Summary
Bacteria employ a variety of mechanisms to promote and control colonization of their respective hosts, including restricting the expression of genes necessary for colonization to distinct situations (i.e. encounter with a prospective host). In the symbiosis between the marine bacterium Vibrio fischeri and its host squid, Euprymna scolopes, colonization proceeds via a transient biofilm formed by the bacterium. The production of this bacterial biofilm depends on a complex regulatory network that controls transcription of the symbiosis polysaccharide (syp) gene locus. In addition to this transcriptional control, biofilm formation is regulated by two proteins, SypA and SypE, which may function in an unusual regulatory mechanism known as partner switching. Best characterized in Bacillus subtilis and other Gram-positive bacteria, partner switching is a signaling mechanism that provides dynamic regulatory control over bacterial gene expression. The involvement of putative partner-switching components within V. fischeri suggests that tight regulatory control over biofilm formation may be important for the lifestyle of this organism.
Introduction
To achieve efficient colonization of their respective hosts, bacteria have evolved complex signaling networks to ensure the proper expression of the genes necessary to respond to the host environment. The result of these regulatory cascades is the induction of key cellular responses required for successful host colonization, such as biofilm formation. Biofilms, or surface-associated communities of cells encapsulated in a matrix, often play an integral role in the attachment of bacterial cells to host or environmental surfaces and in bacterial survival, both within and outside of a host.
The formation of a biofilm is a common strategy utilized among numerous Vibrio species, in which it is predicted to promote bacterial persistence in the environment and/or colonization of eukaryotic hosts (for a recent review see Yildiz and Visick, 2009). Vibrio species are Gram-negative bacteria, typically present in marine environments. Among the Vibrios, several species engage in pathogenic or symbiotic partnerships with specific eukaryotic hosts. This review focuses on the bacterium Vibrio fischeri, as recent work has shown a clear relevance of biofilm formation to the ability of this organism to establish symbiotic colonization of its eukaryotic host, the Hawaiian bobtail squid Euprymna scolopes. Biofilm formation by V. fischeri requires expression of a cluster of polysaccharide biosynthetic genes, termed the symbiosis polysaccharide (syp) locus, that is conserved among several Vibrio species (Yip et al., 2005; Yip, 2006). Importantly, the regulation of this polysaccharide locus involves an intricate network of regulatory proteins, which is predicted to restrict biofilm formation such that it occurs transiently in response to specific host-derived signals (Visick, 2009).
Biofilm formation also appears to be regulated by two largely uncharacterized regulatory proteins encoded within the syp cluster, SypA and SypE. Bioinformatic analyses of these regulatory proteins suggest they contain elements of a regulatory signaling mechanism, termed partner switching. A signaling mechanism most extensively characterized in Gram-positive bacteria, partner switching provides yet another layer of regulatory control over gene expression. In recent years, genome analyses have suggested that potential partner-switching components are present in a wide range of bacteria, including Gram-negative species (Mittenhuber, 2002; Mattoo et al., 2004). Here, following brief overviews of the V. fischeri-squid symbiosis and biofilm formation, we review the partner-switching mechanism, as understood within characterized models, then speculate on the role of this signaling mechanism in the regulation of biofilms by V. fischeri, and potentially, other Vibrio species.
Vibrio fischeri and Euprymna scolopes: Symbiotic initiation depends on biofilm formation
The symbiotic relationship between the marine bacterium V. fischeri and its eukaryotic host, E. scolopes, provides an elegant model of symbiotic bacteria-host interaction (For recent reviews, see Nyholm and McFall-Ngai, 2004; Visick and Ruby, 2006; Stabb, 2006). Newly hatched juvenile squid are aposymbiotic and must acquire their bacterial symbionts from the surrounding seawater. Successful establishment of this symbiotic colonization involves a number of both host- and symbiont-derived responses (Nyholm and McFall-Ngai, 2004). Importantly for this review, exposure to environmental bacteria stimulates newly hatched squid to secrete mucus onto the surface of their symbiotic light organs (Nyholm and McFall-Ngai, 2004; Nyholm et al., 2000). V. fischeri appears particularly adept at adhering to the mucus and forming a biofilm-like aggregate of cells that are poised to enter the light organ (Nyholm and McFall-Ngai, 2003). Subsequently, V. fischeri, but not other bacteria, productively enter and migrate to the crypts, where they establish colonization by multiplying to high cell density.
The formation of a biofilm aggregate outside the squid light organ is essential for efficient initiation of host colonization. Mutants defective in biofilm formation exhibit a significant defect in colonization, while a strain with an enhanced ability to form biofilms exhibits a significant colonization advantage (Yip et al., 2005; Yip et al., 2006). Formation of this biofilm requires the syp locus, consisting of 18 genes predicted to be involved in the synthesis and regulation of a polysaccharide biofilm matrix (Yip et al., 2005; Yip et al., 2006). syp mutants exhibit a significant defect in biofilm formation and host colonization (Yip et al., 2005).
Regulation of biofilm formation: a complex network of regulators
Biofilm formation appears to be under complex regulatory controls. At least four regulators encoded within the syp locus (SypA/E/F/G) and two regulators encoded elsewhere (RscS and VpsR) appear to regulate biofilms at the level of syp transcription or at an unknown level beyond syp activation (recently reviewed in Visick, 2009). Transcription of the syp locus is controlled by the response regulator SypG, which is predicted to be activated via phosphotransfer from an upstream sensor kinase, RscS (Fig. 1)(Hussa et al., 2008). Overexpression of either rscS or sypG promotes substantial biofilm formation, and loss of either gene results in a severe colonization defect similar to syp mutants (Visick and Skoufos, 2001; Hussa et al., 2007; Yip et al., 2006). Two other regulators, the sensor kinase SypF and the response regulator VpsR (a predicted DNA binding protein), also appear to regulate biofilm formation, but it remains unknown how these proteins contribute to the regulatory network (Darnell et al., 2009).
Figure 1.
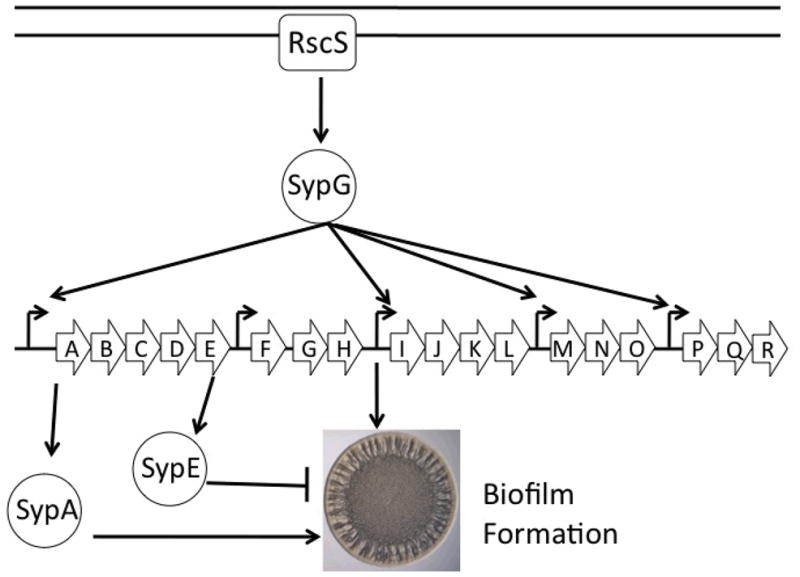
Model of biofilm formation in V. fischeri. The symbiosis polysaccharide (sypA-R) locus is regulated at the transcriptional level via a 2-component regulatory cascade consisting of the sensor kinase, RscS, and the downstream response regulator, SypG. The regulatory proteins, SypA and SypE, exhibit antagonistic regulatory roles, promoting and inhibiting syp-dependent biofilms, respectively. These regulators control biofilm formation via an unknown mechanism that appears to function downstream of syp transcription. Biofilms are represented by the formation of a wrinkled bacterial colony (Yildiz and Visick, 2009).
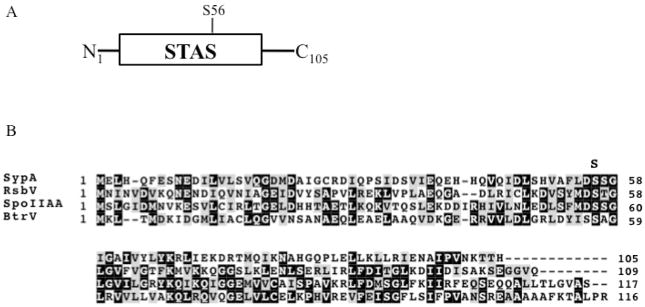
SypA and SypE also contribute to control of biofilm formation, but appear to exert their effects downstream of syp transcription (Hussa et al., 2008; Morris and Visick, unpublished data). Current unpublished data indicates the SypA is required for biofilm formation by V. fischeri (Shibata, Yip and Visick unpublished data). Sequence analysis indicates that sypA codes for a single-domain protein with a predicted sulphate transporter and anti-sigma factor antagonist (STAS) domain (Fig. 2). This domain is conserved among anti-anti-sigma factors, which generally function as positive regulators (Aravind and Koonin, 2000).
Figure 2.
SypA domain structure and multiple sequence alignment. (A) Domain structure of SypA. SypA contains the conserved anti-sigma factor antagonist and sulphate transporter (STAS) domain present in anti-anti-sigma factors. The conserved regulatory serine residue (S56) is indicated. (B) BLAST multiple sequence alignment (Altschul et al., 1997) and identification of conserved residues within V. fischeri SypA and the anti-anti-sigma factors RsbV and SpoIIAA of B. subtilis and BtrV of B. bronchiseptica. Serine 56 of RsbV, serine 58 of SpoIIAA, and serine 59 of BtrU are the phosphorylation targets of the respective serine kinases (Najafi et al., 1995; Yang et al., 1996). This serine residue, indicated by an S, is conserved in SypA (S56). Highlighting of the conserved identical residues (black boxes) and conserved substitutions (grey boxes) was generated using BOXSHADE Server.
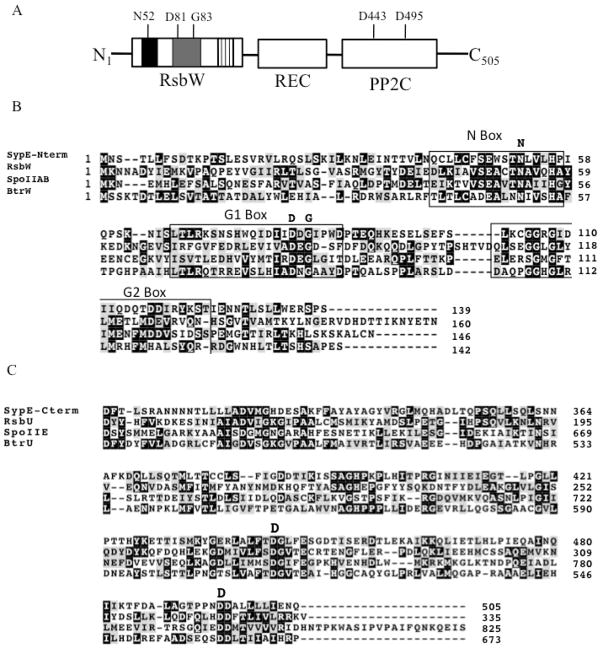
In contrast to SypA, SypE appears to play a dual role in biofilm formation. SypE inhibits SypG-induced biofilm formation, but is required for full biofilm induction under conditions in which the sensor kinase, RscS, is overexpressed (Hussa et al., 2008). Sequence and domain analysis of SypE suggests a unique multi-domain protein. Due to the presence of a conserved receiver (REC) domain, SypE is a predicted response regulator. The activity of response regulators is generally controlled via the phosphorylation of a conserved aspartate residue within the REC domain (Stock, 2009). The phosphorylation state of the REC domain activates an effector domain within the response regulator that results in a response, usually a change in gene transcription (Stock, 2009). Distinct from the typical response regulator, which contains the REC domain in the N-terminus, SypE contains a central REC domain (Fig. 3A). This central REC domain is flanked by two domains of putative opposing functions. The N-terminal domain of SypE exhibits weak sequence similarity to serine kinases of the GHKL (gyrase, Hsp90, histidine kinase, and MutL) family, which includes SpoIIAB B. subtilis (Dutta and Inouye, 2000) (Fig. 3B). The C-terminus of the protein contains a putative serine/threonine phosphatase domain and exhibits strong sequence similarity to the PP2C family of serine phosphatases (Fig. 3C).
Figure 3.
SypE domain structure and multiple sequence alignment. (A) Domain structure of SypE. SypE is a multi-domain protein that contains a central response regulator (REC) domain flanked by an N-terminal serine kinase (RsbW) domain and a C-terminal serine phosphatase (PP2C) domain. The N-terminal RsbW domain of SypE contains the conserved N-, G1-, and G2- boxes important in anti-sigma factor activity, which are indicated by black, gray, and striped boxes, respectively. The conserved residues within the N-terminal RsbW and C-terminal PP2C domains, predicted to be important for serine kinase or serine phosphatase activity, are shown. (B) BLAST multiple sequence alignment (Altschul et al., 1997) of the N-terminal serine kinase domain of SypE with the anti-sigma factors RsbW and SpoIIAB of B. subtilis and BtrW of B. bronchiseptica. The conserved N-, G1-, and G2- boxes are outlined, and the conserved residues required for serine kinase activity are indicated in bold letters above the alignments (Dutta et al., 2000). The SypE serine kinase domain contains the conserved N-box asparagine (N52) and the G1 box aspartate and glycine residues (D81 and G83). (C) BLAST multiple sequence alignment of the C-terminal serine phosphatase domain of SypE with the serine phosphatases RsbU and SpoIIE of B. subtilis and BtrU of B. bronchiseptica. The labeled amino acids indicate the conserved residues required for serine phosphatase activity (Adler et al., 1997) SypE contains the invariant aspartate residues (D443 and D495) predicted to be important in divalent cation binding. For parts B and C, highlighting of the conserved residues (black boxes) and conserved substitutions (grey boxes) was generated using BOXSHADE server.
How these proteins contribute to the biofilm regulatory network remains an area of active research. Although the roles of SypA and SypE in the regulation of biofilm formation remain unknown, bioinformatics suggest that these proteins may constitute a novel partner-switching system.
The partner-switching system
First coined by Alper et al. (1994), the term “partner switch” describes a network of interacting proteins whose interaction with cognate “partners” depends upon a reversible phosphorylation event. Dependent upon which partner proteins interact, the outcome of this partner switching can either negatively or positively regulate a target protein, generally a transcription factor or enzyme. Specifically, the partner-switch mechanism depends upon key regulatory elements, including a serine kinase/anti-sigma factor, a serine phosphatase, an antagonist protein/anti-anti sigma factor, and a target protein (often a sigma factor) (Yang et al., 1996).
The partner-switching mechanism has been best characterized in the Gram-positive bacterium Bacillus subtilis. In B. subtilis and other Gram-positive bacteria, partner-switching systems contribute to the signaling networks that control the activity of sigma factors, the subunit of RNA polymerase that provides promoter specificity. For example, B. subtilis uses partner-switching to control the activity of the sigma factor, sigma B, which regulates the general stress response pathway (Dufour and Haldenwang, 1994). In this system, the anti-sigma factor RsbW negatively regulates the activity of sigma B by directly binding it and preventing interaction with the RNA polymerase core (Dufour and Haldenwang, 1994; Hughes and Mathee, 1998) (Fig. 4A). The binding of an anti-anti-sigma factor, RsbV, to RsbW relieves this negative regulation by sequestering the anti-sigma factor (Dufour and Haldenwang, 1994). This releases sigma B, which can now bind to the RNA polymerase core and promote transcription of sigma B-dependent genes. This results in the induction of proteins involved in protection against multiple bacterial stresses (Price et al., 2001; Yang et al., 1996).
Figure 4.
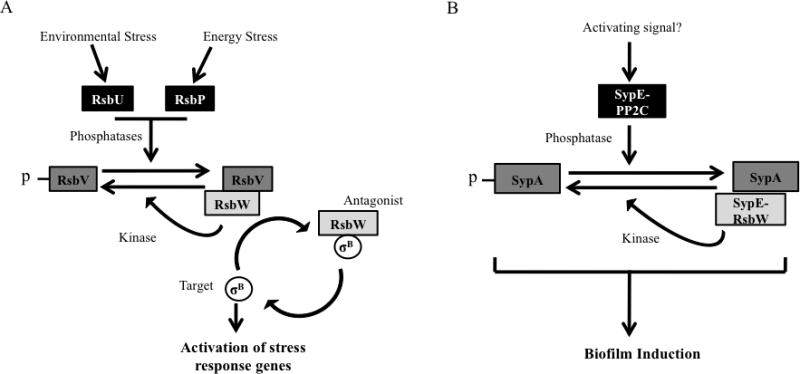
Model of partner-switching pathways. (A) A model of the B. subtilis partner-switching regulatory pathway controlling the activity of the general stress response sigma factor, sigma B. See text for detailed description of the model. (B) A model of the predicted SypA-SypE partner-switching module controlling biofilm formation. SypA and SypE possess the core components of a putative partner-switching signal pathway. The proposed model is constructed from information of the conserved protein domains and current data of biofilm regulation.
Activity of the anti-anti-sigma factor RsbV is controlled via phosphorylation of a conserved serine residue within the STAS domain (Fig. 2B and 4A). In the unphosphorylated state, RsbV can bind and inactivate the anti-sigma RsbW, thus freeing sigma B and upregulating sigma B-dependent genes. However, when this serine residue becomes phosphorylated, RsbV is rendered unable to bind RsbW, which permits RsbW to sequester sigma B and downregulate sigma-B dependent gene expression. The phosphorylation state of this serine residue is controlled by two sets of proteins, one of which is RsbW itself; in addition to its role in sequestering sigma B, RsbW also functions as a serine kinase. Dephosphorylation of this residue is carried out by the serine phosphatases, RsbU and RsbP, which are activated by environmental and energy stress signals, respectively (Voelker et al., 1996). Thus, in this regulatory network, RsbW functions as a regulatory switch, as it reversibly interacts with its cognate sigma factor and anti-anti-sigma factor. Furthermore, the partner switch is regulated by reversible phosphorylation of RsbV.
Conservation of partner-switching: A Gram-positive mechanism?
Since its characterization in B. subtilis, the partner-switching mechanism has been identified as a signaling component in a wide range of Gram-positive bacteria. These include Bacillus cereus (van Schaik et al., 2005), Bacillus anthracis (Fouet et al., 2000), Mycobacterium tuberculosis (Beaucher et al., 2002), Staphylococcus aureus (Miyazaki et al., 1999), and Listeria monocytogenes (Chaturongakul and Boor, 2004; Chaturongakul and Boor, 2006). In these bacterial systems, partner-switch modules are utilized in a manner similar to that observed in B. subtilis: primarily, the regulation of sigma factor activity. However, the output of these modules varies among the individual bacteria. Partner-switching modules have been demonstrated to contribute to the regulation of the general stress response of L. monocytogenes and many other bacteria (Chaturongakul and Boor, 2004), biofilm formation in S. epidermidis (Knobloch et al., 2004), and the expression of virulence-associated genes in M. tuberculosis (Beaucher et al., 2002; Manganelli et al., 2004). Additionally, bacteria may possess multiple partner-switching pathways regulating distinct sets of target proteins. For example, in addition to the RsbU/V/W module that regulates the general stress response, B. subtilis possesses a second set of partner-switching regulators, SpoIIAA/SpoIIAB/SpoIIE. Similar to the sigma B regulatory pathway, these partner-switching proteins control the activity of a sigma, in this case sigma F, which regulates sporulation (Magnin et al., 1997). The partner-switching mechanism, while similar among diverse Gram-positives, has been adapted to respond to specific stimuli and regulate distinct cellular responses.
Partner switching within the Gram-negatives?
Analyses of diverse bacterial genomes suggest that partner-switching orthologs may exist in a wide range of eubacteria (Mittenhuber, 2002; Mattoo et al., 2004). Despite its predicted widespread distribution, partner switching has remained relatively uncharacterized within Gram-negative bacteria. Indeed, partner-switching systems have been characterized in only two Gram-negative bacteria, Bordetella bronchiseptica and Chlamydia trachomatis (Kozak et al., 2005; Hua et al., 2006). Furthermore, only in the case of B. bronchiseptica has a partner-switching module been experimentally demonstrated to regulate a physiological response. This respiratory pathogen utilizes a partner-switching module to control production of a type III secretion system (T3SS)(Mattoo et al., 2004). The T3SS consists of a needle-like secretory apparatus that directly transports virulence proteins into the cytoplasm of host cells. In B. bronchiseptica, the T3SS contributes to persistent colonization of the host trachea and the avoidance of the host immune response (Yuk et al., 2000; Mattoo et al., 2001). The production of the T3SS requires transcription of a gene cluster, the bsc locus, which encodes multiple components of the secretory system (Mattoo et al., 2001). Regulation of the T3SS depends upon a set of genes adjacent to the bsc cluster, the btr locus, which encode orthologs of the B. subtilis RsbU/V/W partner-switching proteins, BtrU/BrtV/BtrW. In vitro and in vivo analyses of the B. bronchiseptica proteins demonstrated that they constitute a regulatory network similar to their B. subtilis counterparts (Kozak et al., 2005). However, this partner-switching system appears to deviate from that of the B. subtilis RsbU/V/W paradigm. First, disruption of any component of the BtrU/V/W partner-switching module results in the loss of Type III secretion (Mattoo et al., 2004), a result that is inconsistent with the B. subtilis model (Fig. 4A). Second, positive regulation of the T3SS requires both the formation of the BtrV/BtrW complex and its dissociation, via phosphorylation of BtrV by BtrW (Kozak et al., 2005). Finally, although the BtrU/V/W module regulates type III secretion, it does not appear to control transcription of the bsc locus (Kozak et al., 2005). Instead, Kozak et al. (2005) suggest that these partner-switching proteins may regulate the T3SS at the posttranscriptional level possibly by interacting with yet unknown regulatory proteins or playing a structural role in the secretory pathway. Thus, although there is conservation of the partner-switching components, the regulatory mechanism appears to vary from that of the Gram-positive paradigm.
Genome analysis of the obligate intracellular pathogen C. trachomatis identified several components of a putative partner-switching module (Hua et al., 2006). In vitro analysis of the candidate genes demonstrated that these proteins could interact. As with B. bronchiseptica, it appears that the C. trachomatis partner-switching system may vary from the B. subtilis paradigm, as in vitro binding assays failed to demonstrate an interaction with any of the three sigma factors encoded in the C. trachomatis genome (Hua et al., 2006). However, the lack of genetic tools and difficulty culturing C. trachomatis have delayed analysis of this potential partner-switching module in vivo.
Together, these studies suggest that the partner-switching mechanism, previously observed only among the Gram-positives, also contributes to regulatory control in Gram-negative bacteria. It remains unknown how these Gram-negative partner-switching proteins regulate downstream targets. Furthermore, it remains unclear how widespread this regulatory mechanism is among Gram-negative bacteria.
SypA and SypE: A potential partner-switching module in V. fischeri?
Several lines of evidence suggest that SypA and SypE may represent a partner-switching module in the Gram-negative bacterium V. fischeri. First, the physical proximity of the sypA and sypE genes and their known roles in biofilm formation suggest a regulatory connection. Second, searches for V. fischeri proteins with sequence similarity to the RsbU/V/W of B. subtilis, and other orthologs, yielded only two candidate proteins, SypA and SypE. Third, these proteins not only contain the conserved domains, but also critical active site residues within these domains.
Specifically, SypA not only contains the conserved STAS domain present among RsbV-like anti-anti-sigma factors, but also retains the conserved serine residue (S56) predicted to be the site of phosphorylation (Fig. 2B). Our preliminary data are consistent with a role for this residue in SypA function (Morris and Visick, unpublished). SypE contains two partner-switching domains: an N-terminal serine kinase (RsbW) and a C-terminal serine phosphatase (PP2C) domain. As shown in Figure 3B, sequence alignment of the N-terminal domain of SypE with RsbW orthologs indicates that SypE contains the conserved N, G1, and G2-boxes present in the Bergerat ATP-binding fold of serine kinases (Dutta and Inouye, 2000). Importantly, SypE possesses the invariant asparagine residue (N52) of the N-box, which is required for Mg2+ ion chelation and coordinates binding of ATP to the nucleotide pocket (Fig. 3B)(Dutta and Inouye, 2000). Sequence alignment also reveals the conserved G1-box aspartate and glycine residues (D81 and G83), predicted to participate in ATP binding and formation of the ATP lid of the nucleotide binding pocket, respectively (Dutta and Inouye, 2000). The conservation of these key residues suggests that, despite poor overall sequence similarity, the N-terminus of SypE may function as an RsbW-like serine kinase. The C-terminus of SypE possesses strong sequence similarity to serine/threonine phosphatases of the PP2C family, including RsbU. As shown in Figure 3C, the C-terminal domain of SypE also possesses the invariant aspartate residues (D306, D323, D443, and D495) that form part of the catalytic core and are predicted to coordinate binding of Mg2+/Mn2+ ions necessary for PP2C catalytic activity (Adler et al., 1997; Rantanen et al., 2007; Shi, 2009).
The antagonistic domains within SypE suggest that this protein may possess both negative and positive regulatory activity, depending on which domain is active (Fig. 4B). Studies of syp-dependent biofilms support this hypothesis: SypE inhibits SypG-induced biofilms, but is required for full expression of biofilms produced by overexpression of the sensor kinase, RscS (Hussa et al., 2008). Furthermore, recent genetic analyses indicate that the antagonistic domains of SypE may be responsible for these apparent dual regulatory activities (Morris and Visick, unpublished data).
Based on bioinformatics and experimental observations, we propose a model in which SypE and SypA constitute part of a partner-switch module (Fig. 4B). In partner-switching systems, RsbW-like serine kinases generally function as antagonists, and inhibit cellular responses via the binding to cognate partner proteins. Thus, potentially the N-terminal (RsbW) domain of SypE regulates biofilm formation by interacting with SypA or a yet unknown regulatory protein. These interactions are possibly dictated by the phosphorylation state of SypA, which may be controlled by the serine kinase and/or the serine phosphatase domains of SypE. The phosphorylation state of the central REC domain may regulate whether SypE functions as a serine kinase or a serine phosphatase. The outcome of this partner-switching network is either the negative or positive regulation of biofilm formation. Work is currently in progress to assess whether these components indeed function as predicted by this model.
Conservation of SypA and SypE among Vibrio species
The syp locus is relatively conserved among several Vibrio species, including both pathogenic and symbiotic bacteria. Although biofilm formation has been investigated in diverse Vibrio species, the role of the syp cluster in species other than V. fischeri is not fully understood. Recently, Kim et al. (2009) demonstrated that the syp locus plays a role in biofilms formed by the pathogenic bacterium Vibrio vulnificus. Specifically, the syp cluster contributes to the production of exopolysaccharides and bacterial attachment to biotic and abiotic surfaces (Kim et al., 2009). Due to the conservation of the syp locus among Vibrio species, we performed a bioinformatic survey of syp-containing Vibrio genomes for SypA and SypE, specifically, or other potential partner-switching proteins. Interestingly, SypA is well-conserved among the syp-containing Vibrio genomes, but SypE appears to be absent in several species. For example, V. vulnificus possesses a SypA ortholog, but lacks any clear SypE-like genes. Intriguingly, the genome of V. vulnificus contains RsbW- and RsbU-like proteins, VVA0582 and VVA1682, which encode a putative RsbW-like anti-sigma factor and an RsbU-like serine phosphatase, respectively. The genomes of Vibrio parahaemolyticus and Aliivibrio salmonicida similarly lack SypE, but encode RsbW and RsbU-like proteins elsewhere. Thus, in several Vibrio species lacking SypE, other putative partner-switching components exist; whether or not they function as predicted or control the activity of SypA remains to be determined.
Concluding Remarks
The symbiotic bacterium V. fischeri employs a variety of regulatory proteins to control biofilm formation and consequently host colonization. While much of the regulation occurs via canonical two-component signaling pathways that control syp transcription, it remains unknown how the syp-encoded regulators SypA and SypE control biofilm formation. Bioinformatics suggest the possibility that SypA and SypE participate in a partner-switching mechanism. However, the downstream target of this potential control mechanism remains unknown. In contrast to the well-established Gram-positive partner-switching paradigms, studies from other Gram-negative bacteria suggest that the downstream target may not necessarily be a sigma factor.
The potential integration of partner-switching components within the V. fischeri biofilm regulatory network would provide yet another layer of signal control. Together, these mechanisms may confine expression of colonization genes to defined conditions (i.e. interaction with juvenile host squid) and/or control the timing of the transient biofilm formation, thus permitting cells to leave the biofilm to enter the symbiotic organ. The presence of the syp locus within multiple Vibrio species exhibiting varied lifestyles (pathogens vs. symbionts) suggests that syp-dependent biofilms may play a role in diverse responses, such as the colonization of respective hosts. The elucidation of the roles of SypA and SypE and the signals to which they respond in V. fischeri thus has the potential to provide a paradigm for understanding partner switching in Vibrios and other Gram-negatives.
Acknowledgments
We thank Valerie Ray, Satoshi Shibata, Justin Eddy, Bruno Lima, Michael Misale, Jon Visick, and Alan Wolfe for their comments on the manuscript. Work on SypA and SypE discussed in this review was supported in part by NIH R01 grant GM59690 awarded to KLV and a grant awarded to ARM by the Conservation Medicine Center of Chicago.
References
- Adler E, Donella-Deana A, Arigoni F, Pinna LA, Stragler P. Structural relationship between a bacterial developmental protein and eukaryotic PP2C protein phosphatases. Mol Microbiol. 1997;23:57–62. doi: 10.1046/j.1365-2958.1997.1801552.x. [DOI] [PubMed] [Google Scholar]
- Alper S, Duncan L, Losick R. An adenosine nucleotide switch controlling the activity of a cell type-specific transcription factor in B. subtilis. Cell. 1994;77:195–205. doi: 10.1016/0092-8674(94)90312-3. [DOI] [PubMed] [Google Scholar]
- Altschul SF, Madden TL, Schaffer AA, Zhang J, Zhang Z, Miller W, Lipman DJ. Gapped BLAST and PSI-BLAST: a new generation of protein database search programs. Nucleic Acids Res. 1997;25:3389–3402. doi: 10.1093/nar/25.17.3389. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Aravind L, Koonin EV. The STAS domain - a link between anion transporters and antisigma-factor antagonists. Curr Biol. 2000;10:R53–5. doi: 10.1016/s0960-9822(00)00335-3. [DOI] [PubMed] [Google Scholar]
- Beaucher J, Rodrigue S, Jacques PE, Smith I, Brzezinski R, Gaudreau L. Novel Mycobacterium tuberculosis anti-sigma factor antagonists control sigmaF activity by distinct mechanisms. Mol Microbiol. 2002;45:1527–1540. doi: 10.1046/j.1365-2958.2002.03135.x. [DOI] [PubMed] [Google Scholar]
- Chaturongakul S, Boor KJ. SigmaB activation under environmental and energy stress conditions in Listeria monocytogenes. Appl Environ Microbiol. 2006;72:5197–5203. doi: 10.1128/AEM.03058-05. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chaturongakul S, Boor KJ. RsbT and RsbV contribute to sigmaB-dependent survival under environmental, energy, and intracellular stress conditions in Listeria monocytogenes. Appl Environ Microbiol. 2004;70:5349–5356. doi: 10.1128/AEM.70.9.5349-5356.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dufour A, Haldenwang WG. Interactions between a Bacillus subtilis anti-sigma factor (RsbW) and its antagonist (RsbV) J Bacteriol. 1994;176:1813–1820. doi: 10.1128/jb.176.7.1813-1820.1994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dutta R, Inouye M. GHKL, an emergent ATPase/kinase superfamily. Trends Biochem Sci. 2000;25:24–28. doi: 10.1016/s0968-0004(99)01503-0. [DOI] [PubMed] [Google Scholar]
- Fouet A, Namy O, Lambert G. Characterization of the operon encoding the alternative sigma(B) factor from Bacillus anthracis and its role in virulence. J Bacteriol. 2000;182:5036–5045. doi: 10.1128/jb.182.18.5036-5045.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hua L, Hefty PS, Lee YJ, Lee YM, Stephens RS, Price CW. Core of the partner switching signalling mechanism is conserved in the obligate intracellular pathogen Chlamydia trachomatis. Mol Microbiol. 2006;59:623–636. doi: 10.1111/j.1365-2958.2005.04962.x. [DOI] [PubMed] [Google Scholar]
- Hughes KT, Mathee K. The anti-sigma factors. Annu Rev Microbiol. 1998;52:231–286. doi: 10.1146/annurev.micro.52.1.231. [DOI] [PubMed] [Google Scholar]
- Hussa EA, Darnell CL, Visick KL. RscS functions upstream of SypG to control the syp locus and biofilm formation in Vibrio fischeri. J Bacteriol. 2008;190:4576–4583. doi: 10.1128/JB.00130-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kim HS, Park SJ, Lee KH. Role of NtrC-regulated exopolysaccharides in the biofilm formation and pathogenic interaction of Vibrio vulnificus. Mol Microbiol. 2009;74:436–453. doi: 10.1111/j.1365-2958.2009.06875.x. [DOI] [PubMed] [Google Scholar]
- Knobloch JK, Jager S, Horstkotte MA, Rohde H, Mack D. RsbU-dependent regulation of Staphylococcus epidermidis biofilm formation is mediated via the alternative sigma factor sigmaB by repression of the negative regulator gene icaR. Infect Immun. 2004;72:3838–3848. doi: 10.1128/IAI.72.7.3838-3848.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kozak NA, Mattoo S, Foreman-Wykert AK, Whitelegge JP, Miller JF. Interactions between partner switcher orthologs BtrW and BtrV regulate type III secretion in Bordetella. J Bacteriol. 2005;187:5665–5676. doi: 10.1128/JB.187.16.5665-5676.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Magnin T, Lord M, Yudkin MD. Contribution of partner switching and SpoIIAA cycling to regulation of sigmaF activity in sporulating Bacillus subtilis. J Bacteriol. 1997;179:3922–3927. doi: 10.1128/jb.179.12.3922-3927.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Manganelli R, Provvedi R, Rodrigue S, Beaucher J, Gaudreau L, Smith I. Sigma factors and global gene regulation in Mycobacterium tuberculosis. J Bacteriol. 2004;186:895–902. doi: 10.1128/JB.186.4.895-902.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mattoo S, Yuk MH, Huang LL, Miller JF. Regulation of type III secretion in Bordetella. Mol Microbiol. 2004;52:1201–1214. doi: 10.1111/j.1365-2958.2004.04053.x. [DOI] [PubMed] [Google Scholar]
- Mattoo S, Foreman-Wykert AK, Cotter PA, Miller JF. Mechanisms of Bordetella pathogenesis. Front Biosci. 2001;6:E168–86. doi: 10.2741/mattoo. [DOI] [PubMed] [Google Scholar]
- Mittenhuber G. A phylogenomic study of the general stress response sigma factor sigmaB of Bacillus subtilis and its regulatory proteins. J Mol Microbiol Biotechnol. 2002;4:427–452. [PubMed] [Google Scholar]
- Miyazaki E, Chen JM, Ko C, Bishai WR. The Staphylococcus aureus rsbW (orf159) gene encodes an anti-sigma factor of SigB. J Bacteriol. 1999;181:2846–2851. doi: 10.1128/jb.181.9.2846-2851.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Najafi SM, Willis AC, Yudkin MD. Site of phosphorylation of SpoIIAA, the anti-anti-sigma factor for sporulation-specific sigma F of Bacillus subtilis. J Bacteriol. 1995;177:2912–2913. doi: 10.1128/jb.177.10.2912-2913.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nyholm SV, Stabb EV, Ruby EG, McFall-Ngai MJ. Establishment of an animal-bacterial association: recruiting symbiotic vibrios from the environment. Proc Natl Acad Sci U S A. 2000;97:10231–10235. doi: 10.1073/pnas.97.18.10231. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nyholm SV, McFall-Ngai MJ. The winnowing: establishing the squid-vibrio symbiosis. Nat Rev Microbiol. 2004;2:632–642. doi: 10.1038/nrmicro957. [DOI] [PubMed] [Google Scholar]
- Nyholm SV, McFall-Ngai MJ. Dominance of Vibrio fischeri in secreted mucus outside the light organ of Euprymna scolopes: the first site of symbiont specificity. Appl Environ Microbiol. 2003;69:3932–3937. doi: 10.1128/AEM.69.7.3932-3937.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Price CW, Fawcett P, Ceremonie H, Su N, Murphy CK, Youngman P. Genome-wide analysis of the general stress response in Bacillus subtilis. Mol Microbiol. 2001;41:757–774. doi: 10.1046/j.1365-2958.2001.02534.x. [DOI] [PubMed] [Google Scholar]
- Rantanen MK, Lehtio L, Rajagopal L, Rubens CE, Goldman A. Structure of Streptococcus agalactiae serine/threonine phosphatase. The subdomain conformation is coupled to the binding of a third metal ion. FEBS J. 2007;274:3128–3137. doi: 10.1111/j.1742-4658.2007.05845.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shi Y. Serine/threonine phosphatases: mechanism through structure. Cell. 2009;139:468–484. doi: 10.1016/j.cell.2009.10.006. [DOI] [PubMed] [Google Scholar]
- Stabb EV. The Vibrio fischeri-Euprymna scolopes light organ symbiosis. In: Thompson FL, Austin B, Swings J, editors. The Biology of Vibrios. Washington, DC: ASM Press; 2006. pp. 204–218. [Google Scholar]
- Stock AM, Robinson VL, Goudreau PN. Two-component signal transduction. Annu Rev Biochem. 2000;69:183–215. doi: 10.1146/annurev.biochem.69.1.183. [DOI] [PubMed] [Google Scholar]
- van Schaik W, Tempelaars MH, Zwietering MH, de Vos WM, Abee T. Analysis of the role of RsbV, RsbW, and RsbY in regulating {sigma}B activity in Bacillus cereus. J Bacteriol. 2005;187:5846–5851. doi: 10.1128/JB.187.16.5846-5851.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Visick KL. An intricate network of regulators controls biofilm formation and colonization by Vibrio fischeri. Mol Microbiol. 2009;74:782–789. doi: 10.1111/j.1365-2958.2009.06899.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Visick KL, Ruby EG. Vibrio fischeri and its host: it takes two to tango. Curr Opin Microbiol. 2006;9:632–638. doi: 10.1016/j.mib.2006.10.001. [DOI] [PubMed] [Google Scholar]
- Voelker U, Voelker A, Haldenwang WG. Reactivation of the Bacillus subtilis anti-sigma B antagonist, RsbV, by stress- or starvation-induced phosphatase activities. J Bacteriol. 1996;178:5456–5463. doi: 10.1128/jb.178.18.5456-5463.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yang X, Kang CM, Brody MS, Price CW. Opposing pairs of serine protein kinases and phosphatases transmit signals of environmental stress to activate a bacterial transcription factor. Genes Dev. 1996;10:2265–2275. doi: 10.1101/gad.10.18.2265. [DOI] [PubMed] [Google Scholar]
- Yildiz FH, Visick KL. Vibrio biofilms: so much the same yet so different. Trends Microbiol. 2009;17:109–118. doi: 10.1016/j.tim.2008.12.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yip ES, Geszvain K, DeLoney-Marino CR, Visick KL. The symbiosis regulator rscS controls the syp gene locus, biofilm formation and symbiotic aggregation by Vibrio fischeri. Mol Microbiol. 2006;62:1586–1600. doi: 10.1111/j.1365-2958.2006.05475.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yip ES, Grublesky BT, Hussa EA, Visick KL. A novel, conserved cluster of genes promotes symbiotic colonization and sigma-dependent biofilm formation by Vibrio fischeri. Mol Microbiol. 2005;57:1485–1498. doi: 10.1111/j.1365-2958.2005.04784.x. [DOI] [PubMed] [Google Scholar]
- Yuk MH, Harvill ET, Cotter PA, Miller JF. Modulation of host immune responses, induction of apoptosis and inhibition of NF-kappaB activation by the Bordetella type III secretion system. Mol Microbiol. 2000;35:991–1004. doi: 10.1046/j.1365-2958.2000.01785.x. [DOI] [PubMed] [Google Scholar]