5′-Single-stranded/duplex DNA junctions are loading sites for E. coli UvrD translocase
Single-molecule analyses of the dual DNA helicase/translocase enzyme UvrD reveal the particular substrate preferences of its monomeric, translocase form, with implications for understanding UvrD's translocase and anti-recombinase roles.
Keywords: DNA junctions, fluorescence, helicase, kinetics, translocase
Abstract
Escherichia coli UvrD is a 3′–5′ superfamily 1A helicase/translocase involved in a variety of DNA metabolic processes. UvrD can function either as a helicase or only as an single-stranded DNA (ssDNA) translocase. The switch between these activities is controlled in vitro by the UvrD oligomeric state; a monomer has ssDNA translocase activity, whereas at least a dimer is needed for helicase activity. Although a 3′-ssDNA partial duplex provides a high-affinity site for a UvrD monomer, here we show that a monomer also binds with specificity to DNA junctions possessing a 5′-ssDNA flanking region and can initiate translocation from this site. Thus, a 5′-ss–duplex DNA junction can serve as a high-affinity loading site for the monomeric UvrD translocase, whereas a 3′-ss–duplex DNA junction inhibits both translocase and helicase activity of the UvrD monomer. Furthermore, the 2B subdomain of UvrD is important for this junction specificity. This highlights a separation of helicase and translocase function for UvrD and suggests that a monomeric UvrD translocase can be loaded at a 5′-ssDNA junction when translocation activity alone is needed.
Introduction
Escherichia coli UvrD is a superfamily 1A (SF1A) DNA helicase/translocase that functions in methyl-directed mismatch repair (Iyer et al, 2006), DNA excision repair (Sancar, 1996), replication restart (Flores et al, 2004, 2005; Michel et al, 2007; Lestini and Michel, 2008) and plasmid replication (Bruand and Ehrlich, 2000). UvrD is also a potent anti-recombinase due to its ability to dismantle RecA protein filaments on single-stranded DNA (ssDNA) (Veaute et al, 2005). The role of UvrD in most of these activities is to function as a helicase to unwind duplex DNA and generate the ssDNA intermediates required for DNA metabolism. However, as the helicase and ssDNA translocase activities of UvrD are separable in vitro (Maluf et al, 2003b; Fischer et al, 2004; Lohman et al, 2008), it is possible that UvrD uses only its ssDNA translocase activity in some of its functions.
Whereas a UvrD monomer is a rapid and highly processive 3′–5′-ssDNA translocase in vitro, a UvrD monomer shows no detectable helicase activity in vitro (Maluf et al, 2003b; Fischer et al, 2004). In fact, activation of the helicase in vitro requires formation of at least a UvrD dimer (Maluf et al, 2003a, 2003b). Hence, UvrD self-assembly can regulate its helicase/translocase activities in vitro (Lohman et al, 2008). A UvrD monomer binds with specificity to a 3′-ss/dsDNA junction and can self-assemble to form an active helicase to unwind the dsDNA (Maluf et al, 2003a, 2003b; Heller and Marians, 2007). UvrD can also bind to different DNA fork structures, and these structures can determine whether the parental or lagging strand duplex is unwound (Cadman et al, 2006).
The anti-recombinase activity of UvrD appears to be due to its ability to dismantle RecA-ssDNA filaments (Flores et al, 2005; Long et al, 2009). Genetic experiments have also suggested that UvrD is required for removal of RecA filaments that can form on the lagging strand after the replisome has encountered DNA damage (Flores et al, 2005). The mechanism by which UvrD initiates disassembly of these RecA-ssDNA filaments is not yet known. UvrD could specifically target these RecA filaments through interactions with specific DNA structures, components of the replisome or the RecA filament itself. Although the yeast Srs2 helicase/translocase shows a similar ability to disrupt Rad51-ssDNA filaments (Krejci et al, 2003; Veaute et al, 2003), this Srs2 activity requires a direct interaction with Rad51 (Antony et al, 2009). However, no direct interaction between UvrD and RecA has yet been demonstrated. In this regard, it is possible that the ability of UvrD to translocate along ssDNA, rather than its DNA-unwinding (helicase) activity per se, is the central property for RecA displacement. We have previously shown that UvrD monomers can bind randomly to ssDNA and initiate directional (3′–5′) translocation along ssDNA in an ATP-dependent reaction (Fischer et al, 2004; Tomko et al, 2007). Here, we show that a UvrD monomer can bind with high specificity to a 5′-ss/dsDNA junction, whether it is part of a partial DNA duplex or a DNA fork structure, and can initiate 3′–5′ directional ssDNA translocation from the junction, while some fraction of the UvrD remains bound to the junction thus forming a transient loop in the ssDNA. This junction specificity is mediated by interactions with the 2B subdomain of UvrD and differs among other SF1 translocases in the UvrD subfamily, such as PcrA and Rep.
Results
UvrD monomers bind with specificity to both 3′- and 5′-ss/dsDNA junctions
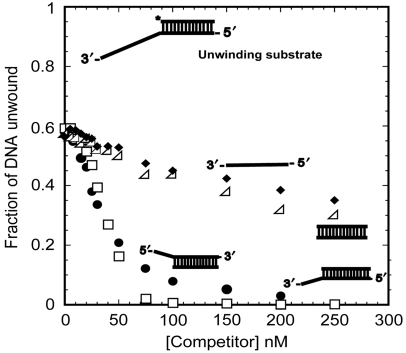
E. coli UvrD monomers bind with specificity to a 3′-ss/dsDNA junction, relative to either ssDNA or dsDNA (Maluf et al, 2003b). In fact, a 3′-ssDNA flanking region as short as four nucleotides provides a high-affinity binding site for a UvrD monomer (Maluf et al, 2003b). However, a UvrD monomer does not initiate DNA unwinding from such a junction. Activation of DNA-unwinding in vitro requires at least a UvrD dimer (Maluf et al, 2003a, 2003b). To test whether UvrD has specificity for a 5′-ss/dsDNA junction, single time point, single-turnover, competition DNA-unwinding experiments were conducted as described (Maluf et al, 2003b), where UvrD (75 nM) was pre-incubated with radiolabelled 3′-dT20-DP18 DNA (25 nM) with increasing concentrations of unlabelled competitor DNA in buffer T20. Unwinding was then initiated by the addition of ATP:Mg2+, the reaction was quenched after 20 s and the extent of unwound, labelled DNA was determined. Figure 1 shows that a 3′-T8-DP18 substrate competes effectively with 3′-dT20-DP18 DNA for UvrD binding while ssDNA (dT20) and dsDNA (DP18) are poor competitors. Interestingly, a 5′-T8-DP18 DNA was also an effective competitor for binding of UvrD, far better than ssDNA and dsDNA, and nearly comparable to a 3′-ss/dsDNA junction. Hence, UvrD shows specificity for binding ss/dsDNA junctions in general. UvrD binding to a 5′- or 3′-ss/dsDNA junction is stoichiometric in buffer T20 (data not shown).
Figure 1.
UvrD has a higher specificity for ss/dsDNA junctions than ssDNA and dsDNA. Single time point, single-turnover competition DNA-unwinding experiments were carried out by incubating UvrD with 32P-labelled unwinding substrate and increasing concentrations of competitor DNAs in T20 buffer at 25°C. Competitor DNAs 3′-dT8-DP18 (□) and 5′-dT8-DP18 (•) strongly compete for UvrD binding as compared with ssDNA (dT20, ♦) or dsDNA (DP18, △).
UvrD monomers can initiate translocation from a 5′-ss/dsDNA junction
UvrD monomers can translocate rapidly and with biased 3′–5′ directionality along ssDNA (Fischer et al, 2004; Tomko et al, 2007). Based on the observation that a UvrD monomer binds with specificity to a 5′-ss/dsDNA junction, we examined whether it can initiate translocation from such a junction. The initial distribution of UvrD monomers bound to an unstructured ssDNA, such as an oligodeoxythymidylate (oligo(dT)), is random (Fischer et al, 2004; Tomko et al, 2007). However, if the ssDNA has a duplex region at one end forming a 5′-ss/dsDNA junction, then the distribution of bound UvrD will be biased to the junction. Such a biased distribution should be detectable from the shape of the UvrD translocation time courses.
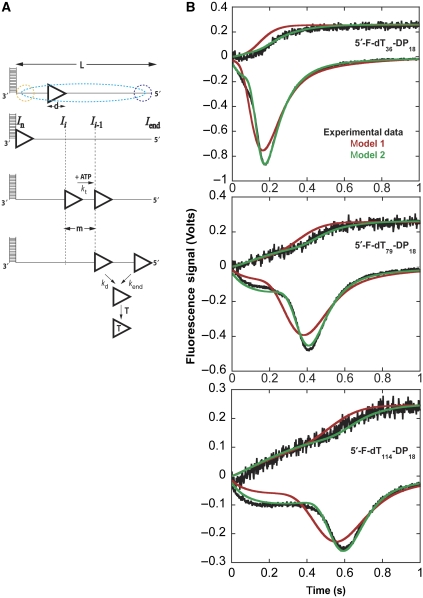
The kinetics of ssDNA translocation by UvrD can be examined by stopped-flow experiments performed using a series of oligodeoxythymidylates of defined length, L (nucleotides), with a fluorophore attached to the 5′-end that undergoes a change in fluorescence intensity when UvrD reaches the 5′-end (Fischer and Lohman, 2004; Fischer et al, 2004; Tomko et al, 2007). Figure 2 shows simulated time courses of arrival of a translocase at the 5′-end of a ssDNA for two cases, differing by the initial distribution of enzyme bound to the DNA. These simulations used an n-step sequential mechanism as described in Tomko et al (2010). In Figure 2A, the translocase initiates from random sites within the ssDNA, whereas in Figure 2B, it initiates at a unique site at the 3′-end. In these simulations, no more than one monomer is bound per DNA molecule and translocation is highly processive so that UvrD only dissociates after reaching the 5′-end of the DNA. Rebinding of UvrD to the DNA is also not allowed. When initiation is from a unique site at the 3′-end, a DNA length dependent lag is observed in the arrival of the translocase at the 5′-end. This kinetic behaviour differs from that observed for initiation at random sites along the ssDNA, which we have observed previously for UvrD translocation along unstructured ssDNA ((dT)L) (Fischer et al, 2004; Tomko et al, 2007).
Figure 2.
UvrD monomers can initiate translocation from a 5′-ss/dsDNA junction. (A, B) N-step sequential model simulation of translocation monitoring arrival of translocase at 5′-end of ssDNA assuming different initial binding distributions, random and 3′-end, respectively. (C) Time courses of UvrD monomer translocation on fluorescein-labelled ssDNA (5′-F-dT54) and 5′-ss/dsDNA (5′-F-dT54-DP18). (D) Time courses of UvrD translocation on 5′-F-dTL-DP18 as a function of 5′-tail length (L=16, 25, 36, 44, 79, 84, 97 and 114). (E) Simulated time courses monitoring UvrD arrival at the 5′-end of 5′-F-dT79-DP18 varying the probability of UvrD initially bound at junction (P(UvrD,junction)). (F) Time-to-peak analysis, see text, of the time courses in (D).
To test whether UvrD can initiate translocation from a 5′-ss/dsDNA junction, we used a series of 5′-ss/dsDNA junction substrates (5′-F-dTL-DP18: L=16, 25, 36, 44, 79, 84, 97 and 114 nts) comprised of an 18-base pair (bp) duplex (DP18) with a 5′-(dT)L-tail labelled covalently at the 5′-end with fluorescein (F). UvrD is pre-incubated with the DNA in buffer T20 then rapidly mixed with ATP, MgCl2 and heparin also in buffer T20 to initiate translocation. Upon reaching the fluorescein at the 5′-end of the DNA, UvrD quenches the fluorescein fluorescence intensity. Heparin is included with the ATP to prevent UvrD from rebinding to the DNA, ensuring single-round kinetics (Fischer et al, 2004). Sedimentation equilibrium experiments confirmed that no more than one UvrD monomer is bound to the 5′-ss/dsDNA when the DNA concentration is in at least a two-fold molar excess over UvrD as is the case in these translocation experiments (Supplementary Figure S1).
Figure 2C compares time courses obtained with a 5′-ss/dsDNA partial duplex (5′-F-dT54-DP18) versus the same length of ssDNA (5′-F-dT54). The time course for the 5′-ss/dsDNA junction shows an initial slow phase/lag, followed by rapid quenching of the fluorescein fluorescence, and then an increase in fluorescence. These results are consistent with UvrD translocation in a 3′–5′ direction and subsequent dissociation upon reaching the 5′-end. The presence of the initial lag phase suggests that UvrD initiation is biased to the ss/dsDNA junction. In contrast, no slow/lag phase is observed in the time course when UvrD is initially bound to 5′-F-(dT)54, consistent with random initiation.
Figure 2D shows the results of experiments performed with a series of junction DNA substrates (5′-F-dTL-DP18), varying in ssDNA length, L. As L increases, the initial slow (lag) phase increases. The time to reach the minimum fluorescence signal (tpeak) increases linearly as a function of the 5′-tail length (Figure 2F), indicating that a UvrD monomer can initiate translocation starting from the 5′-ss/dsDNA junction. The average translocation rate for a UvrD monomer initially bound at the junction, as determined from the inverse of the slope in Figure 2F, is 190±2 nts/s, which is identical to the translocation rate determined for UvrD on 5′-F-(dT)L under the same solution conditions (Fischer et al, 2004; Tomko et al, 2007). Increasing the length of the dsDNA region from 18 to 36 bp or replacing it with a DNA hairpin had no effect on the translocation time courses (Supplementary Figure S2), suggesting an 18-bp duplex is sufficient to maintain the necessary interactions with a UvrD monomer at the junction. The specificity of UvrD for the 5′-ss/dsDNA junction persists for NaCl concentrations up to ∼200 mM NaCl (Supplementary Figure S3C, D and E).
We note that the slow phase observed in the UvrD time courses with the junction DNA is not a strict lag phase, rather there is a slight quenching of the fluorescence signal during this phase and its duration changes with ssDNA tail length. This effect is expected if not all of the UvrD initiates translocation from the junction, but rather some fraction of UvrD also initiates randomly from internal ssDNA sites. This conclusion is supported by kinetic simulations discussed below. We also conducted UvrD translocation experiments with the same junction DNA substrates used above, but monitored the net dissociation of UvrD during translocation, rather than UvrD arrival at the 5′-end of the ssDNA. This was accomplished by monitoring the increase in UvrD Trp fluorescence that accompanies dissociation of UvrD from the DNA as described (Tomko et al, 2007). The resulting time courses (Supplementary Figure S4 and shown later in Figure 3B) are consistent with our hypothesis that a large fraction of UvrD is initiating translocation from the junction.
Figure 3.
Analysis of time courses of UvrD monomer translocation along 5′-ss/dsDNA using a sequential n-step model accounting for a non-random initial binding distribution. (A) Kinetic model for UvrD monomer translocation on 5′-ss/dsDNA as described in the text. UvrD monomers (triangle), high-specificity binding site (junction, yellow circle) and low-specificity binding sites (internal ssDNA sites and 5′-end, blue circle). UvrD is prevented from rebinding the DNA by binding to heparin (protein trap, T). Intermediate positions of UvrD along the ssDNA are shown as Ii, where In represents UvrD bound to the ss/dsDNA junction. (B) Global NLLS analysis of time courses monitoring dissociation of UvrD during translocation and arrival of UvrD at the 5′-end of 5′-F-dTL-DP18 (L=36, 79 and 114) as described in Supplementary data. A subset of the lengths analysed are shown, the rest are shown in Supplementary Figure S8. The coloured lines are simulations of the best-fit parameters (Supplementary Table S1) examining translocation models 1 and 2. The translocation models are summarized in the text and described further in Supplementary data.
Estimate of specificity of UvrD binding to the ss/dsDNA junction
The shapes of the UvrD translocation time courses obtained with the 5′-ss/dsDNA substrates suggest the presence of two populations of DNA-binding sites for UvrD. We hypothesize that UvrD can initiate randomly from internal sites within the ssDNA tail, but also can initiate from the ss/dsDNA junction. To examine this further, we simulated the expected time courses for such a model. We modified our previous model describing UvrD translocation initiating from random sites within a ssDNA (Fischer and Lohman, 2004) to include a second UvrD population that initiates at the duplex junction, as shown in Figure 3A (see Supplementary data for a detailed description of the model). In this model (model 1), a UvrD monomer binds randomly, but with polarity to the ssDNA (L nucleotide long), assuming equal probability (probability r) for protein binding to any stretch of d nucleotides (contact size) within the 5′-ssDNA tail. We also allowed for specific binding of UvrD to the ss/dsDNA junction (with probability q). Upon addition of ATP and MgCl2, all UvrD (regardless of its initial binding site) translocates along the ssDNA with 3′–5′ directional bias in a series of repeated rate-limiting steps with rate constant, kt, until it dissociates from internal ssDNA sites with rate constant, kd, or reaches the 5′-end and then dissociates with rate constant kend. The average number of nucleotides translocated between two consecutive rate-limiting steps is defined as the kinetic step size, m. Upon dissociation from the ssDNA, UvrD is sequestered by a trap, thus preventing rebinding of UvrD to the DNA. We also allow for the possibility that the kinetic parameters (kt, kd and m) describing UvrD that initiates translocation from the ss/dsDNA junction can differ from those for UvrD that initiates from internal ssDNA sites.
Using this model, we simulated time courses in which we varied the probability of UvrD binding to the ss/dsDNA junction versus internal sites within the ssDNA tail. Figure 2E shows simulations for a 5′-ss/dsDNA possessing a 79-nt ssDNA tail. When the probabilities for binding to the junction versus a ssDNA site are the same (q/r=1) (red trace), we recover the expected time course for the random binding case (compare with Figure 2A). Furthermore, when the probability of binding to the junction is 1000-fold greater than to any ssDNA site (q/r=1000) (purple trace) the time course predicted for initial binding only at the 3′-end is recovered (Figure 2B). As the probability for UvrD binding to the junction changes between these limiting cases (q/r=10, 40, 100 in Figure 2E), the time courses reflect initiation from a mixture of both the random binding and 3′-end binding. The initial slow phase reflects translocation by UvrD that initiates randomly from internal ssDNA sites (Figure 2A); the amplitude of this phase decreases as the probability of binding to the junction increases. The second steeper phase reflects translocation by UvrD that initiates at the junction; the amplitude of this rapid phase increases as the probability of binding to the junction increases. The simulations for the mixed populations (q/r=10, 40, 100) capture the characteristics of the experimental time courses using a 5′-ss/dsDNA junction (compare Figure 2E with Figure 2C and D). This supports our hypothesis that these result from two subpopulations of UvrD, one that initiates from the ss/dsDNA junction and one that initiates from internal sites within the ssDNA.
To determine how well this model (model 1) describes the experimental data, we globally fit both the translocation time courses, monitoring fluorescein fluorescence, and the UvrD dissociation time courses, monitoring UvrD Trp fluorescence, to this model using non-linear least squares (NLLS) methods (Supplementary data). From such an analysis, we can also obtain an estimate of the UvrD specificity for binding to the junction as well as values of the kinetic parameters for translocation (Supplementary Table S1; Figure 3B and Supplementary Figure S8). The simplest model (model 1) reproduces the basic characteristics of the experimental time courses (Figure 2D), but it is unable to quantitatively describe the complete experimental time courses, as discussed below. Hence, we introduced a further modification (model 2) that improves the quantitative agreement with the experimental time courses.
Model 1: both populations of UvrD have identical kinetic parameters. Model 1 assumes that the translocation kinetic parameters (kt, kd and m) that we determined previously for UvrD monomer translocation along ssDNA alone (Tomko et al, 2007) also apply to the population of UvrD that initiates from the ss/dsDNA junction. As shown in Figure 3B (red curves), the simulations based on model 1 deviate from the experimental time courses in that the breadth of the peak is wider and the peak position is shifted to earlier times. These shifts suggest that the UvrD initially bound at the junction translocates with a slower rate and/or a smaller kinetic step size (Supplementary Figure S6 and S7). Thus, we next allowed the kinetic parameters to differ for the two different populations of UvrD in model 2.
Model 2: UvrD initiating at the junction has different kinetic properties than UvrD initiating at internal ssDNA sites. In model 2, global NLLS analysis of the translocation and dissociation time courses was performed by constraining the kinetic parameters for UvrD that initiates at ssDNA sites (kt=41.7 step/s, kd=0.84/s and m=4.6 nts/step) to be those determined previously with ssDNA ((dT)L) alone (Tomko et al, 2007), while allowing the kinetic parameters (ktj, kdj and mj) to float for UvrD that initiates at the junction. This resulted in a significantly improved fit to the experimental data (Figure 3B). However, NLLS analysis always returned a negative value for the rate constant for UvrD dissociation from the junction, kdj, hence we constrained kdj to be zero. Using model 2, the best-fit kinetic parameters for UvrD that initiates at the junction are: mj=1.55±0.02 nt/step and a macroscopic translocation rate (mjktj)=192.3±0.3 nt/s (Supplementary Table S1). These should be compared with values of m=4.67±0.13 and mkt=195±3 nts/s for UvrD initiating on ssDNA alone. The analysis using model 2 also yields an estimate of q/r=17±4, indicating a 17-fold higher affinity for UvrD binding to the junction versus an internal site on ssDNA. These kinetic parameters persist for nearly the entire length of the 5′-ssDNA tail, as limiting them for a short distance along the 5′-ssDNA trail (25 nts) yields a poorer fit to the experimental data (Supplementary data).
Some UvrD remains bound to the junction for short times while translocating
Bacillus stearothermophilus PcrA is an SF1A helicase/translocase with significant structural similarity to both UvrD and Rep (Korolev et al, 1997; Velankar et al, 1999; Lee and Yang, 2006). Recent single-molecule fluorescence studies have shown that monomers of PcrA also display specificity for a 5′-ss/dsDNA junction in the presence of ATP and can initiate 3′–5′ directional translocation along ssDNA from the junction (Park et al, 2010). Those studies also show that a PcrA monomer remains in contact with the junction during translocation, resulting in the formation of ssDNA loops as it translocates. Based on this, we considered whether UvrD might also share this property and that this might explain the effect on the translocation kinetic parameters for UvrD initiating from the junction. To test for this we designed a Förster resonance energy transfer (FRET) experiment to detect changes in the relative distance between the 5′-end of the ssDNA and the junction that might occur during translocation while also conducting single-molecule FRET experiments as done with PcrA (Park et al, 2010).
In the ensemble assay, a series of 5′-ss/dsDNA substrates (5′-dTL-DP18: L=25, 35, 44, 54, 65, 79, 97 and 106 nt) were labelled with Cy3 (donor) at the 5′-end of the ssDNA and Cy5 (acceptor) at the ss–dsDNA junction as shown in Figure 4A. Translocation was monitored in the stopped flow as above by exciting the Cy3 donor and monitoring the fluorescence intensities of Cy3 and Cy5 simultaneously. Control experiments with the FRET DNA substrates alone show the expected decrease in the Cy5 signal and concomitant increase in Cy3 signal upon increasing the length of the 5′-tail (Figure 4B) consistent with a prior study (Murphy et al, 2004). As indicated in Figure 4A, if looping of the ssDNA occurs during UvrD translocation, the distance between the 5′-end of the ssDNA and the junction will decrease, resulting in a Cy3 fluorescence decrease and a concomitant Cy5 fluorescence increase. If no looping occurs then no anti-correlated signal changes between Cy3 and Cy5 should occur.
Figure 4.
UvrD translocation time courses for four of the FRET substrates are shown in Figure 4C along with time courses obtained using control substrates labelled with only Cy3 (open circles). Within the first 50 ms of the time courses, anti-correlated Cy3 and Cy5 signal changes (decreasing and increasing, respectively) are observed, consistent with the 5′-end of the ssDNA tail being brought closer to the ss/dsDNA junction. Control experiments confirm the observed FRET is due to ssDNA looping rather than UvrD dissociation from the ssDNA and is not dependent on Cy5 being at the junction (Supplementary Figure S9 and S10). Interestingly, the amplitudes of the signal changes decrease as the length of the 5′-ssDNA tail increases from 25 to 44 nts, with no anti-correlated signal changes observed for lengths >65 nts, unless an internal residue of the 5′-tail (∼25 nts from the junction) is labelled with Cy3 (Supplementary Figure S11). This trend suggests that UvrD-induced ssDNA looping occurs only over a limited distance (<25 nts) after which the junction and loop are released. Indeed, release of the loop is supported by the opposite anti-correlated Cy3 and Cy5 signals (increasing and decreases, respectively) after ∼50 ms, but before UvrD arrival at and dissociation from the 5′-end. To test whether UvrD remained at the ss/dsDNA junction, we repeated these experiments, but excited Cy5 directly. When UvrD is near Cy5, Cy5 fluorescence is enhanced providing a signal to monitor UvrD at the junction. The resulting time courses for FRET substrates with different length ssDNA tails are nearly super-imposable (Figure 4D), indicating that UvrD remains at the junction for the same amount of time regardless of ssDNA length and releases/leaves the junction well before reaching the 5′-end of the ssDNA (compare time courses to dashed line Figure 4D).
These observations suggest that some fraction of UvrD remains in contact with the 5′-ss/dsDNA junction while looping out a short region of ssDNA, but then releases the junction while continuing to translocate along the 5′-tail. This differs from what has been observed for B. stearothermophilus PcrA, which remains in contact with the junction, while repeatedly looping out the entire length of the ssDNA tail (Park et al, 2010). To further investigate UvrD translocation coupled ssDNA looping, we performed single-molecule total internal reflection FRET experiments as was done previously for PcrA (Park et al, 2010). The experimental methods used are summarized in Supplementary data and the results are shown in Figure 5. When UvrD is added to the flow cell with ATP, a series of FRET spikes appear. The asymmetric pattern of gradual FRET increase followed by a rapid drop suggests that ssDNA looping is coupled to UvrD translocation on ssDNA as was observed for PcrA (Park et al, 2010). However, for UvrD, the FRET signal change is irregular and differs from the much more regular persistent cycling as observed for PcrA (compare Figure 5A and B). Hence, the details of the reinitiation of looping differ for UvrD and PcrA. Interestingly, when UvrD is pre-incubated with the DNA substrate in the flow cell and the fluorescence is monitored while flowing in ATP to initiate translocation, as in the protocol used in our stopped-flow experiments, a FRET signal change is observed with a short delay time (Figure 5C and E). In contrast, when the same experiment is performed with PcrA, the delay time to observe the onset of the FRET signal change associated with repetitive looping is much longer (Figure 5D and F), indicating that the kinetics of initiation of repetitive looping by PcrA and UvrD differ. This difference reflects the different relative specificities of UvrD and PcrA for the ss/ds DNA junction in the presence and absence of ATP. As we show below, PcrA has lower specificity for the junction in the absence of ATP than does UvrD. Yet, in the presence of ATP:Mg2+, PcrA shows a higher specificity for the junction. These observations are consistent with the stopped-flow assay results supporting a model in which UvrD can translocate along the ssDNA, by looping out the ssDNA, while bound at the junction, but the activity does not persist over many repetitive cycles as it does for PcrA (Park et al, 2010).
Figure 5.
Single-molecule FRET experiments showing UvrD and PcrA-induced ssDNA looping. Single-molecule time traces are shown with donor intensity in red, acceptor intensity in green and FRET efficiency in blue. (A) Representative single-DNA molecule time trajectories with 400 pM UvrD and 1 mM ATP. (B) Representative single-DNA molecule time trajectories with 400 pM PcrA and 1 mM ATP. (C, D) Representative single-turnover mimic experiments. In all, 500 pM UvrD (or PcrA) is pre-incubated with the surface immobilized DNA in reaction buffer, see Supplementary data, without ATP and MgCl2 for 10 min. Protein-free buffer that contains MgCl2 (5 mM) and ATP (1 mM) is flowed into the chamber to initiate the reaction at the time indicated. (E, F) Histograms of the time delay between the ATP–Mg2+ buffer arrival and commencement of looping, determined from time trajectories such as shown in (C, D).
UvrD monomers bind preferentially to the 3′-ss/duplex junction of a DNA fork, but can transfer to the 5′-ssDNA tail and reinitiate ssDNA translocation
Studies with forked DNA substrates showed that UvrD preferentially unwinds a duplex on the lagging strand of a DNA fork before unwinding the parental duplex (Cadman et al, 2006). Furthermore, single-molecule studies demonstrated that UvrD can switch DNA strands and translocate away from the duplex (Dessinges et al, 2004; Sun et al, 2008). We have shown that a UvrD monomer has specificity for binding to both a 3′-ss/dsDNA and a 5′-ss/dsDNA junction (Figure 1). Hence, we wished to examine the behaviour of a UvrD monomer on a forked DNA structure, where both junctions are present. We added a 3′-tail (3–10 nucleotides) to the 5′-F-dT79-DP18 substrate to form a DNA fork (Figure 6A) and examined the effect on initiation of UvrD translocation along the 5-ssDNA tail. We note that as a UvrD dimer is needed to activate its helicase activity in vitro, a UvrD monomer by itself is unable to unwind the 18-bp duplex even when bound in the correct orientation along the 3′-ssDNA arm of the fork (Maluf et al, 2003b; Fischer et al, 2004). UvrD was pre-incubated with a two-fold excess of the forked DNA and translocation initiated as before. Figure 6A compares the resulting translocation time courses for the 5′-ss/dsDNA junction substrate and three DNA fork substrates with different 3′-ssDNA tail lengths (3, 6 and 10 nt).
Figure 6.
UvrD monomers can switch strands during translocation upon encountering a DNA fork structure. (A) Time courses of UvrD monomer translocation along ssDNA of artificial DNA forks. The black curves are simulations using equation (s22) and the best-fit parameters (Supplementary Table S2) discussed in Supplementary data. The DNA fork substrates were comprised of an 18-bp duplex with a 5′-dT79-tail labelled with fluorescein and different length 3′-dTL-tail (L=3, 6 and 10 nts). (B) UvrD specificity for the ss/dsDNA junctions relative to ssDNA as a function of 3′-tail length. (C) Model for UvrD monomer translocation on ssDNA of fork DNA structures. UvrD binds with high specificity to 3′-ssDNA tail (lengths >10 nts in the presence of 5′-tail). Upon the addition of ATP:Mg2+, the UvrD monomer is unable to unwind the dsDNA but can transfer to the 5′-ssDNA tail with rate constant kswitch. While at the junction, UvrD reels in ssDNA forming a small loop, then releases the loop, continuing translocation along the 5′-ssDNA tail (3′–5′). Upon reaching the 5′-end, UvrD quenches the fluorescein fluorescence then dissociates with rate constant, kend.
The decrease in amplitude of the initial phase as the 3′-tail length increases suggests the presence of a second higher affinity site (3′-ssDNA junction) that competes for UvrD binding to the 5′-ssDNA tail and the 5′-ss/ds DNA junction. We therefore analysed the translocation time courses (black curves in Figure 6A) using a modified translocation model (Supplementary Figure S5), which considers three different populations of UvrD initially bound to the forked DNA: one at the 3′-tail junction, a second at the 5′-tail junction and a third at random sites along the 5′-tail. The model also accounts for the slower recovery of the fluorescence signal as the length of the 3′-tail increases by including an additional step with rate constant kswitch, allowing UvrD monomers initially bound on the 3′-tail to switch to the 5′-tail when encountering the junction as depicted in Figure 6C. UvrD may loop out ssDNA at the junction as shown above after switching ssDNA tails; however, we did not directly test for this with the fork substrates. Figure 6B shows the resulting estimates of UvrD specificity for the 3′-tail junction and 5′-tail junction relative to random binding to the ssDNA tail from our analysis. The relative specificity of UvrD for the 3′-tail junction increases slightly (from ∼75 to ∼100), as the 3′-tail increases from 3 to 10 nts. The highest specificity of UvrD for the 5′-tail junction occurs when the 3′-tail is 6 nts long. The value of kswitch also varied with 3′-tail length (Supplementary Table S2), with values similar to those obtained in single-molecule experiments (Dessinges et al, 2004; Sun et al, 2008).
Mutations in the 2B subdomain alter the specificity for UvrD binding to a 5′-ss/dsDNA junction
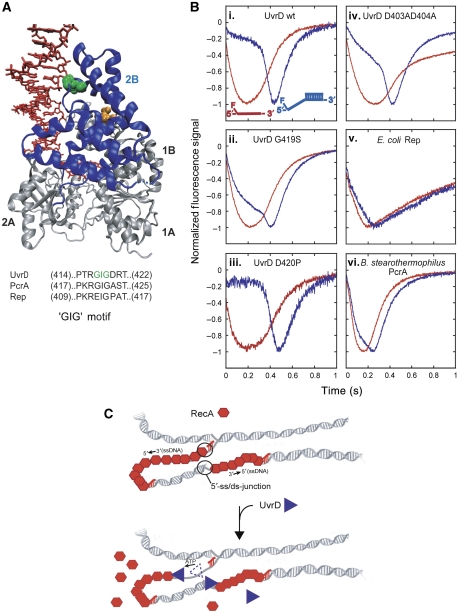
Crystal structures of a UvrD monomer bound to a partial duplex DNA substrate with a short 3′-ssDNA tail show contacts between the 2B subdomain and the dsDNA primarily through a loop region within the 2B subdomain containing the sequence ‘GIG' (Lee and Yang, 2006), consisting of residues G417, I418 and G419 (Figure 7A). Introduction of a G419T mutation within the GIG motif results in a reduction of UvrD affinity for a 3′-ss/dsDNA partial duplex junction, consistent with G419 being important for a UvrD:dsDNA interaction (Lee and Yang, 2006). To determine whether the 2B subdomain has a function in the specificity of UvrD binding to 5′-ss/dsDNA junctions, we examined the effect of several mutations within the 2B subdomain of UvrD on the translocation kinetics on ssDNA versus the 5′-ss/dsDNA junction substrates.
Figure 7.
‘GIG' motif of UvrD 2B domain is important for UvrD specificity for binding 5′-ss/dsDNA junctions. (A) Crystal structure of UvrD bound to a partial duplex with a 3′-ssDNA tail (Lee and Yang, 2006). The green space fill is the GIG (419) of the ‘GIG' motif in the 2B domain (blue ribbon). The orange space fills are residues D403 and D404. The sequence alignment of ‘GIG' motif in UvrD subfamily of SF1 helicase/translocases is shown. (B) Translocation time courses monitoring the arrival UvrD 2B domain mutants and other SF1 helicase/translocases at the 5′-end of ssDNA and 5′-ss/dsDNA substrates. The ssDNA (dT79, red) or 5′-ss/dsDNA (5′-dT79-DP18, blue) is labelled at the 5′-ssDNA end with fluorescein. The time courses are normalized to the peak signal change. (i–iv) wt-UvrD or UvrD 2B domain mutant translocation. (v) E. coli Rep translocation. (vi) B. stearothermophilus PcrA translocation (PcrA (40 nM), 20 mM Tris–HCl pH 7.5, 100 nM DNA, 10 mM NaCl, 10% (v/v) glycerol, 100 nM DNA, 2.5 mM ATP, 3 mM MgCl2 and 4 mg/ml heparin). (C) Proposal for physiological role of UvrD specificity for 5′-ss/dsDNA junctions. UvrD specifically targets RecA filaments that form on the lagging strand of collapsed replication forks. 5′-ss/dsDNA junctions provide a high-affinity site for UvrD to bind and invade the RecA filament, resulting in displacement of RecA by UvrD translocation along the ssDNA.
We examined the translocation behaviour of three ‘2B subdomain' mutants of UvrD. The first was G419S, which changed the second glycine in the ‘GIG' motif to serine. We performed a full ssDNA length dependence of translocation on both ssDNA (5′F-(dT)L) alone as well as the junction substrates (Supplementary Table S3; Supplementary Figure S12); however, in Figure 7B, we show only the data for the substrates with L=79 nucleotides. The comparison for wtUvrD is shown in Figure 7Bi. For UvrD (G419S) (Figure 7Bii), a shift in the peaks of the time courses is observed for the junction DNA versus (dT)79 as was observed for wt-UvrD (Figure 7Bi); however, there is clearly much less of a ‘lag' phase. Analysis of the full-length dependence indicates that the UvrD (G419S) mutant has lower specificity for the 5′-ss/dsDNA junction, which is approximately four-fold lower than wt UvrD (Supplementary Table S3); even though the mutation does not significantly alter the macroscopic rates of ssDNA translocation.
We next examined a double mutation within the 2B subdomain, UvrD (D403AD404A), which, as shown in Figure 7A is at the start of a helix that is adjacent to the ‘GIG' motif (orange residues). It has previously been shown that these mutations enhance the helicase activity of UvrD, although the molecular basis for this has not been determined (Zhang et al, 1998). The translocation time courses indicate that UvrD (D403AD404A) (Figure 7Biv) retains some specificity for the junction, although it is reduced, less than two-fold, compared with wtUvrD, but is higher than for UvrD (G419S). UvrD (D403AD404A) shows a slightly faster rate of translocation (∼248 nts/s) with approximately two-fold higher processivity under the experimental conditions (Supplementary Figure S12; Supplementary Table S3).
Finally, we examined another mutant, UvrD (D420P), as E. coli Rep has a proline at this position (Figure 7A) and Rep displays little specificity for a 5′-ss/dsDNA junction as shown below. The translocation time courses (Figure 7Bii) indicate that UvrD (D420P) has higher specificity as indicated by the more apparent lag phase than observed for wtUvrD. Quantitative analysis of time courses for a series of substrates with different ssDNA tail lengths reveal a nearly six-fold higher specificity for the junction as compared with wtUvrD (Supplementary Table S3; Supplementary Figure S12). The D420P mutant shows a slightly slower rate of translocation of ∼150 nts/s when initiating translocation from ssDNA sites, and ∼176 nts when initiating translocation from the junction (Supplementary Table S3).
Hence, the specificity of UvrD for the 5′-ssDNA–duplex junction is affected by all three mutations within the 2B subdomain, with (G419S) and (D403A, D404A) showing a decrease in junction specificity, whereas D420P shows an increase in junction specificity relative to wtUvrD. These results clearly indicate that the 2B subdomain has a function in the specificity of UvrD binding to the duplex DNA at a 5′-ssDNA junction.
Comparison of UvrD with E. coli Rep and B. stearothermophilus PcrA
E. coli Rep and B. stearothermophilus PcrA are also SF1A helicases and the monomeric forms of these enzymes are rapid and processive ssDNA translocases with 3′–5′ directionality (Dillingham et al, 2000; Brendza et al, 2005; Niedziela-Majka et al, 2007). Rep, UvrD and PcrA are structurally similar and all have 2B subdomains that can undergo substantial rotation about a hinge region connected to the 2A subdomain (Korolev et al, 1997; Velankar et al, 1999). We therefore examined qualitatively the binding specificity of Rep and PcrA monomers for a 5′-ss/dsDNA junction. Translocation of monomers of Rep and PcrA along ssDNA alone and ssDNA with a 5′-ss/dsDNA junction was examined, and the results for substrates with a (dT)79 ssDNA region are shown in Figure 7Bv and Bvi. When compared with UvrD (Figure 7Bi), both Rep and PcrA show more modest differences in their translocation kinetics on ssDNA alone versus a 5′-ssDNA junction substrate. Neither of the time courses for Rep or PcrA on the junction substrates show any clear evidence of a lag phase as is observed for wtUvrD. These results suggest a much lower specificity for junction binding for PcrA and still lower for Rep as compared with wtUvrD. Thus, qualitatively the relative specificities of these translocases for binding to a 5′-ss/dsDNA junction versus ssDNA are UvrD ≫ PcrA > Rep. As shown in Figure 7A, sequence comparisons of the 2B subdomains of UvrD, PcrA and Rep show definite sequence variations within the ‘GIG'-motif region. Based on the results of our mutational studies of the 2B subdomain of UvrD, it is not surprising that there would be differences in junction specificity among these three translocases.
Discussion
Many of the genome maintenance functions of UvrD require its helicase activity to unwind duplex DNA. However, UvrD has other important functions including displacing proteins from DNA, which may require only its translocase activity (Lohman et al, 2008). UvrD displaces the replication termination protein Tus from its specific Ter site (Hiasa and Marians, 1992; Bidnenko et al, 2006). It also promotes DNA replication past transcriptionally active regions of DNA by working in concert with DinG and Rep to displace RNA polymerase ahead of the replisome (Boubakri et al, 2009). UvrD also functions as an anti-recombinase (Zieg et al, 1978), presumably by displacing RecA filaments from ssDNA (Veaute et al, 2005). Whereas displacement of Tus may require UvrD's helicase activity to first unwind the duplex DNA containing the Ter site (Hiasa and Marians, 1992), displacement of RecA from ssDNA may only require the ssDNA translocation activity of UvrD, although this is not yet known. Genetic studies have shown that removal of RecA nucleoprotein filaments that supposedly form on the lagging strand of an inactivated replication fork specifically requires UvrD (Flores et al, 2005; Lestini and Michel, 2008), although it is not known whether direct interactions of UvrD with RecA are required as has been shown for Srs2 displacement of Rad51 filaments (Antony et al, 2009).
The ability of UvrD to translocate along ssDNA is essential for its processive helicase activity and is presumed to be required for its protein displacement activity. However, ssDNA translocation alone is not sufficient to unwind DNA as is evident from the fact that a UvrD monomer is a processive ssDNA translocase, yet is unable to unwind even an 18-bp DNA duplex in vitro (Maluf et al, 2003b; Fischer et al, 2004; Tomko et al, 2007). In fact, a UvrD dimer is the minimal species required to activate DNA-unwinding in vitro (Maluf et al, 2003b; Sun et al, 2008). Thus, the helicase and translocase activities of UvrD are separable where UvrD self-assembly or interactions with other proteins can regulate whether UvrD functions as a helicase or only as a ssDNA translocase (Lohman et al, 2008).
E. coli UvrD can bind a variety of DNA substrates in vitro including DNA forks, nicked DNA, partial DNA duplexes and blunt end DNA, unwinding the DNA with 3′–5′ directionality (Runyon et al, 1990; Runyon and Lohman, 1993; Maluf et al, 2003b; Cadman et al, 2006). Initiation of DNA unwinding by UvrD is more efficient on DNA substrates with a 3′-ss/dsDNA junction and although the 3′-ssDNA provides a loading site for UvrD, it is also possible that the specificity of UvrD for a 3′-ss/dsDNA junction is also important (Maluf et al, 2003b). A UvrD monomer has binding specificity for a 3′-ss/dsDNA junction, despite the inability of a monomer to unwind the dsDNA (Maluf et al, 2003b). Thus, specific binding of a UvrD monomer to a 3′-ss/dsDNA junction may be the first step in the pathway to initiate an active oligomeric helicase at a junction (Maluf et al, 2003a). The fact that a UvrD monomer is unable to initiate DNA unwinding at a 3′-ss/dsDNA junction may prevent a UvrD monomer that is translocating within a ssDNA gap from initiating unwinding of a duplex DNA located at the 5′-end of the ssDNA gap (Lohman et al, 2008).
Our results show that UvrD monomers also possess binding specificity for a 5′-ss/dsDNA junction and that a UvrD monomer can initiate ssDNA translocation from such a junction. In fact, the binding orientation of UvrD for the 5′-ss/dsDNA junction constrains it to translocate away from the dsDNA. Thus, a 5′-ss/dsDNA junction may serve as a high-affinity site for loading UvrD when its ssDNA translocase activity is required rather than its helicase activity. One possibility is that a 5′-ss/dsDNA junction might allow the monomeric UvrD translocase to load onto a ssDNA gap that is coated by RecA as discussed below. Alternatively, structural features of a 5′-ss/dsDNA junction, such as a 3′-hydroxyl, may have a function in UvrD binding to DNA possessing a nick in the phosphate backbone. UvrD can initiate DNA unwinding at a nick in nucleotide excision repair, methyl-directed mismatch repair and replication of plasmids (Sancar, 1996; Bruand and Ehrlich, 2000; Iyer et al, 2006). In these cases, UvrD could bind to the duplex on the 3′-side of the nick and translocate away from the nick along the intact strand in the 3′–5′ direction. This would be consistent with the directionality of Rep (Brendza et al, 2005) and PcrA (Dillingham et al, 2000) when they are recruited to unwind plasmid and phage DNA during rolling circle replication.
The 2B subdomain functions in junction specificity
The monomers of E. coli UvrD (Lee and Yang, 2006), E. coli Rep (Korolev et al, 1997) and B. stearothermophilus PcrA (Velankar et al, 1999) are structurally similar. Each consists of two domains (1 and 2), with each domain composed of two subdomains (1A, 1B and 2A, 2B). The ssDNA-binding site lies across the top of the 1A and 2A subdomains, with a single ATP-binding site at the interface between these subdomains (Figure 7A). The 2B subdomain consists of a nine-helix bundle that is similar in size for UvrD, PcrA and Rep, but has low-sequence conservation and contains none of the conserved SF1 sequence motifs important for nucleic acid stimulated ATP hydrolysis (Gorbalenya et al, 1989; Singleton et al, 2007). In all three proteins, the 2B subdomain can undergo a significant rotation (∼130 deg in the case of Rep bound to ssDNA) about a hinge region connected to the 2A subdomain to go from a ‘closed' form to an ‘open' form (Korolev et al, 1997). The 2B subdomain is in a similar ‘closed' form in all of the crystal structures reported for UvrD and PcrA monomers bound to a ss–dsDNA junction (Velankar et al, 1999; Lee and Yang, 2006), whereas apo PcrA (Subramanya et al, 1996) and apo UvrD (S Korolev, NK Maluf, G Gauss, T Lohman, G Waksman, unpublished data) both crystallize in an ‘open' form.
In the UvrD-3′-ssDNA junction structures, the 2B subdomain contacts the dsDNA backbone through a loop region containing the ‘GIG' motif (Lee and Yang, 2006) (Figure 7A, green residues). Based on the PcrA:DNA and UvrD:DNA crystal structures, it was proposed that interactions between the 2B subdomain and the dsDNA are important for DNA unwinding by a monomer (Velankar et al, 1999; Lee and Yang, 2006); however, biochemical studies have been unable to detect DNA-unwinding activity for Rep, UvrD or PcrA monomers on such DNA substrates in vitro (Cheng et al, 2001; Maluf et al, 2003b; Niedziela-Majka et al, 2007; Sun et al, 2008; Yang et al, 2008). Furthermore, deletion of the 2B subdomain of Rep, to form RepΔ2B, actually activates helicase activity of the monomer in vitro, thus the 2B subdomain is autoinhibitory for Rep monomer helicase activity (Brendza et al, 2005). These studies suggest that the interactions of the 2B subdomain with dsDNA observed in the crystal structures may not be important for its DNA-unwinding activity, but rather may actually inhibit the helicase activity of the monomer and thus have a regulatory function (Brendza et al, 2005) and serve an important biological function (Lohman et al, 2008).
Given the ability of the 2B subdomain to rotate within the monomeric structures of Rep, UvrD and PcrA, it may be possible that the same residues within the 2B subdomain that contact the duplex DNA in the UvrD:3′-ss/dsDNA structures may also contact the duplex region of a 5′-ss/dsDNA junction. Indeed, we have shown here that introducing mutations within the 2B subdomain can have dramatic effects on UvrD specificity for a 5′-ssDNA junction, with some mutations (G419S and D403A, D404A) decreasing specificity, whereas others (D420P) increase specificity. This suggests that the same region of the 2B subdomain may be involved in UvrD monomer binding to a 3′-ss/dsDNA junction as well as to a 5′-ss/dsDNA junction, although how this interaction is made while maintaining the correct 3′–5′ orientation of ssDNA within the binding site is not clear. A similar suggestion has been made for the PcrA monomer (Park et al, 2010).
These observations add to the emerging view that the 2B subdomains of SF1 helicase/translocases are important regulatory domains and may not have direct catalytic functions in destabilizing the dsDNA (Lohman et al, 2008). Here, we show that the 2B subdomain of UvrD regulates the binding specificity of UvrD for DNA junctions and that this specificity may help target this enzyme for particular DNA metabolic processes.
UvrD monomer ssDNA translocation kinetic step size is smaller when UvrD initiates translocation from a ss/dsDNA junction
Another point of interest is that UvrD monomers initiating translocation at a 5′-ss/dsDNA junction appear to have different kinetic properties than when they initiate at an internal ssDNA site. Although the macroscopic translocation rates are similar (Supplementary Table S1), the translocation kinetic step size that we estimate is significantly smaller, ∼1 nt, for UvrD initiating at the junction, compared with ∼4–5 nts when initiating on ssDNA. In fact, a kinetic step size of ∼1 nucleotide has recently been determined from single-molecule studies of PcrA monomers initiating translocation from a 5′-ss/dsDNA junction (Park et al, 2010), whereas a step size of ∼4 nucleotides was estimated from ensemble kinetic experiments (Niedziela-Majka et al, 2007). This difference in apparent step size appears to result from the presence of persistent heterogeneity (static disorder) in the translocation rates of the ensemble population of PcrA monomers. The result of this persistent heterogeneity is that the kinetic step size determined from a variance analysis of the ensemble experiments can be overestimated (Fischer et al, 2010). Unfortunately, the lack of a regular pattern when we performed similar single-molecule experiments with UvrD (Figure 5) does not allow us to determine a step size from the single-molecule experiments. Direct interaction of UvrD with the junction and possibly the resulting looping of ssDNA may result in a smaller kinetic step size. However, it is also possible that the type of initiation site (i.e. ssDNA versus 5′-ss/dsDNA junction) and the different interactions of UvrD with these DNA substrates could influence the extent of any such static disorder in the UvrD population. It is also possible that the simple kinetic models used to analyse these data overlook some aspect of the translocation mechanism.
UvrD monomer loading at 5′-ss/dsDNA junctions may facilitate the anti-recombinase activity of UvrD
Genetic and biochemical studies show that UvrD can disassemble RecA filaments formed on ssDNA in vitro and RecA filaments that have formed on ssDNA at collapsed replication forks (Flores et al, 2005; Veaute et al, 2005); however, the mechanism by which this occurs is not known. The high specificity for UvrD binding to a 5′-ss/dsDNA junction that we report here could facilitate access of UvrD to the RecA filaments. ssDNA gaps formed on the lagging strand during DNA replication will have both 3′- and 5′-ss/dsDNA junctions to which UvrD shows binding specificity (Figure 7C). In the case of a RecA filament formed within such a ssDNA gap, the 5′-ss/dsDNA junction could serve as a high-affinity site to which a UvrD monomer can bind and initiate its ssDNA translocation to carry out disassembly of a RecA filament as depicted in Figure 7C. In the absence of such a site, it might be difficult for UvrD to enter the highly cooperative RecA filament, which is not expected to have many protein-free ssDNA gaps.
Interestingly, the relative specificities of UvrD, PcrA and Rep binding to a 5′-ss/dsDNA junction that we observe in our studies correlate with their abilities to disassemble RecA filaments in vivo. Genetic experiments suggest that both UvrD and PcrA homologues are able to disassemble RecA filaments on ssDNA at damaged replication forks (Flores et al, 2005; Veaute et al, 2005; Lestini and Michel, 2008) and our experiments demonstrate that they both have specificity for binding to 5′-ss/dsDNA junctions in vitro. Under the solution conditions used to pre-incubate UvrD and PcrA with the DNA in our studies (no ATP), the specificity for PcrA binding to the 5′-ss/dsDNA junction is lower than UvrD. However, in the presence of ATP, PcrA can adopt a conformation that shows high specificity for the junction allowing repetitive cycles of ssDNA looping coupled to translocation (Park et al, 2010). In contrast, E. coli Rep shows very little specificity for binding to the 5′-ss/dsDNA junction under our conditions and cannot substitute for UvrD in disassembling RecA filaments at damaged replication forks or disassemble RecA filaments on ssDNA in vitro (Flores et al, 2005; Veaute et al, 2005; Lestini and Michel, 2008).
Materials and methods
Buffers and reagents
Buffers were prepared with reagent grade chemicals using distilled water, further deionized with a Milli-Q purification system (Millipore Corp., Bedford, MA) and were filtered through 0.2 μm filters. Buffer T20 is 10 mM Tris–HCl (pH 8.3 at 25°C), 20 mM NaCl and 20 % (v/v) glycerol (enzyme grade). Other buffers are described in Supplementary data.
Enzymes and DNA
E. coli UvrD was purified and its concentration was determined as described (Runyon et al, 1993) and was stored at −20°C in minimal storage buffer for up to 6 months without loss of translocation activity. E. coli Rep and B. stearothermophilus PcrA were purified as described (Brendza et al, 2005; Niedziela-Majka et al, 2007) and was stored at −20°C. Details of preparation of the UvrD mutants G419S, D420P and D403AD404A are discussed in Supplementary data. The ssDNA substrates used to construct the 5′-ss/dsDNA junction and fork substrates were labelled with fluorescein, Cy3, or Cy5 during synthesis and purified as described (Kozlov and Lohman, 2002) unless noted, dialyzed versus 10 mM Tris–HCl, pH 8.3, stored at −20°C and concentrations were determined spectrophotometrically (Fischer et al, 2004; Lucius et al, 2004). The base composition and annealing of the 5′-ss/dsDNA junction and fork substrates is described in Supplementary Table S4).
Single time point competition DNA-unwinding assays
Single turnover competition DNA-unwinding experiments in which the final extent of DNA unwinding (amplitude) was obtained were performed and analysed as previously (Maluf et al, 2003b). In brief, UvrD was pre-incubated with a mixture of the DNA substrate and competitor DNA (65 μl) in buffer T20 containing 0.2 mg/ml of BSA for 5 min at 25°C and the reaction initiated by adding 65 μl of buffer T20 containing 1 mM ATP:Mg2+ and 4 μM protein trap (10 bp hairpin with a 3′-dT40 ssDNA tail: 5′-GCCTCGCTGC-T5-GCAGCGAGGC-T40-3′). The reactions were quenched after 20 s, with 120 μl of quench buffer, deproteinated with 10% SDS, the reaction products separated by non-denaturing PAGE, and quantified using a STORM 840 Phosphoimager (GE Healthcare, Piscataway, NJ).
Stopped-flow experiments
Experiments were performed in buffer T20 at 25°C using an SX18 MV stopped flow (Applied Photophysics Ltd., Leatherhead, UK) with monochrometer slits set to 2 mm, unless noted otherwise. In translocation experiments, UvrD (25 nM) was pre-incubated with DNA substrates (50 nM) in one syringe and reactions initiated by 1:1 mixing with buffer T20 plus 0.5 mM ATP, 2 mM MgCl2 and 4 mg/ml heparin, unless stated otherwise. All concentrations given are the final concentrations after mixing in the stopped flow.
Kinetics of UvrD monomer translocation. UvrD monomer translocation kinetics were measured under single round conditions (no rebinding of UvrD to ssDNA) using a fluorescent stopped-flow assay to monitor the arrival of UvrD at the 5′-end of the ssDNA region of the DNA substrates, which were labelled with either fluorescein or Cy3 as described (Fischer et al, 2004). Fluorescein fluorescence was excited at 492 nm and emission monitored at >520 nm. Cy3 fluorescence was excited at 515 nm and emission was monitored at 570 nm.
UvrD monomer ssDNA dissociation kinetics. UvrD monomer dissociation kinetics during translocation along 5′-ss/dsDNA and fork substrates were monitored by the increase in UvrD tryptophan fluorescence (Tomko et al, 2007) (λex=280 nm, λem=350 nm) upon dissociation from ssDNA. The monochrometer slits were set 0.8 mm to reduce tryptophan photobleaching.
Monitoring looping of ssDNA during translocation using FRET. UvrD monomer looping of ssDNA during translocation was tested using 5′-ss/dsDNA substrates labelled with both Cy3 (donor, 5′-end of tail) and Cy5 (acceptor, 3′- or 5′-end of top strand) (FRET substrates). Experiments were conducted with dual fluorescence detection, monitoring Cy3 and Cy5 fluorescence on separate channels (Lucius et al, 2004). Cy3 fluorescence was excited at 515 nm and emission monitored at 570 nm. The Cy5 fluorescence due to FRET was monitored at >665 nm. Control experiments using 5′-ss/dsDNA substrates labelled with only Cy3 (donor substrates) were conducted under the same experimental set up as above. Experiments monitoring UvrD translocation away from the junction used the FRET substrates above with Cy5 at the 3′-end of top strand (junction), but the Cy5 fluorescence was excited at 649 nm and emission monitored at >665.
Supplementary Material
Acknowledgments
We thank Thang Ho for synthesis and purification of oligodeoxynucleotides and Dr Anita Niedziela-Majka for expression and purification of UvrD (D403A, D404A). This research was supported by NIH (GM45948 to TML and GM065367 to TH) and by the NSF (0822613 and 0646550 to TH). TH is an investigator with the Howard Hughes Medical Institute. EJT and TML designed the ensemble experiments. EJT purified the wild type and protein, performed the stopped-flow experiments and analysed the data. HJ purified the mutant UvrD proteins, performed the stopped-flow experiments and analysed the data. NKM performed and analysed the DNA-unwinding experiments. TH and JP designed the single-molecule experiments. JP performed and analysed the single-molecule experiments. EJT, JP, TH and TML wrote the paper.
Footnotes
The authors declare that they have no conflict of interest.
References
- Antony E, Tomko EJ, Xiao Q, Krejci L, Lohman TM, Ellenberger T (2009) Srs2 disassembles Rad51 filaments by a protein-protein interaction triggering ATP turnover and dissociation of Rad51 from DNA. Mol Cell 35: 105–115 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bidnenko V, Lestini R, Michel B (2006) The Escherichia coli UvrD helicase is essential for Tus removal during recombination-dependent replication restart from Ter sites. Mol Microbiol 62: 382–396 [DOI] [PubMed] [Google Scholar]
- Boubakri H, de Septenville AL, Viguera E, Michel B (2009) The helicases DinG, Rep and UvrD cooperate to promote replication across transcription units in vivo. EMBO J 29: 145–157 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brendza KM, Cheng W, Fischer CJ, Chesnik MA, Niedziela-Majka A, Lohman TM (2005) Autoinhibition of Escherichia coli Rep monomer helicase activity by its 2B subdomain. Proc Natl Acad Sci USA 102: 10076–10081 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bruand C, Ehrlich SD (2000) UvrD-dependent replication of rolling-circle plasmids in Escherichia coli. Mol Microbiol 35: 204–210 [DOI] [PubMed] [Google Scholar]
- Cadman CJ, Matson SW, McGlynn P (2006) Unwinding of forked DNA structures by UvrD. J Mol Biol 362: 18–25 [DOI] [PubMed] [Google Scholar]
- Cheng W, Hsieh J, Brendza KM, Lohman TM (2001) E. coli Rep oligomers are required to initiate DNA unwinding in vitro. J Mol Biol 310: 327–350 [DOI] [PubMed] [Google Scholar]
- Dessinges MN, Lionnet T, Xi XG, Bensimon D, Croquette V (2004) Single-molecule assay reveals strand switching and enhanced processivity of UvrD. Proc Natl Acad Sci USA 101: 6439–6444 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dillingham MS, Wigley DB, Webb MR (2000) Demonstration of unidirectional single-stranded DNA translocation by PcrA helicase: measurement of step size and translocation speed. Biochemistry 39: 205–212 [DOI] [PubMed] [Google Scholar]
- Fischer CJ, Lohman TM (2004) ATP-dependent translocation of proteins along single-stranded DNA: models and methods of analysis of pre-steady state kinetics. J Mol Biol 344: 1265–1286 [DOI] [PubMed] [Google Scholar]
- Fischer CJ, Maluf NK, Lohman TM (2004) Mechanism of ATP-dependent translocation of E. coli UvrD monomers along single-stranded DNA. J Mol Biol 344: 1287–1309 [DOI] [PubMed] [Google Scholar]
- Fischer CJ, Wooten L, Tomko EJ, Lohman TM (2010) Kinetics of motor protein translocation on Single-Stranded DNA. Methods Mol Biol 587: 45–56 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Flores MJ, Bidnenko V, Michel B (2004) The DNA repair helicase UvrD is essential for replication fork reversal in replication mutants. EMBO Rep 5: 983–988 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Flores MJ, Sanchez N, Michel B (2005) A fork-clearing role for UvrD. Mol Microbiol 57: 1664–1675 [DOI] [PubMed] [Google Scholar]
- Gorbalenya AE, Koonin EV, Donchenko AP, Blinov VM (1989) Two related superfamilies of putative helicases involved in replication, recombination, repair and expression of DNA and RNA genomes. Nucleic Acids Res 17: 4713–4730 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Heller RC, Marians KJ (2007) Non-replicative helicases at the replication fork. DNA Repair (Amst) 6: 945–952 [DOI] [PubMed] [Google Scholar]
- Hiasa H, Marians KJ (1992) Differential inhibition of the DNA translocation and DNA unwinding activities of DNA helicases by the Escherichia coli Tus protein. J Biol Chem 267: 11379–11385 [PubMed] [Google Scholar]
- Iyer RR, Pluciennik A, Burdett V, Modrich PL (2006) DNA mismatch repair: functions and mechanisms. Chem Rev 106: 302–323 [DOI] [PubMed] [Google Scholar]
- Park J, Myong S, Niedziela-Majka A, Lee KS, Yu J, Lohman TM, Ha T (2010) PcrA dismantles RecA filaments by reeling in DNA one nucleotide at a time. Cell 142: 544–555 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Korolev S, Hsieh J, Gauss GH, Lohman TM, Waksman G (1997) Major domain swivelling revealed by the crystal structures of binary and ternary complexes of E. coli rep helicase bound to single-stranded DNA and ADP. Cell 90: 635–647 [DOI] [PubMed] [Google Scholar]
- Kozlov AG, Lohman TM (2002) Stopped-flow studies of the kinetics of single-stranded DNA binding and wrapping around the Escherichia coli SSB tetramer. Biochemistry 41: 6032–6044 [DOI] [PubMed] [Google Scholar]
- Krejci L, Van Komen S, Li Y, Villemain J, Reddy MS, Klein H, Ellenberger T, Sung P (2003) DNA helicase Srs2 disrupts the Rad51 presynaptic filament. Nature 423: 305–309 [DOI] [PubMed] [Google Scholar]
- Lee JY, Yang W (2006) UvrD helicase unwinds DNA one base pair at a time by a two-part power stroke. Cell 127: 1349–1360 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lestini R, Michel B (2008) UvrD and UvrD252 counteract RecQ, RecJ, and RecFOR in a rep mutant of Escherichia coli. J Bacteriol 190: 5995–6001 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lohman TM, Tomko EJ, Wu CG (2008) Non-hexameric DNA helicases and translocases: mechanisms and regulation. Nat Rev Mol Cell Biol 9: 391–401 [DOI] [PubMed] [Google Scholar]
- Long JE, Renzette N, Sandler SJ (2009) Suppression of constitutive SOS expression by recA4162 (I298V) and recA4164 (L126V) requires UvrD and RecX in Escherichia coli K-12. Mol Microbiol 73: 226–239 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lucius AL, Jason Wong C, Lohman TM (2004) Fluorescence stopped-flow studies of single turnover kinetics of E. coli RecBCD helicase-catalyzed DNA unwinding. J Mol Biol 339: 731–750 [DOI] [PubMed] [Google Scholar]
- Maluf NK, Ali JA, Lohman TM (2003a) Kinetic mechanism for formation of the active, dimeric UvrD helicase-DNA complex. J Biol Chem 278: 31930–31940 [DOI] [PubMed] [Google Scholar]
- Maluf NK, Fischer CJ, Lohman TM (2003b) A dimer of Escherichia coli UvrD is the active form of the helicase in vitro. J Mol Biol 325: 913–935 [DOI] [PubMed] [Google Scholar]
- Michel B, Boubakri H, Baharoglu Z, LeMasson M, Lestini R (2007) Recombination proteins and rescue of arrested replication forks. DNA Repair (Amst) 6: 967–980 [DOI] [PubMed] [Google Scholar]
- Murphy MC, Rasnik I, Cheng W, Lohman TM, Ha T (2004) Probing single-stranded DNA conformational flexibility using fluorescence spectroscopy. Biophys J 86: 2530–2537 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Niedziela-Majka A, Chesnik MA, Tomko EJ, Lohman TM (2007) Bacillus stearothermophilus PcrA monomer is a single-stranded DNA translocase but not a processive helicase in vitro. J Biol Chem 282: 27076–27085 [DOI] [PubMed] [Google Scholar]
- Runyon GT, Bear DG, Lohman TM (1990) Escherichia coli Helicase II (UvrD) protein initiates DNA unwinding at nicks and blunt ends. Proc Natl Acad Sci USA 87: 6383–6387 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Runyon GT, Lohman TM (1993) Kinetics of Escherichia coli helicase II-catalyzed unwinding of fully duplex and nicked circular DNA. Biochemistry 32: 4128–4138 [DOI] [PubMed] [Google Scholar]
- Runyon GT, Wong I, Lohman TM (1993) Overexpression, purification, DNA binding, and dimerization of the Escherichia coli uvrD gene product (helicase II). Biochemistry 32: 602–612 [DOI] [PubMed] [Google Scholar]
- Sancar A (1996) DNA excision repair. Annu Rev Biochem 65: 43–81 [DOI] [PubMed] [Google Scholar]
- Singleton MR, Dillingham MS, Wigley DB (2007) Structure and mechanism of helicases and nucleic acid translocases. Annu Rev Biochem 76: 23–50 [DOI] [PubMed] [Google Scholar]
- Subramanya HS, Bird LE, Brannigan JA, Wigley DB (1996) Crystal structure of a DExx box DNA helicase. Nature 384: 379–383 [DOI] [PubMed] [Google Scholar]
- Sun B, Wei KJ, Zhang B, Zhang XH, Dou SX, Li M, Xi XG (2008) Impediment of E. coli UvrD by DNA-destabilizing force reveals a strained-inchworm mechanism of DNA unwinding. EMBO J 27: 3279–3287 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tomko EJ, Fischer CJ, Lohman TM (2010) Ensemble methods for monitoring enzyme translocation along single stranded nucleic acids. Methods 51: 269–276 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tomko EJ, Fischer CJ, Niedziela-Majka A, Lohman TM (2007) A nonuniform stepping mechanism for E. coli UvrD monomer translocation along single-stranded DNA. Mol Cell 26: 335–347 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Veaute X, Delmas S, Selva M, Jeusset J, Le Cam E, Matic I, Fabre F, Petit MA (2005) UvrD helicase, unlike Rep helicase, dismantles RecA nucleoprotein filaments in Escherichia coli. EMBO J 24: 180–189 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Veaute X, Jeusset J, Soustelle C, Kowalczykowski SC, Le Cam E, Fabre F (2003) The Srs2 helicase prevents recombination by disrupting Rad51 nucleoprotein filaments. Nature 423: 309–312 [DOI] [PubMed] [Google Scholar]
- Velankar SS, Soultanas P, Dillingham MS, Subramanya HS, Wigley DB (1999) Crystal structures of complexes of PcrA DNA Helicase with a DNA substrate indicate an inchworm mechanism. Cell 97: 75–84 [DOI] [PubMed] [Google Scholar]
- Yang Y, Dou SX, Ren H, Wang PY, Zhang XD, Qian M, Pan BY, Xi XG (2008) Evidence for a functional dimeric form of the PcrA helicase in DNA unwinding. Nucleic Acids Res 36: 1976–1989 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang G, Deng E, Baugh L, Kushner SR (1998) Identification and characterization of Escherichia coli DNA Helicase II mutants that exhibit increased unwinding efficiency. J Bacteriol 180: 377–387 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zieg J, Maples VF, Kushner SR (1978) Recombination levels of Escherichia coli K-12 mutants deficient in various replication, recombination, or repair genes. J Bacteriol 134: 958–966 [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.