Abstract
The neighbor-exclusion principle is one of the most general and interesting rules describing intercalative DNA binding by small molecules. It suggests that such binding can only occur at every other base-pair site, reflecting a very large negative cooperativity in the binding process. We have carried out molecular mechanics and molecular dynamics simulations to study intercalation complexes between 9-amino acridine and the base-paired heptanucleotide d(CGCGCGC) X d(GCGCGCG), in which the neighbor-exclusion principle was both obeyed and violated. Our studies find no stereochemical preference that favors the neighbor-exclusion-obeying structures over the neighbor-exclusion-violating structures. Alternative explanations for the existence of the neighbor-exclusion principle are vibrational entropy effects that we calculate to favor the more flexible neighbor-exclusion models over the more rigid neighbor-exclusion-violating models and polyelectrolyte (counterion release) effects.
Full text
PDF
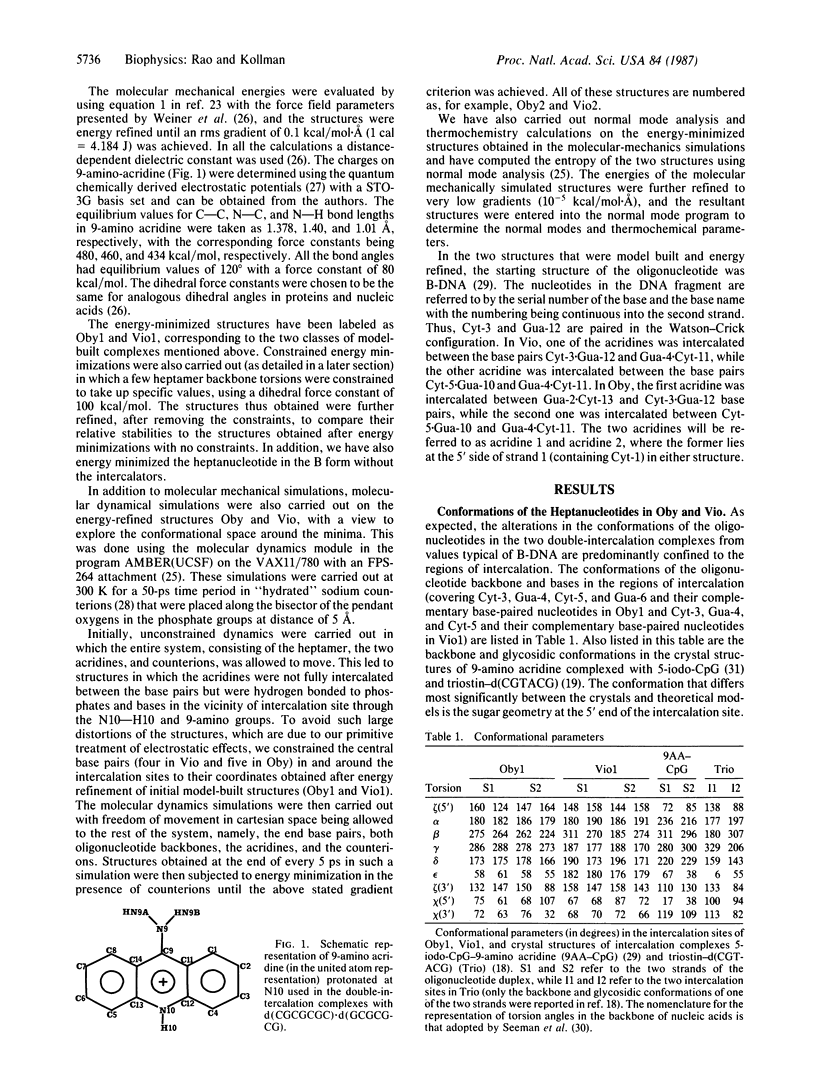
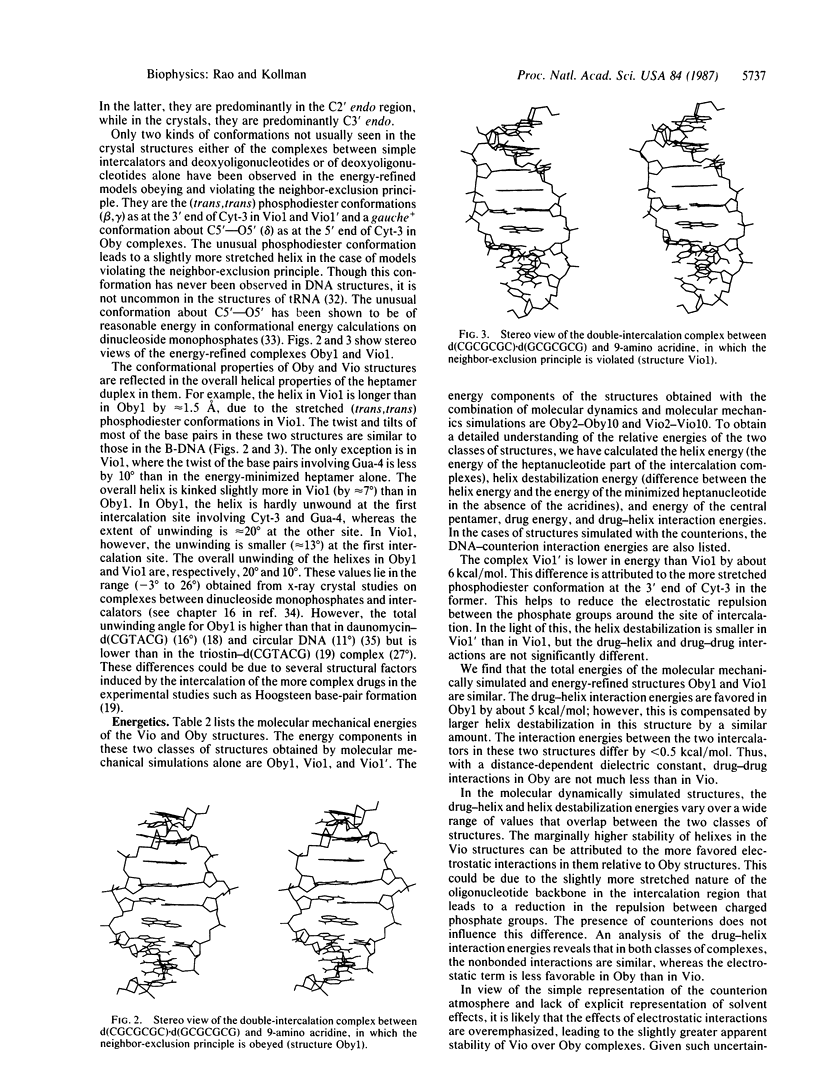


Selected References
These references are in PubMed. This may not be the complete list of references from this article.
- Alden C. J., Arnott S. Stereochemical model for proflavin intercalation in A-DNA. Nucleic Acids Res. 1977 Nov;4(11):3855–3861. doi: 10.1093/nar/4.11.3855. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Alden C. J., Arnott S. Visualization of planar drug intercalations in B-DNA. Nucleic Acids Res. 1975 Oct;2(10):1701–1717. doi: 10.1093/nar/2.10.1701. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Arnott S., Bond P. J., Chandrasekaran R. Visualization of an unwound DNA duplex. Nature. 1980 Oct 9;287(5782):561–563. doi: 10.1038/287561a0. [DOI] [PubMed] [Google Scholar]
- Assa-Munt N., Denny W. A., Leupin W., Kearns D. R. 1H NMR study of the binding of Bis(acridines) to d(AT)5.d(AT)5. 1. Mode of binding. Biochemistry. 1985 Mar 12;24(6):1441–1449. doi: 10.1021/bi00327a024. [DOI] [PubMed] [Google Scholar]
- Assa-Munt N., Leupin W., Denny W. A., Kearns D. R. 1H NMR study of the binding of bis(acridines) to d(AT)5.d(AT)5. 2. Dynamic aspects. Biochemistry. 1985 Mar 12;24(6):1449–1460. doi: 10.1021/bi00327a025. [DOI] [PubMed] [Google Scholar]
- Bond P. J., Langridge R., Jennette K. W., Lippard S. J. X-ray fiber diffraction evidence for neighbor exclusion binding of a platinum metallointercalation reagent to DNA. Proc Natl Acad Sci U S A. 1975 Dec;72(12):4825–4829. doi: 10.1073/pnas.72.12.4825. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Crothers D. M. Calculation of binding isotherms for heterogenous polymers. Biopolymers. 1968 Apr;6(4):575–584. doi: 10.1002/bip.1968.360060411. [DOI] [PubMed] [Google Scholar]
- Denny W. A., Atwell G. J., Baguley B. C., Wakelin L. P. Potential antitumor agents. 44. Synthesis and antitumor activity of new classes of diacridines: importance of linker chain rigidity for DNA binding kinetics and biological activity. J Med Chem. 1985 Nov;28(11):1568–1574. doi: 10.1021/jm00149a005. [DOI] [PubMed] [Google Scholar]
- Friedman R. A., Manning G. S. Polyelectrolyte effects on site-binding equilibria with application to the intercalation of drugs into DNA. Biopolymers. 1984 Dec;23(12):2671–2714. doi: 10.1002/bip.360231202. [DOI] [PubMed] [Google Scholar]
- Jain S. C., Tsai C. C., Sobell H. M. Visualization of drug-nucleic acid interactions at atomic resolution. II. Structure of an ethidium/dinucleoside monophosphate crystalline complex, ethidium:5-iodocytidylyl (3'-5') guanosine. J Mol Biol. 1977 Aug 15;114(3):317–331. doi: 10.1016/0022-2836(77)90253-4. [DOI] [PubMed] [Google Scholar]
- Miller K. J., Pycior J. F. Interaction of molecules with nucleic acids. II. Two pairs of families of intercalation sites, unwinding angles, and the neighbor-exclusion principle. Biopolymers. 1979 Nov;18(11):2683–2719. doi: 10.1002/bip.1979.360181105. [DOI] [PubMed] [Google Scholar]
- Neidle S., Achari A., Taylor G. L., Berman H. M., Carrell H. L., Glusker J. P., Stallings W. C. Structure of a dinucleoside phosphate--drug complex as model for nucleic acid--drug interaction. Nature. 1977 Sep 22;269(5626):304–307. doi: 10.1038/269304a0. [DOI] [PubMed] [Google Scholar]
- Nelson J. W., Tinoco I., Jr Intercalation of ethidium ion into DNA and RNA oligonucleotides. Biopolymers. 1984 Feb;23(2):213–233. doi: 10.1002/bip.360230205. [DOI] [PubMed] [Google Scholar]
- Quigley G. J., Wang A. H., Ughetto G., van der Marel G., van Boom J. H., Rich A. Molecular structure of an anticancer drug-DNA complex: daunomycin plus d(CpGpTpApCpG). Proc Natl Acad Sci U S A. 1980 Dec;77(12):7204–7208. doi: 10.1073/pnas.77.12.7204. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sakore T. D., Reddy B. S., Sobell H. M. Visualization of drug-nucleic acid interactions at atomic resolution. IV. Structure of an aminoacridine--dinucleoside monophosphate crystalline complex, 9-aminoacridine--5-iodocytidylyl (3'--5') guanosine. J Mol Biol. 1979 Dec 25;135(4):763–785. doi: 10.1016/0022-2836(79)90512-6. [DOI] [PubMed] [Google Scholar]
- Seeman N. C., Rosenberg J. M., Suddath F. L., Kim J. J., Rich A. RNA double-helical fragments at atomic resolution. I. The crystal and molecular structure of sodium adenylyl-3',5'-uridine hexahydrate. J Mol Biol. 1976 Jun 14;104(1):109–144. doi: 10.1016/0022-2836(76)90005-x. [DOI] [PubMed] [Google Scholar]
- Singh U. C., Weiner S. J., Kollman P. Molecular dynamics simulations of d(C-G-C-G-A) X d(T-C-G-C-G) with and without "hydrated" counterions. Proc Natl Acad Sci U S A. 1985 Feb;82(3):755–759. doi: 10.1073/pnas.82.3.755. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sobell H. M., Tsai C. C., Jain S. C., Gilbert S. G. Visualization of drug-nucleic acid interactions at atomic resolution. III. Unifying structural concepts in understanding drug-DNA interactions and their broader implications in understanding protein-DNA interactions. J Mol Biol. 1977 Aug 15;114(3):333–365. doi: 10.1016/0022-2836(77)90254-6. [DOI] [PubMed] [Google Scholar]
- Tsai C. C., Jain S. C., Sobell H. M. Visualization of drug-nucleic acid interactions at atomic resolution. I. Structure of an ethidium/dinucleoside monophosphate crystalline complex, ethidium:5-iodouridylyl (3'-5') adenosine. J Mol Biol. 1977 Aug 15;114(3):301–315. doi: 10.1016/0022-2836(77)90252-2. [DOI] [PubMed] [Google Scholar]
- Wakelin L. P., Romanos M., Chen T. K., Glaubiger D., Canellakis E. S., Waring M. J. Structural limitations on the bifunctional intercalation of diacridines into DNA. Biochemistry. 1978 Nov 14;17(23):5057–5063. doi: 10.1021/bi00616a031. [DOI] [PubMed] [Google Scholar]
- Wang A. H., Ughetto G., Quigley G. J., Hakoshima T., van der Marel G. A., van Boom J. H., Rich A. The molecular structure of a DNA-triostin A complex. Science. 1984 Sep 14;225(4667):1115–1121. doi: 10.1126/science.6474168. [DOI] [PubMed] [Google Scholar]
- Wang J. C. The degree of unwinding of the DNA helix by ethidium. I. Titration of twisted PM2 DNA molecules in alkaline cesium chloride density gradients. J Mol Biol. 1974 Nov 15;89(4):783–801. doi: 10.1016/0022-2836(74)90053-9. [DOI] [PubMed] [Google Scholar]



