Abstract
The mechanism by which newly synthesized histones are imported into the nucleus and deposited onto replicating chromatin alongside segregating nucleosomal counterparts is poorly understood, yet this program is expected to bear on the putative epigenetic nature of histone posttranslational modifications. In order to define the events by which naïve pre-deposition histones are imported into the nucleus, we biochemically purified and characterized the gamut of histone H3.1-containing complexes from human cytoplasmic fractions and identified their associated histone PTMs. Through reconstitution assays, biophysical analyses, and live cell manipulations, we describe in detail this series of events, namely the assembly of H3-H4 dimers, the acetylation of histones by the HAT1 holoenzyme, and the transfer of histones between chaperones that culminates with their karyopherin-mediated nuclear import. We further demonstrate the high degree of conservation for this pathway between higher and lower eukaryotes.
Introduction
Canonical nucleosomal histone octamers are formed of a stable (H3-H4)2 tetrameric core flanked by two relatively labile H2A-H2B dimers1. Each histone octamer is enfolded by 147 bp of DNA2 to compact, organize, and regulate access to the underlying genetic material3. There are three major canonical H3 variants in humans, histones H3.1, H3.2, and H3.34. H3.1 and H3.2 differ by a single amino acid substitution (C96S in H3.2), are expressed in S-phase4 and thus termed replication-dependent. While H3.2 is expressed from a single gene, H3.1 levels predominate as it is expressed from ten genes5. H3.3 is replication-independent and expressed at low levels throughout the cell cycle4.
During DNA replication, pre-existing parental histones segregate onto both leading and lagging strands behind the replication fork6, co-depositing alongside newly synthesized counterparts. Early biochemical studies determined that in vitro, nucleosomes predominantly dissociate into a stable (H3-H4)2 tetramer and two H2A-H2B dimers as a function of salt concentration, temperature, and pH1. It was also previously reported that during DNA replication, segregating nucleosomal H3-H4 histones are predominately tetrameric, whereas H2A-H2B histones remain as dimers7, 8. Furthermore, chromatin is thought to be assembled in a sequential manner, through transitory DNA contacts with a central H3-H4 tetramer followed by the addition of two H2A-H2B dimers to complete the nucleosome particle9.
Altogether these findings implied a ‘two-step deposition’ model by which pre-existing nucleosomal histones dissociate ahead of the replication fork to re-distribute on both leading and lagging strands as (H3-H4)2 tetrameric blocks and H2A-H2B dimers. Yet, recent analyses of H3.1 and H3.3 complexes indicated that pre-deposition H3-H4 histone units are handled as dimers in vivo since immunoprecipitation of exogenously expressed epitope-tagged histones would not co-precipitate endogenous counterparts10, 11. Furthermore, biochemical and crystallographic analyses on the anti-silencing factor 1 (ASF1), a major H3-H4 chaperone, indicated that ASF1 binds exclusively an H3-H4 dimer rather than a tetramer12, 13, 14. Since ASF1 co-purifies with subunits of the MCM helicase15, it was proposed that segregating nucleosomal H3-H4 histones dissociate as dimers15, 16. The discrepancy between these recent reports and the earlier ones will only be resolved once the molecular mechanism by which histones are chaperoned and assembled in vivo is thoroughly established. The outcome is important since it may dictate the way cells handle histones as potential carriers of epigenetic information.
Little is known regarding the processing of newly synthesized histones. In humans, newly synthesized histone H4 is acetylated on lysines 5 and 12 by the HAT1-RbAp46 holoenzyme17. Additionally, mass spectrometric analysis of pre-deposition H3.1 histones showed that over a third of this pool contains lysine 9 monomethylation as the sole H3 posttranslational modification18. More recently, the HAT1-RbAp46 holoenzyme and the nuclear autoantigenic sperm protein (NASP) were detected in ASF1 immunoprecipitates from cytosolic fractions16, although the significance of this finding was not clarified. Once in the nucleus, the PCNA-tethered chromatin assembly factor-1 (CAF-1) is essential for the deposition of H3.1-H4 histones onto chromatin during DNA replication19, 10. By interacting with the p60 subunit of CAF-1 (p105 in Drosophila), ASF1 is believed to supply histones to newly replicated or repaired DNA20, 21.
Here, we report the first comprehensive biochemical purification and characterization of cytosolic H3.1 complexes. With this gamut of species, we were able to detect the ongoing transactions whereby histones are transferred from one chaperone to another. These findings bridge the gaps between previous studies and identify the key steps required for the import of newly synthesized histones.
Results
Distinct H3.1 Chaperones in Different Cell Compartments
To ascertain the program whereby naïve pre-deposition histones are handled before incorporation onto chromatin, we purified replication-dependent H3.1 complexes from cytosolic extracts. We employed a HeLa S3 lineage (eH3.1-HeLa)10 that expresses low levels of the replication-dependent histone H3.1 tagged with FLAG and HA epitopes at its C-terminus (eH3.1), from a constitutive viral promoter. Cells were lyzed and fractionated into cytosolic, nuclear, and chromatin-bound fractions. Silver staining of tandem affinity purified eH3.1 samples followed by mass spectrometry allowed the detection of different eH3.1-associated polypeptides, as a function of their subcellular compartments of origin (Fig. 1a). In the cytosolic fraction (S100), six chaperones associated with eH3.1 (HSC70, HSP90, tNASP, sNASP, RbAp46, and ASF1B), in addition to importin-4, histone H4, and the histone acetyltransferase HAT1, among other polypeptides (Fig. 1a). Although the nuclear fraction (NE) also included most of these proteins, western blot analysis revealed stark differences, such as the expected enrichment in CAF1 (Fig. 1b). Similarly, ASF1 was enriched in the cytosolic fraction, and the MCM7 polypeptide was enriched in the chromatin-bound soluble nuclear pellet (SNP) that was devoid of (or drastically reduced in) ASF1 and HAT1 (Fig. 1b). Therefore, histone H3.1 is differentially chaperoned through these different subcellular compartments.
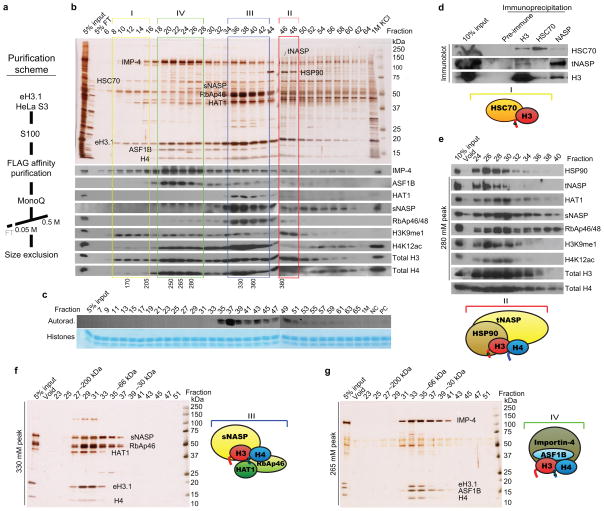
Figure 1.
Subcellular distribution of H3.1-containing complexes. HeLa S3 cells expressing a FLAG-HA tagged histone H3.1 (eH3.1) were fractionated into cytoplasmic (S100), nuclear (NE), and chromatin-bound soluble nuclear pellet (SNP) fractions. (a) Extracts were tandem-affinity purified and resolved by SDS-PAGE for silver staining. (b) Western analysis comparing crude whole cell extracts (WCE), S100, NE, SNP, and total nuclear pellets (TNP) with matching affinity-purified material. (c) Western analysis of endogenous H3.1 co-purifying with exogenous eH3.1 at different cell cycle stages. Arrows mark eH3.1 histones whereas the arrowhead indicates native histone H3.
As per previous reports10, 11, immunoprecipitates containing the epitope tagged histone H3.1 showed a lack of its endogenous counterpart (Fig. 1c). However, while the exogenous eH3.1 gene is constitutively expressed throughout the cell cycle, most cells are normally found outside S-phase. To assess possible association between exogenously and endogenously expressed H3.1 during S-phase, we synchronized the eH3.1-HeLa cells at the G1/S border and verified the level of histones every hour following their release into S-phase (Fig. 1c and Supplementary Fig. 1a). Little change in histone levels was observed in the cytoplasm, highlighting the importance of soluble histone pools outside S-phase. Importantly, barely detectable levels of endogenous histone H3 co-precipitated with exogenous eH3.1 in S-phase, even though endogenous H3 levels surpassed those of the exogenous protein (Fig. 1c). This result corroborates that histones H3-H4 exist as dimeric rather than tetrameric species in the cytoplasm in vivo.
Four Distinct H3.1 Complexes in the Cytoplasm
To evaluate the significance of the proteins associated with cytoplasmic histone H3.1, affinity purified cytoplasmic eH3.1 was first applied to a Superdex 200 gel filtration column. Although all proteins closely eluted, the HAT1 and RbAp46 protein peak was separated by two fractions from the importin-4 protein peak (data not shown). The mass of all proteins combined surpassed that at which they eluted (~200 kDa) suggesting that the eH3.1 immunoprecipitate consists of more than one core complex. We therefore proceeded to further partition the affinity purified eH3.1 material as depicted in Figure 2a.
Figure 2.
Purification of eH3.1 from cytosolic extracts. (a) Purification scheme. (b) Anion exchange chromatography (Mono Q) of affinity-purified cytoplasmic eH3.1 protein complexes. Numbers at the bottom indicate the concentration of potassium chloride (mM) at which peak fractions eluted. Cytoplasmic fractions contain four predominant H3.1 complexes (I–IV). (c) Histone acetyltransferase assay using purified histones, the Mono Q fractions, and tritiated acetyl-CoA as substrate. Top panel, autoradiograph. Bottom panel, Coomassie Blue-stained histone substrates. All HAT activity was specific for histone H4. Note that these fraction numbers are displaced by one (odd numbers), relative to those used for the silver stain and western analyses (even numbers). (d) Endogenous HSC70 co-precipitates with endogenous histone H3. Size exclusion chromatography (S200) of the eH3.1 peaks eluting at (e) 380 mM, (f) 330 mM, and (g) 265 mM KCl from the Mono Q column.
None of the affinity-purified histones bound to a Mono S column (not shown), although pure histones are expected to avidly bind cation exchange resins22. Indeed, eH3.1-containing histone octamers purified to homogeneity from this HeLa lineage tightly bound the cation exchange resin Mono S (not shown), whereas eH3.1 complexes solely bound the anion exchange resin Mono Q (Fig. 2b). This suggested that cellular histones are not present in isolation, but instead are complexed with other proteins that neutralize their positive charge in vivo.
Before proceeding to a large-scale purification, we assessed the effect of cell cycle stage on the assembly and import of cytoplasmic H3.1 histones. To this end, we compared the profile of eH3.1 species obtained after fractionating cells in early or late S-phase (Supplementary Fig. 1b), to that observed in an asynchronous state. No differences were observed among the three cell populations, suggesting a main pathway for the processing and import of newly synthesized histones (compare Fig. 2b with Supplementary Fig. 1b).
Our large-scale purification of cytoplasmic H3.1 complexes revealed four main eH3.1 complexes eluting from the Mono Q column (Fig. 2b). These were numbered (I–IV) to reflect the likely order in the sequence of events during H3.1 transfer, as detailed throughout the text. Fractions were analyzed for suspected enzymatic activity (Fig. 2c) and further purified by gel filtration (Fig. 2e–g).
Fractions 10–16 that eluted at low salt concentrations (170–205 mM KCl) contained the least abundant complex (I) (Fig. 2b), comprising the heat-shock cognate 70 protein (HSC70) and eH3.1, but devoid of histone H4, suggesting that this complex might be the earliest posttranslational eH3.1 species formed at the ribosomal exit. These fractions also contained traces of the C-type HSP40 cofactor DNAJC9 (not shown) that stimulates the ATPase activity of HSC70, thereby accelerating the folding of client proteins23. The co-elution of HSC70 and eH3.1 represented a valid interaction as demonstrated by the co-immunoprecipitation of endogenous HSC70 and histone H3 (Fig. 2d). Due to its low abundance, the complex was barely visible following gel filtration, however the abundant 50 kDa protein that co-eluted within fractions 10–16 from the Mono Q column dissociated from HSP70 on the sizing column (not shown). Interestingly, the eH3.1 histone co-eluting with HSC70 is clearly enriched in H3K9 monomethylation (Fig. 2b), a mark previously detected on newly synthesized histones by mass spectrometry18. Methyltransferase activity, however, was not detected within these fractions (not shown), suggesting that the methylation event is transient and occurs soon after or perhaps concomitant with translation. The co-purification of cytoplasmic HSC70 with histone H3 likely represents a folding rather than a degradation pathway given that the sole detectable PTM was lysine 9 monomethylation, while other normally abundant PTMs (such as trimethylation of lysines 4, 9, or 27) were undetectable by western (not shown). The purified histones migrated as a single sharp band by SDS-PAGE and ubiquitinated species were not detected (not shown).
The Mono Q complex that was next in abundance (II) contained HSP90, tNASP, histone H4, and histone eH3.1 (Fig. 2b). The human NASP gene encodes a full-length transcript that is highly expressed in testes and thus termed ‘testicular’ NASP (tNASP), as well as a splicing variant termed ‘somatic’ NASP (sNASP)24. The tNASP-HSP90 complex (fractions 46–48) shoulders a much more abundant complex (Complex III -fractions 36–42) containing the sNASP variant. To ascertain the interactions between these proteins, fractions 46-48 were analyzed by gel filtration chromatography (Fig. 2e). Indeed, the HSP90-tNASP complex separates by two fractions from the sNASP complex. Complex II remains enriched in H3K9me1, whereas the more abundant complex III is enriched in acetylated histone H4 (Fig. 2b and Fig. 2e). Although the interaction between HSP90 and tNASP has been reported25, the link with histones H3 and H4 was not noted. This complex is likely to be the subsequent step in the processing of histone H3.1 since the H3K9me1 mark remained abundant (Fig. 2b) and importantly, while present, histone H4 was yet to be acetylated (Fig. 2b). Our findings suggest that tNASP is an HSP90 co-chaperone for the assembly of the H3.1-H4 units.
sNASP is a Major H3-H4 Chaperone In Vivo
The most abundant cytoplasmic H3.1 complex (III) consisted of sNASP bound to the eH3.1-H4 histones and co-eluted with the HAT1 holoenzyme (composed of the RbAp46 and HAT1 proteins) (Fig. 2b). All five proteins co-eluted slightly below 200 kDa from size-exclusion chromatography (Fig. 2f and Supplementary Fig. 2a) and appeared to be in a 1:1 stoichiometry based on the size of Complex III. However, this complex eluted as two distinct Mono Q peaks with slight changes in gel filtration elution (but not protein composition) suggesting minor stoichiometric differences or posttranslational modifications. sNASP was one of the most abundant proteins co-eluting with histone eH3.1, as evidenced by western and mass spectrometric analyses (not shown), yet staining by silver was poor when sNASP levels were high (Fig. 2b, fractions 36–42; Fig. 2f, fractions 29–31). This observation has been attributed, in other such cases, to high levels of protein binding silver ions to saturation resulting in a concomitant decrease in the reduction reaction that yields a visible stain on SDS-PAGE gels.
To confirm the composition of Complex III, epitope-tagged sNASP was stably expressed in HEK293F cells, affinity purified, and further fractionated by anion exchange chromatography (Supplementary Fig. 3a). Once again, the main sNASP peak co-eluted with RbAp46, HAT1, and histones H3-H4 (fractions 33–35) with traces of ASF1B. A second minor sNASP peak was evident (fractions 27–29) containing little RbAp46 or HAT1, but most of the co-purified ASF1B. Although sNASP was initially described as a linker histone chaperone, histone H1 was not detected in these cytoplasmic preparations by either western blot or mass spectrometry (not shown). Altogether these results suggest that sNASP is part of a core complex composed of the HAT1 holoenzyme and histones H3-H4 that closely interact with ASF1B.
As expected, the Mono Q fractions containing the cytoplasmic sNASP-HAT1 complex exhibited robust acetyltransferase activity towards histone H4, predominately at lysines 5 and 12 (Fig. 2c). Of note, western analysis revealed a sharp reduction in the levels of H3 lysine 9 monomethylation compared to those within Complexes I and II (Fig. 2b). Mass spectrometric analysis of the histones that co-eluted with Complex III confirmed the overwhelming acetylation of histone H4 and trace amounts of other PTMs (Supplementary Tables 1 and 2). Based on this, we considered Complex III to be the third step in histone H3 processing.
ASF1 Associates with Karyopherins
The last cytoplasmic H3.1 complex (IV) is the one responsible for the nuclear import of the H3.1-H4 histones as it contained the ASF1B chaperone and the importin-4 protein (Fig. 2b, fractions 20–26). Three complexes of identical composition eluted from the Mono Q column, each composed of high levels of ASF1B and importin-4 proteins along with the two histones, and each exhibiting perfect co-elution of these components upon subsequent gel filtration (Fig. 2g; Supplementary Fig. 2b); slight shifts in molecular weights again suggests changes in stoichiometry or posttranslational modifications. Residual peptides from Complex III were sometimes detected, again suggesting a close interaction between the two core complexes. Interestingly, ASF1A was not detected in these fractions by either western blotting or mass spectrometry (not shown).
As in the case of sNASP, stable HEK293F clones expressing FLAG-tagged ASF1B were generated. In agreement with the results above, most of the ASF1B protein eluted with importin-4 and histones H3 and H4 (Supplementary Fig. 3b, fractions 20–22), whereas a less abundant fraction of ASF1B eluted with sNASP, RbAp46, and HAT1 (fractions 26–28). Interestingly, a slower migrating ASF1B species was observed. Mass spectrometric analysis confirmed that the upper band was ASF1B, not ASF1A (not shown). ASF1B posttranslational modifications could account for this migration although it could also be an artifact arising from exogenous expression. The same complex, as well as the dissociation of importin-4 from ASF1B, is also seen in nuclear eH3.1 Mono Q fractions (not shown).
Histone-mediated Chaperone-chaperone Interactions
Since two histone chaperones (sNASP and RbAp46) are in association with histones H3-H4 in Complex III, we next sought to understand their role(s). RbAp46 and the orthologous Drosophila p55 bind a segment of the first alpha helix near the H4 amino-terminal tail26. Since RbAp46 is an integral part of the HAT1 holoenzyme17, we hypothesized that sNASP binding to histones facilitates RbAp46 binding to histone H4, thereby recruiting the associated HAT1 activity. To test this possibility, recombinant FLAG-tagged sNASP was added to recombinant HAT1 holoenzyme (HAT1 and RbAp46) (Fig. 3a). Upon FLAG immunoprecipitation, RbAp46 and HAT1 precipitated with sNASP as a function of the presence of H3-H4. Curiously, the addition of acetyl-CoA to the assembled sNASP-H3-H4-HAT1-RbAp46 complex not only resulted in H4 acetylation, but also stabilized the interaction between sNASP and the HAT1 holoenzyme (Fig. 3a). Recombinant RbAp46 (without HAT1) also co-precipitated with sNASP only in the presence of histones H3-H4 (Supplementary Fig. 4b). A similar finding was observed upon reciprocal immunoprecipitation of the HAT1-RbAp46 holoenzyme (Supplementary Fig. 4c).
Figure 3.
sNASP interacts with HAT1-RbAp46 and ASF1B through histone intermediates. (a) FLAG-tagged sNASP and (b) GST-tagged ASF1B were incubated with reciprocal histone chaperones in the presence or absence of H3-H4 and acetyl-CoA. FLAG immunoprecipitates and GST pull-downs, respectively, were analyzed by western blotting.
That soluble histones are simultaneously bound to sNASP and RbAp46-HAT1 is reminiscent of yeast histones bound to Asf1 and the Vps75-Rtt109 proteins. Although Vps75 is critical for Rtt109 holoenzyme activity, Asf1 has a slight enhancing activity as well27. The same was observed with sNASP. Although RbAp46 has the most stimulating effect on HAT1 activity towards histone H417, we found that sNASP slightly enhanced this effect when pre-bound to histones and purified by gel filtration (Supplementary Fig. 4d).
We next tested if the presence of ASF1B that comprises the final Complex IV would lead to dissociation of sNASP or HAT1. As in the case of sNASP and RpAp46 interaction, the interaction between ASF1B and sNASP was mediated through histones (Fig. 3b). However, ASF1B co-precipitated with histones along with sNASP and the HAT1 holoenzyme. This suggests that other factors or posttranslational modification(s) are required for sNASP and HAT1 dissociation once histones are bound by ASF1B.
sNASP is a Homodimeric Chaperone that Binds H3.1-H4 dimers
Previous analyses suggested that recombinant sNASP homodimerizes28, but the stoichiometry of associatiated histones is unknown. The elution profile of recombinant sNASP from a gel filtration column suggested that it is a homodimer of ~100 kDa (Supplementary Fig. 5a). The tagged recombinant protein has a molecular weight of 53.7 kDa, although the apparent molecular weight by SDS-PAGE is approximately 75 kDa (Fig. 4a). However, while increasing the molar ratio (1:1; 1:2; 1:3) of sNASP:histones, sNASP consistently bound H3-H4 (not H2A-H2B) and consistently eluted within fractions corresponding to a range of ~150 kDa (Supplementary Fig. 5a). The low resolution of gel filtration hampered ascertaining whether one sNASP homodimer binds a single H3-H4 heterodimer (134.1 kDa) or a tetramer (160.9 kDa). To address the makeup of the H3-H4 species in complex with sNASP, we performed the following experiments.
Figure 4.
sNASP homodimerizes but binds H3-H4 heterodimers. (a) Chemical crosslinking of recombinant histones and sNASP or (b) sNASP pre-incubated with H3-H4 at different molar ratios. Crosslinking was performed using wt histones or H3 bearing the indicated point mutations. (c) Sedimentation equilibrium analysis of the complexes between H3-H4 and sNASP. Shown are individual absorbance values at 276 nm recorded at multiple radial positions after equilibrium was reached at 8 krpm (blue), 12 krpm (yellow), and 15 krpm (red). The fitted curves (top graph) are from a single simultaneous fitting of three independent samples at the three speeds. Based on the fixed molecular weights for the postulated species, the data were fitted to the following interaction scheme: (A+A)+B+B⇔A+AB+B⇔(AA)B+B⇔AABB, where A = H3-H4, (AA) is a dimer of A and (A+A) represents self-association, B is the sNASP monomer, AB is the 1:1:1 complex of sNASP:H3:H4, and AABB is its 2:2:2 complex. Bottom graph shows the mean root square deviations.
SDS-PAGE analysis of sNASP that had been crosslinked in vitro corroborated that the protein homodimerizes, considering the apparent 75 kDa molecular weight of HIS-sNASP. This was evidenced by the shift in its migration near 150 kDa, although monomers remained the predominant species (Fig. 4a, Supplementary Fig. 5b–c). Upon crosslinking, wt H3-H4 histones shifted near the 50 kDa mark as bona fide (H3-H4)2 tetramers (Fig. 4a). A 25 kDa dimer population was also detectable. Unlike sNASP, wt (H3-H4)2 exhibited salt-dependency as the tetrameric form predominated over the dimeric form at higher salt concentrations (Supplementary Fig. 5c). The addition of gel-filtration purified wt H3-H4 histones to sNASP followed by crosslinking, altered sNASP migration to just below the 100 kDa and above the 200 kDa marks, consistent with complexes containing 1:1:1 and 2:2:2 (sNASP:H3:H4) stoichiometric ratios, respectively (Fig. 4b, Supplementary Fig. 5b). A third population was at times detected between the 100 and 150 kDa marks, possibly indicating the existence of a complex containing a single sNASP molecule bound to an H3-H4 tetramer (1:2:2 stoichiometry).
The 2:2:2 species might arise from two sNASP molecules each interacting with two H3-H4 dimers or, alternatively, two sNASP molecules binding an (H3-H4)2 tetramer. To differentiate between these two possibilities, we repeated the experiments under conditions non-conducive to tetrameric (H3-H4)2 formation. Five residues are predominantly involved in the H3-H3’ interface responsible for the formation of (H3-H4)2 tetramers2. Interaction through the H113 residue involves hydrogen bonds and its substitution with alanine reduces the tetrameric pool29. We further mutated combinations of C110, L126, and I130 to aspartate to abolish hydrophobic interactions. The H3H113A/C110D/I130D combination abolished the formation of H3-H4 tetramers (Fig. 4a), with weak tetramer formation being visible only under conditions of extensive crosslinking at salt concentrations of 400 mM (Supplementary Fig. 5c–d). In spite of this, these obligate H3-H4 dimers (200 mM NaCl, 100 molar excess BS3) formed the same 1:1:1 and 2:2:2 species as wt histones when increasing amounts of sNASP were added to a fixed amount of histones (Fig. 4b). The addition of increasing amounts of obligate histone dimers onto a fixed concentration of sNASP yielded the same result (Supplementary Fig. 5e). These findings strongly suggest that sNASP dimerizes and forms a complex with two histone H3-H4 dimers.
To corroborate this interpretation, sedimentation equilibrium (SE) analyses were performed by analytical ultracentrifugation (Fig. 4c). For this purpose, we isolated the peak fractions after gel filtration chromatography of HIS-tagged sNASP and (H3-H4)2, as well as pre-mixed HIS-sNASP + wt H3-H4 at 1:1, 1:2 or 2:1 molar ratios. We considered several interaction models using the SEPHAT program30. The SE of our sNASP samples showed self-association with an association constant of about 1× 107 M−1 (data not shown). Based on the data obtained, the most realistic model is depicted in Figure 4c, specifying 1:1:1 (38.5% of sNASP-bound histone species) and 2:2:2 (44.9% of sNASP-bound histone species) complexes, as well as some 1:2:2 (16.6% of sNASP-bound histone species), and small amounts of other species (unbound or aggregated histones or sNASP). This model predicts an association constant of about 3× 107 M−1 for sNASP binding to an H3-H4 dimer to form the 1:1:1 complex. Other models did not account for all crosslinked species that were observed above.
RNAi Analysis of the Replication-dependent Cytosolic Pathway
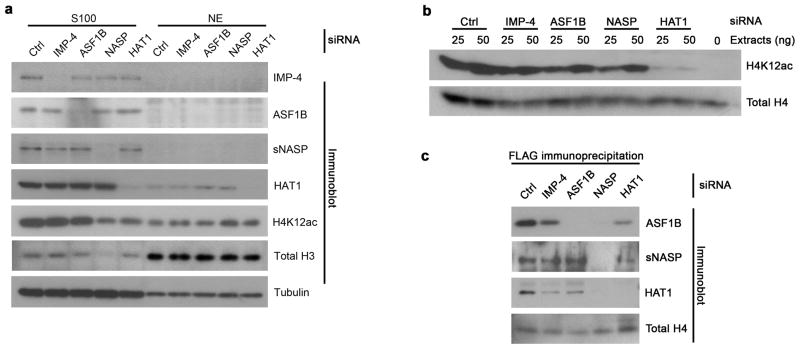
To further scrutinize the pathway delineated through our biochemical analyses, eH3.1 cells were transfected with a scrambled siRNA oligo, or siRNA oligos targeting importin-4, ASF1B, NASP (both isoforms), or HAT1. Compared to the scrambled control, importin-4 and ASF1B knockdowns did not cause an accumulation of cytosolic histones, suggesting some redundancy in the import system (Fig. 5a). Indeed, although importin-4 is the most abundant karyopherin that co-purified with histone eH3.1, mass spectrometry did detect karyopherins β1, β2, and β3 throughout the biochemical purifications (not shown). Similarly, ASF1A may compensate for ASF1B. However, knockdown of NASP proteins resulted in a sharp reduction in cytosolic histone H3 (Fig. 5a), consistent with the role of tNASP in protein folding and sNASP in further processing. Despite the efficient knockdown of HAT1, relatively high levels of acetylated H4 remained 72 hours after transfection, highlighting the importance of low levels of HAT1 activity. By using minute amounts of extracts, HAT1 knockdown did result in far less activity on recombinant histones in vitro (Fig. 5b).
Figure 5.
Importin-4 and ASF1B function downstream of s/tNASP and HAT1 in vivo. (a) Western analysis of H4K12ac levels in cells transfected with siRNA oligos targeting importin-4, ASF1B, NASP (both isoforms), and HAT1. (b) In vitro acetyltransferase activity towards recombinant histones using extracts from cells treated with RNAi against the proteins indicated on top. (c) Immunoprecipitation of histone eH3.1 from cytosolic extracts of RNAi-treated cells.
When histone eH3.1 was immunoprecipitated from extracts of RNAi-treated cells, ASF1B was readily co-precipitated from importin-4 depleted, but not NASP depleted, cells and was greatly reduced in the case of HAT1 depletion (Fig. 5c). This argues that ASF1B functions downstream of sNASP-HAT1 and that ASF1B associates with importin-4 after associating with the fully processed histones. Likewise, sNASP co-precipitated with eH3.1 from cells in which importin-4 or ASF1B was knocked-down, as well as from cells treated with a scrambled control, suggesting that sNASP is not affected by either ASF1 or importin-4 depletion, as it functions upstream. The same is observed with HAT1. However, while NASP RNAi caused a nearly complete loss of HAT1 association with eH3.1, HAT1 RNAi caused only a slight reduction in sNASP co-precipitation with eH3.1, suggesting that sNASP predominantly associates with H3.1 and that HAT1 associates after sNASP has bound the histones.
The sNASP-ASF1 Pathway is Functionally Conserved in Yeast
Despite the low degree of homology between human sNASP and the budding yeast Hif1 protein31, both proteins associate with the HAT1 holoenzyme (yeast Hat1 and Hat2)32. The integrity of the yeast complex requires Hat2 (homologous to RbAp46/48) as its deletion abrogates the interaction of Hat1 with Hif132, 33, but is unaffected by deletion of the asf1, vps75, or rtt109 genes or the importin-4 homologue, kap123 (Fig. 6a, left panel). Asf1 deletion does not affect acetylation of H4K12 but does abrogate H3K56 acetylation (Fig. 6a, right panel), suggesting that H4 acetylation by Hat1 precedes H3 acetylation by Rtt109. The same is observed when Rtt109 is deleted (Supplementary Fig. 6). Again, the loss of H3K56ac is not accompanied by a loss of H4K12ac, although H3K56ac is nearly exclusively nuclear whereas H4 acetylation is well detected in cytoplasmic fractions (not shown). Furthermore, co-immunoprecipitation of Asf1 and the Hat1 complex from the rtt109 null strain suggests that the Hat1-acetylated histones are first transferred to Asf1 for further acetylation by Rtt109 (Supplementary Fig. 6), consistent with Rtt109 being primarily a nuclear protein34.
Figure 6.
Conservation of the H3-H4 pre-deposition pathway in S. cerevisiae. (a) TAP-tagged Hif1 was purified from strains wherein different pre-deposition components were deleted as indicated on top, resolved by SDS-PAGE and silver stained (left panel) or analyzed by western (right panel). (b) TAP-tagged Asf1 was purified from strains lacking Hif1, Hat1, or Hat2 and analyzed by western. (c) Histone dependency for Asf1-Hif1 interactions using TAP-tagged versions of wt Asf1 versus the Asf1 mutant V94R.
To further determine how histones are transferred from Hif1 to Asf1, TAP-tagged Asf1 was immunoprecipitated from strains harboring deletions of hif1, hat1, or hat2. The interaction between Asf1 and the Hat1 complex was dependent on both Hat233 and Hif1, the two histone chaperones in the complex (Fig. 6b). When the endogenous asf1 gene was replaced by the V94R mutant that is deficient in histone binding35, the interactions of Asf1 with Hif1, Hat1, and Hat2 were greatly reduced (Fig. 6c), suggesting that similar to their human counterparts, Asf1 interactions with Hif1 and Hat2 are predominantly mediated through histone intermediates.
Discussion
This study documents the first comprehensive step-by-step transfer of histones from the outset of their posttranslational processing to their nuclear import (Fig. 7). Our studies show that the histones are transferred as dimeric species between cytoplasmic chaperones and that this overall scheme of events is operational in yeast.
Figure 7.
Model for nuclear import of pre-deposition replication-dependent histones. In humans, H3.1 folding is first assisted by the HSC70 chaperone at the ribosomal exit. H3.1 is transferred to HSP90 that, along with the tNASP co-chaperone, assembles it into H3-H4 units. The sNASP chaperone binds H3.1-H4 heterodimers and presents the H4 carboxyl domain to RbAp46 that recruits HAT1 activity. After acetylation of histone H4, the complex is stabilized and the histones transferred to ASF1B. ASF1B associates with importin-4 and the histones are then transported into the nucleus. In budding yeast, Hif1 associates with the H3-H4 histones and the Hat1-Hat2 holoenzyme. The transfer of acetylated histones to Asf1 is mediated through histones.
Assembling Naïve H3-H4 Dimers
To prevent misfolding and aggregation, various protein chaperones congregate near the ribosomal polypeptide exit site36. Although they do not directly bind ribosomes, the Heat-shock Protein 70 family of proteins (of which HSC70 is the major human cytosolic member) are recruited to these sites to adjust hydrophobic segments within a large number of client proteins through binding and release cycles accelerated by ATP hydrolysis36. Upon release by the HSC70 chaperone, a subset of client proteins is transferred to the HSP90 chaperone for further folding. The latter can specialize through associations with a wide-range of co-chaperones, notably a number of proteins bearing the Tetratricopeptide Repeat (TPR) protein-protein interaction domain. NASP is part of the Sim3-Hif1-NASP interrupted TPR (SHNi-TPR) family of proteins31 reported to bind fission yeast H3 and CenH3 histones31, budding yeast H432, human histone H337, and linker histone H124, although these various studies are not always in agreement (discussed below). The association of HSP90 with tNASP suggests that the latter serves as an HSP90 co-chaperone. Interactions between tNASP and HSP90 have been reported, and tNASP has been shown to accelerate the ATPase activity of HSP90 in murine spermatogenic cells25. Based on our results, this step is particularly important since it involves the first posttranslational association between histones H3 and H4, suggesting that HSP90 and tNASP promote the assembly of the heterotypic H3-H4 histone dimer in vivo. Indeed, the knockdown of NASP proteins by RNAi leads to a drastic reduction in overall histone levels in the cytoplasm (Fig. 5a).
sNASP: A Major H3-H4 Chaperone
NASP is a ubiquitous eukaryotic protein sharing a good degree of similarity with the N1/N2-family of H3-H4 histone chaperones38, 24, 28. This chaperone is required for development, as a heterozygous gene deletion results in embryonic lethality in murine models39. Following the cloning of the human NASP gene, both protein isoforms were found to associate with the linker histone H124. The premise that NASP proteins predominantly act as chaperones for linker histones was recently extended by another group, based on the propensity of sNASP to aggregate when mixed with core nucleosomal histones in vitro28. More recently, recombinant sNASP was shown to be capable of binding purified H3-H4 histones in vitro, although with a slightly higher Kd value than histone H137. Interaction with different histone H3 variants is also observed in yeast members of the SHNi-TPR family31, 32. Indeed, we confirmed that sNASP is a major H3-H4 chaperone in human cells. Following the HSP90/tNASP-assisted association of H3-H4, sNASP binds these histones. Since the sNASP complex is the most abundant, it likely keeps soluble pools of histones ready for import and deposition. Yet we find the role of sNASP to be more extensive, given its participation in the processing of naïve histones, such that it presents the histone H4 tail to the HAT1 holoenzyme for acetylation. The entire complex then associates with ASF1B to transfer the histones to this importin-4-associated chaperone. We cannot exclude the possibility that a small proportion of sNASP directly associates with karyopherins, since residual importin-4 was at times seen co-eluting with sNASP after gel filtration. Given the low levels of importin-4 that peaks with the sNASP complex, it is possible that certain conditions may call for sNASP-mediated histone nuclear import as an alternative pathway to ASF1B.
Human RbAp46 and its Drosophila homologue p55 bind to the first helix of the H4 histone fold adjacent to the N-terminal tail26, 40. Structural information for the sNASP chaperone is lacking, but sNASP does preferentially bind histone H3 over histone H4 in vitro37. Indeed, our interaction assays demonstrated that the RbAp46 and sNASP chaperones could simultaneously bind the H3-H4 histone pair. Given that ASF1 binds the carboxyl-terminal regions of both histones13, 14 it is likely that sNASP binds the histone pair predominantly through N-terminal H3 contacts, but structural studies are needed to determine the actual configuration.
Histone modifying enzymes are known to utilize the RbAp46/48 chaperone to catalyze modifications of nucleosomal histones, but the use of two chaperones may be a common strategy for soluble non-nucleosomal intermediates. A similar scenario has been reported in yeast, where two distinct chaperones help catalyze the acetylation of lysine 56 of histone H327. Since sNASP and RbAp46 interact through histone intermediates, the same strategy is likely employed by the sNASP chaperone.
Newly synthesized H3-H4 Histones are Imported as Dimers
Early biochemical studies suggested that during replication, H3-H4 histones segregate onto both leading and lagging strands as intact tetrameric units7. However, this hypothesis has come under scrutiny since epitope-tagged H3 and H4 histones did not co-purify with their more abundant endogenous counterparts from asynchronous cell cultures10, 11, suggesting that new, soluble H3.1-H4 units are predominantly handled as dimers. The chaperoning of histones in the cytoplasm, the nucleus, and on replicating chromatin is likely to differ greatly. The last chaperone that associates with karyopherins to import histone H3-H4 into the nucleus is ASF1, a chaperone that strictly binds histone dimers12, 13, 14. The formation of (H3-H4)2 tetramers upstream of ASF1 in the cytoplasm would therefore seem unlikely. Indeed, our crosslinking and analytical ultracentrifugation experiments demonstrate that each sNASP molecule binds one H3-H4 dimer. As wt (H3-H4)2 tetramers are themselves in equilibrium with H3-H4 dimers, we cannot ascertain if the complexes formed with sNASP in a 1:1:1 stoichiometric ratio result from active breakage of tetramers. However, the relatively high amounts of 1:1:1 complexes as well as the formation of 2:2:2 complexes with obligate H3-H4 dimers does argue that sNASP maintains histones H3 and H4 in a heterodimeric configuration. Therefore, while parental (H3-H4)2 tetramers might not require dissociation during replication, newly synthesized counterparts are assembled from dimers.
Our studies are the first to describe the sequential steps by which the newly synthesized histones H3 and H4 are chaperoned, modified, and transported into the nucleus, a fundamental pathway that is conserved between humans and yeast. As importantly, we have ascertained the enzymatic steps required for the assembly and processing of naïve histones and greatly clarified the roles of cytoplasmic histone chaperones as they help process and transfer their cargo into the nucleus.
Materials and Methods
Constructs and Protein Expression
sNASP was cloned from a HeLa cDNA library into the pET30 plasmid. The ASF1B sequence was subcloned from the pVL1393-ASF1B plasmid into the pGEX4T1 and the pCMV-Tag4 vectors. RBBP7 cDNA was subcloned from the pPK44 vector into the pET28 plasmid. The HAT1 holoenzyme was expressed from baculovirus-infected SF9 cells. Histones were purified as previously described41. Mutations to histone H3 were performed using the Quikchange mutagenesis kit (Stratagene). Recombinant proteins were affinity purified followed by gel filtration.
Cell lines and Tissue Culture
HeLa S3 cells stably expressing eH3.110 were grown as spinner cultures to yield approximately 1 × 1011 cells. To synchronize cultures, cells were grown in the presence of 2 mM thymidine (Sigma) for 19 hrs, washed 3 times with PBS and released for 9 hrs before re-incubating with 2 mM thymidine for another 16 hrs. HEK293F cells stably expressing sNASP or ASF1B were generated by G418 selection of transfected cells.
Biochemical Purification of Cytosolic Complexes
Cells were fractionated as previously described42 41. S100 extracts were affinity purified on an M2 resin (Sigma) and washed twice with 10 resin volumes of BC300 buffer [20 mM Tris, pH 7.6, 300 mM KCl, 0.2 mM EDTA, 10 mM β-mercaptoethanol, 10% Glycerol (v/v), and protease inhibitors] followed by two washes in BC300 + 0.1% NP-40 (v/v) and two washes in BC50 (BC buffer with 50 mM KCl). Complexes were eluted in BC50 containing 125 μg ml−1 FLAG peptide. Eluates were applied to a Mono Q column and eluted through a linear gradient from 50 mM KCl to 750 mM KCl followed by a step elution at 1 M KCl. Peak fractions were applied to a Superdex 200 column and the purified complexes resolved on a 7.5–15% gradient SDS-PAGE. Gels were stained as described43.
Mass Spectrometry
Trypsin in-gel digestion and nano-LC-MSMS were performed as described44. The LC-MSMS data were searched against a human database using a local implementation of X! Tandem45 and confirmed manually. For identification of histone PTMs, gel bands were excised, propionylated, and subjected to a tryptic digest based on Peters et al.46 with modifications. Bands were hydrated in 50 mM NH4HCO3. 2.5 volumes of 30% propionic anhydride:methanol solution (30:70 v/v) were added and the pH adjusted to 7. Samples were incubated at 50°C for 20 min and the reaction stopped with 15 volumes of 25 mM NH4HCO3 and 50% ACN (v/v). Samples were dehydrated with 100% ACN and rehydrated with a trypsin solution. Nano LC-MSMS of trypsin digested histone peptides was based on Trojer et al.47 with modifications using a U-3000 (Dionex) system coupled to LTQ (ThermoFisher). Data was acquired through cyclic series of a full scan followed by zoom scans and MSMS scans of the five most intense ions with a repeat count of two and a dynamic exclusion duration of 60 sec. Relative quantification was performed by determining the peak area of the parent ion in MS full scan or of a unique MSMS fragment.
Enzymatic Activity Assays
An aliquot of each purified fraction was diluted ten fold and one microliter mixed in 25 μl HAT buffer [10 mM Tris, pH 7.6, 250 mM NaCl, 10% Glycerol (v/v), 0.5 mM EDTA, and 1 mM DTT] containing 10 μg core histones and 20 μM [3H] acetyl-CoA at 0.1 μCi ml−1 (Perkin Elmer). Reactions were carried out at 30°C for 30 min and stopped by adding Laemmli buffer. Methyltransferase assays were performed as previously described41.
Yeast Strains and Protein Purification
Strains used in this study are listed in Supplementary Table 3. Strains were constructed using standard yeast media and genetic approaches, and proteins were affinity purified as previously described47.
Antibodies
Polyclonal antibodies against s/tNASP were generated by immunizing two rabbits with the KLH-conjugated Ac-SSSNCVFTDISHLVRK-NH2 peptide. Other antibodies used: ASF1A, HSC70, HSP90, H3, H3K9me1, H4, importin-4, MCM7, p60, RbAp46/48 (Abcam); ASF1B (Cell Signaling); TAP tag (Open Biosystems); FLAG, hemagglutinin tag, β-tubulin (Sigma), HAT1, Histone H1, Myc tag (Santa-Cruz); H3K56ac, H4K5ac, H4K12ac (Upstate); NASP (Proteintech).
Immunoprecipitation and Protein-Protein Interaction Assays
Lysates were pre-cleared with protein A/G beads (Roche) at 4°C for 30 min. 500 μg of cleared cytosolic extracts were incubated with 5 μg antibodies at 4°C overnight. Protein A/G beads were then added and the samples left to rotate for another hour. The beads were washed as described above. For protein-protein interactions, 1 μg of the proteins of interest were mixed in HAT assay buffer with or without [3H] acetyl-CoA at 30°C for 30 min after which either M2 (Sigma), NiNTA (Qiagen), or glutathione (GE Healthcare) beads were added. M2 and glutathione beads were washed as described above and boiled. NiNTA beads were washed twice in the same buffer containing 5 mM, 10 mM, and 30 mM imidazole (Sigma) and eluted with 100 mM imidazole.
Crosslinking Assays
Crosslinking was performed using BS3 (Pierce) in 20 mM HEPES pH 7.9, 10% glycerol (v/v), and 200 mM NaCl, unless otherwise indicated. Protein concentration was 1.5 μM each for sNASP (based on monomeric conformation) and H3-H4 tetramers. Histones and sNASP were pre-incubated at 4°C for 30 min for complexes to form. Crosslinking was allowed to proceed for 1 h at 4°C and then stopped by the addition of Tris pH 7.5 to 50 mM.
Analytical Ultracentrifugation
Sedimentation Equilibrium analyses were performed on a Beckman Coulter ProteomeLab XL-A/XL-I using an 8-place An-50Ti rotor. Samples were prepared as described above in 250 mM NaCl and 10 mM Tris, pH 7.6. Input concentrations for sNASP, (H3-H4)2 tetramers or sNASP-H3-H4 complexes ranged between 0.8–1.2 μM. Absorbance at 276 nm and interference patterns were recorded after reaching equilibrium at 5150 × g, 11600 × g and 18100 × g. All analyses were performed at 4°C. Data was analyzed using the Sedfit and Sedphat softwares30.
RNAi
Cells were transfected using the RNAiMAX reagent (Invitrogen) using 30 μM pooled siRNA oligos (Dharmacon) for 72 hrs. Importin-4 target sequences: GCAUUUCGCUGUACAAGUU, AGUCAGAGGUGCCGGUCAU, CCUCGCAAGUUGUACGCAA, AUGGAGCACCUGCGGGAAU. ASF1B target sequences: GCAGGGAGACACAUGUUUG, AGUGGAAGAUCAUUUAUGU, GACCUGGAGUGGAAGAUCA, CGGACGACCUGGAGUGGAA. NASP target sequences: GGAAAUCACUUCUGGAGUU, GGAAGCAGCUAGUCUUUUA, CCGAAGAAAUGCCAAAUGA, GGAACUGCUACCCGAAAUU. HAT1 target sequences: GCACAAACACGAAUGAUUU, GAAGAUUACCGGCGUGUUA, GCUACAGACUGGAUAUUAA, CUAAUUAGCCCAUAUAAGA.
Supplementary Material
Supplementary Figure 1. Cytoplasmic H3.1 histones are equally processed throughout S-phase. (a) Flow cytometric analysis of HeLa S3 spinner cultures expressing epitope-tagged histone H3.1 and synchronized by a double-thymidine block. (b) Small-scale purification of eH3.1 from cells in early S (3 hr after release) and late S (6 hr after release) show no difference with asynchronous cells expressing epitope tagged histone (Fig. 2).
Supplementary Figure 2. Size exclusion chromatography (S200) of the H3.1 peaks eluting at (a) 360 mM (Complex III) and (b) 250 mM KCl (Complex IV), from the Mono Q column.
Supplementary Figure 3. (a) Purification of sNASP from HEK293F cells stably expressing an epitope tagged version of the protein. After affinity purification from cytoplasmic fractions, sNASP complexes were fractionated by anion exchange chromatography. Most sNASP associates with the HAT1 holoenzyme and contains little ASF1B (blue frame). A minor and distinct sNASP population predominantly associates with ASF1B, but contains little HAT1 holoenzyme (green frame). Importin-4 was undetectable. (b) Purification of cytoplasmic ASF1B from a HEK293F lineage stably expressing an epitope tagged version of the protein. After affinity purification, the ASF1B complexes were fractionated by anion exchange chromatography. Most of ASF1B associates with the importin-4 karyopherin that peaks within fractions that contain lesser amounts of sNASP or HAT1 holoenzyme (green frame). A minor and distinct ASF1B population predominantly associates with sNASP and the HAT1 holoenzyme, but contains relatively little importin-4 (blue frame). Note that the samples used for western versus silver staining are one fraction apart.
Supplementary Figure 4. (a) Coomassie Blue-stained SDS-PAGE of recombinant histones and chaperones used for protein-protein interaction assays. (b) RbAp46 and sNASP interact through histone intermediates. Recombinant FLAG-sNASP and untagged RbAp46 were incubated with or without histones and immunoprecipitated using anti-FLAG antibody and analyzed by western. (c) HIS-tagged HAT1-RbAp46 holoenzyme was incubated with FLAG-tagged sNASP in the presence or absence of H3-H4 and acetyl-CoA and isolated on NiNTA beads prior to western blotting. (d) HAT assay. 5 or 10 μg of recombinant histones or histones pre-bound to sNASP and selected by gel filtration were incubated with increasing amounts of baculovirus-expressed, affinity and gel filtration-purified HAT-RbAp46 holoenzyme.
Supplementary Figure 5. (a) Gel filtration analysis of recombinant sNASP alone (right panel) or pre-incubated with a three-fold molar excess of core histones (left panel). (b) Silver-stained SDS-PAGE of chemically crosslinked recombinant (wt) H3-H4 histones and sNASP at 100 mM NaCl. (c) Western analysis of crosslinked wt, mutant H3-H4 histones, or sNASP at varying salt concentrations for 1 hr with 100-fold excess BS3. (d) Prolonged crosslinking (2 hr) of wt or mutant H3-H4 histones and sNASP at high salt concentration with varying fold excess of BS3. (e) Crosslinking (1 hr, 100-fold excess BS3) of obligate H3-H4 dimers following titration of sNASP.
Supplementary Figure 6. rtt109 deletion does not affect upstream H4 acetylation by Hat1. Asf1 was purified from wt and rtt109 deletion strains. In the latter case, H4 acetylation levels were not disrupted and Asf1 still associated with pre-deposition histones, but H3 was no longer acetylated on lysine 56.
Acknowledgments
Funding for this project was provided by the Howard Hughes Medical Institute and NIH grants GM064844 and R37GM037120 (D.R.), the Natural Sciences and Engineering Research Council of Canada (E.I.C. and W.H.W.K.), the Canadian Cancer Society Research Institute (J.F.G.), and the Deutsche Akademie der Naturforscher Leopoldina (Leopoldina Fellowship Program, LPDS 2009-5, P.V.). The eH3.1 HeLa S3 cells were kindly provided by Drs. Hideaki Tagami and Geneviève Almouzni (Institut Curie). The HAT1-RbAp46, wt ASF1B and V94R Asf1 constructs were kind gifts from Drs. Bruce Stillman (Cold Spring Harbor Laboratory), Herman Silljé (Max Planck Institute) and Carl Mann (CEA/Saclay) respectively. The baculoviruses expressing human HAT1 and RbAp46 were kindly provided by Dr. Robert Kingston (Harvard Medical School). We also thank Shwetal Mehta and Stephanie Kim for technical support with tissue culture, Guoching Zhong for help with yeast TAP purification, and Jin Zhang for a copious amount of histones. We are grateful to Dr. Lynne Vales for comments on the manuscript.
Footnotes
Author Contributions
E.I.C. performed the tissue culture-related work and biochemical purification of eH3.1, sNASP and ASF1B; protein interaction and enzymatic assays, with assistance from Z.G; sub-cloning and mutagenesis; crosslinking experiments, with the assistance of P.V.; AUC runs, with the assistance of L.D.; and RNAi experiments. J.F. performed the work in S. cerevisiae with assistance from W.H.W.K. and H.S. G.L. cloned sNASP and generated stable sNASP clones. H.Z. performed the mass spectrometry analyses. The manuscript was written by E.I.C. and D.R. with assistance from the other authors.
References
- 1.Eickbush TH, Moudrianakis EN. The histone core complex: an octamer assembled by two sets of protein-protein interactions. Biochemistry. 1978;17:4955–64. doi: 10.1021/bi00616a016. [DOI] [PubMed] [Google Scholar]
- 2.Luger K, Mader AW, Richmond RK, Sargent DF, Richmond TJ. Crystal structure of the nucleosome core particle at 2.8 A resolution. Nature. 1997;389:251–60. doi: 10.1038/38444. [DOI] [PubMed] [Google Scholar]
- 3.Campos EI, Reinberg D. Histones: Annotating Chromatin. Annual Review of Genetics. 2009;43:559–599. doi: 10.1146/annurev.genet.032608.103928. [DOI] [PubMed] [Google Scholar]
- 4.Hake SB, Allis CD. Histone H3 variants and their potential role in indexing mammalian genomes: the “H3 barcode hypothesis”. Proc Natl Acad Sci U S A. 2006;103:6428–35. doi: 10.1073/pnas.0600803103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Koessler H, Doenecke D, Albig W. Aberrant expression pattern of replication-dependent histone h3 subtype genes in human tumor cell lines. DNA Cell Biol. 2003;22:233–41. doi: 10.1089/104454903321908629. [DOI] [PubMed] [Google Scholar]
- 6.Sogo JM, Stahl H, Koller T, Knippers R. Structure of replicating simian virus 40 minichromosomes. The replication fork, core histone segregation and terminal structures. J Mol Biol. 1986;189:189–204. doi: 10.1016/0022-2836(86)90390-6. [DOI] [PubMed] [Google Scholar]
- 7.Annunziato AT. Split decision: what happens to nucleosomes during DNA replication? J Biol Chem. 2005;280:12065–8. doi: 10.1074/jbc.R400039200. [DOI] [PubMed] [Google Scholar]
- 8.Xu M, et al. Partitioning of histone H3-H4 tetramers during DNA replication-dependent chromatin assembly. Science. 2010;328:94–8. doi: 10.1126/science.1178994. [DOI] [PubMed] [Google Scholar]
- 9.Gruss C, Wu J, Koller T, Sogo JM. Disruption of the nucleosomes at the replication fork. Embo J. 1993;12:4533–45. doi: 10.1002/j.1460-2075.1993.tb06142.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Tagami H, Ray-Gallet D, Almouzni G, Nakatani Y. Histone H3.1 and H3.3 complexes mediate nucleosome assembly pathways dependent or independent of DNA synthesis. Cell. 2004;116:51–61. doi: 10.1016/s0092-8674(03)01064-x. [DOI] [PubMed] [Google Scholar]
- 11.Benson LJ, et al. Modifications of H3 and H4 during chromatin replication, nucleosome assembly, and histone exchange. J Biol Chem. 2006;281:9287–96. doi: 10.1074/jbc.M512956200. [DOI] [PubMed] [Google Scholar]
- 12.English CM, Maluf NK, Tripet B, Churchill ME, Tyler JK. ASF1 binds to a heterodimer of histones H3 and H4: a two-step mechanism for the assembly of the H3-H4 heterotetramer on DNA. Biochemistry. 2005;44:13673–82. doi: 10.1021/bi051333h. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.English CM, Adkins MW, Carson JJ, Churchill ME, Tyler JK. Structural basis for the histone chaperone activity of Asf1. Cell. 2006;127:495–508. doi: 10.1016/j.cell.2006.08.047. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Natsume R, et al. Structure and function of the histone chaperone CIA/ASF1 complexed with histones H3 and H4. Nature. 2007;446:338–41. doi: 10.1038/nature05613. [DOI] [PubMed] [Google Scholar]
- 15.Groth A, et al. Regulation of replication fork progression through histone supply and demand. Science. 2007;318:1928–31. doi: 10.1126/science.1148992. [DOI] [PubMed] [Google Scholar]
- 16.Jasencakova Z, et al. Replication stress interferes with histone recycling and predeposition marking of new histones. Mol Cell. 2010;37:736–43. doi: 10.1016/j.molcel.2010.01.033. [DOI] [PubMed] [Google Scholar]
- 17.Verreault A, Kaufman PD, Kobayashi R, Stillman B. Nucleosomal DNA regulates the core-histone-binding subunit of the human Hat1 acetyltransferase. Curr Biol. 1998;8:96–108. doi: 10.1016/s0960-9822(98)70040-5. [DOI] [PubMed] [Google Scholar]
- 18.Loyola A, Bonaldi T, Roche D, Imhof A, Almouzni G. PTMs on H3 variants before chromatin assembly potentiate their final epigenetic state. Mol Cell. 2006;24:309–16. doi: 10.1016/j.molcel.2006.08.019. [DOI] [PubMed] [Google Scholar]
- 19.Smith S, Stillman B. Purification and characterization of CAF-I, a human cell factor required for chromatin assembly during DNA replication in vitro. Cell. 1989;58:15–25. doi: 10.1016/0092-8674(89)90398-x. [DOI] [PubMed] [Google Scholar]
- 20.Tyler JK, et al. Interaction between the Drosophila CAF-1 and ASF1 chromatin assembly factors. Mol Cell Biol. 2001;21:6574–84. doi: 10.1128/MCB.21.19.6574-6584.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Mello JA, et al. Human Asf1 and CAF-1 interact and synergize in a repair-coupled nucleosome assembly pathway. EMBO Rep. 2002;3:329–34. doi: 10.1093/embo-reports/kvf068. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Crampton CF, Moore S, Stein WH. Chromatographic fractionation of calf thymus histone. J Biol Chem. 1955;215:787–801. [PubMed] [Google Scholar]
- 23.Han C, et al. HDJC9, a novel human type C DnaJ/HSP40 member interacts with and cochaperones HSP70 through the J domain. Biochem Biophys Res Commun. 2007;353:280–5. doi: 10.1016/j.bbrc.2006.12.013. [DOI] [PubMed] [Google Scholar]
- 24.Richardson RT, et al. Characterization of the histone H1-binding protein, NASP, as a cell cycle-regulated somatic protein. J Biol Chem. 2000;275:30378–86. doi: 10.1074/jbc.M003781200. [DOI] [PubMed] [Google Scholar]
- 25.Alekseev OM, Widgren EE, Richardson RT, O’Rand MG. Association of NASP with HSP90 in mouse spermatogenic cells: stimulation of ATPase activity and transport of linker histones into nuclei. J Biol Chem. 2005;280:2904–11. doi: 10.1074/jbc.M410397200. [DOI] [PubMed] [Google Scholar]
- 26.Song JJ, Garlick JD, Kingston RE. Structural basis of histone H4 recognition by p55. Genes Dev. 2008;22:1313–8. doi: 10.1101/gad.1653308. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Han J, Zhou H, Li Z, Xu RM, Zhang Z. Acetylation of lysine 56 of histone H3 catalyzed by RTT109 and regulated by ASF1 is required for replisome integrity. J Biol Chem. 2007;282:28587–96. doi: 10.1074/jbc.M702496200. [DOI] [PubMed] [Google Scholar]
- 28.Finn RM, Browne K, Hodgson KC, Ausio J. sNASP, a histone H1-specific eukaryotic chaperone dimer that facilitates chromatin assembly. Biophys J. 2008;95:1314–25. doi: 10.1529/biophysj.108.130021. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Andrews AJ, Downing G, Brown K, Park YJ, Luger K. A thermodynamic model for Nap1-histone interactions. J Biol Chem. 2008;283:32412–8. doi: 10.1074/jbc.M805918200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Vistica J, et al. Sedimentation equilibrium analysis of protein interactions with global implicit mass conservation constraints and systematic noise decomposition. Anal Biochem. 2004;326:234–56. doi: 10.1016/j.ab.2003.12.014. [DOI] [PubMed] [Google Scholar]
- 31.Dunleavy EM, et al. A NASP (N1/N2)-related protein, Sim3, binds CENP-A and is required for its deposition at fission yeast centromeres. Mol Cell. 2007;28:1029–44. doi: 10.1016/j.molcel.2007.10.010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Poveda A, et al. Hif1 is a component of yeast histone acetyltransferase B, a complex mainly localized in the nucleus. J Biol Chem. 2004;279:16033–43. doi: 10.1074/jbc.M314228200. [DOI] [PubMed] [Google Scholar]
- 33.Fillingham J, et al. Chaperone control of the activity and specificity of the histone H3 acetyltransferase Rtt109. Mol Cell Biol. 2008;28:4342–53. doi: 10.1128/MCB.00182-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Sundin BA, Chiu CH, Riffle M, Davis TN, Muller EG. Localization of proteins that are coordinately expressed with Cln2 during the cell cycle. Yeast. 2004;21:793–800. doi: 10.1002/yea.1133. [DOI] [PubMed] [Google Scholar]
- 35.Mousson F, et al. Structural basis for the interaction of Asf1 with histone H3 and its functional implications. Proc Natl Acad Sci U S A. 2005;102:5975–80. doi: 10.1073/pnas.0500149102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Hartl FU, Hayer-Hartl M. Converging concepts of protein folding in vitro and in vivo. Nat Struct Mol Biol. 2009;16:574–81. doi: 10.1038/nsmb.1591. [DOI] [PubMed] [Google Scholar]
- 37.Wang H, Walsh ST, Parthun MR. Expanded binding specificity of the human histone chaperone NASP. Nucleic Acids Res. 2008;36:5763–72. doi: 10.1093/nar/gkn574. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Welch JE, Zimmerman LJ, Joseph DR, O’Rand MG. Characterization of a sperm-specific nuclear autoantigenic protein. I. Complete sequence and homology with the Xenopus protein, N1/N2. Biol Reprod. 1990;43:559–68. doi: 10.1095/biolreprod43.4.559. [DOI] [PubMed] [Google Scholar]
- 39.Richardson RT, et al. Nuclear autoantigenic sperm protein (NASP), a linker histone chaperone that is required for cell proliferation. J Biol Chem. 2006;281:21526–34. doi: 10.1074/jbc.M603816200. [DOI] [PubMed] [Google Scholar]
- 40.Murzina NV, et al. Structural basis for the recognition of histone H4 by the histone-chaperone RbAp46. Structure. 2008;16:1077–85. doi: 10.1016/j.str.2008.05.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Nishioka K, Reinberg D. Methods and tips for the purification of human histone methyltransferases. Methods. 2003;31:49–58. doi: 10.1016/s1046-2023(03)00087-2. [DOI] [PubMed] [Google Scholar]
- 42.Dignam JD, Martin PL, Shastry BS, Roeder RG. Eukaryotic gene transcription with purified components. Methods Enzymol. 1983;101:582–98. doi: 10.1016/0076-6879(83)01039-3. [DOI] [PubMed] [Google Scholar]
- 43.Shevchenko A, Wilm M, Vorm O, Mann M. Mass spectrometric sequencing of proteins silver-stained polyacrylamide gels. Anal Chem. 1996;68:850–8. doi: 10.1021/ac950914h. [DOI] [PubMed] [Google Scholar]
- 44.Pungaliya P, et al. TOPORS functions as a SUMO-1 E3 ligase for chromatin-modifying proteins. J Proteome Res. 2007;6:3918–23. doi: 10.1021/pr0703674. [DOI] [PubMed] [Google Scholar]
- 45.Craig R, Cortens JP, Beavis RC. Open source system for analyzing, validating, and storing protein identification data. J Proteome Res. 2004;3:1234–42. doi: 10.1021/pr049882h. [DOI] [PubMed] [Google Scholar]
- 46.Peters AH, et al. Partitioning and plasticity of repressive histone methylation states in mammalian chromatin. Mol Cell. 2003;12:1577–89. doi: 10.1016/s1097-2765(03)00477-5. [DOI] [PubMed] [Google Scholar]
- 47.Trojer P, et al. Dynamic Histone H1 Isotype 4 Methylation and Demethylation by Histone Lysine Methyltransferase G9a/KMT1C and the Jumonji Domain-containing JMJD2/KDM4 Proteins. J Biol Chem. 2009;284:8395–405. doi: 10.1074/jbc.M807818200. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Supplementary Figure 1. Cytoplasmic H3.1 histones are equally processed throughout S-phase. (a) Flow cytometric analysis of HeLa S3 spinner cultures expressing epitope-tagged histone H3.1 and synchronized by a double-thymidine block. (b) Small-scale purification of eH3.1 from cells in early S (3 hr after release) and late S (6 hr after release) show no difference with asynchronous cells expressing epitope tagged histone (Fig. 2).
Supplementary Figure 2. Size exclusion chromatography (S200) of the H3.1 peaks eluting at (a) 360 mM (Complex III) and (b) 250 mM KCl (Complex IV), from the Mono Q column.
Supplementary Figure 3. (a) Purification of sNASP from HEK293F cells stably expressing an epitope tagged version of the protein. After affinity purification from cytoplasmic fractions, sNASP complexes were fractionated by anion exchange chromatography. Most sNASP associates with the HAT1 holoenzyme and contains little ASF1B (blue frame). A minor and distinct sNASP population predominantly associates with ASF1B, but contains little HAT1 holoenzyme (green frame). Importin-4 was undetectable. (b) Purification of cytoplasmic ASF1B from a HEK293F lineage stably expressing an epitope tagged version of the protein. After affinity purification, the ASF1B complexes were fractionated by anion exchange chromatography. Most of ASF1B associates with the importin-4 karyopherin that peaks within fractions that contain lesser amounts of sNASP or HAT1 holoenzyme (green frame). A minor and distinct ASF1B population predominantly associates with sNASP and the HAT1 holoenzyme, but contains relatively little importin-4 (blue frame). Note that the samples used for western versus silver staining are one fraction apart.
Supplementary Figure 4. (a) Coomassie Blue-stained SDS-PAGE of recombinant histones and chaperones used for protein-protein interaction assays. (b) RbAp46 and sNASP interact through histone intermediates. Recombinant FLAG-sNASP and untagged RbAp46 were incubated with or without histones and immunoprecipitated using anti-FLAG antibody and analyzed by western. (c) HIS-tagged HAT1-RbAp46 holoenzyme was incubated with FLAG-tagged sNASP in the presence or absence of H3-H4 and acetyl-CoA and isolated on NiNTA beads prior to western blotting. (d) HAT assay. 5 or 10 μg of recombinant histones or histones pre-bound to sNASP and selected by gel filtration were incubated with increasing amounts of baculovirus-expressed, affinity and gel filtration-purified HAT-RbAp46 holoenzyme.
Supplementary Figure 5. (a) Gel filtration analysis of recombinant sNASP alone (right panel) or pre-incubated with a three-fold molar excess of core histones (left panel). (b) Silver-stained SDS-PAGE of chemically crosslinked recombinant (wt) H3-H4 histones and sNASP at 100 mM NaCl. (c) Western analysis of crosslinked wt, mutant H3-H4 histones, or sNASP at varying salt concentrations for 1 hr with 100-fold excess BS3. (d) Prolonged crosslinking (2 hr) of wt or mutant H3-H4 histones and sNASP at high salt concentration with varying fold excess of BS3. (e) Crosslinking (1 hr, 100-fold excess BS3) of obligate H3-H4 dimers following titration of sNASP.
Supplementary Figure 6. rtt109 deletion does not affect upstream H4 acetylation by Hat1. Asf1 was purified from wt and rtt109 deletion strains. In the latter case, H4 acetylation levels were not disrupted and Asf1 still associated with pre-deposition histones, but H3 was no longer acetylated on lysine 56.