Abstract
The accurate prediction of an RNAs three dimensional structure from its “primary structure” will have a tremendous influence on the experimental design and its interpretation, and ultimately our understanding of the many functions of RNA. This paper presents a general coarse-grained (CG) potential for modeling RNA 3-D structures. Each nucleotide is represented by five pseudo atoms, two for the backbone (one for the phosphate and another for the sugar), and three for the base to represent base-stacking interactions. The CG potential has been parameterized from statistical analysis of 688 RNA experimental structures. Molecular dynamic simulations of 15 RNA molecules with the length of 12 to 27 nucleotides have been performed using the CG potential, with performance comparable to that from all-atom simulations. For ~75% of systems tested, simulated annealing led to native-like structures at least once out of multiple repeated runs. Furthermore, with weak distance restraints based on the knowledge of three to five canonical Watson-Crick pairs, all 15 RNAs tested are successfully folded to within 6.5 Å of native structures using the CG potential and simulated annealing. The results reveal that with a limited secondary structure model, the current CG potential can reliably predict the 3-D structures for small RNA molecules. We also explored an all-atom force field to construct atomic structures from the CG simulations.
Keywords: Coarse-Grained Model, RNA structure, 3-D structure prediction, Molecular Dynamics
Introduction
The importance of RNA has been appreciated since the central dogma was proposed in 1958.1,2 Three RNA molecules, messenger RNA (mRNA), transfer RNA (tRNA), and ribosomal RNA (rRNA) are associated with the cell’s transcription of its DNA into RNA and then translated into proteins.3 While experiments as early as 1971 suggested that RNA is involved in catalysis during protein synthesis,4–6 experiments starting in 1982 confirmed that RNA is directly involved in catalysis, in many different RNA systems with difference chemical reactions.4,7–15 These RNAs form complex three-dimensional structures.16–33
Within the past few years, a large increase in the number of RNAs that form higher-structure, associated with numerous functions in the cell is the foundation for a major paradigm shift in the molecular biology of the cell; RNAs that do not code for proteins are directly associated with the regulation and overall function of the cell 34–44, including different cancers.45–47 Since more than 90% of the human genome is transcribed into RNAs that do not code for proteins,48,49 and the function of an increasing amount of this RNA is now being determined, the prediction of an RNAs higher-order structure and its dynamics will provide great insight into the RNAs contribution to the structure and function of a cell. Towards that end, computational approaches such as molecular modeling have made significant contribution to the understanding of three-dimensional structures and chemical principles of RNA.50–60 The most successful approaches for protein structure prediction so far have been based on comparative analysis or reduced models derived from known structures.61–63
In recent years, increased effort has been devoted to RNA structure prediction as more and more RNA structures have been determined experimentally. A range of models have been developed for nucleic acids, from fully atomistic models to reduced representations.64–67 For example, a knowledge-based atomic energy function has been introduced to predict RNA tertiary structures in the FARNA package.68 Nucleotide cyclic motifs are used in MC-Fold and MC-Sym model to build RNA structure from sequence data.69 These two models seem successful in predicting the tertiary structure of small RNA molecules. In addition, physics-based atomic force fields such as AMBER70–73 and CHARMM74–76 describe the dynamic atomic interaction following traditional molecular mechanics, with parameters derived by fitting to ab initio quantum mechanics calculations and experimental data. It is now feasible with supercomputers to simulate dynamic biological systems as large as an entire virus in atomic detail.77 However, typical applications of the atomistic force fields are usually limited to small oligomers of nucleic acids or routine simulation times on the order of a few nanoseconds.78 On the other hand, CG methods reduce the number of particles and eliminate high-frequency motions in the system. A CG model enlarges the time step in molecular dynamics simulations while also enhancing intrinsically faster dynamics.79–81 Several CG approaches, either knowledge or physics based, have been utilized to study the structures of nucleic acids.68,69,78,82–106
There has been a history of modeling DNA at mesoscale.65,78,82–84,86–89,92–97,99,105 Recently studies such as Knotts IV and co-workers successfully predicted several important DNA behaviors, like salt-dependent melting, bubble formation and rehybridization, with a CG model that uses three interaction sites for phosphate, sugar, and base, respectively.78 For RNA, Malhotra et al. first introduced a reduced potential to refine 3-D structure of low-resolution ribosomal RNAs.52,59 Zhang et al. combined a highly reduced coarse-grained model and a Monte Carlo method to simulate the distribution of viral RNA inside the capsid of cowpea chlorotic mottle virus.100 In these works, each nucleotide was modeled as one bead to reduce the complexity of large RNAs. Cao et al. developed a reduced chain representation model to predict RNA folding thermodynamics based on a statistical mechanical theory.101 The RNA molecules were represented by their backbone and two beads (phosphate and sugar) were used for each nucleotide. Later, Jonikas et al. integrated coarse-grained model with knowledge-based potentials to generate plausible three-dimensional structures.107 Despite the development, automated programs for predicting RNA 3-D structures and their dynamic properties are required to increase their accuracy and robustness.108 In addition, the coarse-grained RNA potentials are mostly aimed at structural refinement rather than predicting dynamics properties.
Aside from the energy or scoring function, the configuration searching and sampling methods are also important components of the structure prediction algorithms. The MC-Fold/MC-Sym and FARNA programs begin by generating structures from small RNA fragments with a few residues. Then, Las Vegas or Monte Carlo sampling is performed to generate a sampling of possible tertiary structures of the entire RNA.68,69 NAST utilizes MD simulations, combined with secondary structure information, to generate 3-D structure candidates for the RNA molecules.107 A coarse-grained (C3’ based one-bead CG model) energy function derived from the structural statistics is applied in the MD simulation. The ranking of structural clusters can be based on the NAST energy function or experimental data (solvent accessibility data from the hydroxyl radical foot printing). Herein we present an “intermediate” coarse-grained potential for modeling RNA 3-D structure using molecular dynamics. Previous CG RNA models typically used one52,100,107 or two101 particles for each nucleotide. To optimize the efficiency and accuracy, we developed a model that represents each nucleotide with five pseudo atoms; two of these represent the backbone–one for the sugar and the other for the phosphate, while three pseudo atoms represent the stacking and base pairing for each base. The analytical potential energy functional forms, parameterization with 3-D structural statistics obtained from experimental structures and initial validation using molecular dynamics simulations of selected RNAs is discussed.
Experimental Methods
Data Collection and Preparation
The CG potential was parameterized using statistics collected from available three dimensional structures of RNA molecules (including both x-ray diffraction structures and nuclear magnetic resonance structures). The RNA structure files were downloaded from The Protein Data Bank (http://www.pdb.org/), Nucleic Acid Database (http://ndbserver.rutgers.edu/index.html), RNA Comparative Analysis Database (rCAD, http://rcat.codeplex.com/, manuscript submitted), and the Comparative RNA Web (CRW) Site109 (http://www.rna.ccbb.utexas.edu/). Only 668 structure files that contained more than 5 base pairs and have the resolution records were analyzed for the statistical calculation. All of the coordinates obtained from nuclear magnetic resonance (NMR) structural files were included in the statistical calculations.
Coarse-Grained RNA Interaction Potential
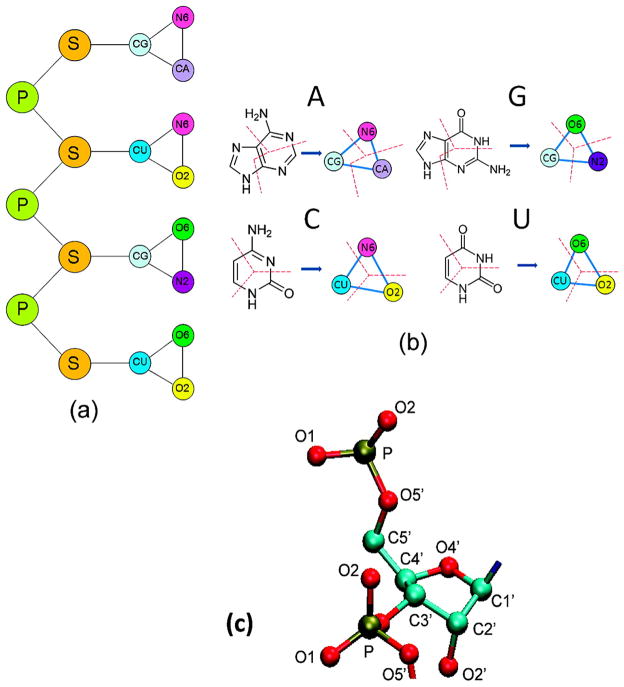
In our CG model, each nucleotide is reduced to five pseudo atoms in RNA (Figure 1). Two of the five pseudo atoms represent phosphate and sugar respectively, which is the minimum requirement to capture the backbone tertiary structures of RNA.110 Each base (A, G, C, and T) is represented by three pseudo atoms, connected by three virtual bonds into a triangle. Compared to earlier models with one particle for each base or each residue,78,107 the use of three pseudo atoms for each base provides us with better ability to capture the stacking and pairing of bases. As the different bases share some common pseudo atoms, nine unique types of pseudo atoms are needed to represent the four canonical RNA component bases in total (Figure 1). The improvement in computational efficiency arises from the reduction of number of particles and larger particle mass that enables greater integration time step in molecular dynamics. The topological and physical properties of the pseudo atoms are listed in Table 1.
Figure 1.
(a) Schematic representation of the CG model for RNA. Phosphate and sugar are represented as one CG particle. The bases A, G, C, and U are represented as three CG particles for each. (b) The components of each CG base. The base is divided by the red dash line. (c) Schematic representation of all-atom RNA backbone.
TABLE 1.
The Properties of Nine CG Particles
| number | CG particle name | mass (amu) | bond connections |
|---|---|---|---|
| 1 | P | 94.970 | 2 |
| 2 | S | 97.054 | 3 |
| 3 | CG | 53.022 | 3 |
| 4 | N6 | 42.030 | 2 |
| 5 | N2 | 54.030 | 2 |
| 6 | O6 | 43.014 | 2 |
| 7 | O2 | 42.006 | 2 |
| 8 | CU | 26.016 | 3 |
| 9 | CA | 39.015 | 2 |
The corresponding CG potential energy is calculated by:
| (1) |
where Ebonded and Enon-bonded are pair-wise bonded and non-bonded energy terms, each representing the sum of contributions of all pairs in the system. The bonded term is further decomposed into:
| (2) |
where Ebond , Eangle and Edihedral are the bond stretching, angle-bending and dihedral energies, respectively.
In classical molecular mechanics, the non-bonded interaction consists of the van der Waals (VDW) and electrostatic contributions. Since our CG model is derived from the 3-D structural statistics of experimental structures, an effective potential is used to represent the potential of mean force of all the non-bonded interactions, including the excluded volume repulsive, the attractive force and the electrostatic force between non-bonded particles, as well as the solvation forces due to the environment. A Buckingham potential is utilized to describe the effective potential (see equation 9).
| (3) |
For each term, the parameterization was performed based on the Boltzmann inversion of the corresponding atomistic distribution functions obtained from the experimental structures. The Boltzmann inversion method performs a potential inversion from a set of known distributions of structural parameters to extract effective CG potentials. In our RNA CG system, the potentials calculated from the Boltzmann inversion method111,112 need to reproduce the distribution of structural parameters including fourteen different bonds, twenty-five types of angles, twenty-eight dihedral angles, and nineteen intermolecular radial distribution functions extracted from statistical results of all available atomistic RNA structures (please see Table 2 to Table 5 for more details). All the parameter-fitting works were performed with the software of Matlab Curve Fitting Tool.
TABLE 2.
The Bond Stretching Interaction Parameters for the CG Model of RNA Fitted by the Gaussian Function and Obtained From Statistical Structures
| bond | b0 | Kb |
|---|---|---|
| 1 - 2 | 3.85 | 11.12 |
| 2 - 3 | 3.74 | 9.79 |
| 2 - 8 | 3.61 | 10.89 |
| 3 - 4 | 4.29 | 57.70 |
| 3 - 5 | 5.66 | 51.66 |
| 3 - 6 | 4.28 | 44.60 |
| 3 - 9 | 4.33 | 109.19 |
| 4 - 7 | 4.55 | 44.00 |
| 4 - 8 | 3.59 | 124.29 |
| 4 - 9 | 3.53 | 93.79 |
| 5 - 6 | 4.57 | 37.14 |
| 6 - 7 | 4.53 | 57.10 |
| 6 - 8 | 3.55 | 89.85 |
| 7 - 8 | 3.52 | 82.87 |
TABLE 5.
The Optimized CG Parameters for the Dihedral Interaction Term Described by Equation (8)
| torsion | V1 | δ1 | V2 | δ2 | V3 | δ3 |
|---|---|---|---|---|---|---|
| 1’-2-3-4a | 3.354 | 120 | −0.606 | 180 | −0.068 | 120 |
| 1’-2-3-9 | 3.801 | 120 | 0.383 | 180 | −0.287 | 120 |
| 1-2-1'-2' | 1.358 | 0 | 0.944 | 180 | 0.574 | 0 |
| 1-2-3-4 | 2.964 | 15 | −0.099 | 180 | −0.247 | 15 |
| 1'-2-3-5 | 3.603 | 120 | 1.167 | 180 | −0.325 | 120 |
| 1-2-3-5 | 3.768 | 0 | 0.52 | 180 | 0.581 | 0 |
| 1'-2-3-6 | 3.409 | 120 | −0.265 | 180 | −0.226 | 120 |
| 1-2-3-6 | 3.077 | 30 | 0.306 | 180 | 0.246 | 30 |
| 1-2-3-9 | 3.299 | 15 | 0.634 | 180 | −0.204 | 15 |
| 1'-2-8-4 | 3.461 | 120 | −0.617 | 180 | 0.294 | 120 |
| 1-2-8-4 | 3.321 | 30 | 1.121 | 180 | −0.156 | 30 |
| 1'-2-8-6 | 2.737 | 120 | −0.666 | 180 | 0.148 | 120 |
| 1-2-8-6 | 2.51 | 30 | 0.518 | 180 | −0.17 | 30 |
| 1'-2-8-7 | 3.304 | 120 | 1.349 | 180 | −0.342 | 120 |
| 1-2-8-7 | 3.844 | 0 | 0.567 | 180 | 0.534 | 0 |
| 2-1-2'-1' | −1.626 | 135 | −0.113 | 180 | −0.246 | 135 |
| 2'-1'-2-3 | −1.661 | 60 | 0.455 | 180 | 0.311 | 60 |
| 2'-1-2-3 | 1.387 | 120 | 0.898 | 180 | −0.516 | 120 |
| 2'-1'-2-8 | −1.531 | 45 | 0.489 | 180 | 0.686 | 45 |
| 2'-1-2-8 | 1.38 | 135 | 0.908 | 180 | −0.691 | 135 |
| 2-3-4-9 | 7.114 | 150 | −2.4 | 180 | 0.516 | 150 |
| 2-3-5-6 | −3.328 | 120 | 0.95 | 180 | 0.101 | 120 |
| 2-3-6-5 | 5.639 | 150 | −2.063 | 180 | −0.009 | 150 |
| 2-3-9-4 | 2.959 | 15 | −1.022 | 180 | 0.666 | 15 |
| 2-8-4-7 | 5.024 | 165 | −1.509 | 180 | −1.807 | 165 |
| 2-8-6-7 | 4.756 | 165 | −1.037 | 180 | −1.455 | 165 |
| 2-8-7-4 | −4.072 | 150 | 0.544 | 180 | −0.144 | 150 |
| 2-8-7-6 | 3.51 | 0 | 0.425 | 180 | 0.457 | 0 |
The prime in the table indicates the atom comes from its neighbor residue
Bond Stretching
The distribution of bond lengths can be represented by the Gaussian function, which is calculated by:
| (4) |
where b, b0 and σ are the parameters obtained through fitting, kB is the Boltzmann factor and T is absolute temperature. Taking the logarithm of both sides of equation (4) and dropping the constant term, after performing Boltzmann inversion, we have:
| (5) |
where the temperature, T, is set to be 298K.
Angle Bending
The distributions of bond angle can be weighted by a factor sin(θ) and renormalized by a factor Zn. The normalized distribution is expressed as:
| (6) |
where θ is the angle between neighboring bonds, while P(θ) and p(θ) are normalized and un-normalized distribution functions of θ. The distributions of bond angles between CG bonds were also fitted with the Gaussian function, and then the Boltzmann inversion were used to calculate Eangle.
| (7) |
Dihedral Rotation
As in atomic force fields, the tensional energy takes the formula of:
| (8) |
where ϕ is the dihedral angle, Kn and δn (n = 1, 2, 3) are force constants and phase angles. The Edihedral were also obtained from performing Boltzmann inversion.
Non-bonded Interactions
A Buckingham potential,113 consisting of a 6 term and an exponential term are used to represents the potential of mean forces between a pair of non-bonded atoms, i and j:
| (9) |
where εij is the depth of the potential well, σij is the radius, and rij is the distance between a pair of atoms. Note that the above equation is used to describe the potential of mean force even though the symbol “E” and the formula are commonly used to represent potential energy. The constants we used here are the same as MM3 force field.85,114–116 We use the pair-specific ε and σ parameters instead of the combing rule for unlike atom pair i and j. The Lennard-Jones (LJ) 6–12 potential and Buckingham potential fitted the non-bonded interactions at the onset were generated. The Buckingham function is “softer” than the LJ 6–12 function in the repulsive region because the exponential term is more suitable to represent the non-bonded potentials in our CG model. However, as shown in Figure S3, even the Buckingham potential is not “soft” enough. In addition, some of the interactions (e.g. N2-N2) clearly show a second or more local minima which are ignored by the Buckingham potential. The complicated shape of the non-bonded potentials can be captured much more accurately by using the spline interpolation functions as in the previous statistical potentials for proteins117. However, in the current study, we would like to explore the capability of the simple Buckingham potential that is implemented in almost all popular molecular modeling packages. In (dihedral) non-bonded potential fitting, we have chosen to primarily reproduce the global energy minima by using a weighted least-square fit. The data points in minimum energy area (0.5 Å within the potential minimum) were assigned a weight of two while the others one. The final fitting results are shown in Figure S3 in Supporting Information. As discussed in the Results and discussion, a nonlinear optimization was later performed on the non-bonded parameters, after all bonded and non-bonded parameters in the CG model were obtained, by minimizing the RMSD between the energy minimized and the experimental structures of selected RNAs. The experimental structures were analyzed to generate the radial distribution functions (RDF) or g(r) for selected pairs of coarse-grained particles (Table 4). The set of 1–2 (directly bonded), 1–3 (separated by 2 bonds), and1–4 (separated by 3 bonds) pairs were not included in RDF calculations. Then the potential of mean force, which corresponds to the Boltzmann inversion of the g(r), is determined from the RDF:
| (10) |
TABLE 4.
The Optimized CG Parameters for Non-Bonded Interaction Described by Equation (9), Including Similar and Unlike Pairs of CG Atoms
| CG atoms | σ (initial) | ε (initial)a | σ (final) | ε (final)b |
|---|---|---|---|---|
| P - P | 11 | 0.287 | 11.2778 | 0.1503 |
| S - S | 11.7 | 0.3827 | 12.1544 | 0.4162 |
| C - C | 4.4 | 0.8851 | 4.1836 | 0.9276 |
| N6-N6 | 3.1 | 1.3875 | 3.4604 | 1.4312 |
| N2-N2 | 4.8 | 1.1004 | 4.7928 | 1.1603 |
| O6-O6 | 3.1 | 1.4354 | 3.7784 | 1.4635 |
| O2-O2 | 4.9 | 1.0526 | 4.8614 | 1.0846 |
| C - N6 | 5.1 | 0.6698 | 5.2158 | 0.3818 |
| O6-O2 | 5.2 | 1.0526 | 5.4321 | 1.2972 |
| N2-O2 | 2.8 | 1.9138 | 2.7974 | 2.0524 |
| C - O6 | 5.2 | 0.8373 | 5.26 | 0.6972 |
| C - O2 | 3.5 | 0.909 | 3.6176 | 0.8886 |
| N6-O6 | 2.85 | 1.866 | 3.0427 | 1.8562 |
| C - N2 | 4.25 | 0.8373 | 4.3342 | 0.8527 |
| N6-O2 | 5.3 | 0.7416 | 5.6477 | 0.7942 |
| N6-N2 | 5.3 | 1.232 | 5.3832 | 1.0547 |
| O6-N2 | 5.4 | 0.7416 | 5.5622 | 0.5273 |
| P - S | 9.4 | 0.5263 | 9.4287 | 0.054 |
| S - C | 5.6 | 0.5502 | 5.615 | 0.5856 |
Columns σ (initial) and ε (initial) are the non-bonded parameters obtained from statistical potential of mean force.
Columns σ (final) and ε (final) are the non-bonded parameters obtained after the optimization
By combining equation (9) and (10), we determine the initial values of ε and σ for each pair. We have also combined certain pairs (the same parameters were used) based on the similarity of the RDF obtained.
Simulation Details
Fifteen different RNA molecules were selected for molecular dynamics simulations using the developed CG potential. For comparison, all-atom simulations were performed on the same set of RNAs using amber99sb force field with explicit and implicit solvent in AMBER10.70,72,118 For the explicit solvent simulations, the TIP3P water model was used.119,120 The Particle Mesh Ewald (PME) method121,122 was applied to treat the long-range electrostatic interactions and a 12 Å cutoff was employed for the van der Waals interactions. After the RNA was solved in the water box, a 150 mM/L Na+ ions were added to the box, and then the Cl− ions were added to neutralize the whole system. All the RNA systems were equilibrated with a 5,000-step energy minimization to remove the bad contacts. Then, the minimized configurations were used as the starting point for a 1 ns NPT molecular dynamics equilibration at 1 atm and 298 K. For the implicit solvent simulations, the Generalized-Born/Surface Area (GB/SA) model was used.123–128 The salt concentration was also 150 mM/L in implicit solvent systems. The time step for all production runs was 1 fs. The CG model molecular dynamics simulations were performed via modified Beeman algorithm with the TINKER software package.129 Different time steps (1 fs to 10 fs) were tested in the CG MD simulations. The normal simulation time for all systems was 10–100 ns. Simulated annealing MD simulations were performed to fold fifteen RNA molecules. In the simulated annealing, after the energy minimization and 2,000 steps of equilibrium, the systems were heated to 1,000 K within the first 2,000 steps and then gradually cooled down to room temperature (298 K) over 100 ns with a linear schedule. For each RNA molecule, we performed 5 independent simulated annealing simulations starting with random seeds. Finally, we performed another 3 independent 100 ns simulated annealing under restraints set by the known secondary structure (Watson-Crick base pairs only). A flat-welled harmonic potential (also known as the Nuclear Overhauser Effect potential) was used to restrain the distance between the pseudo atoms CG and AU (Figure 1) in the canonical Watson-Crick base pairs with a lower and upper bounds of 8.0 Å and 10.0 Å, respectively. The force constant for the harmonic potential is zero when the distance falls within the bounds and 0.5 kcal/Å2 beyond the bounds.
Results and Discussion
Determination of the RNA Coarse-Grained Model Parameters
The probability distribution of all virtual bonds, angles, and torsions (shown in the Supporting Information) were used to fit the valence parameters using equation (4) to (8). The fitted parameters for virtual bonds, angles, and torsions are given in Table 2, Table 3, and Table 5, respectively. The force constants of bonds and angles for pseudo atoms are smaller than those of atomic bonds and angles, meaning a larger time step could be used during the MD simulations in the CG model. As expected, the larger force constants of bonds and angles within the base make the bases fairly stiff (Table 2 and Table 3). However, they are still smaller than those of atomic constants by a factor of 3 to 10.
TABLE 3.
The Bond Angle Interaction Parameters for the CG Model of RNA Fitted by the Gaussian Function and Obtained from Statistical Structures
| angle | θ0 | Ka |
|---|---|---|
| 1-2-1 | 102.78 | 1.356 |
| 1-2-3 | 101.75 | 5.271 |
| 1-2-3'a | 75.89 | 1.864 |
| 1-2-8 | 100.79 | 9.115 |
| 1-2-8' | 74.40 | 2.386 |
| 2-1-2 | 106.18 | 2.040 |
| 2-3-4 | 154.72 | 7.130 |
| 2-3-5 | 104.12 | 12.734 |
| 2-3-6 | 153.94 | 8.162 |
| 2-3-9 | 108.78 | 10.611 |
| 2-8-4 | 163.79 | 6.794 |
| 2-8-6 | 163.79 | 6.794 |
| 2-8-7 | 88.99 | 15.930 |
| 3-4-9 | 66.45 | 35.882 |
| 3-5-6 | 79.38 | 16.156 |
| 3-6-5 | 48.06 | 21.701 |
| 4-3-9 | 48.33 | 49.428 |
| 4-7-8 | 49.44 | 24.490 |
| 4-8-7 | 79.78 | 29.398 |
| 4-9-3 | 65.22 | 17.290 |
| 5-3-6 | 52.57 | 50.065 |
| 6-7-8 | 50.54 | 38.613 |
| 6-8-7 | 79.46 | 31.109 |
| 7-4-8 | 50.84 | 29.033 |
| 7-6-8 | 49.98 | 30.600 |
The prime in the table indicates the atom comes from its neighbor residue
The non-bonded parameters in equation (9) were obtained by mapping the radial distribution functions (RDF) of all the pseudo atoms in existing RNA structures:
| (11) |
where nij(r) is the number of pairs in the given shell from r to r+dr, Ni is the total number of particle I in the system, and d is the mean bulk density of particle j. The reference state here is the expected number of contacts when two pseudo atoms i and j at long distance, which is approximated as the average density of pseudo atoms j.130 Therefore, the g(r) could be normalized to 1 at long distance. The results from g(r) are plugged into equation (10) to get the potential of mean force, which is approximated to be the effective potential function of r. The effective potential functions are shown in Supporting Information. The best fitting results for each type of CG atom are summarized in Table 4.
Optimization of the Non-bonded Parameters
The RNA structural statistics we utilized to derive the non-bonded and bonded parameters effectively include contributions from all energy terms, although to different extents. For example, the actual conformational distribution is affected by both the torsion and non-bonded energy terms in the CG potential. To remove the “redundancy”, we directly compared the structures given by the coarse-grained potential with the experimental structures and adjust the parameters. Since the RNA structure is most sensitive to the non-bonded interaction, we refined the non-bonded parameters using seven RNA molecules with diverse secondary and tertiary structures. The non-bonded parameters were refined by minimizing the difference between the energy-minimized CG structures and their corresponding experimental structures. First, energy minimization was performed on each of the seven RNA molecules, and the structural root mean square deviation (RMSD) from the experimental structure was calculated based on all pseudo atoms. The average of the RMSD over the seven molecules is used as the target function in the optimization of the non-bonded parameters. An optimally conditioned variable metric nonlinear optimization algorithm in TINKER was utilized.129,131–133 The first derivative of the average RMSD with respect to each non-bonded parameter was calculated numerically. In total 38 non-bond parameters were optimized (Table 4). The average RMSD between the experimental and energy-minimized structures dropped from 3.35 Å to 1.75 Å by using the optimized non-bonded parameters.
Validation of the Coarse-Grained Model
We have validated the CG potential in two sets of simulations. First, molecular dynamics simulations of the native RNA structures were performed at room temperature to examine the RMSD fluctuation. Second, simulated annealing molecular dynamics simulations were applied to fold the RNA structures, with and without restraints set by the secondary structure information. Fifteen RNA molecules with high quality experimental structures were chosen for the model validation (Table 6). These RNAs represent common RNA structural motifs, such as a helix with and without non-canonical base pairs, hairpins, internal loops, bulges, and pseudoknots.
TABLE 6.
Comparision of All-Atom Average RMSDs from the Native Crystal Structures for Both the CG Model and the Full-Atom Models. All RMSDs Were Obtained from All CG Atoms (Full-Atom Calculation Using the Same Atom Set as CG Model)
| RMSD (Å) | |||
|---|---|---|---|
| PDB ID | CG model | full-atom implicit water model (amber99sb ff) | full-atom explicit water model (amber99sb ff) |
| 157D | 3.644 | 5.609 | 2.260 |
| 1DQF | 2.478 | 3.274 | 1.935 |
| 1I9X | 3.236 | 3.956 | 1.780 |
| 2JXQ | 2.536 | 2.940 | 1.800 |
| 2K7E | 3.675 | 4.560 | 3.560 |
| 353D | 3.483 | 2.802 | 3.583 |
| 472D | 3.137 | 2.467 | 1.500 |
| 1F5G | 4.425 | 2.950 | 1.800 |
| 1KD3 | 4.570 | 3.740 | 2.560 |
| 1L2X | 2.840 | 2.890 | 2.730 |
| 1LNT | 4.751 | 2.610 | 1.880 |
| 1QCU | 1.669 | 2.560 | 2.210 |
| 1ZIH | 3.532 | 3.180 | 2.010 |
| 2AO5 | 3.326 | 2.090 | 1.350 |
| 1AL5 | 3.080 | 3.770 | 2.210 |
| Average | 3.359 | 3.293 | 2.211 |
All rmsd’s were obtained from all CG atoms (full-atom calculation using the same atom set as the CG model).
The results of the regular room temperature MD simulations are given in Table 6. The RMSDs for both the coarse-grained and all-atom model were calculated as an average over the MD trajectories using the same atom set of five pseudo atoms for better comparison. One base-pair at each of the terminals is ignored in all the RMSD calculations unless specified otherwise. The 15 RNA molecules investigated had an overall average RMSDs with the CG model (3.36 Å). This is similar to the all-atom implicit solvent model (3.30 Å). The all-atom MD simulation with explicit solvent was even closer to the experimental structure, with an average RMSD of 2.21 Å. The RMSD for the CG model (1.66 Å) performed slightly better than the all-atom explicit solvent simulation (2.21 Å) for an A-form helix with only canonical Watson-Crick base pairs. Figure 2a shows the superposition of the native structure (1QCU) and the last frame of 10ns CG MD simulation. The backbone is well preserved by the CG MD in 1QCU, as well as the base-pair and base stacking interactions. As expected, somewhat larger RMSDs have been observed at the 5’ and 3’ terminals of double helix in both CG and all-atom MD simulations of 1QCU, consistent with the assumption that the lack of base stacking interactions and neighboring base-pair connections would decrease the stabilities of the helix terminals
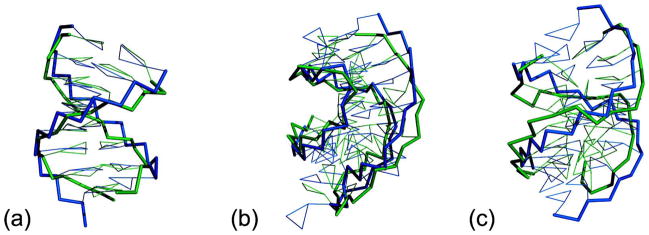
Figure 2.
Superposition of final snapshot from 10 ns CG simulations (colored green) and native structure (colored blue). The backbones are represented as thick sticks and the bases are shown as lines. (a) Superposition of RNA with 12 canonical Watson-Crick base pairs (PDB ID: 1QCU). (b) Superposition of the frameshifting RNA pesudoknot from beet western yellow virus (PDB ID: 1L2X), a single chain with coaxial helices connected by two loops. (c) Superposition of 12 nt dsRNA with 5-bp internal loop (PDB ID: 1LNT).
The frameshifting RNA pesudoknot from beet western yellow virus (PDB ID: 1L2X) is a single chain with coaxial helices connected with two loops.134 The superposition of the native x-ray structure and the last frame of 10ns MD CG simulation are shown in Figure 2b. The backbone is well represented in the CG model. The average RMSD of CG model (2.84 Å) is quite similar to those given by the all-atom simulations using implicit (2.89 Å) and explicit solvent (2.73 Å). Both the all-atom model and the CG model showed a larger fluctuation for non-canonical Watson-Crick base pairs in 1L2X during the MD simulations. The observed unpaired bases which belong to the loops moved back and forth along the direction perpendicular to the backbone, which may play an important role in forming tertiary contacts.
Among the 15 test RNAs, the largest RMSD (4.75 Å) was observed for 1LNT.135 1LNT is a 12-t double-strand helix with a 5-bp interior loop. The superposition of the native x-ray structure and the final snapshot of MD simulation (Figure 2c) show that the large RMSD is mostly due to the base atoms and two terminals. The bases within the interior loop are much more flexible and disordered than base forming Watson-Crick base pairs. It is not surprising that the current CG model has difficulty in capturing them, given that RNA structures in the parameterization set is dominated by canonical Watson-Crick base-pairs.
How well the MD simulations preserve the native structure is only a minimal check of the coarse-grained potential. It is more interesting and challenging to see if the CG potential can be used to “predict” RNA folds. The “folding” of RNA molecules by the CG model was examined by 100 ns simulated annealing molecular dynamics. In this process, the RNA molecule was denatured by being quickly heated to a temperature of 1000 K. With the system temperature gradually (over 100 ns) cooled back down to 298 K, the RNA molecules were expected to fold back to its native or native-like conformations. We have performed five independent simulated annealing runs for each of the 15 RNAs. The final conformations were checked by comparing the structure of the last snapshot of trajectory to its native structure. The minimum and average RMSDs among the 5 repeats are given in Table 7. The results indicated that ~75% of RNAs fold back to their native-like structures (final all-pseudo-atom RMSD < 6.5 Å) at least once among the 5 repeats. Examples of the simulation snapshots and the final annealed structures of 1ZIH (RMSD = 3.8 Å) and 353D (RMSD = 6.3 Å) could be found at Figure 3b and Figure 3d, respectively. Due to the limited sampling capability of simulated annealing method, the RNA molecule is very likely to be trapped into a local minimum. For instance, all five simulated annealing repeats of the 1L2X (contains a pseudoknot), failed to fold back to its right coaxial structures and were stuck in different local minima. Another possible reason is a lack of chemical details in our energy potential function. The stability of some RNA tertiary motifs such as pseudoknots is highly dependent on the solvation environment and metal ion concentration, especially the magnesium hydration effect.136
TABLE 7.
Comparison of All-Atom RMSDs between the Final Structures of 100 ns Simulated-Annealing Simulations to Their Native Structures.
| PDBID | RMSD (Å) without restrains MIN/AVGa | RMSD (Å) with restraints MIN/AVG | number of restrained pairs |
|---|---|---|---|
| 157D | 6.6/7.2 | 5.2/5.5 | 5 |
| 1AL5 | 6.4/8.4 | 5.2/5.8 | 5 |
| 1DQF | 4.2/5.7 | 4.8/5.3 | 4 |
| 1F5G | 5.2/7.0 | 5.8/5.9 | 5 |
| 1I9X | 6.8/8.4 | 3.0/4.7 | 5 |
| 1KD3 | 8.0/9.1 | 5.3/5.6 | 5 |
| 1L2X | 9.7/10.4 | 5.0/5.2 | 5 |
| 1LNT | 6.5/7.7 | 5.0/5.6 | 5 |
| 1QCU | 6.0/8.8 | 4.0/4.4 | 5 |
| 1ZIH | 3.8/4.7 | 3.6/5.0 | 3 |
| 2AO5 | 5.1/6.5 | 2.8/4.6 | 5 |
| 2JXQ | 4.1/6.4 | 3.0/4.8 | 5 |
| 2K7E | 5.9/7.8 | 5.0/5.1 | 5 |
| 353D | 4.8/6.0 | 4.0/5.5 | 5 |
| 472D | 4.6/6.9 | 4.7/5.5 | 3 |
The minimum and average RMSD value among all repeats (the restrained pairs shown in the last column on average correspond to ~40% of each RNA).
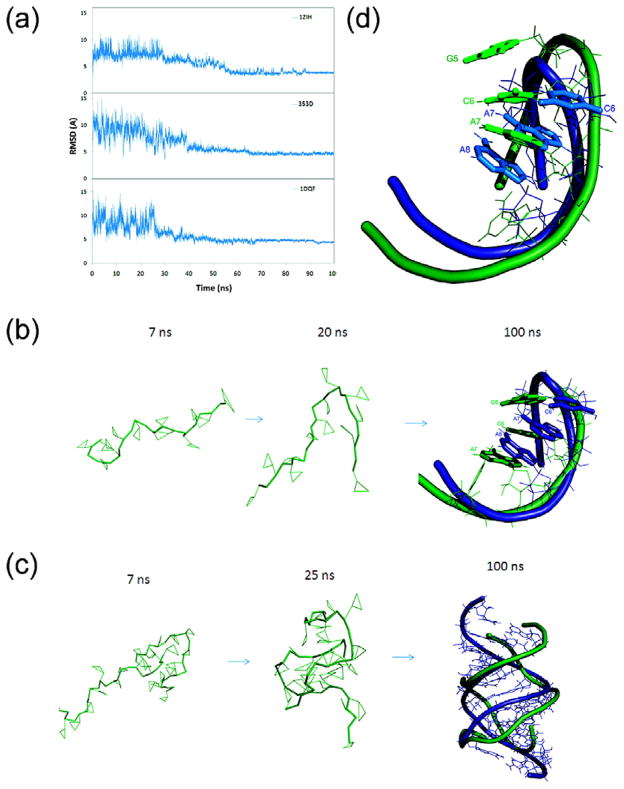
Figure 3.
(a) Comparison of all-atom RMSDs for the structures 1ZIH, 353D and 1DQF during the 100 ns simulated annealing simulations. The simulation temperature was increased to 1,000k within the beginning 2,000 steps and then cooled down to room temperature 298k. The RMSDs were calculated using all of the CG atoms. The figures show how the RNA molecules fold toward to their native structures. (b) Snapshots taken from simulated annealing of 1ZIH. (c) The superposition of final conformation (colored green) and native structure (colored blue) of GCCA tetraloop after 20 ns full-atom MD refinement. The backbone is represented as a ribbon and the base-stacking unit in tetraloop is shown as sticks. (d) Snapshots taken from simulated annealing simulation of 353D.
Here we discuss in detail three RNAs that were successfully folded by the simulated annealing MD simulations, a hairpin 1ZIH,137,138 and two helices: 353D139 and 1DQF.140 1ZIH is a 12-nt single strand RAN hairpin capped by a GCAA tetraloop.138,141 The GCAA tetraloop belongs to the GNRA tetraloop family (N is A, C, G, or U; R is A or G)142 which is a basic building block of RNA structure that often provide sites for tertiary contacts or protein binding.50,142–145 At the beginning of annealing, the RNA was completely denatured due to the heating, showing a RMSD of about 10 Å (Figure 3a and 3b). The structure became stable after 70 ns and the RMSDs dropped to ~3.8 Å when the temperature slowly decreased to 400 K (Figure 3b). Similar behavior was observed in the annealing of two A-form helices, 353D (Figure 3d) and 1DQF. 353D contains two U-G Wobbles and 1DQF contains a bulge residue. Both RMSDs converged to ~4.5 Å at the end of the annealing (Figure 3a). Overall, the successes demonstrate that the CG potential is very promising for ab initio prediction of small RNA tertiary structure from sequence alone.
We have examined the possibility of further refining the CG structures from the simulated annealing by mapping the structures to the corresponding all-atom models and performing all-atom MD simulations. The last frames from the 100-ns simulated annealing simulation of 1ZIH and 353D with RMSDs around 5 Å were taken to generate the initial coordinates for the subsequent all-atom simulations using AMBER10. The all-atom structures were re-constructed from the CG structures via the following steps (please refer to Figure 1c for the atomic labels). 1) The planar all-atom bases were placed based on the three pseudo particles in the CG bases (Figure 1). The C1’ atom, which lies in the base plane, was constructed by extending from the N atom in the bases. Equilibrium bond and angle values from amber99sb force field were used in constructing the atomic coordinates relative to each other. 2) The CG backbone particles P and S were turned into P and C4’ in the all-atom model. The backbone O5’ and C5’ were placed along the vector connecting P and C4’ with the P-O5’ and O5’-C5’ bond lengths to set to be one third of the P-C4’ distance. Similarly, O3’ and C3’ were placed between C4’ and P in the opposite direction. The other two O atoms connecting to P were placed in the plane orthogonal to the O3’-P-O4’. 3) The O4’ in the sugar ring was placed such that it lied in the plane of C3’-C4’-O4’-C1’. Using the C4’-O4’-C1’ as the anchor, the sugar ring (including directly bonded peripheral atoms) in a flat conformation was then constructed. Note that in this new structure, C5’ and C3’ were moved out of the C4’-P vector. 4) The all-atom structure was then relaxed via AMBER energy minimization while the five atoms directly transferred from the CG model (3 base atoms, P and C4’) were constrained to the CG coordinates. The resulting all-atom structures were subjected to a 20,000-step energy minimization without constraints followed by a 20-ns MD simulation with amber99sb force field and GB/SA implicit solvation model. For hairpin 1ZIH, the overall RMSDs dropped slightly from ~5.5 Å to 5 Å. However, the backbone structure of 1ZIH was significantly improved as the RMSD was reduced from 5.4 Å to 3.65 Å. The detailed bases orientations in GCAA loop could be seen in Figure 3c, which shows the superposition of final conformation and native structure of the tetraloop. The GCA bases were stacked on top of each other instead of the CAA base-stacking in native structure with ~60 rotation. The rotation or flipping of the GNRA bases, similar to the U-turn motif 16,33,146 was observed in early studies by NMR analysis and other all-atom molecular dynamic simulations.138,147 The flipping of some of the bases towards the solvent in a tetraloop allows those bases to interact with other RNAs and proteins. For 353D, both all-atom RMSDs and backbone RMSDs remained mostly unchanged after the all-atom refinement. One possible reason is that the final structure of CG simulated annealing may be trapped in a local minimum, and the 20-ns all-atom simulation was not able to allow the helix to relax and re-arrange into the native structure. The results indicate that the multi-stage approach is useful for construction and refinement of all-atom structures after the CG simulation provide the prediction of near-native structures. We also tested the ability of the CG potential to predict 3-D structure when limited restraints are introduced based on the secondary structure information (canonical base pairing) during the simulated annealing simulations. We added restraint by randomly picked 3 to 5 canonical Watson-Crick base pairs in each RNA molecule (Table 7). With these limited restraints, the CG model was able to predict all the 3-D structures for each tested RNA in all 3 independent repeats, with RMSDs in the range of 2.8 Å to 6.5 Å (Table 7). The results are very encouraging as it demonstrates that limited knowledge of canonical base pairing from secondary structure prediction can greatly facilitate the 3-D structure prediction using a CG potential.
Computational Efficiency of the Coarse-Grained Model
The computational efficiency of the CG model is greatly improved when compared to the all-atom model. A nearly two orders of magnitude improvement in MD simulation speed was found with the same time step of 1 fs. This improvement is mainly due to the reduction in the number of bond, angle and torsion calculations. Furthermore, because of the absence of high frequency motions such as bond stretching in the all-atom model, a time step up to 10 fs can be applied to the MD simulation without a noticeable effect on energy and structural stability. In addition, in explicit all-atom simulations using a physical force field, not only is the number of atoms further increased due to the presence of the solvent, the equilibration of water and counter ions distribution is time-consuming. Therefore, the CG model can achieve an improvement of about three orders of magnitude in the simulation speed, which may enable us to study a large system, or extend the simulation time from tens of nanoseconds to the scale of microseconds.
Conclusions
In summary, we developed and applied a new statistical coarse-grained potential to model RNA structures with molecular dynamics. In the coarse-grained potential, each nucleotide is represented by 5 pseudo atoms including 3 in the base ring. The bond, angle, torsion and non-bonded parameters in the CG potential were derived based on the structural statistics sampled from experimental structures of over 600 RNA in PDB. The Boltzmann inversion used to obtain the initial parameters for the CG potential. Subsequently the non-bonded parameters were optimized analytically by comparing the CG minimum energy structures with the experimental structures. The optimization was performed systematically using the optimally conditioned variable metric nonlinear optimization algorithm in TINKER.129,131–133
The resulting potential was validated in molecular dynamics simulations of 15 RNAs, including helices, teraloops, stem loops, bulges and pseudoknots. Room temperature MD simulations starting from the native structures produced very reasonable RMSD (3.36 Å on average), indicating the CG potential is able to maintain the native structure, or the native structures are minima on the CG potential energy surface. The coarse-grained potential was then applied to “predict” the 3-D structures of the 15 RNAs using multiple independent simulated annealing dynamics simulations. For most of the RNAs, at least one out of the five simulated annealing runs, the structure folded into a near native state (RMSD < 6.5 Å). We also noted that the CG potential was able to predict the base stacking behavior in a tetraloop. Furthermore, we also introduced limited distance restraints based on the knowledge of canonical base-pairing in secondary structure. The reliability of the structure prediction using CG potential was drastically improved, and all RNAs were folded into structures with RMSDs less than 6.5 Å. We also investigated the possibility of using an amber all-atom force field to map and refine the CG model into all-atom structures.
Overall, the performance of this simple CG potential is very promising. With limited knowledge of base-pairing from secondary structure prediction, the CG approach can reliably predict the 3-D structure for small RNA molecules of various topologies. The analytical functional form of the CG potential is compatible with existing molecular modeling packages such as NAMD148 and GROMACS,149 so that it can be easily adapted and take advantage of sophisticated simulations algorithms such as replica exchange molecular dynamic method150 that is much more effective than simulated annealing for conformational sampling. Further improvement of the accuracy of the CG potential can be achieved by incorporating more experimental structure and refinement of the parameters. To capture the non-bonded interaction more precisely, spline interpolation functions can be utilized instead of the current simple analytical function,117 In the future, extra types of pseudo atoms will be incorporated to represent the modified bases, especially by methylation, such as inosine (derived from adenine) or pseudouridine (derived from uracil) in tRNA.
Supplementary Material
Acknowledgments
This research is supported by grant from the Robert A. Welch Foundation to PR (F-1691), and RG acknowledges support by Robert A. Welch Foundation (F-1427), NIH (GM067317), Microsoft Research. Z.X is grateful to Yue Shi and Dr. Chunli Yan for their help with the AMBER simulations.
Footnotes
Supporting Information Available. The probability distributions of all virtual bonds, angles, and torsions for parameterization are shown in Figure S1, S2, and S4, respectively. The potential of mean force for each coarse-grained pair could be seen in Figure S3. The RMSDs during 20-ns all-atom simulation refinement are show in Figure S5. This material is available free of charge via the Internet at http://pubs.acs.org.
References
- 1.Crick F. Symp Soc Exp Biol. 1958;XII:139. [PubMed] [Google Scholar]
- 2.Crick F. Nature. 1970;227:561. doi: 10.1038/227561a0. [DOI] [PubMed] [Google Scholar]
- 3.James DW, Tania AB, Stephen PB, Alexander G, Michael L, Richard L CSHLP I. Benjamin Cummings. 2007 [Google Scholar]
- 4.Stark BC, Kole R, Bowman EJ, Altman S. Proceedings of the National Academy of Sciences of the United States of America. 1978;75:3717. doi: 10.1073/pnas.75.8.3717. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Noller HF, Chaires JB. Proc Natl Acad Sci USA. 1972;69:3115. doi: 10.1073/pnas.69.11.3115. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Noller HF, Chang C, Thomas G, Aldridge J. J Mol Biol. 1971;61:669. doi: 10.1016/0022-2836(71)90071-4. [DOI] [PubMed] [Google Scholar]
- 7.Kruger K, Grabowski PJ, Zaug AJ, Sands J, Gottschling DE, Cech TR. Cell. 1982;31:147. doi: 10.1016/0092-8674(82)90414-7. [DOI] [PubMed] [Google Scholar]
- 8.Guerrier-Takada C, Gardiner K, Marsh T, Pace N, Altman S. Cell. 1983;35:849. doi: 10.1016/0092-8674(83)90117-4. [DOI] [PubMed] [Google Scholar]
- 9.Nahvi A, Sudarsan N, Ebert MS, Zou X, Brown KL, Breaker RR. Chemistry & biology. 2002;9:1043. doi: 10.1016/s1074-5521(02)00224-7. [DOI] [PubMed] [Google Scholar]
- 10.Winkler W, Nahvi A, Breaker RR. Nature. 2002;419:952. doi: 10.1038/nature01145. [DOI] [PubMed] [Google Scholar]
- 11.Winkler WC, Cohen-Chalamish S, Breaker RR. Proceedings of the National Academy of Sciences of the United States of America. 2002;99:15908. doi: 10.1073/pnas.212628899. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Rodionov DA, Vitreschak AG, Mironov AA, Gelfand MS. Nucleic Acids Res. 2003;31:6748. doi: 10.1093/nar/gkg900. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Vitreschak AG, Rodionov DA, Mironov AA, Gelfand MS. Trends Genet. 2004;20:44. doi: 10.1016/j.tig.2003.11.008. [DOI] [PubMed] [Google Scholar]
- 14.Doudna JA, Szostak JW. Nature. 1989;339:519. doi: 10.1038/339519a0. [DOI] [PubMed] [Google Scholar]
- 15.Noller HF, Hoffarth V, Zimniak L. Science. 1992;256:1416. doi: 10.1126/science.1604315. [DOI] [PubMed] [Google Scholar]
- 16.Hingerty B, Brown RS, Jack A. Journal of Molecular Biology. 1978;124:523. doi: 10.1016/0022-2836(78)90185-7. [DOI] [PubMed] [Google Scholar]
- 17.Ban N, Nissen P, Hansen J, Moore PB, Steitz TA. Science (New York, NY. 2000;289:905. doi: 10.1126/science.289.5481.905. [DOI] [PubMed] [Google Scholar]
- 18.Wimberly BT, Brodersen DE, Clemons WM, Jr, Morgan-Warren RJ, Carter AP, Vonrhein C, Hartsch T, Ramakrishnan V. Nature. 2000;407:327. doi: 10.1038/35030006. [DOI] [PubMed] [Google Scholar]
- 19.Brodersen DE, Clemons WM, Jr, Carter AP, Wimberly BT, Ramakrishnan V. J Mol Biol. 2002;316:725. doi: 10.1006/jmbi.2001.5359. [DOI] [PubMed] [Google Scholar]
- 20.Kazantsev AV, Krivenko AA, Harrington DJ, Holbrook SR, Adams PD, Pace NR. Proc Natl Acad Sci USA. 2005;102:13392. doi: 10.1073/pnas.0506662102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Torres-Larios A, Swinger KK, Krasilnikov AS, Pan T, Mondragon A. Nature. 2005;437:584. doi: 10.1038/nature04074. [DOI] [PubMed] [Google Scholar]
- 22.Serganov A, Huang L, Patel DJ. Nature. 2009;458:233. doi: 10.1038/nature07642. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Cate JH, Gooding AR, Podell E, Zhou K, Golden BL, Kundrot CE, Cech TR, Doudna JA. Science (New York, NY. 1996;273:1678. doi: 10.1126/science.273.5282.1678. [DOI] [PubMed] [Google Scholar]
- 24.Vidovic I, Nottrott S, Hartmuth K, Luhrmann R, Ficner R. Molecular cell. 2000;6:1331. doi: 10.1016/s1097-2765(00)00131-3. [DOI] [PubMed] [Google Scholar]
- 25.Serganov A, Yuan YR, Pikovskaya O, Polonskaia A, Malinina L, Phan AT, Hobartner C, Micura R, Breaker RR, Patel DJ. Chemistry & biology. 2004;11:1729. doi: 10.1016/j.chembiol.2004.11.018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Bessho Y, Shibata R, Sekine S, Murayama K, Higashijima K, Hori-Takemoto C, Shirouzu M, Kuramitsu S, Yokoyama S. Proceedings of the National Academy of Sciences of the United States of America. 2007;104:8293. doi: 10.1073/pnas.0700402104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Klein DJ, Ferre-D'Amare AR. Science (New York, NY. 2006;313:1752. doi: 10.1126/science.1129666. [DOI] [PubMed] [Google Scholar]
- 28.Serganov A, Huang L, Patel DJ. Nature. 2008;455:1263. doi: 10.1038/nature07326. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Dann CE, 3rd, Wakeman CA, Sieling CL, Baker SC, Irnov I, Winkler WC. Cell. 2007;130:878. doi: 10.1016/j.cell.2007.06.051. [DOI] [PubMed] [Google Scholar]
- 30.Thore S, Leibundgut M, Ban N. Science (New York, NY. 2006;312:1208. doi: 10.1126/science.1128451. [DOI] [PubMed] [Google Scholar]
- 31.Montange RK, Batey RT. Nature. 2006;441:1172. doi: 10.1038/nature04819. [DOI] [PubMed] [Google Scholar]
- 32.Hainzl T, Huang S, Sauer-Eriksson AE. Nature. 2002;417:767. doi: 10.1038/nature00768. [DOI] [PubMed] [Google Scholar]
- 33.Kim SH, Suddath FL, Quigley GJ, McPherson A, Sussman JL, Wang AH, Seeman NC, Rich A. Science (New York, NY. 1974;185:435. doi: 10.1126/science.185.4149.435. [DOI] [PubMed] [Google Scholar]
- 34.Costa FF. Bioessays. 32:599. doi: 10.1002/bies.200900112. [DOI] [PubMed] [Google Scholar]
- 35.Spizzo R, Nicoloso MS, Croce CM, Calin GA. Cell. 2009;137:586. doi: 10.1016/j.cell.2009.04.040. [DOI] [PubMed] [Google Scholar]
- 36.Frohlich KS, Vogel J. Curr Opin Microbiol. 2009;12:674. doi: 10.1016/j.mib.2009.09.009. [DOI] [PubMed] [Google Scholar]
- 37.Georg J, Voss B, Scholz I, Mitschke J, Wilde A, Hess WR. Mol Syst Biol. 2009;5:305. doi: 10.1038/msb.2009.63. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Khraiwesh B, Arif MA, Seumel GI, Ossowski S, Weigel D, Reski R, Frank W. Cell. 2010;140:111. doi: 10.1016/j.cell.2009.12.023. [DOI] [PubMed] [Google Scholar]
- 39.Hale CR, Zhao P, Olson S, Duff MO, Graveley BR, Wells L, Terns RM, PTM Cell. 2009;139:945. doi: 10.1016/j.cell.2009.07.040. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Marraffini LA, Sontheimer EJ. Nat Rev Genet. 2010;11:181. doi: 10.1038/nrg2749. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Hamilton AJ, Baulcombe DC. Science (New York, NY. 1999;286:950. doi: 10.1126/science.286.5441.950. [DOI] [PubMed] [Google Scholar]
- 42.Lecellier CH, Dunoyer P, Arar K, Lehmann-Che J, Eyquem S, Himber C, Saib A, Voinnet O. Science (New York, NY. 2005;308:557. doi: 10.1126/science.1108784. [DOI] [PubMed] [Google Scholar]
- 43.Buchon N, Vaury C. Heredity. 2006;96:195. doi: 10.1038/sj.hdy.6800789. [DOI] [PubMed] [Google Scholar]
- 44.Mattick JS, Taft RJ, Faulkner GJ. Trends Genet. 2010;26:21. doi: 10.1016/j.tig.2009.11.002. [DOI] [PubMed] [Google Scholar]
- 45.Gupta RA, Shah N, Wang KC, Kim J, Horlings HM, Wong DJ, Tsai MC, Hung T, Argani P, Rinn JL, Wang Y, Brzoska P, Kong B, Li R, West RB, van de Vijver MJ, Sukumar S, Chang HY. Nature. 2010;464:1071. doi: 10.1038/nature08975. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Croce CM. Nat Rev Genet. 2009;10:704. doi: 10.1038/nrg2634. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Sassen S, Miska EA, Caldas C. Virchows Arch. 2008;452:1. doi: 10.1007/s00428-007-0532-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Hahn MW, Wray GA. Evolution & development. 2002;4:73. doi: 10.1046/j.1525-142x.2002.01069.x. [DOI] [PubMed] [Google Scholar]
- 49.Taft RJ, Pheasant M, Mattick JS. Bioessays. 2007;29:288. doi: 10.1002/bies.20544. [DOI] [PubMed] [Google Scholar]
- 50.Michel F, Westhof E. J Mol Biol. 1990;216:585. doi: 10.1016/0022-2836(90)90386-Z. [DOI] [PubMed] [Google Scholar]
- 51.Levitt M. Nature. 1969;224:759. doi: 10.1038/224759a0. [DOI] [PubMed] [Google Scholar]
- 52.Malhotra A, Tan RK, Harvey SC. Biophys J. 1994;66:1777. doi: 10.1016/S0006-3495(94)80972-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Fink DL, Chen RO, Noller HF, Altman RB. Rna. 1996;2:851. [PMC free article] [PubMed] [Google Scholar]
- 54.Lehnert V, Jaeger L, Michel F, Westhof E. Chemistry & biology. 1996;3:993. doi: 10.1016/s1074-5521(96)90166-0. [DOI] [PubMed] [Google Scholar]
- 55.Wang R, Alexander RW, VanLoock M, Vladimirov S, Bukhtiyarov Y, Harvey SC, Cooperman BS. J Mol Biol. 1999;286:521. doi: 10.1006/jmbi.1998.2493. [DOI] [PubMed] [Google Scholar]
- 56.Sommer I, Brimacombe R. J Comput Chem. 2001;22:407. [Google Scholar]
- 57.Stagg SM, Mears JA, Harvey SC. J Mol Biol. 2003;328:49. doi: 10.1016/s0022-2836(03)00174-8. [DOI] [PubMed] [Google Scholar]
- 58.Shapiro BA, Yingling YG, Kasprzak W, Bindewald E. Curr Opin Struct Biol. 2007;17:157. doi: 10.1016/j.sbi.2007.03.001. [DOI] [PubMed] [Google Scholar]
- 59.Malhotra A, Harvey SC. J Mol Biol. 1994;240:308. doi: 10.1006/jmbi.1994.1448. [DOI] [PubMed] [Google Scholar]
- 60.Devkota B, Petrov AS, Lemieux S, Boz MB, Tang L, Schneemann A, Johnson JE, Harvey SC. Biopolymers. 2009;91:530. doi: 10.1002/bip.21168. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Meller J, Elber R. Computational Methods for Protein Folding. 2002;120:77. doi: 10.1002/jcc.10014. [DOI] [PubMed] [Google Scholar]
- 62.Kryshtafovych A, Venclovas C, Fidelis K, Moult J. Proteins-Structure Function and Bioinformatics. 2005;61:225. doi: 10.1002/prot.20740. [DOI] [PubMed] [Google Scholar]
- 63.Moult J. Curr Opin Struct Biol. 2005;15:285. doi: 10.1016/j.sbi.2005.05.011. [DOI] [PubMed] [Google Scholar]
- 64.Zwieb C, Muller F. Nucleic Acids Symp Ser. 1997;36:69. [PubMed] [Google Scholar]
- 65.Orozco M, Perez A, Noy A, Luque FJ. Chem Soc Rev. 2003;32:350. doi: 10.1039/b207226m. [DOI] [PubMed] [Google Scholar]
- 66.Jossinet F, Westhof E. Bioinformatics. 2005;21:3320. doi: 10.1093/bioinformatics/bti504. [DOI] [PubMed] [Google Scholar]
- 67.Wu JC, Gardner DP, Ozer S, Gutell RR, Ren P. J Mol Biol. 2009;391:769. doi: 10.1016/j.jmb.2009.06.036. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Das R, Baker D. Proceedings of the National Academy of Sciences of the United States of America. 2007;104:14664. doi: 10.1073/pnas.0703836104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Parisien M, Major F. Nature. 2008;452:51. doi: 10.1038/nature06684. [DOI] [PubMed] [Google Scholar]
- 70.Pearlman DA, Case DA, Caldwell JW, Ross WS, Cheatham TE, Debolt S, Ferguson D, Seibel G, Kollman P. Computer Physics Communications. 1995;91:1. [Google Scholar]
- 71.Cheatham TE, 3rd, Cieplak P, Kollman PA. J Biomol Struct Dyn. 1999;16:845. doi: 10.1080/07391102.1999.10508297. [DOI] [PubMed] [Google Scholar]
- 72.Case DA, Cheatham TE, 3rd, Darden T, Gohlke H, Luo R, Merz KM, Jr, Onufriev A, Simmerling C, Wang B, Woods RJ. J Comput Chem. 2005;26:1668. doi: 10.1002/jcc.20290. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73.Perez A, Marchan I, Svozil D, Sponer J, Cheatham TE, 3rd, Laughton CA, Orozco M. Biophys J. 2007;92:3817. doi: 10.1529/biophysj.106.097782. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74.Brooks BR, Bruccoeri RE, Olafson BD, States DJ, Swaminathan S, Karplus M. J Comput Chem. 1983;4:187. [Google Scholar]
- 75.MacKerell AD, Jr, Banavali N, Foloppe N. Biopolymers. 2000;56:257. doi: 10.1002/1097-0282(2000)56:4<257::AID-BIP10029>3.0.CO;2-W. [DOI] [PubMed] [Google Scholar]
- 76.Brooks BR, Brooks CL, 3rd, Mackerell AD, Jr, Nilsson L, Petrella RJ, Roux B, Won Y, Archontis G, Bartels C, Boresch S, Caflisch A, Caves L, Cui Q, Dinner AR, Feig M, Fischer S, Gao J, Hodoscek M, Im W, Kuczera K, Lazaridis T, Ma J, Ovchinnikov V, Paci E, Pastor RW, Post CB, Pu JZ, Schaefer M, Tidor B, Venable RM, Woodcock HL, Wu X, Yang W, York DM, Karplus M. J Comput Chem. 2009;30:1545. doi: 10.1002/jcc.21287. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 77.Freddolino PL, Arkhipov AS, Larson SB, McPherson A, Schulten K. Structure. 2006;14:437. doi: 10.1016/j.str.2005.11.014. [DOI] [PubMed] [Google Scholar]
- 78.Knotts TA, Rathore N, Schwartz DC, de Pablo JJ. Journal of Chemical Physics. 2007;126:084901. doi: 10.1063/1.2431804. [DOI] [PubMed] [Google Scholar]
- 79.Golubkov PA, Ren PY. Journal of Chemical Physics. 2006;125 doi: 10.1063/1.2244553. [DOI] [PubMed] [Google Scholar]
- 80.Scheraga HA, Khalili M, Liwo A. Annual Review of Physical Chemistry. 2007;58:57. doi: 10.1146/annurev.physchem.58.032806.104614. [DOI] [PubMed] [Google Scholar]
- 81.Golubkov PA, Wu JC, Ren PY. Physical Chemistry Chemical Physics. 2008;10:2050. doi: 10.1039/b715841f. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 82.Maroun RC, Olson WK. Biopolymers. 1988;27:585. doi: 10.1002/bip.360270404. [DOI] [PubMed] [Google Scholar]
- 83.Maroun RC, Olson WK. Biopolymers. 1988;27:561. doi: 10.1002/bip.360270403. [DOI] [PubMed] [Google Scholar]
- 84.Hao MH, Olson WK. Biopolymers. 1989;28:873. doi: 10.1002/bip.360280407. [DOI] [PubMed] [Google Scholar]
- 85.Norman L, Allinger, Young HYuh, Lii J-H. Journal of the American Chemical Society. 1989;111:8851. [Google Scholar]
- 86.Tan RK, Harvey SC. J Mol Biol. 1989;205:573. doi: 10.1016/0022-2836(89)90227-1. [DOI] [PubMed] [Google Scholar]
- 87.Sprous D, Harvey SC. Biophys J. 1996;70:1893. doi: 10.1016/S0006-3495(96)79754-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 88.Sprous D, Tan RK, Harvey SC. Biopolymers. 1996;39:243. doi: 10.1002/(SICI)1097-0282(199608)39:2%3C243::AID-BIP11%3E3.0.CO;2-F. [DOI] [PubMed] [Google Scholar]
- 89.Tan RK, Sprous D, Harvey SC. Biopolymers. 1996;39:259. doi: 10.1002/(sici)1097-0282(199608)39:2<259::aid-bip12>3.0.co;2-9. [DOI] [PubMed] [Google Scholar]
- 90.Massire C, Westhof E. Journal of Molecular Graphics & Modelling. 1998;16:197. doi: 10.1016/s1093-3263(98)80004-1. [DOI] [PubMed] [Google Scholar]
- 91.Tanaka I, Nakagawa A, Hosaka H, Wakatsuki S, Mueller F, Brimacombe R. Rna. 1998;4:542. doi: 10.1017/s1355838298972004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 92.Olson WK, Zhurkin VB. Curr Opin Struct Biol. 2000;10:286. doi: 10.1016/s0959-440x(00)00086-5. [DOI] [PubMed] [Google Scholar]
- 93.Matsumoto A, Olson WK. Biophys J. 2002;83:22. doi: 10.1016/S0006-3495(02)75147-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 94.Coleman BD, Olson WK, Swigon D. Journal of Chemical Physics. 2003;118:7127. [Google Scholar]
- 95.Mergell B, Ejtehadi MR, Everaers R. Physical Review E. 2003;68:021911. doi: 10.1103/PhysRevE.68.021911. [DOI] [PubMed] [Google Scholar]
- 96.Flammini A, Maritan A, Stasiak A. Biophys J. 2004;87:2968. doi: 10.1529/biophysj.104.045864. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 97.LaMarque JC, Le TV, Harvey SC. Biopolymers. 2004;73:348. doi: 10.1002/bip.10529. [DOI] [PubMed] [Google Scholar]
- 98.Nielsen SO, Lopez CF, Srinivas G, Klein ML. Journal of Physics-Condensed Matter. 2004;16:R481. [Google Scholar]
- 99.Peyrard M. Nonlinearity. 2004;17:R1. [Google Scholar]
- 100.Zhang DQ, Konecny R, Baker NA, McCammon JA. Biopolymers. 2004;75:325. doi: 10.1002/bip.20120. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 101.Cao S, Chen SJ. Rna. 2005;11:1884. doi: 10.1261/rna.2109105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 102.Tepper HL, Voth GA. Journal of Chemical Physics. 2005;122:124906. doi: 10.1063/1.1869417. [DOI] [PubMed] [Google Scholar]
- 103.Li XJ, Kou DZ, Rao SL, Liang HJ. Journal of Chemical Physics. 2006:124. doi: 10.1063/1.2200694. [DOI] [PubMed] [Google Scholar]
- 104.Tan RKZ, Petrov AS, Harvey SC. Journal of Chemical Theory and Computation. 2006;2:529. doi: 10.1021/ct050323r. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 105.Vologodskii A. Biophys J. 2006;90:1594. doi: 10.1529/biophysj.105.074682. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 106.Ding F, Sharma S, Chalasani P, Demidov VV, Broude NE, Dokholyan NV. Rna. 2008;14:1164. doi: 10.1261/rna.894608. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 107.Jonikas MA, Radmer RJ, Laederach A, Das R, Pearlman S, Herschlag D, Altman RB. Rna. 2009;15:189. doi: 10.1261/rna.1270809. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 108.Ledford H. Nature. 2010;465:16. doi: 10.1038/465016a. [DOI] [PubMed] [Google Scholar]
- 109.Cannone JJ, Subramanian S, Schnare MN, Collett JR, D'Souza LM, Du Y, Feng B, Lin N, Madabusi LV, Muller KM, Pande N, Shang Z, Yu N, Gutell RR. BMC Bioinformatics. 2002;3:2. doi: 10.1186/1471-2105-3-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 110.Duarte CM, Pyle AM. J Mol Biol. 1998;284:1465. doi: 10.1006/jmbi.1998.2233. [DOI] [PubMed] [Google Scholar]
- 111.Tschop W, Kremer K, Batoulis J, Burger T, Hahn O. Acta Polymerica. 1998;49:61. [Google Scholar]
- 112.Muller-Plathe F. Chemphyschem. 2002;3:754. doi: 10.1002/1439-7641(20020916)3:9<754::aid-cphc754>3.0.co;2-u. [DOI] [PubMed] [Google Scholar]
- 113.Buckingham RA. Proceedings of the Royal Society of London Series A, Mathematical and Physical Sciences. 1938;168:264. [Google Scholar]
- 114.Allinger NL, Yuh YH, Lii JH. Journal of the American Chemical Society. 1989;111:8551. [Google Scholar]
- 115.Lii JH, Allinger NL. Journal of the American Chemical Society. 1989;111:8566. [Google Scholar]
- 116.Lii JH, Allinger NL. Journal of the American Chemical Society. 1989;111:8576. [Google Scholar]
- 117.Summa CM, Levitt M. Proceedings of the National Academy of Sciences of the United States of America. 2007;104:3177. doi: 10.1073/pnas.0611593104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 118.Case DA, Darden TA, Cheatham TEI, Simmerling CL, Wang J, Duke RE, Luo R, Crowley M, Walker RC, Zhang W, Merz KM, Wang B, Hayik S, Roitberg A, Seabra G, Kolossváry I, Wong KF, Paesani F, Vanicek J, Wu X, Brozell SR, Steinbrecher T, Gohlke H, Yang L, Tan C, Mongan J, Hornak V, Cui G, Mathews DH, Seetin MG, Sagui C, Babin V, Kollman PA. University of California; San Francisco: 2008. [Google Scholar]
- 119.Jorgensen WL, Chandrasekhar J, Madura J, Impey RW, Klein ML. Journal of Chemical Physics. 1983;79:926. [Google Scholar]
- 120.Neria E, Karplus M. Journal of Chemical Physics. 1996;105:10812. [Google Scholar]
- 121.Deserno M, Holm C. Journal of Chemical Physics. 1998;109:7678. [Google Scholar]
- 122.Deserno M, Holm C. Journal of Chemical Physics. 1998;109:7694. [Google Scholar]
- 123.Still WC, Tempczyk A, Hawley RC, Hendrickson T. Journal of the American Chemical Society. 1990;112:6127. [Google Scholar]
- 124.Schaefer M, Karplus M. Journal of Physical Chemistry. 1996;100:1578. [Google Scholar]
- 125.Scarsi M, Apostolakis J, Caflisch A. Journal of Physical Chemistry A. 1997;101:8098. [Google Scholar]
- 126.Dominy BN, Brooks CL. Journal of Physical Chemistry B. 1999;103:3765. [Google Scholar]
- 127.Bashford D, Case DA. Annual Review of Physical Chemistry. 2000;51:129. doi: 10.1146/annurev.physchem.51.1.129. [DOI] [PubMed] [Google Scholar]
- 128.Feig M, Im W, Brooks CL. Journal of Chemical Physics. 2004;120:903. doi: 10.1063/1.1631258. [DOI] [PubMed] [Google Scholar]
- 129.Ponder JW. Washington University Medical School. [Google Scholar]
- 130.Skolnick J, Jaroszewski L, Kolinski A, Godzik A. Protein Sci. 1997;6:676. doi: 10.1002/pro.5560060317. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 131.Shanno DF, Phua KH. Mathematical Programming. 1977;25:507. [Google Scholar]
- 132.Davidon WC. Mathematical Programming. 1975;9:1. [Google Scholar]
- 133.Shanno DF, Phua KH. Journal of Optimization Theory and Applications. 1978;25:507. [Google Scholar]
- 134.Egli M, Minasov G, Su L, Rich A. Proceedings of the National Academy ofSciences of the United States of America. 2002;99:4302. doi: 10.1073/pnas.062055599. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 135.Deng J, Xiong Y, Pan B, Sundaralingam M. Acta Crystallogr D Biol Crystallogr. 2003;59:1004. doi: 10.1107/s0907444903006747. [DOI] [PubMed] [Google Scholar]
- 136.Draper DE. Biophys J. 2008;95:5489. doi: 10.1529/biophysj.108.131813. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 137.Heus HA, Pardi A. Science (New York, NY. 1991;253:191. doi: 10.1126/science.1712983. [DOI] [PubMed] [Google Scholar]
- 138.Jucker FM, Heus HA, Yip PF, Moors EHM, Pardi A. J Mol Biol. 1996;264:968. doi: 10.1006/jmbi.1996.0690. [DOI] [PubMed] [Google Scholar]
- 139.Betzel C, Lorenz S, Furste JP, Bald R, Zhang M, Schneider TR, Wilson KS, Erdmann VA. Febs Letters. 1994;351:159. doi: 10.1016/0014-5793(94)00834-5. [DOI] [PubMed] [Google Scholar]
- 140.Xiong Y, Sundaralingam M. Rna. 2000;6:1316. doi: 10.1017/s135583820000090x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 141.Correll CC, Swinger K. Rna. 2003;9:355. doi: 10.1261/rna.2147803. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 142.Woese CR, Winker S, Gutell RR. Proceedings of the National Academy of Sciences of the United States of America. 1990;87:8467. doi: 10.1073/pnas.87.21.8467. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 143.Doherty EA, Batey RT, Masquida B, Doudna JA. Nat Struct Biol. 2001;8:339. doi: 10.1038/86221. [DOI] [PubMed] [Google Scholar]
- 144.Nissen P, Ippolito JA, Ban N, Moore PB, Steitz TA. Proc Natl Acad Sci USA. 2001;98:4899. doi: 10.1073/pnas.081082398. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 145.Cate JH, Gooding AR, Podell E, Zhou K, Golden BL, Szewczak AA, Kundrot CE, Cech TR, Doudna JA. Science. 1996;273:1696. doi: 10.1126/science.273.5282.1696. [DOI] [PubMed] [Google Scholar]
- 146.Jucker FM, Pardi A. RNA. 1995;1:219. [PMC free article] [PubMed] [Google Scholar]
- 147.Sorin EJ, Engelhardt MA, Herschlag D, Pande VS. J Mol Biol. 2002;317:493. doi: 10.1006/jmbi.2002.5447. [DOI] [PubMed] [Google Scholar]
- 148.Phillips JC, Braun R, Wang W, Gumbart J, Tajkhorshid E, Villa E, Chipot C, Skeel RD, Kale L, Schulten K. Journal of Computational Chemistry. 2005;26:1781. doi: 10.1002/jcc.20289. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 149.van der Spoel D, Lindahl E, Hess B, van Buuren AR, Apol E, Meulenhoff PJ, Tieleman DP, Sijbers ALTM, Feenstra KA, Drunen Rv, Berendsen C, HJ 2005 www.gromacs.org.
- 150.Sugita Y, Okamoto Y. Chemical Physics Letters. 1999;314:141. [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.