Abstract
Molecular modelers and cheminformaticians typically analyze experimental data generated by other scientists. Consequently, when it comes to data accuracy, cheminformaticians are always at the mercy of data providers who may inadvertently publish (partially) erroneous data. Thus, dataset curation is crucial for any cheminformatics analysis such as similarity searching, clustering, QSAR modeling, virtual screening, etc., especially nowadays when the availability of chemical datasets in public domain has skyrocketed in recent years. Despite the obvious importance of this preliminary step in the computational analysis of any dataset, there appears to be no commonly accepted guidance or set of procedures for chemical data curation. The main objective of this paper is to emphasize the need for a standardized chemical data curation strategy that should be followed at the onset of any molecular modeling investigation. Herein, we discuss several simple but important steps for cleaning chemical records in a database including the removal of a fraction of the data that cannot be appropriately handled by conventional cheminformatics techniques. Such steps include the removal of inorganic and organometallic compounds, counterions, salts and mixtures; structure validation; ring aromatization; normalization of specific chemotypes; curation of tautomeric forms; and the deletion of duplicates. To emphasize the importance of data curation as a mandatory step in data analysis, we discuss several case studies where chemical curation of the original “raw” database enabled the successful modeling study (specifically, QSAR analysis) or resulted in a significant improvement of model's prediction accuracy. We also demonstrate that in some cases rigorously developed QSAR models could be even used to correct erroneous biological data associated with chemical compounds. We believe that good practices for curation of chemical records outlined in this paper will be of value to all scientists working in the fields of molecular modeling, cheminformatics, and QSAR studies.
1. Introduction
With the recent advent of high throughput technologies for both compound synthesis and biological screening, there is no shortage of publicly or commercially available datasets and databases1 that can be used for computational drug discovery applications (reviewed recently in Williams, et al.2). Rapid growth of large, publicly available databases (such as PubChem3 or ChemSpider4 containing more than 20 million molecular records each) enabled by experimental projects such as NIH's Molecular Libraries and Imaging Initiative5 provides new opportunities for the development of cheminformatics methodologies and their application to knowledge discovery in molecular databases.
A fundamental assumption of any cheminformatics study is the correctness of the input data generated by experimental scientists and available in various datasets. Nevertheless, a recent study6 showed that on average there are two errors per each medicinal chemistry publication with an overall error rate for compounds indexed in the WOMBAT database7 as high as 8%. In another recent study8, the authors investigated several public and commercial databases to calculate their error rates: the latter were ranging from 0.1 to 3.4% depending on the database.
How significant is the problem of accurate structure representation (given that the error rates in current databases may appear relatively low) as it concerns exploratory cheminformatics and molecular modeling research? Recent investigations by a large group of collaborators from six laboratories9, 10 have clearly demonstrated that the type of chemical descriptors has much greater influence on the prediction performance of QSAR models than the nature of model optimization techniques. These findings suggest that having erroneous structures represented by erroneous descriptors should have a detrimental effect on model performance. Indeed, a recent seminal publication8 clearly pointed out the importance of chemical data curation in the context of QSAR modeling. The authors have discussed the error rates in several known databases and evaluated the consequences of both random and systematic errors with respect to the prediction performances of the derivative QSAR models. They also presented several illustrative examples of incorrect structures generated from either correct or incorrect SMILES. The main conclusions of the study were that small structural errors within a dataset could lead to significant losses of predictive ability of QSAR models. The authors further demonstrated that manual curation of structural data leads to substantial increase in the model predictivity. This conclusion becomes especially important in light of the aforementioned study of Oprea et al.6 that cited a significant error rate in medicinal chemistry literature.
Alarmed by these conclusions, we have examined several popular public databases of bioactive molecules to assess possible error rates in structure representation. For instance, the NCI AIDS Antiviral Screen11 (the paper describing this screen12 has been cited 57 times in PubMed Central only) comprises 42687 chemical records with their associated activity. Even quick analysis of this dataset revealed that 4202 (i.e., ca. 10%) compounds should be either treated with care or removed before any cheminformatics investigation: 3350 compounds were mixtures and salts, and we detected 741 pairs of exact duplicates and stereoisomers, possessing different or opposite reported activities. Similar observations can be made for several well known public databases such as NCI Human Tumor Cell Line and PubChem as well as for smaller datasets studied and reported in published literature. For instance, in a study already mentioned above, six research teams each specializing in QSAR modeling (including the authors of this paper!) collaborated on the analysis of the Tetrahymena Pyriformis aquatic toxicity dataset9, 10 comprising 1093 compounds. Later this exact dataset was used by the organizers of CADASTER toxicity challenge13. However, our re-examination of this dataset showed the presence of six pairs of duplicates among 1093 compounds, due to the presence of different metal cations in salts (with different aquatic toxicities measured by pIGC50 ranging from ~0.1 to 1 logarithmic unit). Furthermore, in the new external set compiled by the organizers of the CADASTER challenge to evaluate the comparative performance of competing models, eight out of 120 compounds were found to be structurally identical to modeling set compounds but with different toxicity values (~0.3 pIGC50).
Such disappointing observations may, at least in part, explain why QSAR models may sometimes fail, which is an issue that was brought up in several recent publications14-16 (but none mentioned structure representation or biological annotation errors as possible sources of poor performance of QSAR models). Obviously, cheminformaticians must only use correct chemical structures and biological activities in their studies. Not surprisingly, any structural error translates into either inability to calculate descriptors for incorrect chemical records or erroneous descriptors. As a consequence, models developed using inaccurate data (either structural or biological) will have insignificant or reduced statistical power (cf. Young, et al8) and will be unreliable for prediction. Since the amount of data, the number of models, and the body of cheminformatics publications continue to grow, it becomes increasingly important to address the issue of data quality that inherently affects the quality of models.
Surprisingly, the investigations into how the primary data quality influences the performances of cheminformatics models are almost absent in the published literature. Besides the study by Young et al.8 mentioned above we also found a paper by Southan et al.17, which mentioned briefly some procedures used to determine the number of unique chemical structures in a database. It appears that for the most part cheminformaticians and molecular modelers tend to take published chemical and biological data at their face value and launch calculations without carefully examining the accuracy of the data records. It is indeed difficult to verify the results of biological assays because it is well known that numerical values of bioactivity for the same compounds measured in the same assays frequently disagree between different laboratories. However, there should be much less disagreement concerning the correct representation of a chemical structure for compounds in the databases except in certain difficult cases, such as chemicals with multiple tautomeric forms18. Very often errors in chemical representation are not obvious, and are difficult to identify without special tools and protocols.
Both common sense and the recent investigations described above indicate that chemical record curation should be viewed as a separate and critical component of any cheminformatics research. By comparison, the community of protein X-ray crystallographers has long recognized the importance of structural data curation; indeed the Protein Data Bank (PDB) team includes a large group of curators whose major job is to process and validate primary data submitted to the PDB by crystallographers19. Furthermore, NIH recently awarded a significant Center grant to a group of scientists from the University of Michigan (http://csardock.org/) where one of the major tasks is to curate primary data on protein-ligand complexes deposited to the PDB. Conversely, to the best of our knowledge, even the largest publicly funded cheminformatics project, i.e., PubChem, is considered as a data repository, i.e., no special effort is dedicated to the curation of structural information deposited to PubChem by various contributors. Chemical data curation has been addressed whenever possible within the publicly available ChemSpider project4; however, until now most effort has focused on data collection and database expansion. Thus, it is critical that scientists who build models using data derived from available databases or extracted from publications dedicate their own effort to the task of data curation.
Although there are obvious and compelling reasons to believe that chemical data curation should be given a lot of attention, it is also obvious that, for the most part, the basic steps to curate a dataset of compounds have been either considered trivial or ignored by experts in the field. For instance, several years ago a group of experts in QSAR modeling developed what is now known as OECD QSAR modeling and validation principles16, 20; these are a set of guidelines that the researchers should follow to achieve the regulatory acceptance of QSAR models. There are five stated principles that require QSAR models to be associated with (i) defined endpoint, (ii) unambiguous algorithm, (iii) defined domain of applicability, (iv) appropriate measures of goodness-of-fit, robustness and predictivity and, (v) if possible, mechanistic interpretation. The need to curate primary data used for model development is not even mentioned. Also, in an effort to improve the quality of publications in the QSAR modeling field, the Journal of Chemical Information and Modeling published a special editorial highlighting the requirements to QSAR papers that authors should follow to publish their results in the journal21. Again, no special attention was given to data curation. Finally, there have been several recent publications addressing common mistakes and criticizing various faulty practices in the QSAR modeling field14-16, 22-24; however, none of these papers has explicitly discussed the importance of chemical record curation for developing robust QSAR models. There is an obvious trend within the community of QSAR modelers to establish and follow the standardized guidelines for developing statistically robust and externally predictive QSAR models25. It appears timely to emphasize the importance of and develop best practices for data preparation prior to initiating the modeling process because it is merely senseless to launch massive cheminformatics or molecular modeling investigations if the underlying chemical structures are not correct.
Arguably, each cheminformatics laboratory may have its own protocol to prepare and curate a compound dataset before embarking on a modeling exercise. However, to the best of our knowledge, there is no published compilation of good practices for dataset curation that at least beginners if not established researchers are advised to follow. In our opinion, there is a pressing need to amend the five OECD principles by adding a sixth rule that would request careful data curation prior to model development. Thus, this paper presents an attempt to address the issue of chemical data curation in a systematic way by pursuing the following major goals:
To alert the cheminformatics and molecular modeling community to the fact that a significant fraction of chemical and bioactivity data in the databases used for modeling may be erroneous, which is likely to reduce the quality of derived models.
To develop a set of data curation procedures integrated into a logical functional workflow that would process the input data and correct structural errors whenever possible (sometimes at the expense of removing incomplete or confusing data records).
To share organized protocols for data curation with the scientific community by providing sample case studies and explicit pointers to the sources where procedures discussed in this paper are available (with a bias towards data curation software that is made available free of charge to academic investigators).
To illustrate, with at least a few examples, that rigorously developed QSAR models using well curated primary data may be employed not only for predicting new structures but also to spot and correct errors in biological data reported in databases used either for model development or model validation.
We wish to emphasize the importance of creating and following a standardized data curation strategy applicable to any ensemble of compounds, which should be really viewed as a community exercise. Although this paper targets mainly the beginners in the cheminformatics field, we believe that it may serve as a general reference for best practices for dataset curation that could be useful for all scientists working in the area of cheminformatics, QSAR, and molecular modeling. We point out a general problem in the field that could be responsible for either reducing the quality of published models or preventing the generation of models worthy of publication. Importantly, we do not pretend to provide an ultimate, all-encompassing collection of curation practices covering all types of difficult or ambiguous cases but focus on most frequent and common cases. We hope that other experts will contribute their knowledge and best practices for dealing with both relatively simple as well as complex issues in subsequent publications.
Due to the complexity of modern chemical biology, there is a clear separation between scientists who generate data and those who analyze them. For the latter, data analytical studies are impossible without trusting the original data sources; however, it is important, whenever possible, to verify the accuracy of primary data before developing any model. To emphasize this point, the title of this paper in part repeats the famous proverb that was frequently cited by the late president Ronald Reagan during the cold war era and that traces back to the founder of the Russian KGB Felix Dzerzhinsky who supposedly invented it almost 100 years ago as a founding principle of his organization (cf. http://en.wikipedia.org/wiki/Trust,_but_verify).
2. Main steps for chemical data curation
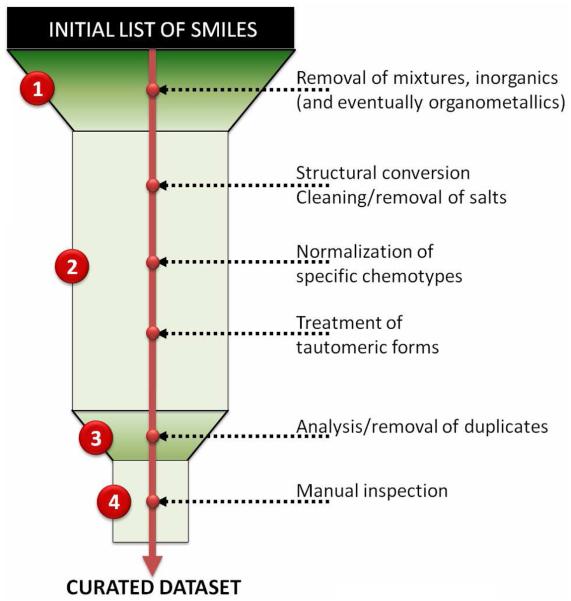
In this section, we discuss the most important steps required to curate a chemical dataset (Figure 1). We specifically focus on chemical structure curation procedures and do not cover the highly relevant but special topic of name to structure conversion, which is often used to create chemical databases (cf. section 3.1); several publications have already addressed the latter subject 7, 26-28. Two main issues are emphasized for each curation procedure: first, we discuss the primary reason why a particular operation should be undertaken, and then provide practical technical advice as to how to do it efficiently. Our goal here is to create a repository of good practices for chemical structure curation, not a software tutorial. The complete technical details concerning the use of each software package employed in our recommended curation protocols can be found on the respective developers' websites and in user manuals. We do not endorse any of the software packages mentioned in this study; however, we are naturally sensitive to the issue of software availability and did tend to select software that is freely available to academic investigators.
Figure 1.
General dataset curation workflow.
It is necessary to note that we focus on the 2D level of molecular structure representation. Such limitation assumes that the topological model (or molecular graph) implicitly contains most of the essential structural information about a given compound. Thus, the curation procedures described herein lead to cleaned 2D representations of compounds. The methods for efficient conversion of 2D molecular graphs to 3D structures are discussed elsewhere29, 30.
2.1. Removal of inorganics and mixtures
Most cheminformatics and QSAR software does not treat inorganic molecules, because the majority of molecular descriptors can be computed for organic compounds only. The inability to model inorganics is an obvious limitation of conventional cheminformatics software. There is a challenging need to develop adequate chemical descriptors for this type of molecules and include them in descriptor calculating software. The fraction of inorganic compounds in most of the available datasets, especially those of relevance to drug discovery is very small. Nevertheless, some datasets generated with the help of automated text-mining approaches extracting data from the literature or electronic sources may contain a significant number of inorganic compounds that are known to have biological effects, e.g., toxic effects (cf. section 3.1). At present, all inorganic compounds must be removed before the descriptors are calculated.
Several approaches can be used to rapidly identify and filter out the inorganic compounds. The following protocol is fast and convenient: assuming that SMILES strings for the original dataset are available and stored in a single SMI file (each line contains the SMILES string for one compound only), one can calculate their empirical formula (e.g., using the cxcalc program included in the ChemAxon JChem package31). Then compounds possessing no carbon atoms (i.e., inorganic molecules) can be easily identified and discarded. A simple Perl or Python script can process the entire database in a few seconds, either by analyzing empirical formula or SMILES strings directly. For non-programmers representing the majority of cheminformaticians, advanced text editors (such as Notepad++32 for Windows) enable a similar automatic treatment with dedicated tools for substring searching and filtering. Once again, in the end a manual inspection of the SMILES list is recommended.
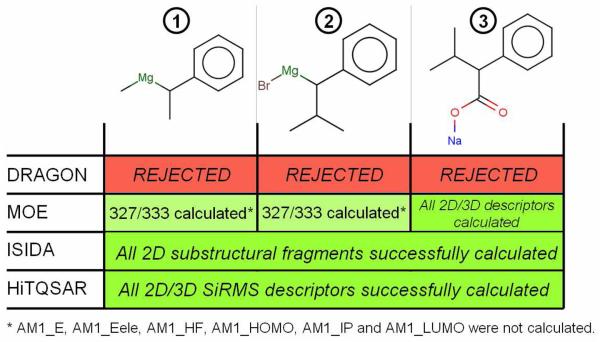
However, it is much more frequent that a dataset includes organic compounds possessing rare elements and organometallics. Dragon molecular descriptors33 can be calculated only for molecules containing the following 38 atoms: H, B, C, N, O, F, Al, Si, P, S, Cl, Cr, Mn, Fe, Co, Ni, Cu, Zn, Ga, Ge, As, Se, Br, Mo, Ag, Cd, In, Sn, Sb, Te, I, Gd, Pt, Au, Hg, Tl, Pb, Bi. As a result, if some compounds in the dataset contain Na, Mg, Ru, etc., they will be rejected by the software (see Figure 2). Meanwhile, such cases could be easily processed by other software, e.g., MOE34. We shall emphasize that we are not highlighting the relative efficiency of different software to compute descriptors for organometallics but merely point out that cheminformaticians should assess whether their modeling tools can handle such compounds and thus decide if they have to remove or keep them in the dataset. There are no available “push-button” solutions for partial data deletion. However, scripts that can identify inorganics by analyzing the compounds' empirical formula and/or SMILES strings can also be utilized to detect organometallics and compounds with rare elements.
Figure 2.
Descriptor calculation for three organometallic compounds using DRAGON, MOE, ISIDA and HiT QSAR software.
Similar to inorganics, most current approaches cannot effectively deal with compound mixtures (although a recent study 35 offered an approach to model such systems). Thus, the second important task of this first curation step concerns the identification and the deletion of mixtures. It is of importance to emphasize for beginners that one single SMILES string can code several molecules: for instance the following SMILES string CC(=N[O-])C(=N[O-])C.CC(=N[O-])C(=N[O-])C.[CH2-]I.C1=CC=NC=C1.[Co] contains one cobalt, two molecules of N,N′-dioxidobutane-2,3-diimine, one iodomethane and one pyridine (CID003290398 in the STITCH database36). Obviously, it is impossible to calculate descriptors using this SMILES string directly.
Treatment of mixtures is not as simple as it appears. The practice of retaining the component with the highest molecular weight or largest number of atoms is common and widely used, but not necessarily the best solution. The best option is first to delete such records prior to descriptor calculation. However, if there is some reason to believe that the experimentally determined biological activity associated with the record is clearly caused by the largest molecule only and not by mixture itself, it is advisable to use the record for the largest molecule in the mixture. Such a simple situation is usually possible only for mixtures formed by a relatively large organic molecule and small inorganic molecule(s), e.g., hydrates, hydrochlorides, etc. In such cases, the molecule with the highest molecular weight or the largest number of atoms (preferably) should be retained for the subsequent analysis. Given that descriptors must be calculated for one molecule only, the most relevant compound in the mixture should be determined and selected. Different situations are possible:
all compounds in the mixture are (or appear to be) identical (e.g., racemic mixtures using 2D representation of molecules): in this case, only one molecule should be kept, and the other(s) should be simply deleted. Certainly, this treatment is only appropriate for 2D QSAR studies when racemic mixture possesses the same activity (property) as corresponding enantiomers;
the mixture contains one large organic compound and several smaller ones, either organic or inorganic. Generally, it is better to delete the entire record. However, if there are some reasons to believe that the experimentally determined biological activity associated with the record is clearly caused by the largest molecule only and not by the entire mixture, one can keep the record: the compound with the highest molecular weight (or the largest number of atoms) can be kept and the others should be deleted;
several similar organic compounds with similar molecular weights: these are the most complicated cases, and usually, the deletion of the entire record is recommended (unless the active ingredient is known and can be selected manually), because it is impossible to determine which compound should be retained for modeling using simple rules and automated software. Manual intervention is required for such cases.
For beginners and non-programmers, the use of ChemAxon Standardizer31 is recommended: the treatment of simple cases is fast and simplified by “drag and drop” graphical tools. Experienced users may prefer to use more advanced tools to determine exactly what kind of mixtures (types i, ii or iii) are present in their datasets.
2.2. Structural conversion and cleaning
The second step of the dataset curation entails the conversion of SMILES strings into 2D molecular graphs. Many programs can accomplish this conversion, e.g., ChemAxon, MOE34, Sybyl37, OpenBabel38, etc. However, the recent study of Young et al.8 emphasized the relatively forgotten problem related to the actual reliability of the conversion from SMILES strings to two-dimensional structures. Indeed they used ChemAxon Marvin to convert a library of SMILES and mined the obtained structures for possible errors. Their results showed that very few compounds (4 out of 2118) were converted incorrectly by Marvin; the other errors were related to the presence of wrong initial SMILES strings (due to manual drawing errors or conversion errors from 2D structures into SMILES strings) in the database. This observation suggests that the direct calculation of descriptors from SMILES using any software is much more risky than using alternative formats (e.g., sdf or Mol2) since SMILES do not allow users to visualize, clean and check chemical structures at 2D level.
Some records in a dataset may correspond to salts that are a common form of many drugs. Although properties of salts can be very different from those of the corresponding neutral molecules 8 and exclusion of salts prior to QSAR analysis is preferred, the removal of metal counterions as well as the neutralization of the remaining carbocations (or carbanions) is still acceptable. Indeed, similar to inorganics, salts are not processed by most of descriptor-generating software and their presence can generate numerous errors in descriptors' calculation. The neutralization of the charged organic molecules is more disputable: it is quite rare to know precisely the experimental conditions under which the compounds have been tested or the physico-chemical environment within cells where the compound is active. In those cases when the pH of the solution and its exact composition are known, it may be possible to evaluate if a compound should have a charge. Making such prediction still requires the knowledge of pKa for each proton donor/acceptor group in the molecule and several available predictors (e.g., from ACDLabs39 or ChemAxon) could help. When reliable estimates are impossible or if descriptors used for the analysis are insensitive to charge, it is simply recommended to neutralize respective compounds, especially in the case of large datasets containing just a few salts. This task can be successfully realized by MOE34, ChemAxon Standardizer or OpenBabel38, which identifies salts, delete counterions and then neutralize the remaining organic compounds. Even zwitterions are successfully treated by most available software. However some difficult cases like compounds where there are covalent bonds between metals and molecules (see compound 3 on Figure 2) can also be encountered. From our experience, such cases are not properly treated by the aforementioned software. Advanced scripts detecting the presence of metals as well as manual curation are thus needed to curate these cases.
Another question concerns the explicit or implicit presence of hydrogen atoms in the structures. In our experience the use of explicit hydrogens for calculating 2D descriptors leads in most cases to QSAR models with higher prediction performances. However, our experience also suggests that sometimes the use of explicit hydrogens may also introduce noise in the descriptor matrix (especially when using fragment-based descriptors) and thus, lead to less reliable models. Many software packages claim to have reliable procedures for adding/removing hydrogens. For instance, our experience indicates that removing hydrogens is not well realized in certain cases, e.g., when hydrogens are attached to nitrogens in rings or in secondary amines. In those cases, hydrogens are not always removed, which leads to error messages in programs calculating descriptors parameterized for the treatment of hydrogen-depleted graphs, as well as incorrect descriptor values.
2.3. Normalization of specific chemotypes
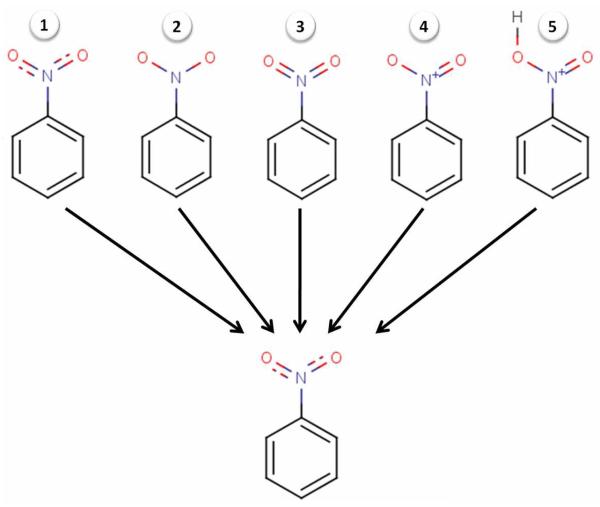
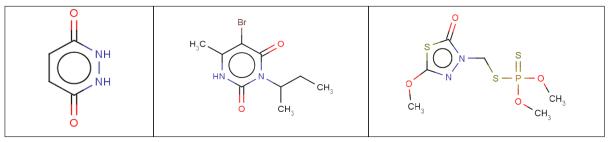
Very often the same functional group may be represented by different structural patterns in a given dataset. For example, nitro groups have multiple mesomers and thus, can be represented using two double bonds between nitrogen and oxygens (neutral form), or one single bond linking the nitrogen and the protonated oxygen, or linking both nitrogen and oxygen atoms that are oppositely charged (Figure 3). For cheminformaticians, these situations may lead to serious inconsistency problems, because molecular descriptors calculated for these different representations of the same chemical group would be significantly different. For example, if two identical compounds contain a nitro group represented by two different patterns, they will not be recognized as identical by conventional similarity metrics because some of their computed descriptors will be different.
Figure 3.
Structure normalization: five types of nitro group representations retrieved in the nitroaromatics dataset for rats and T. pyriformis case studies (see section 3.2 in the text for details).
Manual conversion of all functional groups to some standard forms is too time consuming and could introduce additional human-dependent non-systematic errors. ChemAxon's Standardizer is probably the most well-known tool to rapidly and efficiently realize chemotype normalizations. Users can manually draw the pattern conversion for several functional groups, and store them in a dedicated re-usable xml rule file. Taking into account the specifics of individual research laboratories (modeling software, descriptors, etc.), the users could build a customized database of functional group conversions, which can be applied to every new dataset. Thus, beginners can directly use the library of conversion rules developed by more experienced modelers to treat their datasets in the proper way.
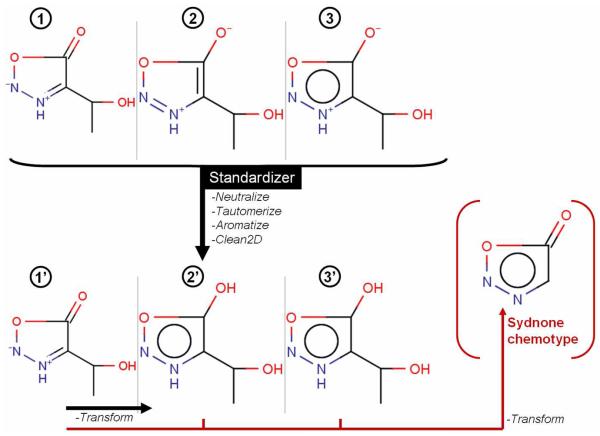
Although ring aromatization and the normalization of carboxyl, nitro and sulfonyl groups are relatively obvious, more complex cases like anionic heterocycles, poly-zwitterions, tautomers, etc., require a deeper analysis and multiple normalization steps. To illustrate this point, we chose three compounds possessing the sydnone chemotype (Figure 4) represented by its different mesomeric forms. The application of the Standardizer with classical settings (neutralize, tautomerize, aromatize and clean2D) was able to normalize two of the three compounds (2′ and 3′) whereas the first compound was not aromatized. A second step using the ‘transform’ function (allowing the conversion of user-defined groups) led to the normalized forms (2′ and 3′) of the compound 1. Depending on the modeled property, the experimental conditions and the other compounds in the dataset, one can transform these compounds into the formal sydnone chemotype (possessing the keto group) like in Figure 4. But, it has to be noted that such notation (aromatic ring and the branched keto group) will be rejected by many descriptor calculating software (Figure 5).
Figure 4.
Use of ChemAxon Standardizer to normalize three compounds possessing the sydnone chemotype (see text for details).
Figure 5.
Examples of misleading structure representations produced by the “general style” option available in ChemAxon Standardizer, which may serve as a potential source of errors for programs calculating molecular descriptors.
In some cases, compounds may exist in several tautomeric forms18, the most common ones being the keto-enolic tautomers. Choosing one form instead of another could have a significant impact on the prediction performances of QSAR models built with such data. As suggested in the study by Young et al.8, discarding one tautomer form may be realized taking into account the compound's mechanism of action, if the latter is known for the studied biological activity. For instance, the choice between keto-enolic tautomeric forms may be influenced by the knowledge about the formation of a specific hydrogen bond with the target receptor, or an aromatic ring. Moreover, experimental conditions of the modeled chemical system (especially, pH) are crucial. A remarkable review addressing different problems related to tautomers was recently published by Dr. Yvonne Martin.18
2.4. Removal of duplicates
Rigorous statistical analysis of any dataset assumes that each compound is unique and thus, structurally different from all other compounds. However, structural duplicates are often present in chemical datasets, especially in large ones. For instance, the same compound could be ordered for a screening campaign from two different sources, given different internal IDs and thus, two corresponding records would be placed in a database (sometimes, with rather different values of the associated experimentally measured property or bioactivity). QSAR models built for such datasets may have artificially skewed predictivity (see section 3.4) even if a dataset contains only a small percentage of duplicates. Duplicates can also affect the observed frequency of a given chemotype in a dataset, the distribution of compounds according to their structural similarity, etc. As a consequence, duplicates must be removed prior to any modeling study. The first step of the procedure requires the detection of identical molecular structures within the set, whereas the second step is dedicated to the comparison of the studied property values for the retrieved duplicates.
A current practice consists of identifying duplicates from SMILES strings which is correct if and only if the latter are canonical SMILES. However, the experience shows that most beginners are not aware of this requirement and often use non-canonical SMILES to identify chemical duplicates. Thus, it is of importance to underline that a given compound can be represented by several SMILES strings: for instance, all the following three formally different SMILES strings, i.e., O=C(OCC)C, C(=O)(OCC)C, and O(C(C)=O)CC are coding the same compound: ethyl acetate. Without the standardization of these SMILES into the CCOC(C)=O, the canonical form (different from the three previous strings), it is impossible to identify them as duplicates in a particular dataset from SMILES strings alone. The calculation of empirical formula from SMILES represents an additional filter to retrieve duplicates.
Once duplicates are identified, the analysis of their properties is mandatory, requiring some manual effort. For a given pair of duplicate structures, if their experimental properties are identical, then one compound should be merely deleted. However, if their experimental properties are numerically different, we shall consider two main scenarios for data curation:
the property value may be wrong for one compound, due to, e.g., a human error when the database was built (these types of errors are often manifested in significant outliers when QSAR models are either built or employed for external predictions). Another frequent case is when the data are compiled from literature sources and the same compound was tested in two or more different laboratories under possibly different experimental conditions, variations in the protocol, etc., leading to the difference in (formally the same) measured property. In this case, supplementary investigations must be done to decide if both entries should be deleted from the dataset, or only one. If the dataset is large enough and there is no obvious explanation of such discrepancies, we recommend to place such cases into a special external test set including all suspicious records and try to reveal the most likely true value by comparing experimental records to the results of consensus predictions from statistically significant and externally validated QSAR models.
both experimental properties are correct but the previous curation tasks (for example, the removal of counterions in salts) have modified the substance records to create such duplicates. For instance, the two records could correspond to two different salts of the same compound (or a neutral compound and its salt). As previously mentioned, their experimental properties can indeed be very different. If both experimental properties are highly similar, the record can be kept associating the structure with the arithmetic average of properties. If they are significantly different, we recommend eliminating both records.
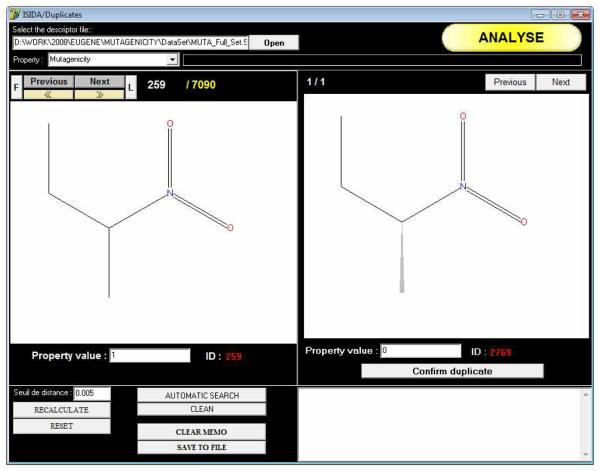
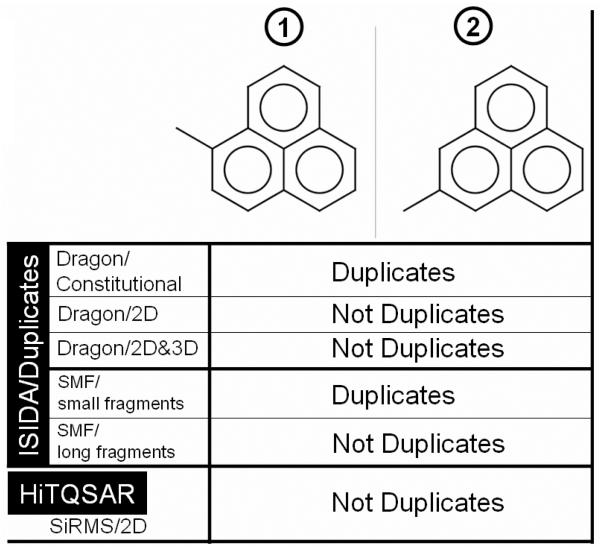
To successfully and rapidly achieve the removal of duplicates, we currently recommend both ISIDA/Duplicates40 (see Figure 6) and HiT QSAR41, 42; these programs are free for academic laboratories and complementary to each other. For each pair of compounds, ISIDA/Duplicates calculates “on the fly” the Euclidean distances between them using the input descriptor matrix of the dataset. Then, all pairs with a distance lower than or equal to a user-defined threshold (zero by default) are considered duplicates. Efficiency depends mainly on the type and the number of descriptors used to represent compounds. The definition of duplicates is strongly associated with the types of descriptors employed to characterize chemicals: for example, two isomers of the same structure would be identified as duplicates if the descriptors used are insensitive to branching in the structures; two stereoisomers will be considered as duplicates in the case where descriptors (e.g., most molecular connectivity indices) do not take into account the chirality. It has to be pointed out that there is no set of descriptors universally recognized to be best for duplicate recognition. An example is given in Figure 7 where two isomers have been processed by ISIDA/Duplicates. If simple descriptors like Dragon/constitutional or ISIDA/small fragments are used, these two compounds are identified as duplicates. One can note that this approach can be useful to rapidly detect pairs of isomers in the database. The use of more complex descriptors such as Dragon/2D and ISIDA/long fragments discriminates the two isomers which are not identified as duplicates anymore. ISIDA/Duplicates natively calculates long fragment descriptors43 that take into account molecular branching and atom connectivity properties, and can also import external sets of descriptors (like Dragon, MOE, etc.) to take into account chirality and other three-dimensional properties. The program can automatically identify duplicates and output the resulting list as well as a curated library of compounds.
Figure 6.
Automatic retrieval of structural duplicates using the ISIDA/Duplicates program: example of stereoisomers (Ames mutagenicity dataset) with opposite mutagenicity properties.
Figure 7.
Two structural isomers retrieved as either duplicates or non-duplicates by ISIDA/Duplicates and HiT QSAR according to different pools of chemical descriptors.
In HiT QSAR the search for duplicates is performed by an innovative one-click tool implementing the CANON algorithm that employs the canonical numeration using the atom connectivity matrix. Each molecule is represented as a string reflecting the empirical formula/order of connectivity for each atom, e.g., benzene would be represented as C6H6/1_2a,1_3a,1_7s,2_4a,2_8s,3_5a,3_9s,4_10s,4_6a,5_11s,5_6a,6_12s/, where atoms 1-6 are carbons and 7-12 are hydrogen atoms. If such strings are similar for different records, the respective compounds are reported as duplicates. However, this approach does have certain limitations: for instance, cis-trans and (R-S) isomers as well as diastereoisomers are considered as duplicates. For this reason the use of both ISIDA/Duplicates and HiT QSAR programs in concert leads to high retrieval rates of real structural duplicates.
2.5. Final manual checking
The last step of the curation entails manual inspection of every molecular structure (as much as possible for the large datasets). We recommend inspecting the curated dataset carefully to establish the types of chemical scaffolds present in the dataset, their relative proportion in the set, etc. Obviously, for large datasets (more than a thousand of compounds), this step is time consuming and extremely laborious. However, several pieces of advice can be formulated to reduce the amount of effort: for instance, to check only compounds with complex structures or having a large number of atoms. Another apparent solution is to generate a representative sample of the set and then, check it for the presence of potential erroneous structures (rechecking of the whole dataset may become unavoidable if significant errors are found). Common errors identified during the manual cleaning procedure may have different origins:
the structure is wrong: a rapid check of both IUPAC compound's name (if available) and its structure is essential to identify possible errors concerning the scaffold and positions of substituents (e.g., due to manual errors or program bugs8 in the conversion of SMILES into 2D structures). Actually, the identification of incorrect structures is the most difficult part of the data curation. The majority of structures are incorrect due to random human errors when the structures have been drawn/converted in an electronic format. Although it is relatively easy for a small dataset to check each individual structure and search for perfect agreement between chemical names and the actual structures, it becomes unfeasible for large datasets. For example, it would take a restrictive amount of time to discover that particular chlorine has to be in position 2 and not in position 1 in the 58,653th compound of a studied dataset. Therefore, we suggest the following protocol: with the development of numerous freely available chemical databases, it is now relatively simple to mine these databases and retrieve chemical structures from a list of names or CAS numbers. Several entries for each name or CAS ID are likely to be retrieved due to known overlaps between databases; some of them are coded as SMILES strings, and some as 2D/3D structures stored in mol, mol2 files, etc. The analysis of these multiple entries for every single compound is then critical: since a given IUPAC name or CAS number is unique for every compound, all structures retrieved using a given query (e.g., 2-chlorobenzylsulfonamide or CAS # 89665-79-2) must represent exactly the same compound. On the other hand, one or several (or all) entries of databases may be wrong. The curation task is then to verify whether the actual structure used in modeling corresponds to the structures retrieved from the database mining exercise. There are no freely available tools specifically dedicated to this task, but we believe that challenges of data integration in bio- and cheminformatics will highlight such tools as being critical for efficient merging between different databases. We also consider that the comparison between different structures retrieved for a given query should be realized at two-dimensional level (three-dimensional if the stereochemistry is specified) but not using SMILES only (there is no guaranty that all SMILES strings in all databases are canonical);
the normalization of bonds is incomplete: common mistakes are related to the presence of different representations of the same functional groups. Despite the normalization procedure, some very specific cases can still be present and thus, the corresponding chemotypes must be corrected manually;
some duplicates may still be present despite the use of automated software to remove them. For instance, some tautomers can still be found. Advanced tools developed internally in private companies or in academic laboratories capable of such fine filtering may exist but they appear to be unavailable in the public domain;
others: wrong charges, presence of explicit hydrogens in a hydrogen depleted structure, incorrect bonds, etc.
2.6. General remarks and disclosure
We would like to stress that the main purpose of this report is not software comparison; this would be simply impossible since the majority of such software is likely hidden within industrial labs. We are mostly concerned with possible sources of inaccurate structures and adequate procedures implemented in available software that should be followed to correct the erroneous data records. Thus, we have attempted to make Table 1, a repository of available software dedicated to data curation, as complete as possible, concerning cases, procedures and listed selected software that is capable of making the requisite corrections. The only bias that we had was towards software that can be obtained free of charge for academic investigators and/or with which we had firsthand experience. Our laboratory is not affiliated in any special way with any of the software vendors and therefore any mentioning of any software should not be regarded as an advertisement.
Table 1.
Summary of major procedures and corresponding relevant software for every step of the data curation process. We invite all interested scientists to enrich this table by adding their preferred procedures and relevant software to the open document available at our web site, http://mml.unc.edu.
| Procedures | Software | Availability |
|---|---|---|
| Inorganics Removal | ChemAxon/Standardizer OpenEye/Filter |
Free for Academia31 Free for Academia67 |
|
Structure Normalization (fragment removal, structural curation, salt neutralization) |
ChemAxon/Standardizer OpenBabel Molecular Networks/ CHECK,TAUTOMER |
Free for Academia31 Free38 Commercial68 |
| Duplicate Removal | ISIDA/Duplicates HiT QSAR CCG/MOE |
Free for Academia40 Free for Academia42 Commercial34 |
|
SDF management/viewer File format converter |
ISIDA/EdiSDF Hyleos/ChemFileBrowser OpenBabel ChemAxon/MarwinView CambridgeSoft/ChemOffice Schrödinger/Canvas ACD/ChemFolder Symix Cheminformatics CCG/MOE Accelrys/Accord Tripos/Benchware Pantheon |
Free40 Free69 Free38 Free for Academia31 Commercial70 Commercial71 Commercial39 Commercial72 Commercial34 Commercial73 Commercial37 |
3. Examples of applications relying on dataset curation
3.1. Cheminformatics analysis of compounds inducing liver injuries
Drug Induced Liver Injury (DILI) is one of the main causes of drug attrition44-46. Elimination of drug candidates likely to cause hepatotoxicity at early stages of drug discovery workflow could significantly increase the rate and reduce the cost of drug development. The ability to predict DILI effects of drug candidates from their chemical structure is critical to help guiding experimental drug discovery projects towards safer medicines. More generally, there is now a great deal of interest both in the US (for instance, with the Toxcast47 program) and Europe (the REACH regulation48) in developing fast and accurate experimental and computational approaches to predict toxic effects of chemicals, e.g., hepatotoxicity.
A large amount of published information that could improve our knowledge about DILI mechanisms is available, but the information is spread over a large body of publications using inconsistent terms. Recently, our group in collaboration with the Biowisdom company49 launched a project concerning the cheminformatics analysis of assertions mined from the biomedical literature that describe DILI effects of chemical compounds. BioWisdom's Sofia™ platform (http://www.biowisdom.com/) was used to generate assertional meta-data, comprising thousands of highly accurate and comprehensive observational statements. These statements are represented in triple constructs: concept_relationship_concept, e.g., Cafestol_suppresses_Bile acid biosynthesis, Azathioprine_induces_Cholestasis, etc. Each assertion is derived from and evidenced by a variety of electronic data sources. More importantly, the assertional meta-data have been collected across different species, i.e., human, rodent and non-rodent animals.
As is often the case for datasets based on literature sources, the initial dataset resulting directly from text mining described compounds by chemical names only. In cases like this, a large number of duplicates is expected because many compounds are described in the literature using many different names; for instance, the ChemSpider 4 search using “aspirin” as a key word indicates that there are more than 35 synonyms that are commonly used for aspirin (or refer to aspirin in literature) and, in total, more than 180 terms (drug names, usual names, etc.) that are related to aspirin. In this recent study 50, after the identifications and curation of obvious chemical synonyms, a set of 1061 compounds (stored as SMILES strings) was retrieved from the analysis of assertions mined from Medline abstracts using text-mining tools. Unlike most traditional QSAR datasets, the compounds were extremely diverse with almost all possible problematic cases in terms of dataset curation: presence of numerous inorganics, mixtures of organics and inorganics, salts, zwitterions and duplicates. No software would be able to compute relevant descriptors for this exotic dataset. As a result no possible cheminformatics analysis (especially QSAR modelling) was possible for this dataset without thorough data curation (see Table 3). Both automatic and manual procedures have been employed to clean this dataset as follows:
- Initially, all inorganic compounds have been removed since our data analysis strategy includes the calculation of molecular descriptors for organic compounds only. We should emphasize again that this is an obvious limitation of many cheminformatics approaches since inorganic molecules are definitely known to induce liver injuries; however, the total fraction of inorganics in our dataset was relatively small. For example, the following compounds have been removed: activated charcoal, cobalt dichloride, ferrous sulphate, zinc chloride, sulphur, cis-diaminedichloroplatinum, manganese chloride, etc. Moreover, additional compounds were removed because (i) their corresponding SMILES strings could not be identified unequivocally due to the inconsistent name or irrelevant labeling code, or (ii) they corresponded to a mixture of compounds (for example, Gramicidin, which involves six antibiotic molecules). Thus, after this step, 993 compounds remained.
- Then, 2D molecular structures (chemical connectivity maps) have been generated from SMILES strings using the ChemAxon's JChem 5.1 program under the control of ISIDA to create a unique SDF file containing both structures and DILI profiles. We also used Standardizer to remove all counterions, clean records including multiple compounds, clean the 2D molecular geometries and normalize bonds (aromatic, nitro groups, etc.) as described in section 2.
- Finally, duplicate molecular structures were detected automatically using the ISIDA/Duplicates 40 program, followed by careful manual inspection of the entire dataset. 951 compounds remained out of the 1061 initial molecules (i.e., as much as ca. 10% of the dataset was eliminated).
Table 3.
Statistical parameters of QSAR models obtained before and after curation.
| ID | Name | R2 | Q2 | R2EF | Sws | Scv | SEF | R2EVS | R2EVS(NM) |
|---|---|---|---|---|---|---|---|---|---|
| 1 | Rat | 0.96 | 0.84-0.93 | 0.89-0.92 | 0.11-0.13 | 0.16-0.24 | 0.20-0.26 | – | – |
| 2 | Rat(NM) | 0.91-0.97 | 0.89-0.95 | 0.45-0.88 | 0.10-0.18 | 0.14-0.28 | 0.28-0.58 | – | – |
| 3 | TP | 0.83 | – | 0.76 | 0.33 | – | 0.38 | 0.54 | −0.58 |
| 4 | TP(NM) | 0.85 | – | 0.54 | 0.31 | – | 0.54 | 0.49 | 0.44 |
| 5 | DILI non-curated | No modeling was possible | |||||||
| 6 | DILI50 | Modeling Set 5-fold external CV Accuracy = 62-68% External sets Accuracy = 56-73% |
|||||||
| 7* |
62Ames non-curated |
SensitivityRF=83%; SensitivitySVM=87%; SpecificityRF=SpecificitySVM=75% AUCGp=88%; AUCSVM=89%; AUCRF=83% |
|||||||
| 8* |
63Ames curated |
SensitivityRF=SensitivitySVM=79%; SpecificityRF=SpecificitySVM=81% AUCGP=86%; AUCSVM=84%; AUCRF=83% |
|||||||
Where:
TP – Tetrahymena pyriformis dataset, (NM) – modeling set with various representations of nitro groups
R2 - determination coefficient, Q2 - cross validation determination coefficient
R2EF- determination coefficient for external folds extracted from the modeling set
Sws - standard error of a prediction for work set
Scv - standard error of prediction for work set in cross validation terms
Sts - standard error of a prediction for external folds extracted from the modeling set
A - number of PLS latent variables, D - number of descriptors, M - number of molecules in the work set
R2EVS - determination coefficient for external validation set
R2EVS(NM) - determination coefficient for external validation set with shuffled nitro groups
AUC – Area Under Curve statistical parameter
RF – Random Forest
SVM – Supporting Vector Machine
GP – Gaussian Processes
Prediction performances are reported for external validation set.
The main objective of the study was to demonstrate the usefulness of classical cheminformatics approaches to analyse assertions of drug-induced liver effects in different species, and more precisely, to explore the relationships between chemical structures and animal DILI toxicity. Thus, our goal was to extract knowledge about the influence of specific scaffolds and chemotypes on DILI. After the critical step of chemical data curation, we have explored the issue of concordance of liver effects across species and found that the concordance values between any two species from the three groups studied, i.e., humans, rodents and non-rodent animals, were relatively low (40-45%), which was in agreement with earlier studies reported in the literature51-53. The subsequent cluster analysis54 of the 951 remaining compounds using 2D fragment descriptors43 allowed us to identify multiple clusters of compounds belonging to structurally congeneric series. Similar liver effect profiles have been observed for most clusters although some compounds appeared as outliers. In several cases of such outliers, additional focused mining of public data sources led to revised assertions that were more in tune with DILI profiles expected on the basis of chemical similarity. Thus, the chemical similarity analysis was helpful in focusing on possible gaps in assertion data for liver effects reported in the literature for different species and correcting the erroneous or missing assertions. In addition, binary QSAR models of liver toxicity were derived and the mean external prediction accuracy in 5-fold external validation study was found to be 65%.
To enable the profile analysis and QSAR modelling using this large dataset extracted from literature using automatic text mining tools, it was essential to utilize various data curation procedures described above. Data cleaning and standardizing was critical since no investigation at all would be possible without having consistent structural representations, correct chemical descriptors and the absence of duplicates. We believe that our study50 presented the first example when rigorous text mining and cheminformatics data analysis were combined towards establishing predictive models of chemical toxicity.
3.2 QSAR modeling of nitroaromatic toxicants
There is a public concern and a strong need in evaluating the potential environmental risks associated with the production, storage, and application of explosive compounds, many of which are nitro- and polynitroaromatics. Recently, a dataset of nitro-aromatic compounds of military interest was compiled from different sources to investigate the relationships between their chemical structure and toxicity. Each compound was manually inspected in order to create a curated dataset. During this process, five different representations of nitro groups were identified (Figure 3). Obviously, the difference in one or two bonds may appear to be insignificant in the context of the entire compound, but in reality, those inconsistencies in the molecular representation of the same functional group could actually lead to different descriptors of the same molecule and, in some cases, to poor QSAR modeling results.
Two sets of nitroaromatic derivatives were compared:
Case Study 1: 28 compounds (2 aromatics and 26 nitroaromatics) tested in rats to evaluate their animal toxicity expressed as log(LD50), mmol/kg.55
Case Study 2: 95 compounds tested against Tetrahymena pyriformis, a ciliated freshwater protozoan to assess their aquatic toxic effects expressed as log(IGC50), mmol/ml.56
In the first case study, five different representations of nitro groups were equally distributed within the modeling set. Initially three different overlapping validation sets were selected according to55, 57. Each external set consisted of one aromatic compound and five nitroaromatics with different types of nitro group representation. The sets with compounds possessing various types of nitro group representations were referred to as “mixed”. 2D simplex descriptors, PLS statistical approach and principles of external test set formation as described in Kuz'min et al.55 were used. Three models were developed using the mixed sets (Table 3). One of them had the same predictivity as original models55 (R2ext ~ 0.9), whereas the prediction accuracy for the two others was much lower (R2ext = 0.45-0.60). Thus, in one case the differences in the nitro group representation had no effect on model predictivity, whereas in the two others the prediction performance decreased significantly. At the same time the goodness-of-fit and robustness of both groups of models for the training set were equal (Tab. 3), confirming the well known Kubinyi paradox 58 and the necessity of external validation59. Thus, one can conclude that simultaneous usage of different types of nitro group representation can significantly influence the predictive ability of the models.
In the second case study, 60% of the modeling set compounds possessed nitro groups represented with aromatic bonds (see Figure 3) whereas the remaining four classes of the nitro group patterns were equally represented (10% each). The same proportions were kept for the external validation set consisting of 63 compounds. The statistical metrics resulting from five-fold external cross-validation of the modeling set and an external validation set were selected as a measure of model predictivity. Similarly to the first case study, the same descriptors and statistical approaches as in the original study56 were used for model generation for the mixed set. Goodness-of-fit and robustness of the original56 and shuffled models were equal (Table 3). Overall comparison of model predictivity in the five-fold external cross-validation experiment shows that the original consensus model is better than the consensus model obtained using the mixed nitro groups. However, their prediction performances estimated using the original external validation set were almost similar. Even more impressive and unexpected results were obtained for the external set when nitro groups were mixed in the same way as for the training set. In the case of a model built on the mixed set, the mixing of the nitro group representation among external validation set compounds did not significantly affect the predictivity (R2ext = 0.49 and R2ext (NM) = 0.44) whereas the difference was dramatic for the original model (R2ext = 0.54 and R2ext (NM) < 0) (Tab. 3). Here, the index “NM” refers to the set with the mixed nitro groups.
The results obtained here with two different nitroaromatics datasets validate the necessity of the nitro group standardization. They show that even small differences in structure representation can lead to significant errors, and even robust and inherently predictive models can fail on non-curated external validation sets.
3.3 ToxRefDB
At the time of the study, the original version of the ToxRef DataBase (http://www.epa.gov/NCCT/toxrefdb/)60 contained 320 compounds tested for their carcinogenicity in both rats and mice (totally 26 panels of experimental data represented as binary results, i.e., toxic or non-toxic). The initial efforts to generate QSAR models for these compounds were unsuccessful: we could not build any statistically significant model based on our standard QSAR modeling workflow25. Thus, we have examined the dataset for possible errors as follows (and we believe this exercise could be illustrative as an example of a training session on the protocol for data examination prior to model building).
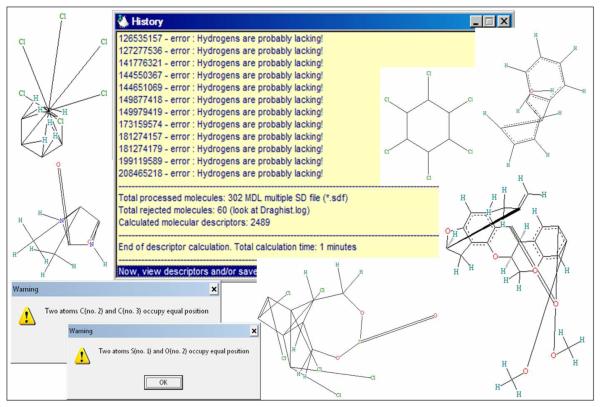
Even a quick examination of the dataset (see Figure 8) revealed that a deep cleaning of 2D structures, addition of explicit hydrogens and standardization of problematic nitro and carboxylic groups was necessary. After applying all of the dataset curation procedures discussed in section 2 as well as aromatization, mixtures and salts removal, standardization of nitro and carboxylic groups, only 293 compounds remained. Subsequent search for duplicates using HiT QSAR and ISIDA software revealed the presence of the (S)-isomer of bioallethrin and its racemate. Since stereochemical information was available for one pair of compounds only, these structures were marked as duplicates on 2D level. Thus, 291 from the original 310 structures were accepted for subsequent modeling (see Table 2).
Figure 8.
Real examples of erroneous structure records in chemical databases leading to Dragon error messages.
Table 2.
Number of investigated compounds in the datasets before and after curation.
| Dataset | Number of compounds | |
|---|---|---|
| Original set | Curated set | |
| Liver toxicants (DILI) | 1061 | 951 (90%) |
| Nitroaromatics (rats) | 28 | 28 (100%) |
| Nitroaromatics (T. pyriformis) | 95 | 95 (100%) |
| ToxRefDB | 320 | 292 (91%) |
| Ames mutagenicity | 7090 | 6542 (92%) |
| Bioavailability (UCSD) | 805 | 734 (91%) |
However, further visual inspection of these structures revealed some misleading representations, especially with respect to unexpected assignment of aromatic bond types. This occurred due to the choice of “General style” instead of the “Basic style” option in the ChemAxon Standardizer. Unlike the “General style”, the ring of 2-pyridone, for instance, is not aromatized under the “Basic style” option. If the “General style” is effectively correct from the chemistry view point, it has to be stressed that many cheminformatics programs will generate errors or simply reject the compound because a carbon atom is not formally tetravalent under this “General style” representation (see Figure 5). Additional information can be found at the following ChemAxon web site (http://www.chemaxon.com/jchem/marvin/help/sci/aromatization-doc.html). This example demonstrates that there is no simple “push-button” solution for chemical data curation and that data inspection and curation including manual involvement is necessary.
Following the chemical record cleaning, the dataset appeared ready for the QSAR modeling. However, the curation procedures should not be limited to structure analysis but should also include the evaluation of the quality of the experimental data (as our earlier example with the analysis of DILI data suggests). Thus, we applied ISIDA/Cluster40 to group similar compounds into clusters. With ISIDA visualization tools, we rapidly identified some suspicious pairs of highly similar compounds (e.g., with only a methyl group in a different position) that nevertheless had large differences in their toxicity profiles. Some of these cases, e.g., ametryn and prometryn could be classical “activity cliffs” but we also found true and suspicious cases of erroneously annotated compounds: for instance, atrazine. At the time of the study and even in a recent version of the ToxRefDB (http://www.epa.gov/NCCT/toxrefdb/files/ToxRefDB_ChronicCancer_2009Apr06.xls), atrazine is annotated as non-tumorigen for both rats and mice. Conversely, two compounds, propazine and simazine, identified by ISIDA/Cluster as structural neighbors of atrazine and different from the latter by presence or absence of only one methyl group, are both annotated as tumorigen agents for rats. After additional investigation we found literature evidence that atrazine has been reported elsewhere as a rat tumorigen 61. We believe that this example suggest a crucial importance of verifying not only molecular structures but also activity data using cheminformatics tools such as clustering by compound similarity and the analysis of property distributions. The results of QSAR modeling of the curated dataset will be published separately.
3.4 Ames Mutagenicity
Recently our group initiated a collaborative modeling project involving many international participants to develop most significant models of the Ames mutagenicity; each research group was expected to use different descriptors and machine learning approaches. The dataset, kindly provided by Dr. K. Hansen62, consists of 7090 compounds classified as mutagenic or non-mutagenic. Briefly, frame-shift mutations or base-pair substitutions can be detected in the Ames test by the exposure of histidine-dependent strains of Salmonella typhimurium to a given compound. Herein, mutagenicity is represented in a binary format: a compound is classified as positive (mutagenic) if it significantly induces revertant colony growth at least in one strain. A compound is labeled negative (non-mutagenic) if it does not induce revertant colony growth in any strain tested.
The original dataset was curated using both HiT QSAR and ISIDA software: (a) all structural duplicates were removed: if both molecules (according to 2D structures) had the same mutagenicity effect, then one of them was removed and if both molecules had different mutagenicity effects, then both were deleted; (b) all inorganic compounds were excluded; (c) the remaining structures were cleaned using the ChemAxon Standardizer and HiT QSAR software; (d) the last step before modeling was the repetition of duplicates search and careful manual checking.
(a) One of the most important steps of the curation procedure is the removal of duplicates: 518 pairs of structural duplicates (at 2D level of structure description) were found by both HiT QSAR and ISIDA software (Figure 6). For the original dataset the situation was as follows: 80% of compounds were represented by 2D structures without any information about stereochemistry and for approximately 20% of the compounds stereochemical information was available. However, in most cases only one of the two enantiomeric forms was characterized. In some cases (about one hundred) the experimental information for both (R)- and (S)- stereoisomers or for two or more diastereoisomers was reported. Moreover, only 7 out of one hundred pairs created by different enantiomers had different mutagenicity. Thus, one can conclude that for the most part the stereochemistry of investigated compounds does not influence their mutagenic effect. As a result, it appeared logical to represent molecular structures at 2D topological level for further QSAR analysis.
Most of the pairs were formed by classical duplicates, i.e., identical (topologically or topologically and stereochemicaly if applicable) structures with the same mutagenicity property; for instance, 2-((4-chlorophenyl)methyl)-oxirane was found twice in the original dataset. Some duplicates represented stereoisomers (R-S or diastereomers) with the same mutagenicity (e.g., (R)- and (S)-penbutolol). In this case, one of the stereoisomers was removed from the dataset. The same procedure was applied for diastereomers with identical mutagenicity (e.g. ursodiol and chenodiol).
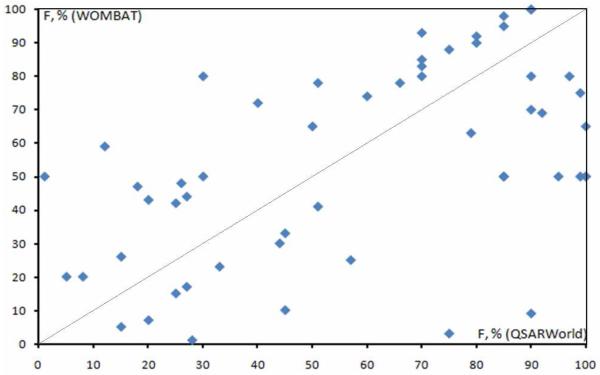
The situation when regular duplicates, similar or different enantiomers ((R)- and (S)-2-nitrobutane) or diastereomers had different mutagenicity values was rare (~ 30 pairs including 7 for different enantiomers) and probably, it could be explained by the presence of errors in the interpretation of experimental data. All such suspicious records were excluded from the subsequent analysis. However, in the absence of data curation, we could easily foresee a situation when identical compounds with identical mutagenicity would be distributed between training and test sets. Should this happen we would expect to observe an artificially enhanced predictive accuracy of the training set models. We shall use this example to illustrate how dataset curation may help design and/or tune more efficiently the modeling parameters such as descriptor types or the machine learning approach. Thus, after duplicates analysis and removal, 6572 compounds remained.
The group that provided us with the Ames dataset has already published the preliminary results of their QSAR studies62; however, the statistical parameters of their models using the non-curated dataset of 7090 compounds were, probably, overestimated since almost 9% of compounds should have been removed. To assess the consequences of their presence in the dataset, we conducted the following study. The dataset was randomly divided into two subsets five times following our standard routine for generating training and validation sets for QSAR modeling and then the content of both sets was analyzed in terms of the presence/absence of duplicates. Results showed that 229-255 out of 518 pairs of duplicates were split between the modeling and external validation sets (it corresponds to the probabilistic distribution). This situation could lead to overestimating the model predictivity, even despite the usage of three-dimensional structures and the whole collection of Dragon-X 1.2 descriptors (including 3D). We would like to emphasize that the overall quality of QSAR models presented in62 is still high despite the presence of duplicates. Nevertheless, in our opinion, their presence significantly biased almost all steps of modeling, from model building to the selection of best models. After the beginning of our collaborative study, the Ames dataset was revised by Dr. Hansen and only 6512 compounds remained in their study63.
(b) In our study, the remaining 6572 compounds were checked for the presence of inorganic compounds using the ISIDA Software. Thirty inorganic compounds such as ammonia and phosphoric acid were excluded.
(c) The remaining 6542 structures were cleaned by the ChemAxon Standardizer (addition of explicit hydrogens, benzene ring aromatization and nitro and carboxylic group standardization) and the HiT QSAR Software (nitro group standardization and connectivity checking).
(d) The last step before the modeling stage was the repetition of search for duplicates and careful manual checking. This step was obviously time-consuming but necessary, because some erroneous structures (lacking hydrogens, having different tautomeric forms or representations of nitro groups, etc.) may become duplicates after structural cleaning. At the end no additional molecule was removed and 6542 compounds still remained for our international QSAR modeling exercise (see Table 2).
The detailed description of the study and its results obtained on the curated dataset will be described in a separate publication because, again, this paper is focusing on the issue of data curation. However, we should mention that in total a group of collaborators has developed as many as 32 predictive QSAR models using different combinations of chemical descriptors and machine learning approaches. It is also worth noting that the results were initially reported by the Hansen group for non-curated datasets62 and the later modeling of the curated data63 showed that the predictivity of models developed on notcurated datasets was indeed somewhat over-estimated (see Table 3) because of the presence of structural duplicates. Unlike small datasets, we should also emphasize that for such a large dataset, the difference in prediction performances of models built before and after curation is statistically significant even when the difference in prediction accuracy is as low as 2%. Moreover, it is clear from Table 3 that models became more balanced (difference between specificity and sensitivity decreased from 8-12% to 2% only). This example illustrates that the use of data curation leads to more predictive and balanced models, along with more objective estimate of their true predictive power.
In addition to statistical aspects of models resulting from the analysis of curated vs non-curated datasets, this study also provides another example of investigation (after the DILI study50), where developed QSAR models were successfully used for the correction of erroneously annotated compounds in the dataset. A compound was considered suspicious and selected for deeper experimental checking if: (i) at least 30 out of 32 models obtained by the different teams failed to predict it accurately (either in modeling or the external validation sets); (ii) two or more structurally similar compounds had different annotations. In total, we have identified 86 suspicious compounds in the external set (51 non-mutagens and 35 mutagens) and 54 compounds in the modeling set (39 non-mutagens and 15 mutagens). Using both manual and automatic literature mining tools, our analysis revealed that 31 compounds (16 from the external set and 15 from the modeling set) were erroneously annotated in the original dataset since we have indeed found published evidence that were in agreement with the model predictions for these compounds. Among them, 29 were originally annotated as non-mutagens, predicted as mutagens and were confirmed by at least one publication to be real Ames mutagens. On the other hand, only two mutagens predicted as non-mutagens were confirmed as non-mutagens. These results are in agreement with earlier observations63 suggesting that the experimental error rate for determining compounds as non-mutagenic is higher than that for mutagens (because compounds tested as negatives in the Ames test in certain bacterial strains may turn out to cause reverse mutations when examined in additional strains). Once again, the identification of 31 mislabeled compounds due to discordance between their stated and predicted properties suggests, perhaps unexpectedly, that predictive QSAR models obtained on carefully curated datasets can be successfully used for experimental biological data curation. Another important impact of experimental data curation applied to these compounds is that, despite an insignificant change in the overall prediction accuracy (see Table 3) for the whole external set (<1%) and the external cross-validation on the modeling set (<0.5%), there is a major improvement in prediction performances for these mislabeled compounds: for the external set, 19% (16 out of 86) of these mislabeled compounds have been predicted correctly (compared to zero percent with the original labels) and 28% (15 out of 54) for the modeling set. Data curation is thus an important criterion for QSAR model improvement in terms of prediction performances and reliability.
3.5 Bioavailability competition
One recent illustration of the famous proverb “the road to hell is paved with good intentions” was the recent “QSARworld Modeling Challenge 2008” organized by QSARWorld (http://www.qsarworld.com). Like other similar challenges, its overall objective was to benchmark modeling techniques from different international teams according to their “blind” prediction performances. The participants were asked to build QSAR models to predict human oral bioavailability using a given training set of compounds, for which both chemical structures and experimental biological data were made available. The best model was supposed to be selected based on the prediction accuracy (RMSE) for an external pre-selected (“blind”) test set, for which only chemical structures were made available.
Our group (and we believe many others) welcomed this Challenge as an excellent opportunity to share the knowledge and experience between QSAR specialists throughout the world. We fully expected to participate in the Challenge that was bound to enable the building of collective QSAR wisdom. Unfortunately, serious concerns regarding the modeling set as well as the method of performance evaluation of QSAR models were rapidly identified. In fact, the concerns were so strong that we concluded that the entire exercise was pointless and withdrew from the participation.
Our decision was based on the following specific reasons: The oral bioavailability data for all test set compounds were supposed to be completely unavailable so that the independent modeling effort could not be compromised in any way (as was indeed the case for another recent Solubility Challenge64, 65). Nevertheless, these “external” data were in fact publicly available through the link placed at the QSARWorld Challenge website [http://modem.ucsd.edu/adme/databases/databases_extend.htm] at the same time when the competition was announced. In fact, the entire dataset (i.e., both training and test sets) was compiled and kindly provided to us upon request by Dr. Tingjun Hou [Dept. of Chemistry and Biochemistry, University of California in San Diego]. In addition, the SDF files provided by QSARWorld contained compound IDs from the Dr. Hou's database66. As a result the blind competitive aspect of the Challenge was obviously compromised because all experimental values for the external dataset were known.
Moreover, it appeared that the quality of the data was low despite the absence of obvious errors in the structure representations. There were only few accidental mistakes, e.g., lack of aromaticity in trapidil or a single (C-O) instead of a double (C=O) bond in ticarcillin. Most likely, some curation procedures were applied by creators of this dataset which is appropriate before launching this kind of challenge. However, the search for duplicates using the HiT QSAR software41, 42 revealed 11 pairs of similar structures. Three of them were different in terms of stereochemical configuration (quinidine and quinine, betamethasone and dexamethasone, and levofloxacin and ofloxacin), and nevertheless different isomers were reported to be associated with identical biological data. The structures in the remaining eight pairs were completely identical. The only difference was in the chemical names of duplicates, e.g. dronabinol and tetrahydrocannabinol or metaprotenerol and orciprenaline. Thus, while the search for duplicates based on names or CAS numbers can be done very rapidly, it is an inefficient tool to discover all chemically identical pairs of compounds. Furthermore, the experimental bioavailability values for similar compounds found in these four pairs differed by 8 to 43%. Such discrepancies are common for datasets compiled from the literature, when experimental values of investigated property are usually averaged. Actually, as we suggested above, activity values with small variations from one source to another could be averaged, but not in the cases such as quetiapine with bioavailability equal to 90% as reported in one source66 and 9% as reported elsewhere 7. Most likely, this difference is caused by simple human error and ‘9’ was intended to be reported as ‘90’. In this case, the removal of a compound with large deviation between experimental values reported in different sources from the training set is highly appropriate. It is also desirable to create a special test set containing such suspicious compounds to help reveal the true value by the means of consensus predictions using QSAR models built on “clean” datasets (cf. our description of both DILI and Ames studies above demonstrating the power of rigorously built QSAR models for correcting false biological data).
In case of any doubt concerning the consistency of the data, the search for analogous information in available sources is also a good solution. Specifically, it was found that some experimental data on human oral bioavailability provided by the Challenge organizers were not always measured on humans. Moreover, the simple comparison of the bioavailability data provided by the Challenge project with those found in WOMBAT-PK7 for a large number of identical compounds revealed a large discrepancy between the two sources for many compounds. Thus, among 220 identical compounds found in both the Challenge and Wombat datasets, 54 compounds showed significant (≥ 10%) differences: ≥ 20% for 25 compounds; ≥ 30% for 15 compounds, and ≥ 50% for 5 compounds (Figure 9). Thus, after the application of the curation procedures to the original dataset, consisting of 805 compounds, only 734 structures remained (see Table 2).
Figure 9.
Experimental bioavailability values (%) from QSARWorld competition (X-axis) vs WOMBAT (Y-axis) for 55 overlapping compounds.
Obviously, we strongly support the idea of organizing QSAR Challenges in general. However, each Challenge should be designed very carefully. We believe that the following simple recommendations may be useful when organizing competitive QSAR studies:
Biological data for the test set should be truly unavailable until the completion of the competition (a good example is the recent 2009 CADASTER Toxicity Challenge13).
Chemical structures should be thoroughly curated, e.g., using procedures outlined in this paper.
Biological data should be of high quality, curated and consistent.
Results of this challenge which are already posted at the QSARWorld website (http://www.qsarworld.com/modleingcompetition08results1.php) also demonstrate the importance of data curation. The best-model is characterized by RMSE of 30%, which is completely unacceptable given that the entire range of data values was 0-100%. Moreover, no additional statistical characteristics of the model quality were reported. In our opinion, the use of RMSE values only to estimate any model performance is insufficient; we always also report R2test (the coefficient of determination is more informative than RMSE in revealing whether or not predicted values reproduce quantitatively the experimental ones) and expect its value to exceed 0.6 to claim that the model is acceptable (as we mentioned the R2test value was not reported but it is highly unlikely that it exceeds 0.6 given the very high RMSE value). This opinion was also validated by the citation of Dr D. Krstajic – the winner of the bioavailability challenge, posted on the same web site: “Unfortunately, for the supplied dataset we were not able to find any good model.” This case study reconfirms that robust and predictive QSAR models cannot be obtained using low-quality data.
4. Conclusions
In this study, the most important steps of dataset cleaning have been described including the removal of inorganics, organometallics, counterions and mixtures; structural cleaning, ring aromatization, normalization of specific chemotypes, curation of tautomeric forms; deletion of duplicates; manual checking of the structures and biological activities. Some general rules following the discussed applications were also formulated:
it is risky to calculate chemical descriptors directly from SMILES. It is preferable to compute descriptors (integral, fragments, etc.) from curated 2D (or 3D if necessary) chemical structures where all chemotypes are strictly normalized;
structural comparison across available databases may facilitate the detection of incorrect structures;
even small differences in functional group representations could cause significant errors in models;
searching for structure-based duplicates and their removal is one of the mandatory steps in QSAR analysis. Such searches based on chemical name or CAS number only are both incomplete and inefficient;
because of the large number of experimental data sources, the search for additional information about investigated property in all available sources is desired to extract valuable knowledge and to compare the data to detect activity cliffs and identify diverse sources of errors;
nothing can replace hands-on participation in the process since some errors obvious to a human are still not obvious for computers. After finishing all the mentioned steps, structures and activities should be checked manually once again.
Table 1 summarizes all proposed procedures (cf. also Figure 1) and provides a list of available software for every step of the curation process. This list is not exhaustive and we invite readers to enrich it via a special link on our website (http://chembench.mml.unc.edu/) by adding references and links to other software that could be of interest in the context of data curation. We consider our paper complimentary to the study by Young et al.8 that provided several examples to illustrate how poor data quality could have detrimental influence on QSAR models.
In conclusion, we believe that there is a clear need for an additional principle that should be added to the five OECD principles for QSAR model development and validation16, 20, and this principle should address the need for data curation before the model development is initiated. We suggest that this additional principle could be formulated as follows: “To ensure the consideration of (Q)SAR models for regulatory purposes, the models must be trained and validated on chemical datasets that have been thoroughly curated with respect to both chemical structure and associated target property values.”
5. Acknowledgements
The authors gratefully acknowledge the financial support from NIH (grant GM66940) and EPA (RD 83382501 and R832720). We also thank Dr. Tudor Oprea and Sunset Molecular, Inc. for the WOMBATPK software license. Finally, the authors should thank the anonymous reviewers of the original version of the manuscript for highly valuable comments and suggestions that helped us significantly improve and enrich this report.
Reference List
- 1.Oprea TI, Tropsha A. Target, Chemical and Bioactivity Databases - Integration is Key. Drug Discov. Today. 2006;3:357–365. [Google Scholar]
- 2.Williams A, Tkachenko V, Lipinski C, Tropsha A, Ekins S. Free online resources enabling crowd-sourced drug discovery. Drug Discovery World. 2010;10:33–39. [Google Scholar]
- 3.PubChem http://pubchem.ncbi.nlm.nih.gov (accessed Feb 1, 2010)
- 4.Chemspider http://www.chemspider.com (accessed Feb 1, 2010)
- 5.Austin CP, Brady LS, Insel TR, Collins FS. NIH Molecular Libraries Initiative. Science. 2004;306:1138–1139. doi: 10.1126/science.1105511. [DOI] [PubMed] [Google Scholar]
- 6.Olah M, Mracec M, Ostopovici L, Rad R, Bora A, Hadaruga N, Olah I, Banda M, Simon Z, Mracec M, Oprea TI. WOMBAT: World of Molecular Bioactivity. In: Oprea TI, editor. Chemoinformatics in Drug Discovery. Wiley-VCH; New York: 2005. pp. 223–239. [Google Scholar]
- 7.Olah M, Rad R, Ostopovici L, Bora A, Hadaruga N, Hadaruga D, Moldovan R, Fulias A, Mracec M, Oprea TI. WOMBAT and WOMBAT-PK: Bioactivity Databases for Lead and Drug Discovery. In: Schreiber SL, Kapoor TM, Wess G, editors. Chemical Biology: From Small Molecules to Systems Biology and Drug Design. Wiley; Weinheim: 2007. pp. 760–786. [Google Scholar]
- 8.Young D, Martin T, Venkatapathy R, Harten P. Are the chemical structures in your QSAR correct? QSAR Comb. Sci. 2008;27:1337–1345. [Google Scholar]
- 9.Tetko IV, Sushko I, Pandey AK, Zhu H, Tropsha A, Papa E, Oberg T, Todeschini R, Fourches D, Varnek A. Critical assessment of QSAR models of environmental toxicity against Tetrahymena pyriformis: focusing on applicability domain and overfitting by variable selection. J. Chem. Inf. Model. 2008;48:1733–1746. doi: 10.1021/ci800151m. [DOI] [PubMed] [Google Scholar]
- 10.Zhu H, Tropsha A, Fourches D, Varnek A, Papa E, Gramatica P, Oberg T, Dao P, Cherkasov A, Tetko IV. Combinatorial QSAR modeling of chemical toxicants tested against Tetrahymena pyriformis. J. Chem. Inf. Model. 2008;48:766–784. doi: 10.1021/ci700443v. [DOI] [PubMed] [Google Scholar]
- 11.NCI AIDS Antiviral Screen dtp.nci.nih.gov/docs/aids/aids_data.html (accessed Feb 1, 2010)
- 12.Weislow OS, Kiser R, Fine DL, Bader J, Shoemaker RH, Boyd MR. New soluble-formazan assay for HIV-1 cytopathic effects: application to high-flux screening of synthetic and natural products for AIDS-antiviral activity. J. Natl. Cancer Inst. 1989;81:577–586. doi: 10.1093/jnci/81.8.577. [DOI] [PubMed] [Google Scholar]
- 13.CADASTER Environmental Toxicity Prediction Challenge http://www.cadaster.eu (accessed Feb 1, 2010)
- 14.Stouch TR, Kenyon JR, Johnson SR, Chen XQ, Doweyko A, Li Y. In silico ADME/Tox: why models fail. J. Comput. Aided Mol. Des. 2003;17:83–92. doi: 10.1023/a:1025358319677. [DOI] [PubMed] [Google Scholar]
- 15.Doweyko AM. QSAR: dead or alive? J. Comput. Aided Mol. Des. 2008;22:81–89. doi: 10.1007/s10822-007-9162-7. [DOI] [PubMed] [Google Scholar]
- 16.Dearden JC, Cronin MT, Kaiser KL. How not to develop a quantitative structure-activity or structure-property relationship (QSAR/QSPR) SAR QSAR Environ. Res. 2009;20:241–266. doi: 10.1080/10629360902949567. [DOI] [PubMed] [Google Scholar]
- 17.Southan C, Varkonyi P, Muresan S. Quantitative assessment of the expanding complementarity between public and commercial databases of bioactive compounds. J. Cheminformatics. 2009;1:1–10. doi: 10.1186/1758-2946-1-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Martin YC. Let's not forget tautomers. J. Comput. Aided Mol. Des. 2009;23:693–704. doi: 10.1007/s10822-009-9303-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Dutta S, Burkhardt K, Young J, Swaminathan GJ, Matsuura T, Henrick K, Nakamura H, Berman HM. Data deposition and annotation at the worldwide protein data bank. Mol. Biotechnol. 2009;42:1–13. doi: 10.1007/s12033-008-9127-7. [DOI] [PubMed] [Google Scholar]
- 20.QSAR Expert Group The report from the expert group on (Quantitative) Structure-Activity Relationships [(Q)SARs] on the principles for the validation of (Q)SARs. ORGANISATION FOR ECONOMIC CO-OPERATION AND DEVELOPMENT: Paris. 2004;49:206. [Google Scholar]
- 21.Jorgensen WL. QSAR/QSPR and Proprietary Data. J Chem. Inf. Model. 2006;46:937. [Google Scholar]
- 22.Maggiora G. On Outliers and Activity Cliffs - Why QSAR Often Disappoints. J. Chem. Inf. Model. 2006;46:1535. doi: 10.1021/ci060117s. [DOI] [PubMed] [Google Scholar]
- 23.Zvinavashe E, Murk AJ, Rietjens IM. Promises and pitfalls of quantitative structure-activity relationship approaches for predicting metabolism and toxicity. Chem. Res. Toxicol. 2008;21:2229–2236. doi: 10.1021/tx800252e. [DOI] [PubMed] [Google Scholar]
- 24.Johnson S. The Trouble with QSAR (or How I Learned To Stop Worrying and Embrace Fallacy) J. Chem. Inf. Model. 2008;48:25–26. doi: 10.1021/ci700332k. [DOI] [PubMed] [Google Scholar]
- 25.Tropsha A, Golbraikh A. Predictive QSAR Modeling Workflow, Model Applicability Domains, and Virtual Screening. Curr. Pharm. Des. 2007;13:3494–3504. doi: 10.2174/138161207782794257. [DOI] [PubMed] [Google Scholar]
- 26.Garfield E. An algorithm for Translating Chemical Name to Chemical Formula. Essays of an Information Scientist. 1984;7:441–513. [Google Scholar]
- 27.Brecher J. From chemical name to structure: finding a noodle in the haystack. Proceedings of the CAS/IUPAC Conference on Chemical Identifiers and XML for Chemistry; CAS/IUPAC Conference on Chemical Identifiers and XML for Chemistry; Columbus (USA). 1-7-2002.2002. [Google Scholar]
- 28.Brecher J. Name=Struct: A Practical Approach to the Sorry State of Real-Life Chemical Nomenclature. J. Chem. Inf. Comput. Sci. 1999;39:943–950. [Google Scholar]
- 29.Takagi T, Amano M, Tomimoto M. Novel Method for the Evaluation of 3D Conformation Generators. J. Chem. Inf. Model. 2009;49:1377–1388. doi: 10.1021/ci800393w. [DOI] [PubMed] [Google Scholar]
- 30.Chen Q, Higgs R, Vieth M. Geometric Accuracy of Three-Dimensional Molecular Overlays. J. Chem. Inf. Model. 2006;46:1996–2002. doi: 10.1021/ci060134h. [DOI] [PubMed] [Google Scholar]
- 31.ChemAxon JChem. http://www.chemaxon.com (accessed Feb 1, 2010)
- 32. Notepad++. http://notepad-plus.sourceforge.net/uk/site.htm (accessed Feb 1, 2010)
- 33.Todeschini R, Consonni V. Handbook of Molecular Descriptors. Wiley-VCH; Weinheim: 2000. p. 667. [Google Scholar]
- 34.MOE Molecular Operating Environment Chemical Computing Group. http://www.chemcomp.com (accessed Feb 1, 2010)
- 35.Kuz'min VE, Muratov EN, Artemenko AG, Varlamova E, Gorb L, Wang J, Leszczynski J. Consensus QSAR Modeling of Phosphor-Containing Chiral AChE Inhibitors. QSAR Comb. Sci. 2009;28:664–677. [Google Scholar]
- 36.Kuhn M, Szklarczyk D, Franceschini A, Campillos M, von MC, Jensen LJ, Beyer A, Bork P. STITCH 2: an interaction network database for small molecules and proteins. Nucleic Acids Res. 2010;38:D552–D556. doi: 10.1093/nar/gkp937. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Tripos, a Certara Company http://tripos.com (accessed Feb 1, 2010)
- 38.OpenBabel: the OpenSource Chemistry Toolbox. http://openbabel.org (accessed Feb 1, 2010)
- 39.ACDLabs Advanced Chemistry Development http://www.acdlabs.com (accessed Feb 1, 2010)
- 40.ISIDA Software . University of Strasbourg; France: http://infochim.u-strasbg.fr (accessed Feb 1, 2010) [Google Scholar]
- 41.Kuz'min VE, Artemenko AG, Muratov EN, Volineckaya IL, Makarov VA, Riabova OB, Wutzler P, Schmidtke M. Quantitative Structure-Activity Relationship studies of [(biphenyloxy)propyl]isoxazole derivatives - human rhinovirus 2 replication inhibitors. J. Med. Chem. 2007;50:4205–4213. doi: 10.1021/jm0704806. [DOI] [PubMed] [Google Scholar]
- 42.Kuz'min VE, Artemenko AG, Muratov EN. Hierarchical QSAR technology on the base of Simplex representation of molecular structure. J. Comp. Aid. Mol. Des. 2008;22:403–421. doi: 10.1007/s10822-008-9179-6. [DOI] [PubMed] [Google Scholar]
- 43.Varnek A, Fourches D, Hoonakker F, Solov'ev V. Substructural fragments: An universal language to encode reactions, molecular and supramolecular structures. J. Comp. -Aided Mol. Des. 2006;19:693–703. doi: 10.1007/s10822-005-9008-0. [DOI] [PubMed] [Google Scholar]
- 44.Fung M, Thornton A, Mybeck K, Wu J, Hornbuckle K, Muniz E. Evaluation of the characteristics of safety withdrawal of prescription drugs from worldwide pharmaceutical markets −1960 to 1999. Drug. Inf. J. 2001;35:293–317. [Google Scholar]
- 45.Watkins P, Seeff L. Drug-induced liver injury: summary of a single topic clinical research conference. Hepatology. 2006;43:618–631. doi: 10.1002/hep.21095. [DOI] [PubMed] [Google Scholar]
- 46.Egan W, Zlokarnik G, Grootenhuis P. In silico prediction of drug safety: despite progress there is abundant room for improvement. Drug Discovery Today: Technologies. 2004;1:381–387. doi: 10.1016/j.ddtec.2004.11.002. [DOI] [PubMed] [Google Scholar]
- 47.US EPA - Environmental Protection Agency ToxCast TM Program : Predicting hazard, characterizing toxicity pathways, and prioritizing the toxicity testing of environmental chemicals. http://www.epa.gov/ncct/toxcast (accessed Feb 1, 2010)
- 48.European Union REACH : Registration, Evaluation, Authorisation and Restriction of Chemical substances. doi: 10.1016/j.chroma.2008.11.074. http://ec.europa.eu/environment/chemicals/reach/reach_intro.htm (accessed Feb 1, 2010) [DOI] [PubMed]
- 49.Biowisdom Ltd http://www.biowisdom.com (accessed Feb 1, 2010)
- 50.Fourches D, Barnes JC, Day NC, Bradley P, Reed JZ, Tropsha A. Cheminformatics analysis of assertions mined from literature that describe drug-induced liver injury in different species. Chem. Res. Toxicol. 2010;23:171–183. doi: 10.1021/tx900326k. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.O'Brien PJ, Irwin W, Diaz D, Howard-Cofield E, Krejsa CM, Slaughter MR, Gao B, Kaludercic N, Angeline A, Bernardi P, Brain P, Hougham C. High concordance of drug-induced human hepatotoxicity with in vitro cytotoxicity measured in a novel cell-based model using high content screening. Arch. Toxicol. 2006;80:580–604. doi: 10.1007/s00204-006-0091-3. [DOI] [PubMed] [Google Scholar]
- 52.Olson H, Betton G, Stritar J, Robinson D. The predictivity of the toxicity of pharmaceuticals in humans from animal data--an interim assessment. Toxicol. Lett. 1998;102-103:535–538. doi: 10.1016/s0378-4274(98)00261-6. [DOI] [PubMed] [Google Scholar]
- 53.Olson H, Betton G, Robinson D, Thomas K, Monro A, Kolaja G, Lilly P, Sanders J, Sipes G, Bracken W, Dorato M, Van DK, Smith P, Berger B, Heller A. Concordance of the toxicity of pharmaceuticals in humans and in animals. Regul. Toxicol. Pharmacol. 2000;32:56–67. doi: 10.1006/rtph.2000.1399. [DOI] [PubMed] [Google Scholar]
- 54.Downs G, Barnard J. Clustering methods and their uses in computational chemistry. Rev. Comp. Chem. 2002;18:1–40. [Google Scholar]
- 55.Kuz'min VE, Muratov EN, Artemenko AG, Gorb LG, Qasim M, Leszczynski J. The effects of characteristics of substituents on toxicity of the nitroaromatics: HiT QSAR study. J. Comp. Aid. Mol. Des. 2008;22:747–759. doi: 10.1007/s10822-008-9211-x. [DOI] [PubMed] [Google Scholar]
- 56.Artemenko AG, Muratov EN, Kuz'min VE, Gorb L, Hill F, Leszczynski J, Tropsha A. QSAR analysis of structural factors and possible modes of nitroaromatics' toxicity on the Tetrahymena Pyriformis. J. Cheminformatics. 2009 doi: 10.1080/1062936X.2011.569950. Submitted. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Kuz'min VE, Muratov EN, Artemenko AG, Gorb LG, Qasim M, Leszczynski J. The effect of nitroaromatics' composition on their toxicity in vivo: Novel, efficient non-additive 1D QSAR analysis. Chemosphere. 2008;72:1373–1380. doi: 10.1016/j.chemosphere.2008.04.045. [DOI] [PubMed] [Google Scholar]
- 58.Kubinyi H, Hamprecht FA, Mietzner T. Three-dimensional quantitative similarity-activity relationships (3D QSiAR) from SEAL similarity matrices. J. Med. Chem. 1998;41:2553–2564. doi: 10.1021/jm970732a. [DOI] [PubMed] [Google Scholar]
- 59.Golbraikh A, Tropsha A. Beware of q2! J. Mol. Graph. Model. 2002;20:269–276. doi: 10.1016/s1093-3263(01)00123-1. [DOI] [PubMed] [Google Scholar]
- 60.Martin MT, Judson RS, Reif DM, Kavlock RJ, Dix DJ. Profiling chemicals based on chronic toxicity results from the U.S. EPA ToxRef Database. Environ. Health Perspect. 2009;117:392–399. doi: 10.1289/ehp.0800074. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Pugh K. Toxicity and Physical Properties of Atrazine and its degradation products: a literature survey. http://www.osti.gov/bridge/purl.cover.jsp?purl=/10190387-0ya2oZ/webviewable/ (accessed Feb 1, 2010)
- 62.Hansen K, Mika S, Schroeter T, Sutter A, Ter Laak A., Steger-Hartmann T, Heinrich N, Muller KR. Benchmark Data Set for in Silico Prediction of Ames Mutagenicity. Proceedings of the 4th German Conference on Chemoinformatics, Goslar; 4th German Conference on Chemoinformatics; CIC; Goslar. 9-11-2008.2008. p. CDD32. [Google Scholar]
- 63.Hansen K, Mika S, Schroeter T, Sutter A, ter Laak A, Steger-Hartmann T, Heinrich N, Muller KR. Benchmark Data Set for in Silico Prediction of Ames Mutagenicity. J. Chem. Inf. Model. 2009;49:2077–2081. doi: 10.1021/ci900161g. [DOI] [PubMed] [Google Scholar]
- 64.Hopfinger AJ, Esposito EX, Llinas A, Glen RC, Goodman JM. Findings of the challenge to predict aqueous solubility. J. Chem. Inf. Model. 2009;49:1–5. doi: 10.1021/ci800436c. [DOI] [PubMed] [Google Scholar]
- 65.Llinas A, Glen RC, Goodman JM. Solubility challenge: can you predict solubilities of 32 molecules using a database of 100 reliable measurements? J. Chem. Inf. Model. 2008;48:1289–1303. doi: 10.1021/ci800058v. [DOI] [PubMed] [Google Scholar]
- 66.Hou T, Wang J, Zhang W, Xu X. ADME evaluation in drug discovery. 6. Can oral bioavailability in humans be effectively predicted by simple molecular property-based rules? J. Chem. Inf. Model. 2007;47:460–463. doi: 10.1021/ci6003515. [DOI] [PubMed] [Google Scholar]
- 67.OpenEye Scientific Software - Filter http://www.eyesopen.com/products/applications/filter.html (accessed Feb 1, 2010)
- 68.Molecular Networks GmbH http://www.molecular-networks.com (accessed Feb 1, 2010)
- 69.Hyleos http://www.hyleos.net (accessed Feb 1, 2010)
- 70.CambridgeSoft http://www.cambridgesoft.com (accessed Feb 1, 2010)
- 71.Schrodinger http://www.schrodinger.com (accessed Feb 1, 2010)
- 72.Symyx http://www.symyx.com (accessed Feb 1, 2010)
- 73.Accelrys http://accelrys.com (accessed Feb 1, 2010)