Abstract
The death inducing signaling complex (DISC) formed by the death receptor Fas, the adapter protein FADD and caspase-8 mediates the extrinsic apoptotic program. Mutations in Fas that disrupt the DISC cause autoimmune lymphoproliferative syndrome (ALPS). Here we show that the Fas–FADD death domain (DD) complex forms an asymmetric oligomeric structure composed of 5–7 Fas DD and 5 FADD DD, whose interfaces harbor ALPS-associated mutations. Structure-based mutations disrupt the Fas–FADD interaction in vitro and in living cells; the severity of a mutation correlates with the number of occurrence of a particular interaction in the structure. The highly oligomeric structure explains the requirement for hexameric or membrane-bound FasL in Fas signaling. It also predicts strong dominant negative effects of Fas mutations, which are confirmed by signaling assays. The structure optimally positions the FADD death effector domain (DED) to interact with the caspase-8 DED for caspase recruitment and higher order aggregation.
Death domains share a common six α-helical bundle structure 1,2 and are versatile modules of protein–protein interactions that mediate the assembly of numerous caspase activating and NF-κB activating complexes in apoptotic and inflammatory signaling including the DISC 3–6. Our current understanding of the Fas DD–FADD DD interaction is both conflicting and confusing. The structures of Fas DD and FADD DD have long been elucidated by the NMR method, with the Fas DD structure determined under an acidic condition that disrupts its capacity to oligomerize and to interact with FADD DD 7–9. Extensive mutagenesis has been performed, but the disruptive mutations map to expanded surfaces with no well-defined patches 7,9–11. The same is true for locations of disease-associated mutations in Fas DD 12–15.
Furthermore, these ALPS-associated disease-causing mutations in Fas DD are not localized at the interfaces with FADD DD in a structure of the complex that was crystallized in 0.95M citric acid and 1.9M ammonium sulfate at pH 4.0 16. Here we present a different structure of the complex that was crystallized in low salt and more neutral pH, which agrees well with electron microscopy, mass spectrometry and multi-angle light scattering studies in shape, size and stoichiometry of the complex in solution. The structure reveals the molecular basis of ALPS mutations, was further validated by structure-based mutagenesis both in vitro and in cells, and provides an elegant framework for optimal interaction with Fas ligand and for caspase recruitment and activation.
RESULTS
Similarity of a 5–7 Fas DD and 5 FADD DD complex to the PIDD–RAIDD complex
To resolve the conflicting data and to elucidate the molecular basis of DD interactions in the DISC, we reconstituted the human Fas (hFas) DD–human FADD (hFADD) DD complex (Supplementary Fig. 1a) and the mouse Fas (mFas) DD–hFADD DD complex and utilized electron microscopy (EM) to visualize the structures of the complexes. EM of negatively stained Fas DD–FADD DD complexes revealed monodisperse and homogeneous particle populations (Supplementary Fig. 1b). Classification of 10,397 particles of the hFas–hFADD complex and 15,271 particles of the mFas–hFADD complex into 100 groups produced class averages that revealed molecules of similar size, about 10 nm in diameter, but with varying structural features (Supplementary Fig. 1c and 1d), indicating different orientations of the complex on the carbon support film. The EM averages of the hFas–hFADD and the mFas–hFADD complexes are visibly indistinguishable.
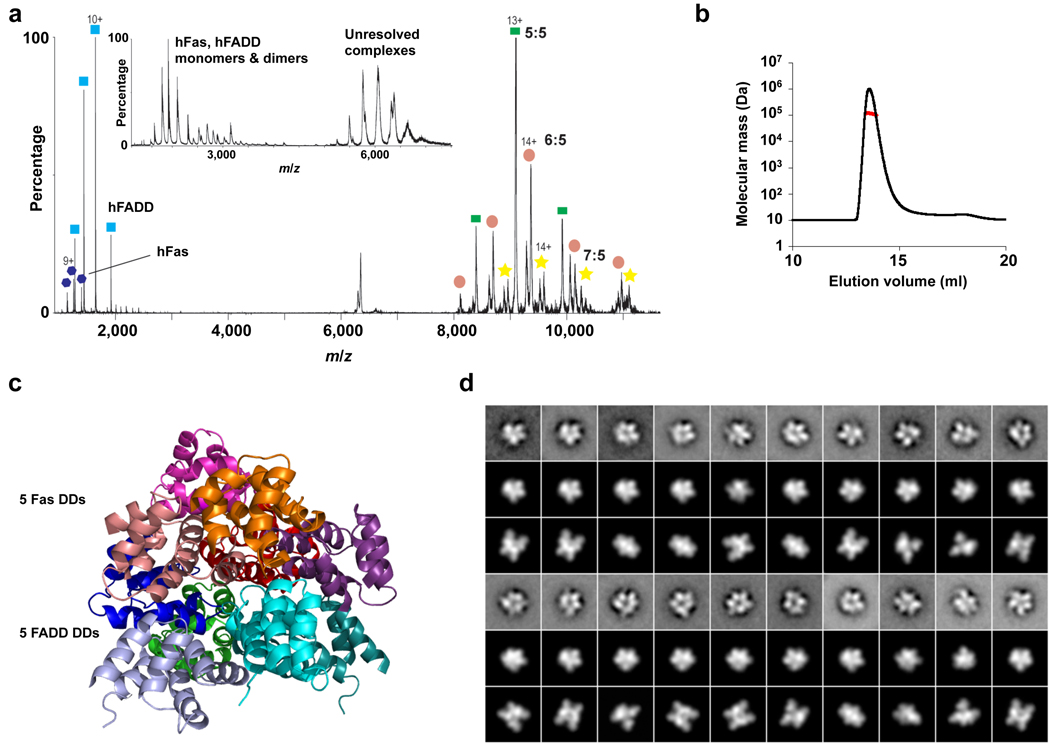
Surprisingly, both the size and the appearance of Fas DD–FADD DD EM projections looked similar to the EM averages of the layered structure of the oligomeric PIDD DD–RAIDD DD complex 17 (Supplementary Fig. 1e). In this complex, a layer of 5 RAIDD molecules is sandwiched between a layer of 5 PIDD molecules on one side and two additional, loosely associated RAIDD molecules on the other side 17. To provide more confidence in this similarity assessment, we used nanoflow electrospray ionization and tandem mass spectrometry to characterize the stoichiometry of the complex. The data showed that the hFas–hFADD complex consists predominantly of a 5:5 species with minor populations of 6:5 and 7:5 species, whereas the mFas–hFADD complex consists predominantly of 5:5 and 6:5 species with a minor population of the 7:5 complex (Fig. 1a, Supplementary Fig. 2a and 2b). We then characterized the size of the complexes quantitatively using multi-angle light scattering (MALS). Among the multiple measurements on multiple samples, a range of molecular masses was obtained, which is consistent with a 5 Fas–5 FADD core complex and loosely bound additional subunits (Fig. 1b, Supplementary Table 1). Therefore, both the mass spectrometry and the MALS measurements support the similarity with the PIDD–RAIDD complex 17. While this similarity was unexpected and suggested that the structure of the Fas DD–FADD DD complex differs from the reported symmetrical tetrameric structure of the complex at high salt and pH 4.0 16, it would agree with the minimal stoichiometry of the extracellular interaction between Fas and its ligand FasL (see below).
Fig. 1.
Biochemical and structural characterization of the Fas DD–FADD DD complexes. (a) Tandem mass spectrum of the hFas–hFADD complex showing the dissociated monomers and the ‘stripped’ complexes at high gas phase collisional activation. The peak centered at 5913 m/z was isolated and collision energy (50–100 V) was applied. Well-resolved peaks at high m/z correspond to the stripped complexes for the 5:5, 6:5 and 7:5 complexes. Inset, mass spectrum of the hFas–hFADD complex showing monomers and dimers of hFas and hFADD at lower m/z and the unresolved charge series at higher m/z. (b) A representative multi-angle light scattering (MALS) measurement of the hFas–hFADD complex. More MALS data are summarized in Supplementary Table 1. (c) Crystal structure of the core 5:5 mFas–hFADD complex. The Fas molecules are shown in warm colors and the FADD DD molecules are shown in cold colors. (d) Class averages of the mFas–hFadd complex (1st and 4th lines) with best matching projections from the current mFas–hFADD crystal structure (2nd and 5th lines) and from a previously published structure of the Fas–FADD complex (3rd and 6th lines) 16.
Crystal structure of the core Fas–FADD DD complex at 6.8 Å resolution
We were able to crystallize the mFas–hFADD complex and obtained its diffraction data at 6.8 Å resolution. We built a layer of 5 Fas DD and a layer of 5 FADD DD using the known structures of hFas and hFADD 7,16, respectively, based on the PIDD–RAIDD complex structure 17. We then solved the structure of the complex by molecular replacement using the Fas DD layer and the FADD DD layer as models (Table 1, Supplementary Table 2). The availability of the individual Fas DD and FADD DD structures effectively provides atomic details to the structure of the complex. The solution revealed a two-layered structure with an upper layer of five Fas DD molecules and a lower layer of five FADD DD molecules (Fig. 1c). This assignment coincides with the greater similarity between Fas and RAIDD, and between FADD and PIDD (Supplementary Fig. 3a). The additional Fas molecules in the complex, which are equivalent to the additional RAIDD molecules in the PIDD–RAIDD complex, are not present in the mFas–hFADD crystal structure due to steric hindrance in crystal packing. The low resolution of the crystals may be reflective of the heterogeneity of the complex. Calculated re-projections of the crystal structure agree well with the experimental class averages (Fig. 1d). Differences in structural details are likely due to staining artifacts.
Table 1.
Data collection and refinement statistics
| mFas DD–hFADD DD complex | |
|---|---|
| Data collection | |
| Space group | C2221 |
| Cell dimensions | |
| a, b, c (Å) | 135.2, 144.5, 131.6 |
| Resolution (Å) | 50.0 − 6.8 (7.0) |
| RSYM (%) | 7.0 (46.2) |
| I / σI | 28.0 (1.3) |
| Completeness (%) | 93.0 (69.0) |
| Redundancy | 4.9 (2.7) |
| Refinement | |
| Resolution (Å) | 20 − 6.8 |
| No. reflections | 1,941 |
| Rwork / Rfree (%) | 35.4 / 34.9 |
| No. atoms | |
| Protein | 7,270 |
| B-factors (Å2) | 451.6 |
| R.m.s. deviations | |
| Bond lengths (Å) | 0.014 |
| Bond angles (°) | 1.7 |
Values in parenthesis are for the highest-resolution shell.
Structure-based mutations have semi-quantitative disruptive effects
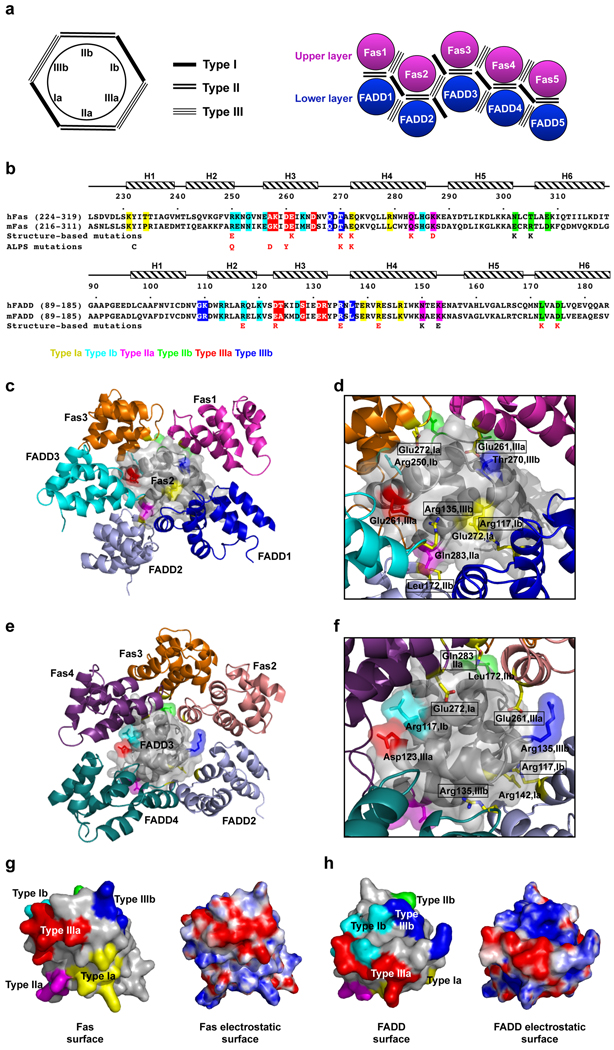
The Fas DD–FADD DD complexes are assembled via an elegant polymerization mechanism involving 3 types of asymmetric interactions at 6 unique interfaces that mediate Fas–FADD, Fas–Fas and FADD–FADD interactions (Fig. 2a, 2b). The type Ia surface composed of residues at the H1 and H4 helices interacts with the type Ib surface composed of residues at H2 and H3. The type IIa surface formed by residues at the H4 helix and the H4–H5 loop interacts with the type IIb surface at the H5–H6 loop and H6 helix. The type IIIa surface from residues at H3 interacts with the type IIIb surface formed by residues at the H1–H2 and the H3–H4 loops. The individual layers are assembled by 5 successive type I and type III interactions. The type II interactions only mediate the interactions between the layers. It may be predicted that type I and type III interfaces of both Fas DD and FADD DD are critical for complex formation. For the type II interaction, only the type IIa surface of the Fas DD and the type IIb surface of the FADD DD should be important.
Fig. 2.
Interactions in the Fas DD–FADD DD complex. (a) Schematic planar diagram showing the construction of the complex. The locations of the three types of contacts are shown. (b) Sequence alignment of human and mouse Fas DD and FADD DD. Major interfacial residues are colored in yellow for type Ia, cyan for type Ib, magenta for type IIa, green for type IIb, red for type IIIa and blue for type IIIb. Single letter codes below the sequences indicate mutations tested with red indicating defective and black indicating non-defective. Locations of ALPS mutations are also shown. (c) Arrangement of molecules around Fas2, viewing into the schematic diagram in (a) from behind the page. (d) Zoom-in of (c) showing residues at the three types of interactions in Fas2 (labeled without a text box) and the surrounding molecules (labeled with a text box). Residues in Fas2 are colored in yellow for type Ia, cyan for type Ib, magenta for type IIa, green for type IIb (residue name not labeled), red for type IIIa and blue for type IIIb. Side chains of residues in the surrounding molecules are shown as stick models colored by atom types. (e) Arrangement of molecules around FADD3, viewing into the schematic diagram in (a) from behind the page. (f) Zoom-in of (e) showing residues at the three types of interactions in FADD3 (labeled without a text box) and the surrounding molecules (labeled with a text box). Residues in FADD3 are colored in yellow for type Ia, cyan for type Ib, magenta for type IIa (residue name not labeled), green for type IIb, red for type IIIa and blue for type IIIb. Side chains of residues in the surrounding molecules are shown as stick models colored by atom types. (g) Side by side surface representations of Fas DD showing the locations of the three types of interactions (left) and the surface charge features (right). (h) Side by side surface representations of FADD DD showing the locations of the three types of interactions (left) and the surface charge features (right).
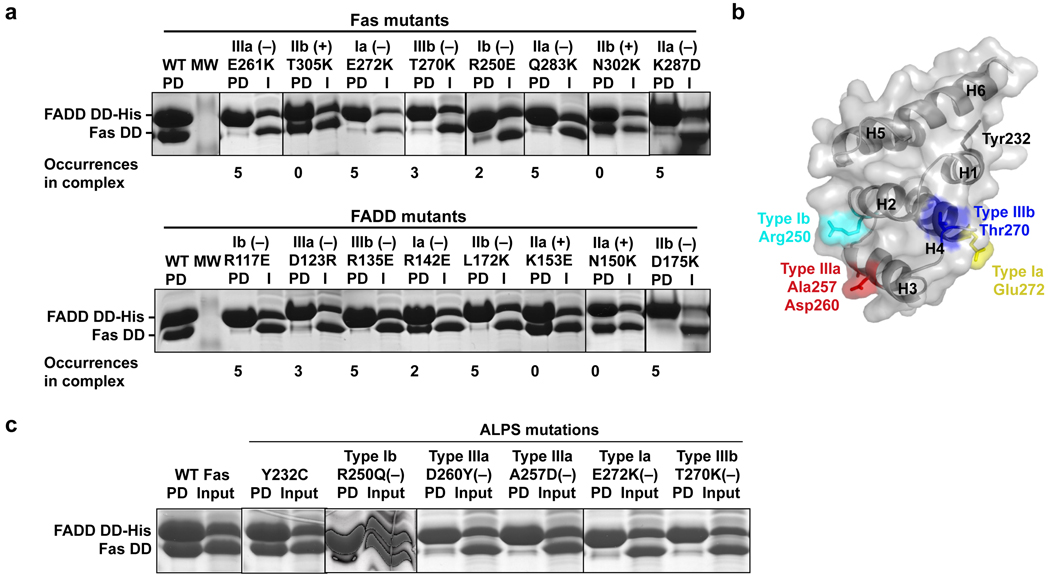
To rigorously confirm the structure, we took an unbiased approach to identify key residues at each of the interfaces in an equivalent hFas DD–hFADD DD complex (Fig. 2b–2f). Residues that bury the most surface area were selected and mutated to charged residues that most likely would disrupt the interaction. One residue each was chosen for each of the type I and type III interfaces in Fas DD and FADD DD. Two residues each were chosen for the type II interface to validate the relative layer positions of Fas DD and FADD DD. These mutations on Fas DD were E272K (type Ia), R250E (type Ib), Q283K (type IIa), K287D (type IIa), T305K (type IIb), N302K (type IIb), E261K (type IIIa) and T270K (type IIIb). The residue numbers are based on the human Fas precursor sequence and 16 should be subtracted for the mature sequence. The mutations on human FADD DD were R142E (type Ia), R117E (type Ib), K153E (type IIa), N150K (type IIa), L172K (type IIb), D175K (type IIb), D123R (type IIIa) and R135E (type IIIb). More than half of the selected residues are either positively or negatively charged, consistent with the salt-dependence of complex formation (Supplementary Fig. 3b) and suggesting that complex formation is driven considerably by electrostatic interactions (Fig. 2g, 2h).
Strikingly, all structure-based mutations at the predicted type I and type III interfaces caused severe disruption of the Fas DD–FADD DD interaction (Fig. 2b, 3a). The type IIa mutations of the Fas DD, but not the type IIb mutations, disrupted the interaction. Conversely, the type IIb mutations of the FADD DD, but not the type IIa mutations, disrupted the interaction. Consistent with their surface locations, none of the disruptive mutations affected the structural integrity of the DDs as shown by circular dichroism (CD) experiments (Supplementary Fig. 3c). The structure was further validated by mutations on conserved surface residues that are not at the observed interface, which retained the Fas–FADD interactions (Supplementary Fig. 3d). Remarkably, there seems to be a semi-quantitative correlation between the severity of the mutational phenotype and the extent of involvement of the interface in complex assembly. For example, the type Ib interface of Fas and the type Ia interface of FADD are only used twice in the complex in comparison with three to five times for the other types of interactions (Fig. 2a), rationalizing the somewhat weaker effects of the Fas R250E type Ib mutation and the FADD R142E type Ia mutation (Fig. 3a).
Fig. 3.
Structure-based mutagenesis and analysis of ALPS mutations. (a) Analysis of structure-based mutations of Fas DD and FADD DD in the in vitro His-tag pulldown assay. (−) indicates a defective interaction. PD: pulldown; I: input. (b) Mapping of ALPS-associated mutation residues onto the surface of Fas DD. (c) Pulldown of ALPS-associated Fas DD mutants by His-tagged FADD DD. (−) indicates a defective interaction. PD: pulldown.
ALPS mutations map to subunit interfaces and disrupt Fas–FADD interaction
Mutations in Fas are the most frequent causes of ALPS and of these, about 70% are mutations in the intracellular DD 12–15. To elucidate whether our Fas DD–FADD DD structure reveals the structural mechanisms of ALPS, we analyzed naturally occurring Fas mutations associated with ALPS (http://research.nhgri.nih.gov/ALPS/fas_tnfrsf6_exon9_mut.shtml). Among the 20 single site ALPS mutations in Fas DD, we selected those that do not involve Gly or Pro and are exposed on the surface of the Fas DD (Supplementary Table 3). These mutations most likely do not influence the structural integrity of the Fas DD. If there are multiple mutations on the same residue, one mutation was chosen for simplicity of the analysis. These criteria selected six mutations, Y232C, R250Q, A257D, D260Y, T270K, and E272K (Fig. 3b). Unexpectedly, three of the residues involved, Arg250, Thr270 and Glu272, are at the centers of type Ib, IIIb and Ia interfaces, respectively. R250E (a mutation related to R250Q), T270K and E272K have already been tested in structure-based mutagenesis and shown to be defective in interacting with the FADD DD. Residues Ala257 and Asp260 reside near the center of the type IIIa interface. In contrast, Tyr232 is exposed near the beginning of helix H1 (Fig. 3b).
Pulldown of wild-type and mutant Fas DD with His-tagged FADD DD showed that ALPS mutants, R250Q, A257D, and D260Y, as well as T270K and E272K, were severely defective in interaction with FADD DD (Fig. 3c). Lack of appreciable structural perturbation had been confirmed by earlier NMR studies for some of these mutations including R250Q and A257D, as well as D260V, a similar mutation to D260Y 11. The structural integrity of T270K and E272K was validated by CD measurements (Supplementary Fig. 3c). In contrast, the Y232C mutant interacted normally with FADD DD. In the family from which it was isolated, the Y232C Fas mutation inhibited apoptosis and caused ALPS only when co-expressed with an extracellular R137W mutation on the other allele, likely by reducing Fas surface expression 13.
The Fas DD–FADD DD structure we present here explains that a majority of the Fas DD ALPS mutations exert their disease phenotypes by failure to form a complex with FADD DD. In contrast, none of the five ALPS mutations, as well as the structurally identified disruptive mutations, are located at the interface in the recently reported Fas DD–FADD DD crystal structure 16 (Supplementary Fig. 4a, 4b, 4c). Instead they map onto the exposed surface of the symmetrical tetrameric complex. In addition, the experimental EM projections of the Fas DD–FADD DD complex are dissimilar to the calculated projections of the tetrameric complex (Fig. 1d, Supplementary Fig. 4d). Because the reported Fas DD–FADD DD complex was crystallized in high salt and low pH 16, factors known to disrupt Fas DD interaction and function 7 (Supplementary Fig. 3b), it is likely that these conditions dramatically altered Fas conformation (Supplementary Fig. 4e), caused a remodeling of the complex, and resulted in the non-physiological oligomer observed in the crystal 16.
In addition to Fas mutations that are associated with ALPS, there are extensive existing mutagenesis data on FADD DD 9,10. Classifying the residues into the three types of interactions allowed us to explain the observed mutational effects of these FADD DD mutants on interactions with Fas DD (Supplementary Table 4). As expected, residues at these interfaces, especially those that are completely buried at the given interface, caused disruption when mutated.
Structure-based mutations disrupt Fas–FADD interaction in living cells
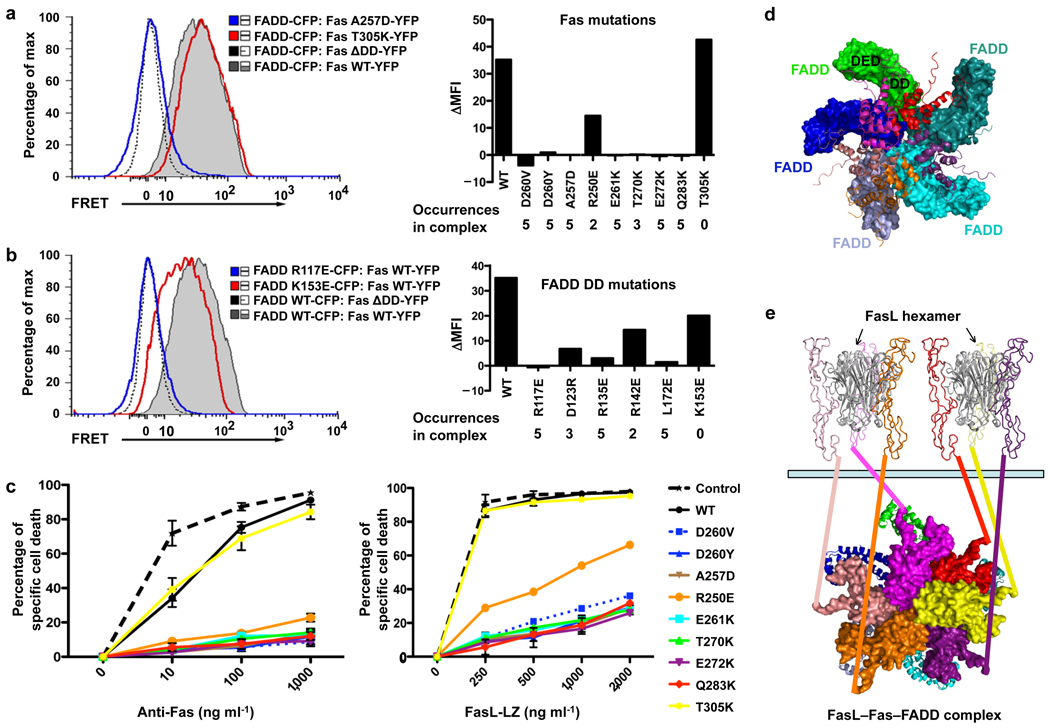
To determine the functional effects of Fas and FADD mutations on signaling complex formation in intact living cells, we performed fluorescence energy transfer (FRET) experiments on full-length Fas and the FADD DD (Fig. 4a, 4b). Full-length Fas and the FADD DD were fused to YFP and CFP, respectively, and co-expressed in 293T cells. FRET using GFP variants is a sensitive measure of protein-protein interactions at distances less than 10 nm 18. Cells co-expressing full-length Fas and the FADD DD produced large FRET signals, indicating complex formation, while cells expressing a Fas mutant lacking the death domain (Fas ΔDD) had reduced FRET signals, indicating that FRET was dependent on DD–DD interactions. Strikingly, all mutant Fas and FADD constructs that were defective in the Fas DD–FADD DD interaction in vitro also had reduced FRET signals compared to the wild-type proteins, whereas the Fas and the FADD mutants that did not affect the Fas DD–FADD DD complex formation in vitro interacted normally with the complementary DD by FRET. The semi-quantitative correlation between the occurrence of a particular interaction in the Fas DD–FADD DD complex and its mutational effect appeared to be more prominent for the interaction in living cells than in vitro (Fig. 4a, 4b).
Fig. 4.
Interactions in living cells and functional effects of Fas DD and FADD DD mutations. (a) 293T cells were transfected with CFP-tagged FADD DD and YFP-tagged wild-type or mutant full-length Fas and analyzed for FRET by flow cytometry. Bi-exponential plots of the FRET signals are shown for cells transfected with the indicated constructs. A plasmid encoding a truncated Fas protein without a death domain (Fas ΔDD) was used as a negative control. Change in geometric mean fluorescence intensity (ΔMFI) was calculated by subtracting the MFI of non-interacting proteins (Fas ΔDD with WT FADD DD) from the MFI in the FRET channel of the indicated interacting pair. At right the mean fluorescence intensity relative to FRET signal from the Fas ΔDD–FADD interaction (ΔMFI) is shown for the indicated Fas DD mutants interacting with wild-type FADD. The occurrences of the interactions in the complex are shown. (b) FRET analysis of 293T cells transfected with either wild-type or mutant FADD DD interacting with full-length Fas or Fas ΔDD. At right the ΔMFI is shown for the indicated FADD DD mutants interacting with wild-type Fas. The occurrences of the interactions in the complex are shown. Results shown in (a) and (b) are representative of three independent experiments. (c) Effect of Fas DD mutants on Fas-induced cell death in transfected Jurkat E6.1 cells. Cells were transfected with the indicated constructs and apoptosis was induced with anti-Fas or FasL-LZ. Cell death was assayed by flow cytometry. Results are the average +/− s.e.m. of specific cell death in three independent experiments. (d) The Fas DD–full-length FADD complex model constructed by superimposing FADD DD with the structure of full-length FADD 20. Fas DDs are shown in ribbon diagrams while FADD molecules are shown in surface representations. The location of the FADD DED for caspase-8 recruitment is shown for one of the FADD molecules. (e) A proposed model for post-receptor DISC formation that utilizes FasL hexamerization. All structures are shown in ribbon diagrams except for the intracellular death domains of Fas, which are shown in surface representations. The two FasL trimers are shown in gray. The six Fas extracellular domains are shown in the same color as their intracellular death domains. A 6th Fas intracellular death domain (yellow) was added to the crystal structure. The extracellular and the intracellular domains are connected via straight lines of the same colors. Full-length FADD molecules are shown in ribbon diagrams. The light green bar represents the cellular membrane.
Dominant negative effects of structure-based and ALPS Fas mutations
Almost all Fas DD mutations in ALPS are dominant and from a heterozygous background, which is consistent with the oligomeric nature and the high cooperativity of assembly in the Fas DD–FADD DD complex. ALPS-associated Fas mutations that cannot bind FADD interfere with Fas-induced apoptosis by dominantly disrupting the wild-type Fas from assembling the DISC 11,14,19. Assuming the complete loss of function of a Fas mutant and a 1:1 mix of wild-type: mutant Fas in a heterozygous patient, the amount of the wild-type signaling complex would be <1/25, which is ~3%, explaining the strong dominant negative phenotype of ALPS-associated Fas DD mutations.
To determine whether structure-based Fas mutants engineered to disrupt the Fas–FADD complex also act dominant negatively, we transfected plasmids encoding these mutants fused to YFP into the Fas-sensitive T cell line Jurkat. As previously described 11, the ALPS-associated mutants A257D, D260Y and D260V almost completely abrogated apoptosis induced by either crosslinked anti-Fas antibody or FasL oligomerized through a leucine zipper tail (FasL-LZ) (Fig. 4c). The structure-based Fas mutants Q283K, E272K, T270K, and E261K were as potent dominant negative mutants as the ALPS-associated Fas mutants. The R250E mutant was somewhat weaker, but still showed more than 50% inhibition at most doses of anti-Fas or FasL-LZ (Fig. 4c). Residue Arg250 of Fas is at the type Ib interface that is less involved in complex formation than other types of interactions (Fig. 2a). The T305K IIb interface mutant that did not interfere with Fas–FADD complex formation had no inhibitory effect on Fas-induced apoptosis. Collectively, these data showed that the Fas DD–FADD DD structure correctly predicted the effects of these mutants in interfering with wild-type Fas signaling complexes.
DISCUSSION
Model of the DISC and requirement for dimers of FasL trimers
Availability of the full-length FADD structure comprised of both the DD and the death effector domain (DED) 20 allowed us to generate a model of the Fas DD in complex with full-length FADD (Fig. 4d). Importantly, the DED domain of FADD does not have any steric clashes with the Fas DD–FADD DD structure and points towards the outside of the complex, poised for interaction with the DED domain of caspase-8.
It is well established that the membrane-bound form of FasL and cross-linked anti-Fas antibodies, but not the proteolytically processed, trimeric soluble form of FasL, trigger apoptosis 21–23. When soluble trimeric FasL is dimerized into an engineered hexameric molecule, it is highly competent to signal apoptosis, suggesting that the minimal signaling competent form of FasL is hexameric 24. Interestingly, a dimer of FasL trimers would bring 6 Fas intracellular DDs into proximity, which is ideal in inducing the formation of the oligomeric complex comprised of 5–7 Fas molecules (Fig. 4e). Because the DED domains of both FADD and caspase-8 are also capable of self-association 25,26, they can mediate further oligomerization of the DISC into microscopically visible clusters 27 to facilitate optimal caspase-8 clustering and activation. Cell death should have an intricately controlled threshold with sensitivity to ligand stimulation and protection from accidental firing. The highly oligomeric nature of the Fas DD–FADD DD complex in the intracellular region and its match in stoichiometry with the Fas–FasL interaction in the extracellular region ensures a highly cooperative and regulated mechanism to control this irreversible cell fate decision.
General DD assembly mechanisms
The current Fas DD–FADD DD complex structure and its similarity to that of the PIDD DD–RAIDD DD complex 17 provide another example of conserved interactions in the DD superfamily which also include the caspase recruitment domain (CARD), the death effector domain (DED) and the Pyrin domain (PYD) 1. The three types of interactions in the Fas DD–FADD DD and the PIDD DD–RAIDD DD complexes have been observed in the 1:1 Pelle DD–Tube DD complex 28, the 1:1 Apaf-1 CARD–caspase-9 CARD complex 29, and the 6:4:4 MyD88–IRAK4–IRAK2 complex 30, but not in the previously published Fas DD–FADD DD complex 16. These interactions form the basis of a polymerization mechanism, which possesses intrinsic helical symmetry 30, in DD assembly. The different stoichiometries in these complexes despite the conservation of interactions establish DD superfamily members as versatile interactors and oligomerizers in signal transduction.
METHODS
Methods and any associated references are available in the online version of the paper at http://www.nature.com/nsmb/.
Supplementary Material
ACKNOWLEDGEMENTS
We thank Dr. Young Chul Park for earlier work on this project and Drs. Kanagalaghatta Rajashankar, Igor Kourinov and Narayanasami Sukumar for data collection at NE-CAT of APS. This work was supported by NIH grant R01-AI50872 (HW), the Post-doctoral Fellowship Program of Korea Science & Engineering Foundation (KOSEF) (JKY), the 2008 Long-term Overseas Dispatch Program for Pusan National University’s Tenure-track Faculty (SBJ), the Biotechnology and Biological Sciences Research Council (AYP), the Royal Society (CVR) and the Walters-Kundert Trust (CVR). SR was a fellow of the German Academy of Sciences Leopoldina (BMBF-LPD 9901/8-163). TW is an investigator of the Howard Hughes Medical Institute.
Footnotes
Accession codes. Protein Data Bank: Coordinates and structure factors for the core mFas–hFADD DD complex have been deposited under accession code 3OQ9. Protein Model Data Base: Coordinates for the 6 hFas–5 hFADD DD complex have been deposited under accession code PM0076550.
AUTHOR CONTRIBUTIONS
H.W. initiated the project and participated in project design and analysis; L.W. provided the samples for EM; L.W. and E.D. performed in vitro mutagenesis experiments; L.W. and Q.Y. performed multi-angle light scattering experiments; J.K.Y., L.W. and S.B.J. grew the crystals and collected the diffraction data; V.K. and H.W. solved the structure; E.D. performed the CD experiments and the salt-dependence experiments; A.J.R., S.R. and T.W. performed the EM experiments; A.C.C. and R.M.S. performed the cell biology experiments; A.Y.P. and C.V.R. performed the mass spectrometry experiments; H.W. made the figures and wrote the manuscript.
COMPETING FINANCIAL INTERESTS
The authors declare no competing financial interests.
REFERENCES
- 1.Park HH, et al. The Death Domain Superfamily in Intracellular Signaling of Apoptosis and Inflammation. Ann Rev Immunology. 2007;25:561–586. doi: 10.1146/annurev.immunol.25.022106.141656. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Kohl A, Grutter MG. Fire and death: the pyrin domain joins the death-domain superfamily. C R Biol. 2004;327:1077–1086. doi: 10.1016/j.crvi.2004.08.006. [DOI] [PubMed] [Google Scholar]
- 3.Chinnaiyan AM, O'Rourke K, Tewari M, Dixit VM. FADD, a novel death domain-containing protein, interacts with the death domain of Fas and initiates apoptosis. Cell. 1995;81:505–512. doi: 10.1016/0092-8674(95)90071-3. [DOI] [PubMed] [Google Scholar]
- 4.Strasser A, Jost PJ, Nagata S. The many roles of FAS receptor signaling in the immune system. Immunity. 2009;30:180–192. doi: 10.1016/j.immuni.2009.01.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Wajant H. The Fas signaling pathway: more than a paradigm. Science. 2002;296:1635–1636. doi: 10.1126/science.1071553. [DOI] [PubMed] [Google Scholar]
- 6.Kischkel FC, et al. Cytotoxicity-dependent APO-1 (Fas/CD95)-associated proteins form a death-inducing signaling complex (DISC) with the receptor. Embo J. 1995;14:5579–5588. doi: 10.1002/j.1460-2075.1995.tb00245.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Huang B, Eberstadt M, Olejniczak ET, Meadows RP, Fesik SW. NMR structure and mutagenesis of the Fas (APO-1/CD95) death domain. Nature. 1996;384:638–641. doi: 10.1038/384638a0. [DOI] [PubMed] [Google Scholar]
- 8.Berglund H, et al. The three-dimensional solution structure and dynamic properties of the human FADD death domain. J Mol Biol. 2000;302:171–188. doi: 10.1006/jmbi.2000.4011. [DOI] [PubMed] [Google Scholar]
- 9.Jeong EJ, et al. The solution structure of FADD death domain. Structural basis of death domain interactions of Fas and FADD. J Biol Chem. 1999;274:16337–16342. doi: 10.1074/jbc.274.23.16337. [DOI] [PubMed] [Google Scholar]
- 10.Hill JM, et al. Identification of an expanded binding surface on the FADD death domain responsible for interaction with CD95/Fas. J Biol Chem. 2004;279:1474–1481. doi: 10.1074/jbc.M304996200. [DOI] [PubMed] [Google Scholar]
- 11.Martin DA, et al. Defective CD95/APO-1/Fas signal complex formation in the human autoimmune lymphoproliferative syndrome, type Ia. Proc Natl Acad Sci U S A. 1999;96:4552–4557. doi: 10.1073/pnas.96.8.4552. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Rieux-Laucat F, Le Deist F, Fischer A. Autoimmune lymphoproliferative syndromes: genetic defects of apoptosis pathways. Cell Death Differ. 2003;10:124–133. doi: 10.1038/sj.cdd.4401190. [DOI] [PubMed] [Google Scholar]
- 13.Bettinardi A, et al. Missense mutations in the Fas gene resulting in autoimmune lymphoproliferative syndrome: a molecular and immunological analysis. Blood. 1997;89:902–909. [PubMed] [Google Scholar]
- 14.Fisher GH, et al. Dominant interfering Fas gene mutations impair apoptosis in a human autoimmune lymphoproliferative syndrome. Cell. 1995;81:935–946. doi: 10.1016/0092-8674(95)90013-6. [DOI] [PubMed] [Google Scholar]
- 15.Oliveira JB, Gupta S. Disorders of apoptosis: mechanisms for autoimmunity in primary immunodeficiency diseases. J Clin Immunol. 2008;28(Suppl 1):S20–S28. doi: 10.1007/s10875-007-9161-4. [DOI] [PubMed] [Google Scholar]
- 16.Scott FL, et al. The Fas-FADD death domain complex structure unravels signalling by receptor clustering. Nature. 2009;457:1019–1022. doi: 10.1038/nature07606. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Park HH, et al. Death domain assembly mechanism revealed by crystal structure of the oligomeric PIDDosome core complex. Cell. 2007;128:533–546. doi: 10.1016/j.cell.2007.01.019. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Siegel RM, et al. Measurement of molecular interactions in living cells by fluorescence resonance energy transfer between variants of the green fluorescent protein. Sci STKE. 2000;2000:PL1. doi: 10.1126/stke.2000.38.pl1. [DOI] [PubMed] [Google Scholar]
- 19.Vaishnaw AK, et al. The molecular basis for apoptotic defects in patients with CD95 (Fas/Apo-1) mutations. J Clin Invest. 1999;103:355–363. doi: 10.1172/JCI5121. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Carrington PE, et al. The structure of FADD and its mode of interaction with procaspase-8. Mol Cell. 2006;22:599–610. doi: 10.1016/j.molcel.2006.04.018. [DOI] [PubMed] [Google Scholar]
- 21.O' Reilly LA, et al. Membrane-bound Fas ligand only is essential for Fas-induced apoptosis. Nature. 2009;461:659–663. doi: 10.1038/nature08402. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Dhein J, et al. Induction of apoptosis by monoclonal antibody anti-APO-1 class switch variants is dependent on cross-linking of APO-1 cell surface antigens. J Immunol. 1992;149:3166–3173. [PubMed] [Google Scholar]
- 23.Muppidi JR, Siegel RM. Ligand-independent redistribution of Fas (CD95) into lipid rafts mediates clonotypic T cell death. Nat Immunol. 2004;5:182–189. doi: 10.1038/ni1024. [DOI] [PubMed] [Google Scholar]
- 24.Holler N, et al. Two adjacent trimeric Fas ligands are required for Fas signaling and formation of a death-inducing signaling complex. Mol Cell Biol. 2003;23:1428–1440. doi: 10.1128/MCB.23.4.1428-1440.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Tibbetts MD, Zheng L, Lenardo MJ. The death effector domain protein family: regulators of cellular homeostasis. Nat Immunol. 2003;4:404–409. doi: 10.1038/ni0503-404. [DOI] [PubMed] [Google Scholar]
- 26.Yang JK, et al. Crystal structure of MC159 reveals molecular mechanism of DISC assembly and FLIP inhibition. Mol Cell. 2005;20:939–949. doi: 10.1016/j.molcel.2005.10.023. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Siegel RM, et al. SPOTS: signaling protein oligomeric transduction structures are early mediators of death receptor-induced apoptosis at the plasma membrane. J Cell Biol. 2004;167:735–744. doi: 10.1083/jcb.200406101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Xiao T, Towb P, Wasserman SA, Sprang SR. Three-dimensional structure of a complex between the death domains of Pelle and Tube. Cell. 1999;99:545–555. doi: 10.1016/s0092-8674(00)81542-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Qin H, et al. Structural basis of procaspase-9 recruitment by the apoptotic protease-activating factor 1. Nature. 1999;399:549–557. doi: 10.1038/21124. [DOI] [PubMed] [Google Scholar]
- 30.Lin SC, Lo YC, Wu H. Helical assembly in the MyD88-IRAK4-IRAK2 complex in TLR/IL-1R signalling. Nature. 2010;465:885–890. doi: 10.1038/nature09121. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Ohi M, Li Y, Cheng Y, Walz T. Negative Staining and Image Classification - Powerful Tools in Modern Electron Microscopy. Biol Proced Online. 2004;6:23–34. doi: 10.1251/bpo70. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Li Z, Hite RK, Cheng Y, Walz T. Evaluation of imaging plates as recording medium for images of negatively stained single particles and electron diffraction patterns of two-dimensional crystals. J Electron Microsc (Tokyo) 59:53–63. doi: 10.1093/jmicro/dfp036. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Frank J, et al. SPIDER and WEB: processing and visualization of images in 3D electron microscopy and related fields. J Struct Biol. 1996;116:190–199. doi: 10.1006/jsbi.1996.0030. [DOI] [PubMed] [Google Scholar]
- 34.Sobott F, Hernandez H, McCammon MG, Tito MA, Robinson CV. A tandem mass spectrometer for improved transmission and analysis of large macromolecular assemblies. Anal Chem. 2002;74:1402–1407. doi: 10.1021/ac0110552. [DOI] [PubMed] [Google Scholar]
- 35.Hernandez H, Robinson CV. Determining the stoichiometry and interactions of macromolecular assemblies from mass spectrometry. Nat Protoc. 2007;2:715–726. doi: 10.1038/nprot.2007.73. [DOI] [PubMed] [Google Scholar]
- 36.Otwinowski Z, Minor W. Processing of X-ray diffraction data collected in oscillation mode. Methods Enzymol. 1997;276:307–326. doi: 10.1016/S0076-6879(97)76066-X. [DOI] [PubMed] [Google Scholar]
- 37.Read RJ. Pushing the boundaries of molecular replacement with maximum likelihood. Acta Cryst. 2001;D57:1373–1382. doi: 10.1107/s0907444901012471. [DOI] [PubMed] [Google Scholar]
- 38.Brunger AT, et al. Crystallography & NMR system: A new software suite for macromolecular structure determination. Acta Crystallogr. 1998;D54:905–921. doi: 10.1107/s0907444998003254. [DOI] [PubMed] [Google Scholar]
- 39.Collaborative Computational Project, N. The CCP4 Suite: Programs for Protein Crystallography. Acta Cryst. 1994;D50:760–763. doi: 10.1107/S0907444994003112. [DOI] [PubMed] [Google Scholar]
- 40.Delano WL. The PyMol Molecular Graphics System. 2002 [Google Scholar]
- 41.Baker NA, Sept D, Joseph S, Holst MJ, McCammon JA. Electrostatics of nanosystems: application to microtubules and the ribosome. Proc Natl Acad Sci U S A. 2001;98:10037–10041. doi: 10.1073/pnas.181342398. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Siegel RM, et al. Fas preassociation required for apoptosis signaling and dominant inhibition by pathogenic mutations. Science. 2000;288:2354–2357. doi: 10.1126/science.288.5475.2354. [DOI] [PubMed] [Google Scholar]
- 43.Herzenberg LA, Tung J, Moore WA, Parks DR. Interpreting flow cytometry data: a guide for the perplexed. Nat Immunol. 2006;7:681–685. doi: 10.1038/ni0706-681. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.