Abstract
Higher throughput thermodynamic measurements can provide value in structure-based drug discovery during fragment screening, hit validation, and lead optimization. Enthalpy can be used to detect and characterize ligand binding, and changes that affect the interaction of protein and ligand can sometimes be detected more readily from changes in the enthalpy of binding than from the corresponding free-energy changes or from protein-ligand structures. Newer, higher throughput calorimeters are being incorporated into the drug discovery process. Improvements in titration calorimeters come from extensions of a mature technology and face limitations in scaling. Conversely, array calorimetry, an emerging technology, shows promise for substantial improvements in throughput and material utilization, but improved sensitivity is needed.
Introduction
As the role of structural biology in drug development continues to grow, the value of related thermodynamic measurements is becoming more widely recognized. Indeed, in a recent review Chaires concludes that “thermodynamic data are an essential complement to structural data in drug development and for the optimization of lead compounds” [1*]. As an increasing body of data makes clear, the formation of isostructural complexes can result in very different thermodynamic consequences, and measuring these consequences can provide additional signatures of binding modes. Thermodynamic measurements can also be used to provide insights into specificity, agonist versus antagonist effects of ligands, and other important properties [1*,2*,3].
Such considerations are of particular value in drug design, where choices made at early stages of the process can have great impact on in vivo efficacy, resistance and toxicity, and where the costs of failure are high. Fragment-based drug discovery (FBDD) is an approach of particular interest and relevance here. Fragments are molecules smaller than typical drugs, and they generally bind with lower affinity than conventional drug screening hits. In FBDD, fragments that bind with high ligand efficiency (binding free energy per atom, not counting hydrogen atoms) are sought as building blocks for constructing larger, higher affinity drug leads [4,5]. Measuring the contributions of enthalpy and entropy to the free energy of binding provides information that can be useful in fragment elaboration and subsequent medicinal chemistry work [6–13].
In this review, we discuss recent progress toward integrating higher throughput calorimetry into structure-based drug discovery, covering both new isothermal titration calorimetry (ITC) instruments and array nanocalorimeters.
Higher throughput ITC instruments
Traditionally, ITC is a low throughput technique, having relatively high material and time requirements (~1mg of protein and 2–3 hours per titration). As a result, ITC has not been used routinely in drug discovery. However, some recent improvements in instrumentation have finally begun to emerge, including the iTC200 and Auto-iTC200 from MicroCal (GE Healthcare) and the Nano ITC from TA Instruments. For example, the iTC200 and Auto-iTC200 provide a 3-fold gain in sensitivity combined with a 7-fold smaller sample cell compared with the Microcal VP-ITC instrument [14**]. These two features reduce the protein requirement by 3 fold and the experiment time to around 30–40 min per titration. The protein concentration must be increased to maintain the same signal: noise ratio, however, which has immediate consequences. Higher titrant concentrations become necessary, so in some cases ligand solubility becomes limiting. In addition, requiring higher protein concentrations may restrict the high affinity limit of the instrument, since accurate Kd’s can only be determined down to ~1000-fold below the protein concentration for standard titrations.
In practice, the increased speed of titration and the ability to automate a series of titrations are the major benefits of the Auto-iTC200. Although retaining a serial titration mechanism, the Auto-iTC200 can, in principle, run ~30–40 titrations per day and up to 384 titrations unattended. Implicit in any consideration of throughput is the question of protein requirements. A typical Auto-iTC200 titration on a 40kDa protein at a 40μM concentration would require ~0.3 mg of protein. This allows ~30 titrations to be run and analyzed using 10mg of protein. This type of throughput could satisfy a typical project chemistry team during hits-to-leads or in lead optimization, providing of course that the target protein was available in sufficient quantity.
ITC in hit validation and lead optimization
In principle, ITC allows the measurement of binding free energies, enthalpies, and stoichiometry from low mM to ~50 pM affinities (see Table 1). Crucially for FBDD, the technique allows solution-based measurement of binding at weak affinities and high ligand concentrations, the region of interest for fragment screening where biochemical assays can suffer from compound interference. Measuring whether a compound binds with a meaningful stoichiometry, reasonable enthalpy, and attractive Kd can be useful for identifying false positives, a matter of high importance in both high throughput screening (HTS) and FBDD. The value of providing this fundamental information early in the drug discovery process has been demonstrated [15] and includes the validation of “tool compounds” taken from the literature. The simplicity and robustness of ITC also makes it valuable during lead optimization.
Table 1.
Summary of capabilities of calorimeters for measuring binding reactions. Ability to be used for indicated application is based on published data demonstrating measurement of the thermodynamic parameters indicated. Kd is the equilibrium dissociation constant, ΔH is the binding enthalpy, n is the stoichiometry coefficient, and ΔS is the binding entropy.
| Calorimeter | Configuration | Availability | Application | ||
|---|---|---|---|---|---|
| Thermodynamic characterization (Kd, ΔH, n, ΔS) | Screen by Kd | Screen by ΔH | |||
| iTC200 (Microcal) and Nano ITC Low Volume (TA Instruments) [14**,41–43] | ~200 μl chamber with stir paddle on spinning injection syringe Thermopile sensors (see online supplement for more detailed information) |
commercial instruments | Broad range of affinities: 20 μM < Kd < ~2 mM for low-C titrations (cannot determine n) 5 nM <Kd < 50 μM for direct titrations ~50 pM <Kd < 10 nM for competitive displacement titrations |
Low throughput (50/day max) | yes |
| Enthalpy array [26,27**,28*] | Open thermally isolated surface, 500 nl reaction, electrostatic drop merging, magnetic mixing Thermistor sensors |
research instrument | Limited range of affinities (Kd>10−5 M) | - | yes, higher throughput |
| [31] | Closed 10 μl chamber, 5 μl/min flow Thermopile sensors |
research instrument | - | - | One component immobilized |
In work at Astex Therapeutics, greater than 400 ITC datasets describing fragments and fragment-derived leads binding to a wide variety of target proteins have been acquired. These datasets complement a similar number available from publically available sources such as the Scorpio [16] (URL: http://scorpio.biophysics.ismb.lon.ac.uk/scorpio.html), PDBcal [17] (URL: http://www.pdbcal.org/) and BindingDB [18] (URL: http://www.bindingdb.org/bind/index.jsp) databases, which compare thermodynamic binding information and structural data for a range of small molecules, natural products and biopolymers interacting with proteins. The data for Astex campaigns clearly show that binding of fragment hits to proteins has been driven predominantly by enthalpy and that this preference has been maintained during X-ray structure-based optimization of fragment hits into nanomolar affinity leads and drug-candidates, as shown in Table 3. With hindsight this trend can be rationalized by invoking molecular interpretations of thermodynamic effects, combined with a biophysical viewpoint of the structure-based drug discovery process.
Table 3.
Average values of binding free energy and enthalpy for fragment-derived compounds binding to Astex protein targets. 5-fold affinity ranges have been selected that cover distinct stages of the drug design process. Our error analysis for ITC measurements is discussed in supplemental information (available online) and indicates that ΔG and ΔH are measured with standard deviations of ± 0.1 kcal/mol and ±0.4 kcal/mol respectively. For replicate titrations which use solutions taken from the same stocks, the variation in ΔH can be reduced to ±0.2 kcal/mol or better.
| Affinity Range (μM) | Description | Number of measurements | <ΔG> kcal/mol | <ΔH> kcal/mol |
|---|---|---|---|---|
| 50–250 | Fragment Screening Hits | 21 | −5.4 | −5.5 |
| 1–5 | Optimized Fragment Hits | 40 | −7.8 | −6.9 |
| 0.02 – 0.10 | Fragment-derived Leads | 40 | −10 | −12 |
| 0.002 – 0.0004 | Optimized Leads | 48 | −11.8 | −10.6 |
Molecular Interpretation of Thermodynamic Effects in FBDD
Binding of an initial fragment hit is often dominated by polar, hydrogen-bonding interactions, while hydrophobic portions of the molecule may limit access of solvent but do not usually make optimal contacts with the protein. Fragments used in screening contain polar heteroatoms because they must be highly soluble in aqueous buffers (solubility > 1mM). Furthermore, fragment scaffolds are usually restricted to simple geometries, typically with one or two aliphatic or aromatic rings decorated with polar functional groups, thus limiting the extent and availability of apolar surface area. In order to bind to a protein, small molecules must first lose a considerable amount of entropy, a disadvantage estimated to be approximately +4 kcal/mol at 25°C [19,20]. These factors mitigate in favor of enthalpically driven binding, at least in the early stages of fragment-based drug design
Hydrogen bonding between ligand and protein usually provides a favorable enthalpy change due to the energy of the bond itself, with a smaller entropic contribution that may be favorable or unfavorable. Large variations occur, though, reflecting the need to desolvate H-bond partners to varying extents and in some cases break intramolecular H-bonds. Also, the formation of multiple hydrogen bonds can severely restrict mobility and thereby reduce entropy significantly. Importantly, once the configurational entropy is lost, additional hydrogen bonds may not have as noticeable an effect.
Removing a hydrophobic area of a ligand from an aqueous environment leads to a loss of solvent order that corresponds to a favorable entropy change, while van der Waals interactions make a smaller, favorable contribution to the binding enthalpy. During later iterations of structure-based design, these hydrophobic contacts are optimized, and there are more opportunities for favorable entropic contributions from the release of ordered water. However, this may be opposed by a further reduction in configurational entropy of the ligand, as its interactions with the protein are strengthened [3]. Astex data suggest that, on average, both entropic and enthalpic contributions to binding affinity can be improved during this phase of structure-based design (Table 3).
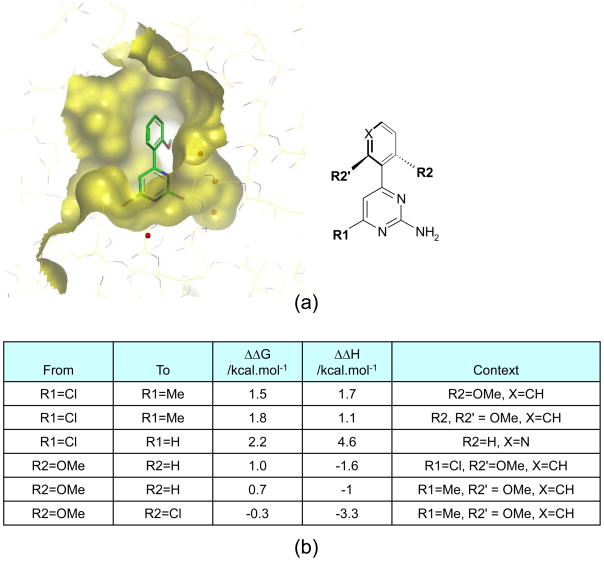
The trend in Table 3 conceals larger, partially compensating changes in the binding enthalpy and entropy that can occur, even though the relationship between ligand structure and free-energy of binding may appear smooth and continuous. One example of such an effect for a series of related amino-pyrimidines binding to HSP90 is evident in Figure 1. Empirically, such effects are usually observed when making changes in regions of the ligand that make close contacts with the protein. Accordingly, the measurement of large enthalpy changes on modification of the ligand can be used to suggest a close contact with the protein, even in the absence of structural data and in cases where the modification leads to little or no change in affinity. As a caution, though, it is difficult to justify such suggestions convincingly, even with high resolution structural data that includes positions of individual water molecules in the active site. The message for FBDD is interpretation of enthalpy and entropy changes needs to be sophisticated to be effective.
Figure 1. Binding thermodynamics for substituted amino-pyrimidine ligands and HSP90.
(a) X-ray structure showing the binding mode of a substituted amino-pyridimidine bound in the ATP-binding site of the N-terminal domain of the chaperone, HSP90.
(b) Changes in free-energy and enthalpy of binding observed on modification of the amino-pyrimidine ligand. At position R1, replacement of Cl by Me leads to a substantial change in ΔH that is accompanied by a similar change in ΔG. Replacing Cl by H, however leads to a much larger change in ΔH than ΔG indicating a high degree of enthalpy-entropy compensation for this particular substitution. Enthalpy-entropy compensation is also evident for both substitutions shown at position R2. For the substitution of OMe by H, entropy effects dominate and ΔΔG and ΔΔH are of opposite sign. For the substitution of OMe by Cl, enthalpy and entropy changes are finely balanced and little change in ΔG results. With the benefit of enthalpy data is it clear that the substitution OMe to Cl at R2 could lead to large improvements in affinity, provided that enthalpy-entropy compensation could somehow be overcome.
Factors such as enthalpies of buffer protonation [21], ligand desolvation, or conformation changes due to binding or ‘unusual’ interactions can complicate the interpretation of thermodynamic data [22]. Repeating measurements in different buffers can elucidate protonation effects, and structural data can be used to identify the presence of new interactions. In contrast, contributions from changes in ligand desolvation remain difficult to probe.
Another thermodynamic parameter of interest in drug design is ΔCP, the change in heat capacity that occurs when a ligand binds to a protein [23,24]. ΔCP is determined from the temperature dependence of the enthalpy change (ΔCp =|∂ΔH/∂T|p) and is dominated by changes in hydration. Desolvation of polar groups is associated with a small increase in heat capacity, while desolvation of apolar groups causes a decrease in heat capacity. Although solvent is ordered around both solvated polar and apolar groups, the ordering is clearly different and, around apolar groups, provides additional vibrational modes that can be populated at normal temperatures. For example, for the burial of an isopropyl group, changes in ΔCp are expected to be ~0.03 kcal/(mol.K), and these should be reflected by changes in ΔH of ~0.6 kcal/mol over 20°C [25].
Array calorimetry
In order to routinely get the full value that thermodynamic measurements can provide in structural biology-based drug discovery, higher throughput instruments will be required. While miniaturization of ITC instruments enables somewhat higher throughput and lower sample use [14**], the measurements are still performed in series and thus have limited throughput relative to the evolving need. Arrays of nanocalorimeters increase throughput by performing measurements in parallel, and they also enable decreased sample use by reducing sample volume to the sub-microliter level.
Tables 1 and 2 list array or array-able nanocalorimeters for biochemical measurements that have been presented in the literature. Table 1 focuses on binding measurements, and Table 2 focuses on enzymatic measurements. For comparison, these tables also list higher throughput ITC instruments.
Table 2.
Summary of capabilities of calorimeters for measuring enzymatic reactions. Ability to be used for indicated application is based on published data demonstrating measurement of the thermodynamic/kinetic parameters indicated. kcat is the turnover number), KM is the Michaelis constant, and KI is the competitive inhibition constant.
| Calorimeter | Configuration | Availability | Application | ||||
|---|---|---|---|---|---|---|---|
| Kinetic characterization | yes/no inhibitor screen | substrate quantitation | |||||
| kcat | KM | KI | |||||
| iTC200 (Microcal) and Nano ITC Low Volume (TA Instruments) [14**,41,44] | ~200 μl chamber with stir paddle on spinning injection syringe Thermopile sensors |
commercial instruments | kcat>0.02 s−1 low throughput (1–2/hour) | 10−8 M to 10−3 M | 10−8 M to 10−3 M | yes | yes |
| Enthalpy array [26,27**,28*] | Open thermally isolated surface, 500 nl reaction, electrostatic drop merging, magnetic mixing Thermistor sensors |
research instrument | kcat>0.5 s−1 higher throughput (10–12/hour, potential for 40/hour) | 10−6 M to 10−3 M | 10−7 Ma to 10−3 M | yes | yes |
| [33,36] | Closed 10 μl chamber, 5 μl/min flow Thermopile sensors |
research instrument | - | - | - | - | yes |
| [34] | Closed 15 nl chamber, 15 nl/s flow Thermopile sensors |
research instrument | - | - | - | - | yes |
| MiDiCal [29,30] | Open 10–20 μl well Thermopile sensors |
research instrument | - | - | - | - | yes |
Lower limit on KI is ≈0.02 [enzyme]
Nanocalorimeters can be classified based on two major design elements: the configuration of the reaction chamber and the type of thermometry. Two types of reaction chambers have been described: open-chamber (batch calorimetry) devices which use syringes or pipettes to place droplets of reactants on the surface of a thermally isolated well or platform [26,27**,28*,29,30] and closed-chamber (typically flow calorimetry) devices which use microfluidic channels to introduce reactants [31,32*,33,34]. Open-chamber designs generally provide better thermal isolation and reduced dead volume compared with microfluidic delivery systems. A limitation of open-chamber systems is evaporation of drops, which must be minimized to achieve reliable measurements. Closed-chamber systems do not suffer from evaporation issues, but their sensitivity is limited by thermal conductance to their surroundings and higher device heat capacity.
The two types of thermometers that have been used in nanocalorimetric devices are thermopiles [29,31,32*,33,34] and thermistors [26,28*]. Thermopiles are composed of thermocouples connected in series, a higher number giving better sensitivity. Thermistors are temperature sensitive resistors which display decreased resistance with increasing temperature. Most of the nanocalorimeters reported in the literature use thermopiles. They have the advantage of not generating heat because current is not required to sense the temperature-dependent voltage. For an array of nanocalorimeters, though, the small size of the device limits the size and number of thermocouple junctions that can be placed in the device, making the fabrication more challenging and increasing the cost substantially compared to thermistors. Sensitive thermistors are easier to fabricate, but they generate heat during a measurement due to the applied current. Larger current improves the electronic signal-to-noise ratio, but the increased resistive heating can amplify unwanted thermal artifacts [35]. In enthalpy array devices [28*], four thermistors are combined in a Wheatstone bridge to directly provide a differential temperature measurement of sample and reference specimens.
A number of closed-chamber microfluidic calorimeters using thermopile heat sensors have been described [31,32*,33,34,36]. In principle, arrays of these devices could be fabricated, but so far the published work only describes the construction and operation of a single calorimetric unit. Flow calorimeters typically require significantly larger sample volumes than batch designs, but microfluidic devices that use as little as 10–20 microliter sample volumes have been described by Lerchner et al [33,37], albeit with a focus on measuring the metabolic heat produced by microorganisms rather than on drug discovery applications. This calorimeter contains a thermopile chip with four measuring sections arranged along a flow path, allowing the transient behavior of a reaction to be resolved. With respect to the potential for application in target-based drug discovery, the device has been used to measure high-activity enzymatic reactions (oxidation of glucose by glucose oxidase, hydrolysis of penicillin G by β-lactamase [33]), but further work appears necessary to enable measurement of specific parameters of interest, such as thermodynamic quantities for binding or kinetic inhibition constants.
The measurement of binding reactions in a flow-through nanocalorimeter has been achieved by immobilizing one binding partner on magnetic streptavidin-coated agarose beads [31]. Using a microfluidic calorimeter of the type described by Lerchner et al [33], streptavidin-coated beads were loaded into the chamber over one or more of the thermopile sensors (~175 μM streptavidin monomer in chamber), and the binding of biotin was measured, yielding a linear relationship between peak area and biotin concentration. In addition, the hybridization of DNA strands was measured by first binding one biotinylated DNA strand to the streptavidin-coated beads and then injecting the complementary DNA strand into the chamber. These are interesting demonstrations, but they are still a long way from measurements relevant to drug design, and immobilization of one binding partner eliminates a major advantage that calorimetry provides.
Lee et al. [32*] also report a closed-chamber calorimeter that can in principle be developed into an array technology. They describe a single unit closed-chamber calorimeter in which the measurement chamber and thermometer are built on a parylene-C membrane. The sensitivity of the closed-chamber device has been increased by surrounding the chamber with a vacuum jacket, providing better thermal isolation of the reaction chamber, albeit at a significant increase in fabrication difficulty. Measurement of the enzymatic reaction of urease was shown, which yielded the expected linear relationship between total energy and initial substrate concentration. In addition, the mixing of methanol with water was shown, but the results indicated some limitations in sample mixing in the device. The device will require further development to achieve good mixing and enable measurement of binding reactions [32*].
Actual array calorimeters have been demonstrated for open chamber designs. For example, Verhaegen, et al [29] developed the MiDiCal array calorimeter using a thermopile-based sensor and published results for the analysis of ascorbic acid concentration in food and pharmaceutical samples [30]. Each array allows up to 48 samples to be measured using reaction volumes of 10 to 20 microliters each. The ascorbic acid content of a sample was determined by measuring the heat evolved by ascorbate oxidase turnover of ascorbic acid. The results were in good agreement with an HPLC reference method, but these reactions gave highly exothermic signals well above the sensitivity limits desired for array calorimeters in target-based drug discovery. The data analysis reveals that diffusion of reagents in the reaction wells has a significant impact on the observed signal compared to the expected sensor readings, indicating that poor sample mixing is an issue.
The array calorimeter with the best published sensitivity is the enthalpy array, an open chamber calorimeter [26,27**,28*]. As shown in Tables 1 and 2, these arrays have been used to measure both binding and enzymatic reactions. For enzymatic reactions, both the total integrated heat and the detailed kinetics have been measured, the former for inhibitor screening or substrate quantitation and the latter for determination of kinetics constants such as kcat, KM, and KI. Of particular interest in FBDD, enthalpy arrays can measure inhibition constants for relatively weak inhibitors at mM compound concentrations [27**], providing a platform for activity-based fragment screening. The arrays were initially used to measure the enthalpy of binding reactions (RNase A-2′-CMP, stretpavidin-biotin), an enzymatic reaction with limited substrate (hexokinase), and mitochondrial respiration [28*]. The thermistor material was improved, yielding lower noise devices, and these were then used to perform titrations to determine Kd of a simple non-biological model system (BaCl2-18-crown-6) and to quantitate the total initial concentration of substrate in an enzymatic reaction [26]. Limitations in the sensitivity of the devices was observed and traced to poor mixing of 500 nanoliter volume samples. Achieving good sensitivity requires rapid and complete mixing of samples, which is hard to achieve with nanoliter sample volumes [26,38,39]. Employing a micro magnetic stir bar, rapid mixing of samples was achieved and enabled measuring enzyme kinetic parameters and determining inhibitor constants for several enzyme systems (trypsin, hexokinase, PKA). In addition, determination of the mode of inhibition (competitive, noncompetitive) was determined from the nonlinear regression of the rate versus substrate concentration data at various inhibitor concentrations. Lineweaver-Burk plots were provided to give visual validation of the inhibition mode [27**].
Concluding remarks
Higher throughput thermodynamic measurements are providing value in structure-based drug discovery, in particular throughout the FBDD process where enthalpy can be used to detect and characterize ligand binding. Critical regions of the ligand that affect its interaction with protein can sometimes be detected more readily from changes in the enthalpy of binding than from the corresponding free-energy changes because of enthalpy-entropy compensation effects. While the interpretation of small enthalpy differences in terms of changes in molecular interactions is difficult, enthalpy-driven binding has been suggested to favor ‘best-in-class’ properties in some cases, such as for HIV protease inhibitors [40]. Experience in the area of protein-folding has also indicated that ΔCP is correlated with the amount of apolar surface area buried on binding, thus providing additional structural information from enthalpy.
Current improvements in conventional titration and array calorimeters indicate the potential for making measurements on hundreds of complexes, using a few milligrams of protein. Improvements in titration calorimeters come from extensions of a mature technology and face limitations in scaling. Conversely, array calorimetry is an emerging technology and shows promise for substantial improvements in throughput and material utilization, but development work is needed to improve the sensitivity. Successful miniaturization and automation could impact not only lead optimization but also primary and secondary screening.
Supplementary Material
Acknowledgments
Support from grants R01EB009191 and R01GM077435 from the National Institutes of Health (USA) for F. Torres, M. Recht, and R. Bruce is gratefully acknowledged. The content is solely the responsibility of the authors and does not necessarily represent the official views of the National Institutes of Health.
Footnotes
Disclosure Statement: F. Torres, M. Recht, and R. Bruce are employed by Palo Alto Research Center Incorporated. G. Williams and J. E. Coyle are employed by Astex Therapeutics Ltd.
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
Contributor Information
Francisco E. Torres, Email: torres@parc.com.
Glyn Williams, Email: G.Williams@astex-therapeutics.com.
References
- *1.Chaires JB. Calorimetry and thermodynamics in drug design. Annu Rev Biophys. 2008;37:135–151. doi: 10.1146/annurev.biophys.36.040306.132812. An excellent review summarizing the value of obtaining complete thermodynamic profiles in drug design. [DOI] [PubMed] [Google Scholar]
- *2.Ladbury JE, Klebe G, Freire E. Adding calorimetric data to decision making in lead discovery: a hot tip. Nat Rev Drug Discov. 2010;9:23–27. doi: 10.1038/nrd3054. The authors highlight the importance of enthalpically-driven versus entropically-driven protein-ligand interactions. They discuss the application of enthalpic efficiency data to complement other data in decision making in lead discovery and optimization. [DOI] [PubMed] [Google Scholar]
- 3.Williams DH, Stephens E, O’Brien DP, Zhou M. Understanding noncovalent interactions: ligand binding energy and catalytic efficiency from ligand-induced reductions in motion within receptors and enzymes. Angew Chem Int Ed Engl. 2004;43:6596–6616. doi: 10.1002/anie.200300644. [DOI] [PubMed] [Google Scholar]
- 4.Carr RA, Congreve M, Murray CW, Rees DC. Fragment-based lead discovery: leads by design. Drug Discov Today. 2005;10:987–992. doi: 10.1016/S1359-6446(05)03511-7. [DOI] [PubMed] [Google Scholar]
- 5.Hopkins AL, Groom CR, Alex A. Ligand efficiency: a useful metric for lead selection. Drug Discov Today. 2004;9:430–431. doi: 10.1016/S1359-6446(04)03069-7. [DOI] [PubMed] [Google Scholar]
- 6.Krishnamurthy VM, Estroff LA, Whitesides GM. Multivalency in Ligand Design. In: Jahnke W, Erlanson D, editors. Fragment-based Approaches in Drug Discovery (vol. 34 of Methods and Principles in Medicinal Chemistry) Wiley-VCH Verlag GmbH & Co; KGaA: 2006. pp. 11–53. [Google Scholar]
- 7.Ladbury JE. Isothermal titration calorimetry: application to structure-based drug design. Thermochimica Acta. 2001;380:209–215. [Google Scholar]
- 8.Mammen M, Choi S-K, Whitesides GM. Polyvalent Interactions in Biological Systems: Implications for Design and Use of Multivalent Ligands and Inhibitors. Angew Chem Int Ed. 1998;37:2754–2794. doi: 10.1002/(SICI)1521-3773(19981102)37:20<2754::AID-ANIE2754>3.0.CO;2-3. [DOI] [PubMed] [Google Scholar]
- 9.Ohtaka H, Muzammil S, Schon A, Velazquez-Campoy A, Vega S, Freire E. Thermodynamic rules for the design of high affinity HIV-1 protease inhibitors with adaptability to mutations and high selectivity towards unwanted targets. Int J Biochem Cell Biol. 2004;36:1787–1799. doi: 10.1016/j.biocel.2004.02.021. [DOI] [PubMed] [Google Scholar]
- 10.Ruben AJ, Kiso Y, Freire E. Overcoming roadblocks in lead optimization: a thermodynamic perspective. Chem Biol Drug Des. 2006;67:2–4. doi: 10.1111/j.1747-0285.2005.00314.x. [DOI] [PubMed] [Google Scholar]
- 11.Taylor JD, Gilbert PJ, Williams MA, Pitt WR, Ladbury JE. Identification of novel fragment compounds targeted against the pY pocket of v-Src SH2 by computational and NMR screening and thermodynamic evaluation. Proteins. 2007;67:981–990. doi: 10.1002/prot.21369. [DOI] [PubMed] [Google Scholar]
- 12.Velazquez-Campoy A, Todd MJ, Freire E. HIV-1 protease inhibitors: enthalpic versus entropic optimization of the binding affinity. Biochemistry. 2000;39:2201–2207. doi: 10.1021/bi992399d. [DOI] [PubMed] [Google Scholar]
- 13.Ward WHJ, Holdgate GA. Isothermal Titration Calorimetry in Drug Discovery. Prog Med Chem. 1999;38:309–376. doi: 10.1016/s0079-6468(08)70097-3. [DOI] [PubMed] [Google Scholar]
- **14.Peters WB, Frasca V, Brown RK. Recent developments in isothermal titration calorimetry label free screening. Comb Chem High Throughput Screen. 2009;12:772–790. doi: 10.2174/138620709789104889. The authors describe a miniaturized ITC microcalorimeter that allows up to 7-fold reduction in protein required and describe its use in higher throughput measurements with a limited injection method and a continuous injection method. [DOI] [PubMed] [Google Scholar]
- 15.Murray CW, Carr MG, Callaghan O, Chessari G, Congreve M, Cowan S, Coyle JE, Downham R, Figueroa E, Frederickson M, et al. Fragment-based drug discovery applied to hsp90. Discovery of two lead series with high ligand efficiency. J Med Chem. 2010;53:5942–5955. doi: 10.1021/jm100059d. [DOI] [PubMed] [Google Scholar]
- 16.Olsson TS, Williams MA, Pitt WR, Ladbury JE. The thermodynamics of protein-ligand interaction and solvation: insights for ligand design. J Mol Biol. 2008;384:1002–1017. doi: 10.1016/j.jmb.2008.09.073. [DOI] [PubMed] [Google Scholar]
- 17.Li L, Dantzer JJ, Nowacki J, O’Callaghan BJ, Meroueh SO. PDBcal: a comprehensive dataset for receptor-ligand interactions with three-dimensional structures and binding thermodynamics from isothermal titration calorimetry. Chem Biol Drug Des. 2008;71:529–532. doi: 10.1111/j.1747-0285.2008.00661.x. [DOI] [PubMed] [Google Scholar]
- 18.Liu T, Lin Y, Wen X, Jorissen RN, Gilson MK. BindingDB: a web-accessible database of experimentally determined protein-ligand binding affinities. Nucleic Acids Res. 2007;35:D198–201. doi: 10.1093/nar/gkl999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Borsi V, Calderone V, Fragai M, Luchinat C, Sarti N. Entropic Contribution to the Linking Coefficient in Fragment Based Drug Design: A Case Study. J Med Chem. 2010 doi: 10.1021/jm901723z. [DOI] [PubMed] [Google Scholar]
- 20.Murray CW, Verdonk ML. Entropic Consequences of Linking Ligands. In: Jahnke W, Erlanson D, editors. Fragment-based Approaches in Drug Discovery (vol. 34 of Methods and Principles in Medicinal Chemistry) Wiley-VCH Verlag GmbH & Co; KGaA: 2006. pp. 55–66. [Google Scholar]
- 21.Gohlke H, Klebe G. Approaches to the description and prediction of the binding affinity of small-molecule ligands to macromolecular receptors. Angew Chem Int Ed Engl. 2002;41:2644–2676. doi: 10.1002/1521-3773(20020802)41:15<2644::AID-ANIE2644>3.0.CO;2-O. [DOI] [PubMed] [Google Scholar]
- 22.Schreyer A, Blundell T. CREDO: a protein-ligand interaction database for drug discovery. Chem Biol Drug Des. 2009;73:157–167. doi: 10.1111/j.1747-0285.2008.00762.x. [DOI] [PubMed] [Google Scholar]
- 23.Cooper A, Johnson CM, Lakey JH, Nollmann M. Heat does not come in different colours: entropy-enthalpy compensation, free energy windows, quantum confinement, pressure perturbation calorimetry, solvation and the multiple causes of heat capacity effects in biomolecular interactions. Biophys Chem. 2001;93:215–230. doi: 10.1016/s0301-4622(01)00222-8. [DOI] [PubMed] [Google Scholar]
- 24.Holdgate GA, Ward WHJ. Measurements of binding thermodynamics in drug discovery. Drug Discovery Today. 2005;10:1543–1550. doi: 10.1016/S1359-6446(05)03610-X. [DOI] [PubMed] [Google Scholar]
- 25.Sturtevant JM. Heat capacity and entropy changes in processes involving proteins. Proc Natl Acad Sci U S A. 1977;74:2236–2240. doi: 10.1073/pnas.74.6.2236. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Recht MI, De Bruyker D, Bell AG, Wolkin MV, Peeters E, Anderson GB, Kolatkar AR, Bern MW, Kuhn P, Bruce RH, et al. Enthalpy array analysis of enzymatic and binding reactions. Anal Biochem. 2008;377:33–39. doi: 10.1016/j.ab.2008.03.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- **27.Recht MI, Torres FE, De Bruyker D, Bell AG, Klumpp M, Bruce RH. Measurement of enzyme kinetics and inhibitor constants using enthalpy arrays. Anal Biochem. 2009;388:204–212. doi: 10.1016/j.ab.2009.02.028. Demonstrates how rapid mixing of samples enables determination of KM, kcat and KI in an array nanocalorimeter. Discusses the limitations for determining these parameters based on the thermal properties of the device (time constant, detector noise, etc) [DOI] [PMC free article] [PubMed] [Google Scholar]
- *28.Torres FE, Kuhn P, De Bruyker D, Bell AG, Wolkin MV, Peeters E, Williamson JR, Anderson GB, Schmitz GP, Recht MI, et al. Enthalpy arrays. Proc Natl Acad Sci U S A. 2004;101:9517–9522. doi: 10.1073/pnas.0403573101. Arrays of nanocalorimeters using thermistor sensors in a Wheatstone bridge configuration. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Verhaegen K, Baert K, Simaels J, Van Driessche W. A high-throughput silicon microphysiometer. Sensors and Actuators A: Physical. 2000;82:186–190. [Google Scholar]
- 30.Vermeir S, Nicolai BM, Verboven P, Van Gerwen P, Baeten B, Hoflack L, Vulsteke V, Lammertyn J. Microplate differential calorimetric biosensor for ascorbic acid analysis in food and pharmaceuticals. Anal Chem. 2007;79:6119–6127. doi: 10.1021/ac070325z. [DOI] [PubMed] [Google Scholar]
- 31.Ahmad LM, Towe B, Wolf A, Mertens F, Lerchner J. Binding event measurement using a chip calorimeter coupled to magnetic beads. Sensors and Actuators B: Chemical. 2010;145:239–245. [Google Scholar]
- *32.Lee W, Fon W, Axelrod BW, Roukes ML. High-sensitivity microfluidic calorimeters for biological and chemical applications. Proc Natl Acad Sci U S A. 2009;106:15225–15230. doi: 10.1073/pnas.0901447106. A closed-chamber nanocalorimeter that achieves increased sensitivity by surrounding the measurement chamber with a vacuum. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Lerchner J, Wolf A, Wolf G, Baier V, Kessler E, Nietzsch M, Krügel M. A new micro-fluid chip calorimeter for biochemical applications. Thermochimica Acta. 2006;445:144–150. [Google Scholar]
- 34.Zhang Y, Tadigadapa S. Calorimetric biosensors with integrated microfluidic channels. Biosens Bioelectron. 2004;19:1733–1743. doi: 10.1016/j.bios.2004.01.009. [DOI] [PubMed] [Google Scholar]
- 35.De Bruyker D, Wolkin MV, Recht MI, Torres FE, Bell AG, Anderson GB, Peeters E, Kolatkar A, Kuhn P, Bruce RH. MEMS-based enthalpy arrays. Transducers ‘07, Lyon & Eurosensors XXI; 14th International Conference on Solid-State Sensors, Actuators, and Microsystems; IEEE; 2007. pp. 1757–1760. [Google Scholar]
- 36.Baier V, Födisch R, Ihring A, Kessler E, Lerchner J, Wolf G, Köhler JM, Nietzsch M, Krügel M. Highly sensitive thermopile heat power sensor for micro-fluid calorimetry of biochemical processes. Sensors and Actuators A: Physical. 2005;123–124:354–359. [Google Scholar]
- 37.Lerchner J, Maskow T, Wolf G. Chip calorimetry and its use for biochemical and cell biological investigations. Chemical Engineering and Processing: Process Intensification. 2008;47:991–999. [Google Scholar]
- 38.Squires TM, Quake SR. Microfluidics: Fluid physics at the nanoliter scale. Review of Modern Physics. 2005;77:977–1026. [Google Scholar]
- 39.Stone HA, Stroock AD, Ajdari A. Engineering Flows in Small Devices: Microfluidics Toward a Lab-on-a-Chip. Annual Rev Fluid Mech. 2004;36:381–411. [Google Scholar]
- 40.Freire E. Do enthalpy and entropy distinguish first in class from best in class? Drug Discov Today. 2008;13:869–874. doi: 10.1016/j.drudis2008.07.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Freyer MW, Lewis EA. Isothermal titration calorimetry: experimental design, data analysis, and probing macromolecule/ligand binding and kinetic interactions. Methods Cell Biol. 2008;84:79–113. doi: 10.1016/S0091-679X(07)84004-0. [DOI] [PubMed] [Google Scholar]
- 42.Sigurskjold BW. Exact analysis of competition ligand binding by displacement isothermal titration calorimetry. Anal Biochem. 2000;277:260–266. doi: 10.1006/abio.1999.4402. [DOI] [PubMed] [Google Scholar]
- 43.Turnbull WB, Daranas AH. On the value of c: can low affinity systems be studied by isothermal titration calorimetry? J Am Chem Soc. 2003;125:14859–14866. doi: 10.1021/ja036166s. [DOI] [PubMed] [Google Scholar]
- 44.Cai L, Cao A, Lai L. An isothermal titration calorimetric method to determine the kinetic parameters of enzyme catalytic reaction by employing the product inhibition as probe. Anal Biochem. 2001;299:19–23. doi: 10.1006/abio.2001.5397. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.