Abstract
Actin has maintained an exquisite degree of sequence conservation over large evolutionary distances for reasons that are not understood. Generating an atomic model of the actin filament (F-actin) has been driven by the desire to explain phenomena from muscle contraction to cytokinesis in mechanistic detail. Here we use electron cryo-microscopy to show that frozen-hydrated actin filaments contain a multiplicity of different structural states. We show (at ~ 10 Å resolution) that subdomain 2 can be disordered, as well as being able to make multiple contacts with the C-terminus of a subunit above it. We link a number of disease-causing mutations in the human ACTA1 gene to the most structurally dynamic elements of actin. Since F-actin is structurally polymorphic it cannot be described using only one atomic model, and must be understood as an ensemble of different states.
INTRODUCTION
Actin was discovered more than 60 years ago1, and has been extensively studied because of its crucial roles in many functions, including cell shape and polarity2 and force generation in muscle3. Under physiological salt concentrations monomeric (globular) actin (G-actin) polymerizes to form filamentous actin (F-actin), a helical filament. Actin is one of the most abundant eukaryotic proteins, and its activities in vivo are modulated by >150 different actin binding proteins (ABPs)4. These ABPs control actin polymerization in the cell, as well as assembly of these filaments into higher order structures. Since the active form of actin is the filament, a significant effort has been devoted to obtaining a high resolution structure of F-actin. The actin filament is not amenable to crystallographic analysis. Actin subunits have been impaired for polymerization by chemical modifications, mutations, or by actin-binding proteins and drugs5–7, and some of the resulting crystal structures have been used to build atomic models of the actin filament by fitting x-ray fiber diffraction patterns from oriented F-actin gels8–11. A recent study using this approach has illuminated the conformational transition from G- to F-actin11, a “flattening” of the actin subunit by a relative rotation of the two major domains.
An extensive literature suggests, however, that F-actin cannot be described by a single structural model. F-actin is known to have a variable twist12 and a variable tilt of subunits13, which suggest a multiplicity of subunit-subunit interactions14. Several key structural elements of actin such as the DNase I-binding loop15 (D-loop), the hydrophobic plug16, and the N-terminus17,18 have been shown to be highly mobile. A single model for F-actin does not appear to be able to explain the crosslinking of residues within the filament19, and these results have suggested a large degree of plasticity and heterogeneity within F-actin.
A remarkable feature of actin is its anomalously low rate of sequence divergence over evolution20. There are no amino acid changes between chicken and human striated skeletal muscle actins21, while human cytoplasmic actin is 87% identitical with yeast actin. In contrast, the bacterial actin homologs that have been identified to data have no such sequence conservation among themselves or with eukaryotic actin22. There have been a number of arguments for this unusual conservation of eukaryotic actin. One explanation is that actin’s interaction with more than 150 ABPs in the cell constrains many residues and imposes a strong selective force4. An analysis of the residues in actin that have diverged23 shows that they are mainly on the surface of the filament where they would interact with actin-binding proteins, suggesting that the divergence of ABPs can explain the divergence of some surface residues in actin, but not the conservation of most buried residues. Since actin requires a specific chaperonin to fold, the interactions with the chaperonin may have placed many constraints on sequence divergence24. Highly expressed proteins evolve slowly25, presumably as a means to prevent protein misfolding, and actin is one of the most highly expressed proteins.
A different explanation for the anomalous sequence conservation involves the multiplicity of F-actin structural states13. In this argument the multiplicity of functions, interactions and higher-ordered structures requires actin to adopt different conformations, and this multiplicity of structural states puts a pressure on most amino acids within the actin molecule. Allosteric interactions have been shown to exist within actin26,27, and allosteric networks must create selection on buried residues that link the distant sites28. F-actin is likely to undergo conformational changes during muscle contraction that are essential for force generation by myosin29. Studies have shown that actin can be changed by mutations29, chemical modifications30,31 or proteolysis32 so as to leave myosin’s binding and actin’s activation of myosin’s ATPase unaffected, but greatly reduce or eliminate force generation.
To understand the intrinsic structural polymorphism of F-actin we used electron microscopy of unmodified frozen-hydrated actin filaments and single-particle approaches to sort heterogeneous segments into more homogeneous classes. Many other structural studies of F-actin have used stabilization of the filaments by phalloidin, a heptapeptide that is tightly bound and likely to change the dynamics of the filament, or nucleation by gelsolin11. The advances that we describe are due to both improved resolution as well as improved means of separating heterogeneous structures into classes. We suggest a relationship between a number of mutations in the human ACTA1 gene that cause disease and the structural dynamics of the actin filament.
RESULTS
Frozen-hydrated actin filaments possess structural heterogeneity
Electron cryo-micrographs of F-actin have been recorded using standard methods (Supplementary Fig. 1a). The approach used involves treating short segments (containing ~ 17 actin subunits) as single particles and using the Iterative Helical Real Space Reconstruction (IHRSR) method that does not involve averaging over long filament lengths33. There is no relationship between the length of the segments that we have used (~ 17 subunits) and the pseudo-repeat of an actin filament (~ 13 subunits), since the single-particle approach to helical reconstruction only requires that the segments contain multiple subunits. The 63,288 segments were divided into six classes using a multi-reference based approach.
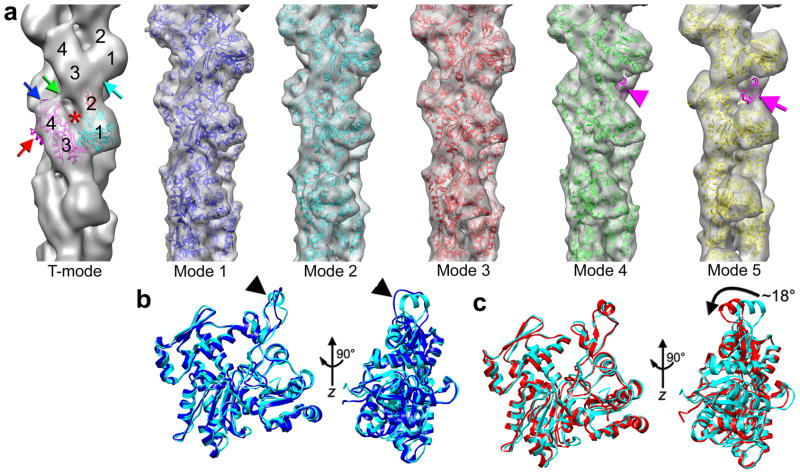
We call a tilted state of the subunit T-mode (Fig. 1a), which accounted for 24% of all segments. The cryo-EM reconstruction of T-mode is consistent with our earlier observations from negatively stained F-actin13 that in this mode SD2 of the lower protomer makes an extensive contact with SD3 of the protomer above it (Fig. 1a, cyan arrow), while the interaction of SD4 of the lower protomer with SD3 of the upper actin subunit weakens compared to “canonical” F-actin (Fig. 1a, green arrow). In the tilted state the nucleotide-binding cleft is open (Fig. 1a, red asterisk) due13 to the rotation around the hinge region of the two domains of actin by ~30°, accompanied by a shift apart of these domains by ~10 Å. The top and the side helix of SD4 in the tilted reconstruction (filtered to 16 Å) (Fig. 1a, blue and red arrows, respectively) stick out of the map suggesting structural variability of SD4. In contrast, SD1 yields an excellent fit at this resolution. The small number of segments in this class (n=15,218) prevented us from further refinement of the structural details of the tilted state.
Figure 1.
Six structural modes of actin found in frozen-hydrated actin filaments. (a) Three-dimensional reconstructions of six structural modes are shown as grey transparent surfaces, while the corresponding atomic models derived using a flexible fitting procedure (Supplementary Methods) are shown as ribbons. The absence of the D-loop (magenta ribbons in mode 4), and SD2 (magenta ribbons in mode 5) are marked with magenta arrowhead and magenta arrow, respectively. (b) The fold of the D-loop in mode1 is a loop, but a helix in mode 2 (black arrowheads). (c) Mode 3 has the D-loop as a helix, which is rotated by ~ 18° from its position in mode 2 (black arrow).
The remaining segments (n=48,070) were sorted according to the structural state of SD2. X-ray crystallography has shown that the D-loop, located at the top of SD2, can fold into a β-strand5, an α-helix6, or be disordered34. We designed model volumes based upon crystal structures and prior knowledge at lower resolution which were then used in a cross-correlation analysis (see Methods). We found five additional structural modes of F-actin (modes 1–5), and the corresponding three-dimensional reconstructions are shown in Fig 1. In contrast to the tilted state, the resolution of the reconstructions for modes 1–4 was determined to be ~ 10 Å (Supplementary Fig 1). In mode 5 the entire density for SD2 is missing, and the overall resolution is poorer (~ 14 Å, Supplementary Fig 1b). The resolution is sufficient to resolve the D-loop in modes 1–3. In mode 1 the density of the D-loop portion of the map is most consistent with being in a loop conformation (Fig. 1a,b, blue ribbons), while in mode 2 the density is more compact and is fit better by a helix (Fig. 1a,b, cyan ribbons). This region has been observed as a helix in one crystal6. The fit as a helix is constrained by both the width and length of the density, given the need to fit the known protein chain. When the density is both thinner and longer, the region is modeled as a loop. An α-helix cannot be extended to fit the longer density without melting of the helix. In the mode 3 the D-loop is also helical (Fig. 1 mode 3, red ribbons), but is rotated towards the exterior of the filament by ~ 18° (Fig. 1c). In the fourth mode (Fig. 1a, mode 4, green ribbons) the D-loop is disordered (Fig. 1a, mode 4, magenta arrowhead). Mode 5 (Fig. 1a, mode 5, yellow ribbons) is marked by the absence of density corresponding to SD2, showing that it must be substantially disordered (Fig. 1a, mode 5, magenta arrow).
At ~10 Å resolution the overall reconstruction yields an excellent match with the recently proposed atomic model of the actin filament11, except that the N-terminus of actin is not visualized in our global averaged map (Supplementary Materials).
The interface between the SD3/SD4 of adjacent actin subunits is similar in four modes
The atomic models of modes 1–4 of F-actin have similar SD3 and SD4 conformations, while being very different in the SD2 portion (Supplementary Fig. 5). This similarity results in a conserved interprotomer SD4-SD3 interface (both lateral and longitudinal) in these four modes. Since the resolution of the fifth mode is significantly worse than that of the other modes, it is hard to conclude whether this SD4-SD3 interface is different in mode 5. In addition to the longitudinal contact (Supplementary Fig. 5c), all modes possess two lateral contacts between the two actin strands (Supplementary Fig. 5d). One lateral contact is formed by residues 110–115 and residues 191–199 from the two neighboring actin protomers across the strand (Supplementary Fig. 5d, blue and yellow ribbons). The other lateral contact is formed by the hydrophobic plug (Supplementary Fig. 5d, residues 263–273 in cyan) forming a contact with residues 171–173 and 285–286 of the actin protomer above it on the opposite strand (Supplementary Fig. 5d, red and black ribbons, respectively), and residues 201–203 of the lower protomer on the opposite strand (Supplementary Fig. 5d, magenta ribbons). Our data are thus in complete agreement with the proposed11 SD3/SD4 lateral and longitudinal interactions.
In mode 5 SD2 is completely disordered (Fig. 1 mode 5, magenta arrow), and the filament is only held together by the interactions between SD3 and SD4 described above. This is consistent with the fact that A204E/P243K actin, with just two point mutations at the longitudinal interface between SD3 and SD4, is non-polymerizable7. We suggest that the ability of the SD3/SD4 interface to hold the filament together allows SD2 of actin to adopt multiple conformations without disrupting the filament.
The D-loop of actin is highly polymorphic
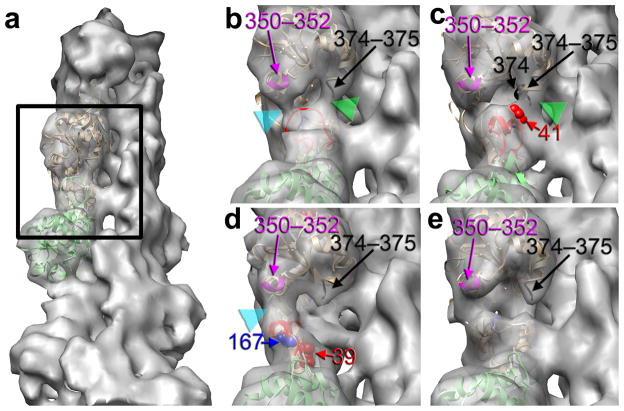
In modes 1–3 (Fig. 1a) we have visualized the entire density expected for SD2, but this density shows a multiplicity of conformations and interactions with SD1 of the subunit above within the same strand (Fig. 2). In mode 1, the D-loop region forms an extended thin ridge of density which is consistent with a loop (Fig. 2b, red ribbons). Because of the extended state, the D-loop is able to make two bridges of density (Fig. 2b, cyan and green arrowheads) with the subunit above – namely with residues 350–352 (Fig. 2b, magenta ribbons), and 374–375 (Fig. 2b, black ribbons) in the C-terminal region. In modes 2 and 3 the density is more consistent with the D-loop being a helix (Fig. 2c,d, red ribbons). Since the D-loop region is more compact in modes 2 and 3, it makes only a single interaction, compared with the two interactions observed in mode 1. In mode 2 this interaction is with the C-terminal residues 374–375 of the protomer above (Fig. 2c, green arrowhead). In mode 3 the D-loop is shifted towards the exterior of the filament (Fig. 1c), making a prominent contact with the 350–352 region of the subunit above (Fig. 2d, cyan arrowhead).
Figure 2.
D-loop of the lower subunit makes polymorphic interactions with the upper protomer. (a) To display details of interaction of the D-loop with the C-terminal portion of the upper protomer the reconstruction is tilted out of plane by 40°. The region of F-actin magnified in (b-e) is marked with a black rectangle. (b) In mode 1 the D-loop (red ribbons) of the lower protomer makes two contacts with the C-terminus of the upper subunit (cyan and green arrowheads), and these are with residues 350–352 (magenta), and 374–375 (black). (c) In mode 2 the D-loop is in the helical conformation, and is capable of making only one interaction with residues 374–375 (black) of the C-terminus of the upper protomer (green arrowhead). In this mode residue 41 (red spheres) is near residue 374 (black spheres). (d) In mode 3 the D-loop is positioned near the exterior of the filament and makes a bridge of density (cyan arrowhead) only with the 350–352 portion (magenta) of the upper subunit. (e) In mode 4 the D-loop is disordered, and does not make any interactions with the C-terminus of the upper protomer.
The multiplicity of interactions between SD1 and SD2 that we observe is consistent with biochemical observations. A cysteine replacing Gln41 can form a disulfide bond with Cys374 of an adjacent actin protomer16. In the recent atomic model of the actin filament11 the distance between the Cα atoms of these two residues is 13.4 Å, which suggests that the two regions of actin must either be shifted from the proposed positions to be able to form a disulfide, or one or both of these residues fluctuates between multiple positions. In our mode 2 (Fig. 2c, red and black spheres) the distance between these two residues is 9.7 Å, which brings the two residues close enough to form a disulfide bond. The ionic bridge between residues 39 and 167 of two neighboring actin protomers is crucial for yeast survival35. In the latest F-actin model11 these two residues are 11.7 Å apart. The position of the D-loop in mode 3 (Fig. 2d) brings the Cα atoms from these residues 6.3 Å apart, making an ionic interaction possible. In mode 4 the D-loop is completely missing in the EM map due to disorder or multiple states (Fig. 2e).
In this mode the C-terminal portion of the upper protomer does not make any contacts with the lower protomer, and the hydrophobic pocket at the base of SD1 and SD3 is open for interactions with ABPs that bind to this site36.
The D-loop conformation is coupled to other parts of actin
We suggest that the D-loop controls the connectivity between the strands. Two parts of the actin filament, the inter-strand contact involving the hydrophobic plug and the N-terminus of actin, both undergo significant structural alterations coupled with the conformation of the D-loop (Fig. 3). Two regions of SD2 contribute to the inter-strand interaction with the hydrophobic plug (Fig. 3a, magenta ribbons) – residues 64–67 (Fig. 3a, blue ribbons), and residues 38–40 (Fig. 3a, red ribbons). In modes 1 and 2 both regions are at the interface with the hydrophobic plug (Fig. 3a). In mode 3 SD2 is closer to the exterior of the filament (Fig. 1c), further from the inter-strand contact than in modes 1 and 2 (Fig. 3a, modes 1–3, cyan arrows). In mode 4 the D-loop is disordered, and thus invisible in the reconstruction (Fig. 1a). Nevertheless, the connectivity between the two actin strands is not compromised (Fig. 3a, mode 4, cyan arrows). This suggests that only the 64–67 region is needed for inter-strand connectivity.
Figure 3.
D-loop may work as an allosteric switch. (a) The two regions of SD2 that are adjacent to the lateral contact (cyan arrows) formed by the hydrophobic plug (263–273 magenta ribbons) are 64–67 (blue ribbons), and 38–40 (red ribbons). In mode 1 and mode 2 these two SD2 regions are positioned similarly, while in mode 3 they are moved away from the hydrophobic plug, which results in the weakening of the lateral contact (compare the lateral bridges of density marked with cyan arrows). In mode 4 the top of SD2 that includes the 38–40 region is missing, but residues 64–67 (blue ribbons) are sufficient to maintain the lateral contact. The N-terminus of actin has different conformations in these four modes (black arrowheads). (b) The orientation of the actin filament used to compare the three states of the N-terminus from the first three modes. (c) The movement of the N-terminus in modes 1–3 is coupled with the fold and orientation of SD2.
At ~ 10 Å resolution fine details of the actin molecule, such as the N-terminus, become visible. In modes 1–4 this portion of actin is very variable (Fig. 3a, black arrowheads). In mode 1 the N-terminus is facing down, while in mode 2 it faces up. In mode 3 the N-terminus has the strongest density among the four modes, presumably because of its stabilization by an interaction with the lower portion of SD1, while in mode 4 the N-terminal density is the weakest. The position of the D-loop seems to be coupled directionally with the position of the N-terminus (Fig. 3b,c).
The polymorphic nature of the N-terminus of actin that we observe is consistent with a proposed mobility of this region from NMR observations17. The coupling of structural states between the N-terminus and SD2 is intriguing. Muscle actin forms filaments that are more stable than those formed by yeast actin, and it has been suggested that due to its different dynamics it cannot support yeast cell growth37. Three N-terminal residues determine the death or survival of yeast cells when SD1 and SD2 of yeast actin are replaced with those from muscle actin38. We speculate that an allosteric coupling between the N-terminus and SD2 explains these observations.
The opening of the ATP-binding cleft and the structural state of SD2 are coupled
A substantial number of segments (31%) were classified as having SD2 completely disordered, arising from two different classes. Only 9% of all segments gave the best correlation with the model having SD2 completely disordered but with the ATP-binding cleft tightly closed, while 22% yielded the best correlation with the model having SD2 disordered but with the cleft open. We were not able to make a reasonable reconstruction of the closed-cleft class because of the small number of segments (n=4,782), while the set with the open cleft (mode 5, n=9,878) yielded a 3D-reconstruction (Fig. 4b) with a measured resolution of ~ 14 Å (Supplementary Fig. 1b).
Figure 4.
The structural states of SD2 and the ATP-binding cleft are coupled. (a) When the D-loop is disordered in mode 4 the ATP-binding cleft (black arrows) remains closed, similar to modes 1–3. Residues 60–64 (red ribbons) from one major domain make a contact with residues 200–205 (magenta ribbons) and residues 284–292 (blue ribbons) of the other major domain, which contribute to the closed state of the cleft. (b) In mode 5 the entire SD2 is disordered and the only longitudinal contact is the one between SD4 and SD3 of the adjacent actin subunits (red arrowhead). Residues 60–64 of SD2 do not make a contact with residues 200–205 (magenta) and 284–292 (blue), and this allows for the opening of the cleft (black arrows). (c) The two protomers from (a) and (b) are aligned to show that when the portion of the SD2 that does not include the D-loop (residues 34–36, 54–69 shown as black ribbons) is disordered, the two major domains of the actin molecule move apart in the direction marked with the black arrow.
In all atomic models of F-actin, including the most recent one11, SD2 of actin contributes to both longitudinal and lateral interactions that hold the filament together. We show that the actin filament can remain intact even though SD2 is completely disordered, and thus absent from the EM density map. In such a filament the only longitudinal contact that maintains the filament is the one between SD4 of the lower protomer and the SD3 of the upper one (Fig. 4b, red arrowhead). An actin filament with SD2 completely disordered should have a decreased torsional and flexural39 rigidity and be more heterogeneous. Thus it is not surprising that the resolution of such a relatively large set is significantly lower than that determined for the other four modes (Supplementary Fig. 1b).
Our data show that the disordering of the D-loop alone does not cause an opening of the nucleotide-binding cleft, since in mode 4 (where the D-loop is completely disordered) the cleft is tightly closed (Fig. 4a, black arrows). Our modeling suggest that residues 60–64 (Fig. 4a, red ribbons) form a contact with residues 200–205 located in SD4 of the same protomer (Fig. 4a, magenta ribbons) and residues 284–292 of SD3 of the upper protomer (Fig. 4a, blue ribbons). This interaction holds the ATP-binding cleft, located between SD3/4 and SD1/2, closed (Fig. 4a, black arrows). When SD2 is disordered, SD1 is able to move away from SD3, which opens the cleft (Fig. 4b, black arrows). This transition between mode 4 and mode 5 is opposite to the flattening of the actin subunit that occurs upon the G to F transition11. This suggests that the interaction of residues 60–64 (Fig. 4a, red ribbons) with residues 200–205 within the same protomer and 284–292 from a different protomer (Fig. 4a, magenta and blue ribbons, respectively) contribute to the flattening of actin upon polymerization.
DISCUSSION
Cooperativity of the observed structural states
As expected from previous work, blocks of different structure are present in the same filament. Our ability to see these separate states requires cooperativity within the actin filament. If adjacent individual protomers were in random states, we would never be able to sort segments (each containing ~ 17 protomers) into separate classes. Our analysis shows that multiple states can be found within the same filament (data not shown), thus the length of the “cooperative unit” is much smaller that that of a typical actin filament (containing hundreds to thousands of protomers). There is an extensive biochemical literature describing cooperativity within the actin filament40–45, so the visualization of these cooperative blocks within the same filament is not surprising. Many of the details that we see are not new features of the actin molecule; rather, they agree with different structural conformations of the actin molecule observed using x-ray crystallography, and are in agreement with biochemical experiments.
The role of structural polymorphism of F-actin in human disease
To date more that 177 mutations in the ACTA1 gene, encoding skeletal striated muscle actin, have been reported to cause disease in humans46. Many of these diseases are clinically severe and lethal within the first year of life. Most of the ACTA1 gene mutations are nonsense, frame-shift, or splice-site mutations that cause premature termination of translation, or the omission of entire exons from the mRNA47,48. We analyzed missense mutations that result in a properly folded and polymerizable actin, and these mutations are shown (Fig. 5a,b colored spheres). The first cluster of mutations is located at the interface between SD4 and SD3 of two adjacent actin subunits within the same helical strand (Fig. 5a, blue spheres). This region is essential, since in mode 5 this is the only contact that contributes to the lateral interactions within the actin filament (Fig. 4b, red arrowhead). Mutations of residue 286 cause severe nemaline myopathy (NM)49, while mutations of residue 292 result in the congenital fiber type disproportion (CFTD)50. Mutation of residue 241 in cytoplasmic gamma actin results in spontaneous bundle formation by the mutant actin and is linked to DFNA20/26 hearing impairment51.
Figure 5.
Mutations in ACTA1 gene causing human disease map to either the inter-subunit contacts or mobile elements of actin. (a,b) Residues that map to the interface between the SD4 and SD3 (241, 286, and 292) are shown as blue spheres. Mutations in SD2 (40, 64, and 66) are marked as red spheres, while residues linked to myopathies and located in regions involved in allosteric interactions are shown in yellow (168) and magenta (357 and 370). Mutations reported in the HP region are cyan spheres (263 and 268). (c) The electron density that corresponds to the N-terminus is marked with red arrows. Modification of the four N-terminal residues (red ribbons) rescues yeast cells harboring the lethal H372R mutation (blue spheres)56. Residue 132 (cyan spheres), linked to nemaline myophathy, resides between the two sites.
The next cluster is located in SD2 of actin (Fig. 5a,b, red spheres). The longitudinal contact formed by the SD2 region is not esssential for filament formation in vitro, since ~35% of all frozen-hydrated actin segments have SD2 partially or completely disordered. We suggest that SD2 works as an allosteric switch, and actins with mutations in the SD2 region will be deficient in their ability to undergo some vital structural alterations. Mutation of residue 40 (D-loop) results in severe NM50, mutation of residue 64 causes typical NM50, while mutations in residue 66 are linked to both actin and nemaline myopathies52. We have shown that SD2 makes multiple interactions with SD1 of the upper protomer, and these observations are consistent with the disease-causing mutations in the ACTA1 gene. Mutation of residue 168 (Fig. 5a, yellow spheres), which neighbors Ala167 involved in ionic bridge formation35, results in severe NM59. This mutation may alter the ability of actin to switch to mode 3, which is very consistent with an Arg39-Ala167 ionic interaction. Residues 357 and 370 reside in the C-terminus of actin (Fig. 5a,b, magenta spheres) which is allosterically linked with the D-loop (Fig. 2). Biochemical studies have described the existence of such coupling in both monomeric and filamentous actin53,54. Mutations at residues 357 and 370 are linked to severe NM46. The hydrophobic plug maintains the lateral interaction within the actin filament, and is involved in allosteric structural interactions with SD2 (Fig. 3) as shown by biochemical studies55. Mutations in two hydrophobic plug residues, 263 and 268 (Fig. 5b, cyan spheres), have been reported in NM cases in humans46.
The H372R mutation (Fig. 5c, blue spheres) suppresses56 the negative effects of substituting the two N-terminal acidic residues of yeast actin with four acidic residues from muscle actin (Fig. 5c, red ribbons). Structural analysis of the first three modes of actin reported here suggests that there is allosteric communication through SD1 within the actin subunit between the N and C termini (Fig. 3). Mutation of residue 132 that is located between the C- and the N-termini of actin (Fig. 5c, cyan spheres) is linked to NM in humans46. Since this residue is largely buried it cannot be interacting with ABPs, and is most likely part of an essential network providing allosteric coupling between the two termini.
F-actin is best understood as an ensemble of states
In the case of multimeric proteins, conformational plasticity can exist within subunits as well as between subunits. We show that under in vitro conditions F-actin does not exist in one structural state, but in a multiplicity of states. Our best expectation in vivo is that no self-nucleated actin filaments exist. Rather, different actin filaments will be nucleated by different ABPs, and this raises the potential for these filaments to be in different structural states42. A single structure for F-actin cannot explain why every buried residue has been under selective pressure for hundreds of millions of years. As long as mutations do not affect the folding, one would expect that such buried residues could be substituted conservatively. The documented allosteric interactions between the hydrophobic plug, the D-loop, and the N- and C-termini must, however, be transmitted through the buried residues in the core of actin, and we suggest that this may have placed the greatest selective pressure on these residues. We have shown that some of these allosteric couplings can be seen in the actin filament at modest (~ 10 Å) resolution, and this is a starting point to understanding how a single protein, actin, can specifically interact with more than 150 other proteins while involved in many different activities.
METHODS
Sample Preparation and Microscopy
Skeletal striated muscle G-actin (2 μM) was polymerized in 10 mM Mops buffer (pH 7.2), 40 mM KCl, 1 mM MgCl2, 0.5 mM DTT, and 0.5 mM ATP for 2–3 h. Droplets (3–4 μl) were applied to glow-discharged grids carbon-coated grids, and both blotted and plunged within a humidified chamber into an ethane slush. All cryo-EM was done on a Tecnai F20 FEG microscope operated at 200 keV at a magnification of 50,000x.
Image Processing
The SPIDER software package57 was used for most image processing, but the EMAN package58 was used to determine the defocus values in the micrographs and to extract filament images from micrographs. A Nikon COOLPIX 8000 scanner was used to digitize 124 cryo-EM micrographs having a defocus range from −1.2 to −4.4 μm at a raster of 2.38 Å per pixel. Initial correction for the contrast transfer function (CTF) was made by multiplying each image by its theoretical CTF. From these CTF-corrected images, 63,288 short (200 pixels long) overlapping segments were extracted. Three model volumes were created by using crystal structures of G-actin6 having actin protomers in the “canonical” state, with missing SD2, and finally in the “tilted” state13. These volumes were scaled to 4.76 Å per pixel and projected into 100 × 100-pixel images with an azimuthal rotational increment of 4°, generating 270 reference projections (3 × 90). The F-actin segments were down-sampled to 4.76 Å per pixel and cross-correlated with the 270 reference projections. A set of 29,970 segments (47%) was selected as similar to the “canonical” state of F-actin, 18,100 segments were assigned to the SD2-disordered class (29%), and 15,218 images (24%) yielded the best match with the tilted reference. Each set (sampled at 2.38 Å per pixel) was reconstructed with the IHRSR method33. After ~30 iterations the canonical set yielded a stable solution of 166.6°/27.5 Å, SD2-disordered set converged to 166.7°/27.6 Å solution, while the tilted set yielded a symmetry of 166.8°/27.6 Å. The canonical and the SD2-disordered sets were combined (n=48,070) and used in subsequent sorting procedures.
A set of models having SD2 of actin in different conformations was generated. An atomic model of the actin filament in the canonical state which has the D-loop in the loop conformation (pdb entry: 2zwh) was used as the first model. In the second model the SD2 of actin (residues 34–69) was replaced with that from the crystal structure where the DB-loop is a helix (pdb entry: 1j6z), while in the last two models the D-loop (residues 37–53), or the whole SD2 (residues 34–69) were removed. The 5 N-terminal residues were removed in each model. These volumes were scaled to 4.76 Å per pixel and projected into 100 × 100-pixel images with an azimuthal rotational increment of 4°, generating 360 reference projections (4 × 90). The 48,070 actin segments were down-sampled to 4.76 Å per pixel and cross-correlated with the 360 reference projections, and each class was reconstructed using IHRSR method. The volumes were corrected for the CTF (because images had been effectively multiplied by the CTF twice, once by the microscope and once by us) by using a Wiener filter assuming that the signal-to-noise ratio in the volume was very large, and a negative B-factor of 1,300 was used to amplify high frequencies in the reconstruction that were damped by the envelope function of the microscope, disorder in the filaments, etc.. Segments that gave the best correlation with the second reference volume (D-loop being a helix) yielded a 3D-reconstruction which had compact but weak density on the top of SD2, and this reflected a substantial heterogeneity in the SD2 portion of the map. Several models having D-loop in helical form, but in different orientations were used for refinement. The quality of the resultant reconstructions especially at their SD2 region was used as a guideline in the sorting process. The best result was achieved when two models having the D-loop in a helical conformation were used – in one model the top of SD2 was left in the “canonical” position, while in the other model it was shifted towards the exterior of the filament. Also, to reveal whether the disordering of the SD2 results in the ATP-binding cleft opening, two models of the actin filament having SD2 absent were generated – one with the cleft closed, another having the cleft opened to the extent observed in one of the crystal structures of actin59. Finally, six models were chosen as giving the best sorting results in terms of the maps quality: model with D-loop being a loop (mode 1), the two models having the top of SD2 as helix in the canonical or shifted conformation (mode 2 and mode 3, respectively), a model missing the D-loop (residues 37–53) (mode 4), and the two models having the whole SD2 (residues 34–69) removed but the cleft open (mode 5) and closed. These volumes were scaled to 4.76 Å per pixel, projected into 100 × 100-pixel images with an azimuthal rotational increment of 4°. The resultant 540 reference projections (6 × 90) were subsequently cross-correlated with the 48,070 actin segments (down-sampled to 4.76 Å per pixel). Each class was reconstructed using IHRSR method, except the class of images sorted as having the cleft closed and the SD2 disordered (n=4,782) which lacked enough segments to generate reasonable volume. Mode1 class (n=8,234) yielded stable solution of 166.6°/27.5 Å, mode2 (n=6,583) yielded a symmetry of 166.6°/27.6 Å, mode3 (n=11,169) converged to 166.6°/27.6 Å, mode4 (n=7,424) resulted in a 166.6°/27.5 Å symmetry, while mode5 (n=9,878) yielded a symmetry of 166.7°/27.6 Å. The resolution for each class was determined as described earlier60 using the Fourier Shell Correlation procedure and a conservative 0.5 criterion (Supplementary Fig. 1). The five reconstructions were corrected for the CTF as described above and filtered to the measured resolution of 10 Å, except the mode 5 reconstruction was filtered to 14 Å. The five filtered volumes were used to generate atomic models (Supplementary Methods).
Supplementary Material
Acknowledgments
This work was supported by NIH GM081303 (to E.H.E.).
Footnotes
Author contributions
A.O. did specimen preparation and EM; V.E.G. did image analysis, classifications, three-dimensional reconstructions, and preliminary model building; R.S. did model building and refinement; V.E.G, A.O. and E.H.E. discussed the data and wrote the paper.
Reference List
- 1.Feuer G, Molnar F, Pettko E, Straub FB. Studies on the composition and polymerization of actin. Hungarica Acta Physiologica. 1948;1:150–163. [PubMed] [Google Scholar]
- 2.Pollard TD, Borisy GG. Cellular motility driven by assembly and disassembly of actin filaments. Cell. 2003;112:453–465. doi: 10.1016/s0092-8674(03)00120-x. [DOI] [PubMed] [Google Scholar]
- 3.Cooke R. The mechanism of muscle contraction. CRC Crit Rev Biochem. 1986;21:53–118. doi: 10.3109/10409238609113609. [DOI] [PubMed] [Google Scholar]
- 4.Salwinski L, et al. The Database of Interacting Proteins: 2004 update. Nucleic Acids Res. 2004;32:D449–D451. doi: 10.1093/nar/gkh086. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Kabsch W, Mannherz HG, Suck D, Pai EF, Holmes KC. Atomic structure of the actin:DNase I complex. Nature. 1990;347:37–44. doi: 10.1038/347037a0. [DOI] [PubMed] [Google Scholar]
- 6.Otterbein LR, Graceffa P, Dominguez R. The crystal structure of uncomplexed actin in the ADP state. Science. 2001;293:708–711. doi: 10.1126/science.1059700. [DOI] [PubMed] [Google Scholar]
- 7.Rould MA, Wan Q, Joel PB, Lowey S, Trybus KM. Crystal structures of expressed non-polymerizable monomeric actin in the ADP and ATP states. J Biol Chem. 2006;281:31909–31919. doi: 10.1074/jbc.M601973200. [DOI] [PubMed] [Google Scholar]
- 8.Holmes KC, Popp D, Gebhard W, Kabsch W. Atomic model of the actin filament. Nature. 1990;347:44–49. doi: 10.1038/347044a0. [DOI] [PubMed] [Google Scholar]
- 9.Lorenz M, Popp D, Holmes KC. Refinement of the F-actin model against x-ray fiber diffraction data by the use of a directed mutation algorithm. J Mol Biol. 1993;234:826–836. doi: 10.1006/jmbi.1993.1628. [DOI] [PubMed] [Google Scholar]
- 10.Tirion MM, ben-Avraham D, Lorenz M, Holmes KC. Normal modes as refinement parameters for the F-actin model. Biophys J. 1995;68:5–12. doi: 10.1016/S0006-3495(95)80156-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Oda T, Iwasa M, Aihara T, Maeda Y, Narita A. The nature of the globular- to fibrous-actin transition. Nature. 2009;457:441–445. doi: 10.1038/nature07685. [DOI] [PubMed] [Google Scholar]
- 12.Egelman EH, Francis N, DeRosier DJ. F-actin is a helix with a random variable twist. Nature. 1982;298:131–135. doi: 10.1038/298131a0. [DOI] [PubMed] [Google Scholar]
- 13.Galkin VE, VanLoock MS, Orlova A, Egelman EH. A new internal mode in F-actin helps explain the remarkable evolutionary conservation of actin’s sequence and structure. Cur Biol. 2002;12:570–575. doi: 10.1016/s0960-9822(02)00742-x. [DOI] [PubMed] [Google Scholar]
- 14.Schmid MF, Sherman MB, Matsudaira P, Chiu W. Structure of the acrosomal bundle. Nature. 2004;431:104–107. doi: 10.1038/nature02881. [DOI] [PubMed] [Google Scholar]
- 15.Kim E, Reisler E. Intermolecular coupling between loop 38–52 and the c-terminus in actin filaments. Biophys J. 1996;71:1914–1919. doi: 10.1016/S0006-3495(96)79390-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Kim E, et al. Cross-linking constraints on F-actin structure. J Mol Biol. 2000;299:421–429. doi: 10.1006/jmbi.2000.3727. [DOI] [PubMed] [Google Scholar]
- 17.Heintz D, Kany H, Kalbitzer HR. Mobility of the N-terminal segment of rabbit skeletal muscle F-actin detected by 1H and 19F nuclear magnetic resonance spectroscopy. Biochemistry. 1996;35:12686–12693. doi: 10.1021/bi961159k. [DOI] [PubMed] [Google Scholar]
- 18.Orlova A, Yu X, Egelman EH. Three-dimensional reconstruction of a co-complex of F-actin with antibody Fab fragments to actin’s amino-terminus. Biophys J. 1994;66:276–285. doi: 10.1016/s0006-3495(94)80791-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Oztug Durer ZA, Diraviyam K, Sept D, Kudryashov DS, Reisler E. F-actin structure destabilization and DNase I binding loop: fluctuations mutational cross-linking and electron microscopy analysis of loop states and effects on F-actin. J Mol Biol. 2010;395:544–557. doi: 10.1016/j.jmb.2009.11.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Doolittle RF. The origins and evolution of eukaryotic proteins. Philos Trans R Soc Lond B Biol Sci. 1995;349:235–240. doi: 10.1098/rstb.1995.0107. [DOI] [PubMed] [Google Scholar]
- 21.Sheterline P, Clayton J, Sparrow J. Actin. Protein Profile. 1995;2:1–103. [PubMed] [Google Scholar]
- 22.Derman AI, et al. Phylogenetic analysis identifies many uncharacterized actin-like proteins (Alps) in bacteria: regulated polymerization, dynamic instability and treadmilling in Alp7A. Mol Microbiol. 2009;73:534–552. doi: 10.1111/j.1365-2958.2009.06771.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Egelman EH. Actin allostery again? Nat Struct Biol. 2001;8:735–736. doi: 10.1038/nsb0901-735. [DOI] [PubMed] [Google Scholar]
- 24.McCormack EA, Llorca O, Carrascosa JL, Valpuesta JM, Willison KR. Point mutations in a hinge linking the small and large domains of beta-actin result in trapped folding intermediates bound to cytosolic chaperonin CCT. J Struct Biol. 2001;135:198–204. doi: 10.1006/jsbi.2001.4385. [DOI] [PubMed] [Google Scholar]
- 25.Drummond DA, Bloom JD, Adami C, Wilke CO, Arnold FH. Why highly expressed proteins evolve slowly. Proc Natl Acad Sci U S A. 2005;102:14338–14343. doi: 10.1073/pnas.0504070102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Strzelecka-Golaszewska H, Mossakowska M, Wozniak A, Moraczewska J, Nakayama H. Long-range conformational effects of proteolytic removal of the last three residues of actin. Biochem J. 1995;307:527–534. doi: 10.1042/bj3070527. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Kim E, Motoki M, Seguro K, Muhlrad A, Reisler E. Conformational changes in subdomain 2 of G-actin: fluorescence probing by dansyl ethylenediamine attached to Gln-41. Biophys J. 1995;69:2024–2032. doi: 10.1016/S0006-3495(95)80072-X. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Suel GM, Lockless SW, Wall MA, Ranganathan R. Evolutionarily conserved networks of residues mediate allosteric communication in proteins. Nat Struct Biol. 2003;10:59–69. doi: 10.1038/nsb881. [DOI] [PubMed] [Google Scholar]
- 29.Drummond DR, Peckham M, Sparrow JC, White DC. Alteration in crossbridge kinetics caused by mutations in actin. Nature. 1990;348:440–442. doi: 10.1038/348440a0. [DOI] [PubMed] [Google Scholar]
- 30.Prochniewicz E, Yanagida T. Inhibition of sliding movement of F-actin by crosslinking emphasizes the role of actin structure in the mechanism of motility. J Mol Biol. 1990;216:761–772. doi: 10.1016/0022-2836(90)90397-5. [DOI] [PubMed] [Google Scholar]
- 31.Kim E, et al. Intrastrand cross-linked actin between Gln-41 and Cys-374. III. Inhibition of motion and force generation with myosin. Biochemistry. 1998;37:17801–17809. doi: 10.1021/bi981286b. [DOI] [PubMed] [Google Scholar]
- 32.Schwyter DH, Kron SJ, Toyoshima YY, Spudich JA, Reisler E. Subtilisin cleavage of actin inhibits In vitro siding movement of actin filaments over myosin. J Cell Biol. 1990;111:465–470. doi: 10.1083/jcb.111.2.465. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Egelman EH. A robust algorithm for the reconstruction of helical filaments using single-particle methods. Ultramicroscopy. 2000;85:225–234. doi: 10.1016/s0304-3991(00)00062-0. [DOI] [PubMed] [Google Scholar]
- 34.Kudryashov DS, et al. The crystal structure of a cross-linked actin dimer suggests a detailed molecular interface in F-actin. Proc Natl Acad Sci U S A. 2005;102:13105–13110. doi: 10.1073/pnas.0506429102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Stokasimov E, McKane M, Rubenstein PA. Role of intermonomer ionic bridges in the stabilization of the actin filament. J Biol Chem. 2008;283:34844–34854. doi: 10.1074/jbc.M804419200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Dominguez R. Actin-binding proteins--a unifying hypothesis. Trends Biochem Sci. 2004;29:572–578. doi: 10.1016/j.tibs.2004.09.004. [DOI] [PubMed] [Google Scholar]
- 37.Kim E, Miller CJ, Reisler E. Polymerization and in vitro motility properties of yeast actin: a comparison with rabbit skeletal α-actin. Biochemistry. 1996;35:16566–16572. doi: 10.1021/bi9623892. [DOI] [PubMed] [Google Scholar]
- 38.McKane M, Wen KK, Meyer A, Rubenstein PA. Effect of the substitution of muscle actin-specific subdomain 1 and 2 residues in yeast actin on actin function. J Biol Chem. 2006;281:29916–29928. doi: 10.1074/jbc.M602251200. [DOI] [PubMed] [Google Scholar]
- 39.Orlova A, Egelman EH. A conformational change in the actin subunit can change the flexibility of the actin filament. J Mol Biol. 1993;232:334–341. doi: 10.1006/jmbi.1993.1393. [DOI] [PubMed] [Google Scholar]
- 40.Prochniewicz E, Katayama E, Yanagida T, Thomas DD. Cooperativity in F-actin: chemical modifications of actin monomers affect the functional interactions of myosin with unmodified monomers in the same actin filament. Biophys J. 1993;65:113–123. doi: 10.1016/S0006-3495(93)81057-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Drewes G, Faulstich H. Cooperative effects on filament stability in actin modified at the C-terminus by substitution or truncation. Eur J Biochem. 1993;212:247–253. doi: 10.1111/j.1432-1033.1993.tb17656.x. [DOI] [PubMed] [Google Scholar]
- 42.Orlova A, Prochniewicz E, Egelman EH. Structural dynamics of F-actin. II. Cooperativity in structural transitions. J Mol Biol. 1995;245:598–607. doi: 10.1006/jmbi.1994.0049. [DOI] [PubMed] [Google Scholar]
- 43.Miki M, Wahl P, Auchet JC. Fluorescence anisotropy of labeled F-actin: influence of divalent cations on the interaction between F-actin and myosin heads. Biochemistry. 1982;21:3661–3665. doi: 10.1021/bi00258a021. [DOI] [PubMed] [Google Scholar]
- 44.Muhlrad A, Cheung P, Phan B, Miller C, Reisler E. Dynamic properties of actin. Structural changes induced by beryllium fluoride. J Biol Chem. 1994;269:11852–11858. [PubMed] [Google Scholar]
- 45.Oosawa F, Fujime S, Ishiwata S, Mihashi K. Dynamic property of F-actin and thin filament. Cold Spring Harbor Symposium of Quantitative Biology. 1972;37:277–285. [Google Scholar]
- 46.Laing NG, et al. Mutations and polymorphisms of the skeletal muscle alpha-actin gene (ACTA1) Hum Mutat. 2009;30:1267–1277. doi: 10.1002/humu.21059. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Nowak KJ, et al. Nemaline myopathy caused by absence of alpha-skeletal muscle actin. Ann Neurol. 2007;61:175–184. doi: 10.1002/ana.21035. [DOI] [PubMed] [Google Scholar]
- 48.Sparrow JC, et al. Muscle disease caused by mutations in the skeletal muscle alpha-actin gene (ACTA1) Neuromuscul Disord. 2003;13:519–531. doi: 10.1016/s0960-8966(03)00101-9. [DOI] [PubMed] [Google Scholar]
- 49.Hennessey ES, Drummond DR, Sparrow JC. Molecular genetics of actin function. Biochem J. 1993;291:657–671. doi: 10.1042/bj2910657. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Feng JJ, Marston S. Genotype-phenotype correlations in ACTA1 mutations that cause congenital myopathies. Neuromuscul Disord. 2009;19:6–16. doi: 10.1016/j.nmd.2008.09.005. [DOI] [PubMed] [Google Scholar]
- 51.Morin M, et al. In vivo and in vitro effects of two novel gamma-actin (ACTG1) mutations that cause DFNA20/26 hearing impairment. Hum Mol Genet. 2009;18:3075–3089. doi: 10.1093/hmg/ddp249. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Ilkovski B, et al. Evidence for a dominant-negative effect in ACTA1 nemaline myopathy caused by abnormal folding, aggregation and altered polymerization of mutant actin isoforms. Hum Mol Genet. 2004;13:1727–1743. doi: 10.1093/hmg/ddh185. [DOI] [PubMed] [Google Scholar]
- 53.Khaitlina SY, Strzelecka-Golaszewska H. Role of the DNase-I-binding loop in dynamic properties of actin filament. Biophys J. 2002;82:321–334. doi: 10.1016/S0006-3495(02)75397-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Kuznetsova I, Antropova O, Turoverov K, Khaitlina S. Conformational changes in subdomain I of actin induced by proteolytic cleavage within the DNase I-binding loop: energy transfer from tryptophan to AEDANS. FEBS Lett. 1996;383:105–108. doi: 10.1016/0014-5793(96)00238-4. [DOI] [PubMed] [Google Scholar]
- 55.Kuang B, Rubenstein PA. The effects of severely decreased hydrophobicity in a subdomain 3/4 loop on the dynamics and stability of yeast G-actin. J Biol Chem. 1997;272:4412–4418. doi: 10.1074/jbc.272.7.4412. [DOI] [PubMed] [Google Scholar]
- 56.McKane M, et al. A mammalian actin substitution in yeast actin (H372R) causes a suppressible mitochondria/vacuole phenotype. J Biol Chem. 2005;280:36494–36501. doi: 10.1074/jbc.M506970200. [DOI] [PubMed] [Google Scholar]
- 57.Frank J, et al. SPIDER and WEB: Processing and visualization of images in 3D electron microscopy and related fields. J Struct Biol. 1996;116:190–199. doi: 10.1006/jsbi.1996.0030. [DOI] [PubMed] [Google Scholar]
- 58.Ludtke SJ, Baldwin PR, Chiu W. EMAN: semiautomated software for high-resolution single-particle reconstructions. J Struct Biol. 1999;128:82–97. doi: 10.1006/jsbi.1999.4174. [DOI] [PubMed] [Google Scholar]
- 59.Chik JK, Lindberg U, Schutt CE. The structure of an open state of β-actin at 2.65 Ångstrom resolution. J Mol Biol. 1996;263:607–623. doi: 10.1006/jmbi.1996.0602. [DOI] [PubMed] [Google Scholar]
- 60.Galkin VE, Orlova A, Cherepanova O, Lebart MC, Egelman EH. High-resolution cryo-EM structure of the F-actin-fimbrin/plastin ABD2 complex. Proc Natl Acad Sci U S A. 2008;105:1494–1498. doi: 10.1073/pnas.0708667105. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.