Abstract
The field of organellar proteomics has emerged as an attempt to minimize the complexity of the proteomics data obtained from whole cell and tissue extracts while maximizing the resolution on the protein composition of a single subcellular compartment. Standard methods involve lengthy density-based gradient and/or immunoaffinity purification steps followed by extraction, one-dimensional or two-dimensional gel electrophoresis, gel staining, in-gel tryptic digestion and protein identification by mass spectrometry. In this paper, we present an alternate approach to purify subcellular organelles containing a fluorescent reporter molecule. The gel-free procedure involves fluorescence-assisted sorting of the secretory granules followed by gentle extraction in a buffer compatible with tryptic digestion and mass-spectrometry. Once the subcellular organelle labeled, this procedure can be done in a single day, requires no major modification to any instrumentation and can be readily adapted to the study of other organelles. When applied to corticotrope secretory granules, it led to a much enriched granular fraction from which numerous proteins could be identified through mass spectrometry.
Keywords: Corticotropes, FACS, FAOS, Protein secretion, Secretory granules
1. INTRODUCTION
As a whole, the field of proteomics aims to identify all proteins present in a tissue or cell type in a given set of conditions at a specific time. However, this endeavor has proven extremely complex due to the thousands of proteins being potentially expressed at a dynamic level varying from a few copies to millions of copies within a single cell.
In an effort to circumvent the ensuing data complexity, a subfield of proteomics has emerged and proposes analysis of the proteome of individual subcellular compartments. In the last few years, numerous studies have been published and led to the identification of anywhere between one hundred to a few thousand proteins specific to many organelles[1, 2], including the cell nucleus in terms of its content in the nuclear envelope [3] and the nucleolus [4], the endoplasmic reticulum [5], the Golgi apparatus [6], the secretory lysosomes [7], the phagosome [8], clathrin-coated vesicles [9] and the mitochondrion [10]. The resulting information has been of the utmost importance in unraveling the specific overlapping and non-overlapping protein composition of cellular compartments as well as their specific roles in biological processes.
In nearly all the subcellular proteomics studies including those above listed, the general methodology relies to a great extent on subcellular fractionation allowing recovery of the organelle through careful selection of physical properties. Hence, following cell or tissue lysis, it is usual to submit the lysate to one or many rounds of differential centrifugation. Velocity and equilibrium centrifugation have established their capability in the past fifty years to separate cellular components differing by density. Once the centrifugation steps are complete, the organellar content is extracted and the proteins are further separated by 1D- or 2D-SDS-PAGE. Upon gel staining, the bands or spots are excised; the proteins are proteolytically digested and are ultimately identified by analyzing their peptides by mass spectrometry. However, in the case of a complex endocrine tissue such as the pituitary gland containing five morphologically distinct endocrine cells, applying this lengthy procedure (48–72 hrs) to analyze the protein content of secretory granules of a single cell type would represent a sizeable task. Hence, in order to do so, it is preferable to rely on using a cell line representative of a given cell type and this, even if the above described method is rendered less efficient when applied to cell cultures [11].
In addition to the difficulty and the duration of the procedure, we aimed at simplifying it for two other main reasons. Firstly, it is still unclear how representative the proteome of an organelle is when taking into perspective the time at which cells are extracted and the time at which MS data is made available following days of preparation, even in the presence of protease inhibitors. Secondly, even after optimization, most steps involved are not very efficient, thereby necessitating large amounts of starting biological material. For example, SDS-PAGE leads unavoidably to staining artifacts, loss of sample and poor separation of very high (over 100 kDa) and very low (under 15 kDa) molecular weight proteins. Consequently, the development of alternative or complementary approaches for subcellular fractionation in organellar proteomics is needed and advocated [2].
Fluorescence assisted cell sorting (FACS) is routinely employed to perform selective enrichment of cell populations bearing specific markers based on fluorescence [12, 13]. In 1985, Murphy’s group first described the concept of single organelle fluorescence analysis (SOFA) and subsequently suggested its application to sort single organelles [14]. The term fluorescence-assisted organelle sorting (FAOS) has been introduced to describe the use of flow cytometry to sort subcellular structures and components. However, this represents a most challenging task as the organelles to be sorted are routinely 10 to 100 times smaller than the original cells. Furthermore, the resistance of the target organelle to the shear and tear fluid forces in the cell sorter is severely limited. Nonetheless, publications have described sorting of organelles such as endosomes [15], mitochondria [16] and phagosomes [17] using various fluorescent probes. However, due to technical constraints, most of these were analytical in nature and none went as far as conducting a proteomic analysis of the sorted organelle although demonstration of significant level of enrichment was accomplished [18].
The dense-core secretory granule is the ultimate compartment in the regulated pathway of secretion. Originally perceived as a simple storage compartment, it is now known to be actively involved in cargo selection, maturation, and secretory processes. However, much remains to be defined in terms of the mechanisms implicated and the proteins involved, and further understanding of this organelle could shed light on several normal and pathological endocrine processes. Proteomics studies of the secretory pathway [5] as well as that of various secretory vesicles and/or granules have been published, some very recently. Relying on classical purification and analysis protocols, the proteomes of atrial secretory granules [19], pancreatic zymogen granules [20, 21], chromaffin granules [22], insulin-containing granules [23] and synaptic vesicles [24, 25] were documented.
The present work focuses on the development and application of a simple subcellular fractionation strategy to the study of the pituitary corticotropes dense-core secretory granules which does not use density gradient centrifugation and gel separation. It involves gentle cell lysis, brief centrifugation, improved FAOS and LC-MS. Using relatively small numbers of cultured cells, it led to identification of more than 100 proteins, including small molecular weight proteins and granule-associated proteins. More importantly, it can be accomplished within a day following proper cell culturing and using readily available facilities.
2. MATERIALS and METHODS
2.1 Cell culture and immunocytochemistry
Materials
Unless otherwise stated, all chemicals, solutions and solvents were of the highest possible grade purchased from Sigma-Aldrich. Primary antibodies (anti-calnexin, anti-giantin, anti-prohibitin, anti-ERp29, anti-PIST, anti-cathepsin B, anti-Cdk9, anti-RPL8, anti-chromogranin A and anti-GFP were purchased from Abcam. Cell culture material was purchased from Invitrogen.
Cell culture and immunocytochemistry
The AtT-20 cells stably expressing the-PHM-mGFP fusion protein were generated as described [26]. Briefly, AtT-20 were transfected with an expression vector encoding for amino acids 1–407 of the peptidyl-glycine α-amidating monooxygenase enzyme fused in frame with monomeric green fluorescent protein. AtT-20 and AtT-20-PHM-mGFP cells were grown in DMEM-F12 containing 10% fetal calf serum, 10% NuSerum, penicillin/streptomycin/glutamine and 0.5 mg/mL G418 in T175 flasks at 37°C in 10% CO2 atmosphere. They were regularly passaged when confluence reached 90–95%. Fresh medium was added to the cells 16–20 hrs before they were collected. Cells were usually kept for no more than 20 generations in order to maintain phenotype and steady levels of fluorescence. WT and PHM-mGFP transfected cells were indistinguishable in appearance and growth properties. Colocalization of ACTH with PHM-mGFP in the secretory granules was seen using a Zeiss LSM510 confocal microscope following procedures previously described [26].
2.2 Cell lysis and fluorescence-assisted organelle sorting (FAOS)
Cell lysis
All the following steps were performed at 37°C. When cells reached 90–95% confluence, they were trypsinized and collected in warm fresh medium. Latrunculin B and nocodazole (Sigma-Aldrich) were added to a final concentration of 5 µM each and the cells were allowed to incubate for 20 min with occasional gentle agitation. The cells from a single T175 flask were pelleted, washed once in PBS and resuspended in 1 mL of sucrose solution (0.34 M sucrose, 10 mM HEPES, 0.3 mM EDTA, protease inhibitor cocktail (Roche), 5 µM latrunculin B, 5 µM nocodazole). From this point on, all steps were carried at 4°C. Two cell lysis procedures were evaluated namely the use of a 2 mL Potter homogenizer (Kontes) and the use of a freeze-thaw procedure. However, submitting them to one freeze (5 min)-thaw (2 min) cycle consistently resulted in over 90% cell lysis as seen under light microscope and thus, this method was used in the current study. The cell extract was further incubated 5 min on ice, and then centrifuged on a bench-top centrifuge at 1 500 × g for 5 min to remove nuclei, unbroken cells and large debris. The 1 500 × g supernatant was collected and diluted with one third volume of cold PBS with EDTA (25 mM final). This final sample was immediately submitted to flow cytometry.
FAOS
Flow cytometry sorting of the secretory granules was performed using a MOFLO ultra high speed cell sorter (Dako) equipped with an Innova 90C ion laser (Coherent) tuned at 488 nm and 200 mW. Threshold was adjusted on the side scatter (SSC) set in linear mode. Green florescence was detected in FL1 using standard filters for GFP detection (530/30). The threshold was adjusted to optimize granules detection, in which case the sorting rate was in the range of 3 000–5 000 events per second, even if over 25 000 events per second were detectable at maximum sensitivity. Since this equipment is usually employed to sort whole cells (6–25 µm), the neutral filter was removed to increase sensitivity in the lower size range and the sample pressure was set to 20 psi (1.38 bars).In order to define reference settings, 450 nm sky blue, 530 nm nile red, 840 nm sky blue and 840 nm nile red control beads (Spherotech) were used to obtain a size and fluorescence estimate of the detected events. A population of event presenting characteristics similar to those expected from secretory granules and known from electron microscopy and literature (relative homogeneity in size, approximately 300–500 nm) was rapidly identified on the density plot representation (X-axis: fluorescence, Y-axis: size, Color-code: density or number of events) using the previously mentioned beads. All sorted events were smaller than 500 nm, as fluorescent as nile red 530 nm beads (positive control for fluorescence), and at least 10 times more fluorescent than sky blue 450 nm beads (negative control for fluorescence). Non-transfected WT AtT-20 cells were also analyzed and results confirmed the absence of endogenous green fluorescence (data not shown). Propidium iodide staining was routinely conducted to confirm the absence of DNA left in the samples after centrifugation and before sorting to exclude intact nuclei. Within the previously detected population of events, the boundaries of the sorting window were set to encompass simultaneously both the peak of fluorescence distribution of events and the peak of size distribution of events (visible on a classical 2D histogram representation, not shown). Data acquisition and processing was performed with the Summit software, v4.3 (Dako). In all cases, the content of the cell lysate was sorted in ice-cold PBS containing 5 µM latrunculin B and the purified granules were centrifuged at 5 000 × g for 20 min at 4°C using a swing-out SW41ti rotor in a L8-80 Ultracentrifuge (Beckman). Following removal of the supernatant, the pellet was washed with cold PBS and centrifuged again at 5 000 × g for 20 min at 4°C.
2.3 Electron microscopy and Western Blot
Electron microscopy
Following sorting and centrifugation, the pellet was post-fixed with 1% osmium tetroxide in 0.1 M cacodylate buffer for 30 min, dehydrated in graded series of ethanol dilutions and embedded in Durcupan (Fluka). After polymerization, serial ultrathin sections from the pellet were cut on an Ultracut ultramicrotome (Reichert-Jung) and collected on Nickel 200 mesh square grids. Sections were counterstained with lead citrate and examined with JEM1200EX electron microscope (Jeol).
Western Blot
1.5 µg each of protein from the total cell lysate, from the 1 500 × g supernatant and from the granule extracts A and B were separated on a 10% SDS-PAGE and analyzed by Western Blot. Anti-GFP (1:5000), anti-ERp29 (1:2500) and anti-PIST (1:2500) were used to reveal the presence of these subcellular markers using enhanced chemiluminescence (GE-Healthcare). Also, 4 µg each of protein from the total cell lysate, from the 1 500 × g supernatant and from the granule extracts A and B were separated on a 10% SDS-PAGE and analyzed by Western Blot. Anti-prohibitin (1:750), anti-Cdk9 (1:750), anti-capthepsin B (1:750), anti-RPL8 (1:750) and anti-chromogranin A (1:750) were used to reveal the presence of these subcellular markers using enhanced chemiluminescence.
2.4 Granule extraction and mass spectrometry
Granule extraction
The pelleted granules were resuspended in a hypoosmotic solution composed of 10 mM EDTA in water and kept for 5 min at 4°C. Following incubation, the pellet was extracted by three consecutive Freeze (2 min) – Boil (1 min) – Sonicate (5 sec) cycles. Following removal of the aqueous solution (extract A), the tube is rinsed with an aqueous solution containing 10 mM EDTA and 0.025% (w/v) SDS and the content submitted to two consecutive Freeze-Boil-Sonicate cycles (extract B). In order to maximize the number and significance of the identified proteins, both successive extracts were submitted to mass spectrometry. The SDS present in the second extract was removed using the SDSAway™ sample preparation kit (Protea Biosciences) according to the manufacturer’s protocol. Extracts A and B obtained from three separate purifications were dried under vacuo.
LC-MS/MS
Protein extracts were reconstituted in 6 M urea, reduced with 1, 4-dithio-DL-threitol and alkylated with iodoacetamide. Proteolytic digestions were performed at 37°C for 5 hrs using a sequencing-grade modified Trypsin (Promega) with a protein-enzyme ratio of 1:25. Chromatographic separation and subsequent analysis were accomplished on a nanoLC system (Eksigent) coupled to a LTQ Orbitrap hybrid mass spectrometer (ThermoFisher). The tryptic peptides were separated using self-pack PicoFrit capillary columns (75 µm i.d. × 10 cm, 15 µm tip) (New Objective) packed with Jupiter C18 reversed-phase stationary phase of 5 µm particle size (Phenomenex). A gradient elution of 4–80 % acetonitrile-water (0.2 % formic acid) in 35 min was used for all separations.
Data analysis
Peaks were generated using Mascot Daemon v.2.1.6 while protein identification was performed with the Mascot software package v.2.1.03 (Matrix Science, London, UK) [27]. The database probed was NCBInr, released on 2006/09/05, restricting the search on the Mus Musculus taxon (106363 sequences) since the AtT-20 cells are of murine origin. The search criteria were as follow: Tryptic digestion; Variable modifications include carbamidomethylation (Cys), di-methylation (Lys), di-methylation (Arg), oxidation (Met) with a peptide mass tolerance of ± 10 ppm and a fragment mass tolerance of ± 0.7 Da. The maximum missed cleavage number was set at 2. Peptide scores were derived from ions scores as a non-probabilistic basis for ranking protein hits and the protein scores as the sum of a series of peptide scores. The score threshold to achieve p<0.05 (95% probability of positive identification per protein, yielding less than 5% false positive identification) is set by Mascot algorithm, and is based on the size of the database and the peptide mass tolerance used for the search. For additional confidence in protein identification, peptides with a score of 10 or less were excluded, while the presence of at least one bold red (high significance) peptide was required per protein. For each protein listed and matching the above-mentioned criteria, the quality of the MS/MS spectra of peptides with scores lower than 50 was manually verified. Proteins identified as “unknown” were usually easily identified by searching the peptide sequence using BLAST algorithm [28] for short, nearly exact matches. When all peptides for a protein hit matched another protein hit, the top scoring hit was selected. Members of the same family of proteins were identified by a sufficient amount of peptides to allow discrimination. In order to be included in our list, a protein must exhibit a minimum score of 50 and be identified by at least two distinct peptides, further enhancing the probability of true identification.
3. Results
3.1 Cell culture and immunocytochemistry
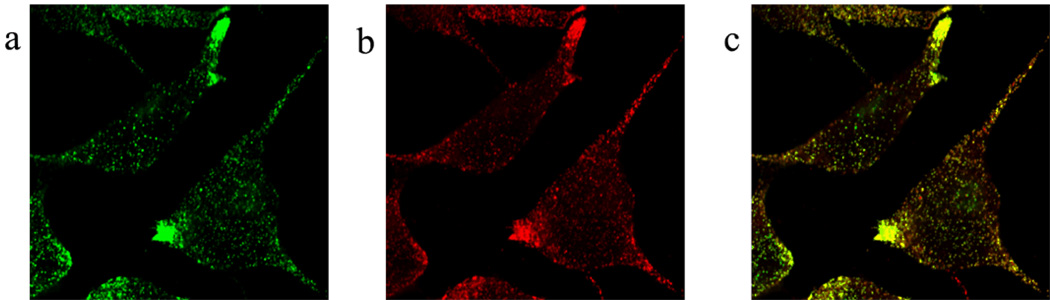
AtT-20 is a tumor-derived, immortalized cell line of murine pituitary corticotropes and a recognized and accepted model for pro-opiomelanocortin (POMC) production and secretion studies. Indeed, as early as 1981, the secretory granules of these cells were isolated, studied and shown to contain mature forms corticotropin and β-lipotropin [29]. For our method to succeed, one has to label the organelle with a fluorescent molecule. In the past, green fluorescent protein (GFP) was shown to be routed to the regulated pathway of secretion and ultimately into secretory granules through linking either to a signal sequence [30] or to the NH2-terminal domain (known as PHM) of the secretory granule resident peptidyl α-amidating monooxygenase (PAM) enzyme [26]. An expression vector, PHM-mGFP (the m refers to the monomeric GFP variant resulting from an A206K mutation [31]), was used to transfect AtT-20 cells and stably expressing cells were obtained. Additional studies [26] and immunocytochemistry experiments confirmed that expression of the fusion protein, although not solely confined to secretory granules, nevertheless is limited to the regulated pathway of secretion. For example (Fig. 1), the PHM-mGFP signal is present in secretory granules which share a pattern of distribution within the cells, of storage and of release in response to stimulation similar to adrenocorticotropin (ACTH), a hormone resulting from POMC processing.
Figure 1. Intracellular localization of the PHM-mGFP fusion protein within AtT-20 transfected cells.
(a) PHM-mGFP (green) (b) ACTH (red) and (c) merging of colors highlights the colocalization of PHM-mGFP and ACTH in the secretory granules at the tips of the cells.
3.2 Cell lysis and fluorescence-assisted organelle sorting
In order to minimize the mechanical stress associated with the use of Potter homogenization and the ensuing damage to organelles and possible formation of microsomes, we used a pre-treatment of the cells with latrunculin B, an actin filament disruptor [32] and nocodazole, a microtubule disruptor [33]. This was done in an effort to inhibit the secretory processes and partially dissolve the cytoskeleton to free the granules from the actin and tubulin meshes within the cells. After treatment, the cells were lysed by one freeze-thaw cycle in an isoosmotic sucrose solution which led to over 90% cell lysis.
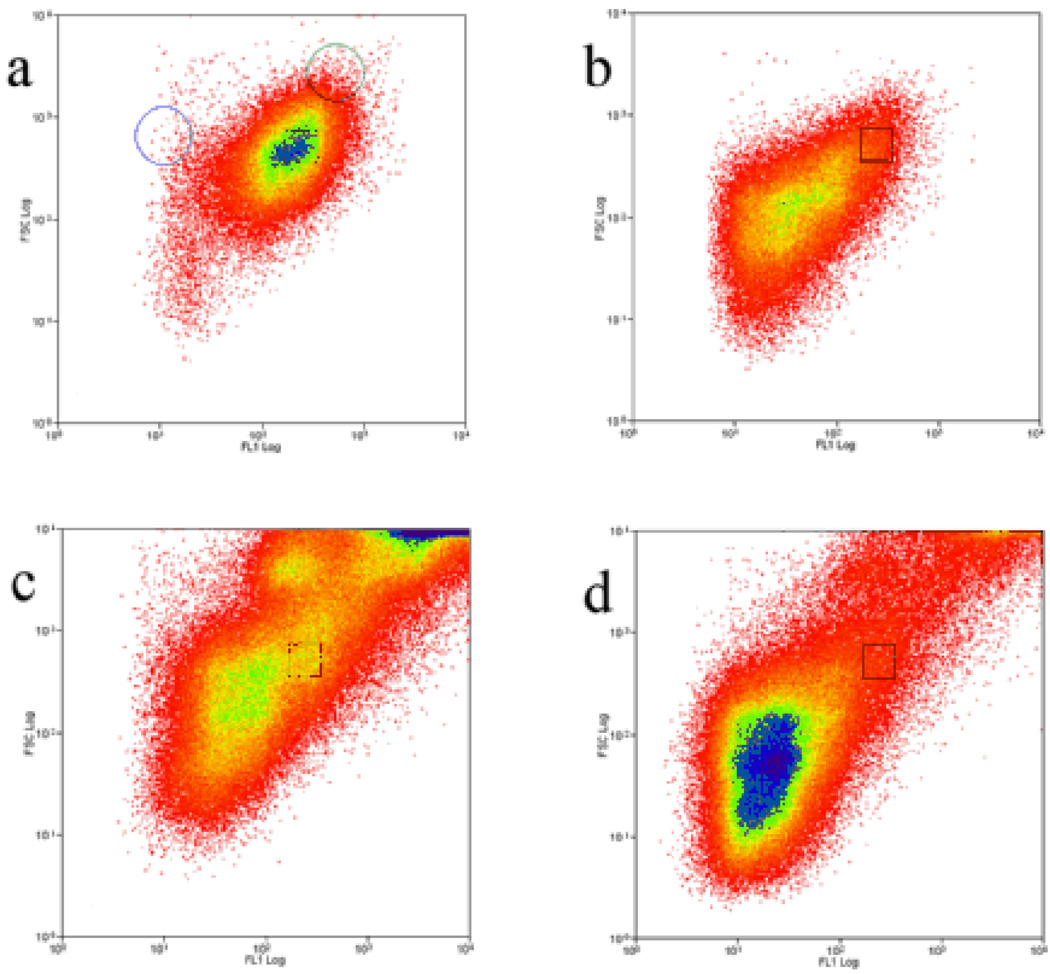
The sample was sorted using a regular cell sorter with the sample pressure lowered in an effort to minimize the shear and tear forces to which the granules are exposed. In addition to reducing the electronic background noise, it also considerably improves the resolution in the small particle size range. A narrow sorting window was selected on the density plot illustrating the fluorescence (X-axis) and size (Y-axis) of the particles in the sample. Commercially available calibration beads (non-fluorescent and fluorescent, 450 nm, 530 nm and 840 nm) were used as reference points to estimate the size and level of fluorescence of the sorted populations. As shown in Fig. 2a, the sorted granule population (black square) was 10 to 50 times more fluorescent than the non fluorescent beads and non-fluorescent events, while being smaller than 500 nm in size. This figure also shows all detectable events in the 1 500 × g supernatant (Fig. 2b) and the total cell lysate (Fig. 2d) with the trigger adjusted to minimum threshold (maximum sensitivity) on the side scatter (SSC) detector. It also illustrates how a brief centrifugation step is necessary to reveal a population (Fig. 2a) that was not clearly distinguishable in the total cell lysate (Fig. 2c). On average, our strict selection parameters (sorting window) allowed sorting of 3–5 million events per cell dish. Evidently, this number can vary depending on the sorting criteria; herein, it corresponds to approximately 20% of detectable events at the optimized sensitivity threshold, or 2–3% of all detectable events at maximum sensitivity.
Figure 2. Fluorescence-assisted organelle sorting of the secretory granules (FAOS).
(a) to (d) are density plots (green fluorescence (X-axis) versus size (Y-axis) of all events detected by the cell sorter under each set of conditions. The black square represents the selected sorting region which corresponds to the secretory granules (see text). (a) 1 500 × g supernatant analysis with the threshold set to optimize the detection of the secretory granule population. The blue circle and green circle indicate the region in which non-fluorescent 450 nm beads and fluorescent 840 nm beads are respectively located when analyzed. (b) 1 500 × g supernatant analysis with the threshold set to the minimum (maximum sensitivity), allowing visualization of the most abundant smaller, low-fluorescent to non-fluorescent events. (c) Total cell lysate analyzed as in (a). The presence of numerous cellular particles of varying fluorescence is visible, masking the granule population. The top right density represents larger fluorescent particles such as intact cells and large ER and Golgi debris. (d) Total cell lysate analyzed as in (b). In order to present informative and representative plots, 75 000 events were pooled in (a) and (b) while 225 000 events were pooled in (c) and (d). Density is color-coded from low (few events) in red to high (many events) in blue.
3.3 Electron microscopy and Western Blot
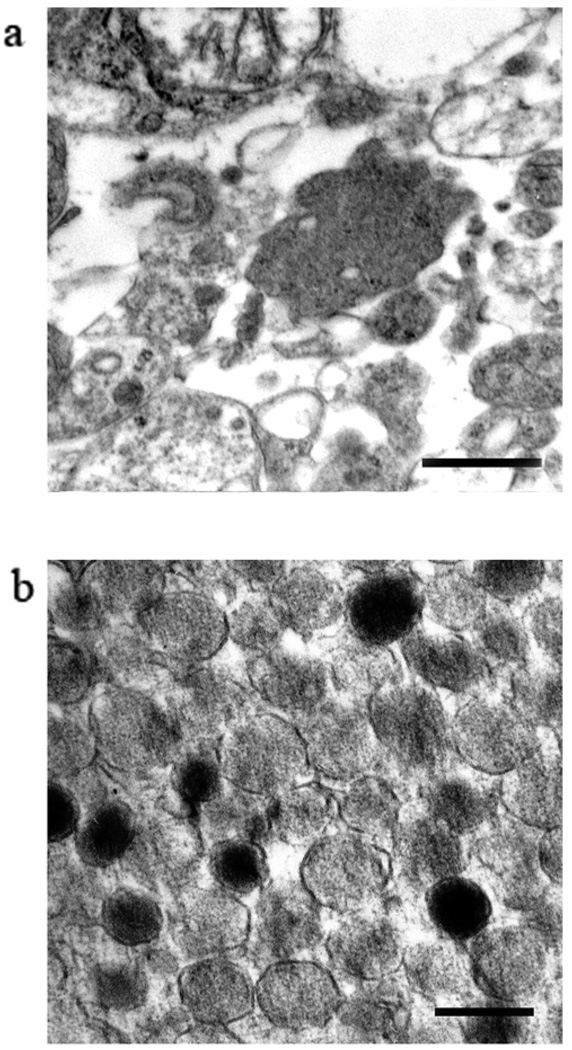
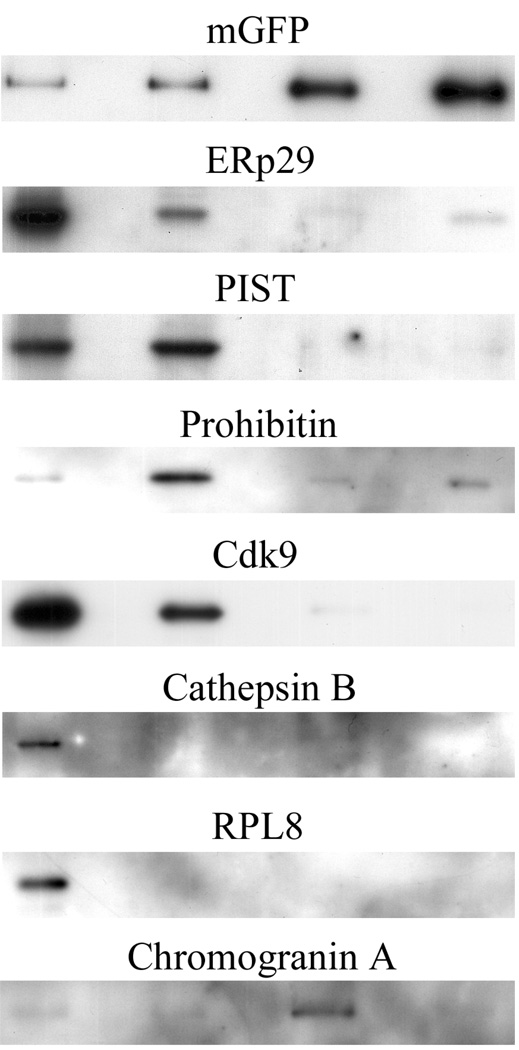
The sorted events were pelleted, fixed and analyzed by electron microscopy. The content of a representative cut through the pelleted total lysate (Fig. 3a) and through the FAOS-purified granules pellet (Fig. 3b) is shown in Fig. 3. It is noteworthy that the granules collected are very homogeneous in size and present an intact morphology. However, most granules appear slightly decondensed, an effect likely resulting from the presence of EDTA in the sorting medium and the passage through the sorter’s lines and nozzle. Samples from the total extract, the 1 500 × g supernatant and the sorted granules were routinely analyzed by SDS-PAGE and Western blotting using a battery of antibodies to assess the enrichment and depletion of specific subcellular markers. As shown in Fig. 4, the FAOS purification leads to a considerable enrichment of the sample in mGFP and chromogranin A (granules) with a concomitant depletion of the sample in ERp29 (ER), PIST (Golgi), prohibitin (mitochondria), Cdk9 (nuclei), cathepsin B (lysosomes) and RPL8 (ribosomes) This illustrates the efficiency of the procedure to selectively sort the granules from the rest of the mGFP-tagged material in the secretory pathway and from non-labeled compartments, a conclusion that is further confirmed by the mass spectrometry data.
Figure 3. Secretory granules visualized by electron microscopy.
(a) A representative cut of the total cell extract; bar: 500 nm. (b) A representative cut thought the purified secretory granules pellet shows a population of intact, membrane bound granules, homogeneous is size. Most of them are slightly decondensed (see text); bar: 200 nm.
Figure 4. Western blots illustrating the granules enrichment.
From left to right, separated by empty lanes are the total cell lysate, the 1 500 × g supernatant, the granule extract A and the granule extract B. 1.5 µg or 4 µg of protein was loaded in each lane (see text). Markers for various subcellular markers and structures were assessed. While the ER and Golgi markers are depleted, the GFP signal, present only in the ER, Golgi and secretory granules, is enriched. The markers for the mitochondria, nucleus, lysosomes and ribosomes are also depleted with a concomitant enrichment in chromogranin A, a recognized granule marker.
3.4 Granule extraction and mass spectrometry
The purified granules were centrifuged, washed and sequentially extracted in water containing 10 mM EDTA, and subsequently in water containing 0.025% SDS (w/v). Extract A was submitted to tryptic digestion and the resulting peptides were separated by liquid chromatography and analyzed with an ESI-LTQ-Orbitrap mass spectrometer. Extract B was treated (see methods) to remove SDS and processed identically. The resulting data was analyzed using the Mascot software [27]. The FAOS-purified material originating from ca. 15 million cells was routinely sufficient to identify over 100 proteins from over 3 000 peptide queries using relatively stringent criteria. Chromogranin A, a recognized secretory granule marker [34], was always and consistently the top scoring protein. Data originating from individual LC-MS/MS analysis of extracts A and B of three distinct granules preparations originating from three separate purification experiments were combined to increase the number of proteins identified and their respective score. With this approach, more than a hundred proteins were identified. Moreover, the reproducibility between the three different granule preparations is very high. Indeed, 96.7% (29/30) of the 30 top scoring proteins were identified in all three preparations and 82.7% (129/156) of all identified proteins were identified in at least two of the three preparations. A complete list (in alphabetical order) of identified proteins is presented in Table 1 while representative MS/MS spectra are presented in supplementary Fig. 1. Furthermore, complete data pertaining to individual proteins and peptides are listed in supplementary Table I.
Table 1. List of all identified proteins.
The table lists the accession number, the number of individual peptides matched, the percentage coverage of the identified protein sequence and the Mascot score of the protein. When the accession number matched to an unknown protein in the database, the identity of the protein was determined by searching, using the BLAST algorithm, the amino acid sequence of the identified peptides in the NCBI databases. The proteins were classified according whether they were previously identified as associated with endocrine secretory granules (ESG), or according to their most often reported localization (PubMed searches) into the following categories: SK: Cytoskeleton, NU: Nucleus, ER: Endoplasmic Reticulum, GO: Golgi Apparatus, EN: Endosome, MI: Mitochondria PR: Proteasome, CY: Cytosol, RI: Ribosome, PM: Plasma Membrane, RI: Ribosome, Variable (proteins having multiple major localization sites or for which not enough information is available). For ESG proteins, a study in which association with endocrine secretory granules was observed, is referenced. When present in ESG as shown by proteomics, it is labeled with a P and referenced.
| Name | Accession | Peptides | Cov% | Score | Localization | Study |
|---|---|---|---|---|---|---|
| 14-3-3 protein epsilon | gi|74189349 | 8 | 35 | 408 | ESG | [45] |
| 14-3-3 protein gamma | gi|3065929 | 7 | 25 | 351 | ESG | P [23] |
| 14-3-3 zeta | gi|1841387 | 11 | 44 | 576 | ESG | P [23] |
| 78 kDa glucose-regulated protein | gi|1304157 | 9 | 16 | 480 | ER | |
| Acrogranin | gi|50852 | 4 | 11 | 230 | Variable | |
| Actin, gamma | gi|74213524 | 21 | 64 | 1107 | ESG | P [23] |
| Actinin, alpha 1 | gi|32766260 | 2 | 2 | 129 | ESG | [46] |
| Adenomatous polyposis coli protein | gi|12643510 | 3 | 1 | 50 | SK | |
| Alpha-NAC | gi|1666692 | 4 | 2 | 252 | NU | |
| Asparagine synthetase B | gi|26345208 | 3 | 6 | 94 | Variable | |
| ATP synthase, beta subunit | gi|23272966 | 4 | 10 | 136 | ESG | P [23] |
| ATPase, Na/K transporting, alpha 1 polypeptide | gi|21595127 | 23 | 27 | 1115 | PM | |
| ATPase, Na/K transporting, beta subunit | gi|54130 | 4 | 18 | 156 | PM | |
| Calmodulin | gi|74143933 | 8 | 47 | 573 | ESG | [47] |
| Calreticulin | gi|26344403 | 3 | 9 | 107 | ER | |
| CAP, adenylate cyclase-associated protein 1 | gi|26343401 | 2 | 7 | 117 | SK | |
| Carnitine deficiency-associated gene 3B | gi|28461294 | 5 | 38 | 203 | Variable | |
| CAST1/ERC2 | gi|38231910 | 3 | 5 | 54 | ESG | [48] |
| Catenin (Ctnnd1) p120 | gi|26006157 | 3 | 4 | 152 | PM | |
| Cellular nucleic acid binding protein | gi|50471 | 2 | 16 | 85 | NU | |
| Chaperonin containing TCP-1 delta subunit | gi|460317 | 2 | 4 | 83 | ESG | [49] |
| Chaperonin containing TCP-1 theta subunit | gi|5295992 | 4 | 8 | 91 | ESG | [49] |
| Chaperonin containing TCP-1, beta subunit | gi|468546 | 3 | 8 | 97 | ESG | [49] |
| Chaperonin subunit 5 | gi|37359776 | 4 | 9 | 243 | Variable | |
| Chaperonin subunit 6a | gi|17391080 | 2 | 9 | 79 | Variable | |
| Charged multivesicular body protein (CHMP4b) | gi|74192743 | 4 | 22 | 117 | EN | |
| Chromogranin A | gi|20071660 | 26 | 38 | 1440 | ESG | P [23] |
| Chromogranin B | gi|50409 | 18 | 29 | 887 | ESG | P [22] |
| Clathrin, heavy polypeptide | gi|33438248 | 6 | 4 | 233 | ESG | P [22] |
| Coatomer protein complex, gamma subunit | gi|19354315 | 2 | 4 | 58 | ER, GO | |
| Cofilin 1 | gi|12861068 | 2 | 9 | 64 | SK | |
| Complexin 2 | gi|62740137 | 2 | 20 | 84 | ESG | [39] |
| Cp27 | gi|3115274 | 2 | 10 | 56 | PM | |
| Cu/Zn superoxide dismutase | gi|226471 | 8 | 50 | 489 | ESG | P [23] |
| Cyclophilin A (Peptidyl-prolyl cis-trans isomerase A) | gi|74146841 | 9 | 66 | 407 | ESG | P [23] |
| Cysteine-rich protein 1 | gi|74212172 | 4 | 71 | 127 | SK | |
| DEAD box polypeptide 3 | gi|18204785 | 3 | 5 | 147 | NU | |
| Desmoplakin | gi|82950147 | 5 | 2 | 111 | SK | |
| Diazepam binding inhibitor | gi|67511482 | 2 | 25 | 110 | ESG | [50] |
| Dynein, heavy chain 1 | gi|13384736 | 4 | 1 | 70 | ESG | [51] |
| Dynein, heavy chain 8 | gi|14335452 | 4 | 1 | 53 | ESG | [51] |
| Dynein, light chain roadblock-type 1 | gi|12842877 | 2 | 10 | 56 | SK | |
| EEF2 | gi|26324898 | 5 | 7 | 165 | NU | |
| EIF4a1 | gi|50820 | 4 | 13 | 146 | NU | |
| EIF4h | gi|39104480 | 4 | 23 | 121 | NU | |
| Elongation factor Tu | gi|26345590 | 17 | 40 | 819 | NU | |
| Enolase 1, alpha | gi|53734652 | 16 | 50 | 1030 | Variable | |
| Fatty acid synthase | gi|74142919 | 3 | 2 | 129 | PM | |
| Fatty acid-binding protein, epidermal | gi|74191310 | 6 | 40 | 246 | ESG | [52] |
| Fructose-bisphosphate aldolase A | gi|7548322 | 17 | 53 | 687 | ESG | P [23] |
| Gag-pro-pol polyprotein | gi|559059 | 8 | 17 | 245 | Variable | |
| Gasdermin 1 | gi|26325036 | 3 | 9 | 102 | Variable | |
| Glucose phosphate isomerase | gi|56554102 | 7 | 18 | 309 | ESG [53] | [53] |
| Glyceraldehyde-3-phosphate dehydrogenase | gi|62201487 | 14 | 49 | 725 | ESG | P [23] |
| Glycyl-tRNA synthetase | gi|18256043 | 2 | 2 | 55 | Variable | |
| GP42/Basigin protein | gi|631683 | 5 | 21 | 212 | PM | |
| Guanine nucleotide binding protein, alpha subunit, inhibitory | gi|309255 | 2 | 4 | 83 | ESG | [54] |
| Guanine nucleotide binding protein, alpha subunit, stimulating | gi|47271396 | 7 | 8 | 232 | ESG | P [22] |
| Guanine nucleotide binding protein, beta 2 subunit | gi|984551 | 7 | 23 | 232 | ESG | P [22],[23] |
| Heat shock protein hsp70 | gi|74220592 | 25 | 43 | 1181 | ESG | [55] |
| Heat-shock protein hsp84/90 | gi|74147026 | 29 | 39 | 1330 | Variable | |
| Heterogeneous nuclear ribonucleoprotein K | gi|12847547 | 2 | 4 | 52 | Variable | |
| HN1 | gi|74220270 | 2 | 27 | 125 | Variable | |
| HN1-like protein | gi|74213941 | 4 | 40 | 192 | Variable | |
| Importin beta 1 | gi|2829480 | 3 | 5 | 144 | Variable | |
| Integrin alpha 6 | gi|110577 | 3 | 19 | 58 | PM | |
| IQ motif containing GTPase activating protein 1 | gi|27370648 | 3 | 4 | 70 | SK | |
| Lactate dehydrogenase 1 | gi|74198692 | 13 | 45 | 636 | Variable | |
| L-type amino acid transporter 1 | gi|6906727 | 2 | 5 | 145 | PM | |
| M2-type pyruvate kinase | gi|74221210 | 23 | 54 | 1389 | Variable | |
| Macrophage migration inhibitory factor | gi|5822094 | 3 | 23 | 69 | ESG | [41] |
| MARCKS-like 1 | gi|13879553 | 2 | 14 | 65 | PM | |
| Microtubule-actin crosslinking factor 1 | gi|94372354 | 16 | 2 | 162 | SK | |
| Monocarboxylate transporter 4 | gi|7688756 | 2 | 5 | 54 | PM | |
| Myosin, heavy polypeptide | gi|20137006 | 19 | 12 | 705 | ESG | P [22] |
| Myosin, light polypeptide | gi|16924329 | 6 | 52 | 345 | ESG | [56] |
| Myosin, regulatory light chain 2 | gi|12851268 | 3 | 20 | 121 | ESG | [56] |
| Myosin-I, beta | gi|1924961 | 2 | 3 | 88 | SK | |
| Myristoylated alanine rich protein kinase C substrate (MARCKS) | gi|6678768 | 2 | 11 | 71 | PM | |
| Nebulin | gi|94366489 | 14 | 1 | 126 | SK | |
| Neural cell adhesion molecule 1 (NCAM) | gi|817984 | 10 | 41 | 480 | ESG | [57] |
| Nucleolin | gi|13529464 | 6 | 7 | 168 | NU | |
| Nucleosome assembly protein 1-like 1 | gi|50722 | 3 | 11 | 109 | NU | |
| Oxidation resistance protein 1 | gi|58047714 | 3 | 4 | 51 | PM | |
| P23 | gi|12843224 | 2 | 16 | 122 | Variable | |
| PDGFA associated protein | gi|74151229 | 5 | 32 | 153 | Variable | |
| Peptidylglycine alpha-amidating monooxygenase | gi|7305367 | 4 | 3 | 193 | ESG | P [23] |
| Peroxiredoxin 1 | gi|12846314 | 3 | 21 | 67 | Variable | |
| Phosphatidylethanolamine binding protein | gi|1517864 | 4 | 31 | 113 | ESG | [40] |
| Phosphofructokinase-1 C isozyme | gi|74224916 | 4 | 10 | 82 | PM | |
| Phosphoglycerate kinase 1 | gi|129903 | 7 | 23 | 438 | Variable | |
| Phosphoglycerate mutase 1 | gi|12844989 | 3 | 13 | 95 | Variable | |
| Plakoglobin | gi|423532 | 2 | 4 | 102 | SK | |
| Profilin 1 | gi|26389590 | 2 | 19 | 64 | SK | |
| Prolyl 4-hydroxylase | gi|54777 | 8 | 17 | 163 | ER | |
| Proopiomelanocortin (POMC) | gi|74227361 | 9 | 45 | 517 | ESG | [58] |
| Proprotein convertase PC1/3 | gi|7305371 | 2 | 2 | 79 | ESG | P [22] |
| Proteasome, 26S non-ATPase subunit 2 | gi|12861131 | 2 | 28 | 96 | PR | |
| Proteasome, delta subunit | gi|74191020 | 2 | 17 | 66 | PR | |
| Protein disulfide isomerase associated 3 | gi|6679687 | 8 | 17 | 247 | ESG | P [23] |
| Rab10 | gi|10435058 | 2 | 11 | 112 | ESG | P [23] |
| Rab14 | gi|10435483 | 2 | 8 | 70 | ESG | P [22],[23] |
| Rab1A | gi|74147521 | 2 | 13 | 109 | ESG | P [23] |
| Radixin | gi|74186081 | 6 | 8 | 106 | SK | |
| Rap1a | gi|53236951 | 3 | 15 | 67 | ESG | P [23] [59] |
| Regulator G-protein signaling 9 | gi|2739458 | 4 | 6 | 53 | PM | |
| Ribosomal protein L10A | gi|6755350 | 3 | 13 | 86 | RI | |
| Ribosomal protein L10E | gi|71051393 | 3 | 11 | 108 | RI | |
| Ribosomal protein L15 | gi|12846287 | 3 | 15 | 82 | RI | |
| Ribosomal protein L17 | gi|12832997 | 3 | 16 | 88 | RI | |
| Ribosomal protein L18 | gi|12840700 | 3 | 19 | 132 | RI | |
| Ribosomal protein L19 | gi|6677773 | 2 | 9 | 66 | RI | |
| Ribosomal protein L21E | gi|94368373 | 4 | 24 | 98 | RI | |
| Ribosomal protein L23 | gi|12849613 | 2 | 25 | 160 | RI | |
| Ribosomal protein L26 | gi|74179650 | 3 | 16 | 94 | RI | |
| Ribosomal protein L3 | gi|74198856 | 3 | 6 | 77 | RI | |
| Ribosomal protein L6 | gi|695638 | 6 | 20 | 133 | RI | |
| Ribosomal protein L8 | gi|74203516 | 3 | 10 | 77 | RI | |
| Ribosomal protein P2 | gi|74140891 | 5 | 70 | 290 | RI | |
| Ribosomal protein S10 | gi|74198792 | 2 | 10 | 72 | RI | |
| Ribosomal protein S16 | gi|7305445 | 3 | 20 | 54 | RI | |
| Ribosomal protein S18 | gi|198578 | 2 | 9 | 85 | RI | |
| Ribosomal protein S2 | gi|12835827 | 2 | 8 | 70 | RI | |
| Ribosomal protein S20 | gi|74181462 | 3 | 22 | 73 | RI | |
| Ribosomal protein S23 | gi|12846275 | 2 | 13 | 68 | RI | |
| ribosomal protein S28 | gi|12833257 | 3 | 46 | 140 | RI | |
| Ribosomal protein S3 | gi|12847921 | 2 | 11 | 57 | RI | |
| Ribosomal protein S4 | gi|12846200 | 4 | 15 | 128 | RI | |
| Ribosomal protein S8 | gi|26353710 | 3 | 17 | 138 | RI | |
| Secretogranin II | gi|54096 | 2 | 2 | 79 | ESG | P [23] |
| Secretogranin III | gi|74211039 | 3 | 11 | 147 | ESG | P [22],[23] |
| Serpin1 | gi|12836024 | 3 | 9 | 81 | ESG | [60] |
| Seryl-aminoacyl-tRNA synthetase 1 | gi|26349967 | 3 | 5 | 89 | MI | |
| Similar to PEST-containing nuclear protein (PCNP) | gi|74141568 | 2 | 21 | 94 | NU | |
| Similar to tumor protein, translationally-controlled 1 (TCTP) | gi|74181622 | 2 | 5 | 55 | PM | |
| Similar to ubiquitin C | gi|94375393 | 3 | 23 | 93 | ESG | P [22] |
| Solute carrier family 3 (CD98) | gi|26354873 | 12 | 28 | 528 | PM | |
| Spectrin, beta 2 | gi|7106421 | 2 | 1 | 54 | ESG | [61] |
| Stathmin | gi|12832714 | 13 | 63 | 463 | SK | |
| Sulfated glycoprotein (prosaposin) | gi|34328185 | 3 | 6 | 126 | ESG | P [23] |
| Synaptosomal-associated protein 25 (SNAP25) | gi|26346913 | 2 | 10 | 88 | ESG | [62] |
| Synuclein, beta | gi|18043841 | 2 | 30 | 103 | SK | |
| Thymosin, beta 10 | gi|74199914 | 2 | 31 | 91 | SK | |
| Titin isoform N2-A | gi|77812697 | 13 | 1 | 139 | SK | |
| Transferrin receptor 2 | gi|15559221 | 3 | 4 | 52 | PM, EN | |
| Triosephosphate isomerase | gi|54855 | 3 | 16 | 140 | Variable | |
| Tropomyosin | gi|74213492 | 22 | 57 | 959 | ESG | [63] |
| Tubulin, alpha 1a | gi|74202338 | 17 | 50 | 991 | ESG | P [22] |
| Tubulin, beta | gi|74141821 | 21 | 65 | 1381 | ESG | [64] |
| Tumor metastatic process-associated protein NM23 | gi|387496 | 2 | 17 | 57 | ER, CY | |
| Ubiquitin carboxy-terminal hydrolase L1 | gi|6755929 | 2 | 12 | 60 | PM | |
| Ubiquitin protein ligase | gi|94399459 | 8 | 2 | 85 | Variable | |
| Ubiquitin-activating enzyme E1 | gi|74228573 | 3 | 4 | 62 | Variable | |
| Vacuolar H+ATPase B2 | gi|74195936 | 3 | 5 | 53 | ESG | P [22] |
| VcpP97 (valosin) | gi|62738728 | 4 | 6 | 94 | ESG | P [22] |
| Y box transcription factor | gi|199821 | 9 | 36 | 491 | NU |
Many other important makers of the secretory granules were identified such as, for example, chromogranin B, POMC, secretogranin III and prohormone convertase PC1/3. A distribution of the identified proteins is presented in Fig. 5 with respect to their known, proposed or putative subcellular localizations. Overall, 27 proteins previously seen in the two above-mentioned studies [22, 23] were also identified herein. Interestingly, we also identified 27 other proteins known from the literature to be present in or associated with the secretory granules but which were never before reported in this type of organellar proteomics studies. Furthermore, 17 cytoskeleton and cytoskeleton-associated proteins involved in transport, docking, membrane fusion and actin/tubulin network remodeling present in our sample are shown to be associated for the first time with the endocrine or neuroendocrine secretory granules. We identified only 5 proteins reported as being ER and Golgi residents, the only other compartments made fluorescent by the transit of our mGFP construct. This highlights the efficiency of our purification approach not only to discriminate between fluorescent and non-fluorescent organelles, but also between fluorescent compartments. Also, only 5 proteins in our list are usually recognized to be restricted to endosomes, mitochondria, lysosomes, or proteasome. This result compares advantageously to those obtained using more conventional methodology [22, 23]. Interestingly, we also identified proteins whose expression and localization is deregulated during metastatic transformation and that are used as tumor markers since they are found in the blood of patients (i.e. M2-pyruvate kinase [35]). If indeed present in secretory granules, such misrouting and misregulation in endocrine and neuroendocrine tumors clearly warrants further investigations.
Figure 5. Pie chart illustrating the distribution of the identified proteins with respect to their known, proposed or putative subcellular localizations.
Pie slices correspond to Cytoskeleton (structure, trafficking, fusion); Granules (protein identified in the literature as being part of or associated with the secretory granules); Nucleus; Other (Endosome, Lysosome, Mitochondria, Proteosome); Plasma membrane; Ribosome; ER/Golgi; Undefined (protein present in more than one major location, or for which there is not enough information available).
Finally, relying on a gel-free approach leads to two important improvements. Firstly, we minimized sample loss frequently associated with gel staining and band extraction while allowing high quality MS data in terms of number and score of peptides from a relatively small sample as illustrated by representative MS/MS spectra (supplementary Fig. 1). Secondly, it contributed to the identification of small molecular weight proteins important to granule function such as complexin 2, phosphatidylethanolamine binding protein and macrophage migration inhibitory factor, which were never previously reported in such proteomic studies.
4. Discussion
Density-based separation techniques have been the tool of choice for decades in cell fractionation experiments. Being readily accessible, equilibrium gradient centrifugation has proven quite effective in enriching a sample for a specific subcellular compartment. However, this method is not without its problems. Indeed, it usually requires large amounts of starting biological material, either from fresh tissues (several bovine adrenal glands [22]) or from large scale cell cultures (8×108 cells [23]) already limiting the experimenter to the study of models for which enough material is available. Also, the procedure is very resource and time-consuming, typically requiring between 48 to 72 hours of centrifugation and electrophoresis. Not only does this delay the time at which mass spectrometry data can be obtained, but it also generates concerns on how complete and accurate is the representation from the proteins extracted from the final gel of the in-vivo state of the organelle.
In order to circumvent this problem, we propose an alternative approach to perform organellar proteomics. Hence, we first used cytoskeleton disruptors in combination with a freeze-thaw cycle to induce cell lysis and ultimately yield an enriched granule preparation. Then, FAOS performed with a cell sorter was efficient in sorting out an abundant, intact and homogeneous population of dense-core secretory granules within one hour. The choice of the appropriate sorting window was rapidly established using readily available calibration beads (controls for size and fluorescence) and known properties of that organelle. What is remarkable is the fact that even if some PHM-mGFP fluorescence is localized throughout the regulated secretory pathway (Fig. 1), we were nevertheless able to isolate a much enriched population of mature secretory granules. It is noteworthy that sorting a very fragile and hypoosmosis-sensitive organelle using a single fluorescent label transiting through multiple subcellular compartments was a demanding task. This method could benefit from using recent multiple simultaneous laser wavelengths cell sorters and/or by combining constructs, fluorescent dyes and/or antibodies since cell sorters do not discriminate the source of the fluorescence and can be employed to sort particles as long as their sizes and fluorescence intensities fall within the detection limits of the instrument. Provided that a specific antibody for a surface protein of the organelle of interest is available, the presented sorting procedure is conceptually identical to that employed for the sorting of cells bearing a specific surface marker, which is what FACS is routinely employed for. FACS identification and/or sorting of several structures presenting a size similar to the secretory granules (subcellular structures, bacteria) using some of the above-mentioned alternate labeling approaches were reported. [18, 36–38]). Doing so, the user can virtually sort and actively exclude any event in a defined window which, in turn, leads to much increased purity of the sorted material. It would be even possible to fractionate an organelle population into specific groups depending on their state at the time of cell lysis as determined, for example, by the presence or absence of a marker. Herein, we optimized the material to be sorted out by selecting stable transfectant but we consider that our method could be of use with transient transfectant as well as with any commercially available organelle-specific dyes or conjugated antibodies.
Even in its simplest form, this method has allowed us to obtain a purified sample very rich in dense-core granules from which more than 150 proteins were identified by mass spectrometry. Many important markers (Table 1) have been identified by numerous high scoring peptides, and some important small molecular weight proteins were identified for the first time in a large scale proteomics study of endocrine granules. Indeed, in the above cited examples, complexin 2 has been suggested to play a role in the exocytosis process [39], phosphatidylethanolamine binding protein (PEBP, also known as Raf-1 kinase inhibitor protein) has been localized to the membrane as well as the intragranular matrix of the chromaffin granules [40] and finally the macrophage migration inhibition factor (MIF), though highly recognized as a soluble lymphokine produced by activated T cells, has been localized to corticotropes and thyrotrope cells secretory granules [41]. Obviously, one ought to expect that the isolated granules contain the appropriate peptides in good amounts. In all studies including ours, this has been the case as can be seen with proANF for atrial granules [19], insulin(s) in β-cell granules [23], neuropeptide Y, enkephalins and adrenomedullin in chromaffin granules [22], various zymogens in pancreatic zymogen granules [20, 21] and corticotropin and proopiomelanocortin in corticotropes (this study). Associated closely with these processing products, all these studies also revealed the presence of the obligatory processing machinery including appropriate convertases, mostly PC1/3 ([22], this study) and PC2 [22, 23], carboxypeptidase-E [19, 22, 23] and PAM ([19, 22, 23]and this study). It is noteworthy, however, that these enzymes do not appear to be routinely observed as, for example, we were not able to detect CP-E nor was the study on chromaffin granules [22] able to detect PAM. Similarly, various recognized granules proteins such as chromogranin A ([22, 23], this study) and B ([19], this study), a variable mixture of secretogranins ([22, 23], and this study), 7B2 [22, 23], proSAAS [22, 23] or granule membrane proteins such as cytB561 [22] and V-ATPase ([19–23], and this study) were reported. All the granules preparations contained characteristic but distinct set of Rab proteins and VAMPs though none of the latter was seen in the present study nor was there any VAMP4 detected in any study. Interestingly, VAMP4 together with a great many number of SNARE and Rab proteins were shown to be abundant in synaptosomes [24, 25]. Furthermore, proteins reported to be associated with secretory granules were observed; these include kalirin [19], myosins ([20] and this study), tubulin and actin ([21] and this study). Actually, prior to our inclusion of actin/tubulin network disruptors, we observed a much larger distribution of fluorescent events likely due to aggregation and/or tethering of granules to actin/tubulin as previously reported [42]. Hence, our observation is much in favor with their close association with the granules as envisioned. Moreover, it is certainly worth noting that each study possesses its own set of unique proteins whose presence and functional role remain to be explored and established. For example, the protein 14-3-3ζ was recently shown to be closely associated with exocytic granules of the human platelet [43]. Also, the CuZn superoxide dismutase was recently reported to be secreted in a calcium and SNARE-dependant manner from somatolactotropes [44]. Other identified proteins of interest include the Na/K-ATPase, MARCKS, various chaperonins, secretory signal-peptide-devoid proteins such as cyclophilin A, serpin-1 and stathmin. The only major unforeseen proteins originated from ribosomes, despite the apparent depletion of these structures based upon RPL8 immunoblotting. It is possible that some PHM-mGFP bound to polyribosomes could have been co-sorted during FAOS. Although ribosomal proteins are present in our list, their significance must also be kept in perspective. Effectively, as a group, they are identified by an average of 3 peptides and a score of 109, which is well below chromogranin A with its 26 peptides for a score of 1440.
In conclusion, we present here a very rapid, simple and adaptable method to purify organelles and obtain their proteome. Requiring relatively low amounts of starting materials, the protocol can be performed within a day. In this presented form, the method can be seen as a complement and/or can be used concomitantly with the conventional protocols for subcellular fractionation. A combination of gel-based and gel-free procedures could certainly maximize the information collected in those studies. Indeed, the former could maximize, through the use of stringent extraction conditions, the recovery of large and poorly soluble proteins such as membrane proteins since clearly, in this study, simple inclusion of low amount of SDS did not permit this. Conversely, the latter could provide information on most soluble proteins in a much more rapid and simple manner. Furthermore, the availability of hundreds of fluorescent molecules, including dyes and antibodies, combined with the speed, power and versatility of flow cytometry could rapidly contribute to overcoming some of the challenges related to sample preparation in organellar proteomics.
Supplementary Material
Acknowledgements
We thank Dany Gauthier for technical expertise and Nadia Rabah for help in cell culture, Annie. Vallée for processing the samples for electron microscopy and Denis Faubert for LC-MS/MS analyses (IRCM). Special thanks to Eric Massicotte and Martine Dupuis for their precious help in optimizing and running the flow cytometry experiments in the IRCM Cytometry Unit. This work was supported by a Canadian Institutes of Health Research operating grant to CL (MT-74479) and by National Institutes of Health grant to REM (DK-32948). D.J. Gauthier is a recipient of a Fonds de la recherche en santé du Québec (FRSQ) studentship, and J.A. Sobota holds a National Research Service Award (DE-017094).
ABBREVIATIONS
- ACTH
corticotropin (adrenocorticotropic hormone)
- BLAST
Basic Local Alignment and Search Tool
- CP-E
carboxypeptidase E or H
- ER
endoplasmic reticulum
- ERp
endoplasmic reticulum protein
- FAOS
fluorescence assisted organelle sorting
- GFP
Aequorea victoria green fluorescent protein
- mGFP
Aequorea victoria monomeric green fluorescent protein
- HRP
horseradish peroxidase
- LTQ
linear quadrupole ion trap
- PAM
peptidyl α-amidating monooxygenase
- PC
proprotein convertase
- PHM
peptidylglycine α-hydroxylating monooxygenase
- PIST
PDZ domain protein interacting specifically with TC10
- POMC
proopiomelanocortin
- Rab
Ras-related in brain
- SNARE
soluble N-ethylmaleimide-sensitive factor (NSF) attachment receptor
- SOFA
single organelle fluorescence analysis
- SSC
side scatter
- TGN
trans-Golgi network
- VAMP
vesicle-associated membrane protein
References
- 1.Ho E, Hayen A, Wilkins MR. Characterisation of organellar proteomes: a guide to subcellular proteomic fractionation and analysis. Proteomics. 2006;6:5746–5757. doi: 10.1002/pmic.200600241. [DOI] [PubMed] [Google Scholar]
- 2.Yates JR, III, Gilchrist A, Howell KE, Bergeron JJ. Proteomics of organelles and large cellular structures. Nat. Rev. Mol. Cell Biol. 2005;6:702–714. doi: 10.1038/nrm1711. [DOI] [PubMed] [Google Scholar]
- 3.Schirmer EC, Florens L, Guan T, Yates JR, III, Gerace L. Nuclear membrane proteins with potential disease links found by subtractive proteomics. Science. 2003;301:1380–1382. doi: 10.1126/science.1088176. [DOI] [PubMed] [Google Scholar]
- 4.Andersen JS, Lam YW, Leung AK, Ong SE, et al. Nucleolar proteome dynamics. Nature. 2005;433:77–83. doi: 10.1038/nature03207. [DOI] [PubMed] [Google Scholar]
- 5.Gilchrist A, Au CE, Hiding J, Bell AW, et al. Quantitative proteomics analysis of the secretory pathway. Cell. 2006;127:1265–1281. doi: 10.1016/j.cell.2006.10.036. [DOI] [PubMed] [Google Scholar]
- 6.Takatalo MS, Kouvonen P, Corthals G, Nyman TA, Ronnholm RH. Identification of new Golgi complex specific proteins by direct organelle proteomic analysis. Proteomics. 2006;6:3502–3508. doi: 10.1002/pmic.200500516. [DOI] [PubMed] [Google Scholar]
- 7.Casey TM, Meade JL, Hewitt EW. Organelle proteomics: identification of the exocytic machinery associated with the natural killer cell secretory lysosome. Mol. Cell. Proteomics. 2007;6:767–780. doi: 10.1074/mcp.M600365-MCP200. [DOI] [PubMed] [Google Scholar]
- 8.Stuart LM, Boulais J, Charriere GM, Hennessy EJ, et al. A systems biology analysis of the Drosophila phagosome. Nature. 2007;445:95–101. doi: 10.1038/nature05380. [DOI] [PubMed] [Google Scholar]
- 9.Borner GH, Harbour M, Hester S, Lilley KS, Robinson MS. Comparative proteomics of clathrin-coated vesicles. J. Cell Biol. 2006;175:571–578. doi: 10.1083/jcb.200607164. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Prokisch H, Scharfe C, Camp DG, Xiao W, et al. Integrative analysis of the mitochondrial proteome in yeast. PLoS. Biol. 2004;2:795–804. doi: 10.1371/journal.pbio.0020160. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Huber LA, Pfaller K, Vietor I. Organelle proteomics: implications for subcellular fractionation in proteomics. Circ. Res. 2003;92:962–968. doi: 10.1161/01.RES.0000071748.48338.25. [DOI] [PubMed] [Google Scholar]
- 12.Ibrahim SF, Van Den EG. Flow cytometry and cell sorting. Adv. Biochem. Eng. Biotechnol. 2007;106:19–39. doi: 10.1007/10_2007_073. [DOI] [PubMed] [Google Scholar]
- 13.Givan AL. Flow cytometry: an introduction. Methods Mol. Biol. 2004;263:1–32. doi: 10.1385/1-59259-773-4:001. [DOI] [PubMed] [Google Scholar]
- 14.Murphy RF. Analysis and isolation of endocytic vesicles by flow cytometry and sorting: demonstration of three kinetically distinct compartments involved in fluid-phase endocytosis. Proc. Natl. Acad. Sci. U.S.A. 1985;82:8523–8526. doi: 10.1073/pnas.82.24.8523. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Wilson RB, Murphy RF. Flow-cytometric analysis of endocytic compartments. Methods Cell Biol. 1989;31:293–317. doi: 10.1016/s0091-679x(08)61616-7. [DOI] [PubMed] [Google Scholar]
- 16.Cossarizza A, Ceccarelli D, Masini A. Functional heterogeneity of an isolated mitochondrial population revealed by cytofluorometric analysis at the single organelle level. Exp. Cell Res. 1996;222:84–94. doi: 10.1006/excr.1996.0011. [DOI] [PubMed] [Google Scholar]
- 17.Dhandayuthapani S, Via LE, Thomas CA, Horowitz PM, et al. Green fluorescent protein as a marker for gene expression and cell biology of mycobacterial interactions with macrophages. Mol. Microbiol. 1995;17:901–912. doi: 10.1111/j.1365-2958.1995.mmi_17050901.x. [DOI] [PubMed] [Google Scholar]
- 18.Bock G, Steinlein P, Huber LA. Cell biologists sort things out: Analysis and purification of intracellular organelles by flow cytometry. Trends Cell Biol. 1997;7:499–503. doi: 10.1016/S0962-8924(97)01160-4. [DOI] [PubMed] [Google Scholar]
- 19.Muth E, Driscoll WJ, Smalstig A, Goping G, Mueller GP. Proteomic analysis of rat atrial secretory granules: a platform for testable hypotheses. Biochim. Biophys. Acta. 2004;1699:263–275. doi: 10.1016/j.bbapap.2004.03.007. [DOI] [PubMed] [Google Scholar]
- 20.Chen X, Walker AK, Strahler JR, Simon ES, et al. Organellar proteomics: analysis of pancreatic zymogen granule membranes. Mol. Cell. Proteomics. 2006;5:306–312. doi: 10.1074/mcp.M500172-MCP200. [DOI] [PubMed] [Google Scholar]
- 21.Rindler MJ, Xu CF, Gumper I, Smith NN, Neubert TA. Proteomic analysis of pancreatic zymogen granules: identification of new granule proteins. J. Proteome. Res. 2007;6:2978–2992. doi: 10.1021/pr0607029. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Wegrzyn J, Lee J, Neveu JM, Lane WS, Hook V. Proteomics of neuroendocrine secretory vesicles reveal distinct functional systems for biosynthesis and exocytosis of peptide hormones and neurotransmitters. J. Proteome. Res. 2007;6:1652–1665. doi: 10.1021/pr060503p. [DOI] [PubMed] [Google Scholar]
- 23.Brunner Y, Coute Y, Iezzi M, Foti M, et al. Proteomics analysis of insulin secretory granules. Mol. Cell. Proteomics. 2007;6:1007–1017. doi: 10.1074/mcp.M600443-MCP200. [DOI] [PubMed] [Google Scholar]
- 24.Burre J, Volknandt W. The synaptic vesicle proteome. J. Neurochem. 2007;101:1448–1462. doi: 10.1111/j.1471-4159.2007.04453.x. [DOI] [PubMed] [Google Scholar]
- 25.Takamori S, Holt M, Stenius K, Lemke EA, et al. Molecular anatomy of a trafficking organelle. Cell. 2006;127:831–846. doi: 10.1016/j.cell.2006.10.030. [DOI] [PubMed] [Google Scholar]
- 26.Sobota JA, Ferraro F, Back N, Eipper BA, Mains RE. Not all secretory granules are created equal: Partitioning of soluble content proteins. Mol. Biol. Cell. 2006;17:5038–5052. doi: 10.1091/mbc.E06-07-0626. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Perkins DN, Pappin DJ, Creasy DM, Cottrell JS. Probability-based protein identification by searching sequence databases using mass spectrometry data. Electrophoresis. 1999;20:3551–3567. doi: 10.1002/(SICI)1522-2683(19991201)20:18<3551::AID-ELPS3551>3.0.CO;2-2. [DOI] [PubMed] [Google Scholar]
- 28.Altschul SF, Gish W, Miller W, Myers EW, Lipman DJ. Basic local alignment search tool. J. Mol. Biol. 1990;215:403–410. doi: 10.1016/S0022-2836(05)80360-2. [DOI] [PubMed] [Google Scholar]
- 29.Gumbiner B, Kelly RB. Secretory granules of an anterior pituitary cell line, AtT-20, contain only mature forms of corticotropin and beta-lipotropin. Proc. Natl. Acad. Sci. U.S.A. 1981;78:318–322. doi: 10.1073/pnas.78.1.318. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.El Meskini R, Jin L, Marx R, Bruzzaniti A, et al. A signal sequence is sufficient for green fluorescent protein to be routed to regulated secretory granules. Endocrinology. 2001;142:864–873. doi: 10.1210/endo.142.2.7929. [DOI] [PubMed] [Google Scholar]
- 31.Zacharias DA, Violin JD, Newton AC, Tsien RY. Partitioning of lipid-modified monomeric GFPs into membrane microdomains of live cells. Science. 2002;296:913–916. doi: 10.1126/science.1068539. [DOI] [PubMed] [Google Scholar]
- 32.de Oliveira CA, Mantovani B. Latrunculin A is a potent inhibitor of phagocytosis by macrophages. Life Sci. 1988;43:1825–1830. doi: 10.1016/0024-3205(88)90282-2. [DOI] [PubMed] [Google Scholar]
- 33.Vasquez RJ, Howell B, Yvon AM, Wadsworth P, Cassimeris L. Nanomolar concentrations of nocodazole alter microtubule dynamic instability in vivo and in vitro. Mol. Biol. Cell. 1997;8:973–985. doi: 10.1091/mbc.8.6.973. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Hendy GN, Bevan S, Mattei MG, Mouland AJ. Chromogranin A. Clin. Invest. Med. 1995;18:47–65. [PubMed] [Google Scholar]
- 35.Oremek GM, Rox S, Mitrou P, Sapoutzis N, Sauer-Eppel H. Tumor M2-PK levels in haematological malignancies. Anticancer Res. 2003;23:1135–1138. [PubMed] [Google Scholar]
- 36.Ashley N, Harris D, Poulton J. Detection of mitochondrial DNA depletion in living human cells using PicoGreen staining. Exp. Cell Res. 2005;303:432–446. doi: 10.1016/j.yexcr.2004.10.013. [DOI] [PubMed] [Google Scholar]
- 37.Koga K, Matsumoto K, Akiyoshi T, Kubo M, et al. Purification, characterization and biological significance of tumor-derived exosomes. Anticancer Res. 2005;25:3703–3707. [PubMed] [Google Scholar]
- 38.Moerch U, Nielsen HS, Lundsgaard D, Oleksiewicz MB. Flow sorting from organ material by intracellular markers. Cytometry A. 2007;71:495–500. doi: 10.1002/cyto.a.20418. [DOI] [PubMed] [Google Scholar]
- 39.Bajohrs M, Darios F, Peak-Chew SY, Davletov B. Promiscuous interaction of SNAP-25 with all plasma membrane syntaxins in a neuroendocrine cell. Biochem. J. 2005;392:283–289. doi: 10.1042/BJ20050583. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Goumon Y, Muller A, Glattard E, Marban C, et al. Identification of morphine-6-glucuronide in chromaffin cell secretory granules. J. Biol. Chem. 2006;281:8082–8089. doi: 10.1074/jbc.M502298200. [DOI] [PubMed] [Google Scholar]
- 41.Nishino T, Bernhagen J, Shiiki H, Calandra T, et al. Localization of macrophage migration inhibitory factor (MIF) to secretory granules within the corticotrophic and thyrotrophic cells of the pituitary gland. Mol. Med. 1995;1:781–788. [PMC free article] [PubMed] [Google Scholar]
- 42.Abu-Hamdah R, Cho WJ, Horber JK, Jena BP. Secretory vesicles in live cells are not free-floating but tethered to filamentous structures: a study using photonic force microscopy. Ultramicroscopy. 2006;106:670–673. doi: 10.1016/j.ultramic.2006.01.013. [DOI] [PubMed] [Google Scholar]
- 43.Hernandez-Ruiz L, Valverde F, Jimenez-Nunez MD, Ocana E, et al. Organellar Proteomics of Human Platelet Dense Granules Reveals That 14-3-3zeta Is a Granule Protein Related to Atherosclerosis. J. Proteome. Res. 2007;6:4449–4457. doi: 10.1021/pr070380o. [DOI] [PubMed] [Google Scholar]
- 44.Santillo MF, Secondo AF, Seru RF, Damiano S, et al. Evidence of calcium- and SNARE-dependent release of CuZn superoxide dismutase from rat pituitary GH3 cells and synaptosomes in response to depolarization. J. Neurochem. 2007;102:679–685. doi: 10.1111/j.1471-4159.2007.04538.x. [DOI] [PubMed] [Google Scholar]
- 45.Roth D, Morgan A, Martin H, Jones D, et al. Characterization of 14-3-3 proteins in adrenal chromaffin cells and demonstration of isoform-specific phospholipid binding. Biochem. J. 1994;301:305–310. doi: 10.1042/bj3010305. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Baldini G, Martelli AM, Tabellini G, Horn C, et al. Rabphilin localizes with the cell actin cytoskeleton and stimulates association of granules with F-actin cross-linked by {alpha}-actinin. J. Biol. Chem. 2005;280:34974–34984. doi: 10.1074/jbc.M502695200. [DOI] [PubMed] [Google Scholar]
- 47.Taylor KA. Regulation and recycling of myosin V. Curr. Opin. Cell Biol. 2007;19:67–74. doi: 10.1016/j.ceb.2006.12.014. [DOI] [PubMed] [Google Scholar]
- 48.Inoue E, Deguchi-Tawarada M, Takao-Rikitsu E, Inoue M, et al. ELKS, a protein structurally related to the active zone protein CAST, is involved in Ca2+dependent exocytosis from PC12 cells. Genes Cells. 2006;11:659–672. doi: 10.1111/j.1365-2443.2006.00970.x. [DOI] [PubMed] [Google Scholar]
- 49.Creutz CE, Liou A, Snyder SL, Brownawell A, Willison K. Identification of the major chromaffin granule-binding protein, chromobindin A, as the cytosolic chaperonin CCT (chaperonin containing TCP-1) J. Biol. Chem. 1994;269:32035–32038. [PubMed] [Google Scholar]
- 50.Metz-Boutigue MH, Goumon Y, Lugardon K, Strub JM, Aunis D. Antibacterial peptides are present in chromaffin cell secretory granules. Cell Mol. Neurobiol. 1998;18:249–266. doi: 10.1023/A:1022573004910. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Kraemer J, Schmitz F, Drenckhahn D. Cytoplasmic dynein and dynactin as likely candidates for microtubule-dependent apical targeting of pancreatic zymogen granules. Eur. J. Cell Biol. 1999;78:265–277. doi: 10.1016/S0171-9335(99)80060-0. [DOI] [PubMed] [Google Scholar]
- 52.Yanase H, Shimizu H, Kanda T, Fujii H, Iwanaga T. Cellular localization of the diazepam binding inhibitor (DBI) in the gastrointestinal tract of mice and its coexistence with the fatty acid binding protein (FABP) Arch. Histol. Cytol. 2001;64:449–460. doi: 10.1679/aohc.64.449. [DOI] [PubMed] [Google Scholar]
- 53.Niinaka Y, Paku S, Haga A, Watanabe H, Raz A. Expression and secretion of neuroleukin/phosphohexose isomerase/maturation factor as autocrine motility factor by tumor cells. Cancer Res. 1998;58:2667–2674. [PubMed] [Google Scholar]
- 54.Sattar AA, Boinpally R, Stromer MH, Jena BP. G(alpha)(i3) in pancreatic zymogen granules participates in vesicular fusion. J. Biochem. 2002;131:815–820. doi: 10.1093/oxfordjournals.jbchem.a003170. [DOI] [PubMed] [Google Scholar]
- 55.Evdonin AL, Martynova MG, Bystrova OA, Guzhova IV, et al. The release of Hsp70 from A431 carcinoma cells is mediated by secretory-like granules. Eur. J. Cell Biol. 2006;85:443–455. doi: 10.1016/j.ejcb.2006.02.008. [DOI] [PubMed] [Google Scholar]
- 56.Neco P, Giner D, Viniegra S, Borges R, et al. New roles of myosin II during vesicle transport and fusion in chromaffin cells. J. Biol. Chem. 2004;279:27450–27457. doi: 10.1074/jbc.M311462200. [DOI] [PubMed] [Google Scholar]
- 57.Chan SA, Polo-Parada L, Landmesser LT, Smith C. Adrenal chromaffin cells exhibit impaired granule trafficking in NCAM knockout mice. J. Neurophysiol. 2005;94:1037–1047. doi: 10.1152/jn.01213.2004. [DOI] [PubMed] [Google Scholar]
- 58.Loh YP, Kim T, Rodriguez YM, Cawley NX. Secretory granule biogenesis and neuropeptide sorting to the regulated secretory pathway in neuroendocrine cells. J. Mol. Neurosci. 2004;22:63–71. doi: 10.1385/JMN:22:1-2:63. [DOI] [PubMed] [Google Scholar]
- 59.Fukai S, Matern HT, Jagath JR, Scheller RH, Brunger AT. Structural basis of the interaction between RalA and Sec5, a subunit of the sec6/8 complex. EMBO J. 2003;22:3267–3278. doi: 10.1093/emboj/cdg329. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Hook V, Metz-Boutigue MH. Protein trafficking to chromaffin granules and proteolytic processing within regulated secretory vesicles of neuroendocrine chromaffin cells. Ann. N.Y. Acad. Sci. 2002;971:397–405. doi: 10.1111/j.1749-6632.2002.tb04502.x. [DOI] [PubMed] [Google Scholar]
- 61.Perrin D, Aunis D. Reorganization of alpha-fodrin induced by stimulation in secretory cells. Nature. 1985;315:589–592. doi: 10.1038/315589a0. [DOI] [PubMed] [Google Scholar]
- 62.Chieregatti E, Witkin JW, Baldini G. SNAP-25 and synaptotagmin 1 function in Ca2+dependent reversible docking of granules to the plasma membrane. Traffic. 2002;3:496–511. doi: 10.1034/j.1600-0854.2002.30706.x. [DOI] [PubMed] [Google Scholar]
- 63.Lin JJ, Warren KS, Wamboldt DD, Wang T, Lin JL. Tropomyosin isoforms in nonmuscle cells. Int. Rev. Cytol. 1997;170:1–38. doi: 10.1016/s0074-7696(08)61619-8. [DOI] [PubMed] [Google Scholar]
- 64.Baltrusch S, Lenzen S. Novel insights into the regulation of the bound and diffusible glucokinase in MIN6 beta-cells. Diabetes. 2007;56:1305–1315. doi: 10.2337/db06-0894. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.