Abstract
Multi-subunit tethering complexes participate in the process of vesicle tethering, the initial interaction between transport vesicles and their acceptor compartments. TRAPPII is a highly conserved tethering complex that functions in the late Golgi and consists of all TRAPPI and three specific subunits. We have purified native yeast TRAPPII and characterized its structure and subunit organization by single-particle electron microscopy. Our data show that the nine TRAPPII components form a core complex that dimerizes into a three-layered, diamond-shaped structure. The TRAPPI subunits assemble into TRAPPI complexes that form the outer layers. The three TRAPPII-specific subunits cap the ends of TRAPPI and form the middle layer responsible for dimerization. TRAPPII binds Ypt1 and likely uses the TRAPPI catalytic core to promote guanine nucleotide exchange. We discuss implications of the TRAPPII structure for coat interaction and TRAPPII-associated human pathologies.
Vesicular trafficking mediates the transport of proteins and lipids between different membrane-bound compartments. Each step along this pathway, from the budding of a transport vesicle at the donor compartment to its fusion with the membrane of the acceptor compartment, must be highly regulated to ensure fidelity of transport. Tethering refers to the initial, long-range interaction between a transport vesicle and its acceptor compartment1. This step is believed to play a crucial role in establishing specificity of vesicle targeting and is mediated by tethering factors that are either long coiled-coil proteins or multi-subunit complexes2. Tethering factors participate in the tethering process, and some of their demonstrated activities include: binding coat proteins of transport vesicles, promoting the organization of SNARE proteins involved in vesicle fusion, and functioning as effectors and/or guanine nucleotide exchange factors (GEFs) for Rab/Ypt GTPases3. The molecular mechanisms of how these proteins and protein complexes coordinate vesicle tethering are poorly understood and may vary considerably between different transport steps.
The budding yeast Saccharomyces cerevisiae possesses eight multi-subunit tethering complexes (TRAPPI, TRAPPII, Dsl1, COG, exocyst, GARP, HOPS/Class C VPS, and CORVET)4. These complexes function in distinct compartments and differ both in the number and primary sequences of their subunits1. TRAPP (Transport protein particle), a highly conserved tethering complex that resides in the Golgi, exists in two forms, TRAPPI and TRAPPII5. TRAPPI has a size of about 300 kDa and was initially thought to contain seven subunits, Bet3, Bet5, Trs20, Trs23, Trs31, Trs33, and Gsg1 (or Trs85), but Gsg1 has recently been shown to be part of a different complex, named TRAPPIII (Table 1)6,7. In yeast, all TRAPPI subunits, except Trs33, are encoded by essential genes. TRAPPI functions as a GEF for Ypt1 (Rab1 ortholog in yeast)8,9, and binds the COPII coat protein Sec23, which has been suggested to mediate the recruitment of incoming ER-derived vesicles to the Golgi10. At the structural level, TRAPPI is the best-characterized tethering complex. An atomic model of TRAPPI was generated based on the crystal structures of two mammalian TRAPPI subcomplexes and an electron microscopy (EM) reconstruction of the yeast complex11. The model revealed that the seven globular subunits arrange side by side to generate a flat and elongated molecule with two extensive surfaces. The subunit organization of this model was confirmed by the recently determined crystal structure of the yeast TRAPPI core in complex with Ypt1, which provided insights into how the assembly of four TRAPPI subunits (Bet3, Bet5, Trs23, and Trs31) creates a catalytic site for promoting GDP/GTP exchange in Ypt1 (ref. 12).
Table 1.
Subunit composition of the TRAPP complexes
| Subunit | Calculated mass (kDa) | Essential gene |
|---|---|---|
| TRAPPI: | ||
| Bet3 (TRAPPC3)* | 22 (20) | Yes |
| Bet5 (TRAPPC1) | 18 (17) | Yes |
| Trs20 (TRAPPC2) | 20 (16) | Yes |
| Trs23 (TRAPPC4) | 25 (24) | Yes |
| Trs31 (TRAPPC5) | 32 (21) | Yes |
| Trs33 (TRAPPC6a,b) | 31 (19,18) | No |
| TRAPPII-specific: | ||
| Tca17 (TRAPPC2L) | 17 (16) | No |
| Kre11 (N/A) | 63 | No |
| Trs120(TRAPPC9) | 148 (139) | Yes |
| Trs130(TRAPPC10) | 128 (142) | Yes |
| TRAPPIII-specific: | ||
| Gsg1 (N/A) | 81 | No |
Mammalian orthologs in parentheses
TRAPPII consists of TRAPPI and three additional large proteins, Kre11 (or Trs65), Trs120, and Trs130, forming a complex with an estimated size of about 1 MDa7. More recently, Tca17 (TRAPPC2L in mammals), a protein with sequence similarity to Trs20, has been shown to be a substoichiometric component of TRAPPII and has been .proposed to play a role in promoting assembly and stability of TRAPPII13,14. TRAPPII binds to coatomer, the heptameric complex that forms the coat of COPI vesicles15,16, and it has been proposed to function in intra-Golgi and endosome-to-Golgi transport. TRAPPII is also believed to function as a GEF for Ypt/Rab GTPases, but the exact identity of its target GTPase remains controversial12,17. Precisely how the three TRAPPII-specific subunits convert the biological function of TRAPPI to that of TRAPPII, in particular how they remodel and/or block the COPII binding site on TRAPPI while creating a new binding site for COPI, is currently not known. It is also unclear whether the TRAPPI components are arranged differently within TRAPPII. TRAPPII appears to be the more dominant TRAPP complex in mammalian cells, and defects in TRAPP complex function are linked to several human pathologies. Notably, point mutations of the Trs20/TRAPPC2 subunit, a shared subunit of TRAPPI and TRAPPII, are associated with the genetic disease spondyloepiphyseal dysplasia tarda (SEDT)18,19, and a truncation mutation of Trs120/TRAPPC9, a TRAPPII-specific subunit, was recently identified to be responsible for autosomal recessive intellectual disability20–22.
To address fundamental questions concerning the function of TRAPPII, we set out to determine its structure. We developed a method to purify native TRAPPII from yeast, which enabled us to characterize the structure and subunit organization of this tethering complex by single-particle EM. With nine components in its fully assembled state, TRAPPII is the most sophisticated tethering complex to be structurally characterized to date. Our structure of TRAPPII reveals substantial divergence in the quaternary structure of multi-subunit tethering complexes and allows us to discuss the functional conversion of TRAPPI to TRAPPII.
Results
Purification of TRAPPII and subunit composition
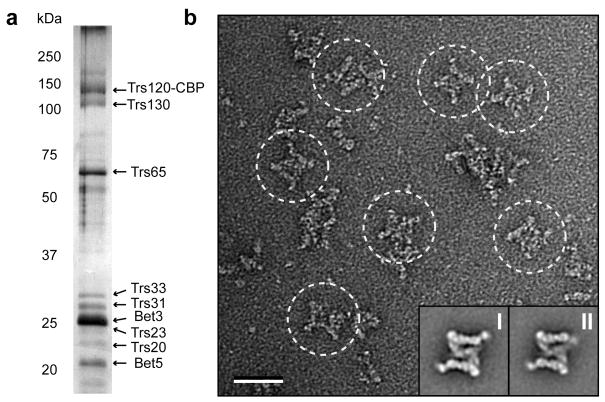
Obtaining sufficient quantities of fully assembled complex is a major impediment in the structural analysis of TRAPPII and other tethering complexes that contain a larger number of subunits. We explored the possibility of isolating native TRAPPII from yeast lysates. Previous reports of TRAPPII purification utilized a yeast strain with epitope-tagged Bet3 (ref. 7). Since Bet3 is a component shared by TRAPPI and TRAPPII, targeting this subunit inevitably results in the purification of a mixture of both TRAPP complexes. We therefore investigated the possibility of isolating TRAPPII by targeting one of the three TRAPPII-specific subunits (Kre11, Trs120, and Trs130). We carried out tandem affinity purification (TAP) from yeast strains, each containing a C-terminal TAP tag on one of the three TRAPPII-specific subunits. SDS-PAGE analysis showed that purifications based on TAP-tagged Trs120 and Trs130 yielded the full TRAPPII complex, whereas the yield of purifications targeting Kre11-TAP was very low, and the subunits were not detectable by silver staining (data not shown). The gel bands from the Trs120-TAP purification (Fig. 1a) were excised and subjected to tandem mass spectrometry. The analysis identified all TRAPPII-specific subunits (Kre11, Trs120, and Trs130) and all shared TRAPPI subunits (Bet3, Bet5, Trs20, Trs23, Trs31, and Trs33) except Gsg1, indicating that Gsg1 is not a stoichiometric component of TRAPPII (Supplementary Table 1). We also did not detect the recently identified Tca17 protein, suggesting that most of the complexes in our preparation are lacking this substoichiometric TRAPPII component (Supplementary Table 1).
Figure 1.
Purification and negative stain EM analysis of yeast TRAPPII. (a) Silver-stained 12.5% SDS-PAGE of TRAPPII purified by the tandem affinity purification (TAP) method. The labeled subunits were identified by mass spectrometry (Supplementary Table 1). Trs120-CBP indicates that the C-terminal calmodulin-binding peptide (CBP) moiety of the TAP tag remains attached after the TEV-protease cleavage step. (b) EM of negatively stained TRAPPII. A raw image of purified TRAPPII (based on TAP-tagged Trs120) shows large, diamond-shaped TRAPPII complexes (circled). The insets show representative class averages of the symmetric (I) and the asymmetric (II) conformer. Scale bar represents 50 nm, and side length of the panels showing the class averages is 59 nm.
While TAP tagging of either Trs120 or Trs130 allowed purification of TRAPPII, the yields were too low for structural studies. We therefore tested different buffer conditions and found that the yield increased by more than five times when we replaced the detergent NP-40 in our buffers by CHAPS. A similar result was obtained in the EM analysis of the Nup84 nucleoporin subcomplex, in which the type of detergent strongly affected the quality and yield of the purified complex23.
TRAPPII is a dimeric complex
With the purification procedure established, we prepared negatively stained specimens of purified TRAPPII and examined its structure by EM. The raw images showed diamond-shaped particles that were much larger than TRAPPI, even accounting for the mass contributed by the three TRAPPII-specific subunits Kre11, Trs120, and Trs130 (Fig. 1b). We manually selected 9,382 particles from 112 images and classified them into 50 classes (Supplementary Fig. 1). The class averages showed that the diamond-shaped TRAPPII, approximately 225 Å × 250 Å in size, has a three-layered architecture, which gives the complex the appearance of a laboratory jack (Fig. 1b, insets). The two outer layers are capped by prominent densities on either end that connect to domains of the middle layer. In many of the averages, the complex showed a clear two-fold symmetry, indicating that TRAPPII is dimeric (Fig. 1b, inset I; Supplementary Fig. 1). This observation supports previous gel filtration studies, which showed that TRAPPII elutes at a peak corresponding to a molecular weight of at least 1 MDa (approximately twice the value of the summed molecular weights of all the individual components)7. While most class averages showed two-fold symmetry, in some classes one half of the complex was different as one of the two capping densities in the outer layer appeared weaker and was at a different position relative to the remainder of the complex (Fig. 1b, inset II). While the physiological relevance of the two TRAPPII conformers remains to be established, they do provide evidence that there is inherent flexibility in the connection of the outer layers to the middle layer.
3D structure of TRAPPII
The well-defined projection averages encouraged us to determine the 3D structure of TRAPPII. The concentrations of our TRAPPII preparations were, however, very low (~0.005 mg ml−1), and we were unable to sufficiently concentrate the TAP-purified samples to prepare vitrified specimens. We therefore prepared TRAPPII by cryo-negative staining and collected pairs of images (50° tilted/untilted) from these specimens (Supplementary Fig. 2). Cryo-negative staining minimizes dehydration-induced sample flattening associated with conventional negative staining and improves the overall quality of the 3D reconstruction24. A total of 8,869 pairs of particles were selected from 104 image pairs, and the particles from the untilted images were classified into 10 classes (Supplementary Fig. 2). The averages showed the same two predominant conformations we observed previously in our negative stain EM analysis (Fig. 2a). We then used the corresponding particles from the tilted images to calculate two separate reconstructions, one for each of the two TRAPPII conformations, using the random conical tilt (RCT) approach25. The particles included in the two density maps represent 22% (conformer I) and 19% (conformer II) of the whole data set respectively. The projection structure of conformer I suggested that it may have two-fold symmetry (Fig. 2a). When we imposed this symmetry on the density map the features became clearer, the resolution improved from 33 Å to 32 Å, and the reprojection from the symmetrized density map matched well with the class average (Supplementary Fig. 2 and 3). By contrast, when we imposed two-fold symmetry on the density map of conformer II, the density mapped showed less features, the resolution dropped from 34 Å to 39 Å, and the reprojection from the symmetrized density map matched the class average less well than the reprojection from the unsymmetrized density map (Supplementary Fig. 2 and 3). Based on these observations, we conclude that although TRAPPII is dimer, only conformation I, but not conformation II, has a symmetric structure. We will therefore base our discussion of the TRAPPII structure on the symmetrized map of conformer I (Fig. 2b) and the unsymmetrized map of conformer II (Fig. 2c).
Figure 2.
3D reconstructions of TRAPPII. (a) Representative class averages of cryo-negatively stained TRAPPII showing the same two conformations observed with negatively stained samples. The side length of the panels is 54 nm. (b) Different views of the 3D reconstruction of TRAPPII corresponding to conformation I in panel a, with an imposed 2-fold symmetry and filtered to a resolution of 32 Å. (c) Different views of the 3D reconstruction of TRAPPII corresponding to conformation II in panel a, with no symmetry imposed and filtered to a resolution of 34 Å. The scale bars shown in b and c represent 5 nm. (d) The main structural features of TRAPPII. Each of the two outer layers consist of a central domain (CD) and two peripheral domains (PD1 and PD2), whereas two copies of the middle domain (MD) form the middle layer.
The 3D reconstruction of conformer I shows that TRAPPII has an apparent thickness of approximately 110 Å (Fig. 2b). This reconstruction resolves distinct features in the three layers (labeled in Fig. 2d). Each of the two outer layers consists of an elongated central domain (CD) that is “capped” by two prominent “rod-like” peripheral domains (PD1 and PD2), which both project into the middle layer. The projection of PD1 interacts intimately with the outer lobe of a middle domain (MD), two of which form the middle layer. The class averages suggest that part of this outer lobe may connect to PD2 of the other half-complex, although the region connecting these two domains is not visible at the contour level at which the map is displayed. The inner lobe of the MD interacts tightly with the inner lobe of the MD of the other half-complex at the two-fold symmetry axis of the full complex. In summary, the MD links the two halves of the complex through PD1 and possibly PD2, thus mediating dimerization of TRAPPII.
The 3D reconstruction of conformer II reveals an asymmetric molecule (Fig. 2c). This asymmetry is caused by an outward rotation of the CD in one half-complex that is coupled to a change in position and a reduction in size of PD2 in the same outer layer. The smaller size of PD2 suggests that loss of the connection to the MD results in PD2 adopting variable orientations, causing the domain to be averaged out and thus to appear smaller. Despite these large conformational changes in one half of TRAPPII, the other half-complex appears largely identical to the structure seen in conformer I.
Subunit organization of TRAPPII
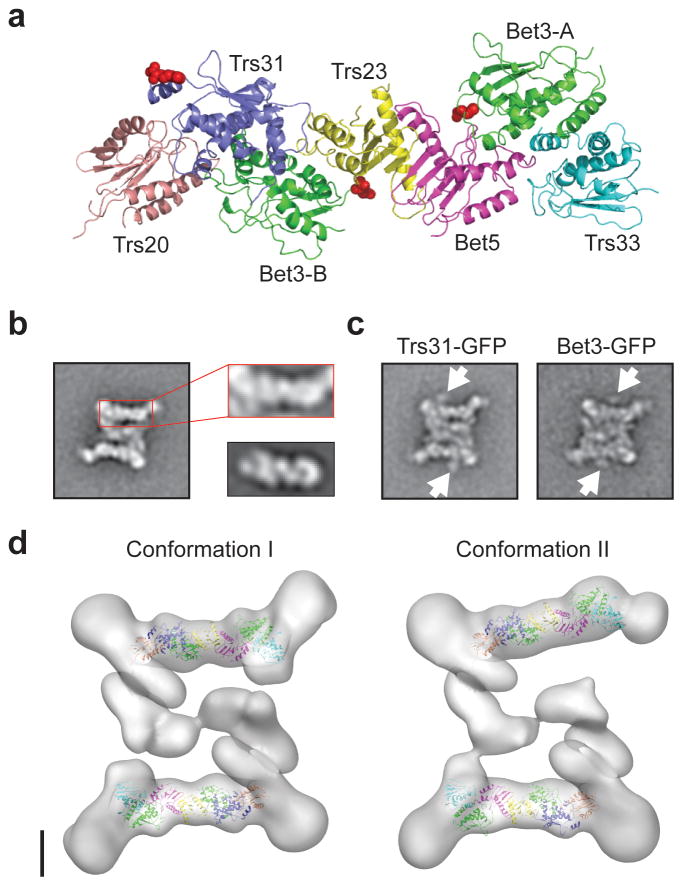
To obtain a better understanding of the intricate TRAPPII structure, we studied the organization of the individual subunits. Closer inspection of the 2D class averages revealed that the projection of the CD resembles the projection of a full seven-protein TRAPPI complex, which was modeled based on the crystal structures of the two mammalian TRAPPI sub-complexes (PDB codes: 2J3T and 2J3W)11 and the crystal structure of the yeast TRAPPI core in complex with Ypt1 (PDB code: 3CUE)12(Fig. 3a,b). To verify that TRAPPI indeed constitutes the CDs of the two outer layers, we generated yeast strains that expressed TRAPPI components, Trs31 or Bet3, fused to a C-terminal GFP tag. Based on the structural model of TRAPPI, C-terminal tagging of these two subunits would not interfere with TRAPPI assembly. We purified labeled Trs31-GFP and Bet3-GFP complexes from these strains based on TAP-tagged Trs120, and examined them by negative stain EM. For each specimen, we selected approximately 3,000 particles and classified them into 100 (Trs31-GFP) or 200 classes (Bet3-GFP) (Supplementary Fig. 4). For both samples, the class averages clearly showed the presence of two globular densities, located in the vicinity of the two outer layers (Fig. 3c). Although each TRAPPI complex contains two Bet3 subunits (Bet3-A and Bet3-B), only one GFP can be seen associated with each TRAPPI complex. The GFP fused to the other Bet3 subunit possibly localizes to a region that is obscured by the TRAPPII density. Based on the distance between the GFPs fused to Trs31 and Bet3, ~6.5 nm, and the locations of these two proteins in the model of TRAPPI (Fig. 3a), Bet3-A is most likely the outward-facing Bet3 subunit. The GFP labeling result thus not only confirms the assignment of TRAPPI to the center of the outer layers, but also suggests that the organization of TRAPPI is conserved in the context of TRAPPII and provides further experimental evidence for the dimeric organization of TRAPPII.
Figure 3.
The TRAPPI subunits localize to the periphery of TRAPPII. (a) Atomic model of TRAPPI generated based on three crystal structures (PDB codes: 2J3T, 2J3W, and 3CUE)11,12. The two copies of Bet3 are labeled Bet3-A and Bet3-B, respectively. The red spheres indicate the C termini of Trs31, Bet3-A, and Bet3-B. (b) The structural features of the TRAPPII CD domain resemble those of TRAPPI. The left panel shows a representative class average of TRAPPII. The side length of the panel is 57 nm. Panels to the right show a larger view of the CD domain of TRAPPII (top) in comparison with the previously published TRAPPI class average11 shown at the same scale (bottom). (c) Representative averages of TRAPPII containing GFP-labeled Trs31 or Bet3. The orange arrows point to the positions of the GFP tags. The side length of the panels is 57 nm. (d) Molecular docking of the TRAPPI model into the EM density maps of the two conformations of TRAPPII. The scale bar represents 5 nm.
Guided by the GFP tagging results, we could unambiguously fit the TRAPPI model into the outer layers of both 3D density maps, further confirming that the structure and organization of TRAPPI is indeed largely preserved within TRAPPII (Fig. 3d). In addition, our docking shows that the TRAPPII-specific components do not interact directly with the flat surfaces of TRAPPI.
With the position of the TRAPPI components determined, we next investigated the localization of the three TRAPPII-specific components. We fused a GFP tag to the C termini of Kre11, Trs120, and Trs130, respectively, and purified TRAPPII from these three strains based on TAP-tagged Trs120 (Kre11-GFP and Trs130-GFP) or Trs130 (Trs120-GFP). Negative stain EM analysis suggested that labeling of Trs120 and Trs130 did not affect the stability of the complex, but C-terminal GFP tagging of Kre11 resulted in very low yields, which prevented further analysis. We manually selected approximately 1,900 Trs130-GFP particles and approximately 5,000 Trs120-GFP particles and classified them into 100 and 200 classes, respectively, but none of the resulting class averages revealed clear densities for the GFPs (Supplementary Fig. 4). A possible explanation is that the GFP tags may be positioned within the particle such that their densities are obscured by those of the three TRAPPII layers. Alternatively, the positions of the GFPs could be too variable, causing them to become blurred out upon averaging. Nevertheless, an appreciable number of class averages of TRAPPII containing GFP-labeled Trs120 showed the complex to be deformed, with an appearance as if the two halves of the complex had been pulled apart (Fig. 4a; Supplementary Fig. 4). This finding may suggest that the C terminus of Trs120 is located near the dimer interface. In these averages, the middle layer was often divided into two parallel densities, clearly defining the dimer interface and demarcating two roughly triangular-shaped half-complexes (Fig. 4a).
Figure 4.
Localization of Trs120 and Trs130. (a) The dimer interface of TRAPPII. Six representative class averages of TRAPII containing GFP-labeled Trs120. The slight deformation and expansion of TRAPPII due to the GFP tag on Trs120 provide a clear view of the dimer interface, which is indicated by a dotted, curved line in the first panel. (b) Antibody labeling of TRAPPII purified from the Trs120 TAP-tagged strain. One representative average and five raw particle images (top) are presented with schematic drawings showing TRAPPII in white and the antibody in grey (bottom). Trs120 localizes to a region around the PD2 domain. (c) Antibody labeling of TRAPPII purified from the Trs130 TAP-tagged strain. One representative average and five raw particle images (top) are presented with schematic drawings showing TRAPPII in white and the antibody in gray (bottom). Trs130 localizes to a region around the PD1 domain. The side length of the panels is 57 nm.
As an alternative approach to GFP tagging, we performed antibody-labeling experiments by incubating purified TRAPPII with an antibody against the C-terminal CBP tag, which remains on the TAP-tagged subunit after the final purification step. While the antibodies were too blurred out in the class averages to precisely identify their positions, the raw particle images collectively showed that Trs120 and Trs130 are located on opposite sides of the TRAPPI complex (Fig. 4b,c). The strong densities capping the outer layers thus appear to represent at least part of the Trs120 and Trs130 subunits. Furthermore, from the docking of TRAPPI into the density map, we can conclude that Trs120 interacts with Trs33 and Bet3-A, and Trs130 with Trs20 and possibly Trs31.
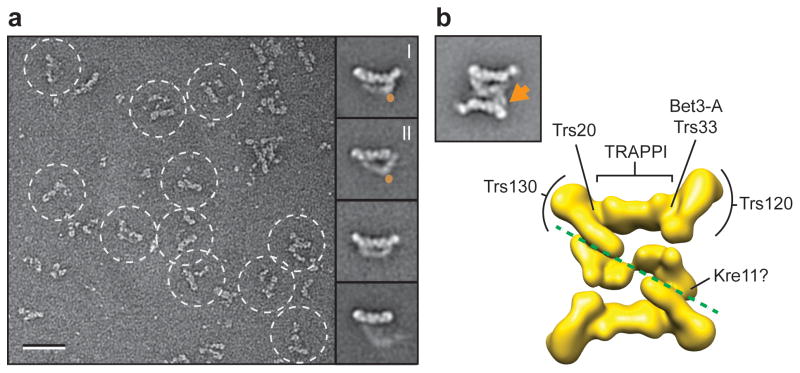
To investigate the localization of Kre11, which could not be tagged, we generated a yeast strain deficient in Kre11. Kre11 is encoded by a non-essential gene in yeast, and deleting the gene does not affect growth in standard culture conditions26. We purified TRAPPII from this strain using TAP-tagged Trs120 and examined the purified complex by negative stain EM. TRAPPII lacking Kre11 was much smaller and appeared to comprise only half of the fully assembled complex (Fig. 5a). We selected a total of 8,627 particles from 115 images and classified them into 100 classes (Supplementary Fig. 5). The individual particle images already showed that the domains usually forming the well-defined middle layer in the fully assembled, “dimeric” TRAPPII adopted a large number of different conformations in the “monomeric” half-complexes. As a result of the structural variability, only a small percentage of the class averages showed well-defined features in this region. These class averages revealed that monomeric TRAPPII could have a rhomboid or triangular shape (Fig. 5a). Interestingly, we could also identify averages that seem to correspond to the two prominent conformations we observed previously with the fully assembled, dimeric TRAPPII (compare Fig. 1a, panels I & II; with Fig. 5a, panels I & II). Although the structural variability in monomeric TRAPPII and the resulting lower quality of the class averages precluded the use of difference mapping to localize Kre11, comparison of the class averages of the fully assembled TRAPPII with those of the monomeric TRAPPII suggests that Kre11 is positioned both at the interface and at the corner of the triangular-shaped dimer (Fig. 5a,b). In summary, our results implicate Kre11 in mediating and/or stabilizing the dimeric assembly of TRAPPII.
Figure 5.
Kre11 mediates dimerization of TRAPPII. (a) EM of negatively stained TRAPPII deficient in Kre11 (Δkre11). A raw image is shown (left) and four representative class averages (right). Labels I and II designate the two dominant conformers. The orange circles designate the potential position of Kre11. The scale bar represents 50 nm, and the side length of the panels showing the class averages is 45 nm. (b) Subunit organization of TRAPP subunits. A representative class average (left) is shown together with the 3D reconstruction in the same orientation (right). The orange arrow marks the possible location of Kre11. The labels indicate the positions of the shared TRAPP subunits Trs20, Bet3-A, and Trs33 and of the TRAPPII-specific subunits Trs120 and Trs130. The green dotted line demarcates the dimer interface.
TRAPPII binds Ypt1
Our docking of the TRAPPI model into the TRAPPII EM density map revealed that one side of each TRAPPI complex is exposed and remains freely accessible for potential binding partners. Comparison of the crystal structure of the yeast TRAPPI core in complex with Ypt1 (Fig. 6a) with the TRAPPI model docked into the TRAPPII EM density map revealed that the accessible side includes the catalytic site of TRAPPI for its Ypt1 GEF activity (Fig. 6b)12. TRAPPI, a well-defined Ypt1 GEF, is unique amongst GEFs in that four distinct subunits have to assemble to form the active site12. To further investigate Ypt binding by TRAPPII, we performed GST pulldown assays with yeast lysates. In repeated experiments, we only observed TRAPPII binding to GST-Ypt1, but not to GST, GST-Ypt31, GST-Ypt32, and GST-Sec4 (Fig. 6c).
Figure 6.
TRAPPII binds Ypt1. (a) Crystal structure of the yeast TRAPPI core in complex with Ypt1 (PDB code: 3CUE)12. (b) The Ypt1 GEF site of TRAPPI is accessible in TRAPPII. Docking of the TRAPPI model into the EM density map of TRAPPII shows that Ypt1 (shown as orange oval) could bind to the same TRAPPI subunits in TRAPPII. (c) GST pulldown assay. The Coomassie blue stained gel (bottom) shows the loading of the GST-fusion proteins. TRAPPII binding was probed by Western blotting using an antibody against the CBP tag (top). GST-Ypt1 binds TRAPPII, but none of the other tested GTPases, GST-Ypt31, GST-Ypt32 and GST-Sec4, pulled down TRAPPII. (d) EM of negatively stained TRAPPII incubated with GST-Ypt1. Raw image (left) showing TRAPPII particles (circled) with an additional density representing the GST tag (orange arrow). Two representative class averages showing that the GST density is associated with the TRAPPI subunits in TRAPPII (right, top panels). Two raw images showing dimeric TRAPPII particles with two labels (right, middle panels) and two monomeric TRAPPII particles with one label (right, bottom panels). Scale bar represents 50 nm, and side length of the panels showing the class averages is 57 nm.
Based on this finding, we attempted to assemble a TRAPPII–Ypt1 complex by incubating GST-Ypt1 with TRAPPII containing TAP-tagged Trs120 for 2 hours prior to the final elution from the calmodulin resin. We prepared negatively stained specimens from the eluted fractions, imaged them, and selected approximately 2,600 particles. About 300 of the particles showed an additional density and were chosen for classification into 10 classes (Supplementary Fig. 6). The averages showed that the additional density, which likely represents the GST fused to Ypt1, localized close to the Ypt1 GEF site of the TRAPPI complex (Fig. 6d). In our raw images, we also found some TRAPPII particles with two additional densities (Fig. 6d) as well as “monomeric” TRAPPII particles with an associated density (Fig. 6d). These observations indicate that the two Ypt1 binding sites in TRAPPII are equally accessible, and that dimerization is not a pre-requisite for the interaction of Ypt1 with TRAPPII. Collectively, our results support recently published work that showed that TRAPPII is a GEF for Ypt1 but not for Ypt31/32 (ref. 12 & 16).
Discussion
Subunit composition and overall architecture of TRAPPII
Our biochemical and mass spectrometric analyses suggest that Gsg1 is unlikely to be a stoichiometric component of yeast TRAPPII. This observation is supported by a recent study showing that Gsg1 nucleates a novel TRAPPIII complex involved in autophagy, and that Gsg1 is not a component of the TRAPPI and TRAPPII complexes6. We were also unable to detect Tca17, suggesting that only a very small percentage of the TRAPPII complexes we purified contained this newly identified substoichiometric component. Even with nine stably associated components, TRAPPII remains the most sophisticated tethering complex in terms of composition. The EM structure of TRAPPII presented here thus represents the largest tethering complex structurally characterized in its fully assembled state.
The overall architecture of TRAPPII is strikingly different from those of the “tower-shaped” Dsl1 tethering complex27 and the “y-shaped” COG1-4 subcomplex (Lees & Yip et al., in press). The divergence in quaternary structures amongst the various tethering complexes is not surprising given the differences in composition and sequences of their constituents. However, at the ultrastructural level, TRAPPII and other tethering complexes all appear to lack a globular central core but instead possess highly extended domains or appendages. It is tempting to speculate that these structural features enable a tethering complex residing at the acceptor compartment to sample a large 3D space to interact with different proteins and protein complexes involved in the tethering process.
The dimeric organization of yeast TRAPPII
The dimerization of TRAPPII is unique amongst the tethering complexes studied to date. A direct consequence of dimerization is the substantial increase in overall dimensions of TRAPPII compared to TRAPPI. According to our EM structure, TRAPPII spans a minimal distance of approximately 225 Å between the two CD domains, and a maximum possible distance of approximately 375 Å, when measured diagonally between the two PD2 domains. Whether adopting this higher ordered structure enables TRAPPII to link remote objects remains to be determined.
Our mutation analysis implicates a role of the TRAPPII-specific subunit Kre11 in mediating TRAPPII dimerization. Precisely how Kre11 performs this function remains to be determined, but the finding that C-terminal tagging (TAP or GFP) of Kre11 destabilizes TRAPPII suggests that an unobstructed C terminus of this protein is critical. The observation that “monomeric” TRAPPII lacking Kre11 is structurally more variable than the wild-type complex indicates that Kre11 may serve to lock the two larger TRAPPII-specific components, Trs120 and Trs130, into more defined conformations required for dimerization.
Despite having a central role in organizing the molecular architecture of TRAPPII, Kre11 is not an essential gene in yeast26, raising the question whether TRAPPII indeed exists as a dimer in vivo. Our EM data on the monomeric and dimeric complexes suggest that apart from Kre11, Trs120 and Trs130 are also involved in constructing the dimer interface. Thus, we cannot rule out the possibility that TRAPPII retains some ability to form dimers in the absence of Kre11 in vivo. It is possible that we were not able to isolate dimeric TRAPPII from the Δkre11 strain, simply because the harsh cell lysis procedure may have disrupted the more labile dimers formed in the absence of Kre11.
Kre11 orthologs are only found in yeast and related fungi28. While TRAPPII in mammals and other organisms may possibly be monomeric, it is also possible that their Trs120 and Trs130 orthologs have evolved the ability to dimerize, without the assistance of Kre11. Different strengths of subunit interactions in tethering complexes from different species are also seen in TRAPPI. Unlike the yeast TRAPPI components, co-expression of the mammalian counterparts did not result in a stable seven-protein complex11. Our structure of TRAPPII shows that Trs120 and Trs130 cap opposite ends of TRAPPI. The mammalian orthologs of Trs120 and Trs130 may thus be required to assemble a stable TRAPP complex. This notion would be in agreement with the observation that mammalian cells likely contain only a single TRAPP complex, the TRAPPII equivalent26,29. An interesting area of future investigation would be to develop strategies to purify mammalian TRAPPII and compare its composition, structure, and subunit organization with that of the yeast complex.
GEF activity and coat interaction of TRAPPII
Our biochemical and EM studies show that TRAPPII preserves and appears to utilize the same catalytic site as TRAPPI, supporting recent data that showed that TRAPPII has GEF activity for Ypt1 (ref. 12 & 16). Ypt1 is a major regulator of trafficking with documented roles in ER-to-Golgi, intra-Golgi, and early endosome-to-late Golgi transport30,31. Future work will undoubtedly focus on further delineating the molecular mechanisms of Ypt1 function, particularly on understanding how and why this small GTPase participates in multiple transport steps, and defining the precise roles of the three TRAPP complexes in modulating and/or coordinating Ypt1 activity in their respective compartments.
A critically important property of tethering complexes is their ability to interact with coat proteins3. Until recently, it was believed that transport vesicles shed all their coat proteins immediately after budding from the donor compartment. It has now been recognized, however, that some coat proteins remain attached to the vesicles and could serve for identification purposes during tethering3. An intriguing question regarding this property of TRAPPII is how the TRAPPII-specific components remodel the Sec23 binding site on TRAPPI, and create a binding site for coatomer, the COPI coat complex. Bet3, the protein that binds Sec23, is present in two copies in TRAPPI (Bet3-A and Bet3-B in Fig. 3a). Our docking of the TRAPPI model into the EM map of TRAPPII shows that the TRAPPII specific subunits Trs120 and Trs130 completely occlude Bet3-B while a large portion of Bet3-A remains accessible (Fig. 6a). While it is tempting to propose that Bet3-A primarily participates in Ypt1 GEF activity and Bet3-B is solely responsible for binding Sec23, we cannot completely exclude the possibility that the structural elements of Bet3-A obstructed by the TRAPPII-specific subunits are also critical for the Sec23 interaction. In terms of interaction with COPI coats, the TRAPPII-specific component Trs130 was shown to interact with the γ subunit of the coatomer16. We suggest that this binding site would be located along the “side” of TRAPPII (Supplementary Fig. 7), as some parts or domains of Trs130 are likely to be accessible in this region. An important area of future research would involve examining the structures of TRAPPI–Sec23 and TRAPPII–coatomer complexes and delineating the precise interfaces and domains involved in these interactions.
Molecular basis for TRAPPII-related diseases
The EM structure of TRAPPII, together with the docking of TRAPPI, provides insights into the molecular basis for two human pathologies resulting from defective TRAPP complex function. Mutations in the Trs20/TRAPPC2 gene are associated SEDT, an X-linked genetic skeletal disorder18,19. Several of these mutations have been shown to reduce stability of Trs20, but how these defects adversely affect TRAPP function remains unknown32,33. Furthermore, it has been shown that Trs20 is dispensable for the GEF activity of TRAPPI12. Our structural data illustrate the pivotal role of Trs20 in bridging the TRAPPI complex to a TRAPPII-specific component, most likely Trs130 (Fig. 5b). The D47Y mutation, which does not affect Trs20 stability, projects into the Trs20-Trs130 interface and could potentially disrupt this critical interaction (Supplementary Fig. 7). Autosomal-recessive intellectual disability and postnatal microcephaly are pathologies recently identified to be associated with truncation mutations of Trs120 (ref. 20–22). Trs120 caps one end of the TRAPPI complex, possibly stabilizing it, and interacts with Trs130. Removal of important domains of Trs120 would undoubtedly abolish TRAPPII assembly and function.
Methods
Yeast strains
We obtained Kre11-TAP, Trs120-TAP, and Trs130-TAP haploid yeast strains from the Yeast TAP-tagged collection (Open Biosystems). Strains with C-terminal GFP-tagged Bet3, Trs31, Kre11, Trs120, and Trs130, and deletion of kre11 were constructed using PCR-based homologous recombination34.
TRAPPII purification
Conventional TAP purification procedure was used35, except that NP-40 in the lysis buffer was replaced by 1% (w/v) CHAPS. The concentration of CHAPS was sequentially lowered with every buffer to reach 0.1% (w/v) at the final elution step. Purified complexes were immediately examined by EM or TCA-precipitated for SDS-PAGE analysis.
Electron microscopy
Negatively stained specimens were prepared as described36. Raw images were recorded at a nominal magnification of 52,000× on a 1K × 1K charge-coupled device (CCD) camera (Gatan) with a Philips CM10 electron microscope operated at 100 kV. Images for digital processing were collected with a Tecnai T12 electron microscope (FEI) operated at 120 kV, on imaging plates at a nominal magnification of 67,000× and a defocus value of −1.5 μm. Imaging plates were read out with a scanner (DITABIS) using a step size of 15 μm, a gain setting of 20,000, and a laser power setting of 30%; 2 × 2 pixels were averaged to yield a pixel size of 4.5 Å at the specimen level37. Cryo-negative staining was performed as described36. Specimens were examined using a CT-3500 cryo-transfer holder (Gatan) and a Tecnai F20 electron microscope (FEI) operated at 200 kV. The same specimen areas were recorded at tilt angles of 50° and 0° on Kodak SO-163 film under low-dose conditions at a nominal magnification of 50,000× and a defocus value of −2 μm. Negatives were developed for 12 min at 20°C using full-strength Kodak D-19 developer. Micrographs were digitized with a Zeiss SCAI scanner using a step size of 7 μm; 3 × 3 pixels were averaged to yield a pixel size of 4.2 Å at the specimen level.
Image processing and 3D reconstruction
For 2D analysis of negatively stained TRAPPII samples, particles were interactively selected using BOXER38. Using SPIDER39, these particles were windowed into 128 × 128-pixel images, rotationally and translationally aligned, and subjected to 10 cycles of multi-reference alignment. Each round of multi-reference alignment was followed by K-means classification specifying 50, 100, or 200 output classes. The references used for the first multi-reference alignment were randomly chosen. For 3D reconstruction, a total of 8,869 particle pairs were selected from both the untilted and 50° tilted images of cryo-negatively stained specimens using WEB39. The selected particles were windowed into 128 × 128-pixel images, and the particles from the untilted specimens were classified into 10 classes. Particles from classes with well-defined features were combined (3 classes for conformation I with a total of 1,993 particles; 2 classes for conformation II with a total of 1,686 particles), and two 3D reconstructions were independently calculated with the particle images from the tilted specimens using the backprojection, backprojection refinement and angular refinement procedures in SPIDER. Two-fold symmetrization in the final angular refinement step improved the resolution and the features of the density map of conformer I but had the opposite effect for that of conformer II (Supplementary Figs. 2 and 3). The symmetrized density map of conformation I was filtered to 32 Å, and the unsymmetrized density map of conformation II was filtered to 34 Å, the resolutions estimated using the Fourier Shell Correlation (FSC) = 0.5 criterion40.
Molecular modeling and docking
A model of the full seven-protein TRAPPI complex was generated by aligning the structures of the two mammalian TRAPPI subcomplexes (PDB codes: 2J3T and 2J3W)11 with the co-crystal structure of Ypt1 bound to the yeast TRAPPI core (PDB code: 3CUE)12. The PDZ domain in the crystal structure of mammalian Trs23 was removed, as it is not present in the yeast ortholog. The resulting model was manually fitted into the TRAPPII EM density map and further refined using the UCSF Chimera package. All molecular graphics images were produced using Pymol (http://www.pymol.org/) and the UCSF Chimera package41.
Antibody labeling
Anti-CBP antibody (Open Biosystems, 0.5 mg ml−1) was added at 1:15 to 1:50 dilutions to purified TRAPPII (Trs120-TAP or Trs130-TAP) and incubated for 15 to 30 minutes at room temperature. The samples were negatively stained and imaged as described above. 909 and 836 labeled particles selected for (Trs120-TAP + anti-CBP) and (Trs130-TAP + anti-CBP), respectively, were subjected to alignment and classification, specifying 100 output classes. In the case of (Trs120-TAP + anti-CBP), 93 particles from the 10 best output classes were combined and subjected to a second classification into 5 classes. In the case of (Trs130-TAP + anti-CBP), 98 particles from the 9 best output classes were combined and subjected to a second classification into 8 classes.
GST pulldown assays
GST-fused Ypt1, Ypt31, Ypt32, and Sec4 were overexpressed in E. coli BL21 (DE3) and purified using immobilized glutathione (Pierce) and gel filtration chromatography. As no GTP was supplemented in the gel filtration buffer (20 mM Tris, pH 8.0, 150 mM NaCl), this procedure results in proteins that are primarily in the GDP-bound state42,43. Yeast cells were harvested at log phase, and resuspended in buffer (50 mM HEPES, pH 7.5, 150 mM NaCl, 1 mM EDTA, 10% (v/v) glycerol, 0.5% (v/v) NP-40, complete protease inhibitor cocktail (Roche), 1 mM PMSF) and lysed by vortexing with glass beads. Equal amounts of lysates were incubated with 15 to 20 μg of GST or the respective GST fusion proteins, and 30 μl of GST mag beads (Genscript) at 4°C for 2 hours. Beads were washed three times with lysis buffer, and resuspended in 1× loading buffer, and analyzed by SDS-PAGE and Western blotting. TRAPPII was detected by using anti-CBP antibodies (Open Biosystems or Genscript).
EM analysis of TRAPPII–GST-Ypt1
GST-Ypt1 (~60 μg) was added to TRAPPII containing Trs130-TAP bound to calmodulin sepharose and incubated at 4°C for 2 hours. Complexes were eluted from the resin and negatively stained specimens were prepared. 2,601 particles were selected, and 379 particles containing a clear label were subjected to alignment and classification, specifying 10 output classes.
Supplementary Material
Acknowledgments
We thank John Nicols for technical assistance with purification and data processing, and Zongli Li for assistance with microscopy and image processing. C.K.Y acknowledges fellowships from the Jane Coffin Childs Memorial Fund and Canadian Institutes of Health Research. T.W. is an Investigator in the Howard Hughes Medical Institute. The molecular electron microscopy facility at Harvard Medical School was established with a generous donation from the Giovanni Armenise Harvard Center for Structural Biology and is maintained with funds from NIH grant PO1 GM62580 (to Stephen C. Harrison). Molecular graphics images were produced using the UCSF Chimera package from the Resource for Biocomputing, Visualization, and Informatics at the University of California, San Francisco (supported by NIH P41 RR-01081).
Footnotes
Accession codes
The 3D maps of TRAPPII have been deposited in the EM Databank (accession codes EMD-5213 and EMD-5214).
Author contributions
C.K.Y. and T.W. conceived the experiments. C.K.Y. performed all experiments. J.B. assisted in generating GFP-labeled yeast strains and data processing. C.K.Y. and T.W. analyzed the data and wrote the manuscript.
References
- 1.Whyte JR, Munro S. Vesicle tethering complexes in membrane traffic. J Cell Sci. 2002;115:2627–2637. doi: 10.1242/jcs.115.13.2627. [DOI] [PubMed] [Google Scholar]
- 2.Sztul E, Lupashin V. Role of vesicle tethering factors in the ER-Golgi membrane traffic. FEBS Lett. 2009;583:3770–3783. doi: 10.1016/j.febslet.2009.10.083. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Cai H, Reinisch K, Ferro-Novick S. Coats, tethers, Rabs, and SNAREs work together to mediate the intracellular destination of a transport vesicle. Dev Cell. 2007;12:671–682. doi: 10.1016/j.devcel.2007.04.005. [DOI] [PubMed] [Google Scholar]
- 4.Kummel D, Heinemann U. Diversity in structure and function of tethering complexes: evidence for different mechanisms in vesicular transport regulation. Curr Protein Pept Sci. 2008;9:197–209. doi: 10.2174/138920308783955252. [DOI] [PubMed] [Google Scholar]
- 5.Sacher M, Kim YG, Lavie A, Oh BH, Segev N. The TRAPP complex: insights into its architecture and function. Traffic. 2008;9:2032–2042. doi: 10.1111/j.1600-0854.2008.00833.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Lynch-Day MA, et al. Trs85 directs a Ypt1 GEF, TRAPPIII, to the phagophore to promote autophagy. Proc Natl Acad Sci U S A. 2010;107:7811–7816. doi: 10.1073/pnas.1000063107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Sacher M, et al. TRAPP I implicated in the specificity of tethering in ER-to-Golgi transport. Mol Cell. 2001;7:433–442. doi: 10.1016/s1097-2765(01)00190-3. [DOI] [PubMed] [Google Scholar]
- 8.Jones S, Newman C, Liu F, Segev N. The TRAPP complex is a nucleotide exchanger for Ypt1 and Ypt31/32. Mol Biol Cell. 2000;11:4403–4411. doi: 10.1091/mbc.11.12.4403. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Wang W, Sacher M, Ferro-Novick S. TRAPP stimulates guanine nucleotide exchange on Ypt1p. J Cell Biol. 2000;151:289–296. doi: 10.1083/jcb.151.2.289. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Cai H, et al. TRAPPI tethers COPII vesicles by binding the coat subunit Sec23. Nature. 2007;445:941–944. doi: 10.1038/nature05527. [DOI] [PubMed] [Google Scholar]
- 11.Kim YG, et al. The architecture of the multisubunit TRAPP I complex suggests a model for vesicle tethering. Cell. 2006;127:817–830. doi: 10.1016/j.cell.2006.09.029. [DOI] [PubMed] [Google Scholar]
- 12.Cai Y, et al. The structural basis for activation of the Rab Ypt1p by the TRAPP membrane-tethering complexes. Cell. 2008;133:1202–1213. doi: 10.1016/j.cell.2008.04.049. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Montpetit B, Conibear E. Identification of the novel TRAPP associated protein Tca17. Traffic. 2009;10:713–723. doi: 10.1111/j.1600-0854.2009.00895.x. [DOI] [PubMed] [Google Scholar]
- 14.Scrivens PJ, et al. TRAPPC2L is a novel, highly conserved TRAPP-interacting protein. Traffic. 2009;10:724–736. doi: 10.1111/j.1600-0854.2009.00906.x. [DOI] [PubMed] [Google Scholar]
- 15.Cai H, Zhang Y, Pypaert M, Walker L, Ferro-Novick S. Mutants in trs120 disrupt traffic from the early endosome to the late Golgi. J Cell Biol. 2005;171:823–833. doi: 10.1083/jcb.200505145. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Yamasaki A, et al. mTrs130 is a component of a mammalian TRAPPII complex, a Rab1 GEF that binds to COPI-coated vesicles. Mol Biol Cell. 2009;20:4205–4215. doi: 10.1091/mbc.E09-05-0387. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Morozova N, et al. TRAPPII subunits are required for the specificity switch of a Ypt-Rab GEF. Nat Cell Biol. 2006;8:1263–1269. doi: 10.1038/ncb1489. [DOI] [PubMed] [Google Scholar]
- 18.Gedeon AK, et al. Identification of the gene (SEDL) causing X-linked spondyloepiphyseal dysplasia tarda. Nat Genet. 1999;22:400–404. doi: 10.1038/11976. [DOI] [PubMed] [Google Scholar]
- 19.Gedeon AK, et al. The molecular basis of X-linked spondyloepiphyseal dysplasia tarda. Am J Hum Genet. 2001;68:1386–1397. doi: 10.1086/320592. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Mir A, et al. Identification of mutations in TRAPPC9, which encodes the NIK-and IKK-beta-binding protein, in nonsyndromic autosomal-recessive mental retardation. Am J Hum Genet. 2009;85:909–915. doi: 10.1016/j.ajhg.2009.11.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Mochida GH, et al. A truncating mutation of TRAPPC9 is associated with autosomal-recessive intellectual disability and postnatal microcephaly. Am J Hum Genet. 2009;85:897–902. doi: 10.1016/j.ajhg.2009.10.027. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Philippe O, et al. Combination of linkage mapping and microarray-expression analysis identifies NF-kappaB signaling defect as a cause of autosomal-recessive mental retardation. Am J Hum Genet. 2009;85:903–908. doi: 10.1016/j.ajhg.2009.11.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Kampmann M, Blobel G. Three-dimensional structure and flexibility of a membrane-coating module of the nuclear pore complex. Nat Struct Mol Biol. 2009;16:782–788. doi: 10.1038/nsmb.1618. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Cheng Y, et al. Single particle reconstructions of the transferrin-transferrin receptor complex obtained with different specimen preparation techniques. J Mol Biol. 2006;355:1048–1065. doi: 10.1016/j.jmb.2005.11.021. [DOI] [PubMed] [Google Scholar]
- 25.Radermacher M. Three-dimensional reconstruction of single particles from random and nonrandom tilt series. J Electron Microsc Tech. 1988;9:359–394. doi: 10.1002/jemt.1060090405. [DOI] [PubMed] [Google Scholar]
- 26.Sacher M, Barrowman J, Schieltz D, Yates JR, 3rd, Ferro-Novick S. Identification and characterization of five new subunits of TRAPP. Eur J Cell Biol. 2000;79:71–80. doi: 10.1078/S0171-9335(04)70009-6. [DOI] [PubMed] [Google Scholar]
- 27.Ren Y, et al. A structure-based mechanism for vesicle capture by the multisubunit tethering complex Dsl1. Cell. 2009;139:1119–1129. doi: 10.1016/j.cell.2009.11.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Cox R, Chen SH, Yoo E, Segev N. Conservation of the TRAPPII-specific subunits of a Ypt/Rab exchanger complex. BMC Evol Biol. 2007;7:12. doi: 10.1186/1471-2148-7-12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Kummel D, Oeckinghaus A, Wang C, Krappmann D, Heinemann U. Distinct isocomplexes of the TRAPP trafficking factor coexist inside human cells. FEBS Lett. 2008;582:3729–3733. doi: 10.1016/j.febslet.2008.09.056. [DOI] [PubMed] [Google Scholar]
- 30.Jedd G, Richardson C, Litt R, Segev N. The Ypt1 GTPase is essential for the first two steps of the yeast secretory pathway. J Cell Biol. 1995;131:583–590. doi: 10.1083/jcb.131.3.583. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Sclafani A, et al. Establishing a role for the GTPase Ypt1p at the late Golgi. Traffic. 2010;11:520–532. doi: 10.1111/j.1600-0854.2010.01031.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Jang SB, et al. Crystal structure of SEDL and its implications for a genetic disease spondyloepiphyseal dysplasia tarda. J Biol Chem. 2002;277:49863–49869. doi: 10.1074/jbc.M207436200. [DOI] [PubMed] [Google Scholar]
- 33.Choi MY, et al. Biochemical consequences of sedlin mutations that cause spondyloepiphyseal dysplasia tarda. Biochem J. 2009;423:233–242. doi: 10.1042/BJ20090541. [DOI] [PubMed] [Google Scholar]
- 34.Longtine MS, et al. Additional modules for versatile and economical PCR-based gene deletion and modification in Saccharomyces cerevisiae. Yeast. 1998;14:953–961. doi: 10.1002/(SICI)1097-0061(199807)14:10<953::AID-YEA293>3.0.CO;2-U. [DOI] [PubMed] [Google Scholar]
- 35.Rigaut G, et al. A generic protein purification method for protein complex characterization and proteome exploration. Nat Biotechnol. 1999;17:1030–1032. doi: 10.1038/13732. [DOI] [PubMed] [Google Scholar]
- 36.Ohi M, Li Y, Cheng Y, Walz T. Negative Staining and Image Classification – Powerful Tools in Modern Electron Microscopy. Biol Proced Online. 2004;6:23–34. doi: 10.1251/bpo70. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Li Z, Hite RK, Cheng Y, Walz T. Evaluation of imaging plates as recording medium for images of negatively stained single particles and electron diffraction patterns of two-dimensional crystals. J Electron Microsc (Tokyo) 2010;59:53–63. doi: 10.1093/jmicro/dfp036. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Ludtke SJ, Baldwin PR, Chiu W. EMAN: semiautomated software for high-resolution single-particle reconstructions. J Struct Biol. 1999;128:82–97. doi: 10.1006/jsbi.1999.4174. [DOI] [PubMed] [Google Scholar]
- 39.Frank J, et al. SPIDER and WEB: processing and visualization of images in 3D electron microscopy and related fields. J Struct Biol. 1996;116:190–199. doi: 10.1006/jsbi.1996.0030. [DOI] [PubMed] [Google Scholar]
- 40.Bottcher B, Wynne SA, Crowther RA. Determination of the fold of the core protein of hepatitis B virus by electron cryomicroscopy. Nature. 1997;386:88–91. doi: 10.1038/386088a0. [DOI] [PubMed] [Google Scholar]
- 41.Pettersen EF, et al. UCSF Chimera – a visualization system for exploratory research and analysis. J Comput Chem. 2004;25:1605–1612. doi: 10.1002/jcc.20084. [DOI] [PubMed] [Google Scholar]
- 42.Christoforidis S, Zerial M. Purification and identification of novel Rab effectors using affinity chromatography. Methods. 2000;20:403–410. doi: 10.1006/meth.2000.0953. [DOI] [PubMed] [Google Scholar]
- 43.Murata T, et al. The Legionella pneumophila effector protein DrrA is a Rab1 guanine nucleotide-exchange factor. Nat Cell Biol. 2006;8:971–977. doi: 10.1038/ncb1463. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.