Abstract
Between 2002 and 2007, travel related cases of Shigella sonnei and S. flexneri in Alberta, Canada were acquired from Central America, the Indian subcontinent and North America. Of this group, resistance to ciprofloxacin and nalidixic acid was identified in isolates from patients who had travelled to the Indian subcontinent. This study provides a Canadian perspective to a growing body of literature linking ciprofloxacin and nalidixic acid resistance to travel to the Indian subcontinent.
Shigella is a common cause of diarrheal illness in North America with a rate of 2.0 per 100,000 in Canada [1] and a rate of 3.2 per 100,000 in the United States [2,3]. Imported cases of Shigella infections have been reported in developed countries following travel to a foreign or developing country [4,5] and may be impacted by factors including socio-economic factors [6], food distribution networks [5] and microbiologic factors [7]. Across multiple geographic regions, high rates of antimicrobial resistance to multiple agents (e.g. sulfonamides, tetracycline, chloramphenicol, ampicillin, and trimethoprim-sulfamethoxazole) have limited the choices for empiric antimicrobial therapy required to manage Shigella infections and reduce fecal excretion of the bacteria [8-10] with descriptions of shifting species dominance and changes in antimicrobial susceptibility [10,11]. Generally, Shigella flexneri and Shigella sonnei are the dominant species and are heavily impacted by changes in antimicrobial susceptibility [12,13].
This study identifies the global regions associated with travel-related cases of S. flexneri and S. sonnei in Alberta, Canada and compares antibiotic resistance patterns of these isolates for 2002 to 2007 inclusive.
Specimens collected 2002-2007 (inclusive) from S. flexneri and S. sonnei infections in Alberta, Canada were included for study. Data collected at time of specimen submission included: date of specimen collection, outbreak association if present, travel history and antibiogram (data source-ProvLab Information Systems; Communicable Disease Report at Alberta Health and Wellness). Outbreaks were defined by public health officials as ≥ 2 epidemiologically related cases. Each outbreak was assigned a unique incident number. Repeat isolates received within six months of original case infections were excluded. Only one representative case for each outbreak was included, unless the isolates had different antibiotic susceptibility patterns. Based on travel history the origin of an isolate was grouped into corresponding regions and continents. Regions included in the study represented major travel destinations for individuals living in Canada. Domestic exposures were defined as "travel within North America."
Isolate confirmation
Presumptive Shigella isolates were confirmed using conventional biochemical tests [14]. Serotyping was done for S. flexneri and phagetyping was done for S. sonnei. Serotyping was performed using commercially available antisera (Denka Seiken USA Inc., Campbell, CA) for S .flexneri and the following serotypes (STs) were determined: 1-4, 6, SH-101, SH-104, and variants × or y [14]. Phage typing was performed on S. sonnei isolates following standard procedures at the National Microbiology Laboratory in Winnipeg, Manitoba [15]. For 2002 and 2003, there were representative but fewer numbers of isolates were available for testing. For example, in 2002 and 2003, only 24% and 58% of representative isolates were available respectively. From 2004-2007, representative isolates for each case of infection were available for susceptibility testing: 2004 (100%), 2005 (100%), 2006 (89%), 2007 (91%).
Susceptibility testing
Susceptibility testing was performed using Sensititre panels (Trek Diagnostic Systems, Cleveland, OH) against the following antimicrobial agents:
• amikacin (AMI)
• amoxicillin/clavulanic acid (AMC)
• ampicillin (AMP)
• cefoxitin (FOX)
• ceftiofur (TIO)
• ceftriaxone (AXO)
• chloramphenicol (CHL)
• ciprofloxacin (CIP)
• gentamicin (GEN)
• kanamycin (KAN)
• nalidixic acid (NAL)
• streptomycin (STR)
• tetracycline (TET)
• sulfisoxazole (SSS)
• trimethoprim/sulfamethoxazole (SXT)
The minimum inhibitory concentrations (MIC) and breakpoints were determined in accordance with guidelines established by the Clinical and Laboratory Standards Institute (CLSI) [16,17].
Data analysis
GraphPad Prism 5 software (GraphPad Software, Inc. La Jolla, CA) was used for statistical analysis.
Between 2002-2007, 578 Shigella isolates were received and confirmed by ProvLab. The overall distribution of species included: S. sonnei 54.7% (n = 316); S. flexneri 33.9% (n = 196); S. boydii 7.6% (n = 44); S. dysenteriae 3.8% (n = 22). Twenty nine S. flexneri and 79 S. sonnei were not archived (stored and cataloged); three S. flexneri could not be cultured; 15 S. sonnei belonged to four outbreaks and were removed as they had the same antibiogram as the index isolate for each outbreak (nine S. sonnei isolates in 2006 and six S. sonnei isolates in 2007). All but four S. flexneri and S. sonnei isolates were isolates from stool specimens; two S. sonnei isolates from blood, and two S. flexneri isolates were from blood and urine. Of the 386 S. flexneri and S. sonnei isolates, 74.9% (n = 289) were associated with international travel; 12.7% (n = 49) associated with domestic exposure within North America; 12.4% (n = 48) unknown travel history or origin of acquisition.
Rate calculations from Alberta population data were utilized to ensure no bias to study. The data set lacks a true denominator for all specimens received and tested. S. flexneri rates ranged from 0.70 to 1.21 per 100,000, and S. sonnei rates ranged from 1.10 to 1.98 per 100,000 per annum. The majority of travel cases for S. flexneri were from Central America (32.3% [53/164]), the Indian subcontinent (22.6% [37/164]) and North America (8.5% [14/164]). The majority of S. sonnei cases were from Central America (39.2% [87/222]), North America (15.8% [35/222]), and the Indian subcontinent (11.3% [25/222]).
Of the 196 S. flexneri isolates, as described above 164 were available for analysis, while 29 were not archived and 3 did not grow. The most common ST for S. flexneri was ST2 (37.8% [62/164]) with 40.3% (25/62) of the ST2 isolates originating from Central America. Of the S. flexneri isolates from the Indian subcontinent the two most common STs were ST2 (40.5% [15/37]) and ST6 (35.1% [13/37]). The most common phage type for S. sonnei was S1 (65.8% [146/222]) with (38.4% [56/146]of S1 isolates from Central America.
Only 1.2% (n = 2) S. flexneri and 8.1% (n = 18) S. sonnei isolates were pan-susceptible to all antibiotics tested. All S. flexneri isolates were susceptible to AMI, GEN, AMC, KAN, FOX, TIO, AXO. All the S. sonnei were resistant to AMP, CHL, NAL, STR, TET and SXT (Table 1).
Table 1.
Travel history and frequency of antimicrobial resistance of Shigella isolates in Alberta, 2002-2007A
| North America | Central America | South America | Africa | Middle East | Indian subcontinent | Far East Asia | Unknown | Western Hemisphere | Eastern Hemisphere | |
|---|---|---|---|---|---|---|---|---|---|---|
| Shigella flexneri | N = 14 | N = 53 | N = 6 | N = 27 | N = 2 | N = 37 | N = 8 | N = 17 | N = 73 | N = 74 |
| n(%) | n(%) | n(%) | n(%) | n(%) | n(%) | n(%) | n(%) | n(%) | n(%) | |
| Streptomycin | 7(50) | 30(57) | 3(50) | 12(44) | 2(100) | 32(86) | 6(75) | 14(82) | 40(55) | 52(70) |
| Ampicillin | 7(50) | 39(74) | 4(67) | 22(81) | 2(100) | 23(62) | 7(88) | 14(82) | 50(68) | 54(73) |
| Trimethoprim-sulfamethoxazole | 7(50) | 21(40) | 3(50) | 17(63) | 2(100) | 26(70) | 6(75) | 14(82) | 31(42) | 51(69) |
| Sulfisoxazole | 7(50) | 25(47) | 3(50) | 21(78) | 2(100) | 26(70) | 8(100) | 14(82) | 35(48) | 57(77) |
| Chloramphenicol | 8(57) | 35(66) | 4(67) | 22(81) | 2(100) | 24(65) | 6(75) | 12(71) | 47(64) | 54(73) |
| Ciprofloxacin | 1(7) | 0(0) | 0(0) | 0(0) | 0(0) | 7(19)1 | 0(0) | 0(0) | 1(1) | 7(9) |
| Nalidixic acid | 1(7) | 0(0) | 0(0) | 0(0) | 0(0) | 21(57) | 0(0) | 3(18) | 1(1) | 21(28) |
| Tetracycline | 13(93) | 51(96) | 6(100) | 25(93) | 2(100) | 37(100) | 6(75) | 17(100) | 70(96) | 70(95) |
| Shigella sonnei | N = 35 | N = 87 | N = 14 | N = 16 | N = 2 | N = 25 | N = 12 | N = 31 | N = 136 | N = 55 |
| Gentamicin | 0(0) | 0(0) | 1(7) | 0(0) | 0(0) | 0(0) | 0(0) | 1(3) | 1(1) | 0(0) |
| Streptomycin | 34(97) | 71(82) | 7(50) | 16(100) | 2(100) | 24(96) | 10(83) | 25(81) | 109(80) | 52(95) |
| Ampicillin | 8(23) | 30(34) | 9(64) | 1(6) | 0(0) | 1(4) | 2(17) | 19(61) | 47(35) | 4(7) |
| Amoxicillin/clavulanic acid | 0(0) | 0(0) | 0(0) | 0(0) | 0(0) | 0(0) | 0(0) | 1(3) | 0(0) | 0(0) |
| Ceftiofur | 0(0) | 0(0) | 0(0) | 0(0) | 0(0) | 0(0) | 0(0) | 2(6) | 0(0) | 0(0) |
| Ceftriaxone | 0(0) | 0(0) | 0(0) | 0(0) | 0(0) | 0(0) | 0(0) | 2(6) | 0(0) | 0(0) |
| Trimethoprim-sulfamethoxazole | 26(74) | 62(71) | 14(100) | 16(100) | 2(100) | 24(96) | 11(82) | 19(61) | 99(73) | 53(96) |
| Sulfisoxazole | 30(86) | 64(74) | 14(100) | 15(94) | 2(100) | 25(100) | 10(83) | 24(77) | 105(77) | 52(95) |
| Chloramphenicol | 1(3) | 0(0) | 8(57) | 0(0) | 0(0) | 1(4) | 1(8) | 0(0) | 9(7) | 2(4) |
| Ciprofloxacin | 0(0) | 0(0) | 0(0) | 0(0) | 0(0) | 0(0) | 0(0) | 0(0) | 0(0) | 0(0) |
| Nalidixic acid | 4(11) | 4(5) | 0(0) | 0(0) | 0(0) | 20(80) | 0(0) | 5(16) | 8(6) | 20(36) |
| Tetracycline | 23(66) | 57(66) | 7(50) | 15(94) | 2(100) | 25(100) | 9(75) | 14(45) | 91(67) | 51(93) |
AData for antimicrobial susceptible isolates are not shown
When median MICs were analyzed for all agents the following changes were identified as in Table 2. For S. flexneri median MICs were within two dilutions for most agents over the study period. Exceptions were for the following agents; AMP (increase), CHL (increase), SXT (increase and following drop), and SSS (decrease). For S. sonnei, median MICs were within two dilutions for most agents over the study period with the following exceptions; exception of AMP (decrease).
Table 2.
Median MICs of antimicrobial agents in S. flexneri and S. sonnei per year.
| AMI | AMP | AMC | AXO | CHL | CIP | SXT | FOX | GEN | KAN | NAL | SSS | STR | TET | TIO | ||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| S. flexneri | Total per year (n) | |||||||||||||||
| 2002 | 10 | 4 | 2 | 2 | < = 0.25 | 0.5 | < = 0.015 | < = 0.12 | 2 | 1 | < = 8 | 1 | > 256 | < = 32 | > 32 | < = 0.12 |
| 2003 | 28 | 2 | > 32 | 8 | < = 0.25 | > 32 | < = 0.015 | > 4 | 2 | 0.5 | < = 8 | 1 | > 256 | > 64 | > 32 | 0.25 |
| 2004 | 38 | 2 | > 32 | 8 | < = 0.25 | 32 | < = 0.015 | > 4 | 2 | 0.5 | < = 8 | 1 | > 256 | > 64 | > 32 | < = 0.12 |
| 2005 | 35 | 2 | > 32 | 8 | < = 0.25 | > 32 | < = 0.015 | > 4 | 4 | 0.5 | < = 8 | 1 | > 256 | 64 | > 32 | 0.25 |
| 2006 | 22 | 2 | > 32 | 8 | < = 0.25 | 32 | < = 0.015 | 0.25 | 2 | 0.5 | < = 8 | 2 | > 256 | 64 | > 32 | < = 0.12 |
| 2007 | 31 | 2 | > 32 | 8 | < = 0.25 | 32 | < = 0.015 | 0.25 | 4 | 0.5 | < = 8 | 2 | < = 16 | 64 | > 32 | < = 0.12 |
| S. sonnei | Total per year (n) | |||||||||||||||
| 2002 | 12 | 2 | 32 | 4 | < = 0.25 | 4 | < = 0.015 | > 4 | 1 | 1 | < = 8 | 1 | > 256 | > 64 | > 32 | 0.25 |
| 2003 | 27 | 2 | 32 | 4 | < = 0.25 | 4 | < = 0.015 | > 4 | 1 | 1 | < = 8 | 1 | > 256 | > 64 | > 32 | 0.25 |
| 2004 | 35 | 2 | 2 | 4 | < = 0.25 | 4 | < = 0.015 | > 4 | 1 | 0.5 | < = 8 | 1 | > 256 | > 64 | > 32 | 0.25 |
| 2005 | 60 | 2 | 2 | 4 | < = 0.25 | 8 | < = 0.015 | > 4 | 2 | 1 | < = 8 | 1 | > 256 | > 64 | > 32 | 0.25 |
| 2006 | 26 | 2 | 2 | 2 | < = 0.25 | 4 | < = 0.015 | > 4 | 2 | 0.5 | < = 8 | 1 | > 256 | > 64 | > 32 | 0.25 |
| 2007 | 19 | 2 | 2 | 2 | < = 0.25 | 4 | < = 0.015 | > 4 | 2 | 0.5 | < = 8 | 2 | > 256 | > 64 | > 32 | 0.25 |
amikacin (AMI), amoxicillin/clavulanic acid (AMC), ampicillin (AMP), cefoxitin (FOX), ceftiofur (TIO), ceftriaxone (AXO), chloramphenicol (CHL), ciprofloxacin (CIP), gentamicin (GEN), kanamycin (KAN), nalidixic acid (NAL), streptomycin (STR), tetracycline (TET), sulfisoxazole (SSS), trimethoprim/sulfamethoxazole (SXT)
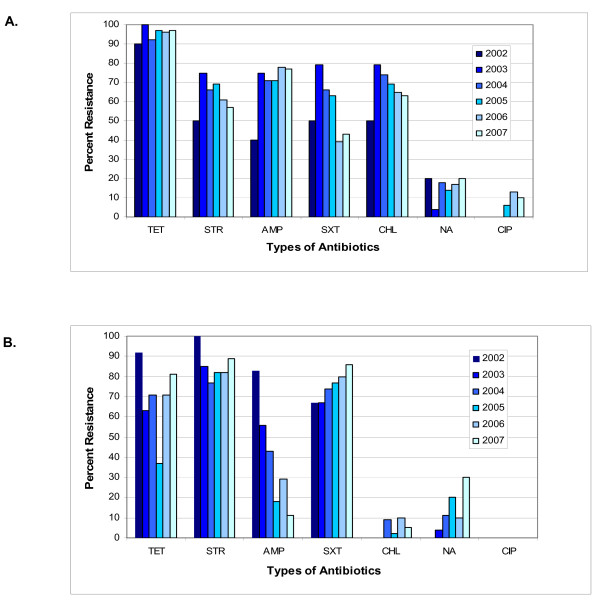
When data was combined for all years, the NAL and CIP resistance was 20.1% (33/164) and 14.9% (33/222) for S. flexneri and S. sonnei respectively. CIP resistance was identified only in S. flexneri isolates (4.9%, 8/164) when averaged over the six-year study period (Fisher's exact test, p = 0.001) (Figure 1a and 1b) CIP resistance in S. flexneri was not steady but instead was most evident in the years 2005, 2006, and 2007 (Figure 1a). Combined CIP and NAL resistance was related to travel to the Indian subcontinent for S. flexneri (84.8%, 28/37) and S. sonnei (80.0%, 20/25) (Fisher's exact test, p < 0.0001). The proportion of antibiotic resistance was constant over six years except for S. sonnei, where AMP resistance decreased from 83% in 2002 to 11% in 2007 (p < 0.0001, χ2 = 36.52, df = 5) and NAL resistance increased from 0% in 2002 to 30% in 2007 (p = 0.0168, χ2 = 13.82, df = 5).
Figure 1.
Frequency of antimicrobial resistance of study isolates from Alberta 2002-2007: 1 a) S. flexneri (n = 164); and 1b) S. sonnei (n = 222).
At the study onset, treatment guidelines suggested a fluoroquinolone for acute traveler's diarrhea regardless of travel location. It is possible that some CIP resistance was underestimated in 2002-2003 due to the smaller number of isolates tested. By 2009, treatment guidelines for acute traveler's diarrhea (outside of Latin America and Africa) suggested azithromycin or a fluoroquinolone [18,19]. Data also suggests that azithromycin resistance may be emerging and resistance rates of 16% have been recently described in Bangladesh [20]. These studies indicate that travel to the Indian subcontinent, in patients returning to Western Canada with traveler's diarrhea should be determined to guide initial empiric treatment options; especially for severe infections because the association of S. flexneri and S. sonnei isolates from this region with fluoroquinolone and potential macrolide resistance [13,21]. Although CIP resistance was described only in S. flexneri, we should remain vigilant for developing gyrA and parC mutations as well as the presence of plasmid mediated quinolone resistance determinants (PMQR) genes that may lead to increasing rates of CIP resistance in travel-related Shigella isolates which are beginning to emerge globally [4,22].
There are multiple factors that may have lead to CIP and NAL resistance in Shigella species originating from the Indian subcontinent [21]. It is possible that part of this emerging resistance may be associated with the increasing dominance of specific STs or clones of Shigella. Both this study and other work have identified a dominance of S. flexneri STs 2 and 6 in isolates of Indian origin and cases of traveler's diarrhea associated with the Indian subcontinent [23]. One factor driving multi-drug resistance in the Indian subcontinent may be the emergence of specific clones within these dominant STs [24]. Therefore, the identification of clonal groups within Alberta strains may be a powerful tool for tracking the development of drug-resistance in Shigella isolates from future cases of traveler's diarrhea.
Competing interests
The authors declare that they have no competing interests.
Authors' contributions
SJD, CL, MA, CF, and ML participated in data analysis and interpretation of susceptibility and travel data, drafted and revised paper, and made follow-up revisions to submission. CL, CF performed susceptibility testing on isolates, and interpreted/analyzed this data. CL collated, analyzed, and interpreted travel history data. LS, BF participated in susceptibility testing on isolates, and reviewed paper. KS, DE collaborated for travel history data, and reviewed/edited paper. ML, CL conceived study design. All authors read and approved the final manuscript draft.
Contributor Information
Steven J Drews, Email: steven.drews@albertahealthservices.ca.
Chris Lau, Email: chris-lau@shaw.ca.
Marnie Andersen, Email: marnie.andersen@albertahealthservices.ca.
Christina Ferrato, Email: Chrsitina.Ferrato@albertahealthservices.ca.
Kim Simmonds, Email: mksimmonds@gmail.com.
Liala Stafford, Email: Liala.Stafford@albertahealthservices.ca.
Bev Fisher, Email: Bev.Fisher@albertahealthservice.ca.
Doug Everett, Email: doug.everett@gov.ab.ca.
Marie Louie, Email: Marie.Louie@albertahealthservices.ca.
Acknowledgements
We thank M. Lovgren, S. Cook, R. Gordon and all the technologists and staff at the Provincial Laboratory for Public Health (Microbiology)(ProvLab) in Calgary and Edmonton, W. Demczuk and the National Microbiology Laboratory in Winnipeg, Manitoba for their support and contributions.
Data from this paper was presented in part as a poster at the 26th International Congress of Chemotherapy and Infection, Toronto, Ontario, 2009.
References
- Demczuk W, Ng LK, Woodward D, Ahmed R, Clark C, Tabor H, Dore K, Ciampa N, Muckle A. Laboratory Surveillance Data for Enteric Pathogens in Canada: Annual Summary 2006. Winnipeg, Manitoba, Canada: Public Health Agency of Canada, National Microbiology Laboratory; 2007. [Google Scholar]
- Centers for Disease Control and Prevention. Shigella Surveillance: Annual Summary, 2004. Atlanta, Georgia, USA: US Department of Health and Human Services; 2005. [Google Scholar]
- Centers for Disease Control and Prevention. National Antimicrobial Resistance Monitoring System for Enteric Bacteria (NARMS): Human Isolates Final Report, 2004. Atlanta, Georgia, USA: U.S. Department of Health and Human Services, CDC; 2007. [Google Scholar]
- Izumiya H, Tada Y, Ito K, Morita-Ishihara T, Ohnishi M, Terajima J, Watanabe H. Characterization of Shigella sonnei isolates from travel-associated cases in Japan. J Med Microbiol. 2009;58:1486–1491. doi: 10.1099/jmm.0.011809-0. [DOI] [PubMed] [Google Scholar]
- Gaynor K, Park SY, Kanenaka R, Colindres R, Mintz E, Ram PK, Kitsutani P, Nakata M, Wedel S, Boxrud D, Jennings D, Yoshida H, Tosaka N, He H, Ching-Lee M, Effler PV. International foodborne outbreak of Shigella sonnei infection in airline passengers. Epidemiol Infect. 2009;137:335–341. doi: 10.1017/S0950268807000064. [DOI] [PubMed] [Google Scholar]
- Simonsen J, Frisch M, Ethelberg S. Socioeconomic risk factors for bacterial gastrointestinal infections. Epidemiology. 2008;19:282–290. doi: 10.1097/EDE.0b013e3181633c19. [DOI] [PubMed] [Google Scholar]
- Todd EC, Greig JD, Bartleson CA, Michaels BS. Outbreaks where food workers have been implicated in the spread of foodborne disease. Part 4. Infective doses and pathogen carriage. J Food Prot. 2008;71:2339–2373. doi: 10.4315/0362-028x-71.11.2339. [DOI] [PubMed] [Google Scholar]
- Niyogi SK. Increasing antimicrobial resistance-an emerging problem in the treatment of shigellosis. Clin Microbiol Infect. 2007;13:1141–1143. doi: 10.1111/j.1469-0691.2007.01829.x. [DOI] [PubMed] [Google Scholar]
- Nelson JD, Kusmiesz H, Jackson LH, Woodman E. Trimethoprim-sulfamethoxazole therapy for shigellosis. JAMA. 1976;235:1239–1243. doi: 10.1001/jama.235.12.1239. [DOI] [PubMed] [Google Scholar]
- Vrints M, Mairiaux E, Van ME, Collard JM, Bertrand S. Surveillance of antibiotic susceptibility patterns among Shigella sonnei strains isolated in Belgium during the 18-year period 1990 to 2007. J Clin Microbiol. 2009;47:1379–1385. doi: 10.1128/JCM.02460-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vinh H, Nhu NT, Nga TV, Duy PT, Campbell JI, Hoang NV, Boni MF, My PV, Parry C, Nga TT, Van Minh P, Thuy CT, Diep TS, Phuong le T, Chinh MT, Loan HT, Tham NT, Lanh MN, Mong BL, Anh VT, Bay PV, Chau NV, Farrar J, Baker S. A changing picture of shigellosis in southern Vietnam: shifting species dominance, antimicrobial susceptibility and clinical presentation. BMC Infect Dis. 2009;9:204. doi: 10.1186/1471-2334-9-204. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Haukka K, Siitonen A. Emerging resistance to newer antimicrobial agents among Shigella isolated from Finnish foreign travellers. Epidemiol Infect. 2008;136:476–482. doi: 10.1017/S0950268807008862. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Srinivasa H, Baijayanti M, Raksha Y. Magnitude of drug resistant Shigellosis: a report from Bangalore. Indian J Med Microbiol. 2009;27:358–360. doi: 10.4103/0255-0857.55460. [DOI] [PubMed] [Google Scholar]
- American Society for Microbiology. Manual of Clinical Microbiology. 8. Washington, DC: American Society for Microbiology; 2003. [Google Scholar]
- Ahmed SF, Riddle MS, Wierzba TF, Messih IA, Monteville MR, Sanders JW, Klena JD. Epidemiology and genetic characterization of Shigella flexneri strains isolated from three paediatric populations in Egypt (2000-2004) Epidemiol Infect. 2006;134:1237–1248. doi: 10.1017/S095026880600642X. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Clinical and Laboratory Standards Institute. Methods for Dilution Antimicrobial Susceptibility Tests for Bacteria that Grow Aerobically; Approved Standard. 7. Wayne, Pennsylvania, USA: Clinical and Laboratory Standards Institute; 2006. [Google Scholar]
- Clinical and Laboratory Standards Institute. Performance Standards for Antimicrobial Susceptibility Testing; 17th Informational Supplement. Wayne, Pennsylvania, USA: Clinical and Laboratory Standards Institute; 2007. [Google Scholar]
- Sanford Guide to Antimicrobial Therapy 2000. 30. Hyde Park, VT: Antimicrobial Therapy Inc; 2000. [Google Scholar]
- Sanford Guide to Antimicrobial Therapy 2009. 39. Sperryville, VT: Antimicrobial Therapy Inc; 2009. [Google Scholar]
- Rahman M, Shoma S, Rashid H, El AS, Baqui AH, Siddique AK, Nair GB, Sack DA. Increasing spectrum in antimicrobial resistance of Shigella isolates in Bangladesh: resistance to azithromycin and ceftriaxone and decreased susceptibility to ciprofloxacin. J Health Popul Nutr. 2007;25:158–167. [PMC free article] [PubMed] [Google Scholar]
- Mensa L, Marco F, Vila J, Gascon J, Ruiz J. Quinolone resistance among Shigella spp. isolated from travellers returning from India. Clin Microbiol Infect. 2008;14:279–281. doi: 10.1111/j.1469-0691.2007.01903.x. [DOI] [PubMed] [Google Scholar]
- Pu XY, Pan JC, Wang HQ, Zhang W, Huang ZC, Gu YM. Characterization of fluoroquinolone-resistant Shigella flexneri in Hangzhou area of China. J Antimicrob Chemother. 2009;63:917–920. doi: 10.1093/jac/dkp087. [DOI] [PubMed] [Google Scholar]
- Dutta S, Rajendran K, Roy S, Chatterjee A, Dutta P, Nair GB, Bhattacharya SK, Yoshida SI. Shifting serotypes, plasmid profile analysis and antimicrobial resistance pattern of Shigellae strains isolated from Kolkata, India during 1995-2000. Epidemiol Infect. 2002;129:235–243. doi: 10.1017/S0950268802007240. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pazhani GP, Niyogi SK, Singh AK, Sen B, Taneja N, Kundu M, Yamasaki S, Ramamurthy T. Molecular characterization of multidrug-resistant Shigella species isolated from epidemic and endemic cases of shigellosis in India. J Med Microbiol. 2008;57:856–863. doi: 10.1099/jmm.0.2008/000521-0. [DOI] [PubMed] [Google Scholar]