Abstract
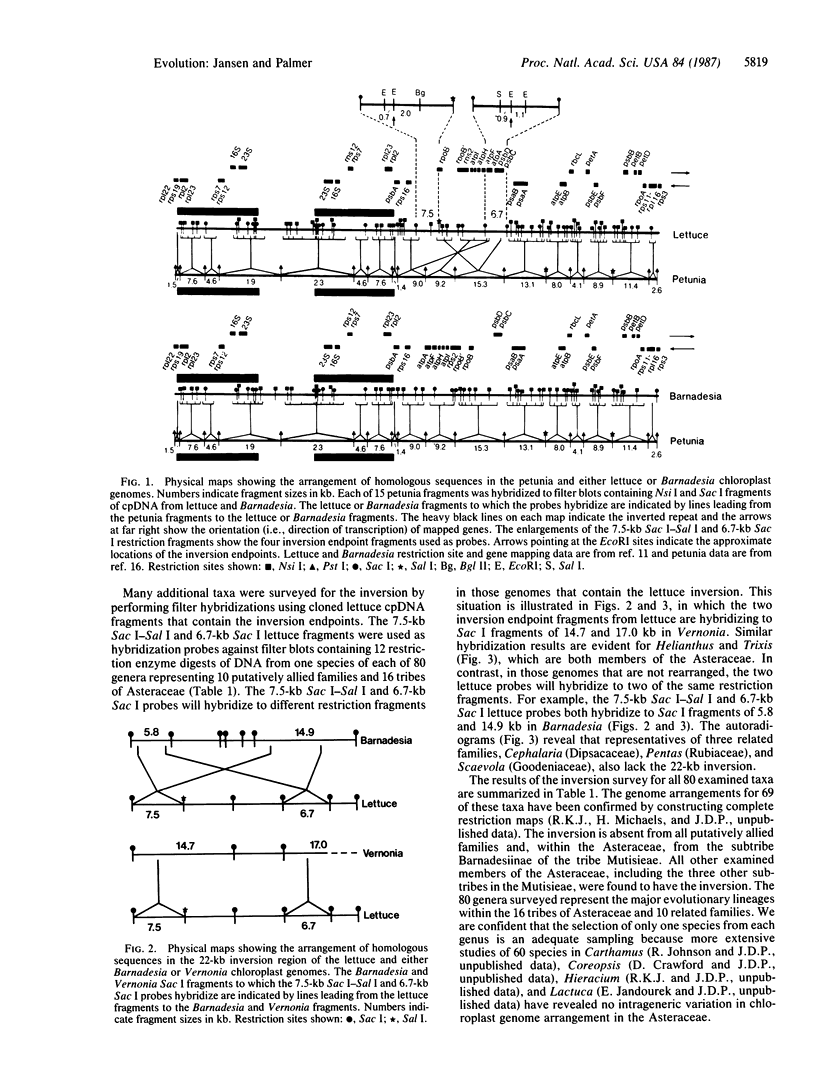
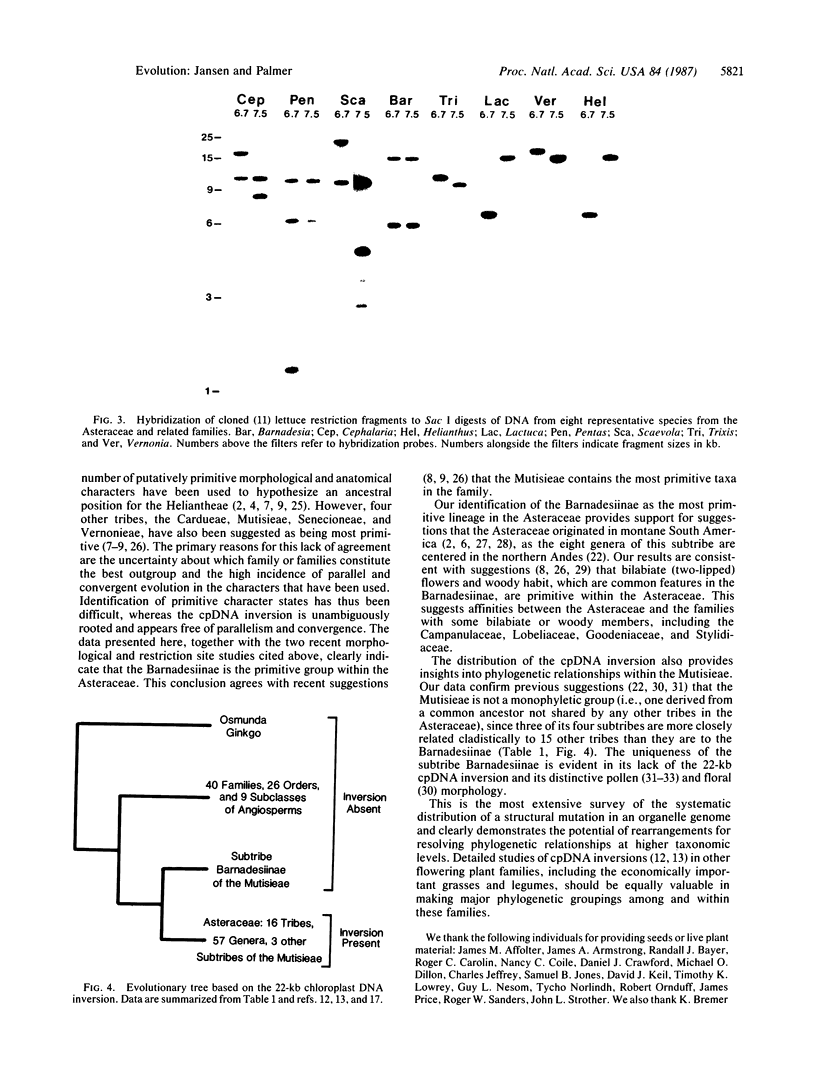
We determined the distribution of a chloroplast DNA inversion among 80 species representing 16 tribes of the Asteraceae and 10 putatively related families. Filter hybridizations using cloned chloroplast DNA restriction fragments of lettuce and petunia revealed that this 22-kilobase-pair inversion is shared by 57 genera, representing all tribes of the Asteraceae, but is absent from the subtribe Barnadesiinae of the tribe Mutisieae, as well as from all families allied to the Asteraceae. The inversion thus defines an ancient evolutionary split within the family and suggests that the Barnadesiinae represents the most primitive lineage in the Asteraceae. These results also indicate that the tribe Mutisieae is not monophyletic, since any common ancestor to its four subtribes is also shared by other tribes in the family. This is the most extensive survey of the systematic distribution of an organelle DNA rearrangement and demonstrates the potential of such mutations for resolving phylogenetic relationships at higher taxonomic levels.
Keywords: angiosperm evolution, molecular systematics, Mutisieae, Barnadesiinae
Full text
PDF


Images in this article
Selected References
These references are in PubMed. This may not be the complete list of references from this article.
- Fluhr R., Edelman M. Conservation of sequence arrangement among higher plant chloroplast DNAs: molecular cross hybridization among the Solanaceae and between Nicotiana and Spinacia. Nucleic Acids Res. 1981 Dec 21;9(24):6841–6853. doi: 10.1093/nar/9.24.6841. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Palmer J. D. Comparative organization of chloroplast genomes. Annu Rev Genet. 1985;19:325–354. doi: 10.1146/annurev.ge.19.120185.001545. [DOI] [PubMed] [Google Scholar]
- Palmer J. D., Thompson W. F. Chloroplast DNA rearrangements are more frequent when a large inverted repeat sequence is lost. Cell. 1982 Jun;29(2):537–550. doi: 10.1016/0092-8674(82)90170-2. [DOI] [PubMed] [Google Scholar]
- Saghai-Maroof M. A., Soliman K. M., Jorgensen R. A., Allard R. W. Ribosomal DNA spacer-length polymorphisms in barley: mendelian inheritance, chromosomal location, and population dynamics. Proc Natl Acad Sci U S A. 1984 Dec;81(24):8014–8018. doi: 10.1073/pnas.81.24.8014. [DOI] [PMC free article] [PubMed] [Google Scholar]