Abstract
Pattern formation in Drosophila embryogenesis has been widely investigated as a developmental and evolutionary model of robustness. To ask whether genetic variation for pattern formation is suppressed in this system artificial selection for divergent egg size was used to challenge the scaling of even-skipped pattern formation in mitotic cycle 14 (stage 5) embryos of Drosophila melanogaster. Three-dimensional confocal imaging revealed shifts in the allometry of eve pair-rule stripes along both A-P and D-V axes as a correlated response to egg size selection, indicating the availability of genetic variation for this buffered trait. Environmental perturbation was not required for the manifestation of this variation. The number of nuclei at the cellular blastoderm stage also changed in response to selection, with large-egg selected lines having more than 1000 additional nuclei relative to small-egg lines. This increase in nuclear number in larger eggs does not scale with egg size, however, as nuclear density is inversely correlated with egg length. Nuclear density varies along the A-P axis but does not correlate with the shift in eve stripe allometry between the selection treatments. Despite its macroevolutionary conservation, both eve stripe patterning and blastoderm cell number vary genetically both within and between closely related species.
Keywords: robustness, segmentation patterning, cell number
Introduction
Many developmental systems buffer the effects of genetic, environmental, and stochastic variation in order to achieve a stereotypical outcome. The early segmentation gene patterning system in Drosophila development is a well-studied example of a robust developmental system. Two properties of robustness have been investigated in this system, precision (positional error) and scaling (relative placement) of expression domains. The anterior-posterior (A-P) axis in D. melanogaster is initially established by maternal mRNAs deposited in the egg (Nusslein-Volhard et al. 1987). The subsequent translation and diffusion of these morphogens, especially Bicoid (Bcd), generates concentration gradients that begin the cascade of developmental events resulting in a segmented morphology. Bcd activates the expression of hunchback (hb) (Tautz 1988) which by mechanisms that remain under debate, produces a precise concentration gradient along the A-P axis during cycles 10–13 (Gregor et al. 2007b). Slightly later in blastoderm development, the pair-rule gene, even-skipped (eve) provides a convenient ‘readout’ of developmental robustness of maternal and gap gene expression. eve is expressed at roughly uniform levels after 11 mitotic divisions (Frasch et al. 1987). After 13 synchronous nuclear divisions (cycle 14 embryos) a periodic pattern appears as seven transverse stripes along the A-P axis of the embryo indicating odd-numbered parasegments (Nusslein-Volhard et al. 1985; Frasch and Levine 1987). Several models to account for precision of gene expression also address the issue of scaling — the placement of domain borders at the same relative A-P position in Drosophila embryos varying in length (Houchmandzadeh et al. 2002; Howard and Rein ten Wolde 2005; Gregor et al. 2007a; Gregor et al. 2007b; He et al. 2008; Manu et al. 2009). Gregor et al. (Gregor et al. 2007b), in particular, proposed that if Bcd is degraded while it resides within the nucleus, this intra-nuclear process could lead to the establishment of a Bcd gradient that scales with egg length. The scaling provided by intra-nuclear degradation, however, requires the additional assumption that the number of nuclei in the blastoderm remain constant with respect to embryo length. Developmental robustness has also been a subject of intense interest to evolutionary biologists, beginning with Waddington (Waddington 1942, 1957). Genetic variation in egg size (length or volume) is abundant in natural populations of D. melanogaster, and varies clinally along latitudinal transects across multiple continents, indicating that this is likely an adaptive response (Azevedo et al. 1996). Lott et al. (Lott et al. 2007) found that the eve stripe pattern in three strains of D. melanogaster was accurately scaled in each relative to embryo length (EL) despite a difference among strains of ~25% EL, even when these strains were crossed to create novel assortments of genomes in differently sized eggs. In contrast, the same authors showed that eve stripe position has evolved between the closely related species D. melanogaster, D. sechellia, and D. simulans (Lott et al. 2007), three species that also differ in egg length.
The contradictory evidence for invariant placement of lateral eve stripes in the face of genetic variation for egg length, but rapid evolution of stripe position between closely related species, motivated us to carry out forced selection for egg size to investigate whether genetic variation for spatial placement of segmentation genes would be revealed as a correlated response to artificial selection for egg size. In order to challenge the strict scaling of eve stripes we generated replicate lines of D. melanogaster selected for large and small egg size by performing divergent selection on a laboratory caged population. With 2-photon microscopy we produced complete 3D representations of mid-blastoderm (cycle 14) embryos. The data allowed us to measure shifts in eve expression in selected lines at a spatial resolution not previously possible, and to investigate variation in cell density, a critical parameter in the nuclear trapping model for scaling of expression patterns in morphogenetic fields.
Materials and Methods
Fly collection and selection
We performed artificial selection for divergent egg volume using population cages of D. melanogaster that originated from a collection of 120 wild-caught females in central Illinois, USA. After 10 generations of random mating, we established three replicate lines for each treatment (large-egg, small-egg, and control) and flies were maintained at 25°C, fed standard fly media, and kept on a 16d life cycle. Freshly laid eggs were collected, digitally photographed and measured using OpenLab 3.1.7. Volume was calculated assuming a prolate spheroid and the 110 largest or smallest embryos (19.1% truncation selection) from each line were retained in the population. Selection continued from January 2007 to June 2008. Final embryo volumes were approximately normally distributed, showing a slightly positive skew. Throughout this work normality was evaluated by visual inspection of data. We tested post-selection egg sizes using a mixed model ANOVA with lines nested within treatments and random effects at the level of line, followed by a Student’s t-test. Statistical analyses of selected embryo size, and all other comparisons discussed here, were performed in JMP 8.0 (SAS Institute) with α = 0.05.
in situ hybridization, image collection, and processing
Embryos were collected, fixed, and fluorescently stained to label the mRNA of the gap gene giant and the pair-rule gene eve using the techniques of Lott et al. (Lott et al. 2007), followed by RNase treatment and mounting as outlined in Luengo Hendriks et al. (Luengo Hendriks et al. 2006). Sytox Green (Invitrogen) was used for the nuclear stain. 3D images of cycle 14 embryos (substages 4–8 on the Fly-Ex website http://flyex.ams.sunysb.edu/flyex) were collected using a Leica SP5 2-photon microscope. Image analysis was performed using PointCloudToolbox (PCT) software (http://bdtnp.lbl.gov/Fly-Net/bioimaging.jsp) (Keranen et al. 2006; Luengo Hendriks et al. 2006; Fowlkes et al. 2008). We input confocal stacks of individual embryos and used PCT to localize and count nuclei, to dorsoventrally (D-V) align all embryos, and to identify locations of the anterior and posterior boundaries of the seven eve stripes in 3 dimensions. Dorsoventral alignment was performed interactively in PCT by visually identifying the ventral eve and gt expression patterns while scrolling through an “unrolled” embryo (http://bdtnp.lbl.gov/Fly-Net/bioimaging.jsp?w=pcmaker). Regarding the stripe border positions, output for each embryo consisted of a 16 ×14 matrix with the 14 columns reporting the relative position of the anterior and posterior of each of the seven eve stripes (in units of % EL). The 16 rows report mean values in 16 regions (strips) around the circumference of the embryo. The circumference of the embryo at 50% EL is divided into 16 equally wide strips beginning at the ventral-most region (row 1 in the matrix) and traveling 360° around the periphery of the ellipsoid embryo. Therefore, each strip or region represents ~22.5° of the whole.
Once matrices were produced describing stripe boundaries for each embryo, we extracted the specific rows from each matrix that represented dorsal, ventral and lateral aspects of the embryo. We used a fully nested mixed model ANOVA to test stripe positions with lines nested within treatments and random effects at the level of line. Stripe position is known to change over the 45–50 minutes of cycle 14 so we included developmental age as a fixed effect to account for this shift according to the method used by Lott et al. (Lott et al. 2007). Since the stripe border positions in this dataset have been normalized for egg length, and therefore are percentages, we performed arcsine transformation of the data and repeated the analysis to check for any influence on the outcome.
Total number and mean density of nuclei
For analysis of total number of nuclei in selected lines we used the PCT data produced from Sytox Green nuclear stain only (n=182). Total number of nuclei was modeled using a fully nested mixed model ANOVA (with lines nested within treatments and random effects at the level of line) followed by a Student’s t-test. In order to extend the range of both nuclear number and embryo size we prepared additional embryos using the identical protocol. We selected D. sechellia-Robertson (n = 20) and D. simulans-FC (n = 20) because they represent large- and small-egg species, respectively. We also included three strains of D. melanogaster, w1118, and two genetically distinct strains fixed for different embryo sizes (small-egg Fra, n = 22 and large-egg Ind, n = 19) (Lott et al. 2007)). We tested for differences in the total number of nuclei among these five groups using a one-way ANOVA followed by a Student’s t-test.
The distribution of mean densities of nuclei (number/μm2) in the artificially selected treatments failed to meet assumptions of normality and were analyzed using a nonparametric Kruskal-Wallis test followed by unplanned comparison testing of the Least Significant Difference (LSD). We considered the possibility that shrinkage due to fixation, in situ hybridization, dehydration, and mounting could affect the estimates of surface area, and therefore density. We compared estimates of length and surface area made from measurements of living embryos with those reported by PCT from all three treatments and found no significant difference in the amount of shrinkage among treatments (Fig. S1).
Next, we used the median distance to neighbor nuclei (an indicator of how closely nuclei were packed) as a proxy for nuclear density. PCT processing of confocal image stacks estimates the surface through the centroid of each nucleus at the periphery. A Voronoi tessellation is then used to determine the “cells” that share an edge. Median distance to neighboring nuclei estimates the distance from the centroid of each nucleus to every nucleus with a shared border, thereby estimating how closely packed nuclei are in each embryo. These estimates are absolute distances between nuclei but our estimates are on fixed and Depex™-mounted embryos and are not intended to represent distances between nuclei in living embryos. These estimates did not meet assumptions of normality and were analyzed using a nonparametric Kruskal-Wallis test followed by unplanned comparison testing of the LSD.
Results
Selection
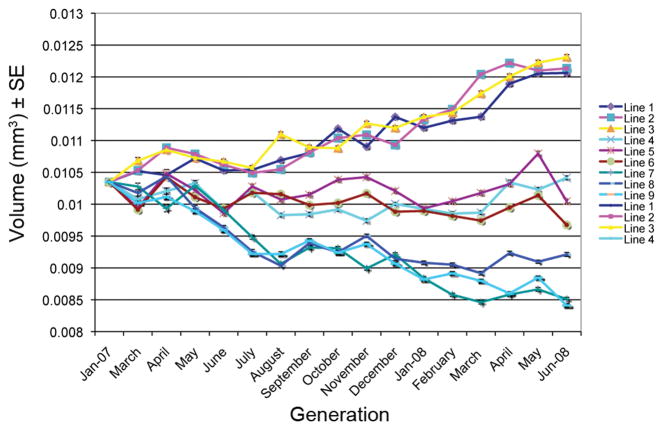
Selection resulted in significant changes in embryo volume relative to control lines (F2,6 = 76.71, P < 0.0001, n = 5169). Means (±SE) of the large-egg, control, and small-egg lines were 12.1 (0.02), 10.0 (0.02), 8.7 (0.02) × 10−3 mm3, respectively. The mean increased 1.5 σP in large-egg lines and decreased 1.2 σP in small-egg lines relative to the base population. Divergent selection for egg volume produced a relatively linear response in egg size in both directions during the course of the experiment (Fig. 1). The response was consistent among replicates across the experiment within both selection treatments, as well as in control lines. After the cessation of direct selection we established inbred sub-lines derived from the large- and small-egg caged populations and these lines have retained the egg size phenotype after 20–21 generations of brother-sister inbreeding (Table S1.). The continuous, consistent response among replicates, indicating that genetic variation for the trait did not appear to be exhausted, and the stability of egg size following the cessation of selection, suggest that the intensity of selection and the size of the selected population prevented inbreeding or genetic drift from being major factors in the response.
Figure 1.
Results of divergent selection on egg volume (mm3) on nine replicate population cages of Drosophila melanogaster (lines 1–3 selected for large eggs, lines 4–6 control lines, lines 7–9 selected for small eggs).
Eve stripe positions
Most previous work has compared only lateral stripe positions. In contrast, by imaging in 3D and using PCT software we were able to examine stripe border positions at 16 different D-V locations, eight on each side of the embryo. We found that, for the ventral-most region, the anterior and posterior borders of stripe 6 and the anterior border of stripe 7 were located on average 1.4% EL more posterior in large-egg lines (P = 0.031, 0.011, and 0.019 respectively, n=108, Table 1, Fig. 2).
Table 1.
ANOVA table for ventral-most eve stripe border positions for anterior and posterior of stripes 6 and 7 using a fully nested mixed model ANOVA (lines nested within treatments, random effects at the level of line, fixed effect term of developmental age added to account for the refinement of stripe position that occurs during mitotic cycle 14).
| Source | df | F | P | |
|---|---|---|---|---|
| Stripe 6 Anterior | Treatment | 2 | 7.24 | 0.031 |
| Line (Treat) & Random | 6 | 0.44 | 0.85 | |
| Developmental Age | 4 | 8.39 | <0.0001 | |
| Error | 95 | |||
| Stripe 6 Posterior | Treatment | 2 | 12.41 | 0.011 |
| Line (Treat) & Random | 6 | 0.44 | 0.85 | |
| Developmental Age | 4 | 8.20 | <0.0001 | |
| Error | 95 | |||
| Stripe 7 Anterior | Treatment | 2 | 9.11 | 0.019 |
| Line (Treat) & Random | 6 | 0.99 | 0.44 | |
| Developmental Age | 4 | 7.03 | <0.0001 | |
| Error | 95 | |||
| Stripe 7 Posterior | Treatment | 2 | 1.87 | 0.24 |
| Line (Treat) & Random | 6 | 1.82 | 0.10 | |
| Developmental Age | 4 | 12.74 | <0.0001 | |
| Error | 95 |
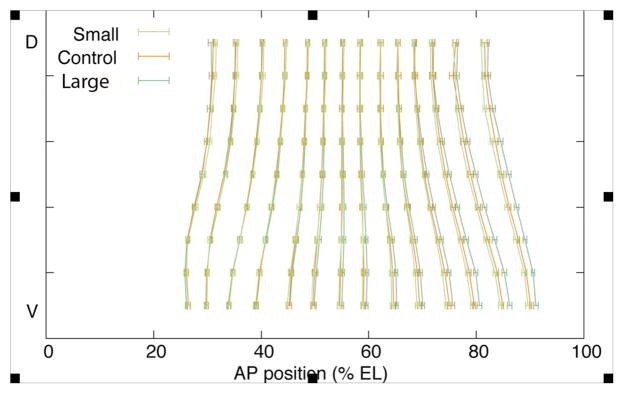
Figure 2.
Mean position for normalized embryos (% EL) for anterior and posterior of seven eve stripes for large-egg, small-egg, and control treatments (± SE) from dorsal (D) to ventral (V) regions.
Specifically, the anterior border of stripe 6 in the large-egg lines was located posteriorly to that in the small-egg lines, but could not be distinguished from controls. The posterior border of stripe 6 and the anterior border of stripe 7 were located more posterior in large-egg lines than either controls or small-egg lines. The posterior border of stripe 7 was not significantly different among treatments (P = 0.24). Dorsal and lateral borders of stripes 6 and 7 also were not significantly different among treatments. Arcsine transformation of stripe border locations (% EL) did not change the results. We also compared the eve stripe border positions for regions adjacent to the ventral-most segment, moving up and around the embryo towards the dorsal region. In the two regions directly adjacent to the ventral-most segment (~45° arc at the surface of the embryo) we also found a significant posterior shift in eve stripe 6 and 7 positions (Table S2).
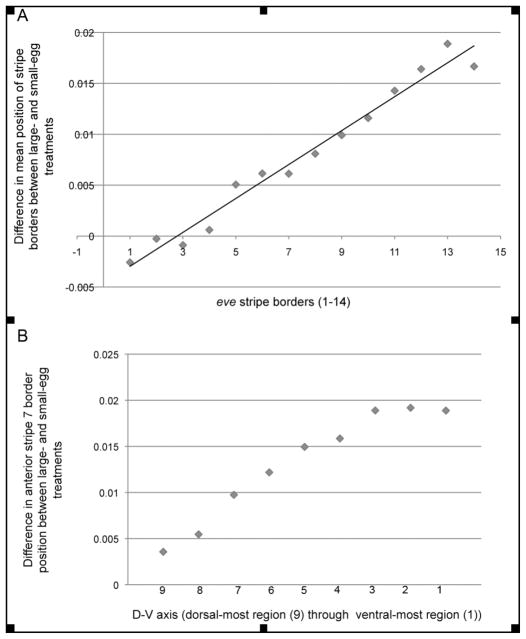
We further investigated whether the shift in stripe position on the ventral side of large-egg selected embryos was restricted to stripes 6 and 7, or whether there was a shift in overall allometry across the embryo, with the cumulative effects only becoming statistically significant at the posterior end (i.e., stripes 6 and 7). By plotting the difference in normalized stripe border locations between the mean large- and small-egg lines (pooled across replicates) for all eve stripes (Fig. 3A), the displacement of the posterior stripes between the treatments clearly initiates anteriorly and increases linearly with successive stripes (r2 = 0.97). Thus, the statistically significant shift of posterior eve stripes in the ventral region of the large-egg selected lines represents a gradual change in the allometry of gene expression and not a disjunct shift of only the two posterior stripes. This increasing displacement of posterior stripes in the ventral aspect of the embryo appears to be anchored at the anterior, however, where eve stripes 1 and 2 can be seen to be in nearly identical locations between the treatments. Stripe width did not differ significantly across treatments.
Figure 3.
eve stripe border displacement in A-P and D-V axes. A) Overall shift in A-P eve stripe pattern illustrated in the linear relationship (r2 = 0.97) of the differences in mean stripe position of all large-egg embryos (pooled across lines) and all small-egg embryos (pooled across lines) plotted for the ventral-most region (1) for anterior and posterior borders of seven eve stripes (1–14). B) Overall shift in D-V eve stripe pattern for anterior border of stripe 7 plotted from dorsal-most region (9) to ventral-most region (1) calculated as the difference in mean stripe position between all large- and small-egg embryos (pooled across lines within treatments
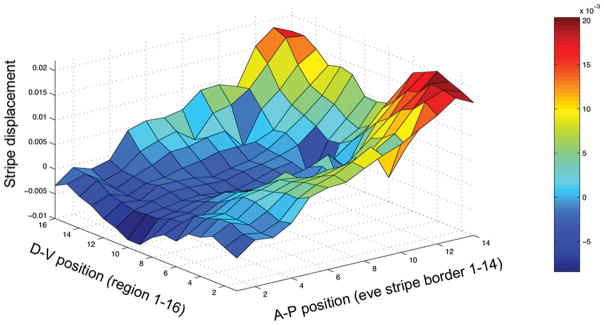
Consistent with our findings of an allometric shift along the length of the A-P axis, we also observed a monotonic displacement of the posterior stripes along the D-V axis converging at a plateau in the ventral regions (Fig. 3B). To further investigate the structure of change between the selection treatments, we calculated the difference in normalized stripe border positions between mean large- and small-egg lines (stripe displacement) for all 14 borders in the 16 regions around the D-V axis (Fig. 4). As the figure clearly shows, stripe displacement changes gradually along both the A-P and the D-V axes, with the least displaced (blue) regions located anterior and dorsally, and the most displaced (red and orange) areas located posterior and ventrally. To summarize, eve stripe border displacement is gradual, both along the A-P and D-V axes, producing what we will subsequently refer to as an “allometric” shift in stripes.
Figure 4.
Three-dimensional representation of stripe border displacement (difference between the mean normalized stripe position of the large- and small-egg lines) plotted against the A-P position (1–14 stripe borders) and D-V position (1–16 regions or strips around the embryo) with blue indicating least (anterior and dorsal), and orange and red indicating most displacement posterior and ventral).
Total number and mean density of nuclei
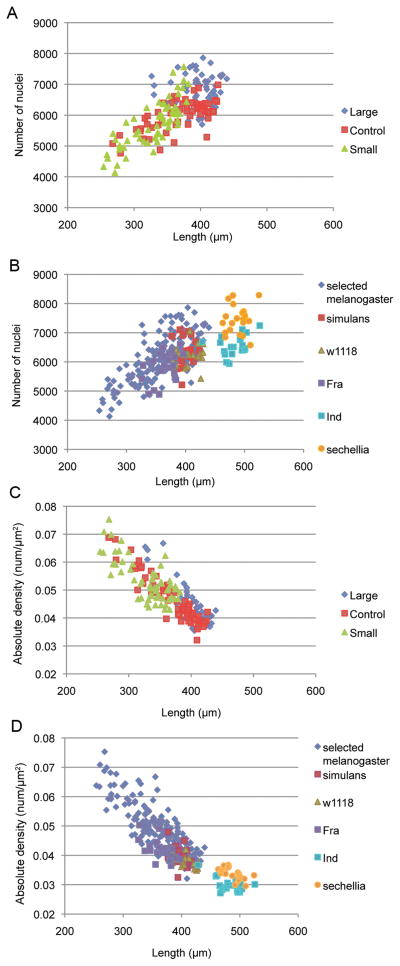
3D representation of embryos using the nuclear stain channel allowed us to count number of nuclei at the periphery of each embryo. The estimates excluded pole cells and yolk nuclei. The large-egg selected lines had significantly more nuclei (6786.9 ± 182.8) than the controls (6014.0 ± 183.0), or the small-egg lines (5728.4 ± 183.2) (F2,6 = 8.93, P = 0.016, n = 182), the latter two not being distinguishable. Counts of nuclei performed by PCT are subject to a small error on the order of a few percent (Luengo Hendriks et al. 2006), but our results are replicated within treatments and any error in counting should not be correlated with embryo size.
To gain further insight into the relationship between egg size and number of nuclei in non-selected lines, we estimated nuclear number using the same methods in five additional isogenic strains and two additional species: Total number of nuclei differed significantly among the unselected embryos D. sechellia-Robertson strain, D. simulans-FC strain, and w1118, Fra, and Ind strains of D. melanogaster (F4 = 38.38, P < 0.0001, n = 100). The large-egg D. sechellia had significantly higher mean total number of nuclei (7448.7 ± 98.6), followed by the Ind strain of D. melanogaster and D. simulans, which could not be distinguished (6569.1 ± 101.2 and 6356.9 ± 98.6, respectively). w1118 had significantly more nuclei than the small-egg Fra strain of D. melanogaster (6181.8 ± 98.6, 5844.6 ± 94.0, respectively). A positive correlation between total number of nuclei and embryo length (P < 0.0001) can be seen in Figure 5 for the selected lines (5A; range of lengths 254–440μm, r2 = 0.50) and all embryos combined (5B; range 254–526μm, r2 = 0.46). Approximately half of all the variation in number of nuclei both within and between species is explained by the variation in embryo length.
Figure 5.
Correlations between nuclear number or mean density and embryo length. A) Positive correlation in total number of nuclei plotted as a function of embryo length for control, large- and small-egg selected D. melanogaster (r2 = 0.50). B) Positive correlation in total number of nuclei plotted as a function of embryo length for D. melanogaster from the selection experiment (treatments pooled), D. sechellia, D. simulans, and W1118, Fra and Ind strains of D. melanogaster (r2 = 0.46). C) Negative correlation between overall nuclear density and length of D. melanogaster embryos within the selection experiment (r2 = 0.74). D) Negative correlation between overall nuclear density and length of embryos including D. melanogaster from the selection experiment (treatments pooled), D. sechellia, D. simulans, and W1118, Fra and Ind strains of D. melanogaster (r2=0.76).
Nuclear density also differed among selected and control treatments (Kruskal–Wallis H=40.77, 2 df, P<0.0001). Least Significant Difference (LSD) comparisons of mean ranks show that small-egg lines had significantly higher mean nuclear density (0.053 ± 0.001 nuclei/μm2) than the controls (0.047 ± 0.001) which could not be distinguished from the large-egg lines (0.045 ± 0.001). Average median distance to neighboring nuclei, another measure of nuclear density, mirrored this result (Kruskal–Wallis, H=43.73, 2 df, P<0.0001). LSD comparisons of mean ranks show that nuclei were more tightly packed in small-egg lines (4.75 ± 0.05 μm median neighbor distance) on average than the controls (5.08 ± 0.04) that could not be distinguished from the large-egg lines (5.19 ± 0.05).
Plots of nuclear density vs. length (Fig. 5C: embryos from selection lines only, r2 = 0.74, 5D: all embryos combined, r2 = 0.76) reveal a significantly negative correlation (P < 0.0001), indicating that although the number of nuclei increase with embryo length, the overall packing of nuclei decreases relative to EL, suggesting the actions of a global mechanism of cell number “counting”.
Is nuclear density correlated with stripe allometry?
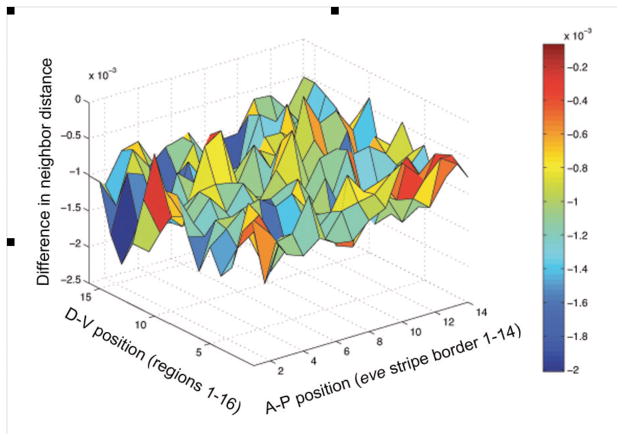
We were interested in exploring whether density along the A-P and/or D-V axes differed between the treatments in any consistent manner, as do stripes, and if so, whether this difference might correlate with stripe allometry differences between treatments. We focused attention on the 14 eve stripe borders x 16 D-V measurements, a total of 224 positions and identified the nuclei closest to the stripe borders in each embryo in both the large- and small-egg lines. We estimated nearest neighbor distances in normalized embryos for each of these nuclei and calculated the difference between the mean of the two selected treatments. A 3D plot of the difference in the means between the large- and small-egg treatments (difference in neighbor distance) as a function of A-P and D-V position (Fig. 6) reveals no apparent spatial structure. To confirm this visual impression, we calculated the correlation between the difference in neighbor distance between treatments and the difference in scaled stripe border positions between treatments (stripe displacement) at the same positions in each embryo (Fig. S2). Although the correlation is significant (P<0.01) the relationship is exceedingly weak (r2 = 0.03), leading us to conclude that local nuclear density (specifically its proxy nuclear neighbor distance) is not causally related to stripe allometry differences between treatments.
Figure 6.
No spatial correlation is apparent in this plot of the difference between mean large and small embryos for normalized median neighbor distance (a proxy for density difference; μm) plotted against the A-P position (1–14 stripe borders) and D-V position (1–16 regions around the embryo).
Evaluation of biophysical model of scaling
Our data on eve stripe position, embryo length, surface area, and number of nuclei from each embryo allowed us to test a general mechanism to produce scaling of expression patterns in morphogenetic fields (Gregor et al. 2007b; Umulis 2009). Recent observations of a dynamic process of nuclear import and export of Bcd have led to a hypothesis that if Bcd degradation occurs exclusively within nuclei, this can produce automatic scaling provided that the number of nuclei does not vary with length (L) (Gregor et al. 2007b). Under this assumption nuclear density will be proportional to 1/L2. The Bcd gradient decays exponentially along the A-P axis (Driever and Nussleinvolhard 1988b; Gregor et al. 2007b) so that its concentration at position x can be reasonably estimated by Aexp(−x/λ), where A is the amplitude, and λ is the length scale. Including nuclear degradation, the length scale of the gradient becomes in relative length units (Umulis 2009). Here ρ is the nuclear density and C is a constant depending on the free diffusion rate, the equilibration constant for nuclear import and export, and the rate of nuclear degradation (Umulis 2009). We assume diffusion and kinetic constants remain fixed across treatments. Using the measured values of mean length and mean nuclear densities from our selected lines we can estimate fate map shifts for the nuclear trapping model under the assumptions that A is constant and that position in the embryo is specified directly by concentration thresholds of the Bcd gradient. A position x along the A-P axis is expected to shift by where L1 and L2 are the mean lengths in two treatments being compared and ρ1 and ρ2 are the mean densities. Using this expression a position in the middle of the embryo (i.e., x =0.50) in the small treatment is expected to shift by 0.05(5% EL) to the anterior in the large treatment. We performed these calculations using other values for x (0.26 and 0.90) and the model consistently predicted an anterior shift in large eggs relative to small eggs (Table S3). The observed difference at x =0.50 between our small- and large-egg lines was 0.007 (< 1% EL) in the opposite direction to the model’s prediction. More specifically, stripe borders in the large–egg lines shifted posterior relative to small-egg lines (and controls), not anterior. Thus, differences in nuclear density in our selection treatments are such that nuclear trapping of Bcd cannot lead to the observed direction in the shift of eve stripes.
Discussion
One of the challenges in studying the evolution of a developmentally buffered trait, such as pattern formation, is to expose otherwise suppressed variation, if it exists. Here we used genetic variation for egg size to challenge the buffering mechanism associated with segmentation patterning and observed changes in eve stripe border positions, nuclear number, and mean nuclear density as correlated responses to selection for egg size.
This study produced five novel findings, which allowed us to address longstanding questions in both evolutionary and developmental biology. [1] There is genetic variation for stripe allometry segregating in natural populations of D. melanogaster; [2] The phenotypic difference in stripe positioning between the selected lines is neither a spatially restricted shift, nor a coordinate shift of all stripes, but rather it is a monotonically increasing (i.e., allometric) shift along both A-P and D-V axes; [3] Genetic variation for this buffered trait was produced in the absence of environmental or genetic perturbation, and therefore must not be entirely suppressed by the developmental canalization process; [4] Cycle 14 nuclear number is not constant, but rather differs by a mean of more than 1000 nuclei as a correlated response to divergent selection for egg size; [5] Nuclear density is inversely related to egg length, but does not differ between selection treatments in a way that can explain the allometric shift between them.
Previous work demonstrating a lack of genetic variation in eve stripe position between strains of D. melanogaster differing in egg length was based on measurements of gene expression at the lateral aspect only (Holloway et al. 2006; Lott et al. 2007; Surkova et al. 2008). We also found a statistically nonsignificant difference in lateral eve stripe border positions between our artificial selection treatments. But examination of each stripe border around the circumference of the selected embryos shows that the significant posterior shift in stripe 6 and 7 borders is not restricted to the ventral-most region only. Rather, they initiate dorsally, where there are no statistically significant differences between selection treatments for stripe borders, and increase monotonically in magnitude to the ventral aspect.
Progressive, i.e., allometric, shifts in eve stripes between the selection treatments along both the A-P and D-V axes suggests that with additional artificial selection, a statistically significant allometric shift could extend even more dorsally to include the lateral aspect of stripe positioning. Therefore, we interpret our results as suggesting that revealed genetic variation in eve stripe position in D. melanogaster can extend across the entirety of the A-P and D-V dimensions of the blastoderm embryo. Indeed, lateral gt and eve stripes do differ significantly among closely related species, offering evolutionary evidence for the existence of such variation (Lott et al. 2007).
We do not know the molecular or cellular mechanisms underlying the shift in eve stripes driven by artificial selection for egg size, but the smooth and monotonic response in both the A-P and D-V dimensions is a significant feature. This may represent an overall stretching of the patterning mechanism, suggesting the involvement of maternal segmentation factors such as Bcd. But genetic analysis (Casanova 1990) and mathematical modeling of the gap gene system indicates that a shift of this type could also result from a more localized perturbation, for example in the terminal system, that has been followed by accommodation or adjustment at the level of gap gene cross-regulation (Vakulenko et al. 2009). Supporting this possibility, mutations in elements of the terminal system, such as tailless, can produce posterior shifts in pair-rule gene expression patterns (Casanova 1990). The allometric shift in eve stripe pattern that we observed, therefore, could represent either type of perturbation.
Although gene expression in the segmentation pathway is one of the most well characterized examples of a robust developmental trait, there was enough genetic variation available segregating in the base population for artificial selection on egg size to produce correlated changes in eve stripe boundaries without intervening mutation or environmental perturbation. Canalized traits are expected to accumulate slightly deleterious cryptic mutations whose full phenotypic manifestation requires genetic or environmental perturbation for release (Masel and Siegal 2009). In yeast, mutational analysis has identified many genes that can act as phenotypic capacitors of cell morphology (Levy and Siegal 2008). Natural populations of D. melanogaster are known to carry large numbers of deleterious mutations (Mukai and Yamaguchi 1974; Watanabe et al. 1976; Mukai and Nagano 1983), some of which might likewise act as phenotypic capacitors for egg-size-related traits. But, if phenotypic capacitors are present, we found no evidence for their increase in frequency through artificial selection, as we failed to observe any increase in the variance of boundary positions for stripes 6 and 7.
After 13 mitotic divisions the maximum possible number of nuclei at the blastoderm stage is 213 or 8192 nuclei, considerably more than the classic value of ~6000 given in the literature (Zalokar and Erk 1976; Turner and Mahowald 1983). We found that cell number is not anywhere near constant at 6000 nuclei in the cycle 14 embryo, but rather is positively correlated with egg size (Fig. 5A). On average, our large-egg selected treatment had more than 1000 additional nuclei than the small-egg selected treatment, a difference that arose as a correlated response to selection for egg size. Yet, in comparing cell numbers in other (nonselected) lines and species, we conclude that cell number must not be entirely a consequence of egg size, but rather must itself be a genetically variable trait. For example, D. sechellia (mean cell number = 7448.7, some embryos approaching the 213 limit) and the Ind strain of D. melanogaster (mean cell number = 6569.1) have nearly non-overlapping distributions of cell number despite having almost identical egg length (Fig. 5B). It would be interesting to investigate whether further selection on egg size is possible without strong deleterious fitness consequences, which would indicate a developmental constraint on evolution. There must also be nongenetic sources of variation in cell number, as isogenic strains of flies can produce embryos varying somewhat in both size and number of nuclei at a given cleavage cycle (Fowlkes et al. 2008).
The positive correlation between nuclear number and embryo length raises the question of whether an embryo is able to “sense” its size and regulate the number of nuclei present in cycle 14. The sensing mechanism, if it exists, does not appear to be influenced by the artificial selection for egg size, as we observe a strong linear correlation extending across the selection treatments. The positive relationship is further extended when we add data from unselected Drosophila lines (Fig. 5B). While nuclear number is positively correlated with egg length, nuclear density does not remain constant, but rather decreases with egg length (Fig. 5C). This decrease in density occurs relatively uniformly across the length (and width) of the embryo, however, and is not likely to be causally involved in the allometric shift in eve stripes in the large-egg treatment. Furthermore, the large-egg lines and the controls cannot be distinguished based on either overall density, or absolute distance to neighboring nuclei, yet they differ in eve stripe border position. Thus, density does not appear to explain the shift in segmentation pattern. Sullivan (Sullivan 1987) also found that expression patterns of the pair-rule gene fushi tarazu were independent of cell density.
Increases in the number of copies of bcd have been shown to result in a posterior shift in pair-rule gene expression (Driever and Nussleinvolhard 1988a), indicating its primary role as a morphogen. Gregor et al. (Gregor et al. 2007b) proposed that the inclusion of degradation of Bcd in nuclei would provide scaling providing that the number of nuclei remains constant. He et al. (He et al. 2008) suggested that the total flux of Bcd protein is not constant, as is commonly assumed, but varies with embryo length as a result of differential maternal provisioning of bcd mRNA. We have shown that the number of nuclei is not constant across embryos of varying size, however, and calculations of predicted shifts in eve stripe borders based on the nuclear trapping model (Gregor et al. 2007b; Umulis 2009) do not correlate with observed shifts in eve stripe border positions. Generally speaking, the automatic scaling provided by nuclear degradation comes at the cost of increased sensitivity to nuclear number. This mechanism could still be operational in much larger embryos with the same number of cleavage cycles as D. melanogaster (Gregor et al. 2005; Gregor et al. 2008) so that the change in nuclear number is negligible compared to the change in length and ρ ~ 1/L2. There must, however, be a different mechanism at work here. One possibility is feedback among the gap genes, which regulate eve expression (Frasch and Levine 1987), and can provide scaling (Manu et al. 2009; Vakulenko et al. 2009). Maternal regulation of the amount of Bcd mRNA deposited during oogenesis could also contribute to scaling, either in concert with gap gene cross-repression or independently (He et al. 2008).
The existence of genetic variation for stripe allometry and the rapid evolutionary change observed in pattern formation both within and between species belies the fact that eve stripe expression is both a highly conserved trait and is subject to strong stabilizing selection (Ludwig et al. 2005). Many features of pattern formation remain remarkably conserved in dipteran evolution, including the shape of the Bcd gradient (Gregor et al. 2005; Gregor et al. 2008), the positioning of eve stripes (Hare et al. 2008), and perhaps even blastoderm cell number (Gregor et al. 2005; Gregor et al. 2008). This suggests to us the presence of developmental constraints on the patterning system preventing components from evolving very far from a global optimum. Short excursions, however, may be permissible, allowing slightly divergent phenotypes in patterning to evolve relatively rapidly. This characteristic of the segmentation system — evolution over short, but not long, evolutionary timescales — may turn out to be an important hallmark of a canalized developmental trait.
Supplementary Material
Acknowledgments
Jeff Birdsley for fly collection and Melissa Hart for help with flies.
LITERATURE CITED
- Azevedo RBR, French V, Partridge L. Thermal evolution of egg size in Drosophila melanogaster. Evolution. 1996;50:2338–2345. doi: 10.1111/j.1558-5646.1996.tb03621.x. [DOI] [PubMed] [Google Scholar]
- Casanova J. Pattern formation under the control of the terminal system in the Drosophila embryo. Development. 1990;110:621–628. doi: 10.1242/dev.110.2.621. [DOI] [PubMed] [Google Scholar]
- Driever W, Nussleinvolhard C. The Bicoid Protein Determines Position in the Drosophila Embryo in a Concentration-Dependent Manner. Cell. 1988a;54:95–104. doi: 10.1016/0092-8674(88)90183-3. [DOI] [PubMed] [Google Scholar]
- Driever W, Nussleinvolhard C. A Gradient of Bicoid Protein in Drosophila Embryos. Cell. 1988b;54:83–93. doi: 10.1016/0092-8674(88)90182-1. [DOI] [PubMed] [Google Scholar]
- Fowlkes CC, Luengo Hendriks CL, Keranen SVE, Weber GH, Rubel O, Huang MY, Chatoor S, DePace AH, Simirenko L, Henriquez C, Beaton A, Weiszmann R, Celniker S, Hamann B, Knowles DW, Biggin MD, Eisen MB, Malik J. A quantitative spatiotemporal atlas of gene expression in the Drosophila blastoderm. Cell. 2008;133:364–374. doi: 10.1016/j.cell.2008.01.053. [DOI] [PubMed] [Google Scholar]
- Frasch M, Hoey T, Rushlow C, Doyle H, Levine M. Characterization and localization of the even-skipped protein of Drosophila. Embo Journal. 1987;6:749–759. doi: 10.1002/j.1460-2075.1987.tb04817.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Frasch M, Levine M. Complementary patterns of even-skipped and fushi-tarazu expression involve their differential regulation by a common set of segmentation genes in Drosophila. Genes & Development. 1987;1:981–995. doi: 10.1101/gad.1.9.981. [DOI] [PubMed] [Google Scholar]
- Gregor T, Bialek W, van Steveninck RRR, Tank DW, Wieschaus EF. Diffusion and scaling during early embryonic pattern formation. Proceedings of the National Academy of Sciences of the United States of America. 2005;102:18403–18407. doi: 10.1073/pnas.0509483102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gregor T, McGregor AP, Wieschaus EF. Shape and function of the Bicoid morphogen gradient in dipteran species with different sized embryos. Developmental Biology. 2008;316:350–358. doi: 10.1016/j.ydbio.2008.01.039. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gregor T, Tank DW, Wieschaus EF, Bialek W. Probing the limits to positional information. Cell. 2007a;130:153–164. doi: 10.1016/j.cell.2007.05.025. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gregor T, Wieschaus EF, McGregor AP, Bialek W, Tank DW. Stability and nuclear dynamics of the bicoid morphogen gradient. Cell. 2007b;130:141–152. doi: 10.1016/j.cell.2007.05.026. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hare EE, Peterson BK, Iyer VN, Meier R, Eisen MB. Sepsid even-skipped Enhancers Are Functionally Conserved in Drosophila Despite Lack of Sequence Conservation. Plos Genetics. 2008;4:e1000106. doi: 10.1371/journal.pgen.1000106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- He F, Wen Y, Deng JY, Lin XD, Lu LJ, Jiao RJ, Ma J. Probing Intrinsic Properties of a Robust Morphogen Gradient in Drosophila. Developmental Cell. 2008;15:558–567. doi: 10.1016/j.devcel.2008.09.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Holloway DM, Harrison LG, Kosman D, Vanario-Alonso CE, Spirov AV. Analysis of pattern precision shows that Drosohila segmentation develops substantial independence from gradients of maternal gene products. Developmental Dynamics. 2006;235:2949–2960. doi: 10.1002/dvdy.20940. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Houchmandzadeh B, Wieschaus E, Leibler S. Establishment of developmental precision and proportions in the early Drosophila embryo. Nature. 2002;415:798–802. doi: 10.1038/415798a. [DOI] [PubMed] [Google Scholar]
- Howard M, Rein ten Wolde P. Finding the Center Reliably: Robust Patterns of Developmental Gene Expression. Physical Review Letters. 2005;95:208103. doi: 10.1103/PhysRevLett.95.208103. [DOI] [PubMed] [Google Scholar]
- Keranen SVE, Fowlkes CC, Luengo Hendriks CL, Sudar D, Knowles DW, Malik J, Biggin MD. Three-dimensional morphology and gene expression in the Drosophila blastoderm at cellular resolution II: dynamics. Genome Biology. 2006;7:R124. doi: 10.1186/gb-2006-7-12-r124. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Levy SF, Siegal ML. Network Hubs Buffer Environmental Variation in Saccharomyces cerevisiae. Plos Biology. 2008;6:2588–2604. doi: 10.1371/journal.pbio.0060264. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lott SE, Kreitman M, Palsson A, Alekseeva E, Ludwig MZ. Canalization of segmentation and its evolution in Drosophila. Proceedings of the National Academy of Sciences of the United States of America. 2007;104:10926–10931. doi: 10.1073/pnas.0701359104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ludwig MZ, Palsson A, Alekseeva E, Bergman CM, Nathan J, Kreitman M. Functional Evolution of a cis-Regulatory Module. Plos Biology. 2005;3:588–598. doi: 10.1371/journal.pbio.0030093. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Luengo Hendriks CL, Keranen SVE, Fowlkes CC, Simirenko L, Weber GH, DePace AH, Henriquez C, Kaszuba DW, Hamann B, Eisen MB, Malik J, Sudar D, Biggin MD, Knowles DW. Three-dimensional morphology and gene expression in the Drosophila blastoderm at cellular resolution I: data acquisition pipeline. Genome Biology. 2006;7:R123. doi: 10.1186/gb-2006-7-12-r123. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Manu, Surkova S, Spirov AV, Gursky VV, Janssens H, Kim AR, Radulescu O, Vanario-Alonso CE, Sharp DH, Samsonova M, Reinitz J. Canalization of Gene Expression in the Drosophila Blastoderm by Gap Gene Cross Regulation. Plos Biology. 2009;7:591–603. doi: 10.1371/journal.pbio.1000049. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Masel J, Siegal ML. Robustness: mechanisms and consequences. Trends in Genetics. 2009;25:395–403. doi: 10.1016/j.tig.2009.07.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mukai T, Nagano S. The genetic structure of natural populations of Drosophila melanogaster. 16. Excess of additive genetic variance of viability. Genetics. 1983;105:115–134. doi: 10.1093/genetics/105.1.115. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mukai T, Yamaguchi O. Genetic structure of natural populations of Drosophila melanogaster. 11. Genetic variability in a local population. Genetics. 1974;76:339–366. doi: 10.1093/genetics/76.2.339. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nusslein-Volhard C, Frohnhofer HG, Lehmann R. Determination of anteroposterior polarity in Drosophila. Science. 1987;238:1675–1681. doi: 10.1126/science.3686007. [DOI] [PubMed] [Google Scholar]
- Nusslein-Volhard C, Kluding H, Jurgens G. Genes affecting the segmental subdivision of the Drosophila embryo. Cold Spring Harbor Symposium Quantitative Biology. 1985;50:145–154. doi: 10.1101/sqb.1985.050.01.020. [DOI] [PubMed] [Google Scholar]
- Sullivan W. Independence of fushi tarazu expression with respect to cellular density in Drosophila embryos. Nature. 1987;327:164–167. doi: 10.1038/327164a0. [DOI] [PubMed] [Google Scholar]
- Surkova S, Kosman D, Kozlov K, Manu, Myasnikova E, Samsonova AA, Spirov A, Vanario-Alonso CE, Samsonova M, Reinitz J. Characterization of the Drosophila segment determination morphome. Developmental Biology. 2008;313:844–862. doi: 10.1016/j.ydbio.2007.10.037. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tautz D. Regulation of the Drosophila segmentation gene hunchback by two maternal morphogenetic centres. Nature. 1988;332:281–284. doi: 10.1038/332281a0. [DOI] [PubMed] [Google Scholar]
- Turner FR, Mahowald AP. Scanning electron microscopy of Drosophila embryogenesis. Dev Biol. 1983;50:95–108. doi: 10.1016/0012-1606(76)90070-1. [DOI] [PubMed] [Google Scholar]
- Umulis D. Analysis of dynamic morphogen scale invariance. Journal of the Royal Society Interface. 2009 doi: 10.1098/rsif.2009.0015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vakulenko S, Manu, Reinitz J, Radulescu O. Size regulation in the segmentation of Drosophila: Interacting interfaces between localized domains of gene expression insure robust spatial patterning. Physical Review Letters. 2009;103(168102):168101–168104. doi: 10.1103/PhysRevLett.103.168102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Waddington CH. Canalization of development and the inheritance of acquired characters. Nature. 1942;150:563–565. doi: 10.1038/1831654a0. [DOI] [PubMed] [Google Scholar]
- Waddington CH. The strategy of the genes. George Allen and Unwin; London: 1957. [Google Scholar]
- Watanabe TK, Yamaguchi O, Mukai T. The genetic variability of third chromosomes in a local population of Drosophila melanogaster. Genetics. 1976;82:63–82. doi: 10.1093/genetics/82.1.63. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zalokar M, Erk I. Division and migration of nuclei during early embryogenesis of Drosophila melanogaster. J Microsc Biol Cell. 1976;25:97–106. [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.