Abstract
Background
BK virus nephropathy (BKVN) may cause renal allograft dysfunction and failure. The gold standard test is kidney biopsy, which is invasive and costly. A noninvasive, accurate biomarker for diagnosis of BKVN and prognostication of allograft function after BKVN infection may improve allograft survival.
Methods
We tested the diagnostic accuracy of our previously reported cutoff of 6.5×105 BKV-VP1 mRNA/ng RNA in urinary cells (Ding et al. Transplantation 2002; 74:987-94) using an independent cohort (N=89). We also examined whether urinary cell mRNA profiles obtained at time of BKVN diagnosis identified patients at risk for subsequent decline of graft function.
Results
BKVN was accurately diagnosed (sensitivity of 100% and specificity of 97%) using our previously reported cutoff value. Levels of granzyme B (GB) mRNA (P=0.002) and proteinase inhibitor-9 (PI-9) mRNA (P=0.01) in urinary cells were higher in BKVN patients with a subsequent decline in renal function (BKVN-DF, N=8) compared to patients with stable function (BKVN-SF, N=10), and were positively associated (GB, P=0.01; PI-9, P=0.04) with rise in serum creatinine from the time of BKVN diagnosis to 12 months post diagnosis. GB levels in the BKVN-DF group were similar to acute rejection(AR) group(N=11, P>0.05), but higher than the normal biopsy group(N= 36, P<0.001); levels in BKVN-SF were lower than the AR group (P<0.01) and not significantly different from the normal biopsy group (P>0.05).
Conclusions
Noninvasive diagnosis of BKVN and prognostication of renal allograft function following BKVN diagnosis are feasible by measurement of transcripts for BKV-VP1, granzyme B, and PI-9 in urine.
INTRODUCTION
Polyomavirus BK-associated nephropathy (BKVN) develops in 1% to 10% of renal transplant recipients (1, 2) and may result in progressive loss of renal allograft function in 16-67% of cases (3-5).
The diagnosis of BKVN is made by demonstrating polyomavirus BK or SV-40 antigen in renal allograft tissue obtained from biopsy specimens (6-8). Several non invasive assays of blood and urine have been developed for the noninvasive diagnosis of BKVN although none have been validated prospectively using a cutpoint from an earlier study and using independent study cohorts (9-14).
A major goal of this study was to develop an independently validated biomarker for the noninvasive diagnosis of BKVN. Towards this goal, we investigated whether our previously reported cutoff value of 6.5×105 BKV-VP1 mRNA /ng RNA from urinary cells (10) can be validated in an independent cohort of renal allograft recipients. An additional objective was to determine whether mRNA profiles, characterized in urine specimens collected at the time of BKVN diagnosis, identified those at risk for renal allograft functional decline following diagnosis. Currently, renal allograft histopathological features such as renal tubular changes, interstitial fibrosis and inflammation are utilized to prognosticate allograft function after BKVN diagnosis (15-17). Previous studies from our laboratory and others have demonstrated that BKV replication is associated with heightened expression of urinary mRNA encoding inflammatory mediators, and that urinary cell mRNA profiles can accurately assess allograft status. Herein we test the hypothesis that this approach will permit accurate diagnosis and prognostication of subsequent allograft function in renal transplant recipients with BKVN. (18), (19).
METHODS
Study populations
The validation study used a cross-sectional study design and included 89 renal allograft recipients who underwent either an indication (for-cause) allograft biopsy (N=45) or a protocol biopsy (N=44) at our institution during 2001-2007. The inclusion criteria were: (1) enrollment in our IRB protocol entitled “Use of PCR to Evaluate Renal Allograft Status”; (2) collection of urine specimen at the time of allograft biopsy; and (3) availability of renal allograft biopsy tissue for SV40 immunostaining. The exclusion criteria were: (1) any BKVN subject used in our earlier study to calculate the BKV VP1 mRNA cutpoint for the noninvasive diagnosis of BKVN (10); (2) patients with inadequate yield of urinary m RNA defined as urinary cell 18S rRNA copies <1×109 copies/per one microgram of total RNA. The 89 subjects included 12 renal allograft recipients with BKVN (allograft biopsy positive for SV40 immunostain) and 77 recipients without BKVN (allograft biopsy negative for SV40 immunostain).
Among the 89 subjects included in the validation study, the 12 with BKVN were 52±10 years old (mean±SD); 3 females and 9 males; 6 deceased donor (DD) grafts and 6 living donor (LD) grafts; mean (±SD) serum creatinine at the time of biopsy was 2.41 ±0.73 mg per deciliter (mg/dl). The non-BKVN group included 77 subjects and was comprised of 44 recipients with stable allograft function and normal protocol biopsy (age 46 ±12 years; 23 females and 21 males; 16 DD grafts and 28 LD grafts; serum creatinine at the time of biopsy: 1.45 ±0.44 mg/dl), and 33 recipients with graft dysfunction and abnormal renal allograft biopsy findings (age 44 ±15 years; 13 females and 20 males; 15 DD grafts and 18 LD grafts; serum creatinine at the time of biopsy: 3.42 ±2.18 mg/dl).
A cohort study design was used for the identification of biomarkers prognostic of renal allograft function following BKVN diagnosis. Eighteen subjects with biopsy confirmed BKVN (allograft biopsy specimen immunostain positive for SV40) who were enrolled in our IRB protocol entitled “Use of PCR to Evaluate Renal Allograft Status between 1999 and 2007 and in whom urine specimens were collected at the time of allograft biopsy prior to changes in management were included. Patients were excluded if: (1) urine specimens yielded inadequate RNA as defined by urinary cell 18S rRNA copies <1×109 copies per one microgram of total RNA; or (2) if urine specimens were collected after changes in immunosuppressive therapy were made following the diagnosis of BKVN. Among the 18 patients included in this study, 11 were also included in the validation study and 7 were patients included in our earlier study of diagnostic criteria of BKVM (10). In the current study, the diagnosis of BKVN was made in all subjects by renal biopsy performed for evaluation of allograft dysfunction (‘for-cause biopsy). The demographics of the prognostic study cohort were: age 51±10 years; 5 women and 13 men; 11 DD grafts and 7 LD grafts; serum creatinine at the time of biopsy: 2.4±0.7 mg/dl.
Among the 18 BKVN patients, 8 had an increase in the level of serum creatinine of ≥0.5mg/dl during the one-year following BKVN diagnosis and were classified as the decline in function group (BKVN-DF, N=8) and the remaining 10 BKVN patients without such an increase were classified as the stable function group (BKVN-SF, N=10). The percentage change in serum creatinine level from the time of BKVN diagnosis to one-year following diagnosis was used to examine the association with urinary cell mRNA levels measured at the time of allograft biopsy. For this analysis, 4 of the18 patients with who had lost renal function within one-year of diagnosis and started on dialysis therapy were assigned a value of 10mg/dl for serum creatinine
Renal Allograft Biopsy Evaluation
Percutaneous core needle biopsies of renal allografts were classified by a single renal pathologist (S.V.S) blinded to the results of urinary cell transcript levels. Biopsy specimens were fixed in 4% paraformaldehyde and embedded in paraffin. Tissue sections were stained with hematoxylin eosin, periodic acid schiff and masson trichrome for light microscopic evaluation. Affinity purified and agarose conjugated IgG2A mouse monoclonal antibody (PAb416, Cat. No. DPO2A, Calbiochem, USA) recognizing the 94 kDa SV40 large T antigen was used to stain the renal allograft biopsies. An allograft nephrectomy specimen with known polyomavirus infection was used as the positive control.
The mean (±SD) number of glomeruli in the biopsy specimens was 9 ± 5 and the renal allograft biopsy specimens were classified using the Banff classification schema for active and chronic tubulo-interstitial parameters (20, 21). Polyomavirus associated nephropathy (PVAN) staging (A, B, C) was based on the criteria developed by Drachenberg et al (15).
Collection of Urine Specimens for Measurement of BKV-VP-1 mRNA Levels and for mRNA Profiling
Urine specimens (about 50ml) were collected at the time of for-cause or protocol biopsies. A single urine specimen was collected from each patient. In the prognostication study, urine specimen was also collected at the time of for-cause biopsy and prior to any alterations in therapy related to BKVN diagnosis.
Total RNA Isolation and Reverse Transcription to cDNA
Urine specimens were centrifuged and total RNA was extracted from the urine cell pellet using RNeasy Minikit (Qiagen), and reverse transcribed to cDNA as described (10).
Real -Time Quantitative PCR Assay for the Measurement of BKV-VP1 mRNA Levels
Absolute quantification of BKV-VP1 mRNA copy numbers was accomplished using the BKV amplicon based standard curve in the real-time quantitative PCR assay (10). BKV specific amplification and detection by the primers and the TaqMan probe used in this study was examined using BKV DNA from stain MM (pBKV 35-1, ATCC number 45026, Rockville MD USA) or JCV DNA (strain 803A, ATCC VRMC-24) as the templates in the real-time quantitative PCR assays. We also determined intra-assay variability and inter-assay variability using BKV DNA, over 7 log concentrations, as the template in the real-time quantitative PCR assay(22).
Pre-amplification Enhanced Real-Time Quantitative PCR Assays for the Measurement of Urinary Cell mRNAs
We have modified the real-time quantitative PCR assays by incorporating a pre-amplification step; this modification allows the measurement of multiple mRNAs (about 50 mRNAs) using as little as 3μL cDNA (23). The pre-amplified cDNA was used as the template in the real-time PCR assays, and levels of mRNA for T cell CD3 epsilon chain (CD3), B cell CD20, granzyme B, proteinase inhibitor-9 (PI-9), CD103, transforming growth factor beta 1 (TGF-β1) and interferon gamma (IFN-γ) were measured using gene specific primer pairs and the TaqMan probe. We measured urinary cell 18S rRNA levels as the reference gene. All PCR assays were performed using 2.5ul of cDNA as the template in a PCR reaction volume of 25ul. Transcript levels were measured using an ABI Prism 7700/7500 system and absolute quantification of transcript levels was accomplished using the BKV amplicon (10) to develop the standard curve (24).
Statistical Analysis
The diagnostic accuracy of urinary cell BKV-VP1 mRNA levels and the prognostic value of urinary cell mRNA profiles were analyzed using the statistical software packages, STATA version 9.2 and GraphPad Prism version 4.0. To ascertain the diagnostic characteristics of a test, we computed sensitivity, specificity and positive and negative predictive values. For two group comparisons, categorical variables were compared using Fisher’s Exact Test, and continuous variables using Mann Whitney Test. Jonckheere-Terpstra Exact Test was used for trend of categorical variables. For more than two group comparisons, Kruskal-Wallis test was used and Dunn’s multiple comparison test was used for accounting for multiple comparisons. mRNA levels deviated significantly from the normal distribution, and the extent of the deviation was substantially reduced through the use of log transformation. We used natural log-transformed mRNA levels in all analyses. Spearman correlation coefficient and partial correlation were computed between natural log-transformed urinary cell mRNA levels and the percentage change in serum creatinine levels with and without controlling for baseline creatinine values. Two sided tests were used for all statistical inferences.
RESULTS
Validation Study: BKV Specific Amplification in the Real-Time Quantitative PCR Assay
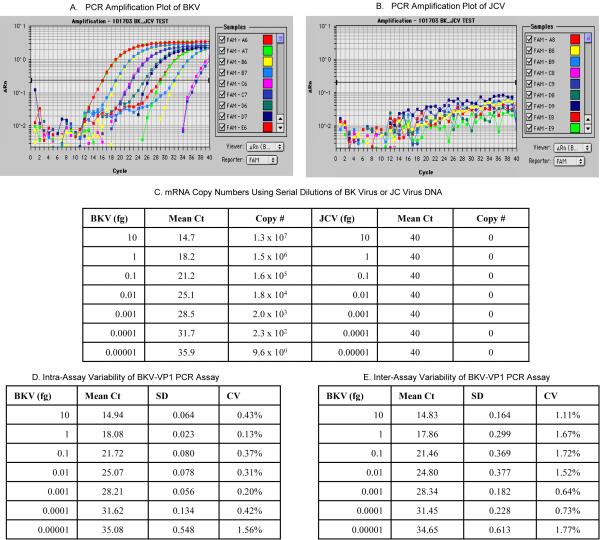
BKV and JCV are evolutionally related and belong to the same polyomavirus group (25). Moreover, BKV and JCV have been identified in the same renal allograft tissue and urine (26, 27). We investigated the specificity of the primers and the TaqMan probe used in this validation study by using BKV DNA or JCV DNA as the template in the real time quantitative PCR assay. Figure1 illustrating the efficient PCR amplification of BKV DNA (Fig.1A) and the lack of amplification of JCV DNA (Fig.1B) demonstrate that the primers and the TaqMan probe used in this investigation detects BKV only and not JCV.
Figure 1. BKV DNA Specific Amplification in the Real Time Quantitative PCR Assay.
PCR amplification plots generated with serial dilutions (10 fg to 0.00001fg) of BKV DNA (BK virus strain MM, ATCC number 45026, pBKV [35-1], Panel A) or JVC DNA (JC virus strain 803A, ATCC number VRMC- 24, Panel B) as the template and using the primers and TaqMan probe designed in our laboratory. The absolute copy numbers calculated using the BKV amplicon are provided in Panel C.
The intra-assay variability is shown in Panel D and the inter-assay variability in Panel E. The mean Ct value, SD and CV of 12 replicates of each concentration within the same PCR run were used to calculate the intra-assay variability. Inter-assay variability was calculated from 2 independent PCR runs of seven different concentrations of BKV DNA.
Messenger RNA abundance, measured with the use of real-time PCR assays, can be reported in relative terms using comparative CT (cycle threshold) method or as absolute copy numbers using the standard curve method (24, 28). Figure 1C shows absolute BKV-VP1 mRNA copy numbers calculated with the use of a BKV amplicon based standard curve method developed in our laboratory (10). Figure 1 also shows the coefficient of variation (CV) found within assays (Fig. 1D) and across assays (Fig. 1E) following PCR amplification of serial dilutions of BKV DNA.
Validation of Urinary cell BKV-VP-1 mRNA Cutpoint for the Noninvasive Diagnosis of BKVN
We measured BKV VP1 mRNA copy numbers in urine specimens collected from the validation study cohort of 89 renal allograft recipients all of whom had undergone allograft biopsy and the presence or absence of BKVN was established using the immunostaining of allograft biopsies for SV40. The median (range) time from transplantation to allograft biopsy was 11 months (2, 36) in the 12 BKVN patients, 4 months (0.2, 13) in the 44 patients with stable graft function and normal biopsies and 8 months (0.13, 151) in the 33 recipients with graft dysfunction and abnormal renal allograft biopsy findings. There was no significant difference in the time from transplantation to biopsy between the BKVN group and the for-cause biopsy group (P>0.05). The time from transplantation to allograft biopsy was significantly different between the BKVN group and the normal biopsy group (P<0.05, Dunn’s Multiple Comparison test).
Table 1 lists the urinary cell BKV VP1 mRNA absolute copy numbers in all 89 subjects. Urinary cell BKV-VP1 mRNA levels were higher than the previously reported cutoff value of 6.5×105 BKV VP1 mRNA copies/ng RNA in all 12 patients with BKVN. In contrast, only 2 of 77 recipients without BKVN patients had levels higher than the previously reported cutoff value. In this validation study, the previously reported cutoff value of value 6.5×105 BKV-VP1 mRNA /ng RNA was diagnostic of BKVN with a sensitivity of 100% and a specificity of 97% (P<0.0001). The negative predictive value was 100% and the positive predictive value was 86% in our cohort with a BKVN prevalence of 13.5% (12 of 89 patients).
Table 1.
Urinary Cell Levels of BKV-VP1 mRNA
| Study Group | Renal Allograft Recipients (N =89) |
BKV Immunostaina |
Urine BKV VP1 mRNA Copies / ng RNA |
Urine BKV VP1 mRNA > 6.5×105copies / ng RNAb |
|---|---|---|---|---|
| I. BKV Nephropathyc (N=12) |
# 1 | Positive | 1.98 × 108 | |
| # 2 | Positive | 1.69 × 108 | ||
| # 3 | Positive | 1.12 × 108 | ||
| # 4 | Positive | 1.04 × 108 | ||
| # 5 | Positive | 9.40 × 107 | 12/12 (100%) | |
| # 6 | Positive | 8.38 × 107 | ||
| # 7 | Positive | 5.71 × 107 | ||
| # 8 | Positive | 5.41 × 107 | ||
| # 9 | Positive | 5.09 × 107 | ||
| # 10 | Positive | 2.41 × 107 | ||
| # 11 | Positive | 9.87 × 106 | ||
| # 12 | Positive | 5.41 × 106 | ||
| II. Acute Cellular Rejectiond (N=13) |
# 1 | Negative | 6.43 × 103 | |
| # 2 | Negative | 2.29 × 103 | 0/13 (0%) | |
| # 3 to #13 | Negative | No Replication e | ||
| III. Borderline Rejectiond (N=5) |
#1 | Negative | 1.47 × 104 | 0/5 (0%) |
| #2 to | #5 | Negative | No Replication e | |
| IV. IF/TAd (N=8) |
#1 to #8 | Negative | No Replication e | 0/8 (0%) |
| V. Otherd (N=7) |
#1 to #7 | Negative | No Replication e | 0/7 (0%) |
| VI. Normal Allograft Biopsyd (N=44) |
#1 | Negative | 5.61 × 106 | |
| #2 | Negative | 2.88 × 106 | 2/44 (4.5%) | |
| #3 | Negative | 2.16 × 105 | ||
| #4 | Negative | 1.78 × 105 | ||
| #5 | Negative | 1.51 × 105 | ||
| #6 | Negative | 1.10 × 105 | ||
| #7 | Negative | 3.70 × 104 | ||
| #8 | Negative | 2.16 × 103 | ||
| #9 to #44 | Negative | No Replication e |
All biopsies were immunostained for SV40 large T antigen using PAb416 mouse mAb.
ROC curve analysis derived cutpoint that yielded the maximum combined specificity and sensitivity for the noninvasive diagnosis of BKVN (10).
BKVN diagnosis was based on positive immunostaining of renal allograft biopsies for SV40 large T antigen. The BKVN biopsies also showed histological changes consistent with BKVN (15).
The renal allograft biopsies were classified using the Banff Classification schema (20). The normal allograft biopsies were from subjects with stable graft function and who underwent protocol biopsies. In the subjects with normal allograft biopsy findings, the serum creatinine levels had not changed by more than 0.2mg/dL during the two weeks before or after the biopsy procedure.
Less than one BKV-VP1 mRNA copy/pg of total RNA.
Prognostication Study: Identification of Parameters Prognostic of Renal Allograft Function Following BKVN Diagnosis
Table 2 lists the demographics and renal allograft biopsy histology findings in the 8 BKVN subjects with a decline in renal allograft function (DF) in the 12 months following BKVN diagnosis and the 10 with stable graft function (SF) during the same period. BKVN-DF patients were more likely to be male (P=0.04) and had higher serum creatinine levels (P=0.02), measured at the time of biopsy diagnosis of BKVN.
Table 2.
Clinical and Histological Parameters of BKVN Patients
| BKVN-DFa N=8 (%) |
BKVN-SFa N=10 (%) |
P Valueb | |
|---|---|---|---|
| Baseline Parameters | |||
| Age (mean± SD) | 52± 6 | 51 ±13 | 0.85 |
| Gender (% male) | 8 (100%) | 5 (50%) | 0.04 |
| Deceased Donor grafts (%) | 4(50%) | 7 (70%) | 0.63 |
| ATG induction (%) | 3 (38%) | 5 (50%) | 0.66 |
| Steroid Maintenance (%) | 4 (50%) | 6 (60%) | 1.0 |
| Biopsy Creatinine (mean±SD) |
2.79±0.5 | 2.03±0.7 | 0.02 |
| Time from Transplant to Biopsy (Months, mean±SD) |
11.6±10.4 | 11.7±6.5 | 0.51 |
|
Histological Grades &
Scores (mean±SD) | |||
| PVAN Grade | 3 A, | 6 A , | 0.41c |
| (Drachenberg Stage [15]) | 2 B, | 2 B, | |
| 3 C | 2 C | ||
| Cytopathic score | 1.4±0.26 | 1.5 ± 0.27 | 0.75 |
| Tubulitis score | 1.3±0.25 | 1.1±0.32 | 0.73 |
| Infiltrate score | 1.9±0.4 | 1.6±0.4 | 0.64 |
| Tubular atrophy score | 1.8±041 | 1.1±0.23 | 0.17 |
| Interstitial Fibrosis score | 1.25±0.31 | 0.8±0.13 | 0.17 |
| BKVN Treatment | |||
| IVIG Therapy (%) | 4 (50%) | 8 (80%) | 0.32 |
| Leflunomide Therapy (%) | 3 (38%) | 6 (60%) | 0.64 |
The BKVN patients with an increase in the level of serum creatinine of ≥0.5mg/dl during the one-year following BKVN diagnosis were classified as the Decline in Function Group (BKVN-DF) and the BKVN patients without such an increase were classified as the Stable Function Group (BKVN-SF).
Categorical variables were compared using Fisher’s Exact Test; Continuous variables were compared using Mann Whitney Test
PVAN grades were compared using Jonckheere-Terpstra Exact test for Trend.
PVAN grade A was more frequent in BKVN-SF group compared to BKVN-DF group, and the histological scores for cytopathic changes, tubulitis, infiltration, tubular atrophy and interstitial fibrosis were all numerically lower in BKVN-SF group compared to BKVN-DF group; none of these differences however were statistically significant (Table 2).
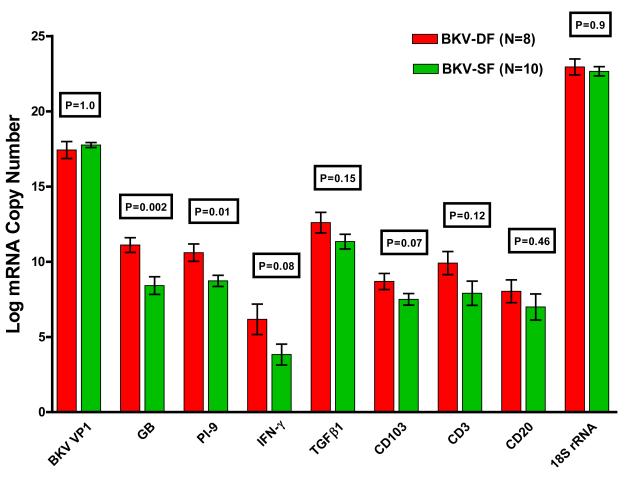
Figure 2 illustrates urinary cell levels of mRNAs in the BKVN-DF group and the BKVN-SF group. The log-transformed mean (±SE) BKV-VP1 copy number was 17.4± 0.6 /ng RNA in the BKVN-DF group, and 17.7±0.2/ng RNA in the BKVN-SF group (P=1.0). Urinary cell levels for granzyme B mRNA and PI-9 mRNA were significantly higher in the BKVN-DF group compared to the BKVN-SF group (Fig. 2). The mean (±SE) granzyme B mRNA copy number was 11.1±0.5/ug RNA in the BKVN-DF group, and 8.4± 0.6 /ug in the BKVN-SF group (P=0.002), and the PI-9 mRNA copy number was 10.6±0.6 /ug RNA in the BKVN-DF group, and 8.7± 0.4 /ug in the BKVN-SF group (P=0.01). The percentage of patients with a decline in allograft function was 0% in the lowest tertile of granzyme B mRNA copy number (log-transformed granzyme B mRNA copy number <9 /ug RNA), 50% in middle tertile (9 to 10.5), and 83% in the highest tertile (>10.5) (P=0.01). The percentage of patients with allograft functional decline was 17% in lowest tertile of PI-9 mRNA copy number (log-transformed PI-9 mRNA copy number <8.6/ug RNA), 33% in the middle tertile (8.6-10.0), and 83% in the highest tertile (>10.0) (P=0.05).
Figure 2. Levels of mRNA in Urinary Cells from Patients with BKVN.
Bar graphs show the log-transformed mean (±SE) of mRNA copies in urine specimens from 8 renal allograft recipient with a decline in renal allograft function following BKVN diagnosis (BKVN-DF) and 10 with stable graft function following BKVN diagnosis (BKVN-SF). Total RNA was isolated from 18 urine specimens collected at the time of allograft biopsy from 18 renal allograft recipients with BKVN and absolute copy numbers of mRNAs were measured using the BKV amplicon in the real time quantitative PCR assays (10). The levels of mRNA for granzyme B and PI-9 were significantly higher in urinary cells from the BKVN-DF subjects compared to the levels in urinary cells from BKVN-SF subjects. The levels of mRNA for BKV VP1, IFN-γ, TGF-β1, CD103, CD3, CD20 and 18S rRNA were not significantly different between the two groups (P>0.05). Two-tailed P-values are based on Mann-Whitney test.
Urinary cell levels of mRNA for IFN-γ (P=0.08), TGF-β1 (P=0.15), CD103 (P=0.07), CD3 (P=0.12), and CD20 (P=0.46) were also higher in the BKVN-DF group compared to the BKVN-SF group (Fig. 2). Tertiles of mRNA for IFN- γ (P=0.25), TGFβ1 (P=0.21), CD103 (P=0.21), CD3 (P=0.21), or CD20 (P=0.41) were not associated with the risk of functional decline post BKVN diagnosis.
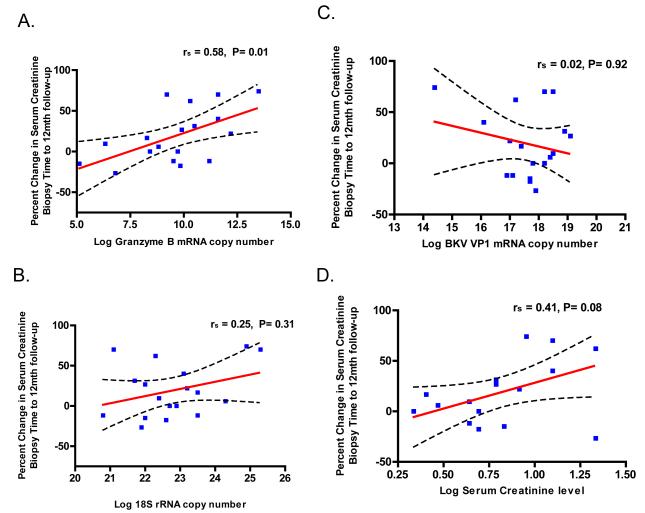
We also examined the linear relationship between urinary cell mRNA levels and the changes in renal allograft function following BKVN diagnosis. Urinary cell levels of granzyme B mRNA (Spearman correlation coefficient rs = 0.58, P=0.01, Fig. 3A) and PI-9 mRNA (rs = 0.48, P=0.04, not shown), both measured at the time of BKVN diagnosis, were correlated with the change in serum creatinine level from the time of allograft biopsy to 12 months post BKVN diagnosis. Levels of 18S rRNA (rs = 0.25, P=0.31, Fig. 3B), BKV-VP1 mRNA (rs =.02, P=0.93, Fig. 3C), IFN-γ (rs =0.19, P=0.52), TGF-β1 (rs =0.32, P=0.2), CD3 (rs=0.19, P=0.44), or CD20 (rs=0.17, P=0.49) were not significantly correlated with the changes in renal allograft function. Serum creatinine levels, measured at the time of BKVN diagnosis, were marginally correlated with the change in serum creatinine levels (rs = 0.41, P=0.08, Fig. 3D). Controlling for the baseline creatinine value did not change correlation coefficients and their P-values materially.
Figure 3. Correlation between the Change in Renal Allograft Function and Transcript Levels or Serum Creatinine.
The relationships between the percent changes in serum creatinine levels 12 months following BKVN diagnosis and the levels of mRNAs in urine specimens or serum creatinine levels, measured at the time of allograft biopsy, are shown, along with Spearman’s rank-order correlation. The percent change in serum creatinine levels were calculated using the formula: 100 × (12 month post biopsy creatinine – creatinine at the time of biopsy / 12 month post biopsy creatinine). A significant positive relationship between the percent change in serum creatinine levels and urinary cell levels of granzyme B mRNA (Panel A). The relationship between the percent change in serum creatinine and levels of mRNA for 18S rRNA (Panel B), BKV-VP1 mRNA (Panel C) and levels of serum creatinine at the time of biopsy (Panel D) are also shown. The graphs display the best-fit line (solid) and the 95% confidence band for the line, indicating the boundaries of all possible straight lines.
Comparison of Urinary Cell mRNA Profiles of BKVN-DF or BKVN-SF Group with Urinary Cell mRNA Profiles of Patients with Acute Rejection or Normal Biopsies
One hypothesis for the differential graft functional outcome (declining vs. stable) following BKVN diagnosis is that BKVN-DF is associated with increased intragraft inflammation, comparable to what is observed in patients with acute rejection. On the other hand, we hypothesize that the degree of intragraft inflammation in the BKVN –SF group is significantly less than that in patients with acute rejection.
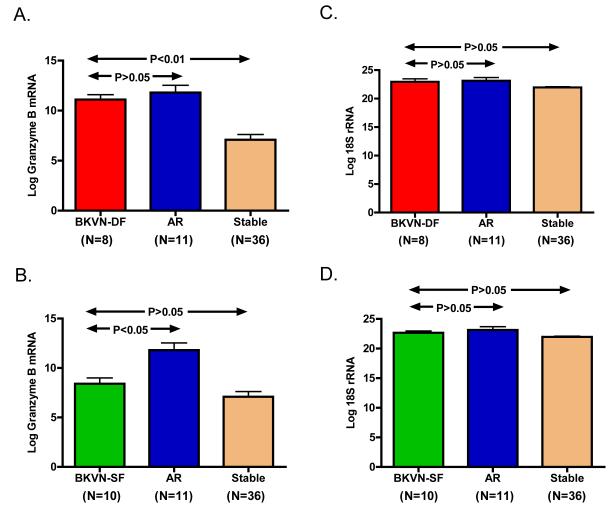
We have reported that urinary cell levels of mRNAs such as granzyme B are diagnostic of intragraft inflammation and AR. Our comparison of urinary cell levels of granzyme B mRNA showed that the levels in the BKVN-DF group (11.1±0.5, n=8) were indeed similar to the levels in the AR group (11.8±0.7, n=11) (P>0.05) and higher compared to the normal biopsy group (7.1±0.5, n=36) P<0.001, Dunn’s Multiple Comparison Test) (Fig. 4A). The urinary cell levels of granzyme B in BKVN-SF group (8.4±0.6, n=10) were significantly lower compared to the AR group (P<0.01) and not significantly different compared to the levels in the normal biopsy group (P>0.05) Fig. 4B). The levels of the reference gene 18S rRNA were not different across the diagnostic categories (Fig. 4 C & D).
Figure 4. Comparison of Urinary Cell mRNA Profiles of BKVN-DF or BKVN-SF Group with Urinary Cell mRNA Profiles of Patients with Acute Rejection or Normal Biopsies.
Bar graphs show the log-transformed mean (±SE) of mRNA copy numbers for granzyme B (GB) and 18S rRNA per one microgram RNA in urine specimens from the BKVN patients with a decline in graft function (BKV-DF, N=8), BKVN patients with stable graft function (BKVN-SF, N=10), the patients with biopsy confirmed acute rejection (AR, N=11) and the patients with stable graft function and normal protocol biopsies (Normal Biopsy, N=36). Urinary cell levels of granzyme B mRNA in the BKVN-DF group (11.1±0.5) were not different compared to the AR group (11.8±0.7) (P>0.05) and higher compared to the Normal Biopsy group (7.1±0.5; P<0.01) (Fig. 4A). Urinary cell levels of granzyme B in BKVN-SF group (8.4±0.6) were significantly lower compared to the AR group (P<0.01) and similar to the levels in the Normal Biopsy group (P>0.05) (Fig. 4B). The levels of the reference gene 18S rRNA were not different across the diagnostic categories (Fig. 4 C & D). Statistical analysis was performed using the log transformed mRNA copy numbers as the dependent variable in the Kruskal-Wallis Test. Dunn’s Multiple Comparison Test was then used to compare the mRNA levels across the diagnoses. Numbers in parenthesis are the number of patients in each diagnostic category.
DISCUSSION
Biomarker development involves sequential phases that include discovery, validation and qualification. The current study serves as a validation study for our earlier report that BKVN can be predicted with a sensitivity of 93.8% and a specificity of 93.9% using a cutoff value of 6.5×105 BKV VP1 mRNA /ng RNA in urinary cells (10). In this validation study using the same cutoff value, BKVN was predicted with 100% sensitivity and 97% specificity. The higher accuracy may be due to study features such as the presence or absence of BKVN being established in each patient by immunostaining the allograft biopsy for SV40 and not using multiple urine samples from the same patient (as was done in our earlier study).
Initial approaches for the noninvasive diagnosis of BKVN focused on the utility of screening for decoy cells in urine, and upon electron microscopic identification of icosahedral viral particles in urine (29-32). Recently, the presence of polyomavirus aggregates in urine, designated as Haufen, has been reported to be diagnostic of BKVN with a sensitivity of 100% and a specificity of 99% (33). PCR based assays utilizing DNA as the template is perhaps the most commonly used assay in the clinic and data exist that plasma/urine BKV DNA viral loads are diagnostic of BKVN (9, 11-14, 34). DNA is more stable and relatively easier to isolate compared to RNA and thus DNA based assays have this technical advantage over the mRNA based assay validated in this study.
We detected the presence of BKV at the mRNA level rather than the DNA level and we made the urinary measurements using RNA isolated from urinary cells. The rationale for our strategy includes: (a) measurement at the mRNA level ensures detection of actively transcribed BKV(35, 36); and (b) measurement using urinary cells as the source of RNA may offer the means of measuring BKV replication within renal tubular cells. The possibility exists however that non renal tubular cells such as bladder epithelial cells as well as any DNA contamination of our RNA preparation contributed to the absolute copy numbers of BKV VP-1 measured in our study. Also, neither blood BKV VP-1 DNA levels nor other diagnostic indicators such as Haufen were measured in our study. Thus, it is not possible to comment on the relative diagnostic accuracy of our assay with the more established test such as the blood BKV-VP-1 DNA test. It is worth noting however that noninvasive diagnosis of BKVN was accomplished in this study with a very high degree of accuracy and that our previously calculated cutpoint for BKV-VP-1 mRNA level in urine was validated using an independent cohort of renal allograft recipients. To the best of our knowledge, validation using a predefined threshold of BKV DNA viral load in blood or urine has not been reported using an independent study cohort.
It is well recognized that BKVN may be associated with a decline in graft function prior to and following the diagnosis. Vasudev et al reported that rate of graft loss in renal allograft recipients with a history of BKVN is significantly higher compared to those without such a history (37). Lipshutz et al reported that 55% of the SPK recipients with BKVN experienced graft loss (38). In our study, 44% of subjects with BKVN diagnosed by renal biopsy performed for allograft dysfunction experienced subsequent graft functional decline, and 22% graft loss. With respect to prognosticators of graft function subsequent to BKVN diagnosis, histological features were indeed more severe in those with a decline in function following the diagnosis compared to those with stable graft function but the differences were not statistically significant. We anticipated urinary cell BKV-VP1 mRNA copy numbers to be informative of subsequent graft function but found that the copy numbers were not prognostic of subsequent graft function. Among the multiple mRNAs measured, only levels of granzyme B mRNA and PI-9 mRNA were prognostic with higher levels in those who developed graft dysfunction in the 12 months post BKVN diagnosis compared to those who did not. Granzyme B is an integral component cytotoxic machinery of T cells and our findings suggest that BKVN patients with cytotoxic T cell activity and not those without such activity are at risk for progressive functional decline (39). PI-9 is an endogenous antagonist of granzyme B and the association of heightened expression with functional decline is more difficult to explain since high levels of PI-9 in renal tubular cells and endothelial cells should help minimize granzyme B dependent cytotoxicity directed at the renal parenchymal cells (40). One intriguing hypothesis is that high PI-9 expression by cytotoxic T cells (CTL) prevents fratricide among them and thereby enhances CTL activity directed at the parenchymal cells (41). Because PI-9 is also expressed by dendritic cells, the professional APCs, another possibility is that high PI-9 expression contributes to more efficient antigen presentation and vigorous CTL activity.
We have reported that urinary cell mRNA profiles are diagnostic of renal allograft status and that the levels of mRNA encoding cytotoxic protein granzyme B are significantly higher in renal allograft recipients with biopsy confirmed acute rejection compared to recipients with normal allograft biopsy findings [reviewed in (18)]. The new findings from current study that the levels of granzyme B mRNA in BKVN patients with a decline in graft function following the diagnosis are indistinguishable from those observed in acute rejection patients and significantly higher compared to stable patients raise the intriguing possibility that some patients with BKVN may have co-existing acute rejection. Because the antigen specificity of the CTLs expressing granzyme B is not established in our study, it is unresolved at this stage whether the increased levels of granzyme B is from alloreactive CTLs, CTLs directed at BKV or both.
Our study has implications relevant for management of renal allograft recipients with allograft dysfunction. We have demonstrated that noninvasive diagnosis of BKVN is feasible by measurement of BKV-VP1 mRNA in urine. Further, we show that urinary cell mRNA levels of granzyme B and PI-9, measured at the time of BKVN diagnosis, may identify patients at risk for continued renal functional decline subsequent to the diagnosis. This interpretation however needs to be validated using an independent cohort of BKVN patients. The third and perhaps most provocative connotation is that immunosuppression reduction following BKVN diagnosis should be undertaken only in those without evidence of ongoing inflammation whereas in those with ongoing inflammation the presence or absence of acute rejection should first be established prior to reduction of immunosuppression. Previous studies suggest several candidate markers present in renal allograft biopsy tissue that may be helpful in distinguishing BKVN from acute rejection, including percentage of perforin positive cells (42), and quantitative differences in the levels of transcripts for CD8, IFN-gamma, perforin, and genes implicated in epithelial-mesenchymal transition and fibrosis (43).
In summary, we have validated the diagnostic accuracy of urinary cell BKV-VP1 mRNA levels for the noninvasive diagnosis BKVN diagnosis. Our findings advance the hypothesis that urinary cell levels of mRNA for granzyme B and PI-9 may identify those at risk renal functional decline following BKVN diagnosis. Our observation that the BKVN patients with a decline in allograft function but not those with stable graft function following BKVN diagnosis share urinary cell gene expression patterns with those with acute rejection raise the possibility that BKVN and acute rejection may co-exist in a subset of patients, and emphasize the urgent need for individualizing immunosuppressive therapy in those diagnosed with BKVN.
ACKNOWLEDGEMENT
The research project was supported by the awards R37AI51652 and R01AI60706 from the National Institute of Allergy and Infectious Disease, National Institutes of Health to M. Suthanthiran. This investigation was also supported by grant ULI RR024996 of the Clinical and Translational Science Center at Weill Cornell Medical College. The research reported in this manuscript is in partial fulfillment of Dr. Darshana Dadhania’s K30 Masters of Science in Clinical Investigation.
Footnotes
This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
REFERENCES
- 1.Hirsch HH, Brennan DC, Drachenberg CB, et al. Polyomavirus-associated nephropathy in renal transplantation: interdisciplinary analyses and recommendations. Transplantation. 2005;79(10):1277. doi: 10.1097/01.tp.0000156165.83160.09. [DOI] [PubMed] [Google Scholar]
- 2.Ramos E, Drachenberg CB, Wali R, Hirsch HH. The decade of polyomavirus BK-associated nephropathy: state of affairs. Transplantation. 2009;87(5):621. doi: 10.1097/TP.0b013e318197c17d. [DOI] [PubMed] [Google Scholar]
- 3.Randhawa PS, Finkelstein S, Scantlebury V, et al. Human polyoma virus-associated interstitial nephritis in the allograft kidney. Transplantation. 1999;67(1):103. doi: 10.1097/00007890-199901150-00018. [DOI] [PubMed] [Google Scholar]
- 4.Nickeleit V, Hirsch HH, Binet IF, et al. Polyomavirus infection of renal allograft recipients: from latent infection to manifest disease. J Am Soc Nephrol. 1999;10(5):1080. doi: 10.1681/ASN.V1051080. [DOI] [PubMed] [Google Scholar]
- 5.Ramos E, Drachenberg CB, Papadimitriou JC, et al. Clinical course of polyoma virus nephropathy in 67 renal transplant patients. J Am Soc Nephrol. 2002;13(8):2145. doi: 10.1097/01.asn.0000023435.07320.81. [DOI] [PubMed] [Google Scholar]
- 6.Purighalla R, Shapiro R, McCauley J, Randhawa P. BK virus infection in a kidney allograft diagnosed by needle biopsy. Am J Kidney Dis. 1995;26(4):671. doi: 10.1016/0272-6386(95)90608-8. [DOI] [PubMed] [Google Scholar]
- 7.Hariharan S. BK virus nephritis after renal transplantation. Kidney Int. 2006;69(4):655. doi: 10.1038/sj.ki.5000040. [DOI] [PubMed] [Google Scholar]
- 8.Drachenberg CB, Papadimitriou JC, Ramos E. Histologic versus molecular diagnosis of BK polyomavirus-associated nephropathy: a shifting paradigm? Clin J Am Soc Nephrol. 2006;1(3):374. doi: 10.2215/CJN.02021205. [DOI] [PubMed] [Google Scholar]
- 9.Nickeleit V, Klimkait T, Binet IF, et al. Testing for polyomavirus type BK DNA in plasma to identify renal-allograft recipients with viral nephropathy. N Engl J Med. 2000;342(18):1309. doi: 10.1056/NEJM200005043421802. [DOI] [PubMed] [Google Scholar]
- 10.Ding R, Medeiros M, Dadhania D, et al. Noninvasive diagnosis of BK virus nephritis by measurement of messenger RNA for BK virus VP1 in urine. Transplantation. 2002;74(7):987. doi: 10.1097/00007890-200210150-00016. [DOI] [PubMed] [Google Scholar]
- 11.Randhawa P, Ho A, Shapiro R, et al. Correlates of quantitative measurement of BK polyomavirus (BKV) DNA with clinical course of BKV infection in renal transplant patients. J Clin Microbiol. 2004;42(3):1176. doi: 10.1128/JCM.42.3.1176-1180.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Viscount HB, Eid AJ, Espy MJ, et al. Polyomavirus polymerase chain reaction as a surrogate marker of polyomavirus-associated nephropathy. Transplantation. 2007;84(3):340. doi: 10.1097/01.tp.0000275205.41078.51. [DOI] [PubMed] [Google Scholar]
- 13.Cross NB, Webster AC, O’Connell PJ, Jeoffreys N, Dwyer DE, Craig JC. Diagnostic accuracy of blood qualitative nucleic acid testing for polyomavirus-associated nephropathy in kidney recipients. Nephrology (Carlton) 2009;14(3):350. doi: 10.1111/j.1440-1797.2009.01118.x. [DOI] [PubMed] [Google Scholar]
- 14.Hirsch HH, Knowles W, Dickenmann M, et al. Prospective study of polyomavirus type BK replication and nephropathy in renal-transplant recipients. N Engl J Med. 2002;347(7):488. doi: 10.1056/NEJMoa020439. [DOI] [PubMed] [Google Scholar]
- 15.Drachenberg CB, Papadimitriou JC, Hirsch HH, et al. Histological patterns of polyomavirus nephropathy: correlation with graft outcome and viral load. Am J Transplant. 2004;4(12):2082. doi: 10.1046/j.1600-6143.2004.00603.x. [DOI] [PubMed] [Google Scholar]
- 16.Gaber LW, Egidi MF, Stratta RJ, Lo A, Moore LW, Gaber AO. Clinical utility of histological features of polyomavirus allograft nephropathy. Transplantation. 2006;82(2):196. doi: 10.1097/01.tp.0000226176.87700.a4. [DOI] [PubMed] [Google Scholar]
- 17.Wadei HM, Rule AD, Lewin M, et al. Kidney transplant function and histological clearance of virus following diagnosis of polyomavirus-associated nephropathy (PVAN) Am J Transplant. 2006;6(5 Pt 1):1025. doi: 10.1111/j.1600-6143.2006.01296.x. [DOI] [PubMed] [Google Scholar]
- 18.Anglicheau D, Suthanthiran M. Noninvasive prediction of organ graft rejection and outcome using gene expression patterns. Transplantation. 2008;86(2):192. doi: 10.1097/TP.0b013e31817eef7b. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Dadhania D, Snopkowski C, Ding R, et al. Epidemiology of BK virus in renal allograft recipients: independent risk factors for BK virus replication. Transplantation. 2008;86(4):521. doi: 10.1097/TP.0b013e31817c6447. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Racusen LC, Solez K, Colvin RB, et al. The Banff 97 working classification of renal allograft pathology. Kidney Int. 1999;55(2):713. doi: 10.1046/j.1523-1755.1999.00299.x. [DOI] [PubMed] [Google Scholar]
- 21.Racusen LC, Halloran PF, Solez K. Banff 2003 meeting report: new diagnostic insights and standards. Am J Transplant. 2004;4(10):1562. doi: 10.1111/j.1600-6143.2004.00585.x. [DOI] [PubMed] [Google Scholar]
- 22.Segura MM, Monfar M, Puig M, Mennechet F, Ibanes S, Chillon M. A real-time PCR assay for quantification of canine adenoviral vectors. J Virol Methods. 163(1):129. doi: 10.1016/j.jviromet.2009.09.010. [DOI] [PubMed] [Google Scholar]
- 23.Muthukumar T, Dadhania D, Ding R, et al. Messenger RNA for FOXP3 in the urine of renal-allograft recipients. N Engl J Med. 2005;353(22):2342. doi: 10.1056/NEJMoa051907. [DOI] [PubMed] [Google Scholar]
- 24.Martell M, Gomez J, Esteban JI, et al. High-throughput real-time reverse transcription-PCR quantitation of hepatitis C virus RNA. J Clin Microbiol. 1999;37(2):327. doi: 10.1128/jcm.37.2.327-332.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Cubitt CL. Molecular genetics of the BK virus. Adv Exp Med Biol. 2006;577:85. doi: 10.1007/0-387-32957-9_6. [DOI] [PubMed] [Google Scholar]
- 26.Randhawa P, Shapiro R, Vats A. Quantitation of DNA of polyomaviruses BK and JC in human kidneys. J Infect Dis. 2005;192(3):504. doi: 10.1086/431522. [DOI] [PubMed] [Google Scholar]
- 27.Drachenberg CB, Hirsch HH, Papadimitriou JC, et al. Polyomavirus BK versus JC replication and nephropathy in renal transplant recipients: a prospective evaluation. Transplantation. 2007;84(3):323. doi: 10.1097/01.tp.0000269706.59977.a5. [DOI] [PubMed] [Google Scholar]
- 28.Livak KJ, Schmittgen TD. Analysis of relative gene expression data using real-time quantitative PCR and the 2(-Delta Delta C(T)) Method. Methods. 2001;25(4):402. doi: 10.1006/meth.2001.1262. [DOI] [PubMed] [Google Scholar]
- 29.Gardner SD, MacKenzie EF, Smith C, Porter AA. Prospective study of the human polyomaviruses BK and JC and cytomegalovirus in renal transplant recipients. J Clin Pathol. 1984;37(5):578. doi: 10.1136/jcp.37.5.578. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Binet I, Nickeleit V, Hirsch HH, et al. Polyomavirus disease under new immunosuppressive drugs: a cause of renal graft dysfunction and graft loss. Transplantation. 1999;67(6):918. doi: 10.1097/00007890-199903270-00022. [DOI] [PubMed] [Google Scholar]
- 31.Howell DN, Smith SR, Butterly DW, et al. Diagnosis and management of BK polyomavirus interstitial nephritis in renal transplant recipients. Transplantation. 1999;68(9):1279. doi: 10.1097/00007890-199911150-00011. [DOI] [PubMed] [Google Scholar]
- 32.Singh HK, Bubendorf L, Mihatsch MJ, Drachenberg CB, Nickeleit V. Urine cytology findings of polyomavirus infections. Adv Exp Med Biol. 2006;577:201. doi: 10.1007/0-387-32957-9_15. [DOI] [PubMed] [Google Scholar]
- 33.Singh HK, Andreoni KA, Madden V, et al. Presence of urinary Haufen accurately predicts polyomavirus nephropathy. J Am Soc Nephrol. 2009;20(2):416. doi: 10.1681/ASN.2008010117. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Hirsch HH, M. M, T K. Prospective Monitoring of BK Virus Load after Discontinuing Sirolimus Treatment in a Renal Transplant Patient with BK Virus Nephropathy. Journal of Infectious Diseases. 2001;184:1494. doi: 10.1086/324425. [DOI] [PubMed] [Google Scholar]
- 35.Cole CN, Conzen S. Polyomaviridae: the viruses and their replication. In: DM K, PM H, editors. Fields’ Virology. vol II. Lippincott Williams & Wilkins; Philadelphia: 2001. p. 2141. [Google Scholar]
- 36.Imperiale M, Major E. Polyomaviruses. In: David Knipe, Peter Howley., editors. Fields Virology. vol 2. Lippincott-Rave; Philadelphia: 2007. p. 2263. [Google Scholar]
- 37.Vasudev B, Hariharan S, Hussain SA, Zhu YR, Bresnahan BA, Cohen EP. BK virus nephritis: risk factors, timing, and outcome in renal transplant recipients. Kidney Int. 2005;68(4):1834. doi: 10.1111/j.1523-1755.2005.00602.x. [DOI] [PubMed] [Google Scholar]
- 38.Lipshutz GS, Mahanty H, Feng S, et al. BKV in simultaneous pancreas-kidney transplant recipients: a leading cause of renal graft loss in first 2 years post-transplant. Am J Transplant. 2005;5(2):366. doi: 10.1111/j.1600-6143.2004.00685.x. [DOI] [PubMed] [Google Scholar]
- 39.Shresta S, Heusel JW, Macivor DM, Wesselschmidt RL, Russell JH, Ley TJ. Granzyme B plays a critical role in cytotoxic lymphocyte-induced apoptosis. Immunol Rev. 1995;146:211. doi: 10.1111/j.1600-065x.1995.tb00690.x. [DOI] [PubMed] [Google Scholar]
- 40.Cunningham TD, Jiang X, Shapiro DJ. Expression of high levels of human proteinase inhibitor 9 blocks both perforin/granzyme and Fas/Fas ligand-mediated cytotoxicity. Cell Immunol. 2007;245(1):32. doi: 10.1016/j.cellimm.2007.03.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Sun J, Bird CH, Sutton V, et al. A cytosolic granzyme B inhibitor related to the viral apoptotic regulator cytokine response modifier A is present in cytotoxic lymphocytes. J Biol Chem. 1996;271(44):27802. doi: 10.1074/jbc.271.44.27802. [DOI] [PubMed] [Google Scholar]
- 42.Rogers NM, Russ GR, Cooper J, Coates PT. Immunophenotyping of interstitial infiltrate does not distinguish between BK virus nephropathy and acute cellular rejection. Nephrology (Carlton) 2009;14(1):118. doi: 10.1111/j.1440-1797.2008.01050.x. [DOI] [PubMed] [Google Scholar]
- 43.Mannon RB, Hoffmann SC, Kampen RL, et al. Molecular evaluation of BK polyomavirus nephropathy. Am J Transplant. 2005;5(12):2883. doi: 10.1111/j.1600-6143.2005.01096.x. [DOI] [PubMed] [Google Scholar]