DNA recognition and the precleavage state during single-stranded DNA transposition in D. radiodurans
In Deinococcus radiodurans, ionizing irradiation activates DNA transposition of ISDra2, which belongs to a recently discovered peculiar class of insertion sequences that uses an ssDNA-based pathway. Various new crystal structures now shed more light onto the action of its respective transposase.
Keywords: Deinococcus, insertion sequence, IS, mobile element, transposition
Abstract
Bacterial insertion sequences (ISs) from the IS200/IS605 family encode the smallest known DNA transposases and mobilize through single-stranded DNA transposition. Transposition by one particular family member, ISDra2 from Deinococcus radiodurans, is dramatically stimulated upon massive γ irradiation. We have determined the crystal structures of four ISDra2 transposase/IS end complexes; combined with in vivo activity assays and fluorescence anisotropy binding measurements, these have revealed the molecular basis of strand discrimination and transposase action. The structures also show that previously established structural rules of target site recognition that allow different specific sequences to be targeted are only partially conserved among family members. Furthermore, we have captured a fully assembled active site including the scissile phosphate bound by a divalent metal ion cofactor (Cd2+) that supports DNA cleavage. Finally, the observed active site rearrangements when the transposase binds a metal ion in which it is inactive provide a clear rationale for metal ion specificity.
Introduction
Exposure to 10 gray (Gy) of ionizing radiation is enough to kill a human yet Deinococcus radiodurans, perhaps the ‘world's toughest bacterium,' has the remarkable ability to survive an instantaneous dose of 5000 Gy without loss of viability (Moseley and Mattingly, 1971). Although this dose effectively shatters the bacterial genome by introducing about 200 double-stranded breaks, within 4 or 5 h following irradiation, D. radiodurans has accurately reassembled its genome. This remarkable task is accomplished mainly using a process termed extended synthesis dependent strand annealing, which involves the extensive synthesis of single-stranded DNA (ssDNA) (Zahradka et al, 2006; Slade et al, 2009).
Like that of essentially all microbes, the genome of D. radiodurans contains a variety of mobile elements, and DNA transposition by insertion sequences (ISs) has been shown to be a major source of spontaneous mutagenesis in this bacterium (Mennecier et al, 2006). ISs are the simplest form of mobile DNA; they can transpose from one genomic location (donor site) to another (target site) assisted by a self-encoded transposase enzyme that specifically recognizes and synapses the ends of the IS. Synapse formation is followed by a regulated set of DNA cutting and joining reactions that first liberate the transposon from donor DNA, and then integrate the transposon ends into target DNA. All of these events are carried out by the transposase, and the exact mechanism by which these are achieved is dependent on the particular IS family.
When IS activity in D. radiodurans was monitored shortly after irradiation, one particular element, ISDra2, stood out as it showed an ∼100-fold increase in activity following 10 kGy of γ irradiation, becoming the predominant transposition event (Mennecier et al, 2006). This is in contrast to non-irradiated cells where ISDra2 insertions are rare. The reason for this increase in ISDra2 transposition during genomic recovery from γ irradiation became clear, as ISDra2 belongs to the recently described IS200/IS605 family (Kersulyte et al, 1998, 2002) that, unlike any other characterized DNA transposition system, uses an asymmetric, ssDNA-dependent pathway for transposition (Guynet et al, 2008; Pasternak et al, 2010). This pathway has several unusual features. Whereas most ISs contain inverted repeat sequences at the two ends, in the IS200/IS605 family, the transposase recognizes its IS ends by binding to subterminal palindromic sequences that form imperfect hairpin structures (designed IPs, for imperfect palindromes; Figures 1A and 2A). Recognition is also strand specific, as only one strand (the top strand) of the transposon is bound and subsequently transposed. Members of the IS200/IS605 family are also unusual from the point of view of target specificity. Whereas most ISs integrate randomly or with minor sequence preferences, ISs in this family always insert just 3′ of a well-defined tetra- or pentanucleotide sequence (Kersulyte et al, 1998; Islam et al, 2003; http://www-is.biotoul.fr/; and P Siguier, personal communication). The exact sequence of this target site is characteristic of the particular family member.
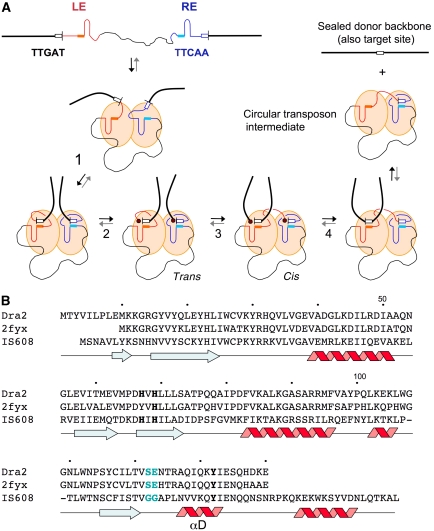
Figure 1.
Model of transposon excision by members of the IS200/IS605 family of insertion sequences (ISs) and sequence alignment of some of their transposases. (A) The IS is shown as a thin black line with its left end (LE) in red and right end (RE) in blue. The imperfect palindromes (IP) are shown schematically as hairpins, which are flanked on their 5′ sides by solid boxes representing the four nucleotides involved in cleavage (or target) site recognition. The transposase, TnpA, is shown as a dimer with orange subunits. Step 1: TnpA synapses the two IS ends by binding the IPs, and brings the cleavage sites (shown as white boxes) at the IS ends into the active sites through DNA–DNA interactions between the nucleotides 5′ of each end (TTGAT at LE, TTCAA at RE in the case of ISDra2) and those at the 5′ sides of the IPs. For clarity, the two IS ends are shown binding in the same orientation to each monomer of the dimer; however, in the determined structures, one IP is rotated by 180° relative to the other (vide infra). Step 2: End cleavage mediated by the active site Tyr on helix αD results in covalent attachment (represented by a red hexagon) of one TnpA subunit to the cleaved LE and the other subunit to the flanking DNA 3′ of the cleaved RE. Step 3: A trans-to-cis movement of the αD helices brings the two ends of the donor DNA together in one active site (monomer on left) and the LE into the proximity of the cleaved RE (monomer on right). Step 4: Nucleophilic attack of the two 3′-OH groups onto the covalent phosphotyrosine intermediates results in sealed donor DNA and a circular transposon intermediate. Reversal of these steps (grey arrows) represents insertion of the transposon intermediate into target DNA. (B) Sequence alignment of the transposases of ISDra2, the IS represented by PDB code 2fyx, and IS608 superimposed on the secondary structure (β-strands in blue, α-helices in red) of IS608 TnpA. The amino acid numbering is for ISDra2 transposase. The active site His (on the fourth β-strand) and Tyr residues (on helix αD) are in bold, and residues putatively involved in the trans-to-cis transition in blue.
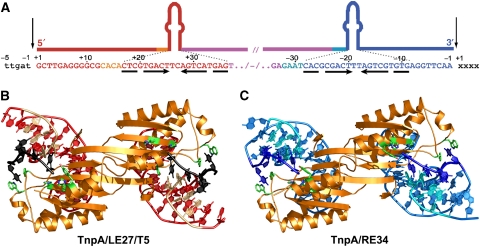
Figure 2.
Structures of TnpADra2 bound to its transposon ends. (A) ISDra2 LE and RE DNA sequences. The overall structures of the TnpADra2 dimer bound to (B) the LE IP (in red and orange) and a short ssDNA (in black) representing the five nucleotides 5′ of the LE cleavage site, which is equivalent to the target site, and (C) the RE IP (in light and dark blue) extended to the transposon 3′ end. The DNA colour scheme corresponds to that shown in Figures 1A and 2A. Residues of TnpADra2 highlighted in green are specifically referred to in the text (Y30, H67, H69, W107, and Y132).
Although most ISs encode transposase enzymes that are members of the retroviral integrase superfamily (Hickman et al, 2010), IS200/IS605 transposases are ssDNA endonucleases belonging to the vast HUH nuclease superfamily. Proteins with HUH nuclease domains are also involved in a diverse variety of other processes such as the initiation of rolling circle replication and conjugative DNA transfer, and replication initiation of certain ssDNA viruses (Koonin and Ilyina, 1993). HUH nucleases contain one or two active site tyrosine residues that act as nucleophiles during DNA cleavage. Upon nucleophilic attack, the transposase becomes covalently attached to DNA through a 5′-phosphotyrosine intermediate, and a free 3′-OH group is released on the other side of the DNA break. DNA cleavage also requires a divalent metal ion cofactor that is coordinated by the two histidine residues that comprise the HUH motif (Datta et al, 2003; Guasch et al, 2003). DNA strand transfer is achieved when a 3′-OH end of a DNA strand attacks the 5′-phosphotyrosine linkage, resulting in a resealed phosphodiester backbone.
A mechanistic model of ssDNA transposition in the IS200/IS605 family has been derived from biochemical experiments and structural data obtained for one of its members, the transposase TnpA of IS608 from Helicobacter pylori (Ronning et al, 2005; Ton-Hoang et al, 2005; Barabas et al, 2008; Guynet et al, 2008). As IS200/IS605 transposases contain only one active site tyrosine (Figure 1B), in order to coordinate cleavage at their left end (LE) and right end (RE), they assemble as dimers; in this way, there are two functional active sites and two binding sites for IP hairpins, one for LE and another for RE (shown schematically in Figure 1A).
Studies on IS608 TnpA revealed two important keys to transposition. The first is that the cleavage sites at both the LE and RE are recognized through non-sequential base-pairing interactions between nucleotides at the cleavage site (shown as a white box at either end in Figure 1A) and four nucleotides at the 5′ base of the IPs (shown as solid orange and light blue boxes). As the sequences of the 4-nt 5′ extensions of the IPs are different at LE and RE, specific recognition of the different LE and RE cleavage site sequences becomes possible. Furthermore, as the circular transposon intermediate to be integrated contains the LE IP and its 4 nt 5′ extension, target site recognition is mechanistically identical to the recognition of the LE cleavage site. The second key is that, after cleavage at LE and RE (step 2 in Figure 1A), a conformational change is believed to occur that swaps the 5′-phosphotyrosine linkages between the two monomers (step 3) in the dimer through the exchange of a mobile α-helix. Resolving these linkages (step 4) yields a circular transposon and a resealed donor DNA strand. Integration occurs when the transposase dimer bound to the circular intermediate captures an ssDNA that contains the target sequence, and the reaction cycle repeats: two cleavages, one swap, and two resolution steps later, the result is an integrated transposon.
Although we now know a great deal about the mechanism of IS608 transposition, some aspects cannot be straightforwardly generalized to other family members. For example, the IS608 target sequence is a tetranucleotide (TTAC) whereas the target sequence of ISDra2 is a pentanucleotide (TTGAT; Islam et al, 2003). It is not clear how the recognition rules derived from the IS608 structures, appropriate for tetranucleotides, would allow recognition of a pentanucleotide. As the mechanism determined for IS608 TnpA can be exploited to redirect insertions to preselected and specified tetranucleotide sites (Guynet et al, 2009), understanding how longer target sequences are recognized may prove extremely useful. Furthermore, the discrimination between the top- and bottom-strand IPs of IS608 TnpA is influenced by nucleotides present at the tip of the hairpin and an extrahelical base in the stem of the IP; there is no such base in the stem of the IPs of ISDra2 leaving open the question of how the two strands are distinguished. Finally, certain amino acids that we concluded should be crucial for IS608 TnpA transposition are simply not found in the ISDra2 transposase.
We describe here a series of crystal structures of the ISDra2 transposase bound to a variety of ssDNA substrates representing either the LE or RE of ISDra2. In conjunction with biochemical and in vivo experiments, these structures expand our understanding of how IS200/IS605 transposases work. In particular, we have captured the transposase with a fully assembled active site including the scissile phosphate, a state that eluded us with IS608 TnpA. We also provide structural explanations for pentanucleotide recognition, strand discrimination, and the metal ion specificity of DNA cleavage.
Results
We have determined crystal structures of four complexes of the ISDra2 transposase (TnpADra2) with various bound DNAs representing several stages of the transposition reaction (Table I). Recent biochemical work has confirmed that TnpADra2 uses ssDNA substrates in in vitro transposition reactions (Pasternak et al, 2010), as does the IS608 transposase. One structure (Figure 2B) is of a ternary complex (designated TnpA/LE27/T5) in which TnpADra2 is complexed with an oligonucleotide containing the LE IP (designated LE27, from LE nt +11 to +37; Figure 2A) and a 5-nt oligonucleotide (T5) comprising the target site, TTGAT. We have also solved a complex (designated TnpA/RE34) between TnpADra2 and a 34-nt oligonucleotide that contains the RE IP and extends to the RE cleavage site (RE nt −34 to −1; Figure 2C). In addition, we have solved two other LE ternary complexes. TnpA/Cd is the complex between TnpADra2 Y132F (the active site Tyr has been mutated to Phe), LE27, and an oligonucleotide in which the target site has been extended by 1 nt, TTGAT↓G, to include the first 5′ nt of the transposon (designated T5↓G where ↓ indicates the cleavage site). TnpA/Cd was crystallized in the presence of Cd2+, a divalent metal ion that robustly supports cleavage. The last complex, TnpA/Zn, contains wild-type TnpA and the same oligonucleotides as TnpA/Cd, but was crystallized in the presence of Zn2+ which supports only a very low level of cleavage. TnpA/LE27/T5 was solved with experimental phasing; the other structures were solved with molecular replacement using TnpA/LE27/T5 as the search model. Representative electron density is shown in Supplementary Figure S1.
Table 1. Crystallographic structure determination and refinement.
| TnpA/LE27/T5 (native) | TnpA/LE27/T5 (I derivative) | TnpA/RE34 | TnpA/Cd | TnpA/Zn | |
|---|---|---|---|---|---|
| Data collection | |||||
| Space group | P212121 | P212121 | P21 | P212121 | P212121 |
| Cell dimensions, a, b, c (Å) | 104.7, 128.2, 140.6 | 104.4, 128.6, 140.1 | 76.8, 89.3, 90.1 | 50.3, 86.9, 128.3 | 49.8, 92.7, 126.1 |
| β (deg) | 111.2 | ||||
| Resolution (Å) | 2.3 | 2.4 | 2.3 | 1.9 | 1.9 |
| Total reflections | 575 978 | 515 046 | 181 638 | 333 244 | 327 763 |
| Unique reflections | 84 171 | 74 480 | 50 130 | 44 818 | 45 279 |
| I/σ(I) | 15.9 (2.4) | 11.2 (2.5) | 16.4 (3.1) | 13.4 (6.91) | 16.4 (4.5) |
| Completeness (%) | 99.5 (97.6) | 100.0 (99.8) | 99.6 (99.0) | 99.8 (99.1) | 96.7 (75.9) |
| Rsym | 0.07 (0.83) | 0.103 (0.716) | 0.049 (0.341) | 0.093 (0.186) | 0.048 (0.244) |
| SIRAS phasing at 3.0 Å | |||||
| RCullis | 0.64 | ||||
| RKraut (iso/ano) | 0.163/0.203 | ||||
| Phasing power (iso/ano) | 1.49/1.9 | ||||
| Combined FOM | 0.402 | ||||
| Molecular replacement at 4.0 Å | |||||
| Rotation function corr. coeff. | 0.209/0.204 | 0.146 | 0.137/0.127 | ||
| Rfactor after rigid body refinement | 0.417 | 0.423 | 0.368 | ||
| Refinement | |||||
| Resolution (Å) | 30.0–2.3 | 30.0–2.3 | 30–1.9 | 30.0–1.9 | |
| No. reflections (F⩾0) | 78 520 | 47 910 | 44 616 | 44 429 | |
| Rwork/Rfree | 0.218/0.249 | 0.215/0.239 | 0.185/0.214 | 0.177/0.210 | |
| No. atoms | |||||
| Protein and nucleic acid | 10 055 | 7236 | 3447 | 3444 | |
| Solvent | 548 | 164 | 401 | 473 | |
| B-factors | |||||
| Protein and nucleic acid | 39.34 | 64.63 | 27.07 | 22.82 | |
| Solvent | 38.18 | 45.30 | 38.76 | 33.49 | |
| R.m.s.d. | |||||
| Bond lengths (Å) | 0.009 | 0.01 | 0.017 | 0.013 | |
| Bond angles (deg) | 4.1 | 3.00 | 1.94 | 4.54 | |
| Dimers per AU | 3 | 2 | 1 | 1 | |
| Ramachandran plot statistics | |||||
| Most favoured | 92.1% | 92.7% | 93.1% | 94.4% | |
| Disallowed | 0 | 0 | 0 | 0 | |
IP recognition by TnpADra2
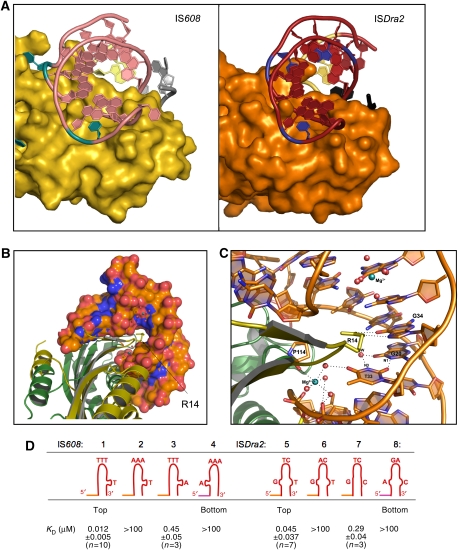
A characteristic feature of transposon DNA sequences in the IS200/IS605 family are subterminal IP sequences located close to the cleavage sites. As only the top strand is cut and subsequently moved to a new genomic location, the transposase must be able to distinguish the top-strand IP hairpins from those on the bottom. The previously determined structures of IS608 TnpA (Ronning et al, 2005) demonstrated that one important discriminating feature is the tip of the hairpin loop, where the 5′-most T at the tip (shown in cyan in Figure 3A) sits in a pocket on the transposase surface which would not be able to accommodate the corresponding adenine that is present at the tip of the bottom-strand IP hairpin. The structures of TnpADra2 bound to the LE IP or the RE IP indicate that this element of discrimination is retained: in all structures, one thymine at the hairpin tip (shown in blue in Figure 3A) is flipped back into a small pocket where it stacks against the peptide plane of Gly89.
Figure 3.
DNA binding by TnpADra2: G:T mismatch recognition and discrimination between the top and bottom strand of the transposon. (A) Comparison of the binding of the tip of the hairpin loop by IS608 TnpA (left; PDB code 2A6O) and TnpADra2 (right). The distinguishing features of the hairpins are shown in shades of blue: the unpaired T in the stem of the IS608 hairpin and the T at the hairpin tip (left), and the mismatched G:T base pair and the T at the hairpin tip of the ISDra2 hairpin (right). (B) Space-filling representation of the complex between TnpADra2 (two subunits shown in shades of green) and LE27 (in orange) highlighting the position of Arg14. (C) A web of hydrogen-bonding interactions and a bound Mg2+ ion mediate the interaction between Arg14 and the G+20:T+33 mismatch on the LE. (D) Dissociation constants for binding between the IS608 and ISDra2 transposases and their IPs and variants determined by fluorescence anisotropy measurements.
The other IP-discriminating feature for IS608 TnpA is an unpaired thymine within the IP stem, which is flipped out and is specifically recognized by the transposase (also shown in cyan in Figure 3A). This structural feature is not conserved in ISDra2. Instead, each IP stem of ISDra2 contains a G:T mismatch (G+20:T+33 on the LE, G−25:T−12 on the RE; Figure 2A) that is recognized and apparently stabilized by TnpADra2.
The key amino acid in the recognition of the G:T mismatch is Arg14, located at the tip of a β-hairpin that dives deep into the major groove and stacks on the top of the mismatched T (shown for T+33 on LE in Figure 3B and C) while forming hydrogen bonds to the immediately adjacent G (G+34 on LE). These interactions twist the DNA such that the Watson–Crick faces of the mismatched nucleotides are turned away from each other thereby avoiding a clash between the protonated N1 of G+34 and the protonated N3 of T+33. A consequence of these DNA distortions is the accumulation of negative charge on one side of T+33 as O4 of T+33 moves toward the sugar-phosphate backbone of the opposite strand. This electrostatic problem is thwarted by the creation of a Mg2+-binding site (Figure 3C). The octahedrally coordinated Mg2+ ion also provides a hydrogen bond to O4 of T+33 through one of its coordinated water molecules. It seems likely that this element of mismatch recognition also contributes to discrimination between the top and bottom strands as such a water-mediated H-bond could not be formed on the bottom-strand IP: with the C:A mismatch of the bottom-strand IP, the Mg2+-bound water molecule cannot act as an H-bond acceptor with the protonated N4 of the cytosine. In all four TnpADra2/DNA complexes, the mode of recognition and stabilization of the G:T mismatch is essentially identical.
Relative to the IP, the location of the G:T mismatch in the ISDra2 stem is exactly where the unpaired base yielding the flipped out thymine is for IS608. The flipped out T is recognized by IS608 TnpA by sequestration between the side chains of Arg52 and Phe75, and by a set of H-bonds (Ronning et al, 2005), whereas the crucial residue for recognizing and stabilizing the G:T mismatch of ISDra2 is Arg14. None of these residues are conserved between the two proteins (Figure 1B).
One final factor that influences IP recognition and strand discrimination by TnpADra2 is the relative ease of hairpin formation by the top- and bottom-strand IPs. Whereas the top-strand IPs on both the LE and RE readily fold into hairpin structures (as assessed by size-exclusion chromatography; data not shown), attempts to form a hairpin with the bottom-strand ISDra2 LE IP sequence were unsuccessful due to its vast preference to form annealed dsDNA products. Even when the small amount of the bottom-strand hairpinned form was chromatographically separated from the dsDNA form, the former rapidly converted to the latter, even at 4°C. Indeed, thermodynamic data suggest that whereas G:T mismatches are well tolerated in dsDNA oligonucleotides, C:A mismatches are less so (SantaLucia and Hicks, 2004).
Both the hairpin tip and the stem IP irregularity are important for IP recognition
The structures of IS608 TnpA and TnpADra2 bound to their IPs suggest that two IP features are important for recognition: the bases at the hairpin tip and an imperfection in the hairpin stem. To investigate the relative influence of these, we determined dissociation constants for IP binding both to IS608 TnpA and TnpADra2 using fluorescence anisotropy with various carboxyfluorescein (FAM)-modified oligonucleotides.
As shown in Figure 3D, modification of either feature resulted in a decrease in binding. The effect was most dramatic when pyrimidine bases at the tip of the IS608 and ISDra2 IPs were changed to purines; for both transposases, this resulted in a complete loss of detectable IP binding (compare IPs #1–#2, and #5–#6). This is consistent with structural observations that the 5′-most T at the hairpin tips sit in a pocket located on each transposase surface too small for a larger purine base.
The imperfections in the IP stems appear to be less important contributors to binding than the bases at the hairpin tips. For IS608 TnpA, changing the flipped out T in the IP stem to A (IP #3) reduced binding ∼40-fold relative to the wild-type IP, whereas correction of the mismatch in the stem of the ISDra2 IP (IP #7) led to an approximately six-fold reduction in binding.
We also tested the ability of the two transposases to bind to their bottom-strand IPs (IP #4 and #8). There was no detectable binding in either case, demonstrating that the discrimination between the top and bottom strand of the transposon occurs at the initial stage of transposase binding.
The role of TnpADra2 Arg14
To evaluate the role of Arg14 in vivo, we used a recently developed genetic system in D. radiodurans (Pasternak et al, 2010) to measure the first stage in IS transposition, excision of the IS from the donor backbone. In this system, the unique active copy of ISDra2 has been replaced by a mutated derivative, ISDra2-113, that is inserted at the unique 5′-TTGAT-3′ target site present in the tetA gene of a TetR cassette; in this way, precise excision of the IS restores a TetR phenotype. In ISDra2-113, both tnpA encoding the transposase and a second gene, tnpB, carried by the IS but which is not necessary for transposition (Pasternak et al, 2010) have been deleted, and the functional ends for transposition have been retained. TnpADra2 is expressed in trans from a plasmid expression vector.
As shown in Table II, precise excision of ISDra2-113 was strictly dependent on the presence of TnpADra2. In strains devoid of plasmid or carrying an empty vector, the frequency of TetR colonies was undetectable, but increased to ∼3 × 10−3 when wild-type TnpADra2 was expressed. When wild-type TnpADra2 was replaced by the R14A mutant, the excision frequency dropped ∼60-fold, supporting a crucial role of Arg14 in IP recognition and subsequent transposon excision.
Table 2. Excision frequencies of the ISDra2-113 derivative in a Deinococcus radiodurans tester straina expressing TnpADra2 variants.
| Resident plasmid | TnpADra2 expressed in trans | Excision frequency |
|---|---|---|
| No | No | <10−8 |
| pGY11559 | No | <10−8 |
| pGY13203 | TnpA wt | 3.2 (±0.6) × 10−3 |
| pGY13521 | TnpAR14A | 5.3 (±1.6) × 10−5 |
| pGY13522 | TnpAS122G/E123G | 1.7 (±0.3) × 10−3 |
| aGY13115 genotype: ΔDR1652ΔDR1651ΩtetAΩISDra2-113 (CamR) (Pasternak et al, 2010). | ||
We also measured the binding affinity of purified TnpADra2 R14A protein to its IP using our fluorescence anisotropy assay. The dissociation constant was ∼1.4 μM, an ∼30-fold loss in binding relative to wild type, which is consistent with the corresponding decrease in excision frequency.
Helix αD is a mobile structural element that can be in cis or trans
A key feature of the molecular model of ssDNA transposition is a large conformational rearrangement that exchanges the location of the two αD helices containing the tyrosine nucleophiles between the two monomers of the transposase dimer (Figure 1A, step 3). Indeed, biochemical data strongly suggest that while a trans-active site arrangement (i.e. the tyrosine of one monomer is close to the HUH motif of the other) is catalytically competent for transposon end cleavage, the resolution steps require a trans/cis isomerization (Barabas et al, 2008). However, we have not yet observed a IS200/IS605 family transposase in the cis configuration. In all the structures of IS608 TnpA determined to date, the active sites are arranged in trans and TnpADra2 is no different. Nevertheless, there does appear to be a precedent for a cis-active site. The coordinates of a high resolution crystal structure of an uncomplexed IS200/IS605 family transposase that is 80.8% identical to TnpADra2 (Figure 1B) have been deposited in the Protein Data Bank under the PDB ID 2fyx, and the protein is in the cis conformation. As the 2fyx transposase is a result of structural genomics efforts, unfortunately no biochemical data are available, and it must be noted that the HUH motif is not intact.
For the trans-to-cis structural transition to take place, modelling suggested that a seven amino acid long loop just upstream of helix αD must change its structure substantially. For IS608 TnpA, this seems feasible as the loop contains two adjacent Gly residues, Gly117 and Gly118, that can act as pivots as the main chain dihedral angle of glycine is less restricted than that of all other amino acids. However, in TnpADra2 and in the protein represented by 2fyx, the equivalently placed residues are Ser and Glu and there are no glycines in the seven-residue loop (Figure 1B), raising the question of how well these residues might permit the conformational change.
We therefore examined the effect in vivo of mutating these two residues to glycine. Expression of the S122G/E123G mutant TnpADra2 gave approximately half the excision frequency observed for wild-type TnpADra2 protein (Table II). Thus, despite their bulkier side chains, Ser122 and Glu123 do not appear to impede a rate-limiting step of transposition and—despite our expectations—mutation of both to Gly does not lead to an increase in activity.
Examination of the main chain torsion angles of the seven-loop residues of the 2fyx protein and TnpADra2 reveals that all the residues are in the most favoured regions of the Ramachandran plot (Supplementary Figure S2). However, it appears that in going from the cis conformation (2fyx) to the trans conformation (all the TnpADra2 structures), the main chain dihedrals of two sequential residues change from one allowed phi/psi region to another, further supporting the notion that a trans/cis isomerization is structurally reasonable.
Both transposase and transposon DNA contribute to TnpADra2 target and cleavage site recognition
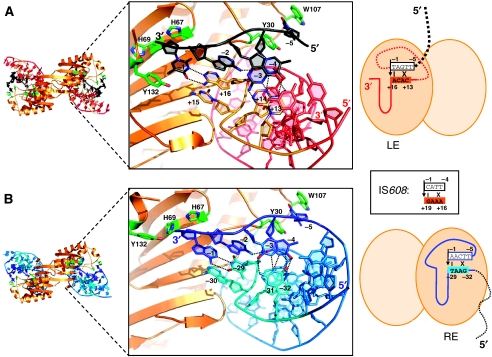
Perhaps the most surprising aspect of the mechanism of ssDNA transposition carried out by IS608 TnpA is that cleavage sites are not recognized by the transposase directly, but rather by non-sequential base pairing between the transposon 5′ extensions of the IPs and sequences just 5′ of the cleavage sites (Barabas et al, 2008). How well are these features of DNA recognition and cleavage recapitulated for ISDra2? Figure 4A and B show cleavage site recognition of the LE and RE by TnpADra2. In contrast to IS608 TnpA, TnpADra2 combines both DNA–DNA and protein–DNA interactions to recognize its cleavage sites. Four of the 5 nt just 5′ of the cleavage site (e.g. T−4, G−3, A−2, T−1 on the LE) are recognized by the 4-nt 5′ extension of the IP, the fifth (T−5) is recognized by TnpADra2 itself. The pattern of recognition echoes that of IS608, that is, the non-linear arrangement of base pairs is the same (see inset in Figure 4). On the other hand, T−5 is sandwiched between Tyr30 and Trp107, and forms hydrogen bonds with surrounding residues including His32 and main chain atoms of Lys106 and Trp107. Neither Trp107 nor His32 is conserved between IS608 TnpA and TnpADra2, and the transposase structures differ in this region due to a shift of 5–6 Å in the position of a short α-helix, which in TnpADra2 is between Gln101 and Trp107 and contains residues that interact with T−5.
Figure 4.
Transposon end and cleavage site recognition by TnpADra2. On the left are the overall TnpADra2 dimer structures corresponding to the complexes TnpA/LE27/T5 (A) and TnpA/RE34 (B). In the middle are close-up views of the active sites. The cartoons on the right show the state during the transposition pathway captured by the crystal structures, corresponding to the model in Figure 1A. In these cartoons, the vertical black arrows correspond to the sites of cleavage and the pattern of base-pairing interactions at the cleavage sites at LE (A) and RE (B) are indicated by the black lines. The inset box shows the corresponding pattern of base-pairing interactions for IS608 (Barabas et al, 2008). The dashed lines indicate portions of DNA not present in the structures.
The first view of a fully assembled active site
To date, none of the structures obtained for IS200/IS605 family transposases have shown the nuclease catalytic site in an active configuration, and we suspect there are two reasons for this. First, helix αD that contains the nucleophilic tyrosine is extremely mobile—as it presumably must be—as it is proposed to move from the trans to cis conformation to complete strand transfer. Correspondingly, in the IS608 TnpA structures helix αD does not pack tightly against the body of the molecule, and small forces such as those resulting from lattice packing appear to influence its position. A similar phenomenon is observed here for TnpADra2. For example, in the TnpA/LE27/T5 ternary complex, there are three dimers in the crystallographic asymmetric unit related to each other mainly by non-crystallographic translations (Supplementary Figure S3). Although the three TnpADra2 transposase dimers and bound oligonucleotides are essentially structurally identical, there are major differences in the positions of the six αD helices. Simulated annealed omit electron density is clear for all of them, indicating that the catalytically important helix αD can be in several distinct positions depending on the crystal packing environment.
The second reason why we have not previously observed a fully assembled active site is that the transposase/DNA structures to date have captured a mechanistically inconsistent state of the system. That is, whereas the product of the cleavage reaction is a free 3′-OH and a phosphotyrosine intermediate, the structures have all contained the 3′-OH group in the active site but without a corresponding phosphotyrosine intermediate or even the scissile phosphate.
Our attempts to obtain ‘mechanistically consistent' complexes with IS608 TnpA have been unsuccessful, largely due to poor biophysical properties of the resulting complexes. However, the ISDra2 TnpA/LE27/T5 structure suggested an alternate approach. As seen in Figure 4A, the 3′-end base of the target sequence, T−1, is base paired to A+16 which in turn is stacked on top of C+15. As the base of C+15 is not engaged in any hydrogen bonding, the structure suggested that if the target sequence were extended by 1 nt in the 3′ direction (corresponding to the first nt of the transposon on LE), a base pair might form between C+15 and this additional nt, thereby perhaps holding the scissile phosphate in the appropriate place in the active site.
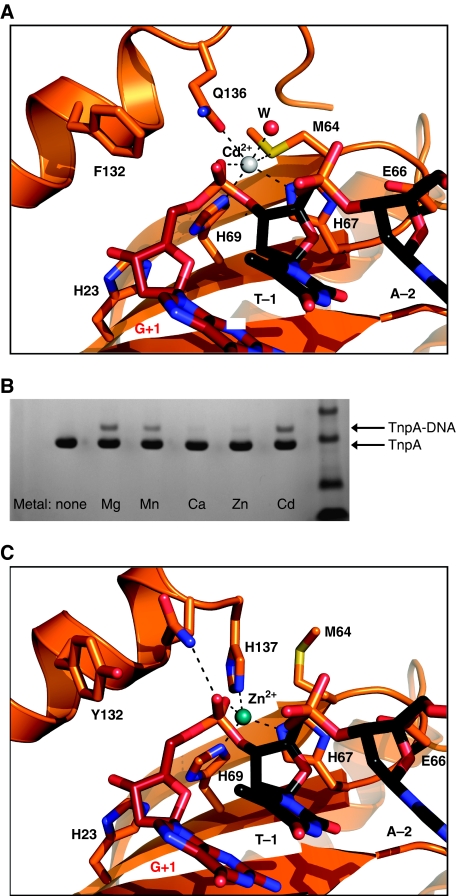
In an attempt to capture the state in the transposition pathway just prior to LE cleavage (which is mechanistically equivalent to target cleavage), we used TnpADra2 with a Y132F mutation bound to LE27 and T5↓G. The best diffracting crystals were obtained in the presence of Cd2+, which was subsequently found to be bound at the active site (Figure 5A). In the structure, the entire T5↓G substrate is visible including G+1 (which is indeed base paired to C+15) and the scissile phosphate. Cd2+ is octahedrally coordinated by His69 and His67 of the HUH motif, Gln136 on helix αD, Sδ of Met64, a water molecule, and one of the non-bridging oxygens of the scissile phosphate. His23 and Glu66 in the second coordination shell of the metal ion presumably keep His69 and His67 in tautomerization states in which the Nδ atoms are protonated so that they can ligand the metal through their Nɛ atoms. The position of His23 is such that it could form a hydrogen bond to the next phosphate group if T5↓G were extended by yet another nt into the transposon end, thus expanding the hydrogen-bonding network.
Figure 5.
The TnpADra2 active site is fully assembled in the presence of Cd2+ but is distorted by Zn2+. (A) The structure of the complex of TnpADra2 Y132F in the presence of Cd2+ (white sphere) bound to LE27 (not visible) and an oligonucleotide, T5↓G, that traverses the LE cleavage site where target nucleotides are shown in black and the first 5′ transposon nucleotide in red. (B) TnpADra2 is fully active for LE cleavage in the presence of Cd2+ but not Zn2+. Cleavage of a 13-mer target sequence results in a covalent complex between the product and TnpADra2, and is detected by Coomassie-stained SDS–PAGE as the formation of a higher molecular weight band. (C) The active site of TnpADra2 is distorted in the presence of Zn2+ (blue sphere), explaining the lack of cleavage activity.
When the position of Tyr132 was modelled based on the observed position of Phe132, it became clear that the oxygen atom of the OH group would be ∼2.6 Å from the phosphorus atom of the scissile phosphate in-line for a nucleophilic attack. To confirm that this structure represents a catalytically relevant state, we evaluated the ability of various divalent metal ions to support LE cleavage activity of the wild-type protein. As shown in Figure 5B, Mg2+, Mn2+ and Cd2+ all support LE cleavage, suggesting that the TnpA/Cd complex represents the precleavage state.
Zn2+ does not support DNA cleavage
We also obtained crystals of TnpADra2 bound to LE27 and T5↓G in the presence of Zn2+. TnpADra2 is only slightly active in 2 mM Zn2+ (Figure 5B) and increasing the zinc concentration to 25 mM does not lead to increased activity (data not shown). Although this divalent metal ion occupies the active site (Figure 5C), we observe a rearrangement of ligands such that Zn2+ is tetrahedrally coordinated, its preferred coordination state (Dudev and Lim, 2003). Three of the four ligands are those that participated in Cd2+ binding: His69 and His67 of the HUH motif and a non-bridging oxygen atom of the scissile phosphate. However, completing the tetrahedral arrangement is His137. Gln136, which in the Cd2+ structure provides a metal ligand, is now hydrogen bonded to the second non-bridging oxygen atom of the scissile phosphate. The net effect of Zn2+'s apparent preference for His137 as a ligand is a positional shift of helix αD such that Tyr132, the active site nucleophile, is pushed ∼6 Å away from the scissile phosphate providing a straightforward explanation for the lack of cleavage activity in the presence of Zn2+.
Discussion
To probe the mechanism of transposition catalysed by members of the prokaryotic IS200/IS605 family, we have been investigating D. radiodurans ISDra2, an element that presents a number of contrasts to the well-characterized H. pylori IS608. We were initially drawn to the IS200/IS605 family as it possesses the smallest known transposases and, as our initial studies revealed, they are single-domain proteins that carry out all of the chemical steps of transposition without the need for any accessory host proteins. TnpADra2 pushes the transposase size requirement even further as it does not have the final C-terminal α-helix found in IS608 TnpA and consists of a mere 140 amino acids. Although the IS608 TnpA and TnpADra2 transposases can be superimposed with a 1.4-Å r.m.s.d. over 123 Cα positions, indicating that they are structurally very similar despite only ∼34% sequence identity, our structural data suggest that amino acid diversity gives this family of proteins the flexibility to recognize and stabilize different DNAs, and to integrate into target sites of differing length.
One of the significant contrasts between these two family members is that ISDra2 specifically integrates into a pentanucleotide target sequence (TTGAT), whereas IS608 is targeted to a different sequence (TTAC). We previously established that the IS608 target site is bound to IS608 TnpA through base-pairing interactions with an internal segment of transposon DNA (Barabas et al, 2008). By analogy, one rather obvious structural solution to selecting a 5-nt target site might have been to use a 5-nt long internal segment of DNA for recognition. However, this is not what occurs for ISDra2. Rather, the tetranucleotide-to-tetranucleotide base-pairing solution is retained as a subset of the basis of recognition, and the fifth nt is specifically recognized by the protein.
This means that for the ISDra2 target sequence 5′-TTGAT, the internal segment of DNA that is used for recognition is 3′-ACAC, and the pattern of base pairing seen for IS608 is maintained. The fifth nt, T−5, is sandwiched in between residues Tyr30 and Trp107, and forms base-specific contacts to three nearby residues with a hydrogen-bonding pattern that suggests a strong preference for T at this position. Satisfyingly, a T is also present at the −5 position at the RE cleavage site.
One consequence of this mode of pentanucleotide recognition is that it indicates that the notion of being able to redirect IS200/IS605 family members to even longer target sequences is unlikely to be as straightforward as expanding the base-pairing complementarity between the target site and the 5′ 4-nt extension. Rather, whereas the complementarity approach works well for redirecting to a 4-nt target sequence (Guynet et al, 2009), incorporating a fifth nucleotide into the ISDra2 target site requires special accommodation in the form of a specific protein-binding pocket. This explains the observation that IS200/IS605 family members appear to be targeted generally only to tetra- and pentanucleotide sequences, and implies that to accommodate an even longer target site would require the dedication of even more of the limited TnpADra2 protein surface to the task.
A second point of contrast between IS608 and ISDra2 is that the IPs at the transposon ends differ in their imperfections. Whereas the IPs at the ends of IS608 have an unpaired nt within the hairpin stem, those of ISDra2 have a mismatched base pair. Our fluorescence anisotropy binding data indicate that these imperfections do not drive DNA end binding but, rather, they enhance binding and presumably account in part for strand discrimination. In two different approaches to selectivity, the two related transposases use non-conserved amino acids for hairpin imperfection recognition. For IS608, the unpaired T is bound within a precisely contoured pocket on the surface of TnpA, whereas the G:T mismatch of ISDra2 is dealt with by a set of interactions organized by an Arg residue of TnpADra2.
Although there are a number of enzymes that recognize mismatched bases as part of their function, the solution used by TnpADra2 appears unique. The G:T mismatch does not form a wobble base pair as seen, for example, in the case of the Vsr endonuclease bound to DNA (Tsutakawa et al, 1999). Instead, a combination of stacking and hydrogen-bonding interactions cause the mismatched T to turn away from the Watson–Crick face of the G. The direction of this motion is opposite to that seen when prokaryotic and eukaryotic MutS bind a G:T mismatch (Lamers et al, 2000; Warren et al, 2007): for TnpADra2, the base turns away from—rather than into—the minor groove of the DNA stem. Furthermore, MutS-bound DNA is sharply bent at the mismatch resulting in severe local narrowing of the major groove, a deformation we do not see for TnpADra2-bound hairpin DNA (although evaluating the DNA geometry is difficult as the stem is relatively short). Another major difference in the mode of G:T mismatch recognition by TnpADra2 and MutS is the nature of the amino acid side chain that stacks against the mismatched T. In MutS, a Phe has sufficient room due to the sharp bend in the DNA to reach into the widened minor groove to facilitate stacking. On the other hand, Arg14 of TnpADra2 forms hydrogen bonds with the base just 3′ of the mismatched T as it reaches into the major groove (G+34 in Figure 3C); this guanine would normally stack against the mismatched T in the standard B form DNA. To our knowledge, this is the first observation of such a geometrical arrangement in the context of mismatch recognition. It is curious that the equivalent of Arg14 is conserved in repetitive extragenic palindrome (REP)-associated tyrosine transposases, which are also members of the IS200/IS605 family (Nunvar et al, 2010; P Siguier, personal communication), suggesting a similar role for this residue in recognizing the imperfect palindromes present in bacterial REP sequences.
In contrast to residues that are involved in recognition and discrimination of the transposon IPs, all of the important active site residues are conserved between IS608 TnpA and TnpADra2. The assembled active site of TnpADra2 was captured structurally in the presence of heavy metal ions and although Cd2+ is certainly not the physiological divalent metal ion used in vivo, its octahedral-coordinating environment produces a cofactor-bound enzyme that is competent for DNA cleavage, and presumably recapitulates the authentic Mg2+-bound form. The structures presented here are consistent with a chemical mechanism proposed for HUH nucleases (Hickman et al, 2002) in which a metal ion both localizes and polarizes the scissile phosphate, setting it up for nucleophilic attack by the catalytic tyrosine. The active site arrangement of TnpADra2 is similar to that seen for TraI, an HUH nuclease involved in the initiation of conjugation (Datta et al, 2003).
We captured a second precleavage state of the enzyme in the presence of Zn2+, in which a different array of ligands was observed tetrahedrally arranged around the metal ion. Although TnpADra2 is not active in the presence of Zn2+, the related HUH nuclease domain of TrwC, the plasmid R388 relaxase, has been structurally characterized in the presence of several divalent metal ions including Zn2+, Cu2+, and Ni2+ (Boer et al, 2006). In all cases, the metal ion-binding environment is tetrahedral, and TrwC was demonstrated to be active in the presence of these three metals. Thus, the lack of activity of TnpADra2 in the presence of Zn2+ is not due to the tetrahedral metal-binding environment per se, nor is there anything untoward about using three His residues at an enzyme active site as metal-coordinating ligands (Dudev and Lim, 2003), as this is precisely what has been observed for the relaxase domains of both TrwC (Guasch et al, 2003) and TraI (Datta et al, 2003; Larkin et al, 2005). Rather, the conformational state of TnpADra2 bound to Zn2+ reflects the fact that the third Zn2+ ligand, His 137, simply happens to be at an inopportune position on helix αD.
The shift in position of the mobile helix αD that accompanies Zn2+ binding concomitantly moves the nucleophilic tyrosine residue away from the scissile phosphate, thus explaining the lack of cleavage activity in the presence of Zn2+. A shift in position of a nucleophile-bearing α-helix has been previously postulated for the prototypical HUH nuclease, the phi X174 gene A protein, this time in the context of a mechanistically critical movement of nucleophiles (van Mansfeld et al, 1986). In that case, two tyrosine residues spaced by three residues were proposed to reside on a helix, which moves during the course of rolling circle replication, alternating in access to different scissile phosphate groups alternatively bound at the active site. It is possible that helix αD mobility is a common feature of HUH nucleases, and that DNA transposases with HUH domains have exploited this inherent structural malleability in the context of transposition to direct strand transfer by allowing a trans/cis-active site isomerization.
Among DNA transposases, there seems to be a very limited number of structurally unrelated enzyme folds capable of carrying out the DNA cleavage and rejoining reactions required to move a segment of DNA from one location to another. The majority of ISs and transposons are believed to have catalytic domains with an RNase H-related fold (i.e. that of retroviral integrase; Nowotny, 2009; Hickman et al, 2010). The wide range of sequences that can be recognized and distinguished is readily understood by the fact that these enzymes are multidomain proteins containing dedicated sequence-specific DNA-binding domains (Davies et al, 2000; Watkins et al, 2004; Rice, 2005; Richardson et al, 2009). Therefore, successfully identifying their own transposon ends does not appear to be a conceptually sophisticated matter. In contrast, IS200/IS605 transposons encode small, single-domain proteins that have the ability to carry out all of the chemical reactions of transposition on their own specific transposon sequences. An additional level of complexity is that, in contrast to most RNase H-like transposases, they insert into specific target sequences. The series of protein/DNA complex structures presented here sheds light on the types of structural differences that account for the ability of a small protein to not only distinguish among transposon DNA sequences but also to identify specific target sites in the absence of any site-specific DNA-binding domains. The variability—an Arg14 here or an Arg52/Phe75 pair there—is subtle yet remarkable in its simplicity.
Materials and methods
Protein purification and crystallization
Oligonucleotides were either obtained desalted from IDT (Coralville, IA) or synthesized using an Applied Biosystems 394 DNA/RNA synthesizer. Iodinated LE27 in which T+18, T+21, T+25, T+30, and T+33 were substituted with 5-I-dU was synthesized using 5-I-dU-CE phosphoramidite (Glen Research, VA); in all cases, oligonucleotides were used without further purification. The gene encoding ISDra2 TnpA was cloned between the NcoI and XhoI sites of pET-15b. R14A and S122G/E123G mutations were introduced by the QuikChange method (Stratagene).
Protein was expressed in BL21 (DE3) cells upon induction by addition of IPTG to a final concentration of 0.5 mM followed by cell growth overnight at 16°C. Cells were harvested, resuspended in 35 mM Tris pH 8.0, 0.5 M NaCl, and 10% (w/v) glycerol, and frozen at −80°C until use. Thawed cells were diluted 1:1 (v/v) with 35 mM Tris pH 8.0, 0.5 M NaCl, 5% glycerol, 5 mM imidazole (Im), and 4 mM β-mercaptoethanol, sonicated, and centrifuged to recover soluble protein. Protein was loaded onto a 5-ml Ni-affinity column and washed sequentially with lysis buffer containing 5 mM Im, then 50 mM Im, followed by gradient elution from 0.05 to 0.5 M Im. Fractions containing TnpADra2 were combined, thrombin (Sigma) added to a final concentration of 2 U/mg protein, and the solution was dialyzed overnight at 4°C against 35 mM Tris pH 8.0, 0.5 M NaCl, 5% glycerol, and 0.5 mM TCEP. Thrombin was removed by passage over benzamidine Sepharose prior to gel filtration on a TSK-SW3000 column. Fractions containing TnpADra2 were concentrated, combined with the appropriate oligonucleotide(s), and dialyzed against 35 mM Tris pH 8.0, 0.15 M NaCl (LE IP) or 0.1 M sodium malonate (RE IP), 10 mM MgCl2, and 0.5 mM TCEP.
The TnpA/LE27/T5 complex was formed by mixing protein at 10 mg/ml with LE27 and T5 at a molar ratio of 1:1.1:1.3 prior to dialysis. Crystals were obtained by the hanging drop method at 19°C by mixing the complex 1:1 with well solution containing 15–20% PEG 6000 and 50 mM sodium acetate pH 4.6. Crystals were cryoprotected by slow soak into 17 mM Tris pH 8.0, 75 mM NaCl, 0.25 mM TCEP, 5 mM MgCl2, 50 mM sodium acetate pH 4.6, 25% PEG 6000 and 15–20% glycerol, and frozen by immersion in liquid propane.
The TnpA/RE34 complex was formed by mixing protein at 10 mg/ml with RE34 DNA at a molar ratio of 1:1.1 prior to dialysis. Crystals were obtained by the hanging drop method at 19°C by mixing the complex 1:1 with well solution containing 0.16 M MgCl2, 0.1 M Tris pH 7.9, and 3.4 M 1,6-hexanediol. Crystals were frozen directly in liquid propane prior to data collection.
The TnpA/Cd complex was formed by mixing protein with a Y132F mutation at 9 mg/ml with LE27 and T5↓G at a molar ratio of 1:1.1:1.3 prior to dialysis. Crystals were obtained overnight at 19°C by hanging drop vapor diffusion upon mixing the complex 1:1 with well solution containing 10 mM CdCl2, 0.1 M sodium acetate pH 4.8, and 14% PEG 4000. Crystals were cryoprotected by serial transfer into solutions containing 17 mM Tris pH 8.0, 75 mM NaCl, 0.25 mM TCEP, 5 mM MgCl2, 5 mM CdCl2, 50 mM sodium acetate pH 4.8, 14% PEG 4000, and increasing glycerol concentration to a final concentration of 20% (v/v). Crystals were frozen by immersion in liquid propane.
The TnpA/Zn complex was formed as described for TnpA/Cd but with the wild-type protein. Crystals were obtained as for TnpA/Cd but with a well solution containing 10 mM ZnCl2, 0.1 M sodium acetate pH 4.2, and 17% PEG 4000.
Crystallographic data collection and structure determination
All diffraction data were collected at 95K on a rotating anode source equipped with multilayer focusing optics using Cu Kα radiation and an Raxis IV image-plate detector. Data were integrated and scaled with Denzo and Scalepack (Otwinowski and Minor, 1997; Table I). Attempts to solve the TnpA/LE27/T5 structure with molecular replacement using IS608 TnpA as a search model were only partially successful, presumably due to the pseudo-translational symmetry between the three dimers in the asymmetric unit. Although two dimers could be located clearly, the orientation and position of the third remained ambiguous. Difference Fourier maps for the third dimer phased with the first two dimers were not of sufficient quality to place it. Therefore, single isomorphous replacement with anomalous scattering experimentally phased maps were calculated based on an iodine derivative obtained using iodinated LE27. Iodine sites were located with Shelxd (Sheldrick, 1998) and their positional and occupancy parameters were refined with Phases-95 (Furey and Swaminathan, 1997) using a phase integrating least squares procedure. The TnpA/RE34 and TnpA/Cd structures were solved with molecular replacement using AMoRe (Navaza, 2001) using the TnpA/LE27/T5 dimer with the lowest average atomic displacement parameters (ADPs) as a search model. TnpA/Zn was solved with a monomeric search model. All molecular models were built with O (Jones et al, 1991) and refined with Cartesian simulated annealing. Non-crystallographic symmetry restraints were used only in the case of TnpA/RE34. Individual ADP refinement and energy minimization was carried out with CNS 1.1 (Brünger et al, 1998). All structures were verified with composite simulated annealed omit maps. Molecular figures were generated using Pymol (DeLano, 2002). Coordinates have been deposited in the Protein Data Bank with the accession codes 2xm3 (TnpA/LE27/T5), 2xma (TnpA/RE34), 2xo6 (TnpA/Cd), and 2xqc (TnpA/Zn).
Fluorescence anisotropy measurements
HPLC-purified 3′ FAM-labelled oligonucleotides were purchased from IDT. For IS608 TnpA, all top-strand oligonucleotides were 27 nt long, and corresponded to LE nt +16 to +42 (5′-AAAGCCCCTAGCTTTTAGCTATGGGGA; numbering as in Barabas et al (2008)), with the modifications shown in Figure 3D. For TnpADra2, the oligonucleotides were 26 nt long, and corresponded to LE nt +13 to +38 (5′-CACACTCGTGACTTCAGTCATGAGTT). For bottom-strand oligonucleotides, the appropriate reverse complement sequences were used.
Oligonucleotides were annealed by resuspension in TE, heating to 95°C followed by rapid cooling on ice or dry ice. The extent of hairpin formation versus double-stranded dimers was monitored by size-exclusion chromatography on Superdex 200 using a Pharmacia SmartSystem. In the case of the ISDra2 bottom-strand hairpin (the only oligonucleotide where the amount of dimer was significant), this also allowed for partial separation of the two species for use in subsequent experiments.
To measure dissociation constants for the interaction between IS608 TnpA or TnpADra2 and various modified IPs, anisotropy changes of FAM-labelled oligonucleotides were monitored upon titration with protein. Each oligonucleotide was added to the highest protein concentration sample (generally 2–5 mg/ml) to a final concentration of either 50 nM DNA (IS608 TnpA) or 40 nM (TnpADra2). Serial dilutions of the protein–DNA mixture were performed in low adsorption 96-well plates (Corning) with buffer (0.2 M NaCl, 20 mM Tris pH 7.5, 20 mM MgCl2, 1 mM DTT) containing the appropriate amount of DNA to keep it constant throughout the experiment. Experiments were performed in triplicate using black, clear flat bottom, 384-well plates (Corning) using a SpectraMax M5 micro-plate reader (Molecular Devices). The excitation wavelength was 493 nm and emission wavelength was 525 nm (cutoff 515 nm).
For each experiment performed in triplicate, fluorescence anisotropy change values were calculated as observed anisotropy minus initial anisotropy (detected in the absence of protein) and plotted as a function of protein concentration. Binding data were fit to a single-binding site model using Microcal Origin 6.0 and the equation A=Af+(Ab−Af) × {(LT+Kd+x)−√[(LT+Kd+x)2−4LTx]}/2LT, where A is the measured anisotropy change, Af is the anisotropy for the free ligand, Ab is the anisotropy for the bound ligand, LT is the total added ligand concentration, and x is the protein concentration. The single-binding site model was tested by analysing Hill plots; no allosteric behaviour was detected. Reported Kd values correspond to the average value for the indicated number of experiments, and the uncertainties correspond to SD values. For Kd values >100 μM in Figure 3D, at the highest protein concentration used, the binding curve did not reach saturation and the Kd was estimated to be over 100 μM (i.e. the apparent inflection point of the sigmoidal-binding curve on a log plot was above 100 μM).
LE cleavage assay
A TnpADra2/LE24 (LE nt +13 to +36) complex was prepared by mixing purified protein and DNA both at 60 μM, followed by overnight dialysis into 0.2 M NaCl, 20 mM Tris pH 7.5, and 0.2 mM TCEP. Cleavage was initiated by addition of 1.1 molar excess of a 13-mer target sequence (5′-TTGATGCTTGAGG) and the appropriate volumes of stock solutions of MgCl2, MnCl2, CaCl2, ZnCl2, or CdCl2 to a final concentration of 2 mM. Reactions were quenched after 2 h at 22°C with SDS–PAGE sample buffer, and the formation of covalently modified TnpADra2 was monitored by SDS–PAGE using 4–12% NuPAGE gels (Invitrogen).
D. radiodurans in vivo excision assay
Bacterial strains, media, and growth conditions have been previously described (Bonacossa de Almeida et al, 2002; Pasternak et al, 2010), as have the construction of the D. radiodurans GY13115 tester strain and the ISDra2-113 mini-transposon (Pasternak et al, 2010).
The D. radiodurans ISDra2 tnpA mutant genes encoding TnpAR14A or TnpAS122G E123G were isolated from the corresponding pET-15b derivatives by NdeI–XhoI restriction and then cloned into plasmid pGY11559 (Mennecier et al, 2004) to generate pGY13521 and pGY13522, respectively. Wild-type tnpA was cloned into pGY11559 to generate pGY13203 (Pasternak et al, 2010) and all constructs were verified by DNA sequencing.
To measure the in vivo excision frequencies of ISDra2-113, 10 individual CamR TetS colonies purified from each derivative of GY13115 strain expressing in trans wild type, mutant, or no TnpADra2 were inoculated into 3 ml of TGY2X supplemented with spectinomycin and grown to an OD650 of 1.3. Determination of the total number of viable cells was performed on TGY plates and excision of ISDra2-113 from the tetA gene was selected on TGY plates containing tetracycline. Colonies were counted after 4 days at 30°C. The frequencies of the excision event per viable cell from these 10 independent measurements were used to calculate mean values and standard deviations.
Supplementary Material
Acknowledgments
We thank A Bailone for fruitful discussions and G Coste for expert technical assistance. This work was supported by the Intramural Program of the National Institute of Diabetes, Digestive, and Kidney Diseases of the National Institutes of Health (FD), the Centre National de la Recherche Scientifique (France, MC and SS), European contract LSHM-CT-2005-019023 (MC), and ANR grant Mobigen (MC and SS). Some data were collected at the Southeast Regional Collaborative Access Team 22-ID beamline at the Advanced Photon Source (APS), Argonne National Laboratory. Use of the APS was supported by the US. Department of Energy, Basic Energy Sciences, Office of Science, under Contract No. W-31-109-Eng-38.
Footnotes
The authors declare that they have no conflict of interest.
References
- Barabas O, Ronning DR, Guynet C, Hickman AB, Ton-Hoang B, Chandler M, Dyda F (2008) Mechanism of IS200/IS605 family DNA transposases: activation and transposon-directed target site selection. Cell 132: 208–220 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Boer R, Russi S, Guasch A, Lucas M, Blanco AG, Pérez-Luque R, Coll M, de la Cruz F (2006) Unveiling the molecular mechanism of a conjugative relaxase: the structure of TrwC complexed with a 27-mer DNA comprising the recognition hairpin and the cleavage site. J Mol Biol 358: 857–869 [DOI] [PubMed] [Google Scholar]
- Bonacossa de Almeida C, Coste G, Sommer S, Bailone A (2002) Quantification of RecA protein in Deinococcus radiodurans reveals involvement of RecA, but not LexA, in its regulation. Mol Genet Genom 268: 28–41 [DOI] [PubMed] [Google Scholar]
- Brünger AT, Adams PD, Clore GM, Delano WL, Gros P, Grosse-kunstleve RW, Jiang JS, Kuszewski J, Nilges M, Pannu NS, Read RJ, Rice LM, Simonson T, Warren GL (1998) Crystallography and Nmr system—a new software suite for macromolecular structure determination. Acta Crystallogr D 54: 905–921 [DOI] [PubMed] [Google Scholar]
- Datta S, Larkin C, Schildbach JF (2003) Structural insights into single-stranded DNA binding and cleavage by F Factor TraI. Structure 11: 1369–1379 [DOI] [PubMed] [Google Scholar]
- Davies DR, Goryshin IY, Reznikoff WS, Rayment I (2000) Three-dimensional structure of the Tn5 synaptic complex transposition intermediate. Science 289: 77–85 [DOI] [PubMed] [Google Scholar]
- DeLano WL (2002) The PyMol molecular graphics system Available at http://www.pymol.org
- Dudev T, Lim C (2003) Principles governing Mg, Ca, and Zn binding and selectivity in proteins. Chem Rev 103: 773–787 [DOI] [PubMed] [Google Scholar]
- Furey W, Swaminathan S (1997) PHASES-95: a program package for processing and analyzing diffraction data from macromolecules. Methods Enzymol 277: 590–620 [DOI] [PubMed] [Google Scholar]
- Guasch A, Lucas M, Moncalian G, Cabezas M, Perez-Luque R, Gomis-Ruth FX, de la Cruz F, Coll M (2003) Recognition and processing of the origin of transfer DNA by conjugative relaxase TrwC. Nat Struct Biol 10: 1002–1010 [DOI] [PubMed] [Google Scholar]
- Guynet C, Achard A, Ton-Hoang B, Barabas O, Hickman AB, Dyda F, Chandler M (2009) Resetting the site: redirecting integration of an insertion sequence in a predictable way. Mol Cell 34: 612–619 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Guynet C, Hickman AB, Barabas O, Dyda F, Chandler M, Ton-Hoang B (2008) In vitro reconstitution of a single-stranded transposition mechanism of IS608. Mol Cell 29: 302–312 [DOI] [PubMed] [Google Scholar]
- Hickman AB, Chandler M, Dyda F (2010) Integrating prokaryotes and eukaryotes: DNA transposases in light of structure. Crit Rev Biochem Mol Biol 45: 50–69 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hickman AB, Ronning DR, Kotin RM, Dyda F (2002) Structural unity among viral origin binding proteins: crystal structure of the nuclease domain of adeno-associated virus Rep. Mol Cell 10: 327–337 [DOI] [PubMed] [Google Scholar]
- Islam MS, Hua Y, Ohba H, Satoh K, Kikuchi M, Yanagisawa T, Narumi I (2003) Characterization and distribution of IS8301 in the radioresistant bacterium Deinococcus radiodurans. Genes Genet Syst 78: 319–327 [DOI] [PubMed] [Google Scholar]
- Jones TA, Zou JY, Cowan SW, Kjeldgaard M (1991) Improved methods for building protein models in electron density maps and the location of errors in these models. Acta Crystallogr A 47: 110–119 [DOI] [PubMed] [Google Scholar]
- Kersulyte D, Akopyants NS, Clifton SW, Roe BA, Berg DE (1998) Novel sequence organization and insertion specificity of IS605 and IS606: chimaeric transposable elements of Helicobacter pylori. Gene 223: 175–186 [DOI] [PubMed] [Google Scholar]
- Kersulyte D, Velapatino B, Dailide G, Mukhopadhyay AK, Ito Y, Cahuayme L, Parkinson AJ, Gilman RH, Berg DE (2002) Transposable element ISHp608 of Helicobacter pylori: nonrandom geographic distribution, functional organization, and insertion specificity. J Bacteriol 184: 992–1002 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Koonin EV, Ilyina TV (1993) Computer-assisted dissection of rolling circle DNA replication. Biosystems 30: 241–268 [DOI] [PubMed] [Google Scholar]
- Lamers MH, Perrakis A, Enzlin JH, Winterwerp HHK, de Wind N, Sixma TK (2000) The crystal structure of DNA mismatch repair protein MutS binding to a G.T mismatch. Nature 407: 711–717 [DOI] [PubMed] [Google Scholar]
- Larkin C, Datta S, Harley MJ, Anderson BJ, Ebie A, Hargreaves V, Schildbach JF (2005) Inter- and intramolecular determinants of the specificity of single-stranded DNA binding and cleavage by the F factor relaxase. Structure 13: 1533–1544 [DOI] [PubMed] [Google Scholar]
- Mennecier S, Coste G, Servant P, Bailone A, Sommer S (2004) Mismatch repair ensures fidelity of replication and recombination in the radioresistant organism Deinococcus radiodurans. Mol Genet Genom 272: 460–469 [DOI] [PubMed] [Google Scholar]
- Mennecier S, Servant P, Coste G, Bailone A, Sommer S (2006) Mutagenesis via IS transposition in Deinococcus radiodurans. Mol Microbiol 59: 317–325 [DOI] [PubMed] [Google Scholar]
- Moseley BEB, Mattingly A (1971) Repair of irradiated transforming deoxyribonucleic acid in wild type and a radiation-sensitive mutant of Micrococcus radiodurans. J Bacteriol 105: 976–983 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Navaza J (2001) Implementation of molecular replacement in AMoRe. Acta Crystallogr D 57: 1367–1372 [DOI] [PubMed] [Google Scholar]
- Nowotny M (2009) Retroviral integrase superfamily: the structural perspective. EMBO Rep 10: 144–151 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nunvar J, Huckova T, Licha I (2010) Identification and characterization of repetitive extragenic palindromes (REP)-associated tyrosine transposases: implications for REP evolution and dynamics in bacterial genomes. BMC Genom 11: 44. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Otwinowski Z, Minor W (1997) Processing of X-ray diffraction data collected in oscillation mode. Meth Enzymol 276: 307–326 [DOI] [PubMed] [Google Scholar]
- Pasternak C, Ton-Hoang B, Coste G, Bailone A, Chandler M, Sommer S (2010) Irradiation-induced Deinococcus radiodurans genome fragmentation triggers transposition of a single resident insertion sequence. PLoS Genet 6: e1000799. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rice PA (2005) Visualizing Mu transposition: assembling the puzzle pieces. Genes Dev 19: 773–775 [DOI] [PubMed] [Google Scholar]
- Richardson JM, Colloms SD, Finnegan DJ, Walkinshaw MD (2009) Molecular architecture of the Mos1 paired-end complex: the structural basis of DNA transposition in a eukaryote. Cell 138: 1096–1108 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ronning DR, Guynet C, Ton-Hoang B, Perez ZN, Ghirlando R, Chandler M, Dyda F (2005) Active site sharing and subterminal hairpin recognition in a new class of DNA transposases. Mol Cell 20: 143–154 [DOI] [PubMed] [Google Scholar]
- SantaLucia J, Hicks D (2004) The thermodynamics of DNA structural motifs. Annu Rev Biophys Biomol Struct 33: 415–440 [DOI] [PubMed] [Google Scholar]
- Sheldrick GM (1998) SHELX: applications to macromolecules. In Direct Methods for Solving Macromolecular Structures, Fortier S (ed) pp 401–411. Dordrecht: Kluwer Academic Publications [Google Scholar]
- Slade D, Lindner AB, Paul G, Radman M (2009) Recombination and replication in DNA repair of heavily irradiated Deinococcus radiodurans. Cell 136: 1044–1055 [DOI] [PubMed] [Google Scholar]
- Ton-Hoang B, Guynet C, Ronning DR, Cointin-Marty B, Dyda F, Chandler M (2005) Transposition of ISHp608, member of an unusual family of bacterial insertion sequences. EMBO J 24: 3325–3338 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tsutakawa SE, Jingami H, Morikawa K (1999) Recognition of a TG mismatch: the crystal structure of very short patch repair endonuclease in complex with a DNA duplex. Cell 99: 615–623 [DOI] [PubMed] [Google Scholar]
- van Mansfeld ADM, van Teeffelen HAAM, Baas PD, Jansz HS (1986) Two juxtaposed tyrosyl-OH groups participate in phiX174 gene A protein catalysed cleavage and ligation of DNA. Nucleic Acids Res 14: 4229–4238 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Warren JJ, Pohlhaus TJ, Changela A, Iyer RR, Modrich PL, Beese LS (2007) Structure of the human MutS alpha DNA lesion recognition complex. Mol Cell 26: 579–592 [DOI] [PubMed] [Google Scholar]
- Watkins S, van Pouderoyen G, Sixma TK (2004) Structural analysis of the bipartite DNA-binding domain of Tc3 transposase bound to transposon DNA. Nucleic Acids Res 32: 4306–4312 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zahradka K, Slade D, Bailone A, Sommer S, Averbeck D, Petranovic M, Lindner AB, Radman M (2006) Reassembly of shattered chromosomes in Deinococcus radiodurans. Nature 443: 569–573 [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.