Abstract
Due to their easy accessibility, proteins outside of the plasma membrane represent an ideal but untapped resource for potential drug targets or disease biomarkers. They constitute the major biochemical class of current therapeutic targets and clinical biomarkers. Recent advances in proteomic technologies have fueled interest in analysis of extracellular proteins such as membrane proteins, cell surface proteins, and secreted proteins. However, unlike the gene expression analyses from a variety of tissues and cells using genomic technologies, quantitative proteomic analysis of proteins from various biological sources is challenging due to the high complexity of different proteomes, and the lack of robust and consistent methods for analyses of different tissue sources, especially for specific enrichment of extracellular proteins. Since most extracellular proteins are modified by oligosaccharides, the population of glycoproteins therefore represents the majority of extracellular proteomes. Here, we quantitatively analyzed glycoproteins and determined the expression patterns of extracellular proteins from 12 mouse tissues using solid-phase extraction of N-linked glycopeptides and liquid chromatography tandem mass spectrometry. We identified peptides enclosing 1231 possible N-linked glycosites from 826 unique proteins. We further determined the expression pattern of formerly N-linked glycopeptides and identified extracellular glycoproteins specifically expressed in each tissue. Furthermore, the tissue specificities of the overexpressed glycoproteins in a mouse skin tumor model were determined by comparing to the quantitative protein expression from the different tissues. These skin tumor-specific extracellular proteins might serve as potential candidates for cell surface drug targets or disease-specific protein markers.
Keywords: Extracellular proteins, glycosylation, solid-phase extraction of glycopeptides, tissue specificity, different tissues, skin tumor, proteomics, and mass spectrometry
INTRODUCTION
Extracellular proteins, such as those associated with the cell membrane, on extracellular cell surface, or secreted to the extracellular environment or body fluids, have important biological functions. These proteins include receptors, growth factors, and regulators for cell communication, channels and transporters for chemicals and solutes, integrins, collagens, and other structural proteins for regulating cell shape, adhesion, and migration. Most importantly, extracellular proteins are easily accessible as potential drug targets for treatments and as surrogate markers for disease detection 1. This explains why extracellular proteins constitute the major biochemical class of therapeutic targets 2 and clinical biomarkers 3. So far, most tumor markers approved by the US Food and Drug Administration (FDA) for clinical usages are extracellular proteins 3. The detection of extracellular proteins in cancer patients, such as HER2 in breast cancer, is also used to select patients for treatment using Herceptin 4.
Extracellular proteins with restricted tissue expression are ideal candidates for new drug targets or protein markers for specific diseases. To reduce the risk of side effects, the drug targets are preferably distributed in few tissue types. Based on information from the Therapeutic Target Database, 53% of the successful drug targets are distributed in no more than two tissues 2,5. In addition, extracellular proteins specific to diseased tissue in body fluids such as prostate specific antigen (PSA) are much more likely to serve as disease specific biomarkers. Therefore, the quantitative analysis of the tissue specificity of extracellular proteins will be particularly useful for the identification of candidate proteins for new drug targets or biomarkers 6–8.
Several proteomic approaches using tandem mass spectrometry have made it possible for quantitative proteomic analyses of certain subclasses of extracellular proteins. These include an improved method to analyze membrane proteins 9–14 and secreted proteins 15, 16. However, due to the complex cellular structures of different tissues, methods for isolating extracellular proteins are often applied to in vitro cultured cells or specific tissue type. These methods have not been able to be applied to the analysis of extracellular proteome of different tissue types for discovery of tissue-specific new drug targets or biomarkers, which requires at least the following properties. First, the method should be uniformly applicable to multiple tissue or cell types. Second, the isolation method should have enough throughputs and reproducibility in order to generate proteomic patterns rapidly and reproducibly. Third, the method should target the extracellular proteome. Fourth, the method should generate distinct and simplified signatures to allow the identification and resolution of different signatures in a limited separation space.
Extracellular proteins are frequently modified by oligosaccharides compared to intracellular proteins 17. An analytic method based on preferential isolation of glycoproteins would greatly enrich extracellular proteins. We have developed, optimized, and automated a method for solid-phase extraction of N-linked glycopeptides (SPEG) from glycoproteins using hydrazide chemistry and tandem mass spectrometry 18–21. In SPEG, glycoproteins from different sources of biological samples are first solubilized by proteolytic digestion. The glycopeptides are then oxidized and specifically immobilized to a hydrazide solid support. After washing away non-immobilized peptides that are not glycosylated, the formerly N-linked glycosylated peptides are then released from the solid support using glycosidase (PNGase F). The isolated peptides are identified and quantified using liquid chromatography-tandem mass spectrometry (LC-MS/MS). Therefore, in a single analysis, the method identifies glycosylated proteins, the site of glycosylation (glycosite) 7, and the relative quantity of the identified formerly N-linked glycopeptides. There are several advantages for extracellular proteome analysis using this method compared to the traditional method. First, there are 1–2 N-linked glycosites for each glycoprotein. The analysis of the reduced number of peptides from each protein translates into favorable limits of detection and accurate quantification of extracellular proteins 22. Second, because peptides generated by this method are specific to peptide sequences containing N-linked glycosites from extracellular glycoproteins, which represent only 3% of the total tryptic peptides from proteins in the entire database, high throughput mass spectrometry-based methods could be used for the specific and sensitive identification and quantification of these peptides in different tissues 22, thus dramatically reducing the challenge of identifying proteins from global proteomic profiles of different tissues. Third, this approach targets proteins with post-translational modifications by glycosylation instead of total protein, it is selective to the mature proteins in their extracellular location 17. In addition, changes in the extent of glycosylation of extracellular proteins shown to correlate with cancer and other disease states can also be captured by this approach 23.
In this manuscript, we describe the quantitative proteomic analysis of the extracellular glycoproteins and determine their tissue specificity from 12 mouse tissues using glycopeptide isolation, liquid chromatography mass spectrometry (LC-MS), and automated tandem mass spectrometry (MS/MS). We further determine the skin tumor associated proteins with limited tissue expression among the 12 tissues. These tissue-specific extracellular glycoproteins may be useful for the development of potential new drug targets and protein markers for disease detection.
METHODS
Materials
Hydrazide resin (Bio-Rad, Hercules, CA), Sodium periodate (Bio-Rad, Hercules, CA), Tris (2-carboxyethyl) phosphine (TCEP) (Pierce, Rockford IL), PNGase F (New England Biolabs, Ipswich, MA), Sequencing grade trypsin (Promega, Madison, WI), C18 columns (Waters, Sep-Pak Vac), CHCA (Agilent, Palo Alto, CA), MALDI 4700 mass calibration standards (Applied Biosystems, Foster City, CA), and other chemicals were purchased from Sigma-Aldrich.
Mouse tissue
Two male and two female adult wildtype NIH01a mice were euthanized by CO2 asphyxiation according to IACUC standard protocol. Tissue samples from 12 different organs were extracted and flash frozen in liquid nitrogen. The 12 tissues included: heart, liver, spleen, stomach, brain, mammary gland, prostate, epidermis, intestine, kidney, ovary and testis.
Solubilization of solid tissues and extraction of peptides
Frozen tissues (20 mg each) were sliced into 1~3mm3 thick and incubated in 100μl of 5mM phosphate buffer and vortexed for 2–3 min. 100μl of trifluoroethanol (TFE) was added to each sample and samples were sonicated for 5 min in an ice-water bath. Samples were then incubated at 60°C for 2 hours followed by sonication for 2 min. Proteins were reduced by 5mM tributylphosphine (TBP) with 30 min incubation at 60°C. Iodoacetamide (10mM) was applied to the mixture and incubated in the dark at room temperature for another 30 min. The samples were diluted 5-fold with 50mM NH4HCO3 (pH 7.8) to reduce the TFE concentration to 10% prior to the addition of Trypsin at a ratio of 1:50 (w/w, enzyme: protein). Samples were digested at 37°C overnight with gentle shaking. The precipitate was discarded by centrifugation. Silver staining was used to test the effect of tryptic digestion and 2mg of total tryptic peptides from each sample were used for N-linked glycopeptide capture in the following steps.
Capture of formerly N-linked glycopeptides from tissue
Formerly N-glycopeptides were isolated from the total tryptic peptides using SPEG 19. The enriched formerly N-linked glycopeptides were concentrated by C18 columns, dried down, and resuspended in 40μl 0.4% acetic acid.
Mass spectrometry analysis
The peptides were identified and quantified by MS/MS analysis using an LTQ ion trap mass spectrometer (Thermo Fisher, San Jose, CA) and LC-MS analysis using an ESI-QTOF mass spectrometer (Waters, Beverly, MA). In both systems, 3μl isolated peptides were injected into a peptide cartridge packed with C18 resin, and then passed through a 10 cm × 75 μm i.d. microcapillary HPLC (μLC) column packed with C18 resin. The effluent from the μLC column entered an electrospray ionization source in which peptides were ionized and passed directly into the respective mass spectrometer. The HPLC mobile phase A and B were 0.1% formic acid (FA) in HPLC grade water and 0.1% formic acid (FA), 5% isopropanol in HPLC grade acetonitrile, respectively. A linear gradient of mobile phase B from 5%–32% over 100 min at flow rate of ~300 nL/min was applied. During LC-MS, data was acquired with a profile mode in the mass range scan between m/z, 400 and 2000 with 3.0 sec scan duration and 0.1 sec interscan. The MS/MS was also turned on to collect MS/MS spectra using data dependent mode. Each sample was analyzed three times to increase the accuracy of quantification.
Data analyses
Quantitative analysis using SpecArray
A suite of software tools were developed to analyze LC-MS data generated by ESI-QTOF analysis of formerly N-linked glycopeptides from each tissue sample 24. The software tools used LC-MS data and sequentially performed the following tasks to determine peptides that were of different abundance in different tissues, respectively: 1) Peptide peak picking: a list of peptide peaks was generated from each LC-MS run; 2) Peptide alignment: Peptides detected in individual LC-MS patterns were aligned based on peptide mass and retention time. 3) Peptide Array: For each peptide peak, an abundance ratio of matched peptides in different samples was determined for each peptide peak using the same method as described in the ASAPRatio software tool developed for LC-ESI-MS/MS data 25.
Peptide identifications
MS/MS spectra were searched with SEQUEST 26 against a mouse protein database (ipi.MOUSE.v3.13.fasta). The peptide mass tolerance is 2.0 Da for QTOF and 3.0 Da for LTQ. Other parameters of database searching are modified as following: oxidized methionines (add 16 Da to methionine), a PNGase F-catalyzed conversion of Asn to Asp and cysteine modification (add 57 Da to cysteine). The output files were evaluated by INTERACT and Peptide Prophet 27, 28. The criterion of Peptide Prophet analysis is a probability score ≥ 0.8 (with error rate of 0.0252) so that low probability peptide identifications were filtered out. For each identified peptide, peptide sequence, protein name, precursor m/z value, peptide mass, charge state, retention time where the MS/MS was acquired, and probability of the peptide identification were recorded and outputted using INTERACT 27.
Mapping the identified peptides to their peaks
Information from the identified peptides in MS/MS spectra was used to map the identified peptides to their corresponding MS peaks of precursor ions in aligned peak array as described previsouly 29. Briefly, the identified peptides from MS/MS spectra were mapped to the MS peaks detected in the same sample in the aligned peptide array using peptide charge state, m/z value of precursor ion, and elution time. The allowable differences for peptide match for charge state, m/z, and retention time were 0, 200 ppm, and 5 minutes, respectively.
Subcellular location of identified proteins
Signal peptides were predicted using SignalP 2.0 30. Transmembrane (TM) regions were predicted using the TMHMM (version 2.0) program 31, which predicts protein topology and the number of TM helices. Information from SignalP and TMHMM were combined to separate proteins into the following categories: i) cell surface - proteins that contained predicted non-cleavable signal peptides and no predicted transmembrane segments; ii) secreted - proteins that contained predicted cleavable signal peptides and no predicted transmembrane segments; iii) transmembrane - proteins that contained predicted transmembrane segments and extracellular loops and intracellular loops; and iv) intracellular - proteins that contained neither predicted signal peptides nor predicted transmembrane regions. All protein sequences were taken from the IPI mouse protein database (version 3.13).
Tissue-specific extracellular proteins
The percentage of signal intensity from each tissue to sum of total signal for all the tissues from peak intensity, number of spectral count, or Affymetrix signal was calculated and used to determine tissue-specific expression. Two criteria were set to select tissue-specific extracellular proteins: (1) the glycosite was only identified in one tissue; (2) the peak intensity from the specific tissue represented over 80% of total peak intensity of the peptide peak from all tissues. The transcriptional expression of the same gene was also explored using Gene atlas 32.
RESULTS
Procedure of quantitative analysis of extracellular proteome
The objective of this study is to apply the glycoproteomic approach to profile extracellular glycoproteins from different tissues and identify tissue-specific expression of extracellular proteins. This requires that the extraction of proteins/peptides can be uniformly applied to different tissues/organs. We started with solubilizing tissues using a combination of homogenization and protease digestion to cleave all proteins to peptides. The formerly N-linked glycopeptides were then captured using SPEG from the total pool of peptides to enrich extracellular glycoproteins.
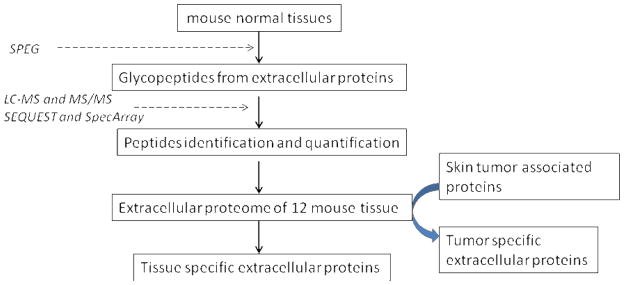
The procedure is schematically illustrated in Figure 1 and consists of three steps. 1) Glycoproteins were isolated and identified from 12 normal mouse tissues including heart, liver, spleen, stomach, brain, mammary gland, prostate, epidermis, intestine, kidney, ovary and testis; 2) The N-linked glycoproteome of 12 normal tissues were generated using quantitative proteomics, and tissue specific proteins were identified; 3) The skin tumor specific glycoproteins were analyzed and compared to the glycoproteome of different normal mouse tissues to determine skin tumor specific proteins 33.
Figure 1.
Diagram of work flow for determination of tissue-specificity of extracellular glycoproteins from normal mouse tissues and skin tumor mouse model.
Isolation of formerly N-linked glycopeptides for specific analysis of extracellular glycoproteins
From 12 normal mouse tissues, a total of 10337 identifications were observed using the MS/MS spectra, database search, and the minimum Peptide Prophet probability of 0.8 (with error rate of 0.0252), and 96% of identifications (9927) contained consensus N-linked glycosylation motif (N-X-S/T, X is any amino acid except proline) 34. This indicated that the majority of identified peptides were formerly N-linked glycosylated and the procedure was specific to peptides with N-linked glycosites. Therefore, we limited our subsequent analysis solely to the identified peptide sequences that contained at least one such N-linked glycosylation motif in order to simplify and to further reduce false positive rates. We were able to identify peptides enclosing a total of 1231 possible N-linked glycosites, which came from 826 unique glycoproteins.
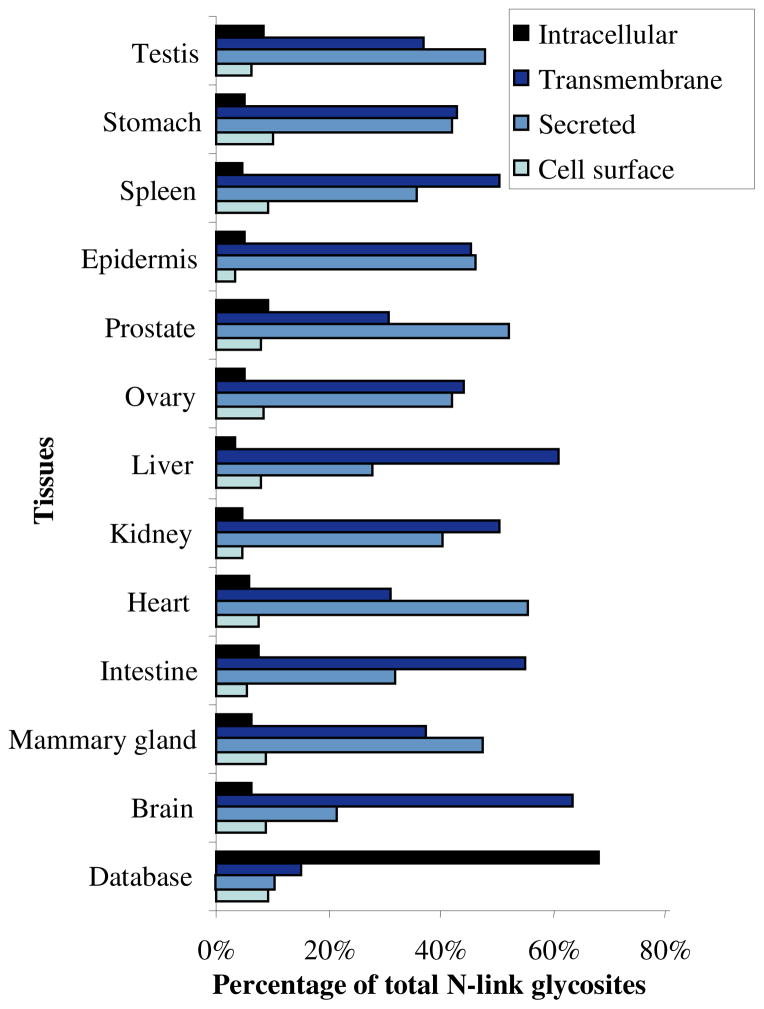
The initial idea of specifically targeting glycoproteins for analysis of extracellular proteins was based on the fact that the vast majority of extracellular proteins are modified by oligosaccharides. To test whether we were indeed sampling the expected extracellular proteome in our analyses, we applied the informatics approach for the prediction of sub-cellular localization for the glycoproteins identified, classifying them into four general classes 31, 35: 1) cell surface proteins, 2) secreted proteins, 3) transmembrane proteins, and 4) intracellular proteins. We would expect glycoproteins to fall into one of the first three of these classes. The cellular distribution of peptides identified is presented in Figure 2. About 88% of possible N-linked glycosites (1087 out of a total of 1231) identified from this study were predicted as extracellular proteins (containing either transmembrane segments or signal peptides or both). In contrast, applying the same informatics methodology to all 50651 entries in the mouse protein sequence database (IPI version 3.13) showed that approximately only one third of all the proteins in database are extracellular proteins. These observations confirmed our initial premise that the targeted isolation and identification of N-linked glycoproteins significantly enriched for the desired classes, i.e., proteins that likely represent good candidates for both markers of disease and therapeutic targets.
Figure 2.
Distribution of cellular locations for proteins identified from different tissues.
Quantitative analysis of glycoproteins from different tissues using spectral count
We then determined whether we could detect quantitative differences in the profiles of the glycoproteins from different tissues. In the above studies, proteins were identified by LC-MS/MS by ESI-LTQ and ESI-QTOF three times each. We used the number of redundant MS/MS spectra (spectral count) of the same peptide in the data set as a crude estimate of the corresponding protein abundance 36. As expected, we observed a wide range of identification frequencies assigned to a specific N-linked glycosite in each tissue type (spectral count from 1 to 319, Supplementary Table 1 online). Formerly N-linked glycopeptides from highly abundant glycoproteins were most often detected in multiple tissue types, suggesting they were housekeeping genes or plasma proteins present in all tissues due to blood circulation. For example, three glycosites were identified from serine protease inhibitor A3K by 610 total MS/MS spectra from all 12 tissues. In contrast to the broad distribution of plasma proteins, the formerly N-linked glycopeptides identified from proteins with a specific function for certain tissue types have limited expression. For example, glycosites, ILQYQPIN#STHELGPLVDLK and LSPYVN#YSFR, were from Splice Isoform 6 of the neuronal cell adhesion molecule and identified by 7 spectra only from brain. This protein is involved in cell adhesion and may play a role in the molecular assembly of the nodes of Ranvier. We then determined the glycoproteins that were uniquely identified in a specific tissue. As shown in supplementary table 1, about 50% of the total identified glycoproteins were only identified from one tissue type. These results indicate that extracellular proteins represent a rich source of tissue-specific proteins and this glycoproteomic approach allows the analysis of expression patterns of extracellular proteins using the robust and universal methodology that could be applied to the analysis of extracellular proteome from different biological sources.
Quantitative analysis of glycoproteins from different tissues using LC-MS
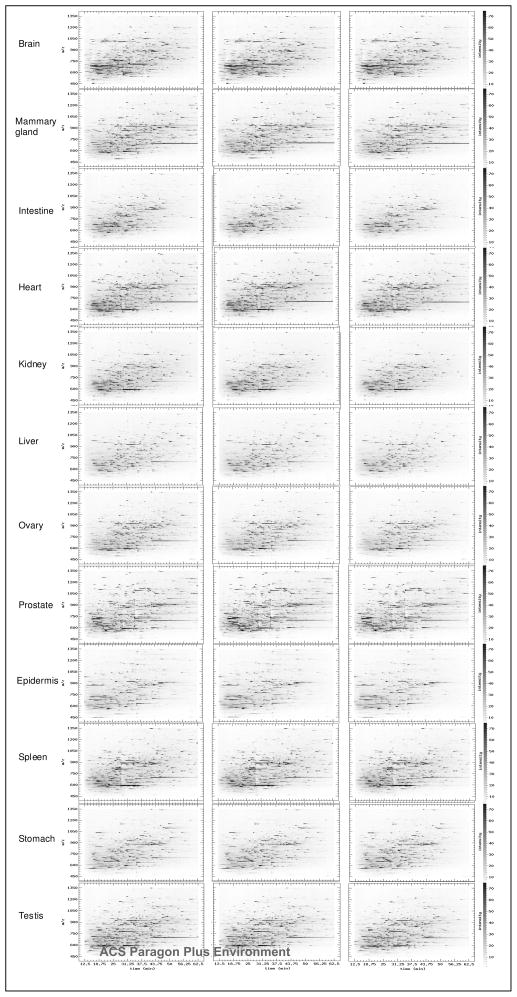
To verify the quantitative results from spectral count, we further determined the relative abundance of formerly N-linked glycopeptides from different mouse tissues using their LC-MS peak intensities of precursor ions. For quantitative study using LC-MS features, peptides need to be separated with a highly reproducible HPLC system and analyzed by the mass spectrometry instrument with high mass accuracy. Peptides analyzed with ESI-QTOF were used to generate LC-MS patterns. To increase the quantification accuracy, Peptides from each tissue were analyzed three times. Figure 3 displays the peptide patterns with three runs of analyses for each tissue using the Pep3D software tool 37. The data showed that the patterns generated from repeated analyses of the same tissue were highly consistent. However, the peptides from different tissues had distinct LC-MS patterns (Figure 3). To quantify the LC-MS patterns, SpecArray was used to detect peaks, to measure peak intensity, and to align corresponding peptide peaks among multiple patterns 24. To increase the quantification confidence, only peptide peaks that appeared at all three repeated analyses within each tissue type were selected. Using this approach, we were able to detect and align 2864 peptide peaks among the 12 tissues for quantitative expression patterns of formerly N-linked glycopeptides. We further mapped each MS peak to their corresponding peptide sequence identified from MS/MS spectrum with the same retention time as the MS peak from the same LC-MS and MS/MS analysis. Four hundred and twenty-five MS peaks can be mapped to their corresponding identified sequences and quantified (Supplementary Table 2).
Figure 3.
LC-MS patterns of formerly N-linked glycopeptides from different tissues. Peptide3D display of peptides detected by LC-MS is shown by analyzing formerly N-linked glycopeptides with three reproducible analyses of the same sample. The detected peptides are indicated as black spots with respected retention time (in minute, X-axis), m/z (in Da, Y-axis), and intensity (darkness of the spots). The peptides from triplicate LC-MS analyses of the same sample are shown for peptides isolated from different tissues (labeled with tissue names).
From these data, we calculated the average intensity and standard deviation for each formerly glycosylated peptide from three repeated analyses (Supplementary Table 2). The mean and median CVs observed in the three repeat LC-MS analyses of the identified peaks were 15.3% and 9.4%, respectively. We next calculated the correlation of different analyses within the three repeated LC-MS analyses. The average correlation of repeated LC-MS analyses within a tissue was 0.9040 with the standard deviation of 0.0573, while the average correlation of peptide intensities from different tissues was 0.3408 with standard deviation of 0.1782.
From the peptide3D, CVs, and correlation studies, the following were apparent. First, the multiple LC-MS analyses of the peptides from the individual mouse tissue were reproducible and quantitative. Second, due to the reduced complexity by glycopeptide capture, peptide peaks were well resolved. Third, the peptide patterns from different tissues were distinct. Collectively, these results suggest that LC-MS analyses of formerly N-linked glycosylated peptides isolated from different tissues are reproducible and the patterns of these peptides can be compared to determine the relative abundance of the peptides.
Extracellular glycoproteins with restricted tissue expression
From our MS/MS analysis, we identified a list of glycoproteins whose expression was dominant in a specific tissue using spectral count by LTQ and verified some of the tissue-specific proteins using precursor peak intensity of LC-MS by QTOF (Supplementary Table 3 online). We determined tissue-specific extracellular proteins in all 12 tissues and found that brain and spleen have the most tissue-specific extracellular glycoproteins, which is consistent with their specialized function. To determine whether similar tissue-specific expression could also be detected in mRNA levels, the Affymetrix data from the mouse gene atlas 32, 38 were examined. For the proteins for which the mRNA data was available, we could conclude that most tissue-specific proteins also had tissue-specific transcription. However, differences in the distribution of mRNA abundance were much less marked in the Affymetrix data compared to the protein expression data, and a number of proteins displayed different tissue expression patterns at the protein level different from their mRNA level. Furthermore, a number of tissue-specific extracellular proteins, for which the mRNA levels were not available, were also identified from this study (Table 1, and Supplementary Table 3 online).
Table 1.
Proteins expressed dominantly in one specific tissue.
| IPI | Protein Name | Organ |
|---|---|---|
| IPI00114279 | Excitatory amino acid transporter 1 | Brian |
| IPI00118385 | Glutamate [NMDA] receptor subunit zeta 1 | Brian |
| IPI00321348 | Immunoglobulin superfamily, member 8 | Brian |
| IPI00120564 | Neuronal cell adhesion molecule | Brain |
| IPI00453537 | 10 days neonate cortex cDNA, RIKEN library, clone:A830029E02 product: weakly similar to BK134P22.1 | Brain |
| IPI00122971 | N-CAM 180 of Neural cell adhesion molecule 1, 180 kDa isoform | Brain |
| IPI00123704 | Sodium/potassium-transporting ATPase beta-2 chain | Brain |
| IPI00125154 | DSD-1-proteoglycan | Brain |
| IPI00221456 | Adult male testis cDNA, synaptic vesicle glycoprotein 2 b | Brain |
| IPI00471176 | Hepatocyte cell adHesion molecule | Brain |
| IPI00465769 | Solute carrier family 12 member 5 | Brain |
| IPI00420554 | Contactin-associated protein-like 2 | Brain |
| IPI00120751 | Adult male brain UNDEFINED_CELL_LINE cDNA, Proton myo-inositol transporter homolog | Brain |
| IPI00131641 | LOC237403 protein | Brain |
| IPI00329927 | Neurofascin | Brain |
| IPI00338983 | Contactin-associated protein 1 | Brain |
| IPI00454159 | Splice Isoform 1 of Chondroitin sulfate proteoglycan 5 | Brain |
| IPI00130389 | Visual cortex cDNA, RIKEN library, clone:K530020M04 product:dipeptidylpeptidase 6, full insert sequence | Brain |
| IPI00131062 | Sodium channel beta-1 subunit precursor | Brain |
| IPI00114063 | Niemann-Pick C1-like protein 1 | Intestine |
| IPI00120907 | Oligopeptide transporter, small intestine isoform | Intestine |
| IPI00153202 | Angiotensin-converting enzyme 2 | Intestine |
| IPI00268184 | Adult male colon cDNA, RIKEN full-length enriched library, membrane-bound aminopeptidase P | Intestine |
| IPI00480532 | NOD-derived CD11c +ve dendritic cells cDNA, hypothetical protein | Intestine |
| IPI00458077 | 4 days neonate male adipose cDNA, N-acylsphingosine amidohydrolase 2 | Intestine |
| IPI00120907 | Oligopeptide transporter, small intestine isoform | Intestine |
| IPI00341098 | Calcium activated chloride channel | Intestine |
| IPI00378366 | N-acetylated-alpha-linked acidic dipeptidase-like protein | Intestine |
| IPI00111366 | Tumor necrosis factor receptor superfamily member 13C | Spleen |
| IPI00113350 | Cannabinoid receptor 2 | Spleen |
| IPI00117413 | Splice Isoform 1 of B-cell receptor CD22 | Spleen |
| IPI00114274 | Semaphorin-4D | Spleen |
| IPI00118413 | Thrombospondin 1 | Spleen |
| IPI00124640 | Osteoclast-like cell cDNA, granulin | Spleen |
| IPI00221418 | NOD-derived CD11c +ve dendritic cells cDNA, hypothetical Phospholipase D/Transphosphatidylase | Spleen |
| IPI00318993 | L-selectin | Spleen |
| IPI00469280 | Bone marrow macrophage cDNA, solute carrier family 30 | Spleen |
| IPI00314355 | B-cell differentiation antigen CD72 | Spleen |
| IPI00311808 | Transmembrane glycoprotein NMB | Spleen |
| IPI00121909 | Class II histocompatibility antigen, M beta 1 chain | Spleen |
| IPI00230509 | Splice Isoform 2 of Sialoadhesin | Spleen |
| IPI00113480 | Myeloperoxidase | Spleen |
| IPI00221911 | Leukocyte surface antigen CD53 | Spleen |
| IPI00343568 | CD180 antigen | Spleen |
| IPI00406609 | Receptor-type tyrosine-protein phosphatase eta | Spleen |
| IPI00318748 | Toll-like receptor 9 | Spleen |
| IPI00265854 | Complement receptor type 2 precursor | Spleen |
| IPI00114361 | Beta-microseminoprotein | Prostate |
| IPI00229083 | Adult male urinary bladder cDNA, hypothetical Kazal-type serine protease inhibitor domain containing protein | Prostate |
| IPI00230295 | Putative polypeptide N-acetylgalactosaminyltransferase-like protein 4 | Prostate |
| IPI00408931 | Carcinoembryonic antigen-related cell adhesion molecule 10 | Prostate |
| IPI00471102 | Adult male tongue cDNA, hypothetical protein | Prostate |
| IPI00133448 | Seminal vesicle antigen | Prostate |
| IPI00308892 | Adult male urinary bladder cDNA, weakly similar to LYSOZYME C, TYPE M | Prostate |
| IPI00396796 | Beta-defensin 50 | Prostate |
| IPI00115482 | Sodium/bile acid cotransporter | Liver |
| IPI00129677 | Asialoglycoprotein receptor major subunit | Liver |
| IPI00226346 | Similar to Rattus norvegicus putative integral membrane transport protein | Liver |
| IPI00355483 | SLC10A5 | Liver |
| IPI00118037 | Adult male testis cDNA, similar to PUTATIVE METALLOPEPTIDASE | Testis |
| IPI00120160 | Zona pellucida sperm-binding protein 3 receptor | Testis |
| IPI00125705 | Testis-specific protein TES101RP | Testis |
| IPI00331487 | Dickkopf-like protein 1 | Testis |
| IPI00120900 | Oviduct-specific glycoprotein | Ovary |
| IPI00123758 | Procollagen-lysine, 2-oxoglutarate 5-dioxygenase 2 | Ovary |
| IPI00128154 | Cathepsin L | Ovary |
| IPI00121337 | Renal sodium-dependent phosphate transport protein 2 | Kidney |
| IPI00122522 | Gamma-glutamyltranspeptidase 1 | Kidney |
| IPI00227906 | Splice Isoform 4 of Ssodium- and chloride-dependent transporter XTRP2 | Kidney |
| IPI00349520 | PREDICTED: similar to low density lipoprotein receptor-related protein 2 | Kidney |
| IPI00124006 | EP1 | Stomach |
| IPI00124269 | Potassium-transporting ATPase beta chain | Stomach |
| IPI00553773 | Secreted gel-forming mucin | Stomach |
| IPI00330068 | MUC6 | Stomach |
| IPI00130342 | Lymphocyte antigen 6 complex locus G6C protein | Epidermis |
| IPI00116056 | Sodium-dependent noradrenaline transporter | Epidermis |
| IPI00263726 | Solute carrier family 2 (facilitated glucoSe tranSporter), member 4 | Heart |
| IPI00469542 | Histidine-rich calcium-binding protein | Heart |
| IPI00123746 | Cadherin-13 | Heart |
| IPI00461384 | 15 days embryo head cDNA, MYELOBLAST KIAA0230 homolog | Mammary Gland |
Skin tumor specific extracellular proteins
From our previous study, 111 proteins were identified with overexpression in skin malignant carcinoma and benign papillomas compared to normal skin tissue 33. Since the proteins with restricted tissue expression are ideal candidates for new drug targets or protein markers for disease detection, we further determined which of the skin tumor overexpressed proteins has tissue-specific expression. By comparing the protein expression pattern from different mouse tissues from this study, 9 skin tumor overexpressed proteins were found expressed in only one normal tissue (Table 2), including extracellular matrix protein 1, lymphocyte antigen 6 complex locus G6C protein, legumain, granulin, myeloperoxidase, etc.
Table 2.
Skin tumor specific extracellular proteins
| IPI | Protein Name | Location | Other Tissue |
|---|---|---|---|
| IPI00122272 | Extracellular matrix protein 1 | Secreted | epidermis |
| IPI00130342 | Lymphocyte antigen 6 complex locus G6C protein | Secreted | epidermis |
| IPI00130627 | Legumain | Secreted | kidney |
| IPI00308971 | Cation-independent mannose-6- phosphate receptor | Transmembrane | ovary |
| IPI00127447 | Lysosome membrane protein II | Transmembrane | spleen |
| IPI00320605 | Integrin beta-2 | Transmembrane | spleen |
| IPI00124640 | Granulin | Secreted | spleen |
| IPI00110810 | Prostate stem cell antigen | Secreted | stomach |
| IPI00113480 | Myeloperoxidase | Secreted | spleen |
Topology of membrane proteins
In this study, we have identified 1231 possible N-linked glycosites from mouse. Since the N-glycosylation occurs on the extracellular domain of the protein, these N-linked glycosites provide experimentally determined evidence for membrane topology of the identified proteins and can be used for drug design. For example, potassium-transporting ATPase β subunit was one of the membrane proteins identified in this study. It is predicted as a single-pass type II membrane protein (http://au.expasy.org/uniprot/P50992). Residues 1-36 were predicted to be cytoplasmic, 37–57 were predicted as transmembrane segment, and residues 58–294 were predicted as extracellular. There are 7 potential N-linked glycosites located in the C-terminus of this protein (Figure 4, N highlighted with red). However, none of them has been proven experimentally. In this study, 5 out of 7 potential N-linked glycosites were identified specifically in stomach (Figure 4, N underlined). Quantification of these five glycosites using peak intensities by LC-MS analysis was consistent with specific expression in stomach (Table 1 and Supplementary Table 3 online). This is also consistent with the mRNA expression data 32, 38. The 5 identified N-linked glycosites at the C-terminal tail of potassium-transporting ATPase β subunit confirmed the orientation of the transmembrane proteins and the extracellular location of the C-terminal tail.
Figure 4.
Sequence, topology, and N-linked glycosites of potassium-transporting ATPase β subunit (ATP4B, 294 residues). ATB4B was predicted as type II membrane protein with residues 1-36 located intracellular (highlighted in yellow), residue 38-57 as transmembrane segment (highlighted with red), and residue 58-294 located extracellular (highlighted with blue). Seven potential N-linked glycosites were predicted in C-terminal region (the Asparagine residues (N) for the N-linked glycosites are highlighted with red. Five of the seven predicted N-linked glycosites were identified in this study (underlined). These experimental evidences also prove the membrane topology with C-terminal tail located in extracellular surface.
DISCUSSION
This paper describes the first quantitative glycoproteomic study to generate tissue-specific profiles of the extracellular glycoproteins from multiple tissues. Genomic approaches for large-scale gene expression analysis using microarrays 32, 38 or other transcriptome analysis methods such as massively parallel signature sequencing 39 have been used to determine gene expression profiles in different tissues. These analyses identified a number of tissue-specific genes, of which less than 20% encoded extracellular proteins. To enrich for genes encoding membrane proteins, the membrane-associated polyribosomal RNA from cell lines was purified and subtracted with RNA extracted from other tissues. 49% of the genes identified using this method encoded membrane or secreted proteins 40. However, due to post-transcriptional regulation, such as translational control, protein modification, subcellular location, and protein stability, protein levels cannot be captured from the genomic data. Determination of candidate proteins expressed on the extracellular surface in a tissue-specific fashion still relies on experimental studies of proteins.
Quantitative proteomic technologies using tandem mass spectrometry have made rapid progress in the analysis of soluble proteins in recent years, but the comprehensive proteomic analysis of extracellular proteins have lagged behind due to technical challenges 41–44. Previous studies to improve the method have mostly focused on strategies to analyze membrane proteins 45. Analysis of membrane proteins using traditional two-dimensional gel electrophoresis has faced the difficulty of insolubility of the membrane proteins in the non-detergent isoelectric focusing buffer. Recent efforts have turned to gel-free methods using tandem mass spectrometry and stable isotope labeling 13. In these approaches, membrane proteins were first isolated from cells or tissues using differential centrifugation and subjected to different strategies to solubilize membrane proteins using either organic acid and cyanogen bromide 9, detergents 10, organic solvents 11, 12, high pH and proteinase K 13, and chloroform extraction 14. For analyzing secreted proteins, such efforts were only reported in studies of the secretome in in vitro cultured cells 15, 16. Although these methods have proved to increase the effectiveness of membrane protein identification, they have not been applied to profiles of extracellular protein expression patterns in multiple cell types and tissues. First, previous methods have reported that at least half of proteins in the analyzed sample were identified as intracellular proteins due to their association with plasma membrane or membrane proteins. Second, the previous studies often use multiple dimensional chromatographic separations and isotopic labeling, thus restricting them to pair wise comparisons to limited sample size. As a result, proteomic studies have not been able to profile multiple tissues to generate expression patterns of extracellular proteins.
In this study, we applied a quantitative glycoproteomic method for high-content analysis of extracellular proteomes from different mouse tissues and identified tissue-specificity of skin tumor associated proteins. This analysis was based on the selective isolation of glycosylated peptides, which is the common feature of extracellular proteins, and subsequent identification and quantification of the complex peptides by LC-MS and MS/MS. However, neither this nor any other method is currently capable of analyzing all extracellular proteins. Extracellular proteins without any N-linked glycosylation site representing ~28% of total extracellular proteins will not be identified by this method. This limitation may be overcome by the analysis of O-linked glycoproteins or other methods that targets the extracellular proteins. By selectively isolating this subset of peptides, the procedure achieved a significant reduction in analytic complexity. We showed that this method was reproducible, achieved increased analytical depth and higher throughput. Furthermore, we demonstrated that, this method could be used to analyze extracellular protein patterns from multiple mouse tissues and identify specific tissue enriched extracellular proteins. For the limited sample source, this study only included 12 mouse organs; however, the platform is now in place, in which additional tissues can be included, such as human tissues in different pathological states. In addition, the data set for tissue-expression profiles of mouse extracellular proteins is useful to determine the tissue-enriched expression of human homologues of these proteins. The expression of tissue-enriched extracellular proteins identified in our study is suitable to normal organs, which may tend to change when cancer or disease occurs.
The possible 1231 N-glycosites reported here were identified using SPEG based on chemical immobilization of glycopeptides to solid support and specific releasing formerly N-linked glycosylated peptides using Peptide: N-Glycosidase F18, 19. The released formerly N-linked glycopeptides were analyzed by tandem mass spectrometry and the identified peptides were filtered with Peptide Prophet probability of 0.8 with the false discovery rate of 0.0252. Using this strategy, 96% of peptide identifications contained the consensus N-linked glycosylation motif. The result was consistent with previous reports that over 90% of identified peptides contained consensus N-linked glycosylation motif 18, 20, 22, 33, 46, 47. This is significant enrichment of formerly N-linked glycosylated peptides comparing to the calculated 7.0% peptides containing N-linked glycosylation motif in the database 46. This indicates that the formerly N-linked glycosylated peptides were specifically enriched and identified, not a coincidence of the falsely identified peptides containing a consensus N-linked glycosylation motif.
Using MS/MS spectra count, not all proteins from each tissue were identified due to the random sampling of peptide precursor ions during the analytical process 22. Therefore, the differences between peptide/protein identifications by MS/MS analyses among different tissue samples might be caused by the fact that only a fraction of the total peptides was identified by MS/MS analyses in the data dependant mode of operation. To increase the coverage of the N-linked glycoproteome and reduce the random sampling of MS/MS spectra, we analyzed the formerly N-linked glycopeptides from each tissue sample three times by LTQ and three times by QTOF. A number of proteins identified are drug targets currently in clinical use or are under research development as targets for new drugs 5. For example, toll-like receptor 9 (TLR9) has been known to participate in the innate immune response to microbial agents. However, TLR 9 also responds to autoantigens from dead cells and leads to systemic autoimmune diseases such as systemic lupus erythematosus (SLE). Inhibitors of TLR9, such as chloroquine and related compounds, have been used to treat autoimmune diseases, and a new drug targeting TLR is a subject of intense development 48.
Protein topology is a description, listing transmembrane segments, and their orientation relative to the membrane 49. Determining the topology is necessary for development of small molecule drugs or antibodies to bind their targets at specific location. Membrane protein topology is generally predicted using topology-prediction algorithms 50. However, the accuracy of the best current topology prediction method is in the range of 55–60% when the entire proteome is analyzed 50. Experimental topology information can be used with prediction methods to increase the prediction accuracy and the current experimental approaches to characterize transmembrane protein topologies include X-ray crystallography, NMR, gene fusion technique, substituted cysteine accessibility method, N-linked glycosylation experiment and other biochemical methods 51. Since the initial steps of N-glycosylation occur on the luminal side of the endoplasmic reticulum, we can expect that only extracytoplasmic portions of membrane proteins can be glycosylated 52 and glycosylation sites can be used to predict the topology of transmembrane proteins. However, only 172 experimentally confirmed human N-linked glycosylation sites 53 were reported prior to the recent glycoproteomic studies on the identification of formerly N-linked glycopeptides in complex biological samples 18, 53. Identification of N-linked glycosylation sites can significantly improve the precision of predicted membrane protein topology. In this study, 1231 possible N-linked glycosites have been identified and many of these sites have not been reported previously. This study identified the largest number of experiential identified N-linked glycosites from mouse comparing to several recent glycoproteomic studies reported by Ghesquiere et al. (identified 127 possible N-linked glycosites) 54, Bernhard et al. (104 possible N-linked glycosites) 55, Tian et al. (463 possible N-linked glycosites) 33, Tian et al. (285 possible N-linked glycosites) 47, and Gundry et al. (235 possible N-linked glycosites) 56. These N-linked glycosites can be used as experimentally determined evidence for membrane topology of the identified proteins and may be useful for drug design. These data are valuable for determining the accuracy of current topology-prediction methods and further development of improved algorithms.
In summary, this method affords one of the most comprehensive routine and high throughput analyses of the extracellular proteome. We therefore believe that it will find broad application in discovery of new drug targets or protein markers for human diseases.
Supplementary Material
Acknowledgments
This work was supported with federal funds from the National Cancer Institute, National Institutes of Health, by Grants R21-CA-114852.
Footnotes
Supporting Information Available: This material is available free at http://pubs.acs.org.
References
- 1.Collins BE, Paulson JC. Cell surface biology mediated by low affinity multivalent protein-glycan interactions. Curr Opin Chem Biol. 2004;8 (6):617–25. doi: 10.1016/j.cbpa.2004.10.004. [DOI] [PubMed] [Google Scholar]
- 2.Zheng CJ, Han LY, Yap CW, Ji ZL, Cao ZW, Chen YZ. Therapeutic targets: progress of their exploration and investigation of their characteristics. Pharmacol Rev. 2006;58 (2):259–79. doi: 10.1124/pr.58.2.4. [DOI] [PubMed] [Google Scholar]
- 3.Sokoll LJ, Chan DW. Biomarkers for Cancer Diagnostics. In: Abeloff MD, Armitage JO, Niederhuber JE, Kastan MB, McKenna WG, editors. Abeloff’s clinical oncology. 4. Elsevier Inc; Philadelphia, PA: 2008. [Google Scholar]
- 4.Hortobagyi GN. Treatment of breast cancer. N Engl J Med. 1998;339 (14):974–84. doi: 10.1056/NEJM199810013391407. [DOI] [PubMed] [Google Scholar]
- 5.Chen X, Ji ZL, Chen YZ. TTD: Therapeutic Target Database. Nucleic Acids Res. 2002;30 (1):412–5. doi: 10.1093/nar/30.1.412. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Zhang H, Chan DW. Cancer Biomarker Discovery in Plasma Using a Tissue-targeted Proteomic Approach. Cancer Epidemiol Biomarkers Prev. 2007;16 (10):1915–7. doi: 10.1158/1055-9965.EPI-07-0420. [DOI] [PubMed] [Google Scholar]
- 7.Zhang H, Loriaux P, Eng J, Campbell D, Keller A, Moss P, Bonneau R, Zhang N, Zhou Y, Wollscheid B, Cooke K, Yi EC, Lee H, Peskind ER, Zhang J, Smith RD, Aebersold R. UniPep, a database for human N-linked glycosites: A Resource for Biomarker Discovery. Genome Biol. 2006;7 (8):R73. doi: 10.1186/gb-2006-7-8-r73. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Zhang H, Liu AY, Loriaux P, Wollscheid B, Zhou Y, Watts JD, Aebersold R. Mass spectrometric detection of tissue proteins in plasma. Mol Cell Proteomics. 2007;6 (1):64–71. doi: 10.1074/mcp.M600160-MCP200. [DOI] [PubMed] [Google Scholar]
- 9.Washburn MP, Wolters D, Yates JR., 3rd Large-scale analysis of the yeast proteome by multidimensional protein identification technology. Nat Biotechnol. 2001;19 (3):242–7. doi: 10.1038/85686. [DOI] [PubMed] [Google Scholar]
- 10.Han D, Eng J, Zhou H, Aebersold R. Quantitative profiling of differentiation-induced microsomal proteins using isotope-coded affinity tags and mass spectrometry. Vol. 19. 2001. pp. 946–951. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Blonder J, Yu LR, Radeva G, Chan KC, Lucas DA, Waybright TJ, Issaq HJ, Sharom FJ, Veenstra TD. Combined chemical and enzymatic stable isotope labeling for quantitative profiling of detergent-insoluble membrane proteins isolated using Triton X-100 and Brij-96. J Proteome Res. 2006;5 (2):349–60. doi: 10.1021/pr050355n. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Blonder J, Conrads TP, Yu LR, Terunuma A, Janini GM, Issaq HJ, Vogel JC, Veenstra TD. A detergent- and cyanogen bromide-free method for integral membrane proteomics: application to Halobacterium purple membranes and the human epidermal membrane proteome. Proteomics. 2004;4 (1):31–45. doi: 10.1002/pmic.200300543. [DOI] [PubMed] [Google Scholar]
- 13.Wu CC, MacCoss MJ, Howell KE, Yates JR. A method for the comprehensive proteomic analysis of membrane proteins. Nat Biotechnol. 2003;21 (5):532–8. doi: 10.1038/nbt819. [DOI] [PubMed] [Google Scholar]
- 14.Mirza SP, Halligan BD, Greene AS, Olivier M. Improved method for the analysis of membrane proteins by mass spectrometry. Physiol Genomics. 2007;30 (1):89–94. doi: 10.1152/physiolgenomics.00279.2006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Pellitteri-Hahn MC, Warren MC, Didier DN, Winkler EL, Mirza SP, Greene AS, Olivier M. Improved mass spectrometric proteomic profiling of the secretome of rat vascular endothelial cells. J Proteome Res. 2006;5 (10):2861–4. doi: 10.1021/pr060287k. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Kratchmarova I, Kalume DE, Blagoev B, Scherer PE, Podtelejnikov AV, Molina H, Bickel PE, Andersen JS, Fernandez MM, Bunkenborg J, Roepstorff P, Kristiansen K, Lodish HF, Mann M, Pandey A. A proteomic approach for identification of secreted proteins during the differentiation of 3T3-L1 preadipocytes to adipocytes. Mol Cell Proteomics. 2002;1 (3):213–22. doi: 10.1074/mcp.m200006-mcp200. [DOI] [PubMed] [Google Scholar]
- 17.Roth J. Protein N-glycosylation along the secretory pathway: relationship to organelle topography and function, protein quality control, and cell interactions. Chem Rev. 2002;102 (2):285–303. doi: 10.1021/cr000423j. [DOI] [PubMed] [Google Scholar]
- 18.Zhang H, Li XJ, Martin DB, Aebersold R. Identification and quantification of N-linked glycoproteins using hydrazide chemistry, stable isotope labeling and mass spectrometry. Nat Biotechnol. 2003;21 (6):660–6. doi: 10.1038/nbt827. [DOI] [PubMed] [Google Scholar]
- 19.Tian Y, Zhou Y, Elliott S, Aebersold R, Zhang H. Solid-phase extraction of N-linked glycopeptides. Nat Protocols. 2007;2:334–339. doi: 10.1038/nprot.2007.42. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Zhou Y, Aebersold R, Zhang H. Isolation of N-linked glycopeptides from plasma. Anal Chem. 2007;79 (15):5826–37. doi: 10.1021/ac0623181. [DOI] [PubMed] [Google Scholar]
- 21.Zou Z, Ibisate M, Zhou Y, Aebersold R, Xia Y, Zhang H. Synthesis and evaluation of superparamagnetic silica particles for extraction of glycopeptides in the microtiter plate format. Anal Chem. 2008;80 (4):1228–34. doi: 10.1021/ac701950h. [DOI] [PubMed] [Google Scholar]
- 22.Zhang H, Yi EC, Li XJ, Mallick P, Kelly-Spratt KS, Masselon CD, Camp DG, 2nd, Smith RD, Kemp CJ, Aebersold R. High throughput quantitative analysis of serum proteins using glycopeptide capture and liquid chromatography mass spectrometry. Mol Cell Proteomics. 2005;4 (2):144–55. doi: 10.1074/mcp.M400090-MCP200. [DOI] [PubMed] [Google Scholar]
- 23.Durand G, Seta N. Protein glycosylation and diseases: blood and urinary oligosaccharides as markers for diagnosis and therapeutic monitoring. Clin Chem. 2000;46 (6 Pt 1):795–805. [PubMed] [Google Scholar]
- 24.Li XJ, Yi EC, Kemp CJ, Zhang H, Aebersold R. A software suite for the generation and comparison of Peptide arrays from sets of data collected by liquid chromatography-mass spectrometry. Mol Cell Proteomics. 2005;4 (9):1328–40. doi: 10.1074/mcp.M500141-MCP200. [DOI] [PubMed] [Google Scholar]
- 25.Li XJ, Zhang H, Ranish JA, Aebersold R. Automated statistical analysis of protein abundance ratios from data generated by stable-isotope dilution and tandem mass spectrometry. Anal Chem. 2003;75 (23):6648–57. doi: 10.1021/ac034633i. [DOI] [PubMed] [Google Scholar]
- 26.Eng J, McCormack AL, Yates JR., 3rd An approach to correlate tandem mass spectral data of peptides with amino acid sequences in a protein database. J Am Soc Mass Spectrom. 1994;5:976–989. doi: 10.1016/1044-0305(94)80016-2. [DOI] [PubMed] [Google Scholar]
- 27.Han DK, Eng J, Zhou H, Aebersold R. Quantitative profiling of differentiation-induced microsomal proteins using isotope-coded affinity tags and mass spectrometry. Nat Biotechnol. 2001;19 (10):946–51. doi: 10.1038/nbt1001-946. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Keller A, Nesvizhskii AI, Kolker E, Aebersold R. Empirical statistical model to estimate the accuracy of peptide identifications made by MS/MS and database search. Anal Chem. 2002;74 (20):5383–92. doi: 10.1021/ac025747h. [DOI] [PubMed] [Google Scholar]
- 29.Tian Y, Tan A, Sun X, Olson MT, Xie Z, Jinawath N, Chan DW, Shih Ie M, Zhang Z, Zhang H. Quantitative Proteomic Analysis of Ovarian Cancer Cells Identified Mitochondrial Proteins Associated with Paclitaxel Resistance. Proteomics - Clinical Applications. 2009;3 (11):1288–1295. doi: 10.1002/prca.200900005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Nielsen H, Engelbrecht J, Brunak S, von Heijne G. A neural network method for identification of prokaryotic and eukaryotic signal peptides and prediction of their cleavage sites. Int J Neural Syst. 1997;8 (5–6):581–99. doi: 10.1142/s0129065797000537. [DOI] [PubMed] [Google Scholar]
- 31.Krogh A, Larsson B, von Heijne G, Sonnhammer EL. Predicting transmembrane protein topology with a hidden Markov model: application to complete genomes. J Mol Biol. 2001;305 (3):567–80. doi: 10.1006/jmbi.2000.4315. [DOI] [PubMed] [Google Scholar]
- 32.Su AI, Wiltshire T, Batalov S, Lapp H, Ching KA, Block D, Zhang J, Soden R, Hayakawa M, Kreiman G, Cooke MP, Walker JR, Hogenesch JB. A gene atlas of the mouse and human protein-encoding transcriptomes. Proc Natl Acad Sci U S A. 2004;101 (16):6062–7. doi: 10.1073/pnas.0400782101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Tian Y, Kelly-Spratt KS, Kemp KS, Zhang H. Identification of glycoproteins from mouse skin tumors and plasma. Clinical Proteomics. 2008;4(2) doi: 10.1007/s12014-008-9014-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Bause E. Structural requirements of N-glycosylation of proteins. Studies with proline peptides as conformational probes. Biochem J. 1983;209 (2):331–6. doi: 10.1042/bj2090331. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Nielsen H, Engelbrecht J, Brunak S, von Heijne G. Identification of prokaryotic and eukaryotic signal peptides and prediction of their cleavage sites. Protein Eng. 1997;10 (1):1–6. doi: 10.1093/protein/10.1.1. [DOI] [PubMed] [Google Scholar]
- 36.Liu H, Sadygov RG, Yates JR., 3rd A model for random sampling and estimation of relative protein abundance in shotgun proteomics. Anal Chem. 2004;76 (14):4193–201. doi: 10.1021/ac0498563. [DOI] [PubMed] [Google Scholar]
- 37.Li X-J, Pedrioli P, Eng J, Martin D, Yi E, Lee H, Aebersold R. A tool to visualize and evaluate data obtained by liquid chromatography/electrospray ionization/mass spectrometry. 2004;76:3856–3860. doi: 10.1021/ac035375s. [DOI] [PubMed] [Google Scholar]
- 38.Su AI, Cooke MP, Ching KA, Hakak Y, Walker JR, Wiltshire T, Orth AP, Vega RG, Sapinoso LM, Moqrich A, Patapoutian A, Hampton GM, Schultz PG, Hogenesch JB. Large-scale analysis of the human and mouse transcriptomes. Proc Natl Acad Sci U S A. 2002;99 (7):4465–70. doi: 10.1073/pnas.012025199. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Jongeneel CV, Delorenzi M, Iseli C, Zhou D, Haudenschild CD, Khrebtukova I, Kuznetsov D, Stevenson BJ, Strausberg RL, Simpson AJ, Vasicek TJ. An atlas of human gene expression from massively parallel signature sequencing (MPSS) Genome Res. 2005;15 (7):1007–14. doi: 10.1101/gr.4041005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Egland KA, Vincent JJ, Strausberg R, Lee B, Pastan I. Discovery of the breast cancer gene BASE using a molecular approach to enrich for genes encoding membrane and secreted proteins. Proc Natl Acad Sci U S A. 2003;100 (3):1099–104. doi: 10.1073/pnas.0337425100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Oda Y, Huang K, Cross FR, Cowburn D, Chait BT. Accurate quantitation of protein expression and site-specific phosphorylation. Proc Natl Acad Sci U S A. 1999;96 (12):6591–6. doi: 10.1073/pnas.96.12.6591. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Veenstra TD, Martinovic S, Anderson GA, Pasa-Tolic L, Smith RD. Proteome analysis using selective incorporation of isotopically labeled amino acids. J Am Soc Mass Spectrom. 2000;11 (1):78–82. doi: 10.1016/S1044-0305(99)00120-8. [DOI] [PubMed] [Google Scholar]
- 43.Gygi SP, Aebersold R. Absolute quantitation of 2-D protein spots. Methods Mol Biol. 1999;112:417–21. doi: 10.1385/1-59259-584-7:417. [DOI] [PubMed] [Google Scholar]
- 44.Link AJ, Eng J, Schieltz DM, Carmack E, Mize GJ, Morris DR, Garvik BM, Yates JR., 3rd Direct analysis of protein complexes using mass spectrometry. Nat Biotechnol. 1999;17 (7):676–82. doi: 10.1038/10890. [DOI] [PubMed] [Google Scholar]
- 45.Wu CC, Yates JR., 3rd The application of mass spectrometry to membrane proteomics. Nat Biotechnol. 2003;21 (3):262–7. doi: 10.1038/nbt0303-262. [DOI] [PubMed] [Google Scholar]
- 46.Zhang H, Loriaux P, Eng J, Campbell D, Keller A, Moss P, Bonneau R, Zhang N, Zhou Y, Wollscheid B, Cooke K, Yi EC, Lee H, Peskind ER, Zhang J, Smith RD, Aebersold R. UniPep--a database for human N-linked glycosites: a resource for biomarker discovery. Genome Biol. 2006;7 (8):R73. doi: 10.1186/gb-2006-7-8-r73. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Tian Y, Gurley K, Meany DL, Kemp CJ, Zhang H. N-linked glycoproteomic analysis of formalin-fixed and paraffin-embedded tissues. J Proteome Res. 2009;8 (4):1657–62. doi: 10.1021/pr800952h. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Marshak-Rothstein A. Toll-like receptors in systemic autoimmune disease. Nat Rev Immunol. 2006;6 (11):823–835. doi: 10.1038/nri1957. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.von Heijne G. Membrane-protein topology. Nat Rev Mol Cell Biol. 2006;7 (12):909–18. doi: 10.1038/nrm2063. [DOI] [PubMed] [Google Scholar]
- 50.Melen K, Krogh A, von Heijne G. Reliability measures for membrane protein topology prediction algorithms. J Mol Biol. 2003;327 (3):735–44. doi: 10.1016/s0022-2836(03)00182-7. [DOI] [PubMed] [Google Scholar]
- 51.Ikeda M, Arai M, Okuno T, Shimizu T. TMPDB: a database of experimentally-characterized transmembrane topologies. Nucleic Acids Res. 2003;31 (1):406–9. doi: 10.1093/nar/gkg020. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Kornfeld R, Kornfeld S. Assembly of asparagine-linked oligosaccharides. Annu Rev Biochem. 1985;54:631–64. doi: 10.1146/annurev.bi.54.070185.003215. [DOI] [PubMed] [Google Scholar]
- 53.Kaji H, Saito H, Yamauchi Y, Shinkawa T, Taoka M, Hirabayashi J, Kasai K, Takahashi N, Isobe T. Lectin affinity capture, isotope-coded tagging and mass spectrometry to identify N-linked glycoproteins. Nat Biotechnol. 2003;21 (6):667–72. doi: 10.1038/nbt829. [DOI] [PubMed] [Google Scholar]
- 54.Ghesquiere B, Van Damme J, Martens L, Vandekerckhove J, Gevaert K. Proteome-wide characterization of N-glycosylation events by diagonal chromatography. J Proteome Res. 2006;5 (9):2438–47. doi: 10.1021/pr060186m. [DOI] [PubMed] [Google Scholar]
- 55.Bernhard OK, Kapp EA, Simpson RJ. Enhanced analysis of the mouse plasma proteome using cysteine-containing tryptic glycopeptides. J Proteome Res. 2007;6 (3):987–95. doi: 10.1021/pr0604559. [DOI] [PubMed] [Google Scholar]
- 56.Gundry RL, Raginski K, Tarasova Y, Tchernyshyov I, Bausch-Fluck D, Elliott ST, Boheler KR, Van Eyk JE, Wollscheid B. The mouse C2C12 myoblast cell surface N-linked glycoproteome: identification, glycosite occupancy, and membrane orientation. Mol Cell Proteomics. 2009;8 (11):2555–69. doi: 10.1074/mcp.M900195-MCP200. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.