | 2.862 | Lyzs | Lysozyme |
| 1438333_at | 2.833 | A230098A12Rik | RIKEN cDNA A230098A12 gene |
| 1424194_at | 2.82 | Rcsd1 | RCSD domain containing 1 |
| 1448000_at | 2.804 | Cdca3 | AV352659 RIKEN full-length enriched, 11 days embryo gonad Mus musculus cDNA clone 7030413D12 3′, mRNA sequence. |
| 1429074_at | 2.771 | Eif5a2 | Eukaryotic translation initiation factor 5A2 |
| 1428097_at | 2.771 | 2510009E07Rik | RIKEN cDNA 2510009E07 gene, mRNA (cDNA clone IMAGE:6491720), partial cds |
| 1426589_at | 2.722 | Gab3 | growth factor receptor bound protein 2-associated protein 3 |
| 1460287_at | 2.712 | Timp2 | Tissue inhibitor of metalloproteinase 2 |
| 1453622_s_at | 2.699 | Mllt3 | Myeloid/lymphoid or mixed lineage-leukemia translocation to 3 homolog (Drosophila); synonyms: Af9, D4Ertd321e, 2210011H10Rik, 2610012I03Rik, 3830408D16Rik; isoform 2 is encoded by transcript variant 2; Mus musculus myeloid/lymphoid or mixed lineage-leukemia translocation to 3 homolog (Drosophila) (Mllt3), transcript variant 2, mRNA. |
| 1424437_s_at | 2.696 | Mtap4 | Microtubule-associated protein 4 |
| 1457829_at | 2.681 | Clgn | Calmegin |
| 1430240_a_at | 2.654 | Clgn | Calmegin |
| 1447326_s_at | 2.638 | Zfp261 | zinc finger protein 261 |
| 1415921_a_at | 2.617 | Tnf | Tumor necrosis factor |
| 1419106_at | 2.611 | Tmem97 | Transmembrane protein 97 |
| 1427195_at | 2.61 | Transcribed sequence with weak similarity to protein ref:NP_081764.1 (M. musculus) RIKEN cDNA 5730493B19 [Mus musculus] | |
| 1419155_a_at | 2.597 | Sox4 | SRY-box containing gene 4 |
| 1429959_at | 2.585 | 6620401D04Rik | 12 days embryo female ovary cDNA, RIKEN full-length enriched library, clone:6620401D04 product:interleukin 10 receptor, beta, full insert sequence |
| 1439734_at | 2.58 | Transcribed sequence with weak similarity to protein pir:S12207 (M. musculus) S12207 hypothetical protein | |
| 1448470_at | 2.578 | Sod2 | Superoxide dismutase 2, mitochondrial |
| 1443882_at | 2.575 | Transcribed sequence with weak similarity to protein ref:NP_081764.1 (M. musculus) RIKEN cDNA 5730493B19 [Mus musculus] | |
| 1448825_at | 2.568 | Pdk2 | Pyruvate dehydrogenase kinase, isoenzyme 2 |
| 1451277_at | 2.551 | Zfp131 | Zinc finger protein 131 |
| 1431602_a_at | 2.547 | 1810064L21Rik | RIKEN cDNA A230065J02 gene |
| 1455958_s_at | 2.538 | 9130017A15Rik | RIKEN cDNA 9130017A15 gene |
| 1420832_at | 2.535 | Klhl22 | Kelch-like 22 (Drosophila) |
| 1452141_a_at | 2.534 | Sepp1 | Selenoprotein P, plasma, 1 |
| 1452302_at | 2.534 | Arhgef10 | ceroid-lipofuscinosis, neuronal 8 |
| 1449450_at | 2.532 | Ptges | Prostaglandin E synthase |
| 1419602_at | 2.524 | Hoxa2 | Homeo box A2 |
| 1458798_at | 2.501 | Transcribed sequences | |
| 1424334_at | 2.497 | Selpl | Selectin, platelet (p-selectin) ligand |
| 1460337_at | 2.454 | Sh3kbp1 | SH3-domain kinase binding protein 1 |
| 1455796_x_at | 2.441 | Olfm1 | olfactomedin 1 |
| 1448024_at | 2.44 | B430320C24Rik | 0 day neonate lung cDNA, RIKEN full-length enriched library, clone:E030020K21 product:weakly similar to HYPOTHETICAL 13.8 KDA PROTEIN [Homo sapiens], full insert sequence |
| 1417605_s_at | 2.411 | St3gal2 | ST3 beta-galactoside alpha-2,3-sialyltransferase 2 |
| 1460206_at | 2.406 | Mybbp1a | MYB binding protein (P160) 1a |
| 1450673_at | 2.398 | Col9a2 | Procollagen, type IX, alpha 2 |
| 1429483_at | 2.395 | Ndp52l1 | H3048G12-3 NIA Mouse 15K cDNA Clone Set Mus musculus cDNA clone H3048G12 3′, mRNA sequence. |
| 1435740_at | 2.377 | E330020C23 | hypothetical protein E330020C23 |
| 1454752_at | 2.371 | AI606861 | Similar to dJ259A10.1 (ssDNA binding protein (SEB4D)) (LOC380843), mRNA |
| 1448147_at | 2.365 | Tnf | Tumor necrosis factor |
| 1433844_a_at | 2.362 | Dusp9 | dual specificity phosphatase 9 |
| 1439810_s_at | 2.358 | Pramel7 | preferentially expressed antigen in melanoma like 7 |
| 1420924_at | 2.353 | Timp2 | Tissue inhibitor of metalloproteinase 2 |
| 1433795_at | 2.352 | Tgfbr3 | 18-day embryo whole body cDNA, RIKEN full-length enriched library, clone:1110036H20 product:unknown EST, full insert sequence |
| 1420774_a_at | 2.343 | 4930570C03Rik | RIKEN cDNA 4930570C03 gene |
| 1434856_at | 2.342 | A130096K20 | hypothetical protein A130096K20 |
| 1432331_a_at | 2.338 | Prrx2 | Paired related homeobox 2 |
| 1418517_at | 2.323 | Gja4 | Gap junction membrane channel protein alpha 4 |
| 1454877_at | 2.315 | Sertad4 | 16 days embryo head cDNA, RIKEN full-length enriched library, clone:C130018M11 product:unclassifiable, full insert sequence |
| 1454727_at | 2.296 | AI173486 | expressed sequence AI173486 |
| 1425212_a_at | 2.289 | Tnf | Tumor necrosis factor |
| 1417625_s_at | 2.281 | Cmkor1 | Chemokine orphan receptor 1 |
| 1433575_at | 2.281 | Sox4 | SRY-box containing gene 4 |
| 1448727_at | 2.277 | Bcl10 | B-cell leukemia/lymphoma 10 |
| 1419276_at | 2.255 | Enpp1 | Ectonucleotide pyrophosphatase/phosphodiesterase 1 |
| 1452861_at | 2.245 | 2010300C02Rik | RIKEN cDNA 2010300C02 gene |
| 1456210_at | 2.241 | Transcribed sequences | |
| 1433845_x_at | 2.226 | Dusp9 | dual specificity phosphatase 9 |
| 1430650_at | 2.223 | Zfp191 | zinc finger protein 191 |
| 1460570_at | 2.217 | 2900019M05Rik | piggyBac transposable element derived 5 |
| 1447537_at | 2.217 | 1500032P08Rik | properdin factor, complement |
| 1419301_at | 2.216 | Fzd4 | 602109129F1 NCI_CGAP_Kid14 Mus musculus cDNA clone IMAGE:4237519 5′, mRNA sequence. |
| 1419309_at | 2.213 | Pdpn | Podoplanin |
| 1417794_at | 2.197 | Casp2 | Caspase 2 |
| 1427401_at | 2.185 | Chrna5 | cholinergic receptor, nicotinic, alpha polypeptide 5 |
| 1458878_at | 2.175 | Yes | Yamaguchi sarcoma viral (v-yes) oncogene homolog |
| 1434277_a_at | 2.171 | 6430570G24 | Adult male olfactory brain cDNA, RIKEN full-length enriched library, clone:6430570G24 product:unknown EST, full insert sequence |
| 1435342_at | 2.17 | Kcnk6 | potassium inwardly-rectifying channel, subfamily K, member 6 |
| 1450839_at | 2.167 | D0H4S114 | DNA segment, human D4S114 |
| 1419157_at | 2.159 | Sox4 | SRY-box containing gene 4 |
| 1434528_at | 2.15 | Aard | alanine and arginine rich domain containing protein |
| 1449110_at | 2.146 | App | Amyloid beta (A4) precursor protein |
| 1454737_at | 2.143 | Dusp9 | dual specificity phosphatase 9 |
| 1419693_at | 2.143 | Colec12 | Collectin sub-family member 12 |
| 1450992_a_at | 2.136 | Meis1 | myeloid ecotropic viral integration site 1 |
| 1426514_at | 2.133 | Agrin | Agrin |
| 1417604_at | 2.13 | St3gal2 | ST3 beta-galactoside alpha-2,3-sialyltransferase 2 |
| 1418709_at | 2.126 | Adarb1 | Adenosine deaminase, RNA-specific, B1 |
| 1426785_s_at | 2.124 | Mgll | monoglyceride lipase |
| 1449449_at | 2.118 | Ptges | Prostaglandin E synthase |
| 1453836_a_at | 2.112 | Mgll | Monoglyceride lipase |
| 1422088_at | 2.102 | Lmyc1 | lung carcinoma myc related oncogene 1 |
| 1424951_at | 2.089 | Cacna1d | Calcium channel, voltage-dependent, L type, alpha 1D subunit |
| 1454082_a_at | 2.076 | Giyd2 | GIY-YIG domain containing 2 |
| 1419156_at | 2.073 | Sox4 | SRY-box containing gene 4 |
| 1452888_at | 2.061 | 1110034G24Rik | RIKEN cDNA 1110034G24 gene |
| 1417129_a_at | 2.059 | Mrg1 | Myeloid ecotropic viral integration site-related gene 1 |
| 1418617_x_at | 2.05 | Clgn | Calmegin |
| 1426869_at | 2.05 | Boc | biregional cell adhesion molecule-related/down-regulated by oncogenes (Cdon) binding protein |
| 1460378_a_at | 2.04 | Myb; Tes | Myeloblastosis oncogene; synonyms: TESS, Tes1, Tes2, testin2, D6Ertd352e; isoform 2 is encoded by transcript variant 2; testin 2; Mus musculus testis derived transcript (Tes), transcript variant 2, mRNA. |
| 1418505_at | 2.036 | Ogdh | Oxoglutarate dehydrogenase (lipoamide) |
| 1434069_at | 2.035 | BC067047 | Adult male testis cDNA, RIKEN full-length enriched library, clone:4930413J11 product:mitochondria located 1 homolog (human), full insert sequence |
| 1442180_at | 2.026 | BC038059 | cDNA sequence BC038059 |
| 1455642_a_at | 2.023 | Fbxo23 | F-box only protein 23 |
| 1426991_at | 2.016 | 1810048J11Rik | RIKEN cDNA 1810048J11 gene |
| 1459818_x_at | 2.014 | Zfp261 | zinc finger protein 261 |
| 1434756_at | 2.013 | 5430421B17 | frizzled homolog 4 (Drosophila) |
| 1456475_s_at | 2.01 | Prkar2b | protein kinase, cAMP dependent regulatory, type II beta |
| 1442308_at | 2.006 | Smyd4 | SET and MYND domain containing 4 |
| 1418872_at | 0.499 | Arsk | Arylsulfatase K |
| 1416967_at | 0.499 | Sox2 | SRY-box containing gene 2 |
| 1458277_at | 0.498 | Ccl25 | chemokine (C-C motif) ligand 25 |
| 1424652_at | 0.498 | Kctd10 | Potassium channel tetramerisation domain containing 10 |
| 1435436_at | 0.497 | Transcribed sequences | |
| 1443822_s_at | 0.497 | D10Ertd214e | DNA segment, Chr 10, ERATO Doi 214, expressed |
| 1450728_at | 0.496 | Fjx1 | AV230815 RIKEN full-length enriched, 0 day neonate skin Mus musculus cDNA clone 4632401J20 3′, mRNA sequence. |
| 1450282_at | 0.496 | Fgf4 | Fibroblast growth factor 4 |
| 1421052_a_at | 0.492 | Sms | Spermine synthase |
| 1430780_a_at | 0.491 | Pmm1 | phosphomannomutase 1 |
| 1428775_at | 0.491 | 1110008L16Rik | RIKEN cDNA 1110008L16 gene |
| 1416605_at | 0.49 | Prkcbp1 | Protein kinase C binding protein 1 |
| 1416505_at | 0.489 | Nr4a1 | Nuclear receptor subfamily 4, group A, member 1 |
| 1447703_x_at | 0.489 | 3110024A21Rik | AV214133 RIKEN full-length enriched, ES cells Mus musculus cDNA clone 2410133J07 3′, mRNA sequence. |
| 1447934_at | 0.488 | 9630033F20Rik | RIKEN cDNA 9630033F20 gene |
| 1420964_at | 0.488 | Enc1 | ectodermal-neural cortex 1 |
| 1451026_at | 0.487 | Ftsj3 | FtsJ homolog 3 (E. coli) |
| 1459658_at | 0.487 | Mcm5 | minichromosome maintenance deficient 5, cell division cycle 46 (S. cerevisiae) |
| 1428869_at | 0.486 | Nolc1 | nucleolar and coiled-body phosphoprotein 1 |
| 1421260_a_at | 0.484 | Pgk1 | Phosphoglycerate kinase 1 |
| 1428069_at | 0.483 | Ubqln1 | Ubiquilin 1 |
| 1444883_at | 0.482 | Tmem19 | L0079C06-3 NIA Mouse E12.5 Female Mesonephros and Gonads cDNA Library Mus musculus cDNA clone L0079C06 3′, mRNA sequence. |
| 1430433_at | 0.481 | 4933406J08Rik | RIKEN cDNA 4933406J08 gene |
| 1450928_at | 0.481 | Idb4 | inhibitor of DNA binding 4 |
| 1445924_at | 0.48 | Transcribed sequence with weak similarity to protein ref:NP_060730.1 (H. sapiens) hypothetical protein FLJ10891 [Homo sapiens] | |
| 1441243_at | 0.478 | Transcribed sequence with weak similarity to protein ref:NP_081764.1 (M. musculus) RIKEN cDNA 5730493B19 [Mus musculus] | |
| 1452609_at | 0.477 | 1190005I06Rik | RIKEN cDNA 1190005I06 gene |
| 1418649_at | 0.477 | Egln3 | EGL nine homolog 3 (C. elegans) |
| 1448272_at | 0.477 | Btg2 | B-cell translocation gene 2, anti-proliferative |
| 1444390_at | 0.476 | Transcribed sequences | |
| 1453251_at | 0.476 | Tyrp1 | Tyrosinase-related protein 1 |
| 1435367_at | 0.476 | Mapk4 | mitogen-activated protein kinase 4 |
| 1417725_a_at | 0.475 | Mtvr2 | Mammary tumor virus receptor 2 |
| 1430208_at | 0.474 | Pir | Pirin |
| 1421113_at | 0.474 | Whrn | Whirlin |
| 1448566_at | 0.469 | Cdc42 | Cell division cycle 42 homolog (S. cerevisiae) |
| 1415938_at | 0.469 | Spink3 | Serine peptidase inhibitor, Kazal type 3 |
| 1422786_at | 0.469 | Slc30a1 | Solute carrier family 30 (zinc transporter), member 1 |
| 1431193_at | 0.464 | 2610524B04Rik | RIKEN cDNA 4932409F03 gene |
| 1454197_a_at | 0.464 | Calb1 | Calbindin-28K |
| 1427640_a_at | 0.463 | Runx1t1 | Runt-related transcription factor 1; translocated to, 1 (cyclin D-related) |
| 1424880_at | 0.463 | Trib1 | tribbles homolog 1 (Drosophila) |
| 1451123_at | 0.462 | D19Wsu12e | DNA segment, Chr 19, Wayne State University 12, expressed |
| 1429937_at | 0.461 | AI662478; BB238373; 1700010I21Rik | RIKEN cDNA D530033C11 gene |
| 1417780_at | 0.459 | Lass4 | longevity assurance homolog 4 (S. cerevisiae) |
| 1428377_at | 0.459 | Btbd11 | BTB (POZ) domain containing 11 |
| 1429123_at | 0.457 | Rab27a | RAB27A, member RAS oncogene family |
| 1419700_a_at | 0.455 | Prom1 | Prominin 1 |
| 1436865_at | 0.454 | F630021I08Rik | RIKEN cDNA F630021I08 gene |
| 1433959_at | 0.454 | 9630048M01Rik | RIKEN cDNA 9630048M01 gene |
| 1456296_at | 0.453 | 5832426L23Rik | RIKEN cDNA 5832426L23 gene |
| 1455890_x_at | 0.453 | Snrpn | small nuclear ribonucleoprotein N |
| 1444292_at | 0.453 | D7Ertd143e | NACHT, LRR and PYD containing protein 12 |
| 1437554_at | 0.452 | Plec1 | plectin 1 |
| 1436319_at | 0.452 | Sulf1 | sulfatase 1 |
| 1423748_at | 0.452 | B830012B01; D530020C15Rik | pyruvate dehydrogenase kinase, isoenzyme 1 |
| 1423259_at | 0.451 | Idb4 | inhibitor of DNA binding 4 |
| 1434815_a_at | 0.45 | AI874665 | mq46b10.x1 Soares_thymus_2NbMT Mus musculus cDNA clone IMAGE:581755 3′, mRNA sequence. |
| 1419708_at | 0.45 | Wnt6 | Wingless-related MMTV integration site 6 |
| 1427023_at | 0.449 | Phyhipl | RIKEN cDNA 4921522K17 gene |
| 1449484_at | 0.449 | Hfe2 | Hemochromatosis type 2 (juvenile) (human homolog) |
| 1449187_at | 0.448 | Pdgfa | BB371842 RIKEN full-length enriched, 16 days embryo head Mus musculus cDNA clone C130061I20 3′, mRNA sequence. |
| 1447181_s_at | 0.446 | Slc7a7 | solute carrier family 7 (cationic amino acid transporter, y+ system), member 7 |
| 1424310_at | 0.446 | Mocs2 | molybdenum cofactor synthesis 2 |
| 1443435_at | 0.445 | Transcribed sequences | |
| 1423786_at | 0.444 | Eif2c3 | Eukaryotic translation initiation factor 2C, 3 |
| 1424643_at | 0.444 | Tcof1 | Treacher Collins Franceschetti syndrome 1, homolog |
| 1428549_at | 0.443 | Ccdc3 | coiled-coil domain containing 3 |
| 1426041_a_at | 0.442 | Bre; Fgd4 | Brain and reproductive organ-expressed protein; synonyms: Frabp, ZFYVE6, 9030023J02Rik, 9330209B17Rik; isoform beta is encoded by transcript variant beta; frabin; Fgd1-related F-actin-binding protein; Mus musculus FYVE, RhoGEF and PH domain containing 4 (Fgd4), transcript variant beta, mRNA.; isoform gamma is encoded by transcript variant gamma; Mus musculus FYVE, RhoGEF and PH domain containing 4 (Fgd4), transcript variant gamma, mRNA. |
| 1449996_a_at | 0.441 | Hspa8 | Heat shock protein 8 |
| 1449031_at | 0.441 | Cited1 | Cbp/p300-interacting transactivator with Glu/Asp-rich carboxy-terminal domain 1 |
| 1420086_x_at | 0.436 | Fgf4 | fibroblast growth factor 4 |
| 1429442_at | 0.433 | 4921503C21Rik | RIKEN cDNA 4921503C21 gene |
| 1422887_a_at | 0.432 | Ctbp2 | |
| 1449146_at | 0.431 | Notch4 | Notch gene homolog 4 (Drosophila) |
| 1427537_at | 0.428 | EPIPL; EPIPL1; 6230424I18Rik | epiplakin 1 |
| 1443241_at | 0.427 | 13 days embryo stomach cDNA, RIKEN full-length enriched library, clone:D530023N15 product:unclassifiable, full insert sequence | |
| 1417392_a_at | 0.426 | My+lat1; AI790233 | solute carrier family 7 (cationic amino acid transporter, y+ system), member 7 |
| 1417061_at | 0.426 | Cdc42 | Cell division cycle 42 homolog (S. cerevisiae) |
| 1427364_a_at | 0.423 | Odc1 | Ornithine decarboxylase, structural 1 |
| 1448931_at | 0.421 | F2rl1 | Coagulation factor II (thrombin) receptor-like 1 |
| 1448568_a_at | 0.418 | Keap1 | Kelch-like ECH-associated protein 1 |
| 1434705_at | 0.417 | D7Wsu87e | TRAF-binding protein |
| 1451527_at | 0.417 | Pcolce2 | Procollagen C-endopeptidase enhancer 2 |
| 1428283_at | 0.412 | Cyp2s1 | Cytochrome P450, family 2, subfamily s, polypeptide 1 |
| 1434239_at | 0.411 | AA408556 | expressed sequence AA408556 |
| 1453072_at | 0.411 | Gpr160 | G protein-coupled receptor 160 |
| 1452384_at | 0.411 | Enpp3 | ectonucleotide pyrophosphatase/phosphodiesterase 3 |
| 1415944_at | 0.411 | Sdc1 | syndecan 1 |
| 1427302_at | 0.409 | Enpp3 | ectonucleotide pyrophosphatase/phosphodiesterase 3 |
| 1423747_a_at | 0.409 | B830012B01; D530020C15Rik | pyruvate dehydrogenase kinase, isoenzyme 1 |
| 1418744_s_at | 0.407 | Musk | Muscle, skeletal, receptor tyrosine kinase |
| 1448688_at | 0.406 | Podxl | Podocalyxin-like |
| 1436227_at | 0.4 | Ebaf | endometrial bleeding associated factor |
| 1431425_a_at | 0.399 | 4930535B03Rik | RIKEN cDNA 4930535B03 gene |
| 1451502_at | 0.394 | Pla2g10 | Phospholipase A2, group X |
| 1444458_at | 0.393 | Transcribed sequence with moderate similarity to protein pir:S12207 (M. musculus) S12207 hypothetical protein | |
| 1422912_at | 0.392 | Bmp4 | Bone morphogenetic protein 4 |
| 1433733_a_at | 0.39 | Cry1 | cryptochrome 1 (photolyase-like) |
| 1434458_at | 0.388 | Fst | follistatin |
| 1438160_x_at | 0.379 | Slco4a1 | AV348121 RIKEN full-length enriched, adult male olfactory bulb Mus musculus cDNA clone 6430631M08 3′, mRNA sequence. |
| 1449455_at | 0.375 | Ehd4 | EH-domain containing 4 |
| 1442379_at | 0.37 | Transcribed sequences | |
| 1455692_x_at | 0.37 | 1700097N02Rik | Adult male testis cDNA, RIKEN full-length enriched library, clone:1700122G02 product:unknown EST, full insert sequence |
| 1449109_at | 0.365 | Socs2 | Suppressor of cytokine signaling 2 |
| 1420085_at | 0.36 | Fgf4 | fibroblast growth factor 4 |
| 1436562_at | 0.359 | Ddx58 | RIKEN cDNA 6430573D20 gene |
| 1416316_at | 0.355 | Slc27a2 | Solute carrier family 27 (fatty acid transporter), member 2 |
| 1438861_at | 0.349 | Bnc2 | RIKEN cDNA 5031434M05 gene |
| 1449231_at | 0.349 | 1110008L16Rik | RIKEN cDNA 1110008L16 gene |
| 1432418_a_at | 0.342 | Ckmt1 | Creatine kinase, mitochondrial 1, ubiquitous |
| 1421385_a_at | 0.339 | Zfp191 | Zinc finger protein 191 |
| 1424531_a_at | 0.331 | Tcea3 | Transcription elongation factor A (SII), 3 |
| 1452004_at | 0.325 | Calca | Calcitonin/calcitonin-related polypeptide, alpha |
| 1443786_at | 0.321 | Utf1 | BB000218 RIKEN full-length enriched, ES cells XhoI/SstI Mus musculus cDNA clone 2410014O14 3′ similar to NM_009482 Mus musculus undifferentiated embryonic cell transcription factor 1 (Utf1), mRNA sequence. |
| 1438084_at | 0.32 | Transcribed sequences | |
| 1450771_at | 0.319 | Dgcr2 | DiGeorge syndrome critical region gene 2 |
| 1417745_at | 0.318 | Hnrpu | Heterogeneous nuclear ribonucleoprotein U |
| 1437004_at | 0.318 | C78977 | Similar to tripartite motif-containing 43 (LOC236469), mRNA |
| 1416899_at | 0.317 | Utf1 | Undifferentiated embryonic cell transcription factor 1 |
| 1426673_at | 0.315 | Cdh3 | Cadherin 3 |
| 1425284_a_at | 0.314 | Ash; 2210402C08Rik; 2410003M20Rik; 4933437C11Rik | RAB27A, member RAS oncogene family |
| 1422952_at | 0.308 | Ng23 | |
| 1440563_at | 0.299 | 15 days embryo head cDNA, RIKEN full-length enriched library, clone:D930010N06 product:unknown EST, full insert sequence | |
| 1426431_at | 0.294 | Jag2 | AV264681 RIKEN full-length enriched, adult male testis (DH10B) Mus musculus cDNA clone 4930502C11 3′ similar to Y14331 Mus musculus partial mRNA for jagged2 protein, clone MB16, mRNA sequence. |
| 1416630_at | 0.289 | Id3 | Inhibitor of DNA binding 3 |
| 1432018_at | 0.287 | Mas1 | MAS1 oncogene |
| 1439260_a_at | 0.286 | Enpp3 | BB039510 RIKEN full-length enriched, 13 days embryo male testis Mus musculus cDNA clone 6030443A19 3′, mRNA sequence. |
| 1459469_at | 0.28 | C78516 | Transcribed sequences |
| 1437588_at | 0.279 | Pou4f2 | POU domain, class 4, transcription factor 2 |
| 1426808_at | 0.278 | Lgals3 | Lectin, galactose binding, soluble 3 |
| 1419930_at | 0.271 | D15Ertd55e | Transcribed sequences |
| 1450194_a_at | 0.27 | c-myb; M16449; AI550390; MGC18531; Myb | myeloblastosis oncogene; synonyms: c-myb, M16449, AI550390, MGC18531; isoform 2 is encoded by transcript variant 2; myb protein; tumor-specific myb protein; Mus musculus myeloblastosis oncogene (Myb), transcript variant 2, mRNA. |
| 1438068_at | 0.269 | BB251859 RIKEN full-length enriched, 7 days neonate cerebellum Mus musculus cDNA clone A730048G08 3′, mRNA sequence. | |
| 1450989_at | 0.269 | Tdgf1 | teratocarcinoma-derived growth factor |
| 1425926_a_at | 0.269 | Otx2 | Orthodenticle homolog 2 (Drosophila) |
| 1423232_at | 0.256 | Map2k5 | Mitogen activated protein kinase kinase 5 |
| 1440867_at | 0.251 | Spry4 | sprouty homolog 4 (Drosophila) |
| 1439746_at | 0.246 | C130085G02Rik | RIKEN cDNA C130085G02 gene |
| 1421317_x_at | 0.245 | c-myb; M16449; AI550390; MGC18531; Myb | myeloblastosis oncogene; synonyms: c-myb, M16449, AI550390, MGC18531; isoform 2 is encoded by transcript variant 2; myb protein; tumor-specific myb protein; Mus musculus myeloblastosis oncogene (Myb), transcript variant 2, mRNA. |
| 1422021_at | 0.244 | Spry4 | sprouty homolog 4 (Drosophila) |
| 1438824_at | 0.234 | Slc20a1 | solute carrier family 20, member 1 |
| 1424942_a_at | 0.219 | Myc | Myelocytomatosis oncogene |
| 1445669_at | 0.217 | Spry4 | sprouty homolog 4 (Drosophila) |
| 1450177_at | 0.217 | Ngfr | nerve growth factor receptor (TNFR superfamily, member 16) |
| 1449064_at | 0.212 | Mab21l2 | Mab-21-like 2 (C. elegans) |
| 1430086_at | 0.202 | Chrna9 | cholinergic receptor, nicotinic, alpha polypeptide 9 |
| 1436657_at | 0.202 | LOC380738 (LOC380738), mRNA | |
| 1453345_at | 0.196 | 3830408G10Rik | RIKEN cDNA 3830408G10 gene |
| 1454974_at | 0.151 | Ntn1 | syntaxin 8 |
| 1423378_at | 0.135 | Adam23 | a disintegrin and metalloprotease domain 23 |
| 1444823_at | 0.131 | Transcribed sequences | |
| 1420463_at | 0.0989 | Clnk | Cytokine-dependent hematopoietic cell linker |
| 1457402_at | 0.0939 | 0 day neonate lung cDNA, RIKEN full-length enriched library, clone:E030031B17 product:unknown EST, full insert sequence | |
| 1431711_a_at | 0.0812 | Apbb2 | Amyloid beta (A4) precursor protein-binding, family B, member 2 |
We identified novel RA-regulated genes which exhibited greater than 2-fold changes compared to the levels of expression in untreated F9 Wt cells, such as Serum/glucocorticoid regulated kinase 3, Topoisomerase (DNA) I, Musculin, plexin A2, Cullin 7, Akt2, serologically defined colon cancer antigen 33, Neuropilin 1, epididymal protein Av381126, Opioid receptor-like 1, Cathepsin B, Matrix metallopeptidase 15, ceroid-lipofuscinosis neuronal 8, vanin 1, endothelin receptor type B, Tuberous sclerosis 2, Chemokine (C-X-C motif) receptor 3, Eukaryotic translation initiation factor 5A2, growth factor receptor bound protein 2-associated protein 3, Microtubule-associated protein 4, and zinc finger protein 261 (Appendix 1).
3.5. Expression Profiles in the F9 Wt and RARγ−/− cell lines following RA addition: RA-regulated genes
To identify RARγ-mediated, RA responsive genes, we compared the differentially expressed genes between the two cell types to the RA-responsive genes in the F9 Wt cells in a Venn diagram (Fig. 4). Among the 345 probe sets which were identified as RA-responsive by at least a two-fold change in expression levels after exposure to RA in F9 Wt cells, 161 probe sets were found to be differentially expressed between F9 Wt and RARγ−/− cells after a 24 h exposure to RA (Appendix 2).
Figure 4. Venn diagram of 1058 genes (Wt vs RARγ−/−), 1348 genes (Wt+RA vs RARγ−/−+RA) and 345 genes (Wt+RA vs Wt).
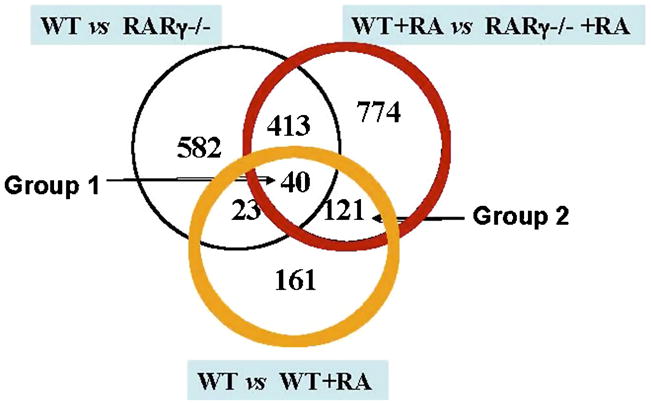
Genes from Figure 3 were analyzed to determine which of the RA-responsive genes were also differentially expressed between the F9 Wt cells and F9 RARγ−/− cells.
Appendix 2.
RA-responsive genes with at least a 2-fold change in expression in F9 Wt cells relative to F9 RARγ−/− cells after RA treatment for 24 h. Fold change = F9 Wt +RA/F9 RARγ−/− +RA.
| Affymetrix ID | Fold Changeab | Gene Symbol | Gene Title |
|---|---|---|---|
| 1455498_at | 122.6 | Transcribed sequence with weak similarity to protein pir:A43932 (H. sapiens) A43932 mucin 2 precursor, intestinal – human | |
| 1423023_at | 24.61 | Sfrp5 | secreted frizzled-related sequence protein 5 |
| 1448926_at | 22.68 | Hoxa5 | Homeo box A5 |
| 1437347_at | 21.85 | Ednrb | endothelin receptor type B |
| 1420603_s_at | 14.36 | Raet1a | retinoic acid early transcript alpha |
| 1433428_x_at | 12.67 | Tgm2 | transglutaminase 2, C polypeptide |
| 1416855_at | 12.61 | Gas1 | BB550400 RIKEN full-length enriched, 2 days pregnant adult female oviduct Mus musculus cDNA clone E230022A16 3′ similar to X65128 M.musculus gas1 mRNA, mRNA sequence. |
| 1450624_at | 10.81 | Bhmt | betaine-homocysteine methyltransferase |
| 1451191_at | 9.949 | Crabp2 | cellular retinoic acid binding protein II |
| 1436075_at | 8.714 | Sfrp5 | secreted frizzled-related sequence protein 5 |
| 1437277_x_at | 8.36 | Tgm2 | transglutaminase 2, C polypeptide |
| 1453501_at | 8.34 | Hoxb1 | Homeo box B1 |
| 1448470_at | 7.705 | Fbp1 | fructose bisphosphatase 1 |
| 1421385_a_at | 7.631 | Myo7a | Myosin VIIa |
| 1430425_at | 6.958 | 5330435L01Rik | RIKEN cDNA 5330435L01 gene |
| 1450839_at | 6.684 | D0H4S114 | DNA segment, human D4S114 |
| 1420924_at | 6.492 | Timp2 | tissue inhibitor of metalloproteinase 2 |
| 1428097_at | 6.483 | 2510009E07Rik | RIKEN cDNA 2510009E07 gene, mRNA (cDNA clone IMAGE:6491720), partial cds |
| 1438512_at | 6.477 | LOC210321 | epididymal protein Av381126 |
| 1426785_s_at | 6.38 | Mgll | monoglyceride lipase |
| 1439810_s_at | 6.263 | Pramel7 | preferentially expressed antigen in melanoma like 7 |
| 1433662_s_at | 5.664 | Timp2 | tissue inhibitor of metalloproteinase 2 |
| 1427433_s_at | 5.661 | 5730446D14Rik | RIKEN cDNA 5730446D14 gene |
| 1454677_at | 5.545 | Timp2 | tissue inhibitor of metalloproteinase 2 |
| 1448494_at | 5.482 | Gas1 | BB550400 RIKEN full-length enriched, 2 days pregnant adult female oviduct Mus musculus cDNA clone E230022A16 3′ similar to X65128 M.musculus gas1 mRNA, mRNA sequence. |
| 1417500_a_at | 5.389 | Tgm2 | transglutaminase 2, C polypeptide |
| 1453836_a_at | 5.373 | Mgll | monoglyceride lipase |
| 1423835_at | 5.279 | Zfp503 | Zinc finger protein 503 |
| 1451481_s_at | 5.22 | D630035O19Rik | RIKEN cDNA D630035O19 gene |
| 1448825_at | 5.09 | Pdk2 | pyruvate dehydrogenase kinase, isoenzyme 2 |
| 1449397_at | 5.047 | Hoxb2 | Homeo box B2 |
| 1450992_a_at | 5.013 | Meis1 | myeloid ecotropic viral integration site 1 |
| 1433795_at | 4.994 | Tgfbr3 | 18-day embryo whole body cDNA, RIKEN full-length enriched library, clone:1110036H20 product:unknown EST, full insert sequence |
| 1419430_at | 4.84 | Cyp26a1 | cytochrome P450, family 26, subfamily a, polypeptide 1 |
| 1447845_s_at | 4.806 | Vnn1 | Vanin 1 |
| 1453286_at | 4.747 | Plxna2 | BB085537 RIKEN full-length enriched, adult male diencephalon Mus musculus cDNA clone 9330200E06 3′, mRNA sequence. |
| 1438682_at | 4.54 | Pik3r1 | Hypothetical protein C530050K14 (C530050K14), mRNA |
| 1420831_at | 4.52 | Qscn6 | quiescin Q6 |
| 1419602_at | 4.455 | Hoxa2 | Homeo box A2 |
| 1417937_at | 4.35 | Dact1 | dapper homolog 1, antagonist of beta-catenin (xenopus) |
| 1449449_at | 4.129 | Ptges | prostaglandin E synthase |
| 1448754_at | 4.118 | Rbp1 | Retinol binding protein 1, cellular |
| 1440143_at | 4.067 | F630022B06Rik | RIKEN cDNA F630022B06 gene |
| 1418486_at | 3.89 | Vnn1 | Vanin 1 |
| 1422674_s_at | 3.854 | Crygc | crystallin, gamma C |
| 1435342_at | 3.836 | Kcnk6 | potassium inwardly-rectifying channel, subfamily K, member 6 |
| 1451277_at | 3.782 | C530046K17Rik | RIKEN cDNA C530046K17 gene |
| 1460287_at | 3.777 | Timp2 | tissue inhibitor of metalloproteinase 2 |
| 1422597_at | 3.726 | Mmp15 | Matrix metalloproteinase 15 |
| 1420565_at | 3.686 | Hoxa1 | Homeo box A1 |
| 1454906_at | 3.557 | Rarb | retinoic acid receptor, beta |
| 1415938_at | 3.518 | Spink3 | serine protease inhibitor, Kazal type 3 |
| 1419309_at | 3.429 | Gp38 | glycoprotein 38 |
| 1435184_at | 3.393 | B430320C24Rik | 0 day neonate lung cDNA, RIKEN full-length enriched library, clone:E030020K21 product:weakly similar to HYPOTHETICAL 13.8 KDA PROTEIN [Homo sapiens], full insert sequence |
| 1452302_at | 3.393 | Arhgef10 | Ceroid-lipofuscinosis, neuronal 8 |
| 1422723_at | 3.387 | Stra6 | Stimulated by retinoic acid gene 6 |
| 1420753_at | 3.375 | Tll | Tolloid-like |
| 1420832_at | 3.279 | Qscn6 | quiescin Q6 |
| 1456481_at | 3.272 | D9Ertd280e | hypothetical protein D930024E11 |
| 1419301_at | 3.228 | Fzd4 | 602109129F1 NCI_CGAP_Kid14 Mus musculus cDNA clone IMAGE:4237519 5′, mRNA sequence. |
| 1418709_at | 3.199 | Cox7a1 | cytochrome c oxidase, subunit VIIa 1 |
| 1424194_at | 3.175 | BC025872 | cDNA sequence BC025872 |
| 1434069_at | 3.041 | BC067047 | Adult male testis cDNA, RIKEN full-length enriched library, clone:4930413J11 product:mitochondria located 1 homolog (human), full insert sequence |
| 1452303_at | 2.966 | Arhgef10 | Ceroid-lipofuscinosis, neuronal 8 |
| 1424334_at | 2.938 | Fbxo23 | F-box only protein 23 |
| 1449450_at | 2.922 | Ptges | prostaglandin E synthase |
| 1420774_a_at | 2.863 | 4930583H14Rik | RIKEN cDNA 4930583H14 gene |
| 1455642_a_at | 2.809 | Fbxo23 | F-box only protein 23 |
| 1419693_at | 2.805 | Colec12 | collectin sub-family member 12 |
| 1434277_a_at | 2.711 | 6430570G24 | Adult male olfactory brain cDNA, RIKEN full-length enriched library, clone:6430570G24 product:unknown EST, full insert sequence |
| 1452141_a_at | 2.71 | Sepp1 | selenoprotein P, plasma, 1 |
| 1435693_at | 2.62 | BC012256 | cDNA sequence BC012256 |
| 1448024_at | 2.559 | B430320C24Rik | 0 day neonate lung cDNA, RIKEN full-length enriched library, clone:E030020K21 product:weakly similar to HYPOTHETICAL 13.8 KDA PROTEIN [Homo sapiens], full insert sequence |
| 1419155_a_at | 2.549 | Sox4 | SRY-box containing gene 4 |
| 1433575_at | 2.506 | Sox4 | SRY-box containing gene 4 |
| 1460605_at | 2.486 | AA606869 | Similar to Homeobox protein goosecoid-like (GSC-2) (LOC381848), mRNA |
| 1418084_at | 2.454 | Nrp | Neuropilin |
| 1419157_at | 2.418 | Sox4 | SRY-box containing gene 4 |
| 1455796_x_at | 2.418 | Olfm1 | olfactomedin 1 |
| 1443882_at | 2.393 | Transcribed sequence with weak similarity to protein ref:NP_081764.1 (M.musculus) RIKEN cDNA 5730493B19 [Mus musculus] | |
| 1427195_at | 2.36 | Transcribed sequence with weak similarity to protein ref:NP_081764.1 (M.musculus) RIKEN cDNA 5730493B19 [Mus musculus] | |
| 1424951_at | 2.347 | 1300006M19Rik | RIKEN cDNA 1300006M19 gene |
| 1417133_at | 2.329 | Pmp22 | peripheral myelin protein |
| 1448727_at | 2.293 | Tle6 | Transducin-like enhancer of split 6, homolog of Drosophila E(spl) |
| 1439341_at | 2.261 | Transcribed sequences | |
| 1435743_at | 2.244 | C130068N17Rik | RIKEN cDNA C130068N17 gene |
| 1416674_at | 2.235 | Ptpru | Protein tyrosine phosphatase, receptor type, U |
| 1418415_at | 2.225 | Hoxb5 | Homeo box B5 |
| 1417605_s_at | 2.209 | Camk1 | calcium/calmodulin-dependent protein kinase I |
| 1456329_at | 2.081 | A230098A12Rik | RIKEN cDNA A230098A12 gene |
| 1417956_at | 2.055 | Cidea | cell death-inducing DNA fragmentation factor, alpha subunit-like effector A |
| 1418445_at | 2.006 | Slc16a2 | solute carrier family 16 (monocarboxylic acid transporters), member 2 |
| 1457829_at | 2.004 | Clgn | Calmegin |
| 1422912_at | 0.497 | Bmp4 | Bone morphogenetic protein 4 |
| 1424643_at | 0.483 | Tcof1 | Treacher Collins Franceschetti syndrome 1, homolog |
| 1438824_at | 0.466 | Slc20a1 | solute carrier family 20, member 1 |
| 1460204_at | 0.465 | Tec | cytoplasmic tyrosine kinase, Dscr28C related (Drosophila) |
| 1444390_at | 0.444 | Transcribed sequences | |
| 1426041_a_at | 0.426 | Fgd4 | FYVE, RhoGEF and PH domain containing 4 |
| 1435436_at | 0.405 | Transcribed sequences | |
| 1430433_at | 0.405 | 4933406J08Rik | RIKEN cDNA 4933406J08 gene |
| 1428283_at | 0.401 | Cyp2s1 | cytochrome P450, family 2, subfamily s, polypeptide 1 |
| 1424310_at | 0.399 | Mocs2 | molybdenum cofactor synthesis 2 |
| 1433733_a_at | 0.396 | Cry1 | cryptochrome 1 (photolyase-like) |
| 1430780_a_at | 0.394 | Pmm1 | Phosphomannomutase 1 |
| 1424942_a_at | 0.391 | Myc | Myelocytomatosis oncogene |
| 1437588_at | 0.391 | Pou4f2 | POU domain, class 4, transcription factor 2 |
| 1419735_at | 0.39 | Csnk | Casein kappa |
| 1429937_at | 0.379 | D530033C11Rik | RIKEN cDNA D530033C11 gene |
| 1416967_at | 0.374 | Sox2 | SRY-box containing gene 2 |
| 1419930_at | 0.372 | D15Ertd55e | Transcribed sequences |
| 1422887_a_at | 0.364 | Ctbp2 | |
| 1438861_at | 0.357 | Bnc2 | RIKEN cDNA 5031434M05 gene |
| 1431425_a_at | 0.354 | 4930535B03Rik | RIKEN cDNA 4930535B03 gene |
| 1451527_at | 0.345 | Pcolce2 | procollagen C-endopeptidase enhancer 2 |
| 1454197_a_at | 0.341 | D19Ertd678e | unnamed protein product; hypothetical protein (evidence: rsCDS,ProCrest,decoder,NCBI CDS Predictor, Longest-ORF) putative; Mus musculus adult male testis cDNA, RIKEN full-length enriched library, clone:4933411H20 product:hypothetical protein, full insert sequence. |
| 1436865_at | 0.335 | F630021I08Rik | RIKEN cDNA F630021I08 gene |
| 1449455_at | 0.333 | Hck | hemopoietic cell kinase |
| 1423259_at | 0.332 | Idb4 | inhibitor of DNA binding 4 |
| 1416899_at | 0.329 | Utf1 | undifferentiated embryonic cell transcription factor 1 |
| 1434705_at | 0.329 | D7Wsu87e | TRAF-binding protein |
| 1416505_at | 0.321 | Nr4a1 | nuclear receptor subfamily 4, group A, member 1 |
| 1421317_x_at | 0.314 | Myb | myeloblastosis oncogene |
| 1443786_at | 0.313 | Utf1 | BB000218 RIKEN full-length enriched, ES cells XhoI/SstI Mus musculus cDNA clone 2410014O14 3′ similar to NM_009482 Mus musculus undifferentiated embryonic cell transcription factor 1 (Utf1), mRNA sequence. |
| 1423748_at | 0.312 | D530020C15Rik | RIKEN cDNA D530020C15 gene |
| 1428775_at | 0.311 | 1110008L16Rik | RIKEN cDNA 1110008L16 gene |
| 1449231_at | 0.31 | Zfp296 | Zinc finger protein 296 |
| 1455692_x_at | 0.308 | 1700097N02Rik | Adult male testis cDNA, RIKEN full-length enriched library, clone:1700122G02 product:unknown EST, full insert sequence |
| 1418649_at | 0.302 | Egln3 | EGL nine homolog 3 (C. elegans) |
| 1438160_x_at | 0.301 | Slco4a1 | AV348121 RIKEN full-length enriched, adult male olfactory bulb Mus musculus cDNA clone 6430631M08 3′, mRNA sequence. |
| 1452609_at | 0.298 | 1190005I06Rik | RIKEN cDNA 1190005I06 gene |
| 1416605_at | 0.292 | Nola2 | nucleolar protein family A, member 2 |
| 1434239_at | 0.288 | AA408556 | expressed sequence AA408556 |
| 1442379_at | 0.287 | Transcribed sequences | |
| 1432018_at | 0.281 | Ascl2 | achaete-scute complex homolog-like 2 (Drosophila) |
| 1420086_x_at | 0.279 | Fgf4 | fibroblast growth factor 4 |
| 1450282_at | 0.277 | Fgf4 | fibroblast growth factor 4 |
| 1449484_at | 0.276 | Stc2 | stanniocalcin 2 |
| 1416316_at | 0.272 | Slc27a2 | solute carrier family 27 (fatty acid transporter), member 2 |
| 1444292_at | 0.27 | D7Ertd143e | NACHT, LRR and PYD containing protein 12 |
| 1440563_at | 0.269 | 15 days embryo head cDNA, RIKEN full-length enriched library, clone:D930010N06 product:unknown EST, full insert sequence | |
| 1451026_at | 0.267 | Ftsj3 | FtsJ homolog 3 (E. coli) |
| 1453345_at | 0.26 | 3830408G10Rik | RIKEN cDNA 3830408G10 gene |
| 1452004_at | 0.259 | Calca | calcitonin/calcitonin-related polypeptide, alpha |
| 1426431_at | 0.252 | Jag2 | AV264681 RIKEN full-length enriched, adult male testis (DH10B) Mus musculus cDNA clone 4930502C11 3′ similar to Y14331 Mus musculus partial mRNA for jagged2 protein, clone MB16, mRNA sequence. |
| 1438084_at | 0.221 | Transcribed sequences | |
| 1418744_s_at | 0.215 | Tesc | Tescalcin |
| 1417745_at | 0.21 | Cpn1 | carboxypeptidase N, polypeptide 1 |
| 1420085_at | 0.208 | Fgf4 | fibroblast growth factor 4 |
| 1453072_at | 0.208 | Gpr160 | G protein-coupled receptor 160 |
| 1449146_at | 0.199 | Notch4 | Notch gene homolog 4 (Drosophila) |
| 1424531_a_at | 0.191 | Tcea3 | transcription elongation factor A (SII), 3 |
| 1436657_at | 0.187 | LOC380738 (LOC380738), mRNA | |
| 1452384_at | 0.187 | Enpp3 | Ectonucleotide pyrophosphatase/phosphodiesterase 3 |
| 1426808_at | 0.181 | Lgals3 | Lectin, galactose binding, soluble 3 |
| 1450989_at | 0.18 | Tdgf1 | teratocarcinoma-derived growth factor |
| 1439746_at | 0.126 | C130085G02Rik | RIKEN cDNA C130085G02 gene |
| 1423378_at | 0.115 | Adam23 | a disintegrin and metalloprotease domain 23 |
| 1425926_a_at | 0.0799 | Otx2 | orthodenticle homolog 2 (Drosophila) |
| 1450177_at | 0.0773 | Ngfr | Nerve growth factor receptor (TNFR superfamily, member 16) |
| 1431711_a_at | 0.07 | 9030409G11Rik | RIKEN cDNA 9030409G11 gene |
Average from three experiments.
Some genes have multiple distinct probes on the GeneChip® resulting in different fold-changes from different probes.
Among the 1348 probe sets which were differentially expressed between the F9 Wt and RARγ−/− cell lines by at least two fold after 24 h-RA exposure, only a total of 161 probe sets were responsive to RA. These results indicate that 88% of these 1348 probe sets are not responsive to RA, or exhibit a less than two-fold change after RA addition to F9 Wt cells. The changes in mRNA levels of these genes between the two cell types indicate that the majority of genes regulated by RARγ are not RA-regulated. However, some of the genes designated as RA non-responsive genes may be partially RA responsive. Their responsiveness to RA may be less than two fold, or the responsiveness to RA may occur at a later time point. Genes in this category include GATA4, GATA6, and laminin B1. Thus, the genes listed (Appendix 2) represent only the putative RARγ-mediated, RA responsive genes at 24 h after RA addition.
3.6. Real-time RT-PCR analyses of selected genes in F9 Wt, RARγ−/−, RARα−/− and RARβ2−/− cell lines in response to RA: Peg1/mest2, tie1, sfrp5, crygc, Zfp503, Runx-1 and Raet1a
To validate the MG-430.2 microarray results and to determine if these RA-regulated genes are specifically regulated through RARγ, the mRNA levels of several of these genes were examined in the F9 Wt, RARγ−/−, RARα−/−, and RARβ2−/− cell lines. The four cell lines were cultured in the presence or absence of RA for 6 h, 24 h, or 48 h. β-actin mRNA was used for comparison. The mRNA levels of Peg1/mest2, sfrp5, crygc, Zfp503, Runx-1 and Raet1a were examined by real-time RT-PCR (Fig. 5). Tie1, a RARγ-regulated gene identified in our earlier gene chip (U74Av2) analyses, was also assessed by real-time quantitative RT-PCR (Fig. 5). These genes were selected for validation because they had relatively high fold changes and/or were found to have interesting biological functions.
Figure 5. Real-time RT-PCR analyses of Tie1, Peg1/mest2, Runx-1, crygc, sfrp5, Zfp503, and Raet1a, in F9 Wt, RARγ−/−, RARα−/−, and RARβ2−/− cell lines in response to RA.
Total RNA was extracted from the F9 Wt, RAR γ−/−, RARα−/−, RARβ2−/− cells cultured in the presence or absence of 1 μM RA for 6, 24, or 48 h. An equivalent amount of RNA (5 μg) was subjected to RT-PCR with primers specific for the indicated genes. Real-time quantitative (SYBR green) PCR of the indicated genes was performed and the mRNA levels were normalized to β-actin mRNA levels. The data are shown as fold increases in mRNA levels relative to the β-actin mRNA levels at each condition. Y axis, relative mRNA expression level. The results shown here are derived from three independent experiments with triplicates. The normalized mRNA levels of F9 RARα−/−, RARβ2−/−, RARγ−/− cell samples were compared to F9 Wt cell samples at the respective time points and drug treatments for statistical significance. The unpaired student’s t-test was used to determine the p values. bars, S.E. *, significant (P<0.05), **, very significant (P<0.001) in comparisons of the RNA values in the mutants with those in F9 Wt treated under the same condition.
Peg1/mest2 encodes “Paternally expressed 1 (Peg1)/mesoderm specific transcript (Mest)”, and is an imprinted gene which is only transcribed from the paternal (father’s) allele [58]. It is expressed predominantly in cells of the mesodermal lineage [59]. Our microarray data indicate that Peg1/mest mRNA levels are not regulated by RA in Wt F9 cells at 24 h (Appendix 2). Peg1/mest2 transcripts exhibit 18.6 ± 3.7-fold and 25.2 ± 6.9-fold higher levels in F9 Wt than in RARγ−/− cells in control and RA treated samples, respectively. Peg1/mest mRNA levels are lower in F9 RARα−/−, RARβ2−/−, and RARγ−/− than in F9 Wt by 45.6-fold (P<0.001), 0.37- fold (P>0.05), and 50.2-fold (P<0.001) at 6 h; 17.9-fold (P<0.001), 0.8-fold (P>0.05), and 77.8-fold (P<0.001) at 24 h; 16.2-fold (P<0.001), 0.92-fold (P>0.05), and 91.9-fold (P<0.001) at 48 h, respectively, by quantitative real time RT-PCR (Fig. 5). Thus, Peg1/mest2 is a putative target gene of both RARγ and RARα.
Runx-1 belongs to the Runt-related (Runx) transcription factor family [60]. Runx1 regulates the differentiation of hematopoietic cells. Mutations in Runx1 are closely associated with human acute myeloid leukemia [61]. Runx1 is also involved in neuronal cell fate specification and the formation of axonal projections [62]. Our microarray data indicate that Runx1 mRNA levels were not responsive to RA in F9 Wt cells at 24 h, and were 11.3 ± 3.2-fold and 15.1 ± 4.5-fold higher in F9 Wt cells than in RARγ−/− cells in control and RA treated samples, respectively. Runx-1 mRNA levels were increased by RA by 2.6 ± 0.6-fold in F9 Wt cells at 48 h (Fig. 4). A RA-associated increase in Runx1 mRNA levels of 4.2 ± 0.5-fold was also seen in RARα−/− cells at 48 h (Fig. 4). In addition, Runx-1 mRNA levels were 5- to 13-fold higher in F9 Wt than in RARγ−/− and RARβ2−/− cells at all of the time points examined (Fig. 5). These results indicate that Runx-1 is a putative target gene of both RARγ and RARβ2.
Crygc encodes gamma C-crystallin, a major component of the eye lens [63]. Mutations in the mouse Crygs gene were demonstrated to cause cataracts [64]. Our microarray data indicate that RA increased Crygc mRNA levels by 15.5 ± 2.2-fold in F9 Wt cells and that Crygc mRNA levels were 3.8 ± 0.9-fold higher in RA-treated F9 Wt cells than in RA-treated F9 RARγ−/− cells at 24 h. Real-time PCR assays confirmed that RA increased Crygc mRNA levels in F9 Wt cells by 16.9 ± 2.4-fold (P<0.001) and 36.3 ± 2.1-fold (P<0.001) at 24 h and 48 h, respectively (Fig. 5). Crygc mRNA levels were higher in F9 Wt than in F9 RARα−/−, RARβ−/−, and RARγ−/− cells by 3.8-fold (P<0.05), 12.7-fold (P<0.001), and 7.4-fold (P<0.001) respectively, after a 24 h- RA treatment (Fig. 5).
Sfrp5 is a member of the Secreted frizzled-related protein (Sfrp) gene family [41]. The secreted Sfrp proteins can bind Wnt molecules and inhibit Wnt signaling [41]. Sfrp5 is expressed in the anterior visceral endoderm (AVE) and the ventral foregut endoderm of the early mouse embryo [65, 66], though genetic evidence indicates that sfrp5 is not essential for axis development [67]. Abnormal down-regulation of Sfrp5 due to abnormal promoter methylation has been suggested in certain tumors [68–70]. The microarray data indicate that RA treatment increased Sfrp5 mRNA levels by 4.0 ± 1.2-fold in Wt F9 cells, and Sfrp5 mRNA levels were 24.6 ± 5.2-fold higher in RA-treated F9 Wt than in RA-treated F9 RARγ−/− cells at 24 h. Real-time RT-PCR data confirmed that RA increased Sfrp5 mRNA levels by 2.2 ± 0.1-fold (P<0.05) and 16.2 ± 2.9-fold (P<0.001), at 24 h and 48 h, respectively, in F9 Wt cells (Fig. 5). Sfrp5 mRNA levels in F9 RARα−/−, RARβ2−/−, and RARγ−/− cells were 2.6-fold (P>0.05), 9.1-fold (P<0.001), and 13.0-fold (P<0.001) lower, respectively, than in F9 Wt cells after a 24 h-RA treatment (Fig. 5) and 1.3-fold (P>0.05), 7.9-fold (P<0.05), and 1.8-fold (P<0.05) lower respectively, than F9 Wt cells after a 48 h- RA treatment (Fig. 5).
Zfp503 (also called Nolz-1) is a member of the nocA/elB/tlp-1 family [71]. Nolz-1 mRNA was dramatically induced upon neural induction of P19 embryonal carcinoma cells by RA [54]. Our microarray data indicate that RA increased Zfp503 mRNA levels by 21.9 ± 3.7-fold in F9 Wt cells, and Zfp503 mRNA levels were 5.2 ± 1.8-fold higher in RA-treated F9 Wt cells than in RA-treated F9 RARγ−/− cells at 24 h. RA increased Zfp503 mRNA levels by 2.2 ± 0.1-fold (P<0.05) and 16.2 ± 2.9-fold (P<0.001) at 24 h and 48 h, respectively, in F9 Wt cells as measured by real-time RT-PCR (Fig. 5). Zfp503 mRNA levels were 2.5-fold (P<0.05) and 4.2-fold (P<0.001) higher in F9 Wt than in F9 RARβ2−/− and RARγ−/− cells, respectively, after a 24 h-RA treatment as measured by real time RT-PCR (Fig. 5). These results indicate that Zfp503 is a putative target gene of both RARγ and RARβ2.
Raet1a encodes a glycoprotein “retinoic acid early inducible protein 1 alpha precursor” (RAE-1alpha). The Rae proteins are anchored on the cell surface by a glycosyl phosphatidylinositol (GPI)-tail [72], and act as ligands for a natural killer cell lectin-like receptor [73]. Rae proteins are not expressed in normal, healthy tissues in adult mice, but are induced by viral infection or cellular transformation, implying a role for Rae proteins in the regulation of immunity [74]. Our microarray data indicate that RA increased Raet1a mRNA levels by 6.4 ± 1.5-fold in F9 Wt cells, and Raet1a mRNA levels were 14.3 ± 2.6-fold higher in RA-treated F9 Wt cells than in RA-treated F9 RARγ−/− cells at 24 h. RA increased Raet1a mRNA levels in F9 Wt cells by 4.9 ± 1.1-fold (P<0.001) and 11.1 ± 2.8-fold (P<0.001) at 24 h and 48 h, respectively, by real-time RT-PCR (Fig. 5). After a 24 h-RA treatment, Raet1a mRNA levels were 6.2-fold (P<0.001), 2.4-fold (P<0.05), and 7.1-fold (P<0.001) lower, respectively, in F9 RARα−/−, RARβ2−/−, and RARγ−/− cells than in F9 Wt cells. These data indicate that all of these RARs are involved in the regulation of Raet1a.
Tie1 mRNA levels were 25.0 ± 8.2-fold higher in F9 Wt cells than in F9 RARγ−/− cells after a 24 h RA treatment (Table 3). Tie1 mRNA levels were also higher in F9 Wt cells than in F9 RARγ−/− cells as assessed by semi-quantitative RT-PCR (Fig. 2). Tie1 mRNA levels increased by 4.1 ± 0.1-fold (p<0.05) and by 18.2 ±3.1-fold (p<0.05) at 24 h and 48 h after RA addition, respectively, in F9 Wt cells. Tie1 mRNA levels were 3.9-fold and 3.3- fold higher in F9 Wt cells than in F9 RARγ−/− cells at 24 h and 48 h after RA addition, respectively (Fig. 5). From these data (Fig. 5), the Tie1 gene has the characteristics of an RARγ specific target gene.
3.7. Further Analysis Using DAVID
For the purpose of understanding how the loss of RARγ affects the biology of F9 cells, we chose to identify the biological processes associated with genes that show reduced or increased expression independently. DAVID identifies categories that are over-represented in the gene list relative to the representation within the genome of a given species. First, the functional annotation clustering tool displays similar annotations together in clusters. The grouping algorithm is based on the hypothesis that similar annotations should have similar gene members. The degree of common genes between two annotations is measured by Kappa statistics, and the Kappa values are then used to classify groups of similar annotations. Next, DAVID calculates the chances of over-representation of the clusters using the Fisher Exact test. The main benefit of over-representation analysis is to order categories associated with a gene list in order to focus on those processes most likely associated with the biological phenomenon under study; in this case, the biological phenomenon is the lack of RARγ function in F9 cells.
The major biological processes that were enriched in either F9 RARγ−/− cells or in F9 Wt cells, based on the differentially expressed genes, are shown (Table 5). Based on the genes that were expressed at higher levels in the control F9 RARγ−/− cells, the biological processes pattern specification, odentogenesis, and organic acid transport were the top terms to be enriched. Four organic acid transporter genes were expressed at higher mRNA levels in F9 RARγ−/− cells than in F9 Wt cells, including Slc43, Slc1a1, Slc38a4, and Riken cDNA 9130023d20. The control F9 RARγ−/− cells were highly enriched in genes involved in pattern specification, such as Otx2, Lef1, Fst, Mib1, and FGF4. The RA treated F9 RARγ−/− cells were highly enriched in genes involved in ribosome biogenesis and cell migration, suggesting that RARγ activity plays a role in regulating ribosome biogenesis and cell migration in F9 cells. Five ribosome genes were expressed at higher mRNA levels in F9 RARγ−/− cells than in F9 Wt cells after a 24 h exposure to RA, including RpL36a, RpL27, mRpL22, RpS24, and RpS20. Genes involved in regulation of ribosome biogenesis including Ftsj3, Gtpb4, and Rrs1, were also expressed at higher mRNA levels in F9 RARγ−/− cells than in F9 Wt cells after a 24 h exposure to RA. The cell migration related genes that had higher mRNA levels in F9 RARγ−/− cells than in F9 Wt cells include Pou4f2, Ngfr, Tdgf, Rtn4, jagged 2, enabled homolog, Ctgf, Tek, and PECAM-1.
Table 5.
Biological processes that are changed in F9 RARγ−/− cells as compared to F9 Wt cells at 24 h in the presence of RA or absence of RA, as indicated by DAVID analyses. “−”, reduced processes; “+”, increased processes.
| Change in F9 RARγ−/− cells | ||||
|---|---|---|---|---|
| Change | # of genes | P* | ||
| 24 h-Control | Morphogenesis | − | 73 | 4.4E-10 |
| Lipid Biosynthesis | − | 22 | 2.7E-6 | |
| Zinc ion homeostasis | + | 3 | 8.5E-4 | |
| Translational initiation | + | 3 | 4.6E-2 | |
| A-P Pattern specification | + | 4 | 4.5E-3 | |
| Odontogenesis | + | 3 | 7.2E-3 | |
| 24 h-RA | Vasculature development | − | 14 | 5.6E-3 |
| Neurogenesis | − | 17 | 1.4E-2 | |
| Ribosome biogenesis and assembly | + | 13 | 7.0E-5 | |
| Cell migration | + | 19 | 1.6E-3 | |
one-tail Fisher Exact Probability Value
The control F9 Wt cells were highly enriched in genes involved in cellular morphogenesis, such as Smo, Gjb3, and Dkk1, suggesting that RARγ activity is important for normal F9 cell morphogenesis. Genes up-regulated in 24 h-RA treated F9 Wt cells were found to be enriched in the biological processes, cell differentiation, nervous system development, vasculature development, and angiogenesis categories, which indicates that F9 Wt cells are induced by RA to differentiate into certain cell lineages, and that these developmental programs require RARγ activity to proceed normally.
4. DISCUSSION
4.1. Microarray analyses of F9 Wt vs. F9 RARγ−/− cells
F9 teratocarcinoma stem cells normally undergo RA-induced differentiation by activating the transcription of genes involved in cellular differentiation and development, and repressing the transcription of stem cell genes [75]. However, how gene transcription on a genome-wide scale is affected when RARγ expression is abolished is poorly understand. The goal of this research was to identify genes which showed altered expression in the absence of RARγ in F9 cells. We chose to study mRNA levels at the genome-wide level by microarray analysis.
Despite the similarity of F9 Wt and F9 RARγ−/− cells in morphology and growth characteristics [76], the gene expression profiles in the two cell lines are distinctive. A total of 1058 probe sets were differentially expressed by greater than 2-fold between the F9 Wt and RARγ−/− cell lines in the absence of exogenously added RA, suggesting that RARγ controls gene transcription in the absence of its ligand. According to the current model of gene regulation by retinoids established by Dilworth et al. [77], unliganded and DNA-bound retinoid receptors repress transcription through the recruitment of the corepressors NCoR and SMRT [78]. However, transient transfection reporter assays have demonstrated that RARγ mediated substantial levels of transcriptional activation in the absence of RA, as did RARβ [79]. Our microarray data demonstrate that a far greater number of genes are expressed at a higher level rather than a lower level in the vehicle-treated, control F9 Wt cells relative to the vehicle-treated F9 RARγ−/− cells (Fig. 3). Our data are consistent with the idea that unliganded RARγ, a nuclear transcription factor, can activate gene transcription. Unliganded RARγ only weakly interacts with SMRT [79]. The intramolecular interactions between helix 3 and helix 12 of the ligand binding domain of RARγ prevent corepressor binding [80]. Unliganded RARα, in contrast, has been shown to interact strongly with the nuclear corepressor SMRT, and RARα does repress transcription [79]. Recent studies indicate that the transcriptional activity of RARγ can also be modified by RARγ phosphorylation by ubiquitin-mediated proteasomal degradation of RARγ [81].
4.2. Comparison of this microarray research in F9 Wt cells with previous studies
There is a high similarity between the RA-responsive gene we identified in F9 Wt cells as compared to those identified by Eifert et al [82], who used 0.5 μM of RA to treat F9 Wt cells for 24 h and used Affymetrix U74AV2 arrays for analysis. However, Eifert et al. detected a greater number of genes changed by 2-fold or more in expression by RA than we did, even though the murine U74Av2 arrays they used contained a lower number of probe sets. One reason is that the data selection criteria for the two experiments were different. We employed a filter first to select genes that gave a “present” call in at least 3 chips out of a total of 12 chips for further analysis. Thus, genes that were not expressed or were weakly expressed across more than nine microarray chips were not selected, even though their mRNA fold changes between RA-treated and vehicle-treated samples were large. An examination of the data analysis protocol described by Eifert et al. suggests that they did not apply such a filter in their analyses; all of the genes that were differentially expressed were subjected to fold change analyses. Second, different concentrations of RA were used in the two studies, which may affect subsets of genes that are sensitive to RA concentration. There is another report describing microarray analysis of F9 RARγ-mediated RA responsive genes [76]. By comparison of our data with this study, there are some overlapping genes. However, because of the differences in the manner in which the microarray analyses were performed, the Parrella et al. report and our experimental design emphasized different aspects of the differentiation process. In our experiments we compared the differentially expressed genes between the F9 Wt and F9 RARγ−/− cells in the presence vs. the absence of RA, while Parrella et al. (2006) only compared differences in gene inducibility by RA between the two cell lines. These researchers also did not emphasize the quantitative differences in gene expression between the two cell lines. Quantitative differences in gene expression have important biological consequences, as demonstrated by the different developmental outcome resulting from a <2-fold variation in the expression of Oct4 in mouse ES cells [83]. Thus, the comparison of quantitative differences in gene expression between F9 Wt and F9 RARγ−/− cells in the presence versus the absence of RA, coupled with bioinformatics analyses concerning gene functions, is much more helpful in understanding the phenotypic differences between the F9 Wt and RARγ−/− cell lines.
4.3. Complex molecular mechanisms of actions of RARs
Over the last twenty years, more than 532 genes have been put forward as either direct or indirect targets of RA in permissive cellular contexts (reviewed by Balmer and Blomhoff, 2002). Our laboratory was the first to clone the Hoxa1 cDNA and show that it was a direct, primary RA target gene [84]. Transcriptional control of Hoxa1 by RA is driven by an RAR/RXR heterodimer bound to a DR5 RARE [85, 86]. Using chromatin immunoprecipitation, Gillespie and Gudas (2007) showed that RA increased the RXRα’s association with the Hoxa1 RARE [87, 88]. In the past few years, additional genes were reported to be regulated by RARs through non-classical RAREs, via coordination of other transcription factors, or through indirect pathways which involved the actions of other transcription factors or through multiple phosphorylation sites of RAR [89–91]. As examined using TESS (Transcription Element Esearching Program, www.cbil.upenn.edu/tess), few of the RARγ-regulated genes identified in our microarray analyses contain classical RAREs in their 5′ or 3′ flanking regions. It is possible that RAREs are located further 5′ or 3′ of the genes, or that some of the genes are “secondary” target genes rather than “primary” targets of the RARs. Thus, more research is required to understand the detailed transcriptional mechanisms by which these genes are regulated by RARγ.
Acknowledgments
We thank members of the Gudas laboratory for scientific discussion. We thank Dr. Xiaohan Tang and Vasundhra Kashyap for reading and reviewing this manuscript and Karl Ecklund for editorial assistance. This research was supported by the National Cancer Institute (LJG) and in part by DOD predoctoral fellowship # W81XWH-04-1-0440 to DS.
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
References
- 1.Hayden LJ, Hawk SN, Sih TR, Satre MA. Metabolic conversion of retinol to retinoic acid mediates the biological responsiveness of human mammary epithelial cells to retinol. J Cell Physiol. 2001;186:437–447. doi: 10.1002/1097-4652(2000)9999:999<000::AID-JCP1043>3.0.CO;2-5. [DOI] [PubMed] [Google Scholar]
- 2.Napoli JL. A gene knockout corroborates the integral function of cellular retinol-binding protein in retinoid metabolism. Nutr Rev. 2000;58:230–236. doi: 10.1111/j.1753-4887.2000.tb01870.x. [DOI] [PubMed] [Google Scholar]
- 3.Blomhoff R, Blomhoff HK. Overview of retinoid metabolism and function. J Neurobiol. 2006;66(7):606–30. doi: 10.1002/neu.20242. [DOI] [PubMed] [Google Scholar]
- 4.Love JM, Gudas LJ. Vitamin A, differentiation and cancer. Curr Op Cell Biol. 1994;6:825–831. doi: 10.1016/0955-0674(94)90051-5. [DOI] [PubMed] [Google Scholar]
- 5.Altucci L, Rossin A, Raffelsberger W, Reitmair A, Chomienne C, Gronemeyer H. Retinoic acid-induced apoptosis in leukemia cells is mediated by paracrine action of tumor-selective death ligand TRAIL. Nat Med. 2001;7(6):680–6. doi: 10.1038/89050. [DOI] [PubMed] [Google Scholar]
- 6.Chambon P. A decade of molecular biology of retinoic acid receptors. FASEB J. 1996;10:940–954. [PubMed] [Google Scholar]
- 7.Kastner P, Mark M, Chambon P. Nonsteroid nuclear receptors: what are genetic studies telling us about their role in real life? Cell. 1995;83:859–869. doi: 10.1016/0092-8674(95)90202-3. [DOI] [PubMed] [Google Scholar]
- 8.Mangelsdorf DJ, Thummel C, Beato M, Herrlich P, Schütz G, Umesono K, et al. The nuclear receptor superfamily: the second decade. Cell. 1995;83:835–840. doi: 10.1016/0092-8674(95)90199-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Gudas LJ. Retinoids and vertebrate development. J Biol Chem. 1994;269:15399–15402. [PubMed] [Google Scholar]
- 10.Means AL, Gudas LJ. The roles of retinoids in vertebrate development. Annu Rev Biochem. 1995;64:201–33. doi: 10.1146/annurev.bi.64.070195.001221. [DOI] [PubMed] [Google Scholar]
- 11.Taneja R, Bouillet P, Boylan JF, Gaub MP, Roy B, Gudas LJ, et al. Reexpression of retinoic acid receptor (RAR)γ or RARβ in RARγ-null F9 cells reveals a partial functional redundancy between the three RAR types. Proc Natl Acad Sci. 1995;92:7854–7858. doi: 10.1073/pnas.92.17.7854. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Mark M, Ghyselinck NB, Chambon P. Function of retinoid nuclear receptors: lessons from genetic and pharmacological dissections of the retinoic acid signaling pathway during mouse embryogenesis. Annu Rev Pharmacol Toxicol. 2006;46:451–80. doi: 10.1146/annurev.pharmtox.46.120604.141156. [DOI] [PubMed] [Google Scholar]
- 13.Allan D, Houle M. Bouchard N, Meyer BI, Gruss P and Lohnes D, RARgamma and Cdx1 interactions in vertebral patterning. Dev Biol. 2001;240(1):46–60. doi: 10.1006/dbio.2001.0455. [DOI] [PubMed] [Google Scholar]
- 14.Wendling O, Ghyselinck NB, Chambon P, Mark M. Roles of retinoic acid receptors in early embryonic morphogenesis and hindbrain patterning. Development. 2001;128:2031–2038. doi: 10.1242/dev.128.11.2031. [DOI] [PubMed] [Google Scholar]
- 15.Ghyselinck NB, Chapellier B, Calleja C, Kumar Indra A, Li M, Messaddeq N, et al. [Genetic dissection of retinoic acid function in epidermis physiology] Ann Dermatol Venereol. 2002;129(5 Pt 2):793–9. [PubMed] [Google Scholar]
- 16.Chapellier B, Mark M, Messaddeq N, Calleja C, Warot X, Brocard J, et al. Physiological and retinoid-induced proliferations of epidermis basal keratinocytes are differently controlled. Embo J. 2002;21(13):3402–13. doi: 10.1093/emboj/cdf331. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Iulianella A, Lohnes D. Chimeric analysis of retinoic acid receptor function during cardiac looping. Dev Biol. 2002;247(1):62–75. doi: 10.1006/dbio.2002.0685. [DOI] [PubMed] [Google Scholar]
- 18.McGowan S, Jackson SK, Jenkins-Moore M, Dai HH, Chambon P, Snyder JM. Mice bearing deletions of retinoic acid receptors demonstrate reduced lung elastin and alveolar numbers. Am J Respir Cell Mol Biol. 2000;23(2):162–7. doi: 10.1165/ajrcmb.23.2.3904. [DOI] [PubMed] [Google Scholar]
- 19.Purton LE, Dworkin S, Olsen GH, Walkley CR, Fabb SA, Collins SJ, et al. RARgamma is critical for maintaining a balance between hematopoietic stem cell self-renewal and differentiation. J Exp Med. 2006;203(5):1283–93. doi: 10.1084/jem.20052105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Dzhagalov I, Chambon P, He YW. Regulation of CD8+ T lymphocyte effector function and macrophage inflammatory cytokine production by retinoic acid receptor gamma. J Immunol. 2007;178(4):2113–21. doi: 10.4049/jimmunol.178.4.2113. [DOI] [PubMed] [Google Scholar]
- 21.Spanjaard RA, Ikeda M, Lee PJ, Charpentier B, Chin WW, Eberlein TJ. Specific activation of retinoic acid receptors (RARs) and retinoid X receptors reveals a unique role for RARgamma in induction of differentiation and apoptosis of S91 melanoma cells. J Biol Chem. 1997;272(30):18990–9. doi: 10.1074/jbc.272.30.18990. [DOI] [PubMed] [Google Scholar]
- 22.Meister B, Fink FM, Hittmair A, Marth C, Widschwendter M. Antiproliferative activity and apoptosis induced by retinoic acid receptor-gamma selectively binding retinoids in neuroblastoma. Anticancer Res. 1998;18(3A):1777–86. [PubMed] [Google Scholar]
- 23.Hatoum A, El-Sabban ME, Khoury J, Yuspa SH, Darwiche N. Overexpression of retinoic acid receptors alpha and gamma into neoplastic epidermal cells causes retinoic acid-induced growth arrest and apoptosis. Carcinogenesis. 2001;22(12):1955–63. doi: 10.1093/carcin/22.12.1955. [DOI] [PubMed] [Google Scholar]
- 24.Pettersson F, Dalgleish AG, Bissonnette RP, Colston KW. Retinoids cause apoptosis in pancreatic cancer cells via activation of RAR-gamma and altered expression of Bcl-2/Bax. Br J Cancer. 2002;87(5):555–61. doi: 10.1038/sj.bjc.6600496. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Siegrist W, Hintermann E, Roggo CN, Apfel CM, Klaus M, Eberle AN. Melanoma cell growth inhibition and melanocortin receptor downregulation induced by selective and non-selective retinoids. Melanoma Res. 1998;8(2):113–22. doi: 10.1097/00008390-199804000-00003. [DOI] [PubMed] [Google Scholar]
- 26.Strickland S, Smith KK, Marotti KR. Hormonal induction of differentiation in teratocarcinoma stem cells: generation of parietal endoderm by retinoic acid and dibutyryl cAMP. Cell. 1980;21(2):347–55. doi: 10.1016/0092-8674(80)90471-7. [DOI] [PubMed] [Google Scholar]
- 27.Wang S-Y, Gudas LJ. Isolation of cDNA clones specific for collagen IV and laminin from mouse teratocarcinoma cells. Proc Natl Acad Sci. 1983;80:5880–5884. doi: 10.1073/pnas.80.19.5880. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Wang SY, LaRosa GJ, Gudas LJ. Molecular cloning of gene sequences transcriptionally regulated by retinoic acid and dibutyryl cyclic AMP in cultured mouse teratocarcinoma cells. Dev Biol. 1985;107(1):75–86. doi: 10.1016/0012-1606(85)90377-x. [DOI] [PubMed] [Google Scholar]
- 29.Hogan BLM, Barlow D, Tilly R. F9 teratocarcinoma cells as a model for the differentiation of parietal and visceral endoderm in the mouse. Cancer Surv. 1983;2:115–140. [Google Scholar]
- 30.Boylan JF, Lohnes D, Taneja R, Chambon P, Gudas LJ. Loss of retinoic acid receptor gamma function in F9 cells by gene disruption results in aberrant Hoxa-1 expression and differentiation upon retinoic acid treatment. Proc Natl Acad Sci U S A. 1993;90(20):9601–5. doi: 10.1073/pnas.90.20.9601. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Faria TN, Mendelsohn C, Chambon P, Gudas LJ. The targeted disruption of both alleles of RARbeta(2) in F9 cells results in the loss of retinoic acid-associated growth arrest. J Biol Chem. 1999;274(38):26783–8. doi: 10.1074/jbc.274.38.26783. [DOI] [PubMed] [Google Scholar]
- 32.Boylan JF, Lufkin T, Achkar CC, Taneja R, Chambon P, Gudas LJ. Targeted disruption of retinoic acid receptor alpha (RAR alpha) and RAR gamma results in receptor-specific alterations in retinoic acid-mediated differentiation and retinoic acid metabolism. Mol Cell Biol. 1995;15(2):843–51. doi: 10.1128/mcb.15.2.843. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Zhuang Y, Faria TN, Chambon P, Gudas LJ. Identification and characterization of retinoic acid receptor beta2 target genes in F9 teratocarcinoma cells. Mol Cancer Res. 2003;1(8):619–30. [PubMed] [Google Scholar]
- 34.Boylan JF, Lufkin T, Achkar CC, Taneja R, Chambon P, Gudas LJ. Targeted disruption of retinoic acid receptor α(RAR α) and RAR γ results in receptor-specific alterations in retinoic acid-mediated differentiation and retinoic acid metabolism. Mol Cell Biol. 1995;15:843–851. doi: 10.1128/mcb.15.2.843. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Abu-Abed SS, Beckett BR, Chiba H, Chithalen JV, Jones G, Metzger D, et al. Mouse P450RAI (CYP26) expression and retinoic acid-inducible retinoic acid metabolism in F9 cells are regulated by retinoic acid receptor gamma and retinoid X receptor alpha. J Biol Chem. 1998;273(4):2409–15. doi: 10.1074/jbc.273.4.2409. [DOI] [PubMed] [Google Scholar]
- 36.Wu C, Khan SA, Peng LJ, Lange AJ. Roles for fructose-2,6-bisphosphate in the control of fuel metabolism: beyond its allosteric effects on glycolytic and gluconeogenic enzymes. Adv Enzyme Regul. 2006;46:72–88. doi: 10.1016/j.advenzreg.2006.01.010. [DOI] [PubMed] [Google Scholar]
- 37.Korhonen J, Lahtinen I, Halmekyto M, Alhonen L, Janne J, Dumont D, et al. Endothelial-specific gene expression directed by the tie gene promoter in vivo. Blood. 1995;86(5):1828–35. [PubMed] [Google Scholar]
- 38.Puri MC, Rossant J, Alitalo K, Bernstein A, Partanen J. The receptor tyrosine kinase TIE is required for integrity and survival of vascular endothelial cells. Embo J. 1995;14(23):5884–91. doi: 10.1002/j.1460-2075.1995.tb00276.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Eklund L, Olsen BR. Tie receptors and their angiopoietin ligands are context-dependent regulators of vascular remodeling. Exp Cell Res. 2006;312(5):630–41. doi: 10.1016/j.yexcr.2005.09.002. [DOI] [PubMed] [Google Scholar]
- 40.Liehr T, Kuhlenbaumer G, Wulf P, Taylor V, Suter U, Van Broeckhoven C, et al. Regional localization of the human epithelial membrane protein genes 1, 2, and 3 (EMP1, EMP2, EMP3) to 12p12.3, 16p13.2, and 19q13.3. Genomics. 1999;58(1):106–8. doi: 10.1006/geno.1999.5803. [DOI] [PubMed] [Google Scholar]
- 41.Jones SE, Jomary C. Secreted Frizzled-related proteins: searching for relationships and patterns. Bioessays. 2002;24(9):811–20. doi: 10.1002/bies.10136. [DOI] [PubMed] [Google Scholar]
- 42.Boulassel MR, Tomasi JP, Deggouj N, Gersdorff M. COCH5B2 is a target antigen of anti-inner ear antibodies in autoimmune inner ear diseases. Otol Neurotol. 2001;22(5):614–8. doi: 10.1097/00129492-200109000-00009. [DOI] [PubMed] [Google Scholar]
- 43.Robertson NG, Skvorak AB, Yin Y, Weremowicz S, Johnson KR, Kovatch KA, et al. Mapping and characterization of a novel cochlear gene in human and in mouse: a positional candidate gene for a deafness disorder, DFNA9. Genomics. 1997;46(3):345–54. doi: 10.1006/geno.1997.5067. [DOI] [PubMed] [Google Scholar]
- 44.Robertson NG, Lu L, Heller S, Merchant SN, Eavey RD, McKenna M, et al. Mutations in a novel cochlear gene cause DFNA9, a human nonsyndromic deafness with vestibular dysfunction. Nat Genet. 1998;20(3):299–303. doi: 10.1038/3118. [DOI] [PubMed] [Google Scholar]
- 45.Eavey RD, Manolis EN, Lubianca J, Merchant S, Seidman JG, Seidman C. Mutations in COCH (formerly Coch5b2) cause DFNA9. Adv Otorhinolaryngol. 2000;56:101–2. doi: 10.1159/000059070. [DOI] [PubMed] [Google Scholar]
- 46.Wang PJ, McCarrey JR, Yang F, Page DC. An abundance of X-linked genes expressed in spermatogonia. Nat Genet. 2001;27(4):422–6. doi: 10.1038/86927. [DOI] [PubMed] [Google Scholar]
- 47.Hiriart E, Gruffat H, Buisson M, Mikaelian I, Keppler S, Meresse P, et al. Interaction of the Epstein-Barr virus mRNA export factor EB2 with human Spen proteins SHARP, OTT1, and a novel member of the family, OTT3, links Spen proteins with splicing regulation and mRNA export. J Biol Chem. 2005;280(44):36935–45. doi: 10.1074/jbc.M501725200. [DOI] [PubMed] [Google Scholar]
- 48.Hirsch D, Stahl A, Lodish HF. A family of fatty acid transporters conserved from mycobacterium to man. Proc Natl Acad Sci U S A. 1998;95(15):8625–9. doi: 10.1073/pnas.95.15.8625. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Raefski AS, O’Neill MJ. Identification of a cluster of X-linked imprinted genes in mice. Nat Genet. 2005;37(6):620–4. doi: 10.1038/ng1567. [DOI] [PubMed] [Google Scholar]
- 50.Davies W, Isles A, Smith R, Karunadasa D, Burrmann D, Humby T, et al. Xlr3b is a new imprinted candidate for X-linked parent-of-origin effects on cognitive function in mice. Nat Genet. 2005;37(6):625–9. doi: 10.1038/ng1577. [DOI] [PubMed] [Google Scholar]
- 51.Banerjee A, Xu HJ, Hu SX, Araujo D, Takahashi R, Stanbridge EJ, et al. Changes in growth and tumorigenicity following reconstitution of retinoblastoma gene function in various human cancer cell types by microcell transfer of chromosome 13. Cancer Res. 1992;52(22):6297–304. [PubMed] [Google Scholar]
- 52.Bastien J, Plassat JL, Payrastre B, Rochette-Egly C. The phosphoinositide 3-kinase/Akt pathway is essential for the retinoic acid-induced differentiation of F9 cells. Oncogene. 2006;25(14):2040–7. doi: 10.1038/sj.onc.1209241. [DOI] [PubMed] [Google Scholar]
- 53.Wang L, Mear JP, Kuan CY, Colbert MC. Retinoic acid induces CDK inhibitors and growth arrest specific (Gas) genes in neural crest cells. Dev Growth Differ. 2005;47(3):119–30. doi: 10.1111/j.1440-169X.2005.00788.x. [DOI] [PubMed] [Google Scholar]
- 54.Chang CW, Tsai CW, Wang HF, Tsai HC, Chen HY, Tsai TF, et al. Identification of a developmentally regulated striatum-enriched zinc-finger gene, Nolz-1, in the mammalian brain. Proc Natl Acad Sci U S A. 2004;101(8):2613–8. doi: 10.1073/pnas.0308645100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Wulf P, Suter U. Embryonic expression of epithelial membrane protein 1 in early neurons. Brain Res Dev Brain Res. 1999;116(2):169–80. doi: 10.1016/s0165-3806(99)00092-9. [DOI] [PubMed] [Google Scholar]
- 56.Kerley JS, Olsen SL, Freemantle SJ, Spinella MJ. Transcriptional activation of the nuclear receptor corepressor RIP140 by retinoic acid: a potential negative-feedback regulatory mechanism. Biochem Biophys Res Commun. 2001;285(4):969–75. doi: 10.1006/bbrc.2001.5274. [DOI] [PubMed] [Google Scholar]
- 57.Scantlebury T, Sniderman AD, Cianflone K. Regulation by retinoic acid of acylation-stimulating protein and complement C3 in human adipocytes. Biochem J. 2001;356(Pt 2):445–52. doi: 10.1042/0264-6021:3560445. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Kaneko-Ishino T, Kuroiwa Y, Miyoshi N, Kohda T, Suzuki R, Yokoyama M, et al. Peg1/Mest imprinted gene on chromosome 6 identified by cDNA subtraction hybridization. Nat Genet. 1995;11(1):52–9. doi: 10.1038/ng0995-52. [DOI] [PubMed] [Google Scholar]
- 59.Reule M, Krause R, Hemberger M, Fundele R. Analysis of Peg1/Mest imprinting in the mouse. Dev Genes Evol. 1998;208(3):161–3. doi: 10.1007/s004270050168. [DOI] [PubMed] [Google Scholar]
- 60.Ito Y. Oncogenic potential of the RUNX gene family: ‘overview’. Oncogene. 2004;23(24):4198–208. doi: 10.1038/sj.onc.1207755. [DOI] [PubMed] [Google Scholar]
- 61.de Bruijn MF, Speck NA. Core-binding factors in hematopoiesis and immune function. Oncogene. 2004;23(24):4238–48. doi: 10.1038/sj.onc.1207763. [DOI] [PubMed] [Google Scholar]
- 62.Yoshikawa M, Senzaki K, Yokomizo T, Takahashi S, Ozaki S, Shiga T. Runx1 selectively regulates cell fate specification and axonal projections of dorsal root ganglion neurons. Dev Biol. 2007;303(2):663–74. doi: 10.1016/j.ydbio.2006.12.007. [DOI] [PubMed] [Google Scholar]
- 63.Graw J. The crystallins: genes, proteins and diseases. Biol Chem. 1997;378(11):1331–48. [PubMed] [Google Scholar]
- 64.Graw J, Neuhauser-Klaus A, Klopp N, Selby PB, Loster J, Favor J. Genetic and allelic heterogeneity of Cryg mutations in eight distinct forms of dominant cataract in the mouse. Invest Ophthalmol Vis Sci. 2004;45(4):1202–13. doi: 10.1167/iovs.03-0811. [DOI] [PubMed] [Google Scholar]
- 65.Finley KR. Tennessen J and Shawlot W, The mouse secreted frizzled-related protein 5 gene is expressed in the anterior visceral endoderm and foregut endoderm during early post-implantation development. Gene Expr Patterns. 2003;3(5):681–4. doi: 10.1016/s1567-133x(03)00091-7. [DOI] [PubMed] [Google Scholar]
- 66.Kemp C, Willems E, Abdo S, Lambiv L, Leyns L. Expression of all Wnt genes and their secreted antagonists during mouse blastocyst and postimplantation development. Dev Dyn. 2005;233(3):1064–75. doi: 10.1002/dvdy.20408. [DOI] [PubMed] [Google Scholar]
- 67.Leaf I, Tennessen J, Mukhopadhyay M, Westphal H, Shawlot W. Sfrp5 is not essential for axis formation in the mouse. Genesis. 2006;44(12):573–8. doi: 10.1002/dvg.20248. [DOI] [PubMed] [Google Scholar]
- 68.Roman-Gomez J, Cordeu L, Agirre X, Jimenez-Velasco A, San Jose-Eneriz E, Garate L, et al. Epigenetic regulation of Wnt-signaling pathway in acute lymphoblastic leukemia. Blood. 2007;109(8):3462–9. doi: 10.1182/blood-2006-09-047043. [DOI] [PubMed] [Google Scholar]
- 69.Nojima M, Suzuki H, Toyota M, Watanabe Y, Maruyama R, Sasaki S, et al. Frequent epigenetic inactivation of SFRP genes and constitutive activation of Wnt signaling in gastric cancer. Oncogene. 2007 doi: 10.1038/sj.onc.1210259. [DOI] [PubMed] [Google Scholar]
- 70.Marsit CJ, McClean MD, Furniss CS, Kelsey KT. Epigenetic inactivation of the SFRP genes is associated with drinking, smoking and HPV in head and neck squamous cell carcinoma. Int J Cancer. 2006;119(8):1761–6. doi: 10.1002/ijc.22051. [DOI] [PubMed] [Google Scholar]
- 71.Cheah PY, Meng YB, Yang X, Kimbrell D, Ashburner M, Chia W. The Drosophila l(2)35Ba/nocA gene encodes a putative Zn finger protein involved in the development of the embryonic brain and the adult ocellar structures. Mol Cell Biol. 1994;14(2):1487–99. doi: 10.1128/mcb.14.2.1487. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.Nomura M, Zou Z, Joh T, Takihara Y, Matsuda Y, Shimada K. Genomic structures and characterization of Rae1 family members encoding GPI-anchored cell surface proteins and expressed predominantly in embryonic mouse brain. J Biochem (Tokyo) 1996;120(5):987–95. doi: 10.1093/oxfordjournals.jbchem.a021517. [DOI] [PubMed] [Google Scholar]
- 73.Cerwenka A, Bakker AB, McClanahan T, Wagner J, Wu J, Phillips JH, et al. Retinoic acid early inducible genes define a ligand family for the activating NKG2D receptor in mice. Immunity. 2000;12(6):721–7. doi: 10.1016/s1074-7613(00)80222-8. [DOI] [PubMed] [Google Scholar]
- 74.Oppenheim DE, Roberts SJ, Clarke SL, Filler R, Lewis JM, Tigelaar RE, et al. Sustained localized expression of ligand for the activating NKG2D receptor impairs natural cytotoxicity in vivo and reduces tumor immunosurveillance. Nat Immunol. 2005;6(9):928–37. doi: 10.1038/ni1239. [DOI] [PubMed] [Google Scholar]
- 75.Gudas LJ, Sporn MB, Roberts AB. Cellular biology and biochemistry of the retinoids. In: Sporn MB, Roberts AB, Goodman DS, editors. The Retinoids: Biology, Chemistry, and Medicine. Raven Press; New York: 1994. pp. 443–520. [Google Scholar]
- 76.Parrella E, Gianni M, Fratelli M, Barzago MM, Raska I, Jr, Diomede L, et al. Antitumor activity of the retinoid-related molecules (E)-3-(4′-hydroxy-3′-adamantylbiphenyl-4-yl)acrylic acid (ST1926) and 6-[3-(1-adamantyl)-4-hydroxyphenyl]-2-naphthalene carboxylic acid (CD437) in F9 teratocarcinoma: Role of retinoic acid receptor gamma and retinoid-independent pathways. Mol Pharmacol. 2006;70(3):909–24. doi: 10.1124/mol.106.023614. [DOI] [PubMed] [Google Scholar]
- 77.Dilworth FJ, Chambon P. Nuclear receptors coordinate the activities of chromatin remodeling complexes and coactivators to facilitate initiation of transcription. Oncogene. 2001;20(24):3047–54. doi: 10.1038/sj.onc.1204329. [DOI] [PubMed] [Google Scholar]
- 78.Bastien J, Rochette-Egly C. Nuclear retinoid receptors and the transcription of retinoid-target genes. Gene. 2004;328:1–16. doi: 10.1016/j.gene.2003.12.005. [DOI] [PubMed] [Google Scholar]
- 79.Hauksdóttir H, Privalsky ML. DNA recognition by the aberrant retinoic acid receptors implicated in human acute promyelocytic leukemia. Cell Growth Diff. 2001;12:85–98. [PMC free article] [PubMed] [Google Scholar]
- 80.Farboud B, Hauksdottir H, Wu Y, Privalsky ML. Isotype-restricted corepressor recruitment: a constitutively closed helix 12 conformation in retinoic acid receptors beta and gamma interferes with corepressor recruitment and prevents transcriptional repression. Mol Cell Biol. 2003;23(8):2844–58. doi: 10.1128/MCB.23.8.2844-2858.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 81.Bruck N, Bastien J, Bour G, Tarrade A, Plassat JL, Bauer A, et al. Phosphorylation of the retinoid X receptor at the omega loop, modulates the expression of retinoic-acid-target genes with a promoter context specificity. Cell Signal. 2005;17(10):1229–39. doi: 10.1016/j.cellsig.2004.12.006. [DOI] [PubMed] [Google Scholar]
- 82.Eifert C, Sangster-Guity N, Yu LM, Chittur SV, Perez AV, Tine JA, et al. Global gene expression profiles associated with retinoic acid-induced differentiation of embryonal carcinoma cells. Mol Reprod Dev. 2006;73(7):796–824. doi: 10.1002/mrd.20444. [DOI] [PubMed] [Google Scholar]
- 83.Niwa H, Miyazaki J, Smith AG. Quantitative expression of Oct-3/4 defines differentiation, dedifferentiation or self-renewal of ES cells. Nat Genet. 2000;24(4):372–6. doi: 10.1038/74199. [DOI] [PubMed] [Google Scholar]
- 84.LaRosa GJ, Gudas LJ. An early effect of retinoic acid: cloning of an mRNA (Era-1) exhibiting rapid and protein synthesis-independent induction during teratocarcinoma stem cell differentiation. Proc Natl Acad Sci U S A. 1988;85(2):329–33. doi: 10.1073/pnas.85.2.329. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 85.Langston AW, Thompson JR, Gudas LJ. Retinoic acid-responsive elements located 3′ of the Hox A and Hox B homeobox gene clusters: functional analysis. J Biol Chem. 1997;272:2167–2175. doi: 10.1074/jbc.272.4.2167. [DOI] [PubMed] [Google Scholar]
- 86.Langston AW, Gudas LJ. Identification of a retinoic acid responsive enhancer 3′ of the murine homeobox gene Hox 1.6. Mech Dev. 1992;38:217–228. doi: 10.1016/0925-4773(92)90055-o. [DOI] [PubMed] [Google Scholar]
- 87.Gillespie RF, Gudas LJ. RAR isotype specificity in F9 teratocarcinoma stem cells results from the differential recruitment of coregulators to retinoic acid response elements. J Biol Chem. 2007 doi: 10.1074/jbc.M704845200. [DOI] [PubMed] [Google Scholar]
- 88.Gillespie RF, Gudas LJ. Retinoid Regulated Association of Transcriptional Co-regulators and the Polycomb Group Protein SUZ12 with the Retinoic Acid Response Elements of Hoxa1, RARbeta(2), and Cyp26A1 in F9 Embryonal Carcinoma Cells. J Mol Biol. 2007 doi: 10.1016/j.jmb.2007.06.079. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 89.Bastien J, Adam-Stitah S, Riedl T, Egly JM, Chambon P, Rochette-Egly C. TFIIH interacts with the retinoic acid receptor gamma and phosphorylates its AF-1-activating domain through cdk7. J Biol Chem. 2000;275(29):21896–904. doi: 10.1074/jbc.M001985200. [DOI] [PubMed] [Google Scholar]
- 90.Cerignoli F, Guo X, Cardinali B, Rinaldi C, Casaletto J, Frati L, et al. retSDR1, a short-chain retinol dehydrogenase/reductase, is retinoic acid-inducible and frequently deleted in human neuroblastoma cell lines. Cancer Res. 2002;62:1196–1204. [PubMed] [Google Scholar]
- 91.Kopf E, Plassat JL, Vivat V, de The H, Chambon P, Rochette-Egly C. Dimerization with retinoid X receptors and phosphorylation modulate the retinoic acid-induced degradation of retinoic acid receptors alpha and gamma through the ubiquitin-proteasome pathway. J Biol Chem. 2000;275(43):33280–8. doi: 10.1074/jbc.M002840200. [DOI] [PubMed] [Google Scholar]