Research highlights
► ChREBP silencing enhances glucose-responsive gene expression in MIN6 β-cells. ► ChREBP modulation of Pdx-1 gene expression might be indirect. ► ChREBP over-expression decreases Pdx-1, MafA, Ins1, Ins2 and GcK mRNA levels in mouse pancreatic islets of Langerhans.
Abbreviations: ARNT, aryl hydrocarbon receptor nuclear translocator; ChREBP, carbohydrate responsive element-binding protein; ChIP, chromatin immunoprecipitation; ChoRE, carbohydrate-responsive element; FAS, fatty acid synthase; GcK, glucokinase; GFP, green fluorescent protein; HIF, hypoxia inducible factor; L-PK, L-type pyruvate kinase; Pdx-1, pancreatic and duodenum homeobox-1; siRNA, small interfering RNA; SREBP-1c, sterol regulatory response-element-binding protein-1c; USF, upstream stimulatory factor
Keywords: ChREBP, Pdx-1, MafA, Insulin, Glucokinase, Gene expression, Pancreatic β-cells, MIN6, Islets of Langerhans
Abstract
Carbohydrate responsive element-binding protein (ChREBP) is a transcription factor whose expression and activity are increased in pancreatic β-cells maintained at elevated glucose concentrations. We show here that ChREBP inactivation in clonal pancreatic MIN6 β-cells results in an increase in Pdx-1 expression at low glucose and to a small, but significant, increase in Ins2, GcK and MafA gene expression at high glucose concentrations. Conversely, adenovirus-mediated over-expression of ChREBP in mouse pancreatic islets results in decreases in Pdx-1, MafA, Ins1, Ins2 and GcK mRNA levels. These data suggest that strategies to reduce ChREBP activity might protect against β-cell dysfunction in type 2 diabetes.
1. Introduction
Pancreatic β-cell glucolipotoxicity [1] is considered to play a significant role in the pathogenesis of type 2 diabetes. Carbohydrate responsive element-binding protein (ChREBP) is a member of the basic helix–loop–helix family of transcription factors and transactivates glucose-responsive genes by binding to DNA as a heterodimer with Max-like protein X1 at a well-defined carbohydrate-responsive element (ChoRE) [2–5]. In the liver, ChREBP is responsible for converting excess carbohydrate to fatty acids for long-term storage [6]. Mice deleted for both alleles of ChREBP display diminished rates of hepatic glycolysis and lipogenesis resulting in high liver glycogen content, low plasma free fatty acid levels and reduced adipose tissue mass [7]. Loss of ChREBP in leptin-null ob/ob mice protects against obesity [7,8].
We, and others, have previously shown that, in pancreatic β-cells, ChREBP is activated by high glucose and is responsible for the induction of the lipogenic genes fatty acid synthase (FAS) and L-type pyruvate kinase (L-PK) [9,10], and the proapoptotic gene Txnip [11,12]. ChREBP also represses aryl hydrocarbon receptor nuclear translocator/hypoxia inducible factor 1-β (ARNT/HIF1-β) [13] shown recently to be diminished in islets [14] and liver [15] of type 2 diabetic humans, and necessary for normal β-cell function and repression of hepatic gluconeogenesis. We sought here to investigate the effects of ChREBP silencing and over-expression on other key glucose-responsive genes in pancreatic islet β-cells, namely pancreatic and duodenal homeobox-1 (Pdx-1), MafA, glucokinase (GcK) and insulin, all critical for normal pancreatic β-cell function.
2. Materials and methods
2.1. Materials
Primers for siRNA construction and PCR were from MWG Biotech (Milton Keynes, UK). Antibodies were described in [9]. Other reagents were from Sigma or Invitrogen.
2.2. Plasmids, adenoviruses and siRNA
pChREBP and ChREBP siRNA have been described in [9]. Adenovirus encoding for ChREBP has been described in [13]. Plasmids and adenoviruses encoding GFP-null and constitutively active and dominant negative forms of SREBP-1c were described in [16]. pPdx1.LucFF, encoding the 5′ flanking region of the mouse pancreatic duodenum homeobox-1 (Pdx-1) gene (−2715 to 0 bp), was generated by PCR from MIN6 cell genomic DNA with the following primers: forward, 5′-ATAT GG TACC CTC CAG TAT CAG GGA GGA-3′ (KpnI site underlined); reverse, 5′-TTT GAGCTC CCA CCC CAG ATC GCT TTG A-3′ (SacI site underlined) and subcloned into pGL3 basic (Promega). Two point mutations [−106C > A; −102T > G] in the Pdx-1 promoter were introduced using Quickchange™ (Stratagene) with the following sense primer: 5′-ATG GCT CCA GGG TAA ACA ACG GGG GGT GCC CCA GAG CCT ATG-3′.
2.3. MIN6 cell culture and islet of Langerhans isolation
MIN6 cells were cultured as in [9]. Mouse islets of Langerhans were isolated and cultured as in [13].
2.4. Single cell reporter gene assay
Intranuclear microinjection of plasmids, antibodies and siRNAs in MIN6 cells were performed at plasmid concentrations of 0.1 (pPdx1.LucFF), and 0.05 (pChREBP, pSREBP-1c, pCMV-RL) mg ml−1, and antibody against ChREBP and SREBP at 1 mg ml−1, before imaging as described in [9].
2.5. Real-time RT-PCR
Total mRNA isolation, cDNA generation and real-time quantitative PCR were performed with primers listed in Table 1, as in [13] and according to the manufacturer’s instructions. Levels of mRNA encoding the indicated genes were normalized compared with cyclophilin mRNA and expressed as the fold change over control (null, 3 mM glucose) and presented as the means ± SEM.
Table 1.
Primers used for real-time RT-PCR.
| mRNA | Forward primer 5′–3′ | Reverse primer 5′–3′ |
|---|---|---|
| MafA | CACCACGTGCGCTTGG | CAGAAAGAAGTCGGGTG |
| Pdx-1 | TGGAGCTGGCAGTGATGTTGA | TCAGAGGCAGATCTGGCCAT |
| Ins1 | GAAGCGTGGCATTGTGGAT | TGGGCCTTAGTTGCAGTAGTTCT |
| Ins2 | AGCCCTAAGTGATCCGCTACAA | CATGTTGAAACAATAACCTGGAAGA |
| GcK | GCTTTTGAGACCCGTTTTGTG | GCCTTCGGTCCCCAGAGT |
| Cyclophilin | TATCTGCACTGCCAAGACTGA | CCACAATGCTCATGCCTTCTTTCA |
2.6. Chromatin immunoprecipitation assay
Chromatin immunoprecipitation was performed essentially as described in [9,13].
2.7. Statistical analysis
Data are given as means ± SEM. Comparisons between means were performed by unpaired two-tailed Student’s t-test with Bonferroni correction as appropriate, using Microsoft Excel.
3. Results
3.1. ChREBP silencing enhances glucose-responsive gene expression in MIN6 pancreatic β-cells
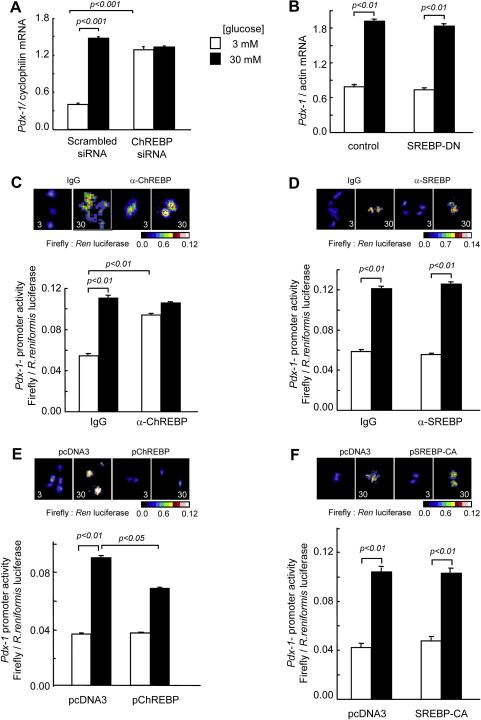
We have previously shown that ChREBP silencing in pancreatic murine insulinoma MIN6 β-cells improves glucose-stimulated insulin secretion, possibly through a decrease in total triglyceride content [9]. Here, we examined the impact of ChREBP silencing by RNA interference on other glucose-responsive genes in MIN6 β-cells. ChREBP knockdown increased the levels of mRNA encoding MafA, GcK and Ins2 at high (30 mM) glucose concentrations, whereas ChREBP silencing increased the expression of the Pdx-1 gene at low (3 mM) glucose concentrations (Table 2 and Fig. 1A). Correspondingly, we observed a similar increase in Pdx-1 promoter activity at low glucose after ChREBP inhibition by microinjection of a specific anti-ChREBP antibody (Fig. 1C), while introduction of a ChREBP expression vector by microinjection suppressed the activity of Pdx-1 promoter at high glucose (Fig. 1E). By contrast, SREBP1-c inactivation or over-expression was without effect on Pdx-1 promoter activity or mRNA levels (Fig. 1B, D and F).
Table 2.
Effects of glucose and ChREBP silencing on mRNA levels in MIN6 cells.
| [Glucose] | Scrambled siRNA |
ChREBP siRNA |
||
|---|---|---|---|---|
| 3 mM | 30 mM | 3 mM | 30 mM | |
| MafA | 0.0294 ± 0.0002 | 0.0332 ± 0.0002*** | 0.0297 ± 0.0002 | 0.0395 ± 0.0002*,¶ |
| GcK | 0.397 ± 0.0002 | 0.927 ± 0.0006*** | 0.386 ± 0.0002 | 1.14 ± 0.0001***,¶ |
| Ins2 | 3.52 ± 0.004 | 9.79 ± 0.004*** | 3.48 ± 0.0007¶ | 10.2 ± 0.004***,¶ |
Culture conditions, total RNA preparation and real-time RT-PCR conditions were as described in Fig. 3. Data are means ± SEM from three independent experiments performed in triplicates, and normalized to cyclophilin mRNA levels. *, *** indicate p < 0.05, 0.0001 for the effect of glucose and ¶ indicates p < 0.05, for the effect of ChREBP siRNA.
Fig. 1.
ChREBP is a repressor of Pdx-1 gene expression in MIN6 cells. (A,B) MIN6 cells were cultured for 48 h in the presence of scrambled or ChREBP siRNA (A), or in the presence of null or SREBP-DN adenoviruses (B), then overnight in 3 mM glucose and finally for 6 h in 3 or 30 mM glucose prior to cell lysis, total RNA extraction and real-time quantitative RT-PCR (see Section 2). (C,D) Pdx-1 promoter activity was monitored via nuclear and cytoplasmic microinjection of Pdx-1 promoter–reporter system and anti-ChREBP (C) or anti-SREBP (D) antibodies (1 mg ml−1), or control IgG as indicated, before culture at the indicated glucose concentrations for 6 h and luciferase imaging as described in Section 2. (E,F) Pdx-1 promoter activity was monitored as above but after co-microinjection of pChREBP (E) or SREBP-CA (F) plasmids. Data are the means ± SEM of three separate experiments.
3.2. ChREBP modulation of Pdx-1 gene expression might be indirect
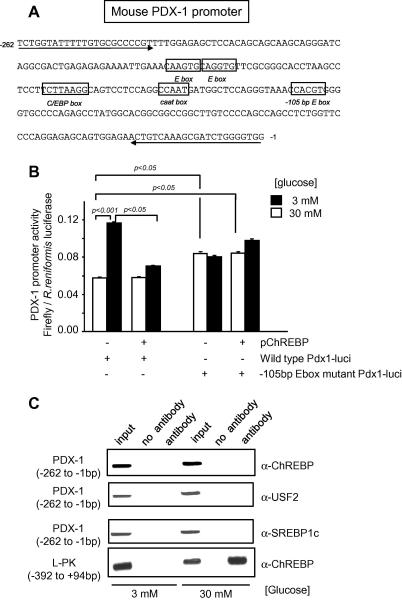
We next sought to identify the region on the Pdx-1 promoter responsive to ChREBP repression. No consensus ChoRE exists on the Pdx-1 promoter, but a proximal E-box, located at −105 bp (Fig. 2A) is highly conserved between species, is protected in DNAse footprints, and has been proposed to confer β-cell specificity to the Pdx-1 promoter [17]. Up to now, it has been thought that this site predominantly binds USF, since mutations abolishing the binding of the latter factor impair the activity of the Pdx-1 promoter, whereas over-expression of a dominant-negative USF2 reduces both Pdx-1 promoter activity as well as Pdx-1 mRNA and protein levels [17,18]. Indeed, mutation of this site abolished both the glucose response and the repressive effect of ChREBP of the Pdx-1 reporter construct (Fig. 2B). However, neither ChREBP, USF2 nor SREBP-1c binding could be detected to the proximal (−260 to +1) region of the promoter by chromatin immunoprecipitation (Fig. 2C). By contrast, and as previously reported [9], ChREBP binding was readily detectable on the proximal L-PK promoter at elevated glucose concentrations (Fig. 2C, bottom panel). We also used this approach to screen a further 11 E-boxes lying in the Pdx-1 promoter region between −2.7 and −0.26 kb (Fig. 3) but could not reveal evidence for ChREBP (not shown).
Fig. 2.
Chromatin immunoprecipitation fails to reveal direct ChREBP binding at the proximal Pdx-1 promoter. (A) DNA sequence of the proximal mouse Pdx-1 promoter showing relevant E-box motifs. Arrows represent the primers used for ChIP assay in (C). (B) Cells were microinjected with either wild-type or mutated pPdx-1.LucFF plus pCMV.RL, plus either empty pcDNA3 or pChREBP as indicated, and incubated and imaged as in Fig. 1. (C) MIN6 cells were cultured in medium containing 3 mM glucose for 16 h prior to stimulation with media containing 3 or 30 mM glucose for 24 h prior to chromatin immunoprecipitation using the antibodies and primers to amplify either Pdx-1 or L-PK promoter as indicated alongside the figure. Data are representative of three independent experiments.
Fig. 3.
Sequences of the primers and E-boxes in the −2.7 kb Pdx-1 promoter probed by ChIP assay. Positions of 5′ and 3′ primers are indicated by arrows and those of E-boxes are highlighted in bold.
3.3. Adenovirus-mediated over-expression of ChREBP inhibits glucose-responsive gene expression in isolated mouse islets of Langerhans
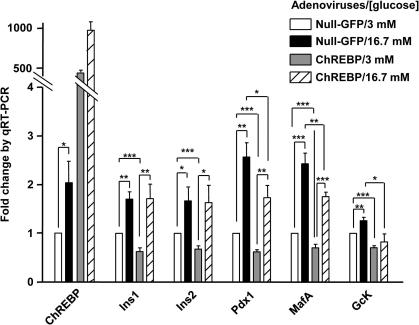
We next examined the effects of ChREBP over-expression on the levels of mRNAs encoding the above glucose-responsive genes in intact islets of Langerhans. Mouse islets were transduced either with a null adenovirus encoding GFP only or with an adenovirus encoding full length wild-type ChREBP [13]. ChREBP over-expression resulted in a decrease in the levels of the mRNA encoding Pdx-1, MafA and GcK at both low and high glucose concentrations, and Ins1 and Ins2 at low glucose concentrations only (Fig. 4).
Fig. 4.
Real-time quantitative PCR analysis of gene expression in mouse islets over-expressing ChREBP. Islets of Langerhans isolated from female CD1 mice aged 12–14 weeks were transduced with either null-GFP or ChREBP adenoviruses (100 MOI) and cultured in 11 mM glucose RPMI media for 48 h and incubation in RPMI media supplemented with 3 or 16.7 mM glucose overnight prior to qRT-PCR as described in methods. Data are means ± SEM from three independent experiments done in triplicates, and normalized to cyclophilin mRNA levels prior to expression as fold change from null infected islets cultured at 3 mM glucose. *p < 0.05; **p < 0.01; ***p < 0.001.
4. Discussion
4.1. Defining a role for ChREBP in glucose-induced pancreatic β-cell dysfunction and diabetes
ChREBP is emerging as an important transcription factor in the pathogenesis of obesity and diabetes and their complications. Indeed, ChREBP is now strongly implicated in the pathogenesis of fatty liver disease and insulin resistance [8,19] acting to induce lipogenic genes. Very recently, ChREBP was proposed to contribute to the development of diabetic nephropathy through the induction of HIF1-α in glomerular mesangial cells [20,21] In the pancreatic β-cell, ChREBP activation by high glucose results in increases in triglyceride content and a reduction in glucose-stimulated insulin secretion [9,13]. We show here that, in addition to the direct effects on L-PK, FAS, Txnip and ARNT promoters previously reported [9,10,12,13] ChREBP also inhibits the expression of several other key β-cell genes, namely Pdx-1, MafA, GcK and insulin.
4.2. Regulation of Pdx-1 gene expression by ChREBP
We show firstly that ChREBP over-expression, both in MIN6 β-cells and pancreatic islets, inhibits Pdx-1 gene expression at high glucose concentrations, and that ChREBP inactivation, achieved either by RNA silencing or antibody microinjection into single cells, increases Pdx-1 expression at low glucose concentrations. Although ChREBP has been shown to possess intrinsic repressive capabilities [13,22], we were unable to demonstrate direct binding of ChREBP in vivo despite scanning the obvious E-boxes, by extensive ChIP assays, at either low or high glucose concentrations (see Fig. 3). We have considered the possibility that an indirect effect of changes in intracellular lipid content could explain the effects of ChREBP on Pdx-1 gene expression, but this would appear to be unlikely given the absence of effects of either inactivation of endogenous SREBP-1c or over-expression of the activated nuclear fragment of SREBP-1c on Pdx-1 promoter activity or mRNA levels (Fig. 1B, D and F), despite well-documented effects of these manoeuvres on cellular triglyceride content [16,23,24]. One possible explanation for the apparent inhibitory effects of ChREBP on Pdx-1 gene expression at low glucose concentrations might be that ChREBP normally serves to sequester another activator(s) of Pdx-1 under these conditions. Translocation of ChREBP to the nucleus at high glucose concentrations, and its activation, either through covalent modification through phosphorylation/dephosphorylation reactions [25], or through an ill-defined mechanism involving the N-terminal LID domain of ChREBP [26,27], may then release the bound transcriptional activator, allowing the latter to bind to the Pdx-1 promoter to stimulate transcription.
It is of note that we were unable here to demonstrate the binding of USF2 at the −105 E-box of the Pdx-1 promoter. Our ChIP assay using both anti-USF and anti-SREBP antibodies has been validated in this cell type on the FAS promoter [9], making a false negative unlikely. The discrepancy between data obtained from in vitro studies and our ChIP assay demonstrates the significance of in vivo (live cell) approaches. Further work will be necessary to identify the key transcription factor binding to the β-cell specific regulatory sequences of the proximal Pdx-1 promoter.
4.3. Regulation of the insulin genes by ChREBP
ChREBP silencing in MIN6 cells resulted in a small, but significant, increase in Ins2 gene expression whereas, in primary islets, ChREBP over-expression only decreased the amount of Ins1 and Ins2 mRNAs at low glucose, but not at elevated glucose concentrations. One possible explanation may lie in the extremely long half-life of insulin mRNAs at high glucose concentrations [28], therefore making a decrease in its transcription rate unnoticeable within the time course of these experiments.
5. Conclusion
We conclude that ChREBP is a key regulator of adult β-cell phenotype, affecting the expression of critical genes. It therefore seems possible that increases in ChREBP expression, prompted by a diabetic milieu, may exacerbate β-cell dysfunction and accelerate β-cell failure in type 2 diabetes. It would be interesting to know whether a β-cell specific ChREBP knockout mouse would be protected against the development of β-cell failure during the course of some forms of diabetes.
Acknowledgments
Supported by grants from Diabetes UK (BDA:RD04/0002895) and Wellcome Trust (WT082366MA) to I.L., and from grants from the Wellcome Trust (Programme Grant 081958/2/07/Z), The European Union (FP6 “Save Beta”; FP7, “IMIDIA”), and the Medical Research Council (G0401641) to G.A.R.
References
- 1.Poitout V., Robertson R.P. Glucolipotoxicity: fuel excess and beta-cell dysfunction. Endocr. Rev. 2008;29:351–366. doi: 10.1210/er.2007-0023. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Shih H., Towle H.C. Definition of the carbohydrate response element of the rat S14 gene. Context of the CACGTG motif determines the specificity of carbohydrate regulation. J. Biol. Chem. 1994;269:9380–9387. [PubMed] [Google Scholar]
- 3.Shih H.M., Liu Z., Towle H.C. Two CACGTG motifs with proper spacing dictate the carbohydrate regulation of hepatic gene transcription. J. Biol. Chem. 1995;270:21991–21997. doi: 10.1074/jbc.270.37.21991. [DOI] [PubMed] [Google Scholar]
- 4.Yamashita H., Takenoshita M., Sakurai M., Bruick R.K., Henzel W.J., Shillinglaw W., Arnot D., Uyeda K. A glucose-responsive transcription factor that regulates carbohydrate metabolism in the liver. Proc. Natl. Acad. Sci. USA. 2001;98:9116–9121. doi: 10.1073/pnas.161284298. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Stoeckman A.K., Ma L., Towle H.C. Mlx is the functional heteromeric partner of the carbohydrate response element-binding protein in glucose regulation of lipogenic enzyme genes. J. Biol. Chem. 2004;279:15662–15669. doi: 10.1074/jbc.M311301200. [DOI] [PubMed] [Google Scholar]
- 6.Uyeda K., Repa J.J. Carbohydrate response element binding protein ChREBP, a transcription factor coupling hepatic glucose utilization and lipid synthesis. Cell Metab. 2006;4:107–110. doi: 10.1016/j.cmet.2006.06.008. [DOI] [PubMed] [Google Scholar]
- 7.Iizuka K., Bruick R.K., Liang G., Horton J.D., Uyeda K. Deficiency of carbohydrate response element-binding protein (ChREBP) reduces lipogenesis as well as glycolysis. Proc. Natl. Acad. Sci. USA. 2004;101:7281–7286. doi: 10.1073/pnas.0401516101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Dentin R., Benhamed F., Hainault I., Fauveau V., Foufelle F., Dyck J.R., Girard J., Postic C. Liver-specific inhibition of ChREBP improves hepatic steatosis and insulin resistance in ob/ob mice. Diabetes. 2006;55:2159–2170. doi: 10.2337/db06-0200. [DOI] [PubMed] [Google Scholar]
- 9.da Silva Xavier G., Rutter G.A., Diraison F., Andreolas C., Leclerc I. ChREBP binding to fatty acid synthase and L-type pyruvate kinase genes is stimulated by glucose in pancreatic beta-cells. J. Lipid Res. 2006;47:2482–2491. doi: 10.1194/jlr.M600289-JLR200. [DOI] [PubMed] [Google Scholar]
- 10.Wang H., Wollheim C.B. ChREBP rather than USF2 regulates glucose stimulation of endogenous L-pyruvate kinase expression in insulin-secreting cells. J. Biol. Chem. 2002;277:32746–32752. doi: 10.1074/jbc.M201635200. [DOI] [PubMed] [Google Scholar]
- 11.Chen J., Hui S.T., Couto F.M., Mungrue I.N., Davis D.B., Attie A.D., Lusis A.J., Davis R.A., Shalev A. Thioredoxin-interacting protein deficiency induces Akt/Bcl-xL signaling and pancreatic beta-cell mass and protects against diabetes. FASEB J. 2008;22:3581–3594. doi: 10.1096/fj.08-111690. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Cha-Molstad H., Saxena G., Chen J., Shalev A. Glucose-stimulated expression of Txnip is mediated by carbohydrate response element-binding protein p300 and histone H4 acetylation in pancreatic beta cells. J. Biol. Chem. 2009;284:16898–16905. doi: 10.1074/jbc.M109.010504. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Noordeen N.A., Khera T.K., Sun G., Longbottom E.R., Pullen T.J., da Silva Xavier G., Rutter G.A., Leclerc I. Carbohydrate-responsive element-binding protein (ChREBP) is a negative regulator of ARNT/HIF-1beta gene expression in pancreatic islet beta-cells. Diabetes. 2010;59:153–160. doi: 10.2337/db08-0868. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Gunton J.E., Kulkarni R.N., Yim S., Okada T., Hawthorne W.J., Tseng Y.H., Roberson R.S., Ricordi C., O’Connell P.J., Gonzalez F.J., Kahn C.R. Loss of ARNT/HIF1beta mediates altered gene expression and pancreatic-islet dysfunction in human type 2 diabetes. Cell. 2005;122:337–349. doi: 10.1016/j.cell.2005.05.027. [DOI] [PubMed] [Google Scholar]
- 15.Wang X.L., Suzuki R., Lee K., Tran T., Gunton J.E., Saha A.K., Patti M.E., Goldfine A., Ruderman N.B., Gonzalez F.J., Kahn C.R. Ablation of ARNT/HIF1beta in liver alters gluconeogenesis lipogenic gene expression and serum ketones. Cell Metab. 2009;9:428–439. doi: 10.1016/j.cmet.2009.04.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Andreolas C., da Silva Xavier G., Diraison F., Zhao C., Varadi A., Lopez-Casillas F., Ferre P., Foufelle F., Rutter G.A. Stimulation of acetyl-CoA carboxylase gene expression by glucose requires insulin release and sterol regulatory element binding protein 1c in pancreatic MIN6 beta-cells. Diabetes. 2002;51:2536–2545. doi: 10.2337/diabetes.51.8.2536. [DOI] [PubMed] [Google Scholar]
- 17.Melloul D., Marshak S., Cerasi E. Regulation of pdx-1 gene expression. Diabetes. 2002;51(Suppl. 3):S320–S325. doi: 10.2337/diabetes.51.2007.s320. [DOI] [PubMed] [Google Scholar]
- 18.Qian J., Kaytor E.N., Towle H.C., Olson L.K. Upstream stimulatory factor regulates Pdx-1 gene expression in differentiated pancreatic beta-cells. Biochem. J. 1999;341(Pt. 2):315–322. [PMC free article] [PubMed] [Google Scholar]
- 19.Iizuka K., Miller B., Uyeda K. Deficiency of carbohydrate-activated transcription factor ChREBP prevents obesity and improves plasma glucose control in leptin-deficient (ob/ob) mice. Am. J. Physiol. Endocrinol. Metab. 2006;291:E358–E364. doi: 10.1152/ajpendo.00027.2006. [DOI] [PubMed] [Google Scholar]
- 20.Isoe T., Makino Y., Mizumoto K., Sakagami H., Fujita Y., Honjo J., Takiyama Y., Itoh H., Haneda M. High glucose activates HIF-1-mediated signal transduction in glomerular mesangial cells through a carbohydrate response element binding protein. Kidney Int. 2010;78:48–59. doi: 10.1038/ki.2010.99. [DOI] [PubMed] [Google Scholar]
- 21.Haase V.H. The sweet side of HIF. Kidney Int. 2010;78:10–13. doi: 10.1038/ki.2010.112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Cairo S., Merla G., Urbinati F., Ballabio A., Reymond A. WBSCR14 a gene mapping to the Williams–Beuren syndrome deleted region is a new member of the Mlx transcription factor network. Hum. Mol. Genet. 2001;10:617–627. doi: 10.1093/hmg/10.6.617. [DOI] [PubMed] [Google Scholar]
- 23.Diraison F., Parton L., Ferre P., Foufelle F., Briscoe C.P., Leclerc I., Rutter G.A. Over-expression of sterol-regulatory-element-binding protein-1c (SREBP1c) in rat pancreatic islets induces lipogenesis and decreases glucose-stimulated insulin release: modulation by 5-aminoimidazole-4-carboxamide ribonucleoside (AICAR) Biochem. J. 2004;378:769–778. doi: 10.1042/BJ20031277. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Wang H., Maechler P., Antinozzi P.A., Herrero L., Hagenfeldt-Johansson K.A., Bjorklund A., Wollheim C.B. The transcription factor SREBP-1c is instrumental in the development of beta-cell dysfunction. J. Biol. Chem. 2003;278:16622–16629. doi: 10.1074/jbc.M212488200. [DOI] [PubMed] [Google Scholar]
- 25.Kawaguchi T., Osatomi K., Yamashita H., Kabashima T., Uyeda K. Mechanism for fatty acid “sparing” effect on glucose-induced transcription: regulation of carbohydrate-responsive element-binding protein by AMP-activated protein kinase. J. Biol. Chem. 2002;277:3829–3835. doi: 10.1074/jbc.M107895200. [DOI] [PubMed] [Google Scholar]
- 26.Li M.V., Chang B., Imamura M., Poungvarin N., Chan L. Glucose-dependent transcriptional regulation by an evolutionarily conserved glucose-sensing module. Diabetes. 2006;55:1179–1189. doi: 10.2337/db05-0822. [DOI] [PubMed] [Google Scholar]
- 27.Davies M.N., O’Callaghan B.L., Towle H.C. Glucose activates ChREBP by increasing its rate of nuclear entry and relieving repression of its transcriptional activity. J. Biol. Chem. 2008;283:24029–24038. doi: 10.1074/jbc.M801539200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Fred R.G., Welsh N. The importance of RNA binding proteins in preproinsulin mRNA stability. Mol. Cell. Endocrinol. 2009;297:28–33. doi: 10.1016/j.mce.2008.06.007. [DOI] [PubMed] [Google Scholar]