Abstract
There now exists a rich set of ontologies that provide detailed semantics for biological entities of interest. However, there is not (nor should there be) a single source ontology that provides all the necessary semantics for describing biological phenomena. In the domain of physiological biosimulation models, researchers use annotations to convey semantics, and many of these annotations require the use of multiple reference ontologies. Therefore, we have developed the idea of composite annotations that access multiple ontologies to capture the physics-based meaning of model variables. These composite annotations provide the semantic expressivity needed to disambiguate the often-complex features of biosimulation models, and can be used to assist with model merging and interoperability. In this paper, we demonstrate the utility of composite annotations for model merging by describing their use within SemGen, our semantics-based model composition software. More broadly, if orthogonal reference ontologies are to meet their full potential, users need tools and methods to connect and link these ontologies. Our composite annotations and the SemGen tool provide one mechanism for leveraging multiple reference ontologies.
Keywords: Biomedical Ontology, Biosimulation, Annotation, Computer Simulation
1. Motivation
Ontology development is often driven by the need to make data and information more sharable and interoperable. In such cases, researchers annotate data and information against a common reference ontology. The ontology provides formal definitions and clarifies the intended semantics for the data, which makes the data more accessible, sharable, and interoperable. As the need for this capability has grown across a variety of biomedical domains, the number and availability of ontologies has grown as well. For many reference ontologies, there has been an intentional design choice to scope their content so that it is orthogonal and non-overlapping—this is a founding principle of the Open Biomedical Ontology (OBO) collection[1]. The vision for this design choice is that ontologies should be interoperable, so that users can access multiple ontologies without conflict.
In biosimulation modeling of physiology or pathology, the use of multiple reference ontologies is essential. To annotate a biosimulation model, one not only needs multiple ontologies for different parts of the model, but one may need multiple ontologies for a single construct in that model. For example, to understand hypertension, one may want to annotate with terms such as fluid pressure, blood, and a specific artery where a pressure is measured. In this paper, we introduce the idea of composite annotations to allow for precise specification of semantics that include more than one ontology.
Our examples and driving use cases are from biosimulation research. This field is maturing to the point where practical, clinical applications for complex, patient-specific models of physiology and pathology are within reach[2]. As evidence of the growth and maturity of this field, there are now many public-domain libraries of biosimulation models available, including libraries with smaller scale models (subcellular or molecular), as well as those with larger scale models (tissue and organ level). [3–5] This growth and maturity has led to great a need for model sharing. Indeed, one purpose for creating libraries of biosimulation models is to allow researchers to find and retrieve models created by others, and then tune, rewrite, or simply incorporate them into their own locally developed biosimulation models. Many biosimulation researchers have recognized the great potential offered by a library of interoperating, plug-and-play biosimulation models—this idea is central to the Physiome vision[6], as well as the recently initiated Virtual Physiological Human project[7]. Unfortunately, there are a host of barriers that inhibit biosimulation model reuse and sharing.
In this paper, we focus on composite annotations as a potential solution to these challenges. We first provide a grammar or specification for our composite annotations, independent of particular syntaxes or implementation details. Next, we describe an implementation of these composite annotations, and an initial demonstration of how our approach and architecture would encourage plug-and-play merging of biosimulation models. As part of this demonstration, we have developed SemGen, a prototype toolset with which researchers can easily create and apply composite annotations, and then use these semantic annotations to merge and integrate biosimulation models. Before we more precisely define composite annotations, we outline the current state of biomedical ontologies and knowledge resources that are relevant to biosimulation, followed by an account of the state-of-the-art in biosimulation modeling research.
1.1 Reference ontologies for biology
In biosimulation, the biological entities of interest depend on the granularity and scale of the model under development. Thus, chemical kinetic models, such as those stored in the BioModels repository[3],* encode the concentrations of chemicals such as proteins, enzymes, and small molecules. In contrast, organ and tissue level models encode larger, anatomic entities such as heart valves, bronchial passages, and muscle tissue. Currently, there are many resources and ontologies available for biological entities—the structures that participate in physiological processes. For example, the Chemicals Entities of Biological Interest resource (ChEBI),[8] UniProt,[9] GO,[10] and KEGG [11] describe molecular entities. For cellular and macroscopic entities (organs and tissues), the Foundational Model of Anatomy (FMA)[12] encodes the canonical anatomy of humans, and other anatomic ontologies apply to other taxonomic species.
However, these entities are insufficient for completely annotating biosimulation models. A defining characteristic of such models is that they represent processes that unfold over time, and not just the physical entities that are participants in those processes. Modelers use variables that represent physical properties of physical entities that change over time; for example, the pressure of aortic blood, or the chemical concentration of thrombin protein in venous blood. Therefore an important resource for biosimulation models must be an ontology of the physical properties that an entity can have. It is exactly this pragmatic need that has driven our development of the Ontology of Physics for Biology (OPB)[13].
Pairing physical properties with physical entities (e.g., pressure of blood), is an idea that is also used for phenotype annotations.[14] More specifically, these authors advocate an “EQ” methodology for entities and qualities. For phenotypes, a quality may be any descriptive term, such as “smaller”, “round”, “increased temperature”, etc. This notion of quality is more formally defined in the Basic Formal Ontology.[15] Our work builds from this idea, but due to our focus on biosimulation models, we restrict qualities to physical properties that can take on values that change over time—properties that might be encoded as variables in a biosimulation model.
As an example that we will use throughout the paper, consider a model of cardiovascular blood circulation and regulation. For such a model, one may have a variable (e.g., “Paorta”) that encodes aortic blood pressure. To annotate this variable, there are (at least) three relevant classes: fluid pressure, the aorta (where pressure is measured) and the blood in the aorta. In our example, fluid pressure is defined in the OPB, whereas blood and aorta are from the FMA. As another example, a variable may encode the concentration of oxygen in the aorta; such a variable would need the same two classes from the FMA (blood and aorta), as well as the class oxygen from ChEBI, and the class chemical concentration from the OPB.
As a straw-man proposal, one might imagine developing a biosimulation ontology that contains terms such as “aortic blood pressure”. However, one would be faced with a combinatoric challenge—every fluid in every bodily compartment could have an associated fluid pressure. Furthermore, blood exists in many spatial compartments (arteries, veins, chambers of the heart, etc), and for each there is a pressure, a volume, and a net flow rate—should any single ontology (e.g., some “biosimulation ontology”) enumerate all such possibilities?
Such an enumeration is conceptually simple, but impractical, as it would require pre-coordinating class names from a cross-product of several large ontologies—an exponentially large number. A more workable solution is to allow such classes to be created on-the-fly, in a post-coordinated manner. With composite annotations, our approach is to allow developers and researchers to synthesize and store post-coordinated annotations, whenever users need multiple ontologies to annotate a single term.
1.2 Biosimulation modeling
Driven by the vision of the Physiome [6], the number of archived, publicly available biosimulation models has exploded, reaching to well over 1000 curated models. In our work, we have drawn primarily from 3 such libraries: The BioModels repository[3], the CellML repository[4], and the NSR JSim model archive[5].
Although libraries of models are an important initial step, archives by themselves are not sufficient to support model interoperation or sharability among researchers. We know both from our own experience[16], and from discussions with colleagues at meetings (such as the 2008 SIAM mini-symposium on “Integrative Modeling: Challenges in Modularity”) that it is extremely time-consuming and difficult to adapt and reuse biosimulation models that were created by others. Current challenges to model sharing and merging include:
Models are written in incompatible modeling languages that differ in semantics and syntax.
Models may have inconsistent and informal annotations with respect to biological knowledge resources and ontologies.
Models frequently have serious errors of unitary imbalance, mathematical inconsistency, and syntax.
As anecdotal evidence, our experiences with two important libraries of biosimulation models (JSim and BioModels) have shown that these sorts of problems are the rule rather than the exception for models.[17] Indeed, it was the frequency of errors in published models that led researchers to design model repositories that emphasize the importance of rigorous curation by third-party scientists.
In spite of these problems and barriers, the biosimulation research community sees great benefit to model integration and reuse. The benefit is the resulting merged model—a model that both has greater functionality than any of its components, and also acts as an internal validity check for each component model[18]. This perceived benefit is also evidenced by current efforts to reduce some of the barriers to reuse, and most notably, to improve annotation efforts.
Even simple annotations can capture at least some of the semantics that underlie a particular term or equation encoded in the syntax of a biosimulation modeling language. At the simplest level, an annotation may be an in-line, natural language comment on a variable. For example, in JSim’s MML code, one may have a line for aortic blood pressure such as:
real Paop(t) mmHg;//Proximal aorta transmural pressure
where text following the “//” is an annotation for the variable “Paop”. This form of annotation does provide some assistance for researchers who may be merging others’ models, but natural language annotations are not amenable to automatic processing. In contrast, annotations that link to unique identifiers in on-line knowledge sources can be compared and processed in a more automatic manner. For example, the MIRIAM guidelines (Minimal Information Requested In the Annotation of biochemical Models;[19]) require that if a model is annotated in terms of external resources, then the annotations must use Unique Resource Identifiers (URIs). Such URIs may be encoded as Uniform Resource Name (URNs) or Uniform Resource Locator (URLs), but must be described by a triple in the following format: {“data-type,” “identifier,” “qualifier”}. As evidence that there is a growing need for these improved annotations, tools have been developed to assist researchers in creating annotations, such as the SemanticSBML tool [20] or the SAINT tool for building MIRIAM annotations[21].
As an example of MIRIAM-compliant annotations, Figure 1 shows a snippet of an SBML model of glycolysis [22] with three annotations to external sources for the entity 2-phospho-D-glycerate (listed inside the “rdf:Bag” structure). In this case, the three sources contain synonymous information about the compound. (Synonymy is not explicit in the RDF code, but is clear in the documentation for SBML, or if one follows the link to pubChem.) Annotations of this form allow for a degree of computational, automatic semantic checks that are useful when merging or understanding different models. For example, other models that include annotations to any of the three URNs (e.g. ChEBI:17835) can automatically be matched as synonymous, regardless of the specific variable or “species” name. This type of capability provides an important step toward model sharing and model merging, and indeed, the SemanticSBML tool uses these annotations to assist researchers with model merging.[20]
Fig 1.
A snippet of SBML code showing three MIRIAM annotations to ChEBI, KEGG & PubChem. The entity (species) is 2-phospho-D-glycerate.
However, although this sort of annotation is more computable than ad hoc annotations, for our purposes, there remain several key limitations. First, all of SBML and BioModels are aimed at the bio-chemical realm, and not other scales nor types of processes beyond bio-chemical reactions. A direct implication of this restriction is that the actual variables whose values change over time (the “properties of interest”) for all SBML species are always chemical amounts or concentrations. Thus, this information is omitted (since it can be implied) in SBML models. For multi-scale applications it is important to make this explicit, so that we can distinguish between (for example) the pressure of blood in the aorta versus its rate of flow.
Second, for multiscale modeling applications, we must be able to annotate a single variable with multiple, orthogonal ontologies, so that we can describe the property of interest (e.g. chemical concentration) as well as the physical entities of interest across multiple scales. The example in Figure 1 annotates a single variable by referencing three non-orthogonal ontologies at a single structural scale to indicate the synonymous semantics for the chemical entity. In contrast, a multi-scale application would require that a cell type be annotated with one ontology, the anatomical organ it is contained-in with another ontology, and the sub-parts of that cell with yet a third ontology. To support this richer annotation ability, we present our more structured, comprehensive approach for composite annotations.
2. Composite annotations
Our goal is to provide a logical construct, the composite annotation, for annotating the physical properties of physical entities of interest for biosimulation models. As an example, imagine a variable that encodes the “concentration of calcium ions in the endoplasmic reticulum of a vascular smooth muscle cell in the wall of a systemic arteriole”. Although “calcium ion” (or any of these components) can be annotated with a single URI, to better capture the semantics of this variable, we need an ordered list of URIs that link to different, orthogonal, reference ontologies. Furthermore, the relations between elements in this list must be explicit, and where possible, described by elements of the OBO Relations Ontology (RO).[23]
Figure 2 shows our schema for composite annotations (top) and the simple example of aortic blood pressure (bottom), rendered in a pseudo-code notation. We view composite annotations as having two parts. The first part (“Fluid_pressure physical_property_of”) identifies the kind of physical property encoded by the biosimulation variable (properties such as flow rate, chemical concentration, pressure, or resistance). Thus, our example has links to the OPB fluid_pressure class, as well as the physical_property_of relationship. The second part of the annotation (“Portion_of_blood contained_in Lumen_of_aorta”) post-coordinates two FMA classes with an RO contained_in relation. Composite annotations for more complex or specific entities are composed simply by extension of the linked list. Thus, the concentration of calcium ion example at the top of this section might include four or five physical entities connected by RO structural relations.
Fig 2.
A schema showing the structure of a composite annotation (top), with the example of aortic blood pressure (bottom).
The top of Figure 2 represents a preliminary grammar for our composite annotations: An ordered list of references to classes in external ontologies, each connected explicitly by a relationship term defined (where possible) by the Relations Ontology. For biosimulation variables, each composite annotation must include exactly one physical property reference, and must contain at least one physical entity reference. More precisely, the first physical entity must be the one which possesses the property being measured.† To guide and accelerate the annotation process, we can include specific constraints on this list when annotating particular types of variables. For example, for any fluid flow-rate annotation, the initial physical entity must be a bodily substance (defined by the FMA as a fluid). As an abstract grammar, our approach to composite annotations is independent of any particular implementation syntax, or of particular choices for reference ontologies.
We do not wish to restrict or require users to annotate terms using only existing reference ontologies. Therefore, we can also accommodate special-purpose physical entities. For example, one may wish to model the blood flow rate through an atrial septal defect, a congenital heart defect that is a pathological conduit between heart chambers. Such an anatomic entity does not belong in the FMA (which represents only normal anatomy), but a user can still compose an annotation such as “Fluid_pressure physical_property_of Portion_of_blood contained_in Lumen_of_atrial_septal_defect”. In such an annotation, the first two terms would have the same URIs as the example in Figure 2 (to the OPB and the FMA), whereas the third term would simply be denoted as custom, and would lack a linkage to any reference ontology.
Of course, if terms in an annotation lack links to ontologies, then these terms cannot be merged or recognized in any automatic manner. Thus, our tools support and encourage a middle ground—users can create custom physical entities, but they should also indicate the relationships, if known, of these entities to known entities in reference ontologies. For example, one could connect the custom term “lumen of atrial septal defect” with knowledge (a) that this is a instance of a biological lumen, (b) that it is connected to the right atrium, and (c) it is connected to the left atrium. While this is still imperfect information, it may provide critical linkages that enable model reuse.
Composite annotations could be implemented and encoded in variety of syntaxes. One could, for example, encode composite annotations in XML or RDF and then use them as in-line annotations in existing languages such as SBML and CellML. However, such a choice has several disadvantages. First, it would be dependent on the syntax of particular biosimulation modeling languages, rather than independent of such choices. Second, we view semantic annotations as “middle-level” knowledge—a set of annotations about a model connects the source code (the bottom level) to reference ontologies (an upper level) that provide more detailed semantics for the entities of interest. Thus, we prefer an implementation that allows these annotations to exist separately from both the modeling source code and the reference ontologies.
Implementation choices for composite annotations depend on how one expects these annotations to be used. Our long-term goal is to demonstrate that composite annotations can support knowledge sharing and model reuse. The remainder of this paper describes our tool, SemGen, that implements our ideas for composite annotations and assists users with model reuse and model merging. For this implementation, we store annotations for any given biosimulation model as a separate OWL file. Figure 3 shows how our implementation relates to the general notion of a composite annotation, and to the example presented in Figure 2. On the left, we reiterate the three selected classes from reference ontologies (FMA and OPB) that provide semantics and a composite annotation for “Paorta”. The snippet of OWL code shows instances that refer to these reference ontology classes, as well as an individual that links to the specific variable name, “Paorta” in the cardiovascular simulation model we have annotated. In our implementation, annotation instances are doubly-linked with properties such as “contained_in” and “contains” (from the Relations Ontology of the OBO). The OWL syntax in this figure is Turtle[24].
Fig 3.
A snippet of the OWL representation for a single composite annotation, Paorta (as in Figure 2). The snippet shows four individuals; the first corresponds to the named variable itself, while the other three correspond to the three reference ontology classes shown on the left. For brevity, we have omitted prefix declarations that provide URIs for ontologies such as OPB, FMA, and RO.
At this time, we do not use the full logical and inferential capabilities of OWL (e.g., we do not require complete semantics of classes via necessary and sufficient conditions, nor use description logic inference). However, as we discuss at the end of this paper, we have chosen OWL because in the longer term we expect to leverage the inferential power of description logic reasoning that this formalism supports.
3. SemGen: A toolset for annotating and merging models
For composite annotations to be effective, we must have tool support for researchers who wish to annotate their models. In particular, to encourage annotation and model sharing, we strive to make it as easy as possible to annotate models, and to hide the complexity of composite annotations and the reference ontologies that they refer to. Indeed, this goal is shared by the designers of SAINT[21] and SemanticSBML[20] for biochemical models. Our SemGen tool is distinct from these efforts in that (a) our composite annotations are designed for multiple scales, rather than just SBML biochemical models, (b) our annotations reference orthogonal ontologies and (c) are stored in a separate middle layer, rather than embedded in the source code.
SemGen is our tool for support of biosimulation model reuse and integration.‡ It includes a number of functions that help to modularize legacy biosimulation models and provide semantics for those models via composite annotations. In this paper, we focus on two capabilities: (1) Sem-Gen’s annotation tool, which helps create annotations, and (2) a merger tool, which integrates annotated models to produce larger models. This implementation is a prototype, and has not yet been formally evaluated with users. However, by working with an implemented tool and real biosimulation models, we are better able to test and validate our ideas for composite annotations and model merging.
Figure 4 shows SemGen’s annotator tool, focused on the variable “Paorta” and its annotation, as described earlier. The top portion of the interface shows a portion of the source code for the biosimulation model—in this case, a cardiovascular model of blood circulation and blood pressure. The syntax shown is MML code for the JSim environment, but our approach can be used with SBML and CellML models as well, as JSim can import models in these languages. As a demonstration of this capability, we have also used the annotator tool to build a set of annotations for a glycolysis SBML model retrieved from BioModels[25].
Fig 4.
A screen from our SemGen annotation tool, showing the composite annotation for Paorta in the CV model. The composite annotation is independent from the source code (shown in top panel).
The bottom panels of Figure 4 show the annotations and their variables—the bottom left is a list of model variables, extracted automatically from the JSim MML code, and the annotation for the selected variable (Paorta) is shown on the bottom right. As we described earlier, this is a composite annotation with reference to terms in the FMA and OPB.
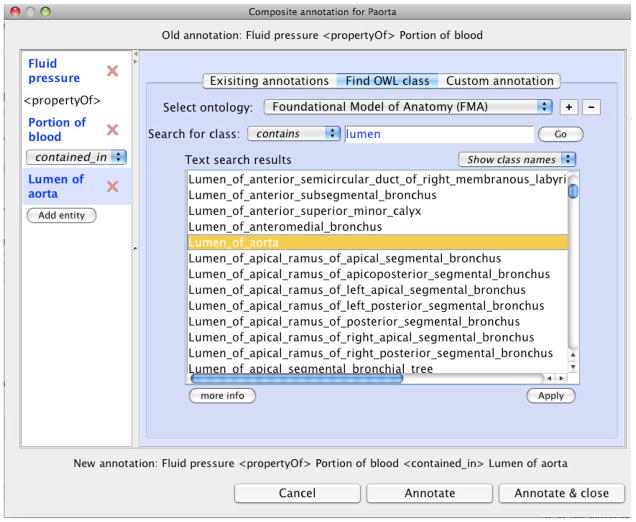
To build such an annotation, users must have access to classes in these external ontologies. Rather than require or expect users to have expertise with these ontologies, or ask them to use external applications for browsing and searching through such ontologies, such as NCBO’s BioPortal system[26], we have designed SemGen to allow for direct browsing of all relevant ontologies. Figure 5 shows a prototype user interface for building a composite annotation, at the point where a user has searched for the classes containing the term “lumen” in the FMA.
Fig 5.
A screen showing how users assemble composite annotations. As highlighted on the left, the user has just selected the FMA term “Lumen of aorta” to fill the third spot in the composite annotation for Paorta.
There are several ways this integrated ontology browsing capability could be implemented. Currently, SemGen provides access and search capabilities for any RDF or OWL syntax ontology on the web, via a locally-developed query interface. However, especially when reference ontologies are large or dynamic, it is important that SemGen does not require a local copy of the reference ontology for retrieving class information. Thus, we would aim for a design such as a web services interface that can process the sorts of queries required for model annotation. An alternative implementation might rely on a BioPortal API that can similarly answer queries about any BioPortal ontology[27]. An advantage of this design would be that both OBO and OWL ontologies would be available; a disadvantage would be that non-BioPortal ontologies would not be available.
4. Demonstration
As a demonstration of the utility of composite annotations, we used SemGen to automate a model-merging task that was previously performed by hand [28]. As described in more detail in our 2008 publication, we wished to combine three legacy JSim models of various aspect of cardiovascular circulation. In particular, we first merged a lumped-parameter model of human cardiovascular circulation (the CV model) with a baroreceptor model that controls heart rate based on aortic blood pressure (the BARO model). We then merged this CV+BARO system with a vascular smooth muscle model (VSM) that alters systemic arteriolar resistance based on intra-cellular calcium dynamics. The goal was to produce a richer, more detailed model that accounts for both heart rate changes (via BARO) and blood pressure changes resulting from calcium fluxes within smooth muscle cells. Unlike any of the individual models, the merged model is multi-scale, and covers a broader range of physiological processes.
In 2008, we developed some initial semantic annotations, and then combined these three models by hand, producing a merged biosimulation model that allows researchers to explore the effects of cellular level processes on heart rate and blood pressure. In this paper, we used our extended composite annotations, and the annotator tool of SemGen to annotate the three models against four reference ontologies: the FMA, the OPB, ChEBI, and the NCI Thesaurus. Next, we used SemGen’s Merger tool to automatically merge the CV, BARO and VSM models.
Our SemGen merger tool works by comparing the semantics of the annotations across a pair of SemSim models. Thus, for our example, we first merged CV and BARO, and then merged the resulting “CV+BARO” model with the VSM model. The SemGen merger tool (a) displays information about the two models in two color-coded panels, (b) provides a set of “suggested” merges where the semantics are similar or identical, and (c) allows the user to create manual connections between any two variables from the two models (this design is partially inspired by the PROMPT tool for merging ontologies[29]). Figure 6 shows the suggested merge points for the first part of our demonstration, when comparing CV and BARO. These are all exact and correct matches—after choosing which data structure (and variable name) to preserve in the merged model, we proceeded to automatically merge the code for these two models.
Fig 6.
A screen from the SemGen merger tool, showing three suggested matches between variables in the CV model and those in the BARO model.
For the second part, when combining CV+BARO with VSM, there are no exact semantic matches between the model variables. To correctly merge the models, the user must choose to make equivalent two variables across these models: “Resistance of systemic arteries and capillaries” from CV+BARO and “Resistance of systemic arterioles” from VSM. This choice is an example of how the model merging process will remain only semi-automatic—individual researchers must make subjective decisions about when and where it is appropriate to link models.
At the end of the process, we found that the semi-automatically merged system generated by SemGen reproduced the numerical results of the original hand-merged model (from our 2008 work). Figure 7 shows a trace of the running, merged biosimulation model, showing aortic blood pressure, the variable we have discussed throughout this paper, under two different conditions of calcium concentration.
Fig 7.

A trace of the aortic blood pressure under two conditions—with and without a stimulated level of calcium in the smooth muscle of the arterioles.
5. Discussion
In this paper, we have developed the idea of composite annotations, and demonstrated their value for biosimulation models. Composite annotations are a theoretical construct that can be implemented in a variety of syntaxes wherever multiple ontologies are needed for annotation. In addition to the theory, we have also demonstrated pragmatic use of composite annotations, via the SemGen toolset with a specific example task of merging three biosimulation models. In this final section, we discuss some open challenges and opportunities for (a) biosimulation model sharing and integration, and (b) the use of semantic-web style inferential capabilities to further leverage collections of composite annotations.
5.1 Knowledge sharing
Broadly speaking, sharing and reuse works only if members of the community see a benefit to sharing, and the community has a common understanding of the problem being tackled. For biosimulation research, we claim that there is ample evidence that the community sees a strong benefit to model sharing and reuse. As we have described, researchers have already built numerous model libraries, and are already sharing and reuse model code, in spite of the high cost and challenge of understanding and adapting code that is poorly or incompletely annotated. In addition, there are initiatives, such as MIRIAM, to improve the quality and completeness of current annotations. As another example, a group of CellML researchers have recently published an effort to improve the scope and semantics of CellML model annotations.[30]
However, for sharing to work, researchers must also share a common understanding of some ground truth to which different models can be compared and then shared. For biosimulation research, these truths are best specified by ontologies that capture the underlying physical and mathematical theory for biosimulation modeling. Unfortunately, developing consensus on a set of orthogonal reference ontologies for biological entities and biological processes remains a significant challenge. This challenge is well-acknowledged, and a number of current efforts aim to tackle this problem (e.g., MIREOT for combining ontologies[31], and the work of BioPortal to encourage ontology mappings[26].)
An underlying assumption of our work is that researchers can come to a consensus on a set of “sufficient” ontologies and/or that ontologists can work together to appropriately connect that set of ontologies together. For example, as shown in Figure 2, PATO, the “phenotypic quality” ontology[32] overlaps in part with the OPB, and both can currently be used to annotate physical properties such as pressure. If one set of annotators use PATO, while another group uses OPB, then it will be challenging to merge models or share knowledge across these two groups. In this particular case, we have recently begun a collaboration with the developers of PATO to address this problem head-on: We will either define a set of mappings or linkages to connect terms across the two ontologies, or to partition the entities of interest across the two sources, so that they are more orthogonal or at least compatible.
If developing consensus for the theory and ontologies of physical entities and properties is hard, then capturing an appropriate theory for processes and the equations in biosimulation models may be even harder. The composite annotation schema we have described is for model variables, and is not sufficient for annotating the model equations that capture mathematical relationships between variables. Of course, variables must be annotated before equations, since a critical aspect to capture is which model variables play which roles in a particular equation. In the bio-chemical realm, the Systems Biology Ontology[33] has done a fairly comprehensive job of characterizing many rate equations for chemical kinetics (e.g. Michaelis–Menten equations, etc.). This ontology provides a good start, but it does not by itself indicate how to associate particular variables in a particular model with the variables in a specific equation.
5.2 Semantic web technologies
Our work to date also leaves open a number of interesting challenges for semantic web knowledge representation and inference. As described in section 4, our current tool only suggests exact semantic matches, where all parts of the composite annotation match. However, if the reference ontologies include sufficient information, an inference system or a set of rules could also suggest potential matches as being relevant to the merge process. For example, the FMA includes a rich variety of partonomy information about anatomic entities. If two composite annotations both refer to the FMA, and there is a part_of relationship between those entities, then the SemGen merger tool could suggest this as an additional, partial match. In fact, the merger tool could be improved to use a variety of information present in reference ontologies to suggest potential matches. For example, it could also use the basic “subclass-of” information in a hierarchical organization to suggest matches, in the case where one annotation may be more specific than another.
In a similar manner, the SemGen annotator tool would be improved if it could constrain the construction of composite annotations to follow a set of rules. For example, if the property selected is “fluid pressure” (from the OPB), then we could use inference to only allow for the selection of a physical entity that is a bodily fluid. Likewise, once that fluid is selected, and the “contained-in” relation is selected, then we could check to make sure the next entity is a lumen or container (spaces that provide boundaries for fluids). These sorts of capabilities require some level of inference. Our choice of OWL as a representation language is partially due to the availability of inference engines that might enable such “smarter” annotation and in turn, more automated model merging.
We view composite annotations themselves as a reusable resource. For example, different biosimulation models may contain the same notion of “aortic blood pressure.” Rather than require an annotator to repeat the construction of a composite annotation for this concept, we envision a system that allows annotators to search for and retrieve composite annotations for reuse. This kind of reuse facilitates a modular approach to biosimulation design. For example, a researcher may want to link their model with a model that simulates aortic blood pressure. With reusable composite annotations, they will be able to search for all models that contain aortic blood pressure, identify those models that are at their desired level of granularity and ultimately merge one of the models with their own. To facilitate such searches, annotations themselves need to be viewed as first-class entities that can be indexed and reused. Thus, each composite annotation will need a Unique Resource Identifier (URI), following standards such as provided by MIRIAM[19].
As described by the OBO foundry, the vision of orthogonal ontologies is that they can then be interoperable, so that users can access multiple ontologies without conflict. The work we have presented here provides some theoretical details and an concrete example of how to achieve this vision, where researchers can use multiple ontologies in a scalable, computable manner. The theory is our description or grammar for composite annotations. For our example, we first developed a tool, SemGen, for creating these composite annotations for biosimulation models and then merging such models. Finally, we demonstrated the potential value of both composite annotations and the use of multiple reference ontologies, by showing how they can assist with a specific task of biosimulation model merging.
Acknowledgments
This work was partially funded by the American Heart Association, and by NIH grants #R01 HL087706-01 and #T15 LM007442-06. We thank our reviewers for many constructive ideas that improved the manuscript. We also thank Todd Detwiler for considerable help with implementation of queries from SemGen to reference ontologies.
Footnotes
The authors of this resource use the title “BioModels Database”. However, we prefer using a more expansive word such as “library” or “repository”, as we are not concerned with database issues such as retrieval efficiency. In addition, the BioModels organization provides a suite of additional functionality beyond that of databases, such as easy browsing, programmatic web services, and curation services.
As defined by the Basic Formal Ontology, the physical property (a type of quality) must inhere in the first physical entity.
SemGen is a research prototype tool, under constant modification and development. It is not yet available for downloading by the wider research community, although this is certainly our goal.
References
- 1.Smith B, Ashburner M, et al. The OBO Foundry: coordinated evolution of ontologies to support biomedical data integration. Nat Biotechnol. 2007;25(11):1251–5. doi: 10.1038/nbt1346. http://obofoundry.org/ [DOI] [PMC free article] [PubMed]
- 2.Neal ML, Kerckhoffs RC. Current progress in patient-specific modeling. Briefings in Bioinformatics. 2010;11:111–126. doi: 10.1093/bib/bbp049. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Le Novere N, Bornstein B, et al. BioModels Database: a free, centralized database of curated, published, quantitative kinetic models of biochemical and cellular systems. Nucleic Acids Res. 2006;34(Database issue):D689–91. doi: 10.1093/nar/gkj092. http://www.ebi.ac.uk/biomodels/ [DOI] [PMC free article] [PubMed]
- 4.Lloyd CM, Lawson JR, Hunter PJ, Nielsen PF. The CellML Model Repository. Bioinformatics. 2008;24(18):2122–3. doi: 10.1093/bioinformatics/btn390. http://www.cellml.org/models. [DOI] [PubMed]
- 5.JSim Model Archive. http://physiome.org/jsim/models/
- 6.Hunter PJ, Borg TK. Integration from proteins to organs: the Physiome Project. Nat Rev Mol Cell Biol. 2003;4(3):237–43. doi: 10.1038/nrm1054. [DOI] [PubMed] [Google Scholar]
- 7.Virtual Physiological Human - Network of Excellence home page. http://www.vph-noe.eu/home/
- 8.de Matos P, Alcantara R, et al. Chemical Entities of Biological Interest: an update. Nucleic Acids Res. 2010;38(Database issue):D249–54. doi: 10.1093/nar/gkp886. http://www.ebi.ac.uk/chebi/ [DOI] [PMC free article] [PubMed]
- 9.The Universal Protein Resource (UniProt) Nucleic Acids Res, 2010. 2010;38(Database issue):D142–8. doi: 10.1093/nar/gkp846. http://www.uniprot.org. [DOI] [PMC free article] [PubMed]
- 10.Ashburner M, Ball CA, et al. Gene ontology: tool for the unification of biology. The Gene Ontology Consortium. Nat Genet. 2000;25(1):25–9. doi: 10.1038/75556. http://www.geneontology.org/ [DOI] [PMC free article] [PubMed]
- 11.Kanehisa M, Goto S. KEGG: kyoto encyclopedia of genes and genomes. Nucleic Acids Res. 2000;28(1):27–30. doi: 10.1093/nar/28.1.27. http://www.genome.ad.jp/kegg/ [DOI] [PMC free article] [PubMed]
- 12.Rosse C, Mejino JLV. A Reference Ontology for Bioinformatics: The Foundational Model of Anatomy. Journal of Biomedical Informatics. 2003;36:478–500. doi: 10.1016/j.jbi.2003.11.007. [DOI] [PubMed] [Google Scholar]
- 13.Cook DL, Mejino JV, Jr, Neal ML, Gennari JH. Bridging Biological Ontologies and Biosimulation: The Ontology of Physics for Biology. AMIA Annual Symposium; 2008; Washington, D.C. [PMC free article] [PubMed] [Google Scholar]
- 14.Washington NL, Haendel MA, et al. Linking human diseases to animal models using ontology-based phenotype annotation. PLoS Biol. 2009;7(11):e1000247. doi: 10.1371/journal.pbio.1000247. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Basic Formal Ontology (BFO) http://www.ifomis.org/bfo.
- 16.Neal ML, Gennari JH, Arts T, Cook DL. Advances in semantic representation for multiscale biosimulation: A case study in merging models. Pac Symp Biocomput. 2009 [PMC free article] [PubMed] [Google Scholar]
- 17.Sauro H, Butterworth E. Personal communication. 2009.
- 18.Kerckhoffs RC, Neal ML, et al. Coupling of a 3D finite element model of cardiac ventricular mechanics to lumped systems models of the systemic and pulmonic circulation. Ann Biomed Eng. 2007;35(1):1–18. doi: 10.1007/s10439-006-9212-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Le Novere N, Finney A, et al. Minimum information requested in the annotation of biochemical models (MIRIAM) Nat Biotechnol. 2005;23(12):1509–15. doi: 10.1038/nbt1156. [DOI] [PubMed] [Google Scholar]
- 20.Krause F, Uhlendorf J, et al. Annotation and merging of SBML models with semanticSBML. Bioinformatics. 2010;26(3):421–2. doi: 10.1093/bioinformatics/btp642. http://www.semanticsbml.org. [DOI] [PubMed]
- 21.Lister A, Pocock M, Taschuk M, Wipat A. Saint: A Lightweight Integration Environment for Model. Bioinformatics. doi: 10.1093/bioinformatics/btp523. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Albert MA, Haanstra JR, et al. Experimental and in silico analyses of glycolytic flux control in bloodstream form Trypanosoma brucei. J Biol Chem. 2005;280(31):28306–15. doi: 10.1074/jbc.M502403200. BioModels link: http://www.ebi.ac.uk/biomodels-main/BIOMD0000000211. [DOI] [PubMed]
- 23.OBO Relationship Ontology. http://www.obofoundry.org/ro/
- 24.Turtle: Terse RDF Triple Language. http://www.w3.org/TeamSubmission/turtle/
- 25.BioModels Database: A Database of Annotated Published Models. http://www.ebi.ac.uk/biomodels/
- 26.NCBO BioPortal. http://bioportal.bioontology.org/
- 27.Noy N. Personal communication.
- 28.Gennari JH, Neal ML, Carlson BE, Cook DL. Integration of multi-scale biosimulation models via light-weight semantics. Pac Symp Biocomput. 2008;13:414–25. [PMC free article] [PubMed] [Google Scholar]
- 29.Noy NF, Musen MA. The PROMPT suite: Interactive tools for ontology merging and mapping. Int J of Human-Computer Studies. 2003;59(6):983–1024. [Google Scholar]
- 30.Wimalaratne SM, Halstead MDB, et al. Biophysical annotation and representation of CellML models. Bioinformatics. 2009;25(17):2263. doi: 10.1093/bioinformatics/btp391. [DOI] [PubMed] [Google Scholar]
- 31.Courtot M, Gibson F, et al. MIREOT: The Minimum Information to Reference an External Ontology Term. Proceedings of the International Conference on Biomedical Ontology; 2009. pp. 87–90. [Google Scholar]
- 32.Gkoutos GV, Green EC, et al. Using ontologies to describe mouse phenotypes. Genome Biol. 2005;6(1):R8. doi: 10.1186/gb-2004-6-1-r8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Le Novere N, Courtot M, Laibe C. Adding Semantics in Kinetics Models of Biochemical Pathways. 2nd International ESCEC Symposium on Experimental Standard Conditions on Enzyme Characterizations; 2006. http://www.ebi.ac.uk/sbo/ [Google Scholar]