Abstract
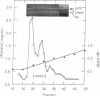
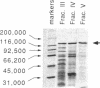
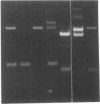
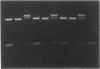
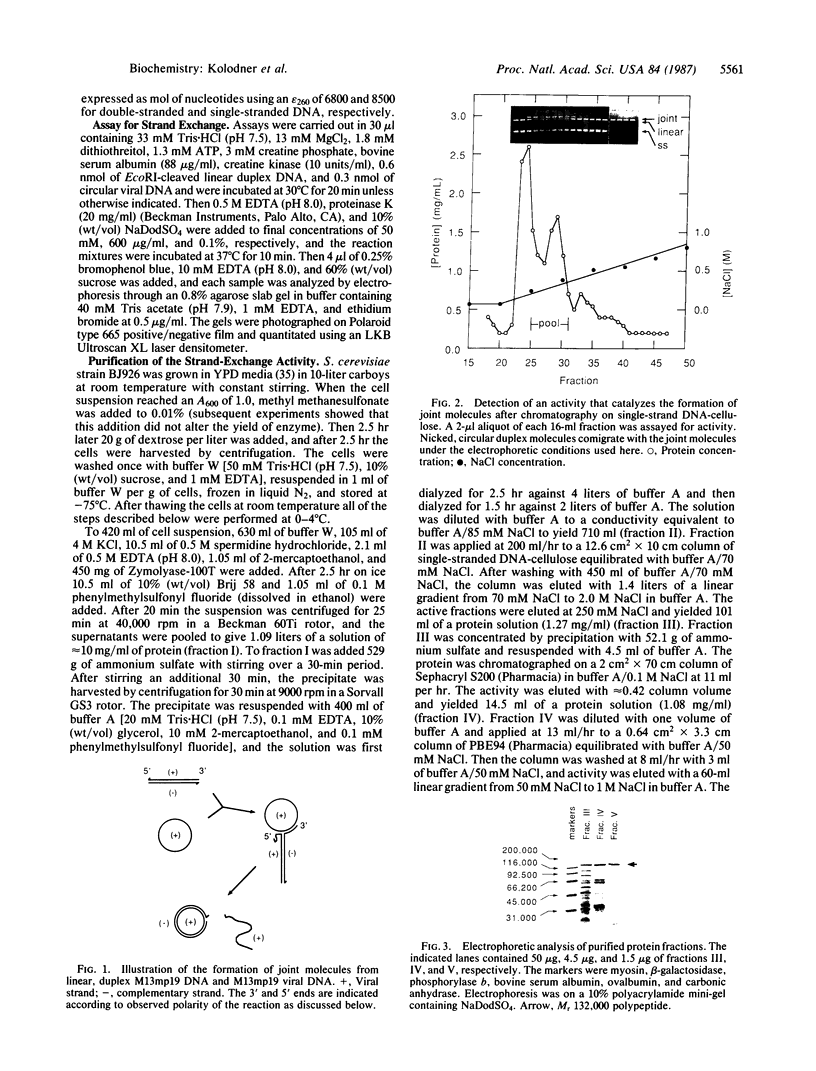
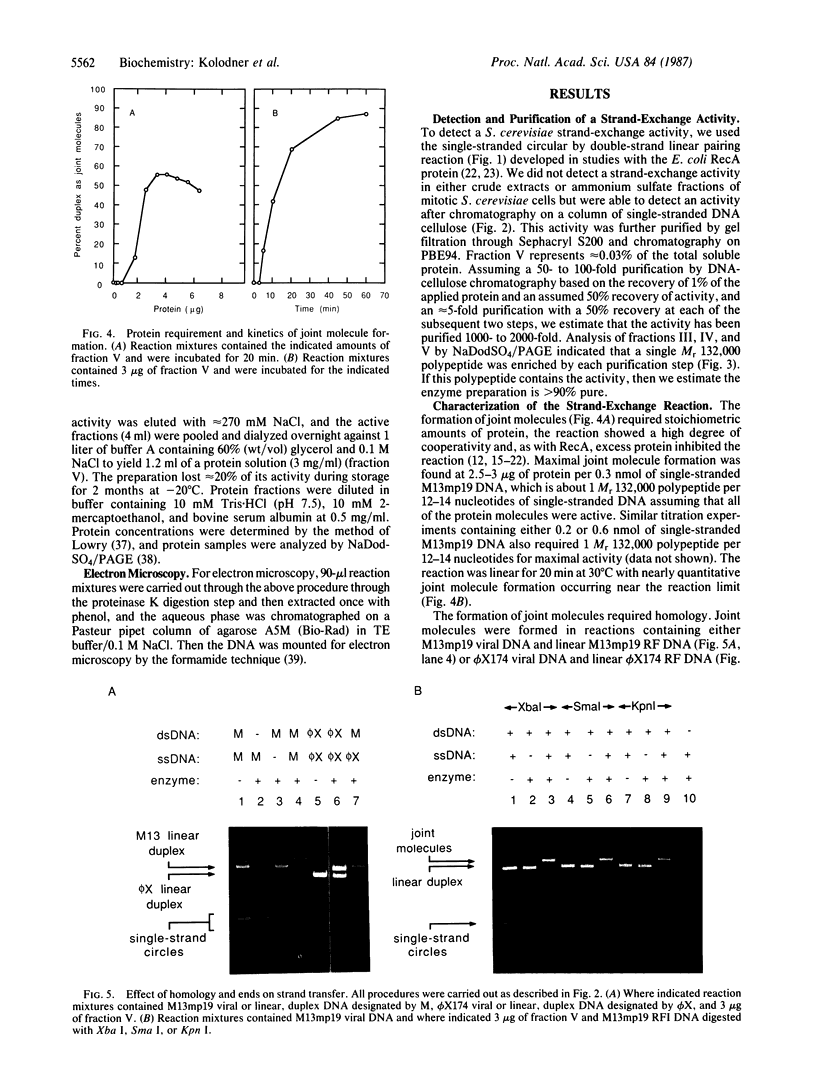
An activity that catalyzes the formation of joint molecules from linear M13mp19 replicative form DNA and circular M13mp19 viral DNA was purified 1000- to 2000-fold from mitotic Saccharomyces cerevisiae cells. The activity appeared to reside in a Mr 132,000 polypeptide. The reaction required that the substrates be homologous and also required Mg2+. There was no requirement for ATP. The reaction required stoichiometric amounts of protein and showed a cooperative dependence on protein concentration. Electron microscopic analysis of the joint molecules indicated they were formed by displacement of one strand of the linear duplex by the single-stranded circular molecule. This analysis also showed that heteroduplex formation started at the 3'-homologous end of the linear duplex strand followed by extension of the hybrid region toward the 5'-homologous end of the linear duplex strand (3'-to-5' direction).
Full text
PDF


Images in this article
Selected References
These references are in PubMed. This may not be the complete list of references from this article.
- Birnboim H. C., Doly J. A rapid alkaline extraction procedure for screening recombinant plasmid DNA. Nucleic Acids Res. 1979 Nov 24;7(6):1513–1523. doi: 10.1093/nar/7.6.1513. [DOI] [PMC free article] [PubMed] [Google Scholar]
- CLARK A. J., MARGULIES A. D. ISOLATION AND CHARACTERIZATION OF RECOMBINATION-DEFICIENT MUTANTS OF ESCHERICHIA COLI K12. Proc Natl Acad Sci U S A. 1965 Feb;53:451–459. doi: 10.1073/pnas.53.2.451. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cassuto E., West S. C., Mursalim J., Conlon S., Howard-Flanders P. Initiation of genetic recombination: homologous pairing between duplex DNA molecules promoted by recA protein. Proc Natl Acad Sci U S A. 1980 Jul;77(7):3962–3966. doi: 10.1073/pnas.77.7.3962. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Clark A. J. Recombination deficient mutants of E. coli and other bacteria. Annu Rev Genet. 1973;7:67–86. doi: 10.1146/annurev.ge.07.120173.000435. [DOI] [PubMed] [Google Scholar]
- Cox M. M., Lehman I. R. Directionality and polarity in recA protein-promoted branch migration. Proc Natl Acad Sci U S A. 1981 Oct;78(10):6018–6022. doi: 10.1073/pnas.78.10.6018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cunningham R. P., Shibata T., DasGupta C., Radding C. M. Single strands induce recA protein to unwind duplex DNA for homologous pairing. Nature. 1979 Sep 20;281(5728):191–195. doi: 10.1038/281191a0. [DOI] [PubMed] [Google Scholar]
- DasGupta C., Wu A. M., Kahn R., Cunningham R. P., Radding C. M. Concerted strand exchange and formation of Holliday structures by E. coli RecA protein. Cell. 1981 Aug;25(2):507–516. doi: 10.1016/0092-8674(81)90069-6. [DOI] [PubMed] [Google Scholar]
- Dunn K., Chrysogelos S., Griffith J. Electron microscopic visualization of recA-DNA filaments: evidence for a cyclic extension of duplex DNA. Cell. 1982 Apr;28(4):757–765. doi: 10.1016/0092-8674(82)90055-1. [DOI] [PubMed] [Google Scholar]
- Flory J., Radding C. M. Visualization of recA protein and its association with DNA: a priming effect of single-strand-binding protein. Cell. 1982 Apr;28(4):747–756. doi: 10.1016/0092-8674(82)90054-x. [DOI] [PubMed] [Google Scholar]
- Formosa T., Alberts B. M. Purification and characterization of the T4 bacteriophage uvsX protein. J Biol Chem. 1986 May 5;261(13):6107–6118. [PubMed] [Google Scholar]
- Griffith J., Formosa T. The uvsX protein of bacteriophage T4 arranges single-stranded and double-stranded DNA into similar helical nucleoprotein filaments. J Biol Chem. 1985 Apr 10;260(7):4484–4491. [PubMed] [Google Scholar]
- Hinton D. M., Nossal N. G. Cloning of the bacteriophage T4 uvsX gene and purification and characterization of the T4 uvsX recombination protein. J Biol Chem. 1986 Apr 25;261(12):5663–5673. [PubMed] [Google Scholar]
- Hotta Y., Tabata S., Bouchard R. A., Piñon R., Stern H. General recombination mechanisms in extracts of meiotic cells. Chromosoma. 1985;93(2):140–151. doi: 10.1007/BF00293161. [DOI] [PubMed] [Google Scholar]
- Hsieh P., Meyn M. S., Camerini-Otero R. D. Partial purification and characterization of a recombinase from human cells. Cell. 1986 Mar 28;44(6):885–894. doi: 10.1016/0092-8674(86)90011-5. [DOI] [PubMed] [Google Scholar]
- Kahn R., Cunningham R. P., DasGupta C., Radding C. M. Polarity of heteroduplex formation promoted by Escherichia coli recA protein. Proc Natl Acad Sci U S A. 1981 Aug;78(8):4786–4790. doi: 10.1073/pnas.78.8.4786. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kahn R., Radding C. M. Separation of the presynaptic and synaptic phases of homologous pairing promoted by recA protein. J Biol Chem. 1984 Jun 25;259(12):7495–7503. [PubMed] [Google Scholar]
- Kmiec E. B., Holloman W. K. Homologous pairing of DNA molecules by Ustilago rec1 protein is promoted by sequences of Z-DNA. Cell. 1986 Feb 28;44(4):545–554. doi: 10.1016/0092-8674(86)90264-3. [DOI] [PubMed] [Google Scholar]
- Kmiec E., Kroeger P., Holliday R., Holloman W. Homologous pairing promoted by Ustilago RecI protein. Cold Spring Harb Symp Quant Biol. 1984;49:675–682. doi: 10.1101/sqb.1984.049.01.076. [DOI] [PubMed] [Google Scholar]
- LOWRY O. H., ROSEBROUGH N. J., FARR A. L., RANDALL R. J. Protein measurement with the Folin phenol reagent. J Biol Chem. 1951 Nov;193(1):265–275. [PubMed] [Google Scholar]
- Manservigi R., Spear P. G., Buchan A. Cell fusion induced by herpes simplex virus is promoted and suppressed by different viral glycoproteins. Proc Natl Acad Sci U S A. 1977 Sep;74(9):3913–3917. doi: 10.1073/pnas.74.9.3913. [DOI] [PMC free article] [PubMed] [Google Scholar]
- McEntee K., Weinstock G. M., Lehman I. R. Binding of the recA protein of Escherichia coli to single- and double-stranded DNA. J Biol Chem. 1981 Aug 25;256(16):8835–8844. [PubMed] [Google Scholar]
- McEntee K., Weinstock G. M., Lehman I. R. Initiation of general recombination catalyzed in vitro by the recA protein of Escherichia coli. Proc Natl Acad Sci U S A. 1979 Jun;76(6):2615–2619. doi: 10.1073/pnas.76.6.2615. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Meselson M. S., Radding C. M. A general model for genetic recombination. Proc Natl Acad Sci U S A. 1975 Jan;72(1):358–361. doi: 10.1073/pnas.72.1.358. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pugh B. F., Cox M. M. recA protein binding to the heteroduplex product of DNA strand exchange. J Biol Chem. 1987 Jan 25;262(3):1337–1343. [PubMed] [Google Scholar]
- Radding C. M. Genetic recombination: strand transfer and mismatch repair. Annu Rev Biochem. 1978;47:847–880. doi: 10.1146/annurev.bi.47.070178.004215. [DOI] [PubMed] [Google Scholar]
- Radding C. M. Homologous pairing and strand exchange in genetic recombination. Annu Rev Genet. 1982;16:405–437. doi: 10.1146/annurev.ge.16.120182.002201. [DOI] [PubMed] [Google Scholar]
- Radding C. M. Recombination activities of E. coli recA protein. Cell. 1981 Jul;25(1):3–4. doi: 10.1016/0092-8674(81)90224-5. [DOI] [PubMed] [Google Scholar]
- Rush M. G., Warner R. C. Alkali denaturation of covalently closed circular duplex deoxyribonucleic acid. J Biol Chem. 1970 May 25;245(10):2704–2708. [PubMed] [Google Scholar]
- Shibata T., Cunningham R. P., DasGupta C., Radding C. M. Homologous pairing in genetic recombination: complexes of recA protein and DNA. Proc Natl Acad Sci U S A. 1979 Oct;76(10):5100–5104. doi: 10.1073/pnas.76.10.5100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stasiak A., Stasiak A. Z., Koller T. Visualization of RecA-DNA complexes involved in consecutive stages of an in vitro strand exchange reaction. Cold Spring Harb Symp Quant Biol. 1984;49:561–570. doi: 10.1101/sqb.1984.049.01.063. [DOI] [PubMed] [Google Scholar]
- Symington L. S., Morrison P., Kolodner R. Plasmid recombination intermediates generated in a Saccharomyces cerevisiae cell-free recombination system. Mol Cell Biol. 1985 Sep;5(9):2361–2368. doi: 10.1128/mcb.5.9.2361. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Szostak J. W., Orr-Weaver T. L., Rothstein R. J., Stahl F. W. The double-strand-break repair model for recombination. Cell. 1983 May;33(1):25–35. doi: 10.1016/0092-8674(83)90331-8. [DOI] [PubMed] [Google Scholar]
- Witkin E. M. Ultraviolet mutagenesis and inducible DNA repair in Escherichia coli. Bacteriol Rev. 1976 Dec;40(4):869–907. doi: 10.1128/br.40.4.869-907.1976. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yonesaki T., Minagawa T. T4 phage gene uvsX product catalyzes homologous DNA pairing. EMBO J. 1985 Dec 1;4(12):3321–3327. doi: 10.1002/j.1460-2075.1985.tb04083.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zimmerman S. B., Oshinsky C. K. Enzymatic joining of deoxyribonucleic acid strands. 3. Further purification of the deoxyribonucleic acid ligase from Escherichia coli and multiple forms of the purified enzyme. J Biol Chem. 1969 Sep 10;244(17):4689–4695. [PubMed] [Google Scholar]