Abstract
T-cell vaccination may prevent or treat cancer and infectious diseases, but further progress is required to increase clinical efficacy. Step-by-step improvements of T-cell vaccination in phase I/II clinical studies combined with very detailed analysis of T-cell responses at the single cell level are the strategy of choice for the identification of the most promising vaccine candidates for testing in subsequent large-scale phase III clinical trials. Major aims are to fully identify the most efficient T-cells in anticancer therapy, to characterize their TCRs, and to pinpoint the mechanisms of T-cell recruitment and function in well-defined clinical situations. Here we discuss novel strategies for the assessment of human T-cell responses, revealing in part unprecedented insight into T-cell biology and novel structural principles that govern TCR-pMHC recognition. Together, the described approaches advance our knowledge of T-cell mediated-protection from human diseases.
1. Introduction
Despite major advancements in the fields of molecular and cellular biology, and the improved understanding of cancer formation and progression, the surgical removal of tumors remains the most effective therapeutic strategy against cancer. While radiation therapy and chemotoxic drugs are often employed to successfully prolong the disease-free survival or to slow down tumor progression, their limited specificity in targeting neoplastic cells is often responsible for a wide spectrum of common clinical side effects. In this respect, immunotherapy is a promising therapeutic alternative to avoid such side effects by activating the patient's own immune system against tumor cells. In this Paper we focused on selected aspects of current vaccination strategies against melanoma, as well as new and sophisticated tools employed by immunologists to analyze cellular immune responses by “zooming in” on single tumor-specific lymphocytes.
1.1. Melanoma
Melanoma arises from the pigment-producing melanocytes. It is the major cause of mortality among skin malignancies. The incidence is steadily increasing at rates over 3% per year, with many hundreds of new cases per 100 000 inhabitants, and mortality rates ranging from 0.1 to >10% [1]. These figures, however, differ largely depending on risk factors such as sun exposure depending on the local climate. Melanoma is one of the most antigenic and immunogenic cancers with a high percentage of tumors expressing well-characterized tumor-associated antigens. Immunotherapy targeting one or several of these tumor-expressed antigens has shown promising results over the past years in enhancing antitumor immune responses. It is now well established that spontaneous tumor antigen-specific T-cell responses are generated in melanoma patients that can be detected both in the circulation as well as at the tumor sites. While spontaneous T-cell responses have been reported against cancer-germline antigens encoded by the MAGE family, and against NY-ESO-1, T-cell frequencies are generally very low (10−7 to 10−4) [2]. An exception to this is the natural immune response observed in most melanoma patients against the Melan-A/MART1 differentiation antigen presented in the context of the MHC class I molecule HLA-A2. In untreated patients, frequencies of T-cells specific for this antigen usually range from 0.01% to 0.1% of total circulating CD8pos cells [3, 4]. These frequencies are often much higher in metastatic lymph nodes and other metastases of melanoma patients [5, 6]. In fact unusually high frequencies (10−3 to 10−4) of A2/Melan-A-specific T lymphocytes are already found in the blood of newborns and healthy A2+ individuals. This population of self-peptide specific T-cells is preferentially selected in the thymus, presumably due to cross reactivity to unknown self-peptides. Consequently, large numbers of such cells are released from the thymus into the periphery as mature, naive precursor T-cells [7, 8]. Thus, the activation and expansion of this population of Melan-A-specific CD8 T lymphocytes to induce clinically relevant tumor cell lysis represents an important target of immunotherapy.
1.2. Therapeutic Immunization Strategies
The aim of an efficient cancer vaccine is to activate de novo the immune system against tumor cells and/or to enhance the preexisting tumor-specific response. An ideal vaccine would induce expansion of large populations of cytotoxic T-cells, with potent antitumor effector functions, both at the tumor-site but also as a systemic immune surveillance for long periods of time. The choice of adjuvants and antigenic peptides used, their combination, and timing are important factors. Currently there are three major approaches of immunotherapy: antigen-based vaccines, adoptive cell transfer of efficient antitumor T-cells, and stimulation of the immune system in an antigen-nonspecific manner.
Optimal vaccines consist of live or attenuated microbes. However, for many infectious diseases, and for cancers in general, such vaccines are not available. Therefore, synthetic vaccines are developed generally following the rational-based microbe-induced immune mechanisms. Synthetic vaccines are composed of at least two basic components: antigen and adjuvant. The rational of using antigens for cancer immunotherapy is based on the relatively large consensus that immune protection against malignant disease requires antigen-specific (adaptive) immune responses including T-cells. Some experts argue that stimulation of the innate immune system alone may be sufficient to generate tumor-specific immunity, since cancer tissue often produces tumor antigens allowing some activation of antigen-specific immune responses. Therefore, an increasing number of novel immune therapies are developed without taking advantage of (synthetic or recombinant) tumor antigens, essentially because this approach simplifies drug production and application. However, tumor cells produce only low amounts of antigen, which is often not present at the optimal location and/or time. Therefore, immune responses triggered by naturally expressed antigens are not sufficiently timed, neither strong nor anatomically focused to protect from tumor progression. In addition, immunotherapy without antigen often requires high and in part toxic drug doses, in contrast to vaccines containing synthetic antigens that can have powerful biological effects even at low doses. For these reasons we propose that synthetic cancer vaccines should include tumor antigens.
Besides antigen, the second essential vaccine component is the so-called immunological “adjuvant.” Adjuvants are immune stimulating agents, which are important because immune responses remain poor when antigens are administered alone. For many years, adjuvants have been developed empirically, without significant progress in the understanding of their molecular nature and mechanisms of action. The discovery of dendritic cells (DCs) and of their central role in linking innate with adaptive immune responses was key for progress. Besides regulating central mechanisms of the innate immune system, DCs are the most effective antigen-presenting cells for enabling antigen-specific T- and B-cell responses. But how are they put in action? Only about 15 years ago it was discovered that DCs become activated due to triggering of pathogen recognition receptors (PRRs). These receptors enable the innate immune system to sense microbes. The best-characterized families of PRRs are the Toll-like receptors (TLRs) that bind microbial products [9–12]. Drugs that stimulate TLRs are promising adjuvants, particularly CpGs that bind to TLR9. CpGs are oligonucleotides containing CpG motifs similar to those observed in bacterial DNA. Our group has shown that vaccination with emulsions with IFA (Incomplete Freund's Adjuvant), containing antigenic peptides and CpG, rapidly induces strong human CD8pos T-cell responses (0.5%–8% of circulating CD8pos T-cells) in melanoma patients [13]. Moreover, it is the first synthetic vaccine formulation to consistently induce ex vivo detectable T-cell responses even when using a natural tumor peptide, that is, the natural Melan-A/MART-1 sequence [14].
A novel concept of lymphodepletion was recently introduced in the field of immunotherapy, with the aim of making “immunological space” and freeing up access to cytokines such as IL-7 and IL-15 for the tumor-specific T-cells to optimally expand. The rational for this is based on observations made from viral systems indicating that protective immune responses require large frequencies of antigen-specific T-cells. Recently, Rosenberg and colleagues [15, 16] have shown promising results with adoptive cell transfer of autologous melanoma-specific T-cells, combined with high-dose IL-2, in late stage melanoma patients, following transient lymphodepleting chemotherapy regimen. Although highly successful (objective antitumor response of 50% for stage IV melanoma), such a treatment is not easily available for a large number of patients, due to the requirement of highly specialized laboratories to support the delicate procedures of isolation and in vitro expansion of autologous T-cells, and the intensive care units necessary for the clinical management of the side effects caused by high-dose IL-2.
1.3. Insight into the Efficiency of a Vaccine Requires Detailed Analysis of the Immune Response
An increase in the number and frequency of antigen-specific T lymphocytes is often observed following peptide vaccination alone, thus achieving the first aim of candidate cancer vaccines. Unfortunately, the majority of these patients still experience tumor progression, despite increased numbers of tumor-specific T lymphocytes. It is believed that in the small number of patients showing disease stabilization or even tumor regression following immunotherapy, tumor-specific T-cells may succeed in infiltrating tumor tissue, destroying some of the tumor cells and temporarily reversing the local immunosuppressive environment. They would thus act like a spark for the activation, proliferation, and antitumor effector function of already infiltrated or newly activated tumor-specific T-cells.
It can thus be implied that a successful vaccine may not solely depend on the induction of large numbers of tumor-specific T lymphocytes but rather on the activation of a population of effector T-cells with potent antitumor activities and resistance to the local immunosuppressive environment (reviewed in [2]). In order to improve the clinical efficacy of such vaccines, a better characterization of the tumor-specific immune responses is required. Specifically each vaccine-induced immune response should be analyzed in terms of phenotype, function, selection of specific T-cells from the repertoire, and their affinity/avidity for the MHC-peptide complex. Importantly, one also needs to examine the changes and the degree of stability of these parameters over time and following repeated rounds of vaccination. Furthermore, a better understanding of the mechanisms that allow long-lasting memory T-cell survival and of strategies to increase T-cell migration and effector functions at the tumor site is needed.
1.4. Cancer versus Infection Models
From an immunological perspective, tumors can be compared with persistent viral infections, since in both situations T-cells may successfully protect from disease. The constant presence of antigen stimulates the immune system by continuously triggering antigen-specific T-cells. The major difference between cancer and infection is that, in the latter, the immune response is highly efficient in controlling the virus and in providing long-lasting protection. Contrarily, antitumor responses show less robustness compared with antiviral responses. Some of the explanations for this may be (i) immune tolerance against self-antigens, (ii) local immune suppression induced by tumors, and (iii) tumor escape mechanisms which render the existing immune responses less efficient. Nevertheless, chronic infections with genetically stable viruses such as Epstein-Barr virus (EBV) and cytomegalovirus (CMV) provide useful model situations for studying the generation and maintenance of highly efficient T-cell memory responses and for identification of T-cell correlates of protection in humans.
We divided this paper into six sections, each describing a particular aspect of current methodological approaches and technical tools used to dissect antigen-specific CD8 T-cell responses. Our strategy combining both ex vivo and in vitro analyses, and utilizing molecular and cellular approaches is schematically depicted in Figure 1. Each major component of this outline corresponds to one section of the text, thus providing a guideline for the reader throughout this paper. The observed similarities and differences between viral- and tumor-specific T-cell responses will be discussed, since they not only reveal the progress of novel vaccine formulations against cancers like melanoma, but also the obstacles which need to be overcome in order to further improve clinical efficacy.
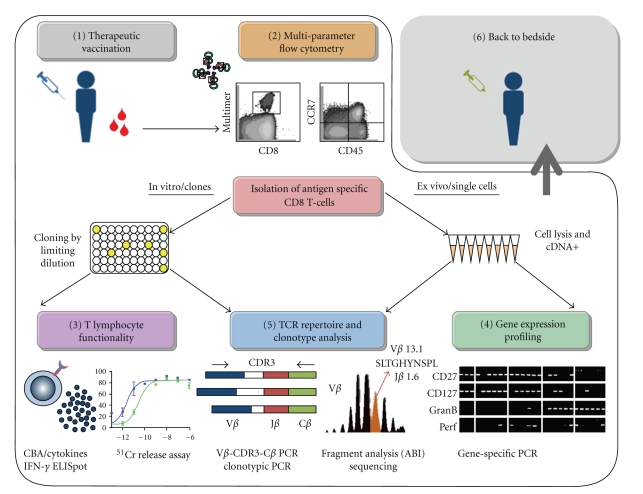
Figure 1.
Cellular and molecular approaches for the characterization of cytotoxic T-cells following therapeutic vaccination. Each step (1 to 6) in the strategy bears the same name and section number as described in the body of text. (1) Patients undergoing therapeutic vaccination are monitored closely throughout the clinical study, and blood and tissue samples are collected at numerous time points. (2) Antigen-specific CD8 T-cell populations are first visualized by the use of fluorescent peptide-MHC multimers then analyzed for their phenotype and subset composition using multiparameter flow cytometry. T-cell populations of interest can subsequently be isolated for further in vitro [3, 5] or ex vivo [4, 5] studies. (3) In vitro generated T-cell clones can be subjected to a series of assays to determine their functionality, including target cell lysis (51C release assay) and cytokine production (ELISpot). (4) Ex vivo sorted single cells are lysed and cDNA purified for gene-expression and TCR repertoire analysis. (5) Single cell samples or individual T-cell clones can be subjected to spectratyping, for the study of TCR repertoire diversity, selection, and clonotype composition. The unique signature of each T-cell can be identified, and its frequency determined. Furthermore, individual T-cell clonotypes can be followed across different T-cell compartments, and over time following therapeutic vaccination. (6) Conclusions drawn from this complete analysis of phenotype and functionality of vaccine-induced immune responses in melanoma patients can be taken back to the bedside. These results can be translated into improved therapeutic vaccination regimens aiming for more powerful immune activation and more efficient and specific antitumor responses.
2. Multiparameter Flow Cytometry Analysis of Antigen-Primed T Lymphocytes
The development of designed fluorescent peptide/MHC multimers (formerly called tetramers), which bind stably to specific TCR molecules on the surface of T-cells [5, 17], has revolutionized the detection of viral- and tumor-specific T-cells. This approach makes it possible to directly carry out subset analysis at the antigen-specific T-cell level ex vivo without preliminary in vitro rounds of antigen stimulation (reviewed in [18]). Multimer technology combined with multiparametric flow cytometry analysis has allowed gaining a better understanding of the T-cell specific immune responses against viruses and tumors. Furthermore, the ex vivo analysis of antigen-specific populations can be accompanied by the isolation of T-cell populations of interest, as well as the use of peptide-MHC multimers as a stimulus in in vitro functional assays. Briefly, PBMCs from vaccinated melanoma patients are first enriched for CD8pos T lymphocytes using anti-CD8-coated magnetic microbeads. Alternatively, T lymphocytes can be extracted from resected metastatic lymph nodes (TILNs) or distant soft/visceral metastases (TILs) [19]. Purified CD8 T-cells are then incubated using PE-labeled HLA-A*0201/peptide multimers [5, 17], in combination with fluorescent antibodies (i) for extracellular differentiation markers (e.g., CCR7, CD45RA, CD28, CD27), (ii) for cytolytic molecules (e.g., perforin, granzyme B), and/or (iii) for cytokines (IFNγ, TNFα) (Figure 1).
2.1. Defining Human CD8 T Lymphocyte Subpopulations
Major efforts have been made in recent years to understand the relationship between different T-cell subpopulations. An important task has been to define molecular markers that readily identify and isolate T-cells sharing discrete stages of cell differentiation. Circulating naive T lymphocytes form a relatively homogenous population expressing a well-defined set of surface glycoproteins and are characterized by the null expression of effector mediators (e.g., IFN-γ, TNF-α, granzyme B, perforin, FAS/CD95) and by proliferative potential (e.g., long telomeres, high detectable levels of TREC copies). During the past decades, primed antigen-experienced T lymphocytes have mostly been classified into two distinct subsets, for example, effector and memory cells [20]. Effectors are presumably rather short-lived, produce cytolytic effector molecules, and are capable of migrating to the site of infection and of killing target cells directly ex vivo. In contrast, memory cells are long lived, persist after pathogen clearance, and have increased survival properties and cell division capacities (reviewed in [21]). In line with this concept, human CD8 T-cells have been delineated with the help of two cell surface markers, the high isoform of the common lymphocyte antigen CD45RA and the chemokine receptor CCR7, and based on their anatomical location [22–24]. Central memory (TCM) T-cells are characterized by the ability to repeatedly circulate into lymph nodes and eventually encounter antigen presented by incoming CCR7pos mature dendritic cells, in contrast to effector memory (TEM) cells which downregulate CCR7 and appear specialized in migrating to peripheral nonlymphoid tissues.
Although this two-marker procedure to identify functionally distinct CD8 T-cell subsets has proven popular, the recent technical improvements in the ability to dissect the immune response using multiparameter flow cytometry have indicated the existence of highly heterogeneous antigen-primed CD8 T subpopulations [25–38]. Importantly, the term of “effector” and “memory” CD8poscell may only apply for situations of acute and resolved infections after which the pathogen is cleared from the host, and not in situations of persistent latent or chronic active infections where it becomes more difficult to use this simplified view of defining primed T-cells (reviewed in [39, 40]). Indeed, in recent years, a variety of other phenotypical and functional markers have been added allowing at present the discrimination of a wide spectrum of antigen-experienced CD8 T-cells with different phenotypes, functions, and half-lives and that may account for the heterogeneity of “memory” and “effector” cells observed following persistent chronic infections (reviewed in [40, 41]).
2.2. Phenotype of Tumor Antigen-Specific T-Cells following Therapeutic Vaccination
Since the fast advancing flow cytometry technology allows us to analyze as many as 20 fluorochromes simultaneously using 7 lasers, the difficult task now becomes the choice of the appropriate extracellular and intracellular markers, which would be the most informative in terms of the phenotypic and functional characteristics. Therefore, our own strategy of monitoring antitumor T-cell responses following peptide vaccination of melanoma patients relies on studies that we perform using well-established surface markers that allow to best correlate a particular phenotype of a T-cell with its function in vivo.
First, we uncovered additional heterogeneity among effector memory and effector CD8 T-cell subsets by studying the functional attributes of such T-cells distinguished on the basis of expression of the costimulatory receptors CD27 and CD28 [32, 34, 42]. Our studies show that these subsets can be subdivided into early differentiated (CCR7−CD45RA−CD27+CD28+) or late differentiated (CCR7−CD45RA+/-CD27+/-CD28−) T-cells. Early differentiated cells express low levels of effector mediators such as granzyme B and perforin and high levels of CD127/IL-7Rα, have a relatively short replicative history (long telomeres, detectable copies of TRECs), and display strong ex vivo telomerase activity. Therefore, these cells are closely related to central memory T-cells (CCR7+CD45RA−CD27+CD28+). Conversely, late differentiated cells have undergone additional rounds of in vivo cell division and share effector-type properties with increased expression of effector mediators and strong ex vivo cytolytic activity (Figure 2). In line with these data, our group more recently reported on the detailed analysis of CD8 T-cell responses specific for EBV and CMV viruses [43]. During chronic infection with EBV and CMV, CD8pos T-cell responses to A2/GLC and A2/NLV antigens, respectively, are composed of heterogeneous populations of T-cells of various differentiation stages, with EBV-specific responses being less differentiated than CMV-specific ones [25–38]. Although the sizes/proportions of these subsets varied between EBV and CMV responses, our recent observations revealed that all subsets, from early differentiated to late differentiated stages, were found in both viral-specific responses [43]. Remarkably, this finding was also observed for CD8 T-cells specific for the influenza matrix protein peptide (Flu-MA), where up to 20% of these cells were composed of effector T-cells, alongside the predominant memory cells [44].
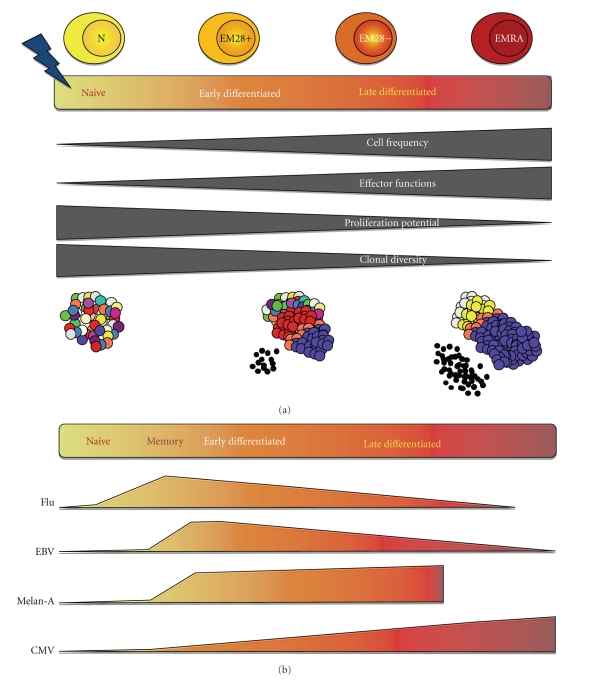
Figure 2.
Model of CD8 T-cell differentiation. (a) The highly diverse naive T-cell compartment contains T-cell clonotypes of very low frequency (represented by one color), and null expression of effector mediators but high potential for proliferation in response to antigen. Following encounter with cognate antigen, CD8 T-cells are activated and begin a process of cellular differentiation characterized by a gradient of early- and late-differentiated cellular states, that can be described by changes within four major parameters as depicted by dark grey blocks. As a result of the massive cellular proliferation that ensues activation, there is an increase in the frequency of T-cells and a restriction in the TCR diversity, since the pool of primed differentiated T-cells is composed of a small number of clonally expanded T-cells (blue, white, yellow, and orange T-cells). Concomitantly, there is a decrease in proliferative potential from naive and early differentiated T-cell subsets towards highly differentiated subsets. Moreover the accumulation of effector functions (high production of effector molecules, small black dots) in these expanded populations of T-cells defines further maturation of differentiated T-cells. (b) The degree of differentiation of T-cells along this spectrum has been shown to vary with antigen specificity. As such, the antigen-specific responses against influenza virus and against persistent EBV and CMV viruses are compared with the tumor-specific response against the Melan-A antigen, while taking into account the four parameters described in (a). EBV-specific cells are more differentiated than influenza-specific cells but less so than CMV-specific cells which are composed primarily of highly differentiated CD8 T-cells. While both early and late differentiated subsets have also been identified in Melan-A specific responses, their proportions are highly variable between different patients following vaccination. In addition, tumor-specific responses contain a much lower fraction of highly differentiated EMRA (effector memory CD45RA+) cells compared with viral-specific responses.
Over the past decade, our group performed extensive work on antitumor responses against melanoma expressed antigens before as well as upon peptide vaccination [3, 13, 45–52], and we showed that vaccination is often found to increase the number of tumor antigen-specific T-cells and to induce their cell differentiation. Phenotypic analysis of Melan-A/MART-1 specific T-cells in vaccinated patients showed similarities to that of virus-specific (EBV and CMV) T-cells, in that they were comprised primarily of effector memory (TEM or EM) cells containing both early (EM CD28+ defined as EM28pos) and late differentiated (EM CD28− defined as EM28neg) T-cells. Compared with the viral system where we observed almost identical phenotypes among individuals (i.e., proportions of EM28+ and EM28− for EBV-specific T-cells), the overall phenotype of Melan-A/MART-1 specific T-cells, specifically the proportion of EM28pos versus EM28neg T-cells, seems to be more variable and to depend on each patient and his/her vaccination history. Nevertheless, peptide vaccination has been shown to drive the Melan-A/MART-1 response towards cell differentiation with progressive upregulation of effector mediators and cytolytic activity. From this point of view, current vaccination strategies emerge as progressively successful in inducing tumor-specific T-cells at high frequencies and with similar phenotypic and functional characteristics as those associated with long-lasting protective responses (Rufer, Speiser, et al., unpublished observations).
2.3. Model of CD8 T-Cell Differentiation
Altogether, these in-depth analyses [32, 34, 45, 48] are in agreement with a model according to which there is a differentiation pathway with the stepwise loss of homing (CCR7), costimulatory (CD27, CD28), and cytokine (IL-7Rα) receptors, as well as concomitant upregulation of molecules involved in cell-cell adhesion and target cell destruction (Figure 2). Importantly, influenza-, EBV-, and CMV-specific T-cells follow the same pathway of cell differentiation, although CMV-specific cells are more frequently late differentiated than influenza- and EBV-specific cells [43, 44].
The pathway of T-cell differentiation described here (Figure 2) appears also to apply to self-/tumor-specific T-cells such as Melan-A/MART-1 and NY-ESO-1 in melanoma patients [45, 48], aside from being found in T-cells specific for persistent viruses [29, 43], in T-cells responding to acute viral infections like influenza [44], and in other types of T-cells (γδpos, CD4pos) [53]. Essentially, following antigen stimulation, less differentiated cells like naive and central memory T-cells expressing CCR7 would initially differentiate into effector memory cells, then ultimately into highly differentiated effector type of cells (Figure 2). Sallusto and colleagues suggested that this differentiation process will depend upon the signal-strength of interaction between the T-cell and the antigen-presenting cell during the initial priming and expansion phase, specifically on factors such as the affinity of the TCR-MHC-peptide interaction, the concentration of antigen, and costimulatory molecules and cytokines [23, 54, 55]. In this model, T-cells that receive a weak signal will be unfit to differentiate and will rapidly die [56]. An intermediate signal or a strong signal that is followed by the clearance of antigen will result in the development of fit memory cells which can eventually give rise to precursors for rapid effector cells generation upon antigen restimulation. When T-cells receive a strong signal, they differentiate towards effectors.
Thus, the presence and concentration of antigen would be one of the main factors determining at what point along this differentiation spectrum, each memory response would be found. In other words, the relative proportions of “memory-like” T-cell-subsets compared with the “effector-like” T-cell subsets would account for the observed differences between various immune responses. Specifically, in the presence of persistent antigen, EBV-specific memory cells will be driven towards an effector memory phenotype. Contrarily, several weeks or months after clearance of pathogens causing short-term acute infections, that is, in absence of antigen, the central memory compartment would be dominant, while a lower fraction of T-cells would exhibit an effector memory phenotype. We also propose that early (memory-like) and late differentiated (effector-like) cells serve important but distinct purposes to the overall antigen-specific immune response and in providing protection against different viruses. Such observations are in line with those made in the mouse model (reviewed in [40]), as it was shown that while immediate protection is better provided by differentiated effector-type T-cells present in peripheral tissues [57], early differentiated (TCM-like) cells are most potent at protecting against systemic infection with lymphocytic choriomeningitis virus and respiratory challenge infections [57–59].
2.4. Recently Improved Strategies to Multiparameter Flow-Cytometry Analysis
The ex vivo characterization of T-cells is limited to T-cell populations with a frequency higher than 0.01% among total CD8 T-cells [3, 4]. This is more problematic for T-cell specific responses of rather low frequencies such as those generated against tumor antigens, in contrast to the relatively frequent viral specific T-cell responses. To address such problems, two groups have recently developed techniques for simultaneous analysis of T-cells with multiple different antigen specificity. The first consists in the combination of 4 fluorochromes for each T-cell specificity instead of a single one, allowing the detection of 15 different specificities in parallel [60]. The other strategy is to increase the number of peptide-MHC multimers labeled with a two-dimensional combination of fluorochromes using six different quantum dots (QD565, QD585, QD605, QD655,QD705, and QD800) and the two most intense fluorochromes PE and APC, which allows the visualization of T-cells with up to 28 different specificities [61]. While both strategies make it possible to simultaneously analyze numerous antigen-specific responses in a single step, they require careful setup of experimental controls. Furthermore, these approaches are currently limited on one hand by the development and application of sophisticated reagents and on the other by the thorough analysis of the data. Nevertheless, aside from decreasing the time of the experimental procedure, one of the most important advantages is that they allow the investigator to save precious material from patients and donors, since the sample does not have to be subdivided for multiple separate analyses [61].
The development of MHC class II multimers has been much less successful compared with MHC class I multimers due to several reasons. First, human MHC class II molecules are highly polymorphic. Second, peptide binding affinity is generally lower for class II as opposed to class I molecules, which is particularly evident for peptides derived from tumors. One of the most successful strategies has proven to be the initial production of HLA-DR molecules loaded with a “placeholder” peptide followed by a peptide exchange step using the peptide of interest. The efficient replacement by the peptide of interest remains one of the major limitations. Recently, Ayyoub and colleagues reported DR52b/NY-ESO-1 multimers using the strategy of His-tagged peptides that allows isolation of folded MHC/peptide monomers by affinity purification before tetramerization [62]. Using these molecules, they could detect ex vivo CD4 T-cell responses to the NY-ESO-1 peptide in patients undergoing a vaccination trial with recombinant NY-ESO-1 protein, Montanide ISA 51 (Seppic, France), and CpG ODN 7909 (Pfizer, USA). The application of this strategy to other tumor- and self-antigen-derived peptides may significantly accelerate the development of reliable MHC class II multimers to monitor antigen-specific CD4 T-cells.
2.5. Advantages and Limitations of the Multiparameter Flow Cytometry Analysis
The major advantage of the multiparameter flow-cytometric analysis is that it allows to ex vivo visualize the antigen-specific pool of lymphocytes and to divide it into numerous subpopulations. As discussed above, antigen-experienced T lymphocytes are highly heterogeneous and are composed of a wide range of discrete subsets. It is thus of high importance to elucidate the roles of these different subsets and to identify how they are regulated. In that regard, in vivo/ex vivo analyses remain the gold standard, and flow cytometry represents an instrumental technology to study antigen-specific T-cell biology ex vivo. Moreover, differences between various T-cell populations within the same response can only be revealed in such studies. On the contrary, the phenotypic analysis of a T-cell response as a whole can lead to false conclusions and biases induced by the predominant subset, while missing the presence of a less frequent yet biologically important population. Multiparameter flow cytometry technology allows the parallel detection of multiple activation markers, such as the expression of cytokines and chemokines by specific populations of cells allowing the fine characterization of particular profiles. Thereby it is possible to determine which parameters define distinct functional subsets in different antigen-specific T-cell contexts. For instance, Makedonas and colleagues recently showed that a polyfunctional response led by IL-2 upregulation was necessary for the control of EBV and the clearance of influenza, while conversely a strong perforin production was rather important in the clearance of CMV and adenovirus infection [63]. Lastly the technology allows to sort each of these populations of interest into various forms (tubes, plates), cell numbers (single cells), and for different purposes (in vitro cloning, ex vivo functional assays, and molecular assays) as will be discussed in the following sections.
3. Assessing Functionality of Antigen-Specific T Lymphocytes
It is evident that the analysis of phenotype of T lymphocytes using multiparameter flow cytometry can reveal a high degree of heterogeneity, especially among the antigen primed T-cell population. However, one should not only judge a book by its cover, not a T-cell solely on its phenotype since a high degree of heterogeneity exists even for T lymphocytes of similar phenotypes or alternatively, similar functions for T lymphocytes of distinct phenotypes. Thus, characterizing the functional capacities of each individual T lymphocyte is crucial to the understanding of their roles in vivo and to designing future strategies to enhance and prolong these functions through therapeutic vaccination. This section will discuss current methods to study T-cell function either directly ex vivo or following expansion in vitro and outline the lessons learned about the functionality of viral and tumor-specific T lymphocytes.
3.1. Current Strategies for Measuring T Lymphocyte Function
Helper CD4 T-cells have been shown to be important in the generation of efficient and long-lasting memory responses; however, cytotoxic CD8 T-cells remain the key effectors in the fight against most viral infections and tumors. The functionality of CD8 T-cells can be studied in vitro or ex vivo, whereby three parameters are often measured: cytotoxicity, cytokine secretion, and proliferation.
The first pathway of cytotoxicity involves the engagement of death receptors (TNF-α and Fas/FasL). The flow-cytometric analysis of the expression of these receptors by various T-cell subsets is possible; however, current in vitro assays are not sufficiently sensitive to correlate this with differences in cytotoxic capacities by the same cells (reviewed in [41]). The second pathway employed by CD8 T-cells is the granzyme-perforin-dependent cytotoxicity. This is a highly complex mechanism, which can be very difficult to assess using currently available approaches and extrapolated to the true capacities of a T-cell in vivo. Specifically, the cytotoxic potential of a T-cell in vivo will depend upon several factors, such as the level of constitutive expression of granzyme and perforin and storage into lytic organelles, the quantity released following activation, and the rate of regeneration of these molecules. These parameters may already differ significantly from cell to cell in vivo and may be further altered following in vitro stimulation [41].
Nevertheless, numerous assays exist to study CD8 T-cell cytolysis, the first being the degree of target cell death induced by CD8 T-cells in an antigen-specific manner. In vitro generated clones expanded in medium supplemented with human serum and recombinant human IL-2 are commonly used in such assays. One of the major advantages of this approach is that the periodic restimulation of these clones using phytohemagglutinin (PHA) and irradiated allogeneic PBMC as feeder cells allows the generation of a large number of cells and for extended periods of time. This opens the possibility for experiments to be repeated, as well as the simultaneous testing of several conditions (antigenic peptide concentrations, effector to target ratios). The classical method of determining the in vitro lytic activity of antigen-specific T-cell clones is through the 4 h 51Cr release assays using antigenic peptide-pulsed T2 target cells (HLA-A2+/TAP−/−) [64]. In the case of tumor-reactive T-cells, melanoma tumor cell lines such as Me 275 and Me 290 (HLA-A2+/Melan-A+) and NA8-MEL (HLA-A2+/Melan-A−) can also be used as target cells in the presence or absence of exogenous Melan-A peptide [8]. The percentage of specific lysis is then calculated in terms of the observed level of 51Cr released compared with the spontaneous and total possible release by the target cells. To quantify the efficiency of T-cell—target cell recognition, results can be expressed in terms of the amount of peptide required to reach 50% of the maximal lysis (EC50). Alternatively the lytic activity of a T-cell can be measured in a CD8 coreceptor independent setting, specifically using C1R target cells transfected with mutant HLA-A2 molecules that abrogate the docking on the CD8 molecule [43, 65]. When these results are compared with the level of lysis using targets expressing HLA-A2 wild type, such assays allow the determination of the degree of CD8 dependency, which can in turn be an indicator of the level of binding avidity of the TCR for the peptide-MHC molecule. While the 51Cr-release assay has several practical advantages, it is not the most sensitive approach to detect small differences in lytic abilities of different clones, particularly since the prior in vitro expansion of T-cells may have dramatically altered their intrinsic cytotoxic machinery.
The alternative approach to study cell-mediated cytotoxicity is through the multiparameter flow cytometry analysis of markers associated with cellular death by the target cell or with the cytolytic potential of the effector T-cell. The most popular approaches for the former are based on the detection of caspase activation, annexin V binding to apoptotic cells, and uptake of propidium iodine (PI) or 7-amino-actinomycin D (7-AAD) by dead or dying target cells (reviewed in [66]). On the other hand, the most commonly used markers to detect potential killer T-cells versus non killers are granzymes A, B, and perforin. To overcome the constraint of having to fix the cells to allow intracellular staining of these proteins, Betts et al. described a novel strategy allowing the visualization of antigen-specific T-cells expressing the cell surface CD107 degranulation marker [67]. Accumulating evidence now shows that the expression of CD107 by CD8 T-cells correlates well with their cytotoxic potential and as a result CD107 is becoming routinely used in clinical trial monitoring alongside markers of activation and subset classification (discussed in [66]).
The Live Count Assay (LCA) was first described by Devêvre et al. It combines several much-needed parameters in T-cell functionality measurements. The LCA is a highly sensitive, ex vivo method of analyzing low numbers of antigen-specific T-cells for their cytolytic potential. Briefly a 1 : 1 mixture of specific and control target cells labeled with CMTMR is added to fluorescently sorted antigen-specific CD8+ T-cells and incubated for 4 h in the presence of degranulation marker CD107a- and CD8-specific mAbs. Samples are harvested, resuspended in staining buffer containing DAPI and analyzed by flow-cytometry [68]. The optimization of the existing LCA protocol in order to ensure minimal sample consumption during fluorescence sorting process is ongoing (Mahnke, Devevre, Speiser et al.; manuscript in submission).
Experimental approaches, which allow the determination of T-cell function along with T-cell quantification, continue to be of high interest. For instance the ELISpot assay allows the detection of T-cells having the capacity to produce IFN-γ, a major cytokine secreted by CD8 T-cells and having important actions in increasing CTL-mediated cytotoxicity. The high sensitivity of this method makes it suitable for ex vivo analyses of populations with low frequencies of antigen-specific T-cells and of precious samples (PBMC, TIL, TILN). Moreover it has been used for the characterization of fine antigen-specificity and cross-reactivity on Melan-A-specific T-cell clones [69] using a library of peptides predicted to be potentially cross-reactive with the Melan-A peptide [70]. The authors showed that increased differentiation of derived clones was associated with a decrease in TCR cross-reactivity and an increased specificity against the native Melan-A peptide [69].
The simultaneous detection of multiple cytokines using approaches such as the cytometric bead array (CBA from BD) or Luminex has significantly advanced the field beyond the commonly used ELISA technique. While these approaches are highly sensitive in detecting low amounts of expressed cytokines, they require prior stimulation using specific peptides and stimulating cells (DC or tumor cells). Alternatively the intracellular cytokine staining (ICS) allows the detection of cytokines expressed ex vivo by T-cells and their subsets. The major drawback of the ICS is that it requires fixation of the cells, meaning that viable cells expressing a cytokine of interest cannot be isolated and placed in culture to further determine its functional properties. Nevertheless this flow cytometry-based technique allows the simultaneous analysis of other intracellularly and extracellularly expressed markers, which has recently improved our understanding of the polyfunctionality of T-cells. Specifically, recent data suggest that cells capable of secreting multiple cytokines may play important roles in the control of HIV infection, since an immune response comprising such multifunctional cells was more frequently observed in individuals who maintained a low level of the virus and normal levels of the CD4+ T-cells for many years (reviewed in [41]).
Last but not least, a proliferative potential of fully functional cytotoxic T-cells is an equally important parameter to an efficient immune response. As with many other methods used to study functionality, the limitations of some of the approaches used to study proliferation (BrdU staining, CFSE dilution) involve an in vitro stimulation step prior to the analysis. These assays can be informative in making the distinction between cells with a high capacity for proliferation such as central memory cells versus cells with a lower capacity like highly differentiated cells, but may be less sensitive to small differences between more functionally similar subsets. Furthermore, a more accurate estimate of the in vivo proliferative potential and history of a T-cell may be provided by ex vivo strategies. Currently it is possible to determine proliferative activity using Ki67 staining of T-cells ex vivo, although this is also limited by an inability to keep such cells viable for additional tests. The degree of telomere shortening is another parameter, which can provide information about the extent of in vivo proliferation of a T-cell. Two of the most precise techniques currently used to measure telomere length are flow-FISH, based on the hybridization of telomere repeats using fluorescently labeled probes [71, 72], and STELA (single telomere length analysis) [73], a PCR-based technique of amplification of telomeric and subtelomeric regions in individual chromosomes. Future studies are thus also needed to get a more precise measurement of telomere length in small number of cells, and even single cells. However, since telomere dynamics does not seem to provide the only explanation for the persistence of T-cell clonotypes (discussed in [74]), other mechanisms of long-term survival in vivo should be investigated. Specifically, the roles of IL-7 and IL-15 survival cytokines should be further investigated in various T-cell populations across the differentiation spectrum, as are molecules involved in increased survival and resistance to apoptosis such as Bcl2. Finally, the replicative history of T lymphocytes can also be investigated by quantifying their content of TRECs, which are stable DNA episomes formed during TCR-α rearrangement and are diluted out with each cell division [75].
3.2. Future Targets for the Study of T Lymphocyte Functionality
The fast advancing field of multiparameter flow cytometry will continue to open many opportunities for the analysis of the functional profiles of phenotypically distinct subsets of antigen-specific T-cells in an ex vivo fashion. It is now even possible to observe the events of the downstream signaling cascades to the T-cell receptor or chemokine receptors at the single cell level. For example, the level of Phospho-Stat1 (pY701) and phospho-Stat5 (pY694) can be observed after 15-minute stimulation with either interferon alpha and gamma or IL-2/IL-15 on tumor antigen-specific CD8 T-cells [76]. In addition one of the downstream events of T-cell activation via the T-cell receptor is the phosphorylation of ERK1/2 (pYr202/pY204). Contact of antigen-specific peptide-pulsed antigen-presenting cells (APCs) leads to the phosphorylation of ERK1/2 which can be detected as a shift of the activated antigen-specific T-cells compared to unstimulated control cells [77]. Another parameter which can be useful in obtaining information about the capacity of tumor antigen-specific T-cells to react to an appropriate signal is the expression of the CD3zeta chain by intracellular staining which can be compared with the level expressed by total CD8 T-cells and viral antigen-specific T-cells [78, 79]. Importantly, additional markers allowing to select viable T-cells for further ex vivo cellular and molecular assays of T lymphocyte functionality are needed in the future, in order to enhance our understanding of the functional properties that correlate with immune protection.
3.3. The Functional Capacities of Tumor Reactive T Lymphocytes
It is now well established that an efficient antigen-specific CD8 T-cell response is one characterized by effector cells with a strong ability to lyse antigen-specific target cells. This is highly important in viral disease, both for acute (e.g., influenza) as well as for chronic or persistent (e.g., herpes viruses) infections. It is now commonly accepted that T-cells specific for foreign (e.g., viral) antigens can give rise to strong protective immune responses, whereas self/tumor antigen-specific T-cells are thought to be less efficient. Therefore, it is of great importance to study the functional capacity, in particular the multifunctionality, of tumor-reactive T lymphocytes before and following therapeutic vaccination (Figure 1). We previously described a study on the functional and proliferative potential of a dominant CD8 T-cell response directed against NY-ESO-1 [48]. We could identify two functionally distinct populations of tumor-specific T-cells. The major population making up to 90% of the cells (EM28neg) displayed the hallmarks of highly differentiated and active effector T-cells. These cells were able to mediate efficient ex vivo killing, produced IFN-γ, and still retained proliferative capacity upon antigenic stimulation, while up-regulating NK-like receptors such as CD57, CD94 as well as PD-1, an inhibitory receptor. The other relatively minor population (EM28pos), representing between 5 to 10% of NY-ESO-1 reactive CD8 T-cells, was also differentiated with features consistent with a resting memory state (CD28+CD27+CD127+PD-1−), suggesting that they may serve to maintain effector cells [48]. These findings are in line with another study, in which we identified a single naturally primed T-cell clone that dominated the CD8 T-cell response to the Melan-A/MART-1 antigen [45]. The dominant clone expressed a high avidity TCR to cognate tumor antigen, efficiently killed tumor cells, and prevailed in the differentiated effector memory (EM28neg) T-cell compartment. Other outstanding properties of this monoclonal T-cell population were the in vivo high frequencies, long-term persistence (>3.5 years), and efficient homing to metastases. Remarkably, during concomitant vaccination, we observed progressive enhancement of effector attributes, thus showing that repeated peptide vaccination together with IFA and CpG allows the induction of functionally competent tumor-reactive T-cells [45]. More recently, we took advantage of CpG oligodeoxynucleotides as a powerful vaccine adjuvant to induce and characterize T-cell responses upon vaccination with the natural Melan-A (EAAGIGILTV, “EAA”) versus the analog Melan-A (ELAGIGILTV, “ELA”) peptide antigen [14]. Compared with vaccination with analog peptide, natural peptide-induced T-cell frequencies were approximately twofold lower. However, T-cells showed superior tumor reactivity because of (i) increased functional avidity for natural antigen and (ii) enhancement of T-cell activation and effector function. Altogether, such studies are essential to characterize the functional requirements of tumor-specific T-cells upon vaccination. Moreover, they support further development of vaccine formulations with CpG and peptides in large-scale phase III trials.
It is essential that effector-type T-cells, once activated upon antigenic challenge, reach (“home to”) affected tissues for the elimination of infected or malignant cells. In human cancers, current knowledge of the in vivo functions of tumor-specific T-cells has been until recently restricted to peripheral blood T-cells; however, studies on the functional activities of such cells at the site of tumor lesions are becoming possible and popular [80, 81]. At present, several reports have described that while circulating Melan-A-specific CD8 T-cells exhibited characteristics in common with the effector T-cells, tumor-specific T-cells that reside in metastatic lymph nodes and soft tissue and visceral metastases appeared functionally attenuated as compared with circulating T-cells, despite their accumulation at high frequencies in tumor lesions [50, 82, 83]. This coincided with a significant increase of FoxP3+ regulatory T-cell activity within the tumor [50, 84]. Moreover, Melan-A-specific T-cells failed to produce IFNγ in both metastatic lymph nodes and nonlymphoid tissue metastases, but this defect was reversible in the presence of low dose IL-2/IL-7 cytokines in vitro [82], in line with gene expression profiling revealing downregulation of the interferon signaling pathway in T-cells from patients with metastatic melanoma [76]. Finally, Beyer and colleagues recently described that these nontumor reactive CD8 T-cells are characterized by a molecular program associated with the hallmarks of “division arrest anergy” [85]. There is still only limited data available regarding T-cell clonotypes, likely because this approach is labor intensive (see coming sections). Yet, clonotypic analyses provide promising results, mostly because T-cell clonotypes can be followed in a straightforward manner at any time and body location using the TCR as a clonotypic marker (reviewed in [74, 86]). For instance, further comparisons of the phenotype and T-cell clonotype composition between viral- and tumor-specific responses should be carried out between the blood and tumor site in patients where samples are available. Such analyses could extent previous studies while focusing on changes in the proportions of T-cell subsets and the potential selection of TCR clonotypes following migration to metastatic lesions [48]. This would also allow determining whether the expression of the tumor antigen at these sites negatively influences selection of tumor-specific TCR clonotypes compared with “bystander” viral-specific T-cells, which are not stimulated locally by their specific antigen.
4. Each T-Cell Is Unique: Gene Expression Profiling of Single Cells
An alternative strategy of avoiding the need for in vitro cultured T-cell clones is to analyze the gene-expression profile of ex vivo sorted cells. The standard microarray approach can be used for gene-expression profiling of T-cells of particular phenotypes; however, it is limited to the study of at least 100–1000 cells [87]. Nevertheless, the technique has evolved over the years to allow gene-expression profiling of coding and noncoding regions of the genome, which still makes it the goldstandard approach for identifying novel differentially expressed genes. Once these genes of interest have been identified, the ultimate goal is to perform detailed analyses on a limited number of antigen-specific T-cells and subpopulations. For this purpose, our group has developed a strategy consisting of cell lysis and cDNA synthesis in a single-step procedure, followed by a modified RT-PCR protocol that relies on the detection of specific cDNAs after global amplification of expressed mRNAs [32, 88]. This method yields sufficient cDNA from as few as five cells. Thus, a major advantage is that it allows the analysis of gene expression and TCR repertoire analysis in small-purified subpopulations of antigen-specific cells even when a limited amount of material is available. Furthermore, since the global amplification is not selective and the amplified cDNA remains stable for long periods of time at −80°C, this approach allows the amplification of genes of interest even at later time-points. We have previously employed this strategy to study the heterogeneity of CD8 T subpopulations. Five-cell aliquots isolated following multiparameter flow cytometric analysis were subjected to global amplification of cDNA, followed by the specific amplification by PCR of genes known to be differentially expressed at distinct stages of cellular differentiation (Figure 1). As such, the expression patterns of the CD27 costimulatory receptor, the IL-7Rα, and the expression of the cytotoxic molecules granzyme B and perforin have permitted the identification of a number of T-cell subpopulations, including pre-effector and effector memory subsets [32, 34, 88], as well as the fine characterization of tumor-specific CD8 T-cell responses in the context of therapeutic peptide vaccination (see [45, 48]; Rufer, Speiser et al., unpublished observations).
4.1. Towards Single Cell Profiling
During T-cell differentiation, the stochastic events become more prominent involving a set of modifications of multiple gene expressions inducing subtle or dramatic changes in the cell. Such changes incur variability among the antigen-primed T-cell populations and even among “naive” cells. CD8 T lymphocytes are a well-defined example of such a phenomenon. Such inevitable heterogeneity in biological systems emphasizes the need to determine this variation by analyzing single cells. Recently, we have optimized the above-described strategy of global cDNA amplification at the single cell level. This provides us with the added advantage of being able to determine the gene-expression profile of a single T-cell clone defined by its unique CDR3 sequence (i.e., a TCR clonotype). One drawback of this method is that low frequency TCR clonotypes require the isolation of numerous single cell samples for its detection. Nevertheless, it is highly powerful in that one can follow single TCR clonotypes over time before and after vaccination and compare their gene-expression profiles between (i) vaccination protocols, (ii) patients, (iii) T-cell subpopulations, (iv) anatomical localizations, and (v) over time. Furthermore, one can correlate the information gathered on the ex vivo gene-expression profile of a TCR clonotype of interest, with the functionality of a T-cell clone bearing the same unique receptor in vitro. Currently, our group is interested in understanding how the differentiation of melanoma-specific T-cells is affected by factors such as the type and dose of antigenic peptide used for vaccinating patients. The profiles of these cells will be subsequently compared against EBV- or CMV-specific T-cells, of which we know that they ensure efficient viral control in vivo, in spite of differences in phenotype, functionality, and TCR clonotype composition [43].
The cDNA global amplification technique provides information on the expression of genes at the mRNA level, and not at the protein level. However, the combination of the single cell gene-expression profiling and the multiparameter flow cytometry analysis using antibodies against the respective proteins can overcome this limitation. The disadvantage of the nonspecific amplification technique is that it is not quantitative, since the main principle of the technique relies on the nonspecific amplification of all expressed mRNAs present in the cell. Furthermore, particularly at the single cell level, this technique remains limited to the study of genes expressed at moderate and high levels within the cell, since genes expressed at very low levels may not be detected even following the global amplification procedure. Thus, overcoming this limitation may be a challenging task, although working with cDNA material of high quality and purity is indispensable. This requires a very clean and sterile working environment. All steps preceding the global amplification must be performed in an RNAse and DNAse free environment, free from potential sources of contaminant material.
4.2. Single Cell Profiling Strategies by Other Groups
Although for some time the direct quantification of gene expression a single cell was considered impossible since the amount of mRNA extracted from a cell is minute, there is now strong evidence of its feasibility, allowing efficient and reliable assessment of gene expression of individual cells [89–91]. Peixoto and colleagues reported of a method wherein they successfully quantified the expression of 20 different genes simultaneously from a single cell using reverse transcription of single cell mRNA with 3′-specific primer of each gene of interest [89, 90]. This cDNA was then amplified by a multiplex PCR of all 20 gene-specific 3′ and 5′ primers, followed by a seminested real-time PCR for each individual gene. Using this approach, Monteiro et al. investigated the heterogeneity of the CD8pos T-cell compartment, by first identifying several T-cell subpopulations using four extra-cellular markers (CCR7, CD45RA, CD27, CD28), followed by the analysis of mRNAs coding for chemokines, cytotoxic molecules, or effector cytokines expressed in single cells from these subsets [33]. This in-depth single cell profiling could demonstrate that certain T-cell subsets belonging to the memory compartment such as the EM T-cells expressing CD27 at high levels could exhibit similar functionality profiles as naive T-cells. Additionally CCR7− cells expressing both CD27 and CD28 showed similar profiles as central memory cells, in line with our own observations [34]. On the other hand, highly differentiated CD27 and CD28 double negative cells were highly similar in their gene-expression profiles regardless of CD45RA expression. The results obtained by this quantitative single-cell multiplex RT-PCR approach clearly depicted a high degree of heterogeneity with each cellular subset displaying characteristic patterns of gene expression, pointing to a hierarchical order of T-cell differentiation stages among the antigen-experienced cells [33]. Furthermore, even for individual cells belonging to the same subset, the gene-expression profiles showed variability, outlining the necessity to study multiple parameters in order to characterize the uniqueness of each single T-cell. The main drawback of this strategy is that it can quantify multiplex PCR applied to only 20 preselected genes of interest expressed in a cell and thus fails to amplify high amounts of cDNA that can eventually be used to individually quantify any gene of interest, for example, TCR BV clonotype expression.
Since the study of single T lymphocytes remains one of the ultimate goals, other groups have developed approaches for this highly specific type of analysis. For instance, the Ampligrid platform by Beckman Coulter can be employed to sort single cells on unique hydrophilic/hydrophobic structured glass slides containing individual wells, which can then be directly used for PCR amplification in a very small volume allowing higher sensitivity. The chemically modified glass slide wells can be used as single reaction centers for cell growth and PCR all in one place [92] in which cells are stimulated, washed, and dried in situ, avoiding numerous handling steps and cellular stress [93]. This platform has already been used for the genetic analysis of circulating tumor cells in small-scale clinical trials [94] and is particularly advantageous for multiplex real-time PCR [95], robotic micromanipulations, and laser microdissection [96]. This method has a high sensitivity with a detection level threshold of 32 pg of purified DNA, allowing the minisequencing of mitochondrial DNA down to a single lymphocyte [97]. Yet, it also requires further optimization at the single cell level, since the efficiency may sometimes vary drastically from 20 to 80% [98] and a second seminested amplification may still be necessary for certain genes [99]. Furthermore, the optimal handling of minute volumes of numerous single cell samples should require robotized approaches.
5. T-Cell Receptor (TCR) Repertoire and TCR Clonotype Analysis
A better understanding of the structural principles that govern TCR-pMHC binding is essential to promote research and clinical applications. The quality of TCRs recruited during disease, or by vaccination, dominantly influences the potency of immune responses. To improve therapeutic immune interventions, TCRs need to be fully characterized, and the mechanisms for their recruitment and function must be elucidated. Results from such studies have a strong impact on the design of antigens for vaccination as well as on the choice of optimal TCRs for adoptive T-cell therapy, with or without TCR gene transfer.
Several strategies exist to assess the TCR repertoire diversity and clonal composition of various pools of antigen-specific T-cells. A straightforward approach is to combine the staining with peptide-MHC multimers with a panel of monoclonal antibodies (mAb) directed against the variable region of the TCR β chain (anti-BV). While this approach allows the direct and quantitative assessment of BV usage by tumor- and viral-specific CD8 T lymphocytes, it does not reveal whether the BV-restricted T-cell response analyzed is of monoclonal or of polyclonal nature (reviewed in [86]). The immunoscope or CDR3 spectratyping technique measures the size heterogeneity of the CDR3 region of the TCR and provides a powerful tool to characterize in-depth the level of the TCR repertoire complexity (reviewed in [86, 100]). Since most human TRBV segments can be amplified using 22 BV-specific primers as previously reported [101], fluorescently labeled primers specific for each TRBV gene segment and a primer specific for the constant region of the β chain of the TCR are used to amplify specifically the expressed CDR3β gene segment. The labeled PCR products are then run on an automated sequencer in the presence of fluorescent size markers to obtain the precise size of the CDR3 region (reviewed in [86]). Expansion of particular antigen-specific T-cell clonotypes is visualized as single dominant PCR peaks of a given size and PCR products are sequenced to obtain the nucleotide sequence of the CDR3 region. Analyses are then focused on determining the dominant clonotypes (defined as the presence of the same TCR BV-CDR3-BJ sequence in two separate subsets or time-points). They can be further divided into highly dominant or subdominant clonotypes, based on their frequencies in an immune response. Subsequently a comparison of the frequencies of a given clonotype can be made between different T-cell subsets, between different time-points, and even among different individuals (Figure 1).
5.1. Advantages and Limitations of the Spectratyping/Clonotyping Technique
Today, the spectratyping/clonotyping approach has proven to be a strong analytical technique, allowing both detecting oligoclonal T-cell expansion ex vivo and assessing the presence and frequencies of dominant T-cell clonotypes among virus- and tumor-specific CD8 T-cell subpopulations [43–45, 48, 49, 51, 52]. Yet, this approach has some limitations. The first one stems from the cross reactivity of the different TRBV oligonucleotide primers used. Due to the homology between several TRBV gene segments and the existence of several subtypes for numerous TRBV gene families (e.g., 14 variations of TRBV13 and 25 for TRBV6), the primer sequences utilized have a certain degree of cross-reactivity. In fact, the likelihood of unspecific recognition can be theoretically calculated for each primer set against all BV gene segments. Nevertheless, performing individual TRBV PCR (in contrast to multiplexing), sequencing of doubtful PCR amplicons, and careful analysis of the upstream sequences allows to accurately determine whether the PCR product corresponds to the correct amplified TCR BV family or is the result of cross-reactivity against another BV family. Another limitation is that there are instances where the TCR of one sample or clone cannot be identified despite repeated efforts. The more likely reason for this observation is that the 22 pairs of primers used detect an estimated >90% of all known TRBV families and subfamilies. Thus, although detection of the majority of all TRBV subfamilies is theoretically ensured, some TCR clonotypes among antigen-specific T-cell responses could express rare and particular BV segments and thus remain undetected by the spectratyping technique (own unpublished observations).
The spectratyping approach of analyzing T-cell clonotype composition requires a high number of samples to be sequenced. The strategy developed by our group involves the analysis of CDR3 sizes among T-cells population as an early step, followed by the analysis of potential candidates for dominant clonotypes. This reduces the number of sequencing reactions of unique BV and unique CDR3 lengths. When the sequencing of several PCR products has yielded the same TCR sequence for a given BV, clonotypic primers can be designed to allow faster identification of these TCR clonotypes among the remaining samples. The forward and reverse clonotypic primers are designed to specifically recognize the unique CDR3 sequence of each clonotype and are used in combination with the BC primer, and BV primer respectively. In certain instances, clonotypic primers cannot be designed, because of lack of specificity or because CDR3β sequences are too similar. This is the case for most EBV-specific TCR clonotypes of the TRBV2 and TRBV4 families as they share highly similar CDR3 sequence motifs, sometimes differing by only one amino acid, and in these situations, all samples are being sequenced [43].
Finally, as the spectratyping technique is only semiquantitative, the amplification of cDNA from antigen-specific T-cell pools might introduce biases, leading to the nondetection of variations present within a particular CDR3β length (reviewed in [102]). Sequencing of antigen-specific T-cells at the single cell level should overcome most of these restrictions [103–105]. This aspect as well as the limitation in the determination of precise frequencies from individual clonotypes has prompted us to recently redefine our spectratyping/clonotyping approach towards the single cell level.
5.2. Clonotype Selection and Composition of Antigen-Specific T Lymphocyte Subsets
It has been proposed that the large heterogeneity that is observed among T-cell subsets may be influenced by the clonotype composition found within these subpopulations. Indeed, different TCRs, which vary in mechanism and strength of peptide-MHC binding, may lead to distinct activation and differentiation. However, it is evident that cellular heterogeneity exists even within the same T-cell clonotype (reviewed in [106]), as demonstrated in TCR-transgenic mice [106, 107], and in the elegant study by Stemberger and coworkers [108], in which it was shown that a single naive CD8 T-cell precursor can differentiate into different effector and memory subsets. Studies of the human TCR repertoire are somewhat conflicting, with some groups reporting that effector and memory are derived from identical, naive precursor cells [109, 110], while others suggest that they are recruited from distinct precursor cells [111].
Our group recently performed an in-depth analysis of the clonal TCR repertoire diversity and selection among healthy donors infected with persistent herpes viruses [43] or following influenza infection [44]. Sequencing of the TCR beta chains showed that the clonal composition was highly restricted for both influenza- and CMV-specific cells (between 1–3 dominant clonotypes), whereas it was relatively diverse for EBV-specific cells (on average 15 clonotypes). Importantly, these analyses revealed for the first time in a steady state of human T-cell memory that individual clonotypes can differentiate and be maintained within both the early- (memory) and late-differentiated (effector) T-cell subsets [44]. Data also indicate, in the EBV model, that the early differentiated memory subset is the most diverse and contains virtually all clonotypes found in the highly differentiated effector subset [43]. Yet, striking differences in the patterns of dominance can be observed among both subsets, since some clonotypes are selected with differentiation while others are not. Remarkably, clonotype selection and composition of EBV- and CMV-specific subsets upon differentiation are highly preserved over time, with the presence of the same dominant clonotypes at specific differentiation stages over a period of 4 years. Altogether, these observations demonstrate that T-cell clonotypes segregate with differentiation, but the clonal composition once established is kept constant for at least several years [43] (Figure 3).
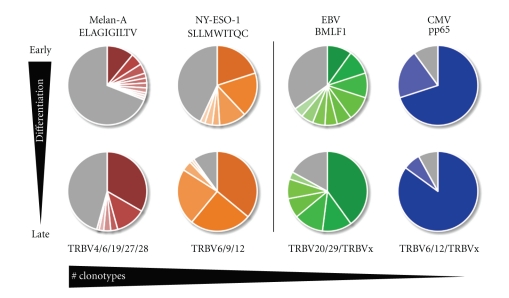
Figure 3.
TCR repertoire restriction and TCR clonotype selection are driven by differentiation. Analysis of TCR repertoire diversity and clonotype composition between tumor-specific responses (left side of the figure) and viral-specific responses (right side of the figure) suggest that all antigen-specific T-cell responses undergo selection along with progressive cellular differentiation (top to bottom of figure). As such, early differentiated subsets show a higher degree of clonal diversity relative to their more differentiated subsets. Nevertheless, dominant clonotypes are also found in the former but make up significantly larger proportions of the more differentiated compartments (defined as TCR clonotype selection) supporting a linear model of differentiation. In addition, viral-specific responses which are composed of highly differentiated cells (EMRA T-cells) also show a higher degree of TCR repertoire restriction compared with tumor-specific responses, evident not only in the late but also already in the early differentiated subsets (indicated by a decrease in numbers of BV families and numbers of clonotypes from left to right). Each colored slice of pie represents a unique TCR clonotype, while the grey slices represent nondominant TCRs, that is, the sequences identified only once within the antigen-specific TCR repertoire. Melan-A (ELAGIGILTV) and NY-ESO-1 (SLLMWITQC) peptide sequences are indicated for the tumor-specific responses; virus specific T-cells were analyzed for the epitope EBV BMLF1280–288 (GLCTLVAML) and for the CMV epitope pp65495–503 (NLVPMVATV). All responses are HLA-A2 restricted. Preferential TRBV family usage is depicted based on IMGT's nomenclature [112].
Therapeutic vaccines against cancer aim to induce effective immune responses similar to protective antiviral responses. Previously, we showed that a potent vaccine formulation composed of low doses of Melan-A/MART-1 peptide, IFA, and low doses of CpG oligonucleotides induced ex vivo detectable T-cell expansions in virtually all melanoma patients reaching on average more than 1% of specific CD8 T-cells [13, 14]. Extensive analyses of the tumor-specific responses among peptide vaccinated patients revealed that the Melan-A/MART-1-specific T-cell responses were also relatively diverse comprising between 10 to 15 clonotypes, depending on individual patients (Rufer, Speiser et al., unpublished observations). These data are consistent with another report [49], in which we found that the number of dominant NY-ESO-1-specific T-cell clonotypes among patients was between 6 and 10 clonotypes (Figure 3). A striking observation is that tumor-specific T-cell responses can, in some rare patients, be dominated by single dominant clones [45, 49]. Collectively, a general observation is that, similarly to viral-specific T-cell responses, responses to Melan-A and NY-ESO-1 epitopes are also caused by selection and amplification of particular T-cell clonotypes, following T-cell differentiation (Figure 3). Specifically, the majority of the patients showed progressive TCR repertoire restrictions, from early differentiated to late differentiated T-cells, indicating the occurrence of oligoclonal T-cell expansions in the latter compartment. Thus, the EM28neg (“effector-like”) subset consists primarily of codominant T-cell clonotypes, whereas the EM28pos (“memory-like”) contains large polyclonal TCR repertoires with the presence of numerous nonclonotypic sequences (see [45, 48], Rufer, Speiser et al., unpublished observations). Finally, most tumor-specific clonotypes identified in the late differentiated EM28neg subset were also found within the EM28pos T-cells, in agreement with our recent observation made in CD8 T-cell responses against influenza [44] and herpes viruses [43]. Altogether, these results provide further evidence for a linear model of T-cell differentiation, in which a small number of clonotypes are selected to differentiate from a larger pool of less differentiated “memory” cells. Figure 3 summarizes the TCR diversity observed for tumor-specific CD8 T-cell responses compared with viral-specific responses. While larger numbers of codominant clonotypes are found for Melan-A/MART-1-specific T-cells, CMV responses involve low numbers of clonotypes. Nevertheless in all instances a clear selection is observed with T-cell differentiation, such that only a small number of TCR clonotypes are found to dominate the highly differentiated subsets.
5.3. Alternative Strategies to Study TCR Repertoire Diversity and Composition of Antigen-Specific CD8 T-Cell Responses
Koup, Douek, and coworkers have developed over the past decade an alternative approach to address the complexity of the TCR repertoires in several antigenic systems [113]. Its principle is based on the use of a template switch-anchored RT-PCR with primers specific for the 3′ TCR constant region to amplify all expressed TRBV or TRAV gene products. Since no primers specific for the TRBV- or TRAV-gene segment are applied, this strategy does not introduce bias associated to particular Vβ or Vα families (see the above discussion). The resulting PCR amplicons are then ligated into a plasmid vector, and cloned and selected colonies are sequenced. This powerful strategy allows the analysis of TCRs at the single cell level and provides an absolute frequency of particular CDR3β usage. One of the major disadvantages of this technique is the relative high cost associated with the need to sequence thousands of individual CDR3 gene segments. Moreover, biases related to the efficiency of the ligation and transformation of bacterial cells may still be introduced. Finally, the design of clonotypic primers specific to identified dominant T-cell clones when combined to quantitative real-time PCR allows the quantification of clonotypes at any given time-point during longitudinal studies [113]. This approach has led several research groups to achieve major advances in the fine characterization of CD8 T-cell responses during SIV [114–117], HIV [118–123], CMV and EBV [124, 125] infections as well as in patients suffering from melanoma [126, 127] or of hematological malignancies [128]. Describing in detail these publications is beyond the scope of this paper and, therefore we have chosen to discuss some aspects of this extensive work that is more closely related to ours.
An important question concerns the frequency and the functional properties of public versus private clonotypes and the bona fide clinical benefit obtained after their in vivo expansion following viral infection or therapeutic vaccination. Public clonotypes are defined by the presence of the same identical TRBV-CDR3-BJ and/or TRAV-CDR3-AJ sequences found in two unrelated individuals or animals. Recently, Price et al. have described the protective activity mediated by public Gag- specific TRBV clonotypes in vaccine-induced SIV-specific CD8 T-cell responses, and observed that the number of public clonotypes correlated inversely with the virological outcome after SIV challenge [114]. Compared to private vaccine-induced SIV-specific T-cells, T-cell populations with public clonotypes exhibited greater levels of cross-reactivity to epitope variants, thus indicating the need to preserve some degree of cross-reactivity to contain viral escape [114]. Similarly, public TRBV sequences that are shared between HLA-matched individuals have also been described within the TCR repertoire specific for viral epitopes that do not mutate as in the case of EBV and CMV [43, 125, 129]. Venturi and colleagues have proposed that the sharing of TCR-beta chain between healthy individuals is strongly associated with TCR-beta production frequency and that a process of convergent recombination facilitates the more efficient production of those TRBV sequences [115, 130, 131]. We recently identified public TRBV sequences isolated from 15 melanoma patients, and unlike viral-specific TCR repertoires, such public TRBVs were primarily expressed by nondominant and infrequent T-cell clonotypes [51]. This highly contrasted with private CDR3β signatures frequently found in T-cell clonotypes that dominated repertoires of individual patients. Clearly, common TRBV motifs have been selected after vaccination in different patients, but at much lower frequency compared to distinct and private TCR structures. Yet, both public and private antigen-specific TCRs recognized tumor cells with comparable avidities suggesting that neither had a clear functional advantage [51]. Future directions involve understanding the structural and biophysical impact of public and private TCRs in the process of recognition of the pMHC complex, T-cell activation, and subsequent T-cell functionality.
The characterization of T-cell clonotypes bearing particular TCR CDR3 motifs, of their frequencies in well-defined phenotypic populations, as well as of their related cytolytic functions has revealed the need to understand the basis of the structure-function relationship of a given TCR to its pMHC molecule. This requires detailed studies performed at the molecular level of the TCR and the delineation of multiple parameters involved in the TCR signaling (e.g., CD8 coreceptor), combined with the characterization of intrinsic biophysical factors of the TCR molecule (e.g., CDR structural loops and binding kinetic rates). In that regard, the recent crystal structure of the Melan-A-MHC-TCR complex by Cole and colleagues brought important insights into the key elements within the TRAV and TRBV gene segments that are involved in the recognition of the Melan-A antigen [127]. They showed that only CDR1α, CDR3α, and CDR3β loops make contacts with the antigen and propose that this unusual pattern of antigen recognition may explain the unique characteristics and extraordinary frequencies of CD8pos T-cell responses to this epitope. Moreover, Asn92 is the only residue from the CDR3α that interacts with the antigen [127], and since it is commonly expressed by TRAV12-2 (or TCR Vα2.1) positive T-cell clonotypes [51], this finding may provide some explanation for the preferential segment usage of the TRAV12-2 in Melan-A-specific T-cell responses [132, 133].
Peptide-based cancer vaccines are often prepared with altered “analog” peptide antigens that have been optimized for HLA class I binding, in order to enhance their immunogenicity. An obvious but crucial point is that structural modifications of peptides should not alter TCRαβ repertoires or TCR binding properties, to ensure that vaccination-primed T-cell clonotypes remain highly specific for the natural antigen and efficiently recognize tumor cells. Recently, we demonstrated fine specificity differences and enhanced recognition of naturally presented antigen by T-cells after vaccination with natural Melan-A/MART-1 peptide as compared with analog peptide [14]. These data highlight the importance of careful re-evaluation of studies using analog antigens with regard to the risk of activating T-cells with “imprecise” antigen specificity or low TCR affinity. We recently addressed the question whether the observed fine peptide antigen specificity could be explained by structurally different TCRs recruited and promoted by vaccination with natural versus analog Melan-A/MART-1 peptide. In this study [51], we compared the TCR primary structures of 1489 HLA-A*0201/Melan-A26–35 specific CD8 T-cells derived from patients after vaccination with the natural or the analog peptide. Collectively, our data indicate that T-cell repertoires generated against natural or analog Melan-A peptide exhibited slightly different but otherwise structurally conserved TCR features, strongly suggesting that the differences in binding affinity/avidity of TCRs towards pMHC observed between both cohorts of vaccinated patients are caused by subtle structural TCR variations [51]. Very recently, Cole and colleagues [126] evaluated the intraindividual clonotypic responses to both analog and native Melan-A peptide following in vitro peptide stimulation, thus allowing comparison between samples derived from the same naive T-cell pool. They found that despite a clear bias towards particular TRBV gene usages and the presence of the GXG public motif in the CDR3β loop, similarly to previous reports [51, 134], the majority of the clonotypes were nonoverlapping between the two TCR repertoires [126]. These data further illustrate (i) the importance to monitor T-cell responses at the clonotype level and (ii) emphasize the need to the careful evaluation of such heteroclitic peptide-based immune interventions to ensure efficacy in the clinic.
6. Applying Improved Knowledge to Clinical Studies
Progress in basic immunology has led to a better understanding of immune protection against cancer. Although there is still much more to learn, we now know how to activate the immune system and which cellular and molecular components are key for immune protection. The knowledge is currently applied in clinical studies, with the aim of introducing immunotherapy against solid cancers in clinical routine in a few years from now (Figure 1).
The design of potent antitumor vaccines currently faces several obstacles, which will need to be addressed in order to improve their efficacy. One of them is the choice of optimal tumor antigens, as reviewed by Boon et al. [2]. For instance, mutated antigens, although unique to tumor cells, may not be shared among different individuals, thus raising the problem of individualized versus universal tumor-vaccines. In contrast, differentiation antigens (such as Melan-A/MART-1) are expressed by tumors from many individuals. However, their expression in normal cells poses targeting problems thus toxicity, and of breaking immune tolerance. Since these concerns are found on opposite sides of the balance, addressing both of them will prove difficult. The use of potent adjuvants may be successful in stimulating immune responses at a cost of autoimmune effects, as for example observed in melanoma patients who developed vitiligo following vaccination [135].
Recent technological advancements now allow dissecting immune responses and compare the phenotypic and functional characteristics of various antigen-specific T-cell populations. An increased understanding of the generation and maintenance of memory T-cells in virus-specific responses will also help to better determine T-cell correlates of protection. This is important in the design of tumor-specific vaccines, as well as in determining which T-cell subset may be most optimal for adoptive cell transfer. Furthermore, recent studies have shown that vaccination with peptide analogues [14] with enhanced binding to MHC may result in the selection of unfavorable T-cells [51, 126]. Therefore, the stability of the peptide-MHC complex should always be taken into account when assessing affinity and avidity of TCR-mediated recognition.
Aside from the fact that tumor-/self-antigen-specific T-cells are less frequent than T-cells specific for pathogens, their capacity to recognize and interact with cognate antigen bearing cells is inferior. This is at least in part explained by the lower TCR affinity, resulting in reduced functional avidity. With a few exceptions, the so far best human cancer-specific TCRs have been isolated from patients with extraordinary strong spontaneous CD8 T-cell responses, often associated with unusually favorable clinical histories [45, 48]. Interestingly, T-cells isolated from metastases, particularly from tumor infiltrated lymph nodes, usually bear better TCRs than T-cells from peripheral blood, reflecting enrichment of specific T-cells in the tumor microenvironment. Unfortunately, TCRs isolated after vaccination with tumor antigens are often of lower avidity [45, 136–138], indicating that current generations of tumor vaccines are suboptimal for the selection of the best T-cells. It may even be possible that many of the TCRs isolated after immunotherapy have been primed earlier and were thus not primed but only boosted by the applied therapy. This issue needs to be clarified, for a full characterization of vaccination driven selection of T-cell clonotypes. It is necessary to improve vaccines in this regard, for example, by reducing vaccine antigenicity (with e.g., lower antigen doses or weaker antigens), or by limiting antigen presentation to professional antigen-presenting cells. However, it remains possible that patient's tumor specific TCR repertoires are fundamentally deficient. In this case, gene therapy with improved TCRs may be a valid approach (reviewed in [139]). Current and future developments in this regard [140, 141] will show whether this is a critical point. Finally, competent TCRs may exist in cancer patients but T-cell precursor frequencies may be too low, limiting the potential to generate strong and long-lasting T-cell responses. If so, gene therapy with naturally occurring TCRs, introduced in larger numbers of (naive) T-cells may be beneficial for improved clinical outcome.
In this paper we have focused on CD8 T-cell responses, which are important for cancer patients, since CD8 T cells can counteract tumor progression. However, tumors and their microenvironment include many more components. Tumors have been described as “wounds that never heal, that regularly reorganize their strategies for persistence and progression. Instead of stopping the disease, some components of the immune system can actually promote cancer, by provoking chronic inflammation and elaborating factors that may support tumor cell growth, survival, and angiogenesis. Furthermore, established tumors use immune-regulatory circuits to generate an immune-suppressive environment, which can act as substantial barriers to protective T-cell responses. Progress in our understanding of the molecular mechanisms governing the interaction between tumors and the immune system has been the basis for improvements of cancer therapy (reviewed in [142]). In view of the formidable challenge to fully understand the plethora of processes employed by tumors, it is evident that further work is needed to optimize cancer therapy, by targeting principal mechanisms of malignant diseases.
Acknowledgments
The authors are thankful to patients and their families, healthy donors and clinical collaborators. They also thank all members of Their research groups, and all Their external collaborators and supporters for their dedicated contributions. They also apologize for those scientists whose studies could not be mentioned due to space limitations. The authors' studies are supported by the Ludwig Institute for Cancer Research Ltd., the Cancer Research Institute (USA), the Cancer Vaccine Collaborative, Atlantic Philanthropies (USA), the Wilhelm Sander-Foundation (Germany), the Swiss Cancer League/Oncosuisse, the Swiss National Science Foundation, and the Swiss National Center of Competence in Research (NCCR) Molecular Oncology.
Abbreviations
- CpG:
Deoxycytidylate-phosphate-deoxyguanylate oligodeoxynucleotide
- EBV:
Epstein-Barr virus
- CMV:
Cytomegalovirus
- Influenza:
Flu
- IFA:
Incomplete Freund's Adjuvant
- FISH:
Fluorescence in situ hybridization.
References
- 1.Lasithiotakis KG, Petrakis IE, Garbe C. Cutaneous melanoma in the elderly: epidemiology, prognosis and treatment. Melanoma Research. 2010;20(3):163–170. doi: 10.1097/CMR.0b013e328335a8dd. [DOI] [PubMed] [Google Scholar]
- 2.Boon T, Coulie PG, Van Den Eynde BJ, Van Der Bruggen P. Human T cell responses against melanoma. Annual Review of Immunology. 2006;24:175–208. doi: 10.1146/annurev.immunol.24.021605.090733. [DOI] [PubMed] [Google Scholar]
- 3.Speiser DE, Liénard D, Pittet MJ, et al. In vivo activation of melanoma-specific CD8+ T cells by endogenous tumor antigen and peptide vaccines. A comparison to virus-specific T cells. European Journal of Immunology. 2002;32(3):731–741. doi: 10.1002/1521-4141(200203)32:3<731::AID-IMMU731>3.0.CO;2-H. [DOI] [PubMed] [Google Scholar]
- 4.Liénard D, Rimoldi D, Marchand M, et al. Ex vivo detectable activation of Melan-A-specific T cells correlating with inflammatory skin reactions in melanoma patients vaccinated with peptides in IFA. Cancer Immunity. 2004;4:p. 4. [PubMed] [Google Scholar]
- 5.Romero P, Dunbar PR, Valmori D, et al. Ex vivo staining of metastatic lymph nodes by class I major histocompatibility complex tetramers reveals high numbers of antigen- experienced tumor-specific cytolytic T lymphocytes. Journal of Experimental Medicine. 1998;188(9):1641–1650. doi: 10.1084/jem.188.9.1641. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Romero P, Valmori D, Pittet MJ, et al. Antigenicity and immunogenicity of Melan-A/MART-1 derived peptides as targets for tumor reactive CTL in human melanoma. Immunological Reviews. 2002;188:81–96. doi: 10.1034/j.1600-065x.2002.18808.x. [DOI] [PubMed] [Google Scholar]
- 7.Dutoit V, Rubio-Godoy V, Pittet MJ, et al. Degeneracy of antigen recognition as the molecular basis for the high frequency of naive A2/Melan-A peptide multimer+CD8+ T cells in humans. Journal of Experimental Medicine. 2002;196(2):207–216. doi: 10.1084/jem.20020242. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Zippelius A, Pittet MJ, Batard P, et al. Thymic selection generates a large T cell pool recognizing a self-peptide in humans. Journal of Experimental Medicine. 2002;195(4):485–494. doi: 10.1084/jem.20011658. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Beutler B. Inferences, questions and possibilities in Toll-like receptor signalling. Nature. 2004;430(6996):257–263. doi: 10.1038/nature02761. [DOI] [PubMed] [Google Scholar]
- 10.Germain RN. An innately interesting decade of research in immunology. Nature Medicine. 2004;10(12):1307–1320. doi: 10.1038/nm1159. [DOI] [PubMed] [Google Scholar]
- 11.Janeway CA, Jr., Medzhitov R. Innate immune recognition. Annual Review of Immunology. 2002;20:197–216. doi: 10.1146/annurev.immunol.20.083001.084359. [DOI] [PubMed] [Google Scholar]
- 12.Takeda K, Kaisho T, Akira S. Toll-like receptors. Annual Review of Immunology. 2003;21:335–376. doi: 10.1146/annurev.immunol.21.120601.141126. [DOI] [PubMed] [Google Scholar]
- 13.Speiser DE, Liénard D, Rufer N, et al. Rapid and strong human CD8+ T cell responses to vaccination with peptide, IFA, and CpG oligodeoxynucleotide 7909. Journal of Clinical Investigation. 2005;115(3):739–746. doi: 10.1172/JCI23373. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Speiser DE, Baumgaertner P, Voelter V, et al. Unmodified self antigen triggers human CD8 T cells with stronger tumor reactivity than altered antigen. Proceedings of the National Academy of Sciences of the United States of America. 2008;105(10):3849–3854. doi: 10.1073/pnas.0800080105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Dudley ME, Wunderlich JR, Robbins PF, et al. Cancer regression and autoimmunity in patients after clonal repopulation with antitumor lymphocytes. Science. 2002;298(5594):850–854. doi: 10.1126/science.1076514. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Dudley ME, Wunderlich JR, Yang JC, et al. Adoptive cell transfer therapy following non-myeloablative but lymphodepleting chemotherapy for the treatment of patients with refractory metastatic melanoma. Journal of Clinical Oncology. 2005;23(10):2346–2357. doi: 10.1200/JCO.2005.00.240. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Altman JD, Moss PAH, Goulder PJR, et al. Phenotypic analysis of antigen-specific T lymphocytes. Science. 1996;274(5284):94–96. doi: 10.1126/science.274.5284.94. [DOI] [PubMed] [Google Scholar]
- 18.Klenerman P, Cerundolo V, Dunbar PR. Tracking T cells with tetramers: new tales from new tools. Nature Reviews Immunology. 2002;2(4):263–272. doi: 10.1038/nri777. [DOI] [PubMed] [Google Scholar]
- 19.Derré L, Jandus C, Baumgaertner P, et al. Quantitative multiparameter assays to measure the effect of adjuvants on human antigen-specific CD8 T-cell responses. Methods in Molecular Biology. 2010;626:231–249. doi: 10.1007/978-1-60761-585-9_16. [DOI] [PubMed] [Google Scholar]
- 20.Volkert M, Marker O, Bro-Jorgensen K. Twp populations of T lymphocytes immune to the lymphocytic choriomeningitis virus. Journal of Experimental Medicine. 1974;139(5):1329–1343. doi: 10.1084/jem.139.5.1329. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Ahmed R, Gray D. Immunological memory and protective immunity: understanding their relation. Science. 1996;272(5258):54–60. doi: 10.1126/science.272.5258.54. [DOI] [PubMed] [Google Scholar]
- 22.Sallusto F, Lenig D, Förster R, Lipp M, Lanzavecchia A. Two subsets of memory T lymphocytes with distinct homing potentials and effector functions. Nature. 1999;401(6754):708–712. doi: 10.1038/44385. [DOI] [PubMed] [Google Scholar]
- 23.Sallusto F, Geginat J, Lanzavecchia A. Central memory and effector memory T cell subsets: function, generation, and maintenance. Annual Review of Immunology. 2004;22:745–763. doi: 10.1146/annurev.immunol.22.012703.104702. [DOI] [PubMed] [Google Scholar]
- 24.Sallusto F, Palermo B, Hoy A, Lanzavecchia A. The role of chemokine receptors in directing traffic of naive, type 1 and type 2 T cells. Current Topics in Microbiology and Immunology. 1999;246:123–129. doi: 10.1007/978-3-642-60162-0_16. [DOI] [PubMed] [Google Scholar]
- 25.Gillespie GMA, Wills MR, Appay V, et al. Functional heterogeneity and high frequencies of cytomegalovirus-specific CD8+ T lymphocytes in healthy seropositive donors. Journal of Virology. 2000;74(17):8140–8150. doi: 10.1128/jvi.74.17.8140-8150.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Hislop AD, Gudgeon NH, Callan MFC, et al. EBV-specific CD8+ T cell memory: relationships between epitope specificity, cell phenotype, and immediate effector function. Journal of Immunology. 2001;167(4):2019–2029. doi: 10.4049/jimmunol.167.4.2019. [DOI] [PubMed] [Google Scholar]
- 27.Champagne P, Ogg GS, King AS, et al. Skewed maturation of memory HIV-specific CD8 T lymphocytes. Nature. 2001;410(6824):106–111. doi: 10.1038/35065118. [DOI] [PubMed] [Google Scholar]
- 28.Gamadia LE, Rentenaar RJ, Baars PA, et al. Differentiation of cytomegalovirus-specific CD8+ T cells in healthy and immunosuppressed virus carriers. Blood. 2001;98(3):754–761. doi: 10.1182/blood.v98.3.754. [DOI] [PubMed] [Google Scholar]
- 29.Appay V, Dunbar PR, Callan M, et al. Memory CD8+ T cells vary in differentiation phenotype in different persistent virus infections. Nature Medicine. 2002;8(4):379–385. doi: 10.1038/nm0402-379. [DOI] [PubMed] [Google Scholar]
- 30.Papagno L, Spina CA, Marchant A, et al. Immune activation and CD8+ T-cell differentiation towards senescence in HIV-1 infection. PLoS Biology. 2004;2(2, article no. E20) doi: 10.1371/journal.pbio.0020020. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Tomiyama H, Takata H, Matsuda T, Takiguchi M. Phenotypic classification of human CD8+ T cells reflecting their function: inverse correlation between quantitative expression of CD27 and cytotoxic effector function. European Journal of Immunology. 2004;34(4):999–1010. doi: 10.1002/eji.200324478. [DOI] [PubMed] [Google Scholar]
- 32.Rufer N, Zippelius A, Batard P, et al. Ex vivo characterization of human CD8+ T subsets with distinct replicative history and partial effector functions. Blood. 2003;102(5):1779–1787. doi: 10.1182/blood-2003-02-0420. [DOI] [PubMed] [Google Scholar]
- 33.Monteiro M, Evaristo C, Legrand A, Nicoletti A, Rocha B. Cartography of gene expression in CD8 single cells: novel CCR7- subsets suggest differentiation independent of CD45RA expression. Blood. 2007;109(7):2863–2870. doi: 10.1182/blood-2006-06-027060. [DOI] [PubMed] [Google Scholar]
- 34.Romero P, Zippelius A, Kurth I, et al. Four functionally distinct populations of human effector-memory CD8+ T lymphocytes. Journal of Immunology. 2007;178(7):4112–4119. doi: 10.4049/jimmunol.178.7.4112. [DOI] [PubMed] [Google Scholar]
- 35.Hislop AD, Annels NE, Gudgeon NH, Leese AM, Rickinson AB. Epitope-specific evolution of human CD8+ T cell responses from primary to persistent phases of Epstein-Barr virus infection. Journal of Experimental Medicine. 2002;195(7):893–905. doi: 10.1084/jem.20011692. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Chen G, Shankar P, Lange C, et al. CD8 T cells specific for human immunodeficiency virus, Epstein-Barr virus, and cytomegalovirus lack molecules for homing to lymphoid sites of infection. Blood. 2001;98(1):156–164. doi: 10.1182/blood.v98.1.156. [DOI] [PubMed] [Google Scholar]
- 37.Khan N, Cobbold M, Keenan R, Moss PAH. Comparative analysis of CD8+ T cell responses against human cytomegalovirus proteins pp65 and immediate early 1 shows similarities in precursor frequency, oligoclonality, and phenotype. Journal of Infectious Diseases. 2002;185(8):1025–1034. doi: 10.1086/339963. [DOI] [PubMed] [Google Scholar]
- 38.Wills MR, Okecha G, Weekes MP, Gandhi MK, Sissons PJG, Carmichael AJ. Identification of naive or antigen-experienced human CD8+ T cells by expression of costimulation and chemokine receptors: analysis of the human cytomegalovirus-specific CD8+ T cell response. Journal of Immunology. 2002;168(11):5455–5464. doi: 10.4049/jimmunol.168.11.5455. [DOI] [PubMed] [Google Scholar]
- 39.Rocha B, Tanchot C. The Tower of Babel of CD8+ T-cell memory: known facts, deserted roads, muddy waters, and possible dead ends. Immunological Reviews. 2006;211:182–196. doi: 10.1111/j.0105-2896.2006.00378.x. [DOI] [PubMed] [Google Scholar]
- 40.Wiesel M, Walton S, Richter K, Oxenius A. Virus-specific CD8 T cells: activation, differentiation and memory formation. Acta Pathologica, Microbiologica. et Immunologica Scandinavica. 2009;117(5-6):356–381. doi: 10.1111/j.1600-0463.2009.02459.x. [DOI] [PubMed] [Google Scholar]
- 41.Chattopadhyay PK, Roederer M. Good cell, bad cell: flow cytometry reveals T-cell subsets important in HIV disease. Cytometry Part A. 2010;77(7):614–622. doi: 10.1002/cyto.a.20905. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Speiser DE, Migliaccio M, Pittet MJ, et al. Human CD8+ T cells expressing HLA-DR and CD28 show telomerase activity and are distinct from cytolytic effector T cells. European Journal of Immunology. 2001;31(2):459–466. doi: 10.1002/1521-4141(200102)31:2<459::aid-immu459>3.0.co;2-y. [DOI] [PubMed] [Google Scholar]
- 43.Iancu EM, Corthesy P, Baumgaertner P, et al. Clonotype selection and composition of human CD8 T cells specific for persistent herpes viruses varies with differentiation but is stable over time. Journal of Immunology. 2009;183(1):319–331. doi: 10.4049/jimmunol.0803647. [DOI] [PubMed] [Google Scholar]
- 44.Touvrey C, Derré L, Devevre E, et al. Dominant human CD8 T cell clonotypes persist simultaneously as memory and effector cells in memory phase. Journal of Immunology. 2009;182(11):6718–6726. doi: 10.4049/jimmunol.0803095. [DOI] [PubMed] [Google Scholar]
- 45.Speiser DE, Baumgaertner P, Barbey C, et al. A novel approach to characterize clonality and differentiation of human melanoma-specific T cell responses: spontaneous priming and efficient boosting by vaccination. Journal of Immunology. 2006;177(2):1338–1348. doi: 10.4049/jimmunol.177.2.1338. [DOI] [PubMed] [Google Scholar]
- 46.Derré L, Ferber M, Touvrey C, et al. A novel population of human melanoma-specific CD8 T cells recognizes melan-AMART-1 immunodominant nonapeptide but not the corresponding decapeptide. Journal of Immunology. 2007;179(11):7635–7645. doi: 10.4049/jimmunol.179.11.7635. [DOI] [PubMed] [Google Scholar]
- 47.Barbey C, Baumgaertner P, Devevre E, et al. IL-12 controls cytotoxicity of a novel subset of self-antigen-specific human CD28+ cytolytic T cells. Journal of Immunology. 2007;178(6):3566–3574. doi: 10.4049/jimmunol.178.6.3566. [DOI] [PubMed] [Google Scholar]
- 48.Derré L, Bruyninx M, Baumgaertner P, et al. In vivo persistence of codominant human CD8+ T cell clonotypes is not limited by replicative senescence or functional alteration. Journal of Immunology. 2007;179(4):2368–2379. doi: 10.4049/jimmunol.179.4.2368. [DOI] [PubMed] [Google Scholar]
- 49.Derré L, Bruyninx M, Baumgaertner P, et al. Distinct sets of αβ TCRs confer similar recognition of tumor antigen NY-ESO-1157-165 by interacting with its central Met/Trp residues. Proceedings of the National Academy of Sciences of the United States of America. 2008;105(39):15010–15015. doi: 10.1073/pnas.0807954105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Appay V, Jandus C, Voelter V, et al. New generation vaccine induces effective melanoma-specific CD8+ T cells in the circulation but not in the tumor site. Journal of Immunology. 2006;177(3):1670–1678. doi: 10.4049/jimmunol.177.3.1670. [DOI] [PubMed] [Google Scholar]
- 51.Wieckowski S, Baumgaertner P, Corthesy P, et al. Fine structural variations of alphabetaTCRs selected by vaccination with natural versus altered self-antigen in melanoma patients. The Journal of Immunology. 2009;183:5397–5406. doi: 10.4049/jimmunol.0901460. [DOI] [PubMed] [Google Scholar]
- 52.Voelter V, Pica A, Laurent J, et al. An unusual case of metastatic melanoma sensitive to chemotherapy and immunotherapy, with late immune escape in the brain. Cancer Immunity. 2008;8:p. 6. [PMC free article] [PubMed] [Google Scholar]
- 53.Chattopadhyay PK, Betts MR, Price DA, et al. The cytolytic enzymes granyzme A, granzyme B, and perforin: expression patterns, cell distribution, and their relationship to cell maturity and bright CD57 expression. Journal of Leukocyte Biology. 2009;85(1):88–97. doi: 10.1189/jlb.0208107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Sallusto F, Lanzavecchia A. Exploring pathways for memory T cell generation. Journal of Clinical Investigation. 2001;108(6):805–806. doi: 10.1172/JCI14005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Lanzavecchia A, Sallusto F. Understanding the generation and function of memory T cell subsets. Current Opinion in Immunology. 2005;17(3):326–332. doi: 10.1016/j.coi.2005.04.010. [DOI] [PubMed] [Google Scholar]
- 56.Gett AV, Sallusto F, Lanzavecchia A, Geginat J. T cell fitness determined by signal strength. Nature Immunology. 2003;4(4):355–360. doi: 10.1038/ni908. [DOI] [PubMed] [Google Scholar]
- 57.Bachmann MF, Wolint P, Schwarz K, Oxenius A. Recall proliferation potential of memory CD8+ T cells and antiviral protection. Journal of Immunology. 2005;175(7):4677–4685. doi: 10.4049/jimmunol.175.7.4677. [DOI] [PubMed] [Google Scholar]
- 58.Roberts AD, Ely KH, Woodland DL. Differential contributions of central and effector memory T cells to recall responses. Journal of Experimental Medicine. 2005;202(1):123–133. doi: 10.1084/jem.20050137. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Wherry EJ, Teichgräber V, Becker TC, et al. Lineage relationship and protective immunity of memory CD8T cell subsets. Nature Immunology. 2003;4(3):225–234. doi: 10.1038/ni889. [DOI] [PubMed] [Google Scholar]
- 60.Newell EW, Klein LO, Yu W, Davis MM. Simultaneous detection of many T-cell specificities using combinatorial tetramer staining. Nature Methods. 2009;6(7):497–499. doi: 10.1038/nmeth.1344. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Hadrup SR, Bakker AH, Shu CJ, et al. Parallel detection of antigen-specific T-cell responses by multidimensional encoding of MHC multimers. Nature Methods. 2009;6(7):520–526. doi: 10.1038/nmeth.1345. [DOI] [PubMed] [Google Scholar]
- 62.Ayyoub M, Dojcinovic D, Pignon P, et al. Monitoring of NY-ESO-1 specific CD4+ T cells using molecularly defined MHC class II/His-tag-peptide tetramers. Proceedings of the National Academy of Sciences of the United States of America. 2010;107(16):7437–7442. doi: 10.1073/pnas.1001322107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Makedonas G, Hutnick N, Haney D, et al. Perforin and IL-2 upregulation define qualitative differences among highly functional virus-specific human CD8+ T cells. PLoS Pathogens. 2010;6(3, article no. e1000798) doi: 10.1371/journal.ppat.1000798. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Brunner KT, Mauel J, Cerottini JC, Chapuis B. Quantitative assay of the lytic action of immune lymphoid cells on 51-Cr-labelled allogeneic target cells in vitro; inhibition by isoantibody and by drugs. Immunology. 1968;14(2):181–196. [PMC free article] [PubMed] [Google Scholar]
- 65.Pittet MJ, Rubio-Godoy V, Bioley G, et al. α3 domain mutants of peptide/MHC class I multimers allow the selective isolation of high avidity tumor-reactive CD8 T cells. Journal of Immunology. 2003;171(4):1844–1849. doi: 10.4049/jimmunol.171.4.1844. [DOI] [PubMed] [Google Scholar]
- 66.Zaritskaya L, Shurin MR, Sayers TJ, Malyguine AM. New flow cytometric assays for monitoring cell-mediated cytotoxicity. Expert Review of Vaccines. 2010;9(6):601–616. doi: 10.1586/erv.10.49. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Betts MR, Brenchley JM, Price DA, et al. Sensitive and viable identification of antigen-specific CD8+ T cells by a flow cytometric assay for degranulation. Journal of Immunological Methods. 2003;281(1-2):65–78. doi: 10.1016/s0022-1759(03)00265-5. [DOI] [PubMed] [Google Scholar]
- 68.Devêvre E, Romero P, Mahnke YD. LiveCount Assay: concomitant measurement of cytolytic activity and phenotypic characterisation of CD8+ T-cells by flow cytometry. Journal of Immunological Methods. 2006;311(1-2):31–46. doi: 10.1016/j.jim.2006.01.011. [DOI] [PubMed] [Google Scholar]
- 69.Appay V, Speiser DE, Rufer N, et al. Decreased specific CD8+ T cell cross-reactivity of antigen recognition following vaccination with Melan-A peptide. European Journal of Immunology. 2006;36(7):1805–1814. doi: 10.1002/eji.200535805. [DOI] [PubMed] [Google Scholar]
- 70.Rubio-Godoy V, Dutoit V, Zhao Y, et al. Positional scanning-synthetic peptide library-based analysis of self- and pathogen-derived peptide cross-reactivity with tumor-reactive Melan-A-specific CTL. Journal of Immunology. 2002;169(10):5696–5707. doi: 10.4049/jimmunol.169.10.5696. [DOI] [PubMed] [Google Scholar]
- 71.Rufer N, Dragowska W, Thornbury G, Roosnek E, Lansdorp PM. Telomere length dynamics in human lymphocyte subpopulations measured by flow cytometry. Nature Biotechnology. 1998;16(8):743–747. doi: 10.1038/nbt0898-743. [DOI] [PubMed] [Google Scholar]
- 72.Rufer N, Brümmendorf TH, Kolvraa S, et al. Telomere fluorescence measurements in granulocytes and T lymphocyte subsets point to a high turnover of hematopoietic stem cells and memory T cells in early childhood. Journal of Experimental Medicine. 1999;190(2):157–167. doi: 10.1084/jem.190.2.157. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73.Baird DM, Rowson J, Wynford-Thomas D, Kipling D. Extensive allelic variation and ultrashort telomeres in senescent human cells. Nature Genetics. 2003;33(2):203–207. doi: 10.1038/ng1084. [DOI] [PubMed] [Google Scholar]
- 74.Iancu EM, Speiser DE, Rufer N. Assessing ageing of individual T lymphocytes: mission impossible? Mechanisms of Ageing and Development. 2008;129(1-2):67–78. doi: 10.1016/j.mad.2007.10.005. [DOI] [PubMed] [Google Scholar]
- 75.Douek DC, McFarland RD, Keiser PH, et al. Changes in thymic function with age and during the treatment of HIV infection. Nature. 1998;396(6712):690–695. doi: 10.1038/25374. [DOI] [PubMed] [Google Scholar]
- 76.Critchley-Thorne RJ, Yan N, Nacu S, Weber J, Holmes SP, Lee PP. Down-regulation of the interferon signaling pathway in T lymphocytes from patients with metastatic melanoma. PLoS Medicine. 2007;4(5, article no. e176) doi: 10.1371/journal.pmed.0040176. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 77.Perez OD, Mitchell D, Jager GC, et al. Leukocyte functional antigen 1 lowers T cell activation thresholds and signaling through cytohesin-1 and Jun-activating binding protein 1. Nature Immunology. 2003;4(11):1083–1092. doi: 10.1038/ni984. [DOI] [PubMed] [Google Scholar]
- 78.Kuss I, Rabinowich H, Gooding W, Edwards R, Whiteside TL. Expression of ζ in T cells prior to interleukin-2 therapy as a predictor of response and survival in patients with ovarian carcinoma. Cancer Biotherapy and Radiopharmaceuticals. 2002;17(6):631–640. doi: 10.1089/108497802320970235. [DOI] [PubMed] [Google Scholar]
- 79.Whiteside TL. Down-regulation of ζ-chain expression in T cells: a biomarker of prognosis in cancer? Cancer Immunology, Immunotherapy. 2004;53(10):865–878. doi: 10.1007/s00262-004-0521-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 80.Dunbar PR, Smith CL, Chao D, et al. A shift in the phenotype of melan-A-specific CTL identifies melanoma patients with an active tumor-specific immune response. Journal of Immunology. 2000;165(11):6644–6652. doi: 10.4049/jimmunol.165.11.6644. [DOI] [PubMed] [Google Scholar]
- 81.Mortarini R, Piris A, Maurichi A, et al. Lack of terminally differentiated tumor-specific CD8+ T cells at tumor site in spite of antitumor immunity to self-antigens in human metastatic melanoma. Cancer Research. 2003;63(10):2535–2545. [PubMed] [Google Scholar]
- 82.Zippelius A, Batard P, Rubio-Godoy V, et al. Effector function of human tumor-specific CD8 T cells in melanoma lesions: a state of local functional tolerance. Cancer Research. 2004;64(8):2865–2873. doi: 10.1158/0008-5472.can-03-3066. [DOI] [PubMed] [Google Scholar]
- 83.Ahmadzadeh M, Johnson LA, Heemskerk B, et al. Tumor antigen-specific CD8 T cells infiltrating the tumor express high levels of PD-1 and are functionally impaired. Blood. 2009;114(8):1537–1544. doi: 10.1182/blood-2008-12-195792. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 84.Ahmadzadeh M, Felipe-Silva A, Heemskerk B, et al. FOXP3 expression accurately defines the population of intratumoral regulatory T cells that selectively accumulate in metastatic melanoma lesions. Blood. 2008;112(13):4953–4960. doi: 10.1182/blood-2008-06-163048. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 85.Beyer M, Karbach J, Mallmann MR, et al. Cancer vaccine enhanced, non-tumor-reactive CD8+ T cells exhibit a distinct molecular program associated with "division arrest anergy". Cancer Research. 2009;69(10):4346–4354. doi: 10.1158/0008-5472.CAN-08-3796. [DOI] [PubMed] [Google Scholar]
- 86.Rufer N. Molecular tracking of antigen-specific T-cell clones during immune responses. Current Opinion in Immunology. 2005;17(4):441–447. doi: 10.1016/j.coi.2005.06.003. [DOI] [PubMed] [Google Scholar]
- 87.Appay V, Bosio A, Lokan S, et al. Sensitive gene expression profiling of human T cell subsets reveals parallel post-thymic differentiation for CD4+ and CD8+ lineages. Journal of Immunology. 2007;179(11):7406–7414. doi: 10.4049/jimmunol.179.11.7406. [DOI] [PubMed] [Google Scholar]
- 88.Rufer N, Reichenbach P, Romero P. Methods for the ex vivo characterization of human CD8+ T subsets based on gene expression and replicative history analysis. Methods in Molecular Medicine. 2005;109:265–284. doi: 10.1385/1-59259-862-5:265. [DOI] [PubMed] [Google Scholar]
- 89.Peixoto A, Monteiro M, Rocha B, Veiga-Fernandes H. Quantification of multiple gene expression in individual cells. Genome Research. 2004;14(10):1938–1947. doi: 10.1101/gr.2890204. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 90.Peixoto A, Evaristo C, Munitic I, et al. CD8 single-cell gene coexpression reveals three different effector types present at distinct phases of the immune response. Journal of Experimental Medicine. 2007;204(5):1193–1205. doi: 10.1084/jem.20062349. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 91.Esumi S, Wu S-X, Yanagawa Y, Obata K, Sugimoto Y, Tamamaki N. Method for single-cell microarray analysis and application to gene-expression profiling of GABAergic neuron progenitors. Neuroscience Research. 2008;60(4):439–451. doi: 10.1016/j.neures.2007.12.011. [DOI] [PubMed] [Google Scholar]
- 92.Fiesel FC, Voigt A, Weber SS, et al. Knockdown of transactive response DNA-binding protein (TDP-43) downregulates histone deacetylase 6. The EMBO Journal. 2010;29(1):209–221. doi: 10.1038/emboj.2009.324. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 93.Waak J, Weber SS, Waldenmaier A, et al. Regulation of astrocyte inflammatory responses by the Parkinson’s disease-associated gene DJ-1. FASEB Journal. 2009;23(8):2478–2489. doi: 10.1096/fj.08-125153. [DOI] [PubMed] [Google Scholar]
- 94.Alunni-Fabbroni M, Sandri MT. Circulating tumour cells in clinical practice: methods of detection and possible characterization. Methods. 2010;50(4):289–297. doi: 10.1016/j.ymeth.2010.01.027. [DOI] [PubMed] [Google Scholar]
- 95.Mayer V, Schoen U, Holinski-Feder E, Koehler U, Thalhammer S. Single cell analysis of mutations in the APC gene. Fetal Diagnosis and Therapy. 2009;26:148–156. doi: 10.1159/000248721. [DOI] [PubMed] [Google Scholar]
- 96.Woide D, Mayer V, Wachtmeister T, et al. Single particle adsorbing transfer system. Biomedical Microdevices. 2009;11(3):609–614. doi: 10.1007/s10544-008-9270-8. [DOI] [PubMed] [Google Scholar]
- 97.Lutz-Bonengel S, Sänger T, Parson W, et al. Single lymphocytes from two healthy individuals with mitochondrial point heteroplasmy are mainly homoplasmic. International Journal of Legal Medicine. 2008;122(3):189–197. doi: 10.1007/s00414-007-0190-6. [DOI] [PubMed] [Google Scholar]
- 98.Kroneis T, Gutstein-Abo L, Kofler K, et al. Automatic retrieval of single microchimeric cells and verification of identity by on-chip multiplex PCR. Journal of Cellular and Molecular Medicine. 2010;14(4):954–969. doi: 10.1111/j.1582-4934.2009.00784.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 99.Holodick NE, Repetny K, Zhong X, Rothstein TL. Adult BM generates CD5+ B1 cells containing abundant N-region additions. European Journal of Immunology. 2009;39(9):2383–2394. doi: 10.1002/eji.200838920. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 100.Pannetier C, Even J, Kourilsky P. T-cell repertoire diversity and clonal expansions in normal and clinical samples. Immunology Today. 1995;16(4):176–181. doi: 10.1016/0167-5699(95)80117-0. [DOI] [PubMed] [Google Scholar]
- 101.Roux E, Helg C, Dumont-Girard F, Chapuis B, Jeannet M, Roosnek E. Analysis of T-cell repopulation after allogeneic bone marrow transplantation: significant differences between recipients of T-cell depleted and unmanipulated grafts. Blood. 1996;87(9):3984–3992. [PubMed] [Google Scholar]
- 102.Turner SJ, Kedzierska K, La Gruta NL, Webby R, Doherty PC. Characterization of CD8+ T cell repertoire diversity and persistence in the influenza a virus model of localized, transient infection. Seminars in Immunology. 2004;16(3):179–184. doi: 10.1016/j.smim.2004.02.005. [DOI] [PubMed] [Google Scholar]
- 103.Maryanski JL, Jongeneel CV, Bucher P, Casanova J-L, Walker PR. Single-cell PCR analysis of TCR repertoires selected by antigen in vivo: a high magnitude CD8 response is comprised of very few clones. Immunity. 1996;4(1):47–55. doi: 10.1016/s1074-7613(00)80297-6. [DOI] [PubMed] [Google Scholar]
- 104.Turner SJ, Diaz G, Cross R, Doherty PC. Analysis of clonotype distribution and persistence for an influenza virus-specific CD8+ T cell response. Immunity. 2003;18(4):549–559. doi: 10.1016/s1074-7613(03)00087-6. [DOI] [PubMed] [Google Scholar]
- 105.Kedzierska K, Turner SJ, Doherty PC. Conserved T cell receptor usage in primary and recall responses to an immunodominant influenza virus nucleoprotein epitope. Proceedings of the National Academy of Sciences of the United States of America. 2004;101(14):4942–4947. doi: 10.1073/pnas.0401279101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 106.Kaech SM, Wherry EJ. Heterogeneity and cell-fate decisions in effector and memory CD8+ T cell differentiation during viral infection. Immunity. 2007;27(3):393–405. doi: 10.1016/j.immuni.2007.08.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 107.Wherry EJ, Ha S-J, Kaech SM, et al. Molecular signature of CD8+ T cell exhaustion during chronic viral infection. Immunity. 2007;27(4):670–684. doi: 10.1016/j.immuni.2007.09.006. [DOI] [PubMed] [Google Scholar]
- 108.Stemberger C, Huster KM, Koffler M, et al. A single naive CD8+ T cell precursor can develop into diverse effector and memory subsets. Immunity. 2007;27(6):985–997. doi: 10.1016/j.immuni.2007.10.012. [DOI] [PubMed] [Google Scholar]
- 109.Kedzierska K, Venturi V, Field K, Davenport MP, Turner SJ, Doherty PC. Early establishment of diverse T cell receptor profiles for influenza-specific CD8+CD62Lhi memory T cells. Proceedings of the National Academy of Sciences of the United States of America. 2006;103(24):9184–9189. doi: 10.1073/pnas.0603289103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 110.Bouneaud C, Garcia Z, Kourilsky P, Pannetier C. Lineage relationships, homeostasis, and recall capacities of central- and effector-memory CD8 T cells in vivo. Journal of Experimental Medicine. 2005;201(4):579–590. doi: 10.1084/jem.20040876. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 111.Baron V, Bouneaud C, Cumano A, et al. The repertoires of circulating human CD8+ central and effector memory T cell subsets are largely distinct. Immunity. 2003;18(2):193–204. doi: 10.1016/s1074-7613(03)00020-7. [DOI] [PubMed] [Google Scholar]
- 112.Lefranc M-P, Giudicelli V, Ginestoux C, et al. IMGT, the international ImMunoGeneTics database. Nucleic Acids Research. 1999;27(1):209–212. doi: 10.1093/nar/27.1.209. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 113.Douek DC, Betts MR, Brenchley JM, et al. A novel approach to the analysis of specificity, clonality, and frequency of HIV-specific T cell responses reveals a potential mechanism for control of viral escape. Journal of Immunology. 2002;168(6):3099–3104. doi: 10.4049/jimmunol.168.6.3099. [DOI] [PubMed] [Google Scholar]
- 114.Price DA, Asher TE, Wilson NA, et al. Public clonotype usage identifies protective gag-specific CD8+ t cell responses in SIV infection. Journal of Experimental Medicine. 2009;206(4):923–936. doi: 10.1084/jem.20081127. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 115.Venturi V, Hui YC, Price DA, Douek DC, Davenport MP. The role of production frequency in the sharing of simian immunodeficiency virus-specific CD8+ TCRs between macaques. Journal of Immunology. 2008;181(4):2597–2609. doi: 10.4049/jimmunol.181.4.2597. [DOI] [PubMed] [Google Scholar]
- 116.Price DA, West SM, Betts MR, et al. T cell receptor recognition motifs govern immune escape patterns in acute SIV infection. Immunity. 2004;21(6):793–803. doi: 10.1016/j.immuni.2004.10.010. [DOI] [PubMed] [Google Scholar]
- 117.Smith MZ, Asher TE, Venturi V, et al. Limited maintenance of vaccine-induced simian immunodeficiency virus-specific CD8 T-cell receptor clonotypes after virus challenge. Journal of Virology. 2008;82(15):7357–7368. doi: 10.1128/JVI.00607-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 118.Almeida JR, Price DA, Papagno L, et al. Superior control of HIV-1 replication by CD8+ T cells is reflected by their avidity, polyfunctionality, and clonal turnover. Journal of Experimental Medicine. 2007;204(10):2473–2485. doi: 10.1084/jem.20070784. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 119.Schaubert KL, Price DA, Salkowitz JR, et al. Generation of robust CD8+ T-cell responses against subdominant epitopes in conserved regions of HIV-1 by repertoire mining with mimotopes. European Journal of Immunology. 2010;40(7):1950–1962. doi: 10.1002/eji.200940079. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 120.Almeida JR, Sauce D, Price DA, et al. Antigen sensitivity is a major determinant of CD8+ T-cell polyfunctionality and HIV-suppressive activity. Blood. 2009;113(25):6351–6360. doi: 10.1182/blood-2009-02-206557. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 121.Geldmacher C, Metzler IS, Tovanabutra S, et al. Minor viral and host genetic polymorphisms can dramatically impact the biologic outcome of an epitope-specific CD8 T-cell response. Blood. 2009;114(8):1553–1562. doi: 10.1182/blood-2009-02-206193. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 122.Schaubert KL, Price DA, Frahm N, et al. Availability of a diversely avid CD8+ T cell repertoire specific for the subdominant HLA-A2-restricted HIV-1 Gag p2419-27 epitope. Journal of Immunology. 2007;178(12):7756–7766. doi: 10.4049/jimmunol.178.12.7756. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 123.Kan-Mitchell J, Bajcz M, Schaubert KL, et al. Degeneracy and repertoire of the human HIV-1 Gag p1777-85 CTL response. Journal of Immunology. 2006;176(11):6690–6701. doi: 10.4049/jimmunol.176.11.6690. [DOI] [PubMed] [Google Scholar]
- 124.Venturi V, Chin HY, Asher TE, et al. TCR beta-chain sharing in human CD8+ T cell responses to cytomegalovirus and EBV. Journal of Immunology. 2008;181(11):7853–7862. doi: 10.4049/jimmunol.181.11.7853. [DOI] [PubMed] [Google Scholar]
- 125.Price DA, Brenchley JM, Ruff LE, et al. Avidity for antigen shapes clonal dominance in CD8+ T cell populations specific for persistent DNA viruses. Journal of Experimental Medicine. 2005;202(10):1349–1361. doi: 10.1084/jem.20051357. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 126.Cole DK, Edwards ES, Wynn KK, et al. Modification of MHC anchor residues generates heteroclitic peptides that alter TCR binding and T cell recognition. The Journal of Immunology. 2010;185:2600–2610. doi: 10.4049/jimmunol.1000629. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 127.Cole DK, Yuan F, Rizkallah PJ, et al. Germ line-governed recognition of a cancer epitope by an immunodominant human T-cell receptor. Journal of Biological Chemistry. 2009;284(40):27281–27289. doi: 10.1074/jbc.M109.022509. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 128.Melenhorst JJ, Scheinberg P, Chattopadhyay PK, et al. High avidity myeloid leukemia-associated antigen-specific CD8+ T cells preferentially reside in the bone marrow. Blood. 2009;113(10):2238–2244. doi: 10.1182/blood-2008-04-151969. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 129.Trautmann L, Rimbert M, Echasserieau K, et al. Selection of T cell clones expressing high-affinity public TCRs within human cytomegalovirus-specific CD8 T cell responses. Journal of Immunology. 2005;175(9):6123–6132. doi: 10.4049/jimmunol.175.9.6123. [DOI] [PubMed] [Google Scholar]
- 130.Venturi V, Kedzierska K, Price DA, et al. Sharing of T cell receptors in antigen-specific responses is driven by convergent recombination. Proceedings of the National Academy of Sciences of the United States of America. 2006;103(49):18691–18696. doi: 10.1073/pnas.0608907103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 131.Venturi V, Price DA, Douek DC, Davenport MP. The molecular basis for public T-cell responses? Nature Reviews Immunology. 2008;8(3):231–238. doi: 10.1038/nri2260. [DOI] [PubMed] [Google Scholar]
- 132.Dietrich P-Y, Le Gal F-A, Dutoit V, et al. Prevalent role of TCR α-chain in the selection of the preimmune repertoire specific for a human tumor-associated self-antigen. Journal of Immunology. 2003;170(10):5103–5109. doi: 10.4049/jimmunol.170.10.5103. [DOI] [PubMed] [Google Scholar]
- 133.Trautmann L, Labarrière N, Jotereau F, et al. Dominant TCR Vα usage by virus and tumor-reactive T cells with wide affinity ranges for their specific antigens. European Journal of Immunology. 2002;32(11):3181–3190. doi: 10.1002/1521-4141(200211)32:11<3181::AID-IMMU3181>3.0.CO;2-2. [DOI] [PubMed] [Google Scholar]
- 134.Serana F, Sottini A, Caimi L, et al. Identification of a public CDR3 motif and a biased utilization of T-cell receptor V beta and J beta chains in HLA-A2/Melan-A-specific T-cell clonotypes of melanoma patients. Journal of Translational Medicine. 2009;7, article no. 21 doi: 10.1186/1479-5876-7-21. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 135.Uchi H, Stan R, Turk MJ, et al. Unraveling the complex relationship between cancer immunity and autoimmunity: lessons from melanoma and vitiligo. Advances in Immunology. 2006;90:215–241. doi: 10.1016/S0065-2776(06)90006-6. [DOI] [PubMed] [Google Scholar]
- 136.Clay TM, Custer MC, McKee MD, et al. Changes in the fine specificity of gp100((209–217))-reactive T cells in patients following vaccination with a peptide modified at an HLA-A2.1 anchor residue. Journal of Immunology. 1999;162(3):1749–1755. [PubMed] [Google Scholar]
- 137.Stuge TB, Holmes SP, Saharan S, et al. Diversity and recognition efficiency of T cell responses to cancer. PLoS Medicine. 2004;1(2, article no. e28) doi: 10.1371/journal.pmed.0010028. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 138.Le Gal F-A, Ayyoub M, Dutoit V, et al. Distinct structural TCR repertoires in naturally occurring versus vaccine-induced CD8+ T-cell responses to the tumor-specific antigen NY-ESO-1. Journal of Immunotherapy. 2005;28(3):252–257. doi: 10.1097/01.cji.0000161398.34701.26. [DOI] [PubMed] [Google Scholar]
- 139.Bendle GM, Haanen JB, Schumacher TN. Preclinical development of T cell receptor gene therapy. Current Opinion in Immunology. 2009;21(2):209–214. doi: 10.1016/j.coi.2009.02.007. [DOI] [PubMed] [Google Scholar]
- 140.Johnson LA, Morgan RA, Dudley ME, et al. Gene therapy with human and mouse T-cell receptors mediates cancer regression and targets normal tissues expressing cognate antigen. Blood. 2009;114(3):535–546. doi: 10.1182/blood-2009-03-211714. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 141.Schmid DA, Irving MB, Posevitz V, et al. Evidence for a TCR affinity threshold delimiting maximal CD8 T cell function. Journal of Immunology. 2010;184(9):4936–4946. doi: 10.4049/jimmunol.1000173. [DOI] [PubMed] [Google Scholar]
- 142.Dougan M, Dranoff G. Immune therapy for cancer. Annual Review of Immunology. 2009;27:83–117. doi: 10.1146/annurev.immunol.021908.132544. [DOI] [PubMed] [Google Scholar]