Redox regulation of the stability of the SUMO protease SENP3 via interactions with CHIP and Hsp90
SENP3 was recently implicated as redox sensor affecting hypoxia-inducible factor-1-dependent transcription under conditions of mild oxidative stress. In a novel mechanism, the chaperone Hsp90 selectively stabilizes oxidized SENP3 by protecting it from ubiquitination mediated by the co-chaperone CHIP.
Keywords: CHIP, Hsp90, ROS, SENP3, ubiquitin
Abstract
The molecular chaperone heat shock protein 90 (Hsp90) and the co-chaperone/ubiquitin ligase carboxyl terminus of Hsc70-interacting protein (CHIP) control the turnover of client proteins. How this system decides to stabilize or degrade the client proteins under particular physiological or pathological conditions is unclear. We report here a novel client protein, the SUMO2/3 protease SENP3, that is sophisticatedly regulated by CHIP and Hsp90. SENP3 is maintained at a low basal level under non-stress condition due to Hsp90-independent CHIP-mediated ubiquitination. Upon mild oxidative stress, SENP3 undergoes thiol modification, which recruits Hsp90. Hsp90/SENP3 association protects SENP3 from CHIP-mediated ubiquitination and subsequent degradation, but this effect of Hsp90 requires the presence of CHIP. Our data demonstrate for the first time that CHIP and Hsp90 interplay with a client alternately under non-stress and stress conditions, and the choice between stabilization and degradation is made by the redox state of the client. In addition, enhanced SENP3/Hsp90 association is found in cancer. These findings provide new mechanistic insight into how cells regulate the SUMO protease in response to oxidative stress.
Introduction
Molecular chaperones and the related degradation machinery constitute two mutually exclusive but collaborative pathways in cells (Cyr et al, 2002). Although the molecular chaperones heat shock proteins Hsp70 and Hsp90 buffer misfolded proteins and help refold them, they can also assist in the delivery of fatally damaged proteins to the ubiquitin–proteasome protein degradation machinery (Wegele et al, 2004). The induction of Hsp70, Hsp90 and other molecular chaperones is among the most important responses to environmental challenge, and these molecules confer protection against stresses by refolding or stabilizing certain important client proteins (Kampinga, 2006). On the other hand, C-terminus of heat shock cognate protein 70 (Hsc70)-interacting protein (CHIP), a dual-function co-chaperone/ubiquitin ligase, mediates the ubiquitination and subsequent proteasome-dependent degradation of several chaperone client proteins (Rosser et al, 2007), but is also involved in protein folding and stabilization via stimulation of heat shock transcription factor 1 and induction of the chaperones (Dai et al, 2003). The balance between protein folding/stabilization and degradation can be disturbed under some physiological and/or pathological processes, such as aging, disease, or stress, in which the proteins face a choice between the two pathways. CHIP seems to participate in protein folding/stabilization and degradation decisions (McDonough and Patterson, 2003). However, the characteristics of the client proteins that decide their fate, stabilization or degradation, remain obscure under most circumstances.
CHIP, through its three tandem tetratricopeptide repeat (TPR) domain, binds to the TPR acceptor sites on Hsp90 and Hsp70 proteins, and it serves as an E3 ubiquitin ligase through its Ubox domain, which facilitates the processing of chaperone client proteins (Zhang et al, 2005). CHIP is implicated in various neurodegenerative diseases characterized by protein misfolding and aggregation (Muchowski and Wacker, 2005). CHIP not only has a critical function in the quality control of cellular proteins, but also has an essential function in stress recovery systems by removing excess or unneeded proteins (Qian et al, 2006). However, it remains unknown why CHIP-mediated degradation sometimes occur to ‘normal' proteins under unstressed conditions, and use a chaperone-independent manner (Xu et al, 2002; Shang et al, 2009).
Recently, leucine-rich repeat kinase-2 (LRRK2), the mutation of which is the most common cause of late-onset Parkinson's disease, was found to be regulated by both CHIP and Hsp90 (Ding and Goldberg, 2009; Ko et al, 2009). Overexpression of Hsp90 stabilizes LRRK2 and prevents CHIP-mediated LRRK2 degradation (Ding and Goldberg, 2009). The CHIP–Hsp90 complex also executes similar regulation of tau (Dickey et al, 2007). These findings provide unusual examples that reciprocal actions by CHIP and Hsp90 maintain the level of a protein. Hsp90 can stabilize many proteins under stresses such as ischemia, cold, heat, and/or inflammation (Mosser and Morimoto, 2004). The clients of Hsp90 include the cytoskeletal proteins, membrane receptor tyrosine kinases, and androgen receptors as well as various other transcription factors and signalling proteins (Goetz et al, 2003). Wild-type (WT) p53 and its mutant can be stabilized by Hsp90 under heat stress conditions and in cancer cells (Wang and Chen, 2003). Under these circumstances, whether CHIP participates in the stress recovery process, and what contribution it makes to Hsp90, have not been elucidated.
Redox imbalance, mainly resulting from increased production of reactive oxygen species (ROS), is the most common insult to cellular homeostasis. We previously found that SENP3, a SUMO/sentrin protease, is rapidly induced upon mild oxidative stress due to a reduction in ubiquitination and 26S proteasome-mediated degradation. SENP3 accumulation in the nucleoplasm allows it to have a function in controlling various nuclear events (Huang et al, 2009; Han et al, 2010). In an attempt to determine the regulatory mechanisms by which ROS modulate the turnover of SENP3, we investigated the molecules that stabilize and destabilize SENP3 and discovered a sophisticated interaction of SENP3 with both CHIP and Hsp90. In the present study, we report that SENP3 is constitutively degraded by CHIP, which serves as an ubiquitin E3 ligase under resting conditions, and SENP3 is thus maintained at a low basal level. Upon oxidative stress, Hsp90 is recruited to bind with SENP3, impeding ubiquitination by CHIP, and SENP3 is thus stabilized. Remarkably, the recruitment and binding of Hsp90 to SENP3 is triggered by oxidative modification of two cysteine residues on the SENP3 molecule. These findings demonstrate for the first time that CHIP and Hsp90 can interplay with a client protein under unstressed and stressed conditions, and that the decision to choose stabilization or degradation is made by the redox state of the client protein. Moreover, our findings expand understanding of the roles of CHIP and Hsp90 in their mutual relationship, as we show surprisingly that CHIP degrades SENP3 independently of Hsp90, whereas Hsp90 stabilizes SENP3 dependently of CHIP.
Results
SENP3 stability is regulated by CHIP
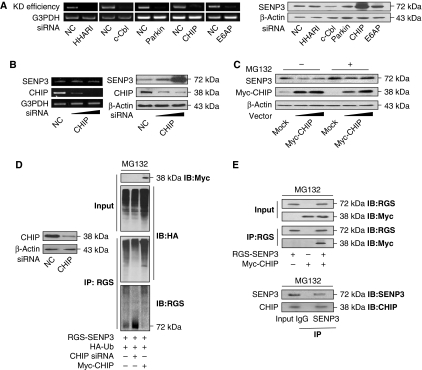
As our previous study indicated that SENP3 might be constantly degraded through the ubiquitin–proteasome pathway under resting conditions, we screened for the molecule that mediates the ubiquitination of SENP3. The abundance of SENP3 protein in HeLa cells was assessed after knocking down a series of ubiquitin E3 ligases. The knockdown efficiency by siRNA for human homologue of Ariadne (HHARI), c-Cbl, parkin, E6-associated protein (E6AP), and CHIP was examined by RT–PCR (Figure 1A, left). The results of immunoblotting (IB) for SENP3 showed that depletion of CHIP led to a robust increase of endogenous SENP3, whereas the other ubiquitin E3 ligases appeared to be irrelevant (Figure 1A, right). The knockdown of endogenous CHIP did not change the mRNA level of SENP3 (Figure 1B, left) but increased its protein level in a dose-dependent manner (right). By contrast, overexpression of exogenous CHIP decreased SENP3 protein dose-dependently in the absence of the 26S proteasome inhibitor MG132 (Figure 1C). As SENP3 could barely be detected after CHIP overexpression in the absence of MG132, all the following co-immunoprecipitation (co-IP) experiments where CHIP was ectopically expressed were performed in the presence of MG132. Co-IP was then carried out in HEK293T cells co-transfected with RGS-SENP3, HA-ubiquitin, CHIP siRNA, or tagged CHIP. The ubiquitin conjugation of SENP3 was readily detected in the presence of MG132; it was largely eliminated by CHIP silencing but was enhanced by overexpression of CHIP (Figure 1D). We further examined whether CHIP was associated with SENP3 in cells. A physical interaction between CHIP and SENP3 was revealed by IP in both exogenous (Figure 1E, upper) and endogenous settings (lower). As SENP3 is a nucleolar protein, to assess the co-localization of SENP3 with CHIP in vivo, we observed cells transfected with red fluorescent protein-tagged SENP3 with and without MG132 treatment, and visualized CHIP by fluorescent isothiocyanate-labelled antibody. In the absence of MG132, CHIP was localized in the cytoplasm and, in particular, the nucleoplasm, showing no overlap with SENP3 that resided within the nucleoli. However, the nucleoplasmic co-localization of CHIP and SENP3 occurred in the presence of MG132 (Supplementary Figure S1; Supplementary data). This result, while revealing the nucleoplasmic co-localization of these two molecules, also implies that their interaction accounts for the degradation of SENP3 outside of nucleoli in resting cells. Taken together, these data indicate that CHIP regulates SENP3 stability through the ubiquitin–proteasome pathway.
Figure 1.
SENP3 stability is regulated by CHIP. (A) HeLa cells were transfected with 50 nM non-specific siRNA (NC) or specific siRNA for five genes, respectively, for 48 h. The efficiency of knockdown (KD efficiency) was determined by RT–PCR (left). SENP3 protein level was evaluated by IB using anti-SENP3 (right). (B) HeLa cells were transfected with 50 nM NC or CHIP siRNA (20 or 50 nM) for 48 h. SENP3 level and the efficiency of siRNA were determined by RT–PCR (left). SENP3 abundance was examined by IB after cells were transfected with 50 nM NC or CHIP siRNA (20 or 50 nM) for 48 h. The expression of CHIP protein was monitored by anti-CHIP (right). (C) SENP3 abundance was examined by IB after HeLa cells were transfected with mock DNA (40 ng) or Myc-CHIP (20 or 40 ng) for 48 h in the absence/presence of MG132 (10 μM) for the last 12 h. The expression of CHIP protein was monitored by anti-Myc. (D) The efficiency of CHIP knockdown was determined by IB after HEK293T cells were transfected with 50 nM NC and CHIP siRNA for 48 h (left). Cells were co-transfected with CHIP siRNA, Myc-CHIP, and HA-ubiquitin (HA-Ub) for 48 h, and MG132 (10 μM) was added for the later 12 h. Co-IP using anti-RGS and IB using the indicated antibodies were performed to determine the ubiquitin conjugation of SENP3 (right). (E) Cells were co-transfected with Myc-CHIP and RGS-SENP3 for 48 h and incubated with MG132 (10 μM) for the later 12 h. The exogenous proteins were co-immunoprecipitated by anti-RGS and immunoblotted using the indicated antibodies (upper). Cells were incubated with MG132 (10 μM) for 12 h. Co-IP using IgG or anti-SENP3 antibodies were performed, and precipitation of endogenous CHIP was determined by IB with anti-CHIP (lower).
SENP3 is degraded by CHIP in an Hsp90-independent manner
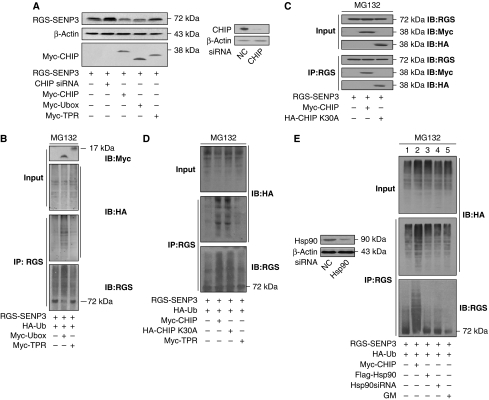
Overexpression of the E3 ubiquitin ligase domain, that is, the Ubox domain, of CHIP, similarly to full-length (FL) CHIP, reduced exogenous SENP3 levels, but TPR domain overexpression had no effect (Figure 2A). Consistent with this result, the ubiquitin conjugation of SENP3 was enhanced by overexpression of the Ubox domain but not the TPR domain (Figure 2B). As CHIP usually functions in association with Hsp90 (Murata et al, 2003), we then assessed whether Hsp90 is involved in the interaction of CHIP with SENP3. Although the fact that the Ubox domain alone could execute ubiquitination already implied that the function of CHIP does not depend on its association with a chaperone, CHIP K30A, a mutant lacking the ability to interact with its chaperone proteins (Yang et al, 2006), was used to provide further evidence. We did an IP assay after overexpressing WT or K30A mutant CHIP together with tagged SENP3 in cells. The CHIP–SENP3 interaction did not rely on the association of CHIP with Hsp90, as both WT and K30A mutant CHIP could be immunoprecipitated by an antibody against the SENP3 tag (Figure 2C). The ubiquitin-conjugation assay showed that both WT and K30A mutant CHIP mediated ubiquitination of SENP3, whereas the TPR domain had no effect (Figure 2D). Neither overexpression nor knockdown of Hsp90 changed the basal level of SENP3 ubiquitination and, likewise, an inhibitor of Hsp90 (geldanamycin (GM)) did not have an effect (Figure 2E). These data suggest that CHIP, through its Ubox domain, mediates the ubiquitination/proteasome-dependent degradation of SENP3, thus maintaining SENP3 protein at a low basal level, and this effect does not require the chaperone Hsp90.
Figure 2.
SENP3 is degraded by CHIP in an Hsp90-independent manner. (A) HEK293T cells were co-transfected with CHIP siRNA, Myc-CHIP, or its domains (Myc-TPR or Myc-Ubox) and RGS-SENP3 for 48 h. The abundance of SENP3 was examined, and the efficiencies for Myc-CHIPs transfection and endogenous CHIP knockdown were monitored by IB. (B) Cells were co-transfected with TPR and Ubox domains of CHIP (Myc-TPR or Myc-Ubox), RGS-SENP3, and HA-ubiquitin (HA-Ub) for 48 h, and MG132 (10 μM) was added for the later 12 h. Co-IP using anti-RGS and IB using the indicated antibodies were carried out to determine the ubiquitination of SENP3. (C) Cells were co-transfected with Myc-CHIP, HA-CHIP (K30A), and RGS-SENP3 for 48 h and incubated with MG132 (10 μM) for the later 12 h. The exogenous proteins were co-immunoprecipitated by anti-RGS and immunoblotted using the indicated antibodies. (D) Cells were co-transfected with Myc-CHIP, HA-CHIP (K30A), TPR domain of CHIP (Myc-TPR), RGS-SENP3, and HA-Ub for 48 h, and MG132 (10 μM) was added for the later 12 h. Co-IP using anti-RGS and IB using the indicated antibodies were carried out to determine the ubiquitination of SENP3. (E) Cells were co-transfected with 50 nM NC or Hsp90 siRNA for 48 h, and the efficiency of the Hsp90 knockdown was determined by IB (left). Cells were co-transfected with RGS-SENP3 and HA-Ub with or without Myc-CHIP, Flag-Hsp90, or Hsp90siRNA for 48 h and MG132 was added for the later 12 h. Geldanamycin (GM, 5 μM) was added as indicated for the last 1 h. Co-IP using anti-RGS and IB using the indicated antibodies were performed to determine the ubiquitination (right).
SENP3 is stabilized upon oxidative stress, whereas the protein level of CHIP, its association with SENP3 and its redox state remain unchanged
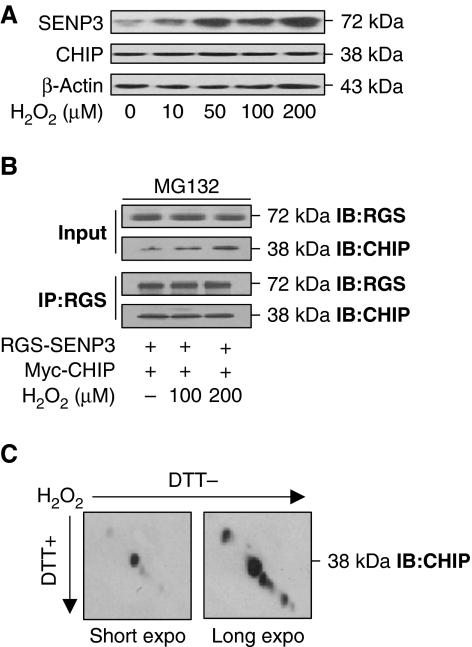
Our previous data showed that hydrogen peroxide (H2O2)-induced increase in SENP3 protein is due to inhibition of the ubiquitin–proteasome pathway (Huang et al, 2009), and the above data in the present study indicate that ubiquitination of SENP3 is mediated by CHIP. We therefore suspected that H2O2 might regulate CHIP protein expression or its binding with SENP3. We thus evaluated the quantity of CHIP and its interaction with SENP3 when cells were exposed to the various low concentrations of H2O2 used previously. After cells were exposed to H2O2, endogenous SENP3 rapidly accumulated in a dose-dependent manner. The amount of endogenous CHIP, however, remained unchanged (Figure 3A). Co-IP assays showed that in cells overexpressing tagged SENP3 and CHIP, the binding of the two molecules was not attenuated upon H2O2 exposure (Figure 3B). As CHIP is a cysteine-rich E3 ligase, and its enzymatic activity can be inactivated under severe oxidative stress (LaVoie et al, 2007), we reasoned that the oxidative effects of H2O2 were likely to be manifested by the formation of disulphide bridges that might disable the catalytic activity of CHIP. To determine whether intra- or intermolecular disulphide bonds formed in CHIP after it was exposed to this low dose of H2O2, redox diagonal electrophoresis was performed. The results excluded this possibility: CHIP migrated well on the diagonal line (Figure 3C), indicating no intra- or intermolecular disulphides. Therefore, we conclude that SENP3 becomes stabilized under mild oxidative stress and that CHIP is not the target of ROS.
Figure 3.
The protein level of CHIP, its association with SENP3, and its redox state remain unchanged upon oxidative stress. (A) HeLa cells were treated with the indicated concentrations of H2O2 for 1 h. SENP3 and CHIP protein levels were evaluated, respectively, by IB. (B) Myc-CHIP and RGS-SENP3 were co-transfected into HEK293T cells for 48 h with MG132 (10 μM) incubation for the later 12 h. H2O2 was added at the indicated concentrations for the last 1 h. Exogenous proteins were co-immunoprecipitated using anti-RGS and immunoblotted using the indicated antibodies. (C) HeLa cells were treated with 100 μM H2O2 for 1 h. CHIP protein was examined by redox diagonal electrophoresis and IB using CHIP antibody. The blots with short- and long-time exposure were displayed to show the position of CHIP relative to the diagonal line.
SENP3 degradation is repressed by Hsp90 upon oxidative stress
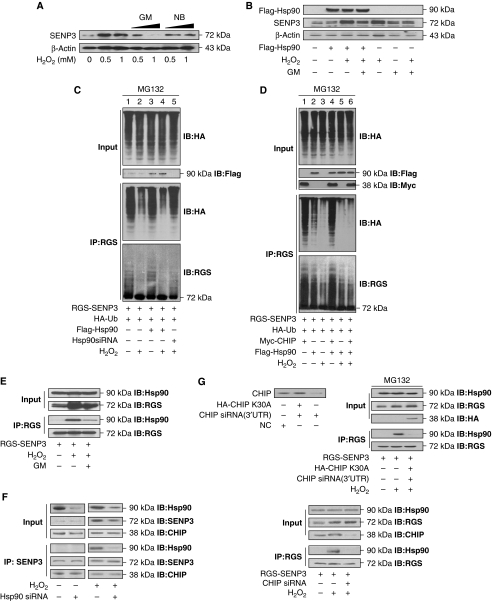
Given the close association of CHIP with Hsp90 and the crucial role of Hsp90 in stabilization of various signalling proteins (Richter and Buchner, 2001), we assumed that Hsp90 is involved in the process wherein mild oxidative stress induces the stabilization of SENP3 protein. There are two reagents, novobiocin (NB) and GM, that by binding to the C-terminal domain and N-terminal ATP/ADP pockets of Hsp90, respectively, block the association of Hsp90 with its client proteins (Chiosis et al, 2004). Indeed, the H2O2-induced accumulation of SENP3 was reversed by GM but not NB (Figure 4A), suggesting that Hsp90 may mediate the process of H2O2-induced SENP3 stabilization and that the N-terminal domain of Hsp90 is involved. Strikingly, overexpression of Hsp90 alone did not lead to the accumulation of SENP3. However, significant accumulation of SENP3 was induced by overexpression of Hsp90 in combination with H2O2 treatment or H2O2 treatment alone, and GM blocked these effects to some extent (Figure 4B). To investigate whether Hsp90 protects SENP3 from ubiquitination, we performed ubiquitination assays in HEK293T cells co-transfected with tagged SENP3, ubiquitin, Hsp90 or siRNA for Hsp90 in the presence/absence of H2O2. As predicted, the ubiquitin conjugation of SENP3 was attenuated by H2O2, but not by overexpressing Hsp90 alone (Figure 4C, lanes 2 and 3). Overexpression of Hsp90 in combination with H2O2 treatment did decrease SENP3 ubiquitination (lane 4). These results are in accordance with the data on SENP3 accumulation. When Hsp90 was depleted by siRNA, H2O2-induced elimination of ubiquitin conjugation was reversed (lane 5). Moreover, these effects were assessed under conditions of CHIP overexpression where SENP3 was intensively ubiquitin conjugated (Figure 4D, lane 1). Consistent with the above findings, Hsp90 blocked the CHIP-mediated ubiquitination of SENP3 only in the presence of H2O2 (compare lanes 4 and 6). Taken together, our findings clearly indicate that Hsp90 stabilizes SENP3 protein upon H2O2-induced mild oxidative stress by blocking ubiquitin-mediated proteasomal degradation through impairing the E3 ligase function of CHIP.
Figure 4.
Hsp90 repress SENP3 degradation upon oxidative stress. (A) HeLa cells were pre-treated with geldanamycin (GM, 1 or 5 μM) for 1 h or novobiocin (NB, 0.1 or 0.5 mM) for 12 h. H2O2 was added at the indicated concentrations for the last 1 h. SENP3 protein levels were evaluated by IB. (B) HEK293T cells were transfected with Flag-Hsp90 for 48 h and exposed to 100 μM H2O2 or/and 5 μM GM as indicated for the last 1 h. SENP3 protein levels were evaluated and Hsp90 transfection efficiency was monitored by IB. (C) HEK293T cells were co-transfected with RGS-SENP3 and HA-Ub with or without Flag-Hsp90 or Hsp90siRNA (50 nM) for 48 h, and MG132 was then added for the later 12 h. H2O2 was added as indicated for the last 1 h. Co-IP using anti-RGS and IB using the indicated antibodies were performed to determine the ubiquitination of SENP3. (D) HEK293T cells were co-transfected with RGS-SENP3 and HA-Ub with or without Myc-CHIP or/and Flag-Hsp90 for 48 h, and MG132 was added for the later 12 h. H2O2 was added as indicated for the last 1 h. Co-IP using anti-RGS and IB using the indicated antibodies were performed to determine the ubiquitination of SENP3. (E) HEK293T cells were transfected with RGS-SENP3 for 48 h and exposed to 100 μM H2O2 or/and 5 μM GM for the last 1 h. The proteins were co-immunoprecipitated using anti-RGS, and binding of endogenous Hsp90 with RGS-SENP3 was determined by IB using the indicated antibodies. (F) HEK293T cells were transfected with Hsp90siRNA (50 nM) for 48 h and exposed to 100 μM H2O2 as indicated for the last 1 h. The endogenous proteins were co-immunoprecipitated using anti-SENP3, and binding of endogenous CHIP or Hsp90 with SENP3 was determined, respectively, by IB using the indicated antibodies for re-blotting. (G) HEK293T cells were co-transfected with HA-CHIP K30A and non-specific siRNA or siRNA (3′UTR) that specifically targeted to endogenous CHIP, respectively. CHIP protein level was evaluated by IB using anti-CHIP to determine the efficiency and specificity of the CHIP siRNA (upper panel, left). RGS-SENP3 with or without CHIP siRNA (3′UTR) plus HA-CHIP K30A were co-transfected into HEK293T cells for 48 h, and MG132 was added for the later 12 h. H2O2 (100 μM) was added as indicated for the last 1 h. The proteins were co-immunoprecipitated using anti-RGS and immunoblotted using the indicated antibodies to determine the binding of endogenous Hsp90 with SENP3 in the presence of wild-type or mutant CHIP (upper, right). HEK293T cells were co-transfected with RGS-SENP3 or/and CHIP siRNA for 48 h. H2O2 was added for the last 1 h. The proteins were co-immunoprecipitated using anti-RGS and immunoblotted using the indicated antibodies to determine the binding of endogenous Hsp90 with SENP3 in the presence/absence of endogenous CHIP (bottom).
We next sought to determine how Hsp90 and SENP3 interact under mild oxidative stress. Fluorescent images showed that the two proteins co-localize in the nucleoplasm upon H2O2 treatment (Supplementary Figure S2). These data confirm the spatial association of the two proteins, but they might merely reflect a redistribution of SENP3 into the nucleoplasm upon H2O2 treatment. To gain evidence of a physical interaction between them, co-IP was carried out in the presence/absence of H2O2 and/or GM. Interestingly, Hsp90 associated with SENP3 only upon H2O2 exposure. Their binding was abolished in the absence of H2O2 or by GM (Figure 4E). We therefore suspected that the binding of Hsp90 with SENP3 upon oxidative stress changed the mode of CHIP–SENP3 binding which was otherwise maintained under non-stress conditions. As we knew that the association between SENP3 and CHIP was not lost under stress (see Figure 3B), we assumed that Hsp90 might bind to SENP3 by means of intervening between SENP3 and CHIP, making CHIP indirectly bound to SENP3. To address this possibility, we assessed the endogenous SENP3–CHIP association after knockdown of Hsp90. The results demonstrated that the SENP3–CHIP association was maintained regardless of the presence or absence of Hsp90, which was true not only under non-stressed but also under oxidative stress conditions (Figure 4F). We then became curious about the role of CHIP in the SENP3–Hsp90 association under oxidative stress. We conducted a rescue assay by overexpressing WT CHIP and the CHIP K30A mutant in cells in which endogenous CHIP was knocked down by an siRNA irrelevant to the exogenous CHIP. Unexpectedly, the results showed that the SENP3–Hsp90 association upon stress was abolished when WT CHIP was replaced by the mutant unable to interact with Hsp90 (Figure 4G, upper panel). Knockdown of WT CHIP also led to a failure of the SENP3–Hsp90 association (Figure 4G, lower panel). These results reveal that the SENP3–Hsp90 association under stress requires the presence of CHIP, implying that CHIP may help to ensure Hsp90 binding to SENP3.
SENP3 interplays with CHIP and Hsp90 in a sophisticated way
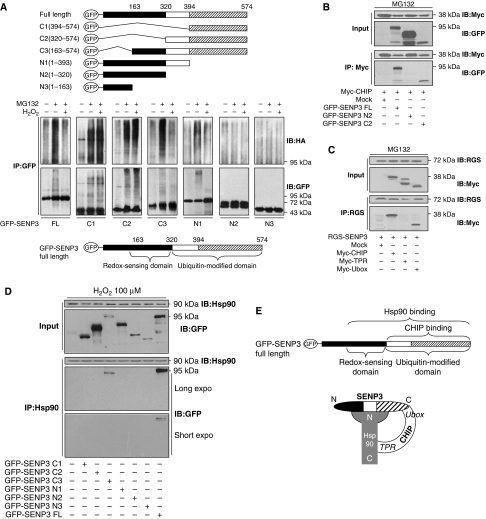
The above data proposed a complex interrelationship among these three molecules that needed clarification. We constructed various truncations of SENP3 based on its sequence (Gong and Yeh, 2006) to map the domains that are subject to ubiquitin ligation and that are responsible for sensing H2O2. The truncates, as illustrated in Figure 5A, upper panel, were transfected into HEK293T cells before the cells were exposed to MG132 and H2O2 treatments. The ubiquitin-conjugation patterns of FL and truncated SENP3 were examined by co-IP. The results showed that the extent and the pattern of ubiquitination varied among the truncates (Figure 5A, middle panel, see the middle lanes, MG132+). Conjugation on the three C-terminal truncates was considerably similar to that on the FL SENP3. However, among the three N-terminal truncates, only N1 was able to be ubiquitinated to a small extent, and no ubiquitination was detectable with N2 or N3 (Figure 5A, middle panel). Therefore, the region at the C-terminus with concentrated lysine residues, ranging from residues 320 to 574, is viewed as the ubiquitin-modified domain (Figure 5A, bottom panel). H2O2-treatment attenuated ubiquitin conjugation to FL SENP3 (Figure 5A, middle panel, see the right lanes, MG132+ plus H2O2+). Strikingly, in all the truncates but C3 and N1, ubiquitination was no longer inhibited by ROS (Figure 5A, middle panel). Thus, we found that the region overlapped by C3 and N1, minus the region overlapped by C3 and C2, ranging from amino acids 163 to 320, is the domain that senses ROS and in turn blocks ubiquitination. We refer to this region as the ‘redox-sensing domain' (Figure 5A, bottom panel).
Figure 5.
SENP3 interplays with CHIP and Hsp90 in a sophisticated way. (A) SENP3 truncates were illustrated (top). HEK293T cells were co-transfected with HA-Ub and GFP-SENP3 full length (FL) or truncates for 48 h. Cells were treated with MG132 (10 μM) for the later 12 h and H2O2 (100 μM) for the last 1 h. Co-IP using anti-GFP and IB using anti-HA were performed to determine the ubiquitination of SENP3, and GFP-SENP3 was detected with anti-GFP (middle). The domains of SENP3 were identified (bottom). (B) Myc-CHIP was co-transfected into HEK293T cells with FL or two truncates of GFP-SENP3 (N2 and C2) for 48 h, respectively. Cells were cultured in the presence of MG132 (10 μM) for the later 12 h. The proteins were co-immunoprecipitated using anti-Myc and immunoblotted using the indicated antibodies. (C) RGS-SENP3 was co-transfected into HEK293T cells with FL or two domains of Myc-CHIP (TPR and Ubox) for 48 h, respectively. Cells were cultured in the presence of MG132 (10 μM) for the later 12 h. The proteins were co-immunoprecipitated using anti-RGS and immunoblotted using the indicated antibodies. (D) HEK293T cells were transfected with GFP-SENP3 FL and truncates (N1, N2, N3, C1, C2, or C3), respectively, for 48 h. H2O2 was added for the last 1 h. Co-IP using anti-Hsp90 and IB using the indicated antibodies were performed to determine the binding of endogenous Hsp90 with SENP3 and its truncates. (E) A speculative model depicting the interplay among SENP3, Hsp90, and CHIP upon oxidative stress.
To dissect the domains that mediate the interaction between SENP3 and CHIP, SENP3 and its truncates were overexpressed and co-IP was performed using an antibody against the tag on CHIP. As shown in Figure 5B, CHIP co-precipitated with FL SENP3 and the SENP3-C2 truncate but not the N2 truncate. Reciprocally, co-IP was performed after overexpression of CHIP and its truncates using an antibody against the tag on SENP3. We found that SENP3 co-precipitated with FL CHIP and the Ubox domain but not the TPR domain (Figure 5C). These results suggest that the binding of CHIP and SENP3 is mediated through an interaction between the Ubox domain of CHIP and the C-terminal domain (C2, 320–574) of SENP3, where CHIP may execute the ubiquitination of SENP3.
We next dissected the interaction domains between SENP3 and Hsp90. Detailed mapping using all the SENP3 truncates was performed. Intriguingly, in addition to FL SENP3, Hsp90 could also bind to the C3 truncate, but not the other truncates (Figure 5D). This result clearly demonstrates that Hsp90 associates to a region on SENP3 that corresponds exactly to the redox-sensing domain plus the ubiquitin-modified domain. We hence concluded that an intact redox-sensing domain and the ubiquitin-modified domain are both required for Hsp90 binding with SENP3. Although the ubiquitin-modified domain is also bound by CHIP, two lines of evidence indicate that Hsp90–SENP3 association under oxidative stress does not repel, but rather, requires CHIP–SENP3 association: one is that CHIP–SENP3 association is not affected by Hsp90 (Figure 4F), and another is that Hsp90–SENP3 interaction needs CHIP (Figure 4G). As the Hsp90–SENP3 association could be abolished by GM, a reagent acting at the N-terminus of Hsp90, the interacting domain of Hsp90 must be its N-terminus. Therefore, we propose a model for interplay of the three molecules under oxidative stress (Figure 5E). Importantly, this information strongly suggests that SENP3 does not become a client of Hsp90 until some intrinsic change specifically appears within the redox-sensing domain under oxidative stress.
Blockade of SENP3 ubiquitination is triggered by oxidative modification of cysteines on SENP3, which recruits Hsp90
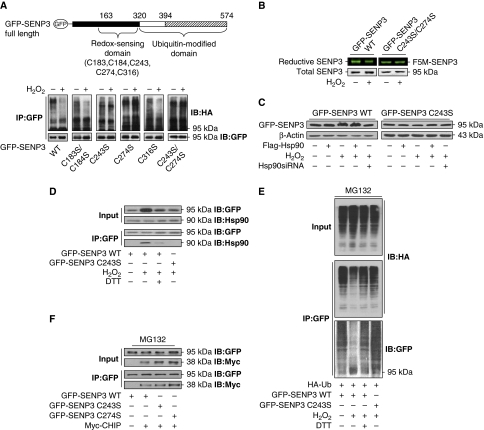
We next sought to characterize the intrinsic feature in the redox-sensing domain to make SENP3 a client of Hsp90. As cysteine residues are major target of oxidative modifications, all five cysteines within the redox-sensing domain were potentially involved in the regulation of ubiquitination. We individually mutated these cysteines to serines (Figure 6A, upper). These SENP3 mutants were transfected into cells followed by H2O2 treatment. The results of ubiquitination assays showed that the C243S and C274S mutants lost their responsiveness to ROS for the repression of ubiquitination. The double mutant showed a pattern similar to that of the single mutant, suggesting that both cysteine residues are essential (Figure 6A).
Figure 6.
Blockade of SENP3 ubiquitination is triggered by oxidative modification of cysteines on SENP3, which recruits Hsp90. (A) The identified domains of SENP3 and the sites for mutagenesis with cysteines replaced by serines in the redox-sensing domain (upper). HEK293T cells were co-transfected with HA-Ub and GFP-SENP3 wild-type (WT) or C/S mutants for 48 h. Cells were treated with MG132 (10 μM) for the later 12 h and H2O2 (100 μM) for the last 1 h. Co-IP using anti-GFP and IB using the anti-HA were performed to determine the ubiquitination of SENP3, and GFP-SENP3 was detected with anti-GFP (bottom). (B) HEK293T cells were transfected with GFP-SENP3 WT or C243/274S mutant for 48 h. Cells were treated with H2O2 (100 μM) for 30 min before incubation with F5M for another 30 min. IP with anti-GFP was performed. Total GFP-SENP3 protein levels were evaluated by IB with anti-GFP. Reductive GFP-SENP3 with F5M fluorescence in the same bands were visualized and photographed. (C) HEK293T cells were transfected with GFP-SENP3 WT or mutant C243S with or without Flag-Hsp90 or Hsp90 siRNA for 48 h, and 100 μM H2O2 was added for the last 1 h. SENP3 protein levels were evaluated by IB. (D) HEK293T cells were transfected with GFP-SENP3 WT or mutant C243S for 48 h. A measure of 100 μM H2O2 and 5 mM DTT were added for the last 1 h. The proteins were co-immunoprecipitated using anti-GFP and immunoblotted using the indicated antibodies to determine the binding of endogenous Hsp90 with WT or mutant SENP3. (E) HEK293T cells were co-transfected with HA-Ub and GFP-SENP3 WT or mutant C243S for 48 h and MG132 was added for the later 12 h. H2O2 and DTT were added for the last 1 h. The proteins were co-immunoprecipitated using anti-GFP and immunoblotted using the indicated antibodies to determine the ubiquitination of SENP3. (F) HEK293T cells were co-transfected with GFP-SENP3 WT or mutants C243S and C274S, and Myc-CHIP for 48 h. MG132 was added for the later 12 h. The proteins were co-immunoprecipitated using anti-GFP and immunoblotted using the indicated antibodies to determine the binding of Myc-CHIP with SENP3 WT or mutants.
The dye fluorescein-5-maleimide (F5M) labels cysteines of which the thiol is in reduced form. F5M fluorescence intensity thereby reflects, and is proportional to, the reductive degree of a protein (Zuo et al, 2009). For WT SENP3, upon exposure to H2O2, the quantity of F5M-labelled SENP3 decreased, whereas the total level of SENP3 protein increased, revealing that some thiols of cysteines in SENP3 were oxidized (Figure 6B). If cysteines 243 and 274 truly sense subtle increases in ROS, the mutated SENP3 would be unable to be oxidized upon exposure to a low dose of H2O2. Indeed, the C243/274S mutant showed neither the accumulation of total protein nor the attenuation in F5M fluorescent intensity displayed by WT SENP3 (Figure 6B). The C243S mutant completely lost the accumulation in response to the presence of H2O2 and Hsp90 (Figure 6C), indicating that cysteine 243 (as well as 274) is required for the stabilization of SENP3. Co-IP assays further demonstrated that although H2O2 induced binding of WT SENP3 with Hsp90 and the thiol-reducing agent dithiothreitol (DTT) reversed this binding, the C243S mutant could no longer be recognized and bound by Hsp90 under the same H2O2 exposure (Figure 6D). In agreement with this result, ubiquitination of C243S was not attenuated by H2O2 treatment (Figure 6E). These data suggest that either cysteine 243 or 274 undergoes oxidative modification under mild oxidative stress, and this functions as a signal to recruit Hsp90. The binding of Hsp90 following recognition of oxidized cysteine(s) consequently protects SENP3 from ubiquitination by CHIP.
It was next necessary to determine whether the association of CHIP with SENP3 was affected by mutation of the redox-sensing cysteines. Co-IP was performed, and the data showed that CHIP could bind to the C243S and C274S mutants as well as to WT SENP3 (Figure 6F). This result confirmed the idea that the physical association between CHIP and SENP3 is not redox responsive.
SENP3 interacts with CHIP and Hsp90 in differential modes under non-stress and oxidative stress conditions
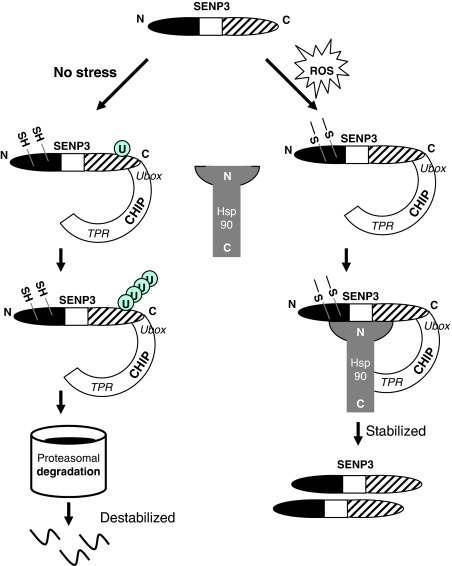
Taken together, the multiple lines of evidence presented above delineate a regulatory mechanism for SENP3 turnover under non-stress and oxidative stress conditions (Figure 7). CHIP, independent on chaperones, binds to the ubiquitin-modified domain of SENP3 and mediates its ubiquitination, causing SENP3 to be degraded outside of the nucleolus and generally maintained at a low basal level. When the cell encounters a mild oxidative stress, the thiols of cysteines 243 and/or 274 in SENP3 undergo oxidative modification, which releases a signal for the recruitment of Hsp90. Hsp90 does not compete with CHIP for binding to the ubiquitin-modified domain, but rather, it binds to a region containing both the redox-sensing domain and the ubiquitin-modified domain of SENP3 and leads to abrogation of CHIP-mediated ubiquitin ligation. The physical association of CHIP and SENP3 is not replaced or altered by Hsp90 binding to SENP3; instead, the CHIP–Hsp90 interaction are required under this context, serving as a supporter for the Hsp90–SENP3 association.
Figure 7.
SENP3 interacts with CHIP and Hsp90 in differential modes under non-stress and oxidative stress conditions. A model for the mechanisms underlying stabilization and degradation of SENP3.
Accumulation of SENP3 in cancer cells correlates with enhanced Hsp90–SENP3 association and increased ROS
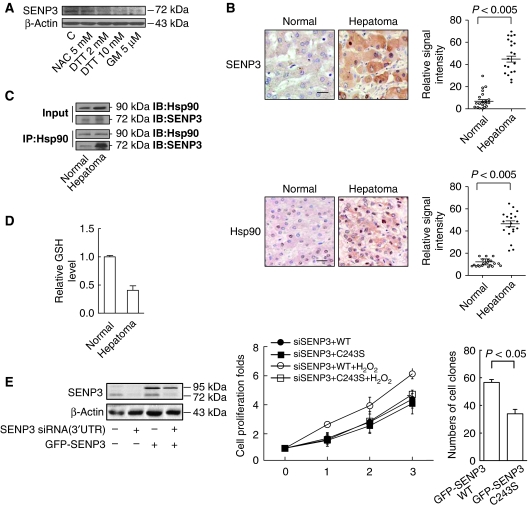
HepG2 is a perpetual cell line, which was derived from human hepatocarcinoma (Pan et al, 2004). Endogenous SENP3 was readily detected in these cancerous cells, implying that it might accumulate due to the binding of Hsp90 in response to high ROS levels. This assumption was corroborated by the fact that the level of SENP3 was markedly attenuated when HepG2 cells were exposed to the anti-oxidant N-acetyl cysteine (NAC), DTT, or the Hsp90 inhibitor GM (Figure 8A). To evaluate the situation in real tissues, SENP3 and Hsp90 immunohistochemistry were conducted on 20 hepatocarcinoma and normal liver autopsy tissue samples. Positive staining for SENP3 was undetectable in normal liver, but it was readily visible in the most of the cancerous hepatocytes, localizing in the nucleus and the cytoplasm with predominant accumulation in the nucleus (Figure 8B, left, upper). By contrast, positive Hsp90 staining was visible in normal hepatocytes, mainly in the nucleus, but was much stronger in the cancerous hepatocytes, both in the nucleus and the cytoplasm (Figure 8B, left, lower). Quantitative image analysis demonstrated that the average intensities of SENP3 and Hsp90 were significantly higher in hepatocarcinoma cells (Figure 8B, right). Furthermore, co-IP assays were performed using tissue extracts derived from two pairs of freshly collected hepatocarcinoma and adjacent tissues. The results showed that binding of endogenous Hsp90 with endogenous SENP3 was robustly enhanced in hepatocarcinoma (Figure 8C). The redox state of these paired tissues was compared by measurement of the reduced glutathione (GSH) level. The results showed that hepatocarcinoma was more prone to an oxidative state than normal liver tissue (Figure 8D). Collectively, these data suggest that SENP3 accumulation in cancer cells might correlate with its enhanced interaction with Hsp90 under oxidative stress.
Figure 8.
Accumulation of SENP3 in cancer cells correlates with enhanced Hsp90–SENP3 association and increased ROS. (A) HepG2 cells were pre-treated with NAC for 4 h, DTT or GM for 1 h. SENP3 protein level was evaluated by IB using anti-SENP3. (B) Immunohistochemistry for SENP3 and Hsp90 were performed in 20 hepatoma and adjacent normal tissues. Scale bar=10 μM (left). The relative signal intensity was analysed (right). (C) IP was performed in 100 mg of tissues evenly derived from two pairs of fresh hepatoma and adjacent normal tissues using anti-Hsp90, and IB was performed with 1/2 loading of hepatoma sample. (D) The GSH level was assessed in the tissues similar to (C). Quantification showed the means±s.d. of the relative GSH levels. (E) HeLa cells were co-transfected with NC or SENP3 3′UTR siRNA and the construct of GFP-SENP3 WT for 72 h. The endogenous SENP3 and the exogenous GFP-SENP3 were determined with anti-SENP3 and anti-GFP (left). HeLa cells were co-transfected with SENP3 3′UTR siRNA and the constructs of GFP-SENP3 WT or C243S. After 48 h, cells were re-seeded and treated with 100 μM H2O2 for 1 h once a day from the second day. Cell proliferation was assessed and indicated by a fold increase. The values are expressed as the means±s.d. of two independent experiments (middle). HepG2 cells were stably transfected with GFP-SENP3 WT or C243S and retroviral vector expressing shRNA for SENP3. Colony formation assay was performed and indicated by the numbers of colonies in soft agar per 1000 seeded cells (n=3 dishes) (right).
Finally, to evaluate the biological and pathological significance of the redox-sensing function of SENP3, we tested cell proliferation and soft agar colonegenesis in cells expressing WT or mutant SENP3. Endogenous SENP3 was depleted by siRNA that did not target exogenous SENP3 (Figure 8E, left). The results of cell proliferation demonstrated that cells with the SENP3 C243S mutant lost responsiveness to ROS-induced proliferation (Figure 8E, middle). Moreover, soft agar colonegenesis assay was conducted in cells stably expressing WT/mutant SENP3 and shRNA for SENP3. The results demonstrated a weaker colonegenetic potential in cells expressing the C243S SENP3 (Figure 8E, right). Taken together, these data strongly indicate that SENP3 overaccumulates in liver cancer and has a function in tumour enhancement by responding to aberrant redox environments.
Discussion
Oxidative stress occurs as a consequence of an imbalance between the production and scavenging of ROS. Severe oxidative stress leads to oxidation and aggregation of vital proteins and DNA. The expression of Hsps may increase dramatically following such stress (Swindell et al, 2007). Some Hsps, mainly members of the Hsp70 family and its co-chaperones, select and direct aberrant proteins to the proteasome or lysosome for degradation (Whitesell and Lindquist, 2005). In some cases when proteins can be rescued, the same system can refold damaged proteins (Broadley and Hartl, 2009). The Hsp and co-chaperone system thus has a crucial function as a cleanser under these circumstances. However, when mild oxidative stress occurs, cells may modify their signalling and gene expression profiles to adjust their behaviour. Diverse post-translational modifications on proteins may be involved in these processes, during which ROS lead to oxidative modification, rather than typical oxidative damage, of proteins (Kumsta and Jakob, 2009). Whether and how the Hsp and co-chaperone system participate in these adaptive responses remain largely unknown. The present study provides an example of the role of Hsp90 and its co-chaperone CHIP in the stabilization of a normal protein under mild oxidative stress induced by low doses of H2O2.
We found that the binding of Hsp90 to SENP3, a SUMO2/3-specific protease, leads to SENP3 stabilization under mild oxidative stress and that this is achieved via blockage of the CHIP-mediated ubiquitination that otherwise constantly occurs under non-stressed conditions. We previously demonstrated that following nuclear accumulation of SENP3, it re-localizes from the nucleoli to all over the nucleoplasm, which provides this SUMO protease with a brand-new spectrum of substrates. The important transcription co-activator p300 and the nuclear body component PML, among others, become substrates of SENP3 under mild oxidative stress. De-conjugation of SUMO2/3 from p300 and PML leads to changes in their functions; the transcriptional activity of hypoxia-inducible factor-1 is enhanced, and the role of negative regulator of PML for cell proliferation is impaired (Huang et al, 2009; Han et al, 2010), which all contribute to cellular adaptation to mild oxidative stress. Therefore, Hsp90 and CHIP cooperate as critical modulators that ensure the function of a protein in this process.
Hsp90 is an abundant and essential protein in eukaryotic cells as well as in bacteria. As one of the most important chaperones, Hsp90 stabilizes, refolds, and activates its client proteins (Young et al, 2001). Hsp90 stabilizes mutant oncogenic proteins that are prone to misfolding, thereby enabling malignant transformation (Cowen, 2009). Compromising Hsp90 function can reverse oncogenic cellular phenotypes (Neckers, 2007). Hsp90 also stabilizes unmutated regulators of signalling in fungal cells, conferring drug resistance (Cowen, 2009). In neuronal cells, phosphorylated tau is a client of the Hsp90 chaperone network. In addition, abnormal tau (hyperphosphorylated) displays enhanced binding to Hsp90 (Dickey et al, 2007). Although an increasing body of research has revealed many in vivo Hsp90 client proteins and proven that the association of client proteins with Hsp90 complexes is important for their activity, the interaction of Hsp90 with client proteins is currently still poorly understood. Among the most intriguing puzzles are the specific mechanism of recognition of non-native substrates by Hsp90 and the coordination of Hsp90 with the CHIP degradation machinery (Richter and Buchner, 2001).
Being in a complex together with the client, CHIP and Hsp90 can have cooperative or antagonistic effects on the client (Qian et al, 2006; Xia et al, 2007). For instance, dephosphorylation and refolding of p-tau is initially facilitated by an Hsp90-containing complex that prevents degradation; however, when refolding is subverted by Hsp90 inhibition, p-tau is transferred to the Hsp70/CHIP complex and degraded via polyubiquitination (Dickey et al, 2007). Our present study revealed that CHIP mediates constitutive ubiquitination and degradation of SENP3 under non-stress condition, whereas by contrast, Hsp90 binds to SENP3 under mild oxidative stress and mediates its stabilization. This is the first demonstration to our knowledge that CHIP and Hsp90 are responsible, respectively, for controlling the levels of a protein under non-stressed and stressed conditions. Moreover, our data provide new insight into the mutual relationship of these two molecules. CHIP is believed to mediate degradation of the clients in a chaperone-dependent manner, until recently it is noticed that CHIP can be independent of Hsp90 or other chaperones (Parsons et al, 2008; Shang et al, 2009) Our findings here highlight this novel concept by confirming that Hsp90 is dispensable for CHIP-mediated SENP3 degradation under non-stressed condition. On the contrary, CHIP is known to be indispensable for the chaperones under various circumstances (McDonough and Patterson, 2003; Rosser et al, 2007), although the precise contributions of CHIP as a co-chaperone to Hsp90 remain unclear. Our study demonstrates that Hsp90 abrogates the ubiquitin ligase function of CHIP, but Hsp90's action requires the presence of CHIP and the CHIP–Hsp90 interaction. This previously unperceived contribution suggests an alternative co-chaperone function of CHIP: a molecule supporting the complex for the Hsp90-mediated client stabilization.
Our present data show that, upon oxidative stress, Hsp90 binds to a region of SENP3 comprising the redox-sensing domain and the ubiquitin-modified domain. More interestingly, a prerequisite for this Hsp90–SENP3 association is that Hsp90 recognizes specific oxidative modifications on cysteines 243 and/or 274 in the redox-sensing domain of SENP3. This study is the first demonstration that oxidation of a protein diverts its fate from degradation to stabilization. A study of structure biology in the future may provide better elucidation to whether the site for Hsp90 binding is spatially close to the site for CHIP binding, and how the Hsp90–SENP3 binding affects the function of CHIP but simultaneously requires the presence of CHIP.
Reversibility, a crucial aspect of all regulatory mechanisms, is one of the most important characteristics of thiol oxidative modification (Kumsta and Jakob, 2009). We showed that stabilization of SENP3 is reversed by an anti-oxidant thiol-reducing agent as well as by an Hsp90 inhibitor. This result implies that Hsp90 binding to SENP3 and the consequent SENP3 stabilization are subject to physiological modulation along with environmental ROS fluctuations.
Redox regulation of chaperones has been illustrated in two molecules, prokaryotic Hsp33 and eukaryotic 2-Cys peroxiredoxin, which use ROS to activate their chaperone function (Jang et al, 2004; Winter et al, 2008; Kumsta and Jakob, 2009). However, the client selectivity of the chaperones under these circumstances has not been explained. By contrast, our report unveils an intrinsic signal formed by oxidative modification of the client protein instead of the chaperone. This unique mechanism may better explain the high specificity of SENP3 stabilization, as we have determined that only this SUMO protease in the SENP family is immediately stabilized by the low concentrations of H2O2 we used (Han et al, 2010).
The role of Hsp90 in cancer has received much attention in the past decade, because it is upregulated in various primary cancers and functions as an integral part of the machinery that allows cancer cells to escape normal regulation (Pick et al, 2007). Recently, CHIP was found to suppress tumour progression in human breast cancer by inhibiting oncogenic pathways, and CHIP levels are negatively correlated with the malignancy of human breast tumour tissues (Kajiro et al, 2009). Our previous and current data demonstrate an accumulation of SENP3 in a series of primary cancers. The present data have further revealed that binding between Hsp90 and SENP3 is enhanced in cancerous hepatocytes in which ubiquitination of SENP3 is attenuated. Therefore, this study, in line with the ideas that Hsp90 is oncogenic whereas CHIP is a tumour suppressor, provides a novel redox-sensitive client protein that is aberrantly regulated by the CHIP–Hsp90 machinery in cancer.
Materials and methods
Cell culture and treatments
HeLa and HEK293T cells were cultured in Dulbecco's modified Eagle's medium (GibcoBRL, Gaithersburg, MD). HepG2 cells were cultured in a 1:1 mixture of DMEM and 1640 (GibcoBRL). All media were supplemented with 10% newborn calf serum (Biochrom AG, Germany). Cells were maintained at 37°C in a humidified atmosphere with 5% CO2. MG132 (Merck KGaA, Germany) was used to block proteasome activity. To examine the association with ROS, cells were treated with H2O2, NAC, and DTT (Sigma-Aldrich, St Louis, MO). GM (ALEXIS Biochemicals, Switzerland) and NB (Merck) were used to block Hsp90 activity.
Constructs and transient transfections
The constructs were described in Supplementary data. The constructs were transiently transfected or co-transfected into cells using Lipofectamine 2000 (Invitrogen, Carlsbad, CA) according to the manufacturer's instructions.
Reverse transcription and polymerase chain reaction (RT–PCR)
RT–PCR and quantitative PCR were carried out by standard procedures. The primers and the conditions were described in Supplementary data.
Immunoblotting
IB was performed using the routine methods and antibodies are described in Supplementary data.
Co-IP and denaturing co-IP
Co-IP and denaturing co-IP were carried out for both endogenous and ectopically expressed proteins using the routine methods described in Supplementary data.
Immunohistochemistry
Twenty normal liver specimens and 20 tumour tissues were derived from archived autopsic and pathological tissues processed as paraformaldehyde-fixed and paraffin-embedded specimens. Sections were incubated overnight at 4°C with primary antibodies against SENP3 or Hsp90 (all 1:100) followed by biotinylated secondary antibody. Immunohistochemical reactions were visualized using peroxidase-conjugated streptavidin, in which diaminobenzidine was used as a chromogen. Sections were finally counterstained with hematoxylin. The relative intensities of signal positive for SENP3 and Hsp90 were estimated in five fields under × 40 by Zeiss KS400 Version 2.2 software.
siRNA and shRNA
siRNA specific for c-Cbl, E6AP, HHARI, CHIP, parkin, SENP3, SENP3 (3′UTR), Hsp90, CHIP (3′UTR), and non-specific control (NC) siRNA were synthesized (RIBOBIO, China), and transfected using Lipofectamine 2000. The viral vector for SENP3 shRNA was constructed and introduced into cells by the previously used methods (Wang et al, 2009; Han et al, 2010). The sequences of siRNA and shRNA were described in Supplementary data.
Sequential two-dimensional non-reducing/reducing SDS–PAGE
This method has been referred as redox diagonal electrophoresis (Zuo et al, 2009) and was briefly described in Supplementary data.
Assessment of thiol reduction by F5M
Thiol reduction of the SENP3 WT/mutant was assessed by the previously used method (Zuo et al, 2009). The detailed procedure was described in Supplementary data.
Reduced GSH assay
Tissues using two pairs of fresh hepatocarcinoma and the adjacent normal liver were prepared and analysed for GSH level according to the manufacturer's instructions (Jiancheng Bioengineering Institute, China) as described previously (Han et al, 2010) and in Supplementary data.
Cell proliferation assay
As HeLa cells are sensitive to H2O2-induced cell proliferation, they were used for assessment of responsiveness of SENP3 redox-sensing domain to ROS. Cells were seeded at 50% confluence and co-transfected with SENP3 3′UTR siRNA and the constructs of GFP-SENP3-WT or GFP-SENP3-C243/274S. After 48 h, cells were re-seeded in 24-well plates at 30% confluence and were treated with 100 μM H2O2 once a day. Proliferation was assessed using previously described methods (Han et al, 2010) for 3 days.
Colony formation assay
The colony formation capability of HepG2 cells in soft agar was investigated in cells stably expressing GFP-SENP3-WT or -C243/274S followed by infection of SENP3 shRNA. The constructs of WT and mutant SENP3 were mutated to become resistant to shRNA. The procedures for stable transfection and retroviral introduction of shRNA were used as previously (Han et al, 2010) and described in Supplementary data. Colony formation assay was conducted using the previous method (Cai et al, 2008). The plates were incubated for 15 days until colonies were counted.
Statistical analysis
SPSS11.5 software was used for statistical analysis. ANOVA was applied for comparison of the means of two groups, in which Student–Newman–Kewls was further used for the comparison of each of the two groups. A value of P<0.05 was considered significant.
Supplementary Material
Acknowledgments
We thank Dr Xiao-Dan Yu of Institute of Basic Medical Sciences, Beijing, for the CHIP constructs, Dr Shubing Qian of the University of Cornell for Hsp90 constructs, and Dr Chen Wang of The Institute of Biochemistry and Cell Biology, Chinese Academy of Science, Shanghai, for the CHIP K30A mutant constructs. This work was supported by grants from the National Natural Science Foundation of China (30971437, J Yi), Shanghai Municipal Science and Technology Commission (08JC1413800 and 10410704000, J Yi), Shanghai Municipal Education Commission (J50201, J Yi) and Ren Ji Collaboration Project (XR Wang and J Yi).
Footnotes
The authors declare that they have no conflict of interest.
References
- Broadley SA, Hartl FU (2009) The role of molecular chaperones in human misfolding diseases. FEBS Lett 583: 2647–2653 [DOI] [PubMed] [Google Scholar]
- Cai J, Niu X, Chen Y, Hu Q, Shi G, Wu H, Wang J, Yi J (2008) Emodin-induced generation of reactive oxygen species inhibits RhoA activation to sensitize gastric carcinoma cells to anoikis. Neoplasia 10: 41–51 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chiosis G, Vilenchik M, Kim J, Solit D (2004) Hsp90: the vulnerable chaperone. Drug Discov Today 9: 881–888 [DOI] [PubMed] [Google Scholar]
- Cowen LE (2009) Hsp90 orchestrates stress response signaling governing fungal drug resistance. PLoS Pathog 5: e1000471. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cyr DM, Hohfeld J, Patterson C (2002) Protein quality control: U-box-containing E3 ubiquitin ligases join the fold. Trends Biochem Sci 27: 368–375 [DOI] [PubMed] [Google Scholar]
- Dai Q, Zhang C, Wu Y, McDonough H, Whaley RA, Godfrey V, Li HH, Madamanchi N, Xu W, Neckers L, Cyr D, Patterson C (2003) CHIP activates HSF1 and confers protection against apoptosis and cellular stress. EMBO J 22: 5446–5458 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dickey CA, Kamal A, Lundgren K, Klosak N, Bailey RM, Dunmore J, Ash P, Shoraka S, Zlatkovic J, Eckman CB, Patterson C, Dickson DW, Nahman NS Jr, Hutton M, Burrows F, Petrucelli L (2007) The high-affinity HSP90-CHIP complex recognizes and selectively degrades phosphorylated tau client proteins. J Clin Invest 117: 648–658 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ding X, Goldberg MS (2009) Regulation of LRRK2 stability by the E3 ubiquitin ligase CHIP. PLoS One 4: e5949. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Goetz MP, Toft DO, Ames MM, Erlichman C (2003) The Hsp90 chaperone complex as a novel target for cancer therapy. Ann Oncol 14: 1169–1176 [DOI] [PubMed] [Google Scholar]
- Gong L, Yeh ET (2006) Characterization of a family of nucleolar SUMO-specific proteases with preference for SUMO-2 or SUMO-3. J Biol Chem 281: 15869–15877 [DOI] [PubMed] [Google Scholar]
- Han Y, Huang C, Sun X, Xiang B, Wang M, Yeh ET, Chen Y, Li H, Shi G, Cang H, Sun Y, Wang J, Wang W, Gao F, Yi J (2010) SENP3-mediated de-conjugation of SUMO2/3 from PML is correlated with accelerated cell proliferation under mild oxidative stress. J Biol Chem 285: 12906–12915 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Huang C, Han Y, Wang Y, Sun X, Yan S, Yeh ET, Chen Y, Cang H, Li H, Shi G, Cheng J, Tang X, Yi J (2009) SENP3 is responsible for HIF-1 transactivation under mild oxidative stress via p300 de-SUMOylation. EMBO J 28: 2748–2762 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jang HH, Lee KO, Chi YH, Jung BG, Park SK, Park JH, Lee JR, Lee SS, Moon JC, Yun JW, Choi YO, Kim WY, Kang JS, Cheong GW, Yun DJ, Rhee SG, Cho MJ, Lee SY (2004) Two enzymes in one; two yeast peroxiredoxins display oxidative stress-dependent switching from a peroxidase to a molecular chaperone function. Cell 117: 625–635 [DOI] [PubMed] [Google Scholar]
- Kajiro M, Hirota R, Nakajima Y, Kawanowa K, So-ma K, Ito I, Yamaguchi Y, Ohie SH, Kobayashi Y, Seino Y, Kawano M, Kawabe Y, Takei H, Hayashi S, Kurosumi M, Murayama A, Kimura K, Yanagisawa J (2009) The ubiquitin ligase CHIP acts as an upstream regulator of oncogenic pathways. Nat Cell Biol 11: 312–319 [DOI] [PubMed] [Google Scholar]
- Kampinga HH (2006) Chaperones in preventing protein denaturation in living cells and protecting against cellular stress. Handb Exp Pharmacol 172: 1–42 [DOI] [PubMed] [Google Scholar]
- Ko HS, Bailey R, Smith WW, Liu Z, Shin JH, Lee YI, Zhang YJ, Jiang H, Ross CA, Moore DJ, Patterson C, Petrucelli L, Dawson TM, Dawson VL (2009) CHIP regulates leucine-rich repeat kinase-2 ubiquitination, degradation, and toxicity. Proc Natl Acad Sci USA 106: 2897–2902 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kumsta C, Jakob U (2009) Redox-regulated chaperones. Biochemistry 48: 4666–4676 [DOI] [PMC free article] [PubMed] [Google Scholar]
- LaVoie MJ, Cortese GP, Ostaszewski BL, Schlossmacher MG (2007) The effects of oxidative stress on parkin and other E3 ligases. J Neurochem 103: 2354–2368 [DOI] [PubMed] [Google Scholar]
- McDonough H, Patterson C (2003) CHIP: a link between the chaperone and proteasome systems. Cell Stress Chaperones 8: 303–308 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mosser DD, Morimoto RI (2004) Molecular chaperones and the stress of oncogenesis. Oncogene 23: 2907–2918 [DOI] [PubMed] [Google Scholar]
- Muchowski PJ, Wacker JL (2005) Modulation of neurodegeneration by molecular chaperones. Nat Rev Neurosci 6: 11–22 [DOI] [PubMed] [Google Scholar]
- Murata S, Chiba T, Tanaka K (2003) CHIP: a quality-control E3 ligase collaborating with molecular chaperones. Int J Biochem Cell Biol 35: 572–578 [DOI] [PubMed] [Google Scholar]
- Neckers L (2007) Heat shock protein 90: the cancer chaperone. J Biosci 32: 517–530 [DOI] [PubMed] [Google Scholar]
- Pan X, Wang Y, Zhang M, Pan W, Qi ZT, Cao GW (2004) Effects of endostatin-vascular endothelial growth inhibitor chimeric recombinant adenoviruses on antiangiogenesis. World J Gastroenterol 10: 1409–1414 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Parsons JL, Tait PS, Finch D, Dianova II, Allinson SL, Dianov GL (2008) CHIP-mediated degradation and DNA damage-dependent stabilization regulate base excision repair proteins. Mol Cell 29: 477–487 [DOI] [PubMed] [Google Scholar]
- Pick E, Kluger Y, Giltnane JM, Moeder C, Camp RL, Rimm DL, Kluger HM (2007) High HSP90 expression is associated with decreased survival in breast cancer. Cancer Res 67: 2932–2937 [DOI] [PubMed] [Google Scholar]
- Qian SB, McDonough H, Boellmann F, Cyr DM, Patterson C (2006) CHIP-mediated stress recovery by sequential ubiquitination of substrates and Hsp70. Nature 440: 551–555 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Richter K, Buchner J (2001) Hsp90: chaperoning signal transduction. J Cell Physiol 188: 281–290 [DOI] [PubMed] [Google Scholar]
- Rosser MF, Washburn E, Muchowski PJ, Patterson C, Cyr DM (2007) Chaperone functions of the E3 ubiquitin ligase CHIP. J Biol Chem 282: 22267–22277 [DOI] [PubMed] [Google Scholar]
- Shang Y, Zhao X, Xu X, Xin H, Li X, Zhai Y, He D, Jia B, Chen W, Chang Z (2009) CHIP functions an E3 ubiquitin ligase of Runx1. Biochem Biophys Res Commun 386: 242–246 [DOI] [PubMed] [Google Scholar]
- Swindell WR, Huebner M, Weber AP (2007) Transcriptional profiling of Arabidopsis heat shock proteins and transcription factors reveals extensive overlap between heat and non-heat stress response pathways. BMC Genomics 8: 125. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang C, Chen J (2003) Phosphorylation and hsp90 binding mediate heat shock stabilization of p53. J Biol Chem 278: 2066–2071 [DOI] [PubMed] [Google Scholar]
- Wang R, Zou Y, Yuan Z, Wang Y, Chen Y, Mao Y, Zhu ZA, Li H, Tang X, Lu J, Yi J (2009) Autografts and xenografts of skin fibroblasts delivering BMP-2 effectively promote orthotopic and ectopic osteogenesis. Anat Rec (Hoboken) 292: 777–786 [DOI] [PubMed] [Google Scholar]
- Wegele H, Muller L, Buchner J (2004) Hsp70 and Hsp90—a relay team for protein folding. Rev Physiol Biochem Pharmacol 151: 1–44 [DOI] [PubMed] [Google Scholar]
- Whitesell L, Lindquist SL (2005) HSP90 and the chaperoning of cancer. Nat Rev Cancer 5: 761–772 [DOI] [PubMed] [Google Scholar]
- Winter J, Ilbert M, Graf PC, Ozcelik D, Jakob U (2008) Bleach activates a redox-regulated chaperone by oxidative protein unfolding. Cell 135: 691–701 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xia T, Dimitropoulou C, Zeng J, Antonova GN, Snead C, Venema RC, Fulton D, Qian S, Patterson C, Papapetropoulos A, Catravas JD (2007) Chaperone-dependent E3 ligase CHIP ubiquitinates and mediates proteasomal degradation of soluble guanylyl cyclase. Am J Physiol Heart Circ Physiol 293: H3080–H3087 [DOI] [PubMed] [Google Scholar]
- Xu W, Marcu M, Yuan X, Mimnaugh E, Patterson C, Neckers L (2002) Chaperone-dependent E3 ubiquitin ligase CHIP mediates a degradative pathway for c-ErbB2/Neu. Proc Natl Acad Sci USA 99: 12847–12852 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yang K, Shi H, Qi R, Sun S, Tang Y, Zhang B, Wang C (2006) Hsp90 regulates activation of interferon regulatory factor 3 and TBK-1 stabilization in Sendai virus-infected cells. Mol Biol Cell 17: 1461–1471 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Young JC, Moarefi I, Hartl FU (2001) Hsp90: a specialized but essential protein-folding tool. J Cell Biol 154: 267–273 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang M, Windheim M, Roe SM, Peggie M, Cohen P, Prodromou C, Pearl LH (2005) Chaperoned ubiquitylation--crystal structures of the CHIP U box E3 ubiquitin ligase and a CHIP-Ubc13-Uev1a complex. Mol Cell 20: 525–538 [DOI] [PubMed] [Google Scholar]
- Zuo Y, Xiang B, Yang J, Sun X, Wang Y, Cang H, Yi J (2009) Oxidative modification of caspase-9 facilitates its activation via disulfide-mediated interaction with Apaf-1. Cell Res 19: 449–457 [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.