Abstract
Mating is fundamental to most organisms, although the physiological and transcriptional changes associated with this process have been largely characterized only in Drosophila. In this study, we use honey bees as a model system since their queens undergo massive and permanent physiological and behavioral changes following mating. Previous studies have identified changes associated with the transition from a virgin queen to a fully-mated, egg-laying queen. Here, we further uncouple the mating process to examine the effects of natural mating vs. instrumental insemination and saline vs. semen insemination. We observed effects on flight behavior, vitellogenin expression, and significant overlap in transcriptional profiles between our study and analogous studies in Drosophila, suggesting that some post-mating mechanisms are conserved across insect orders.
Introduction
Mating is fundamental to the success and reproduction of any sexual species. In animals ranging from invertebrates to mammals, this process results in massive behavioral and physiological changes in females. Among insects, these changes have been studied in most detail in Drosophila melanogaster, and include production of mature eggs, a rise in oviposition rates, increased feeding, a reduction in receptivity to additional mating (McGraw et al. 2008; 2004; Wolfner 1997; 2002), and activation of the immune system (Lawniczak et al. 2007). Mating may occur only once or several times throughout the course of a female’s life, and as such, the changes induced by mating may be some of the most dramatic and plastic physiological processes observed in animals. Several studies have been conducted in Drosophila to identify the transcriptional and physiological changes associated with the mating process in female flies (Lawniczak and Begun 2004; Mack et al. 2006; McGraw et al. 2008; 2004). However, in order to truly understand how general these changes might be, it is important to conduct similar studies in a variety of organisms across multiple insect orders. Here, we use the honey bee (Apis mellifera) as a model system to study post-mating changes in females.
A honey bee queen mates during a brief period early in her lifetime, and following this process she undergoes massive and permanent physiological and behavioral changes(Haydak 1949; Keeling et al. 2003; Plettner et al. 1997; Tanaka and Hartfelder 2004; Tarpy and Page 2000). A young virgin queen emerges into a colony of sterile workers and usually initiates several orientation flights prior to her first mating flight, which occurs when she is approximately one week old (Winston 1991). Throughout the process, the queen will mate with a large number of malesor ‘drones’ (12 on average; Tarpy et al. 2004). Once mating is completed, the queens’ ovaries become fully activated and she initiates egg-laying, at a rate upwards of 1500 eggs per day. The queen will store the sperm in her spermathecae for the remainder of her life (on average 1–3 years), and will only fly again if the colony swarms. Her pheromone profiles also change, signaling her reproductive state to the workers (Keeling et al. 2003; Kocher et al. 2009; Plettner et al. 1993).
Previous research has demonstrated that there are large-scale transcriptional changes in the brain and ovaries that are associated with the mating process in honey bee queens (Kocher et al. 2008). It appears that physiological changes in the ovaries and pheromone production in the mandibular glands may be initiated immediately as the mating process is initiated, but that changes in flight and egg-laying behavior seem to require the completion of the entire mating processor operate on a slower timescale. Consistent with these physiological and behavioral changes, transcriptional changes in the brains and the ovaries appear to be uncoupledas well.
There are still several important questions related to honey bee reproductive biology that remain to be elucidated. First, queens mate with several males before they stop taking mating flights and initiate egg-laying(Roberts 1944; Schluns 2005; Tarpy and Page 2001). However, we still do not fully understand what cues a queen uses to determine that she has successfully completed the mating process. In Drosophila females, seminal proteins trigger specific behavioral and physiological changes (Wolfner 2002). In some polyandrous butterflies and in Dipetalogaster maximus, insemination volume appears to trigger stretch receptors in the abdomens of the females that influence post-mating behavioral changes (Nijhout 1984; Sugawara 1979). In honey bees, both sperm/seminal fluid and the volume transferred are critical for causing post-mating changes in mating-flight behavior (Tarpy 2000) and pheromone production (Richard et. al. unpublished data). Furthermore, instrumental insemination (II) techniques are commonly employed in breeding programs in honey bees (Laidlaw and Page 1997). It is often the case that queens undergoing II (as opposed to natural mating) have an increased latency to egg-laying and differences in pheromone profiles(Kaftanoglu and Peng 1982; Lodesani and Vecchi 1996). Exposure to carbon dioxide before and during this II process appears to improve these disadvantages, but it does not fully correct the discrepancy (Engels et al. 1976; Mackensen 1947). It remains unclear exactly why the II process has these effects on queens.
Here, we approach these questions by monitoring behavioral, physiological, and brain transcriptional changes in queens collected 2 days after mating or II in order to capture changes that are occurring early in the post-mating transition. We used four groups of queens: virgins, queens allowed to take a single mating flight (naturally mated), and queens instrumentally inseminated with either saline or semen. We then used a comparative genomics approach to search for common transcriptional changes associated with the mating process in insects. This approach will help us to understand 1) how quickly post-mating changes occur in the brain, and whether we can detect transcriptional changes in the brain even if the mating process has not been completed, 2) what transcriptional differences are associated with different aspects of the mating process, and 3) how conserved these pathways are with Drosophila post-mating transcriptional responses.
Results
Flight Behavior
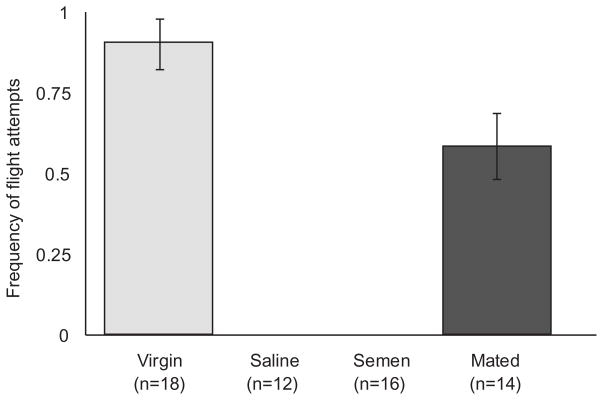
The cues a queen uses to determine when she has completed mating are not well characterized. In order to address this question, we monitored the flight attempts for all queens in each of our treatment groups before and after mating or II. As expected, reproductive status had a significant effect on flight attempts (Figure 1; Chi-square, p<0.0001). Virgin queens and queens limited to a single mating flight both continued to attempt to take mating flights, though the naturally mated queens attempted less frequently (Student’s t-test, p=0.01). None of the inseminated queens attempted to fly following II.
Figure 1. Flight behavior.
Reproductive state significantly affected flight attempts (Chi-square, p<0.0001). None of the inseminated queens attempted to fly two days following instrumental insemination, and fewer mated queens attempted to fly than virgin queens (Student’s t-test, p=0.01). Error bars represent the standard error.
Vitellogenin transcript abundance
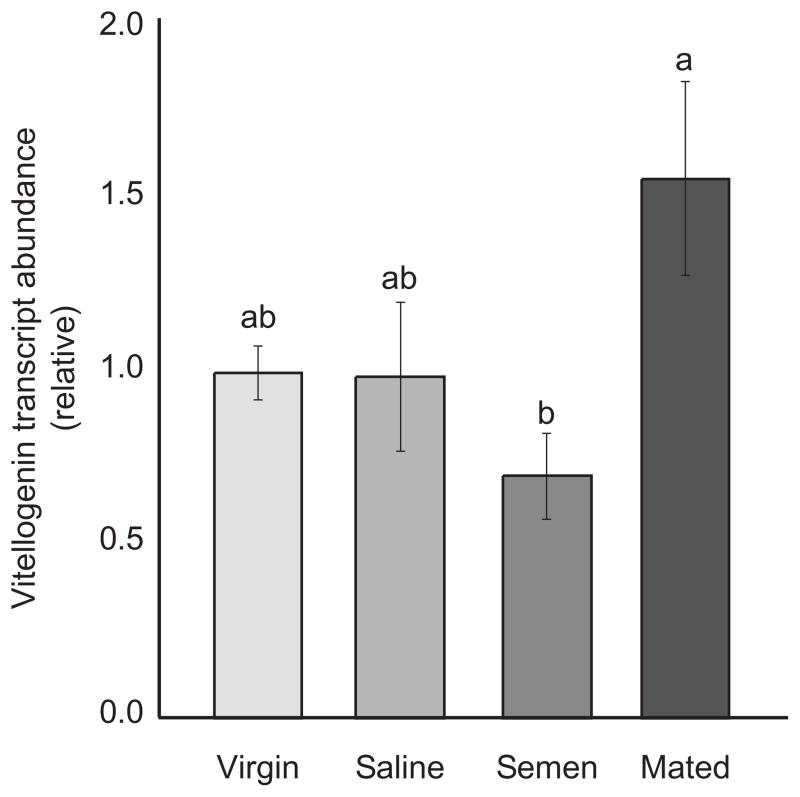
Vitellogenin (Vg) is a protein that is produced in the fat bodies and sequestered in developing oocytes, and it plays a humoral role in ovary development in most insects (Valle 1993). In honey bees, this protein also plays an important role in behavior and division of labor, and several theories have been proposed suggesting that this protein has been co-opted to function in task-specialization among worker bees (Amdam et al. 2006). However, the role of Vg in the mating process still remains to be elucidated in the honey bee. In this study, we used quantitative, real-time PCR to monitor the expression levels of Vg transcripts in the abdominal fat bodies of our queens. We found that mated queens had the highest levels of Vg transcript, virgin and saline-inseminated queens were intermediate, and semen-inseminated queens had the least amount of Vg expressed in the fat bodies (Figure 2, one-way ANOVA, p=0.04, Tukey LSD).
Figure 2. Abdominal Vitellogenin transcript abundance.
Using quantitative, real-time PCR (n=6, each group), we identified significant variation in abdominal transcript abundance of Vg mRNA (ANOVA, p=0.04, Tukey LSD). Mated queens had the highest level of Vg transcripts, while semen-inseminated queens had the lowest level. Virgin and saline-inseminated queens were intermediate. Error bars represent the standard error.
Brain transcriptional profiling
We used whole-genome microarrays to identify transcriptional changes in the brain that were associated with post-mating behavioral and physiological changes. 9,850/13,439 transcripts on the whole-genome microarrays were expressed in our samples and included in the data analysis. There were 631 transcripts (corresponding to 434 predicted genes with fly orthologs) that were differentially expressed among groups (FDR<0.001). The top 10 Gene Ontology (GO) biological processes are summarized in Table 1. 364 of these 631 transcripts were significantly different between naturally mated and saline-/semen-inseminated queens (F-test, p<0.05); genes associated with ‘macromolecule biosynthesis’ were overrepresented in mated queens relative to the inseminated groups (p<0.0001), and genes associated with ‘organic acid metabolism’ were up in the inseminated groups relative to the naturally mated queens (Fisher’s exact test, p<0.02) There were 361/631 transcripts that were significantly different between naturally mated and semen-inseminated queens (F-test, p<0.05), with genes associated with ‘response to stimulus’ overrepresented in mated- relative to inseminated queens (Fisher’s exact test, p<0.04), and genes associated with ‘amine metabolism’ up in semen-inseminated queens relative to mated queens (Fisher’s exact test, p<0.04). 308/631 transcripts were differentially expressed between saline- and semen-inseminated queens (F-test, p<0.05); genes associated with ‘cell communication’ were up in saline relative to semen-inseminated groups (p=0.04), and genes associated with ‘regulation of cell shape’ were overrepresented in semen relative to saline-inseminated queens (p<0.04). Finally, there were 529/631 transcripts differentially expressed between virgins relative and all of the other groups (F-test, p<0.05); genes expressed at higher levels in virgins were associated with the GO biological process ‘response to heat’ (Fisher’s exact test, p<0.0001), and genes expressed at higher levels in the mated or inseminated groups were associated with ‘protein biosynthesis’ (Fisher’s exact test, p<0.0001). A complete list of genes and GO terms associated with the differences between each group is provided in Table S1.
Table 1.
Gene Ontology biological processes
| GO Biological Process | Count | % | p-value |
|---|---|---|---|
| translation | 43 | 9.4 | <0.0001 |
| biosynthetic process | 67 | 14.6 | 0.005 |
| peripheral nervous system development | 11 | 2.4 | 0.009 |
| response to temperature stimulus | 7 | 1.5 | 0.010 |
| vitamin transport | 3 | 6.5 | 0.019 |
| response to stimulus | 57 | 12.4 | 0.020 |
| response to heat | 6 | 1.3 | 0.023 |
| behavior | 22 | 4.8 | 0.023 |
| protein folding | 13 | 2.8 | 0.023 |
| cellular protein metabolic process | 105 | 22.8 | 0.039 |
The 631 transcripts were used in a Gene Ontology analysis. The list of the top ten biological processes associated with these transcripts is summarized in this table. A complete list of terms can be found in supplemental table S1.
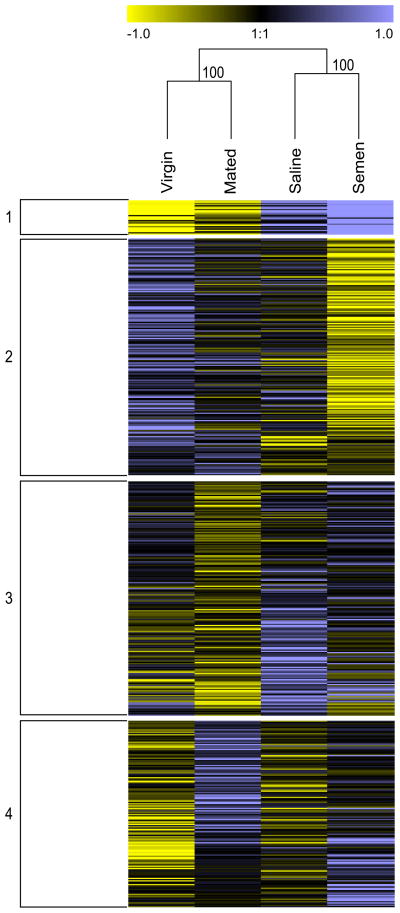
Hierarchical clustering (Figure 3) suggests that the major factor influencing brain gene expression among our groups is the effect of II. This clustering pattern matches the behavioral pattern we observed for flight attempts. K-means clustering generated 4 clusters of genes (Figure 3). The first cluster represents transcripts upregulated by the II process and contains an overrepresentation of genes associated with exonuclease activity (Fisher’s exact test, p<0.0005) and peptidase activity (Fisher’s exact test, p<0.02). The second cluster depicts transcripts downregulated by the II process and includes an overrepresentation of genes associated with phagocytosis (Fisher’s exact test, p<0.0001), cellular biosynthetic processes (Fisher’s exact test, p<0.001), vesicle-mediated transport (Fisher’s exact test, p<0.008), and oxidative phosphorylation (Fisher’s exact test, p=0.03). Clusters 3 and 4 represent transcripts associated with exposure to semen. In cluster 3, these transcripts are down-regulated by semen exposure and include genes associated with translation (Fisher’s exact test, p<0.0001), ion transport (Fisher’s exact test, p<0.006), cell redox homeostasis (Fisher’s exact test, p<0.03), carboxylic acid transport (Fisher’s exact test, p=0.03), and eye-antennal disc development (Fisher’s exact test, p<0.04). Cluster 4 contains transcripts upregulated by semen exposure and includes genes involved in regulation of transport (Fisher’s exact test, p<0.001), establishment of protein localization (Fisher’s exact test, p=0.04), and response to abiotic stimulus (Fisher’s exact test, p<0.05). A complete list of the GO terms associated with each of these clusters is provided in Table S2.
Figure 3. Hierarchical and k-means clustering.
Clustering analysis of the 631 transcripts significantly associated with mating demonstrates that instrumental insemination has a strong effect on brain gene expression. These results match the behavioral patterns that we observed for flight attempts. K-means clustering revealed 4 main clusters of genes. Cluster 1 identifies transcripts that are upregulated by the instrumental insemination process, while Cluster 2 depicts transcripts downregulated by the instrumental insemination process. Clusters 3 and 4 identify transcripts associated with the effects of semen exposure. Cluster 3 seems to contain a set of transcripts that are downregulated by semen exposure in mated queens relative to virgins, and in semen- relative to saline-inseminated queens. Cluster 4 depicts transcripts upregulated by exposure to semen and seminal proteins, with these transcripts upregulated in mated and semen-inseminated queens relative to the other groups. Values are zero-centered transcriptional data in the log2 scale; a value of 1 indicates a 2-fold change in expression levels for a given transcript.
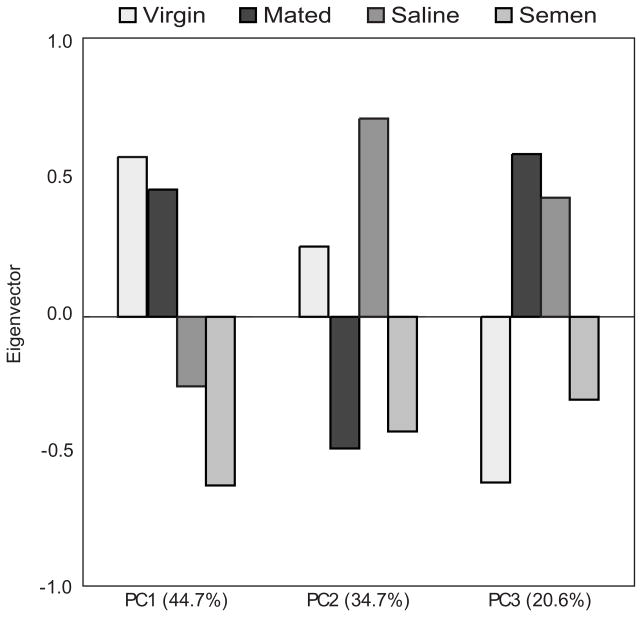
A principal component analysis on the significant transcripts revealed transcriptional differences associated with II effects and the effects of semen exposure on brain gene expression (Figure 4). The first principal component demonstrates that 44.7% of the variation in gene expression is associated with the effects of II. Principal component 2 encapsulates 34.7% of the variation in gene expression and appears to be associated with the effects of semen on brain gene expression. Principal component 3 probably represents the interaction between the II procedure and the effects of semen-exposure, with inseminated groups showing opposite patterns of variation associated with semen-exposure compared to the mated and virgin groups.
Figure 4. Principal components analysis.
PCA based on the 631 significant transcripts identified 3 principal components. PC1 demonstrates that 44.7% of the variation in gene expression is associated with the effects of instrumental insemination. Principal component 2 encapsulates 34.7% of the variation in gene expression, and appears to be associated with the effects of semen on brain gene expression. Principal component 3, and it probably represents the interaction between the instrumental insemination procedure and the effects of semen-exposure, with inseminated groups showing opposite patterns of variation associated with semen-exposure compared to the mated and virgin groups.
Comparative studies
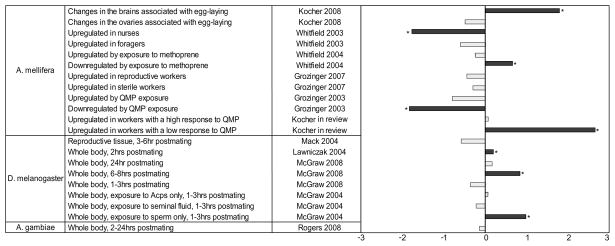
We compared our list of 631 transcripts associated with 2-day post-mating changes to previous studies that identified candidate genes associated with mating in Drosophila (Lawniczak and Begun 2004; Mack et al. 2006; McGraw et al. 2008; 2004), Anopheles (Rogers et al. 2008), and Apis mellifera (Kocher et al. 2008). Significant over-or under-representation was determined using a two-tailed Fisher’s Exact Test, and the results are summarized in Figure 5. The genes associated with 2 day post-mating changes overlapped significantly with the genes associated with 5-day post-mating changes in a study comparing virgin queens, mated and laying queens. Furthermore, there was a significant overlap with multiple studies in Drosophila which characterized post-mating changes in this species, but we observed no significant overlap between our results and the Anopheles gambiae study.
Figure 5. Comparison of our results to previous transcriptional studies.
We compared the results from this study to a set of transcriptional studies in Drosophila, Anopheles, and Apis mellifera. To facilitate comparisons, the deviation of observed overlap from the expected number of overlapping genes observed was converted to a z-score. The x-axis represents the standardized value of these deviations, with 0 representing no deviation, a score of 2 representing a difference that is two standard deviations greater than expected, and −2 representing a difference two standard deviations less than expected. Stars indicate a significant deviation based on a two-tailed Fisher Exact test (p<0.05).
Table 2 lists the Gene Ontology terms associated with the genes overlapping between honey bees and Drosophila. Notably, this list includes several genes associated with defense response (heat shock protein cognate 5, defensin, superoxide dismutase, transferrin 1, lethal (2) essential for life, and glutathione s transferase d1), antioxidant activity (superoxide dismutase, glutathione s-transferase 27, and peroxiredoxin 2540), and oxidoreductase activity (cg8665-pa, cg3560-pa, superoxide dismutase, glutathione s-transferase 27, gh21316p, cg3523-pa, cypixf2, and peroxiredoxin 2540).
Table 2.
Significant GO Terms for all genes overlapping between this study and the Drosophila mating studies.
| Category | Term | Count | % | P-Value |
|---|---|---|---|---|
| BP | defense response | 6 | 12 | 3.10E-03 |
| BP | response to biotic stimulus | 4 | 8 | 1.60E-02 |
| BP | determination of muscle attachment site | 2 | 4 | 3.20E-02 |
| CC | cytosolic ribosome (sensu Eukaryota) | 4 | 8 | 4.70E-03 |
| CC | cytosolic part | 4 | 8 | 1.10E-02 |
| CC | cytoplasm | 13 | 26 | 1.40E-02 |
| CC | ribosomal subunit | 4 | 8 | 2.70E-02 |
| CC | ribosome | 4 | 8 | 4.00E-02 |
| CC | ribonucleoprotein complex | 5 | 10 | 4.70E-02 |
| MF | antioxidant activity | 3 | 6 | 9.90E-03 |
| MF | oxidoreductase activity | 8 | 16 | 1.10E-02 |
| MF | voltage-gated potassium channel activity | 3 | 6 | 1.80E-02 |
| MF | metal ion transmembrane transporter activity | 4 | 8 | 2.60E-02 |
| MF | potassium channel activity | 3 | 6 | 3.00E-02 |
| MF | voltage-gated cation channel activity | 3 | 6 | 3.20E-02 |
| MF | voltage-gated channel activity | 3 | 6 | 3.60E-02 |
| MF | voltage-gated ion channel activity | 3 | 6 | 3.60E-02 |
| MF | glutathione peroxidase activity | 2 | 4 | 3.60E-02 |
The results from this study showed a significant overlap with the results from three previous mating studies in Drosophila (Lawniczak 2004, McGraw 2004, McGraw 2008). The resulting genes that overlapped in one or more of these studies were input into DAVID for Gene Ontology analysis. The GO Terms associated with these genes are listed in the above table.
We also compared our gene list to previously published transcriptional studies in honey bees, including: QMP exposure (Grozinger et al. 2003b), nursing/foraging behavior (Whitfield et al. 2003), methoprene treatment (Whitfield et al. 2006), and worker ovary activation (Grozinger et al. 2007), as well as genes associated with pollen hoarding identified from quantitative trait loci mapping studies(Hunt et al. 2007). We found a significant overlap between genes associated with retinue response behavior and exposure to methoprene (a juvenile hormone analog). We found no significant positive associations between post-mating changes and division of labor, and in fact there was less overlap than expected by chance. There was also significantly less overlap than expected by chance with genes associated with queen pheromone exposure. These comparisons are summarized in Figure 5.
Discussion
This study uncouples the mating process to examine the effects of natural mating vs. instrumental insemination (II) and saline vs. semen insemination in honey bee queens. The results of this study overlapped well with our previous study that examined behavioral, physiological, and transcriptional changes associated with the full mating transition (from virgin to egg-laying queens; Kocher et al. 2008), suggesting that some of these genes are consistently regulated throughout the entire mating process. Furthermore, we found similar results to studies in Drosophila melanogaster, suggesting that these post-mating changes may be conserved across insect orders.
In general, we found that the II process has large-scale effects on brain gene expression and flight behavior, and that our brain gene expression patterns match the behavioral patterns that we observed in the field. One possible reason that the II groups may have exhibited the strongest behavioral and transcriptional changes is that both the saline- and semen-inseminated groups received the same volume (10 μL) during the II procedure. This may indicate that honey bee queens use volume as a cue for flight attempts (perhaps by receiving input from stretch receptors in the lateral oviducts; see Tarpy 2000). Alternatively, both groups were also exposed to carbon dioxide (CO2) treatment and physical manipulation with the II device, and it is also possible that this exposure inhibits further flight attempts by honey bee queens and causes dramatic transcriptional changes. Previous studies found that exposure of virgin queens to CO2 accelerates ovarian development and initiation of egg-laying (Engels et al. 1976; Engels and Imperatriz-Fonseca 1990; Mackensen 1947). However, CO2 does not appear to affect the levels of biogenic amines in their brains (Harris et al. 1996). Additional studies have shown that CO2 treatment alone will inhibit flight attempts, and CO2 treatment with physical manipulation has a greater effect on brain gene expression than CO2 treatment alone (Niño et al, unpublished data). Again, these results suggest that the mating process is quite complex and regulated by a number of factors.
Interestingly, while II appears to trigger immediate and complete behavioral changes (e.g., the cessation of mating flight attempts) and large-scale brain transcriptional changes, it does not have the same effects on ovary development or pheromone production (Kocher et al. 2009). In the case of these physiological post-mating changes, the naturally mated queens are significantly different from virgins, while the instrumentally inseminated queens are intermediate. These results reaffirm the results from Kocher et al. (2008) and suggest that changes associated with flight behavior and brain gene expression are uncoupled—or at least operating at a different timescale—from changes associated with ovary development and pheromone production. Combined with our previous studies, it appears that honey bee queens may use volume (and/or carbon dioxide exposure) as the primary cue to cease mating flights, and that exposure to semen and/or seminal proteins may be less critical to this process.
In the present study, Vg transcripts were the greatest in mated queens (which matches ovary development; Kocher et al 2009), but levels were the lowest in semen-inseminated queens, with virgins and saline-inseminated queens intermediate and not significantly different from either mated or semen-inseminated queens. These results are surprising, since mating and ovary development are expected to be correlated with an increase in Vg expression. It is possible that there are negative effects of II that inhibit this process, or that Vg RNA and protein levels in the hemolymph are not tightly linked in this system.
We identified 631 transcripts that are associated with flight behavior and II. There were significant expression differences between virgins and inseminated or mated queens, inseminated queens and naturally mated queens, and queens inseminated with semen vs saline, suggesting that the transcriptional changes associated with post-mating changes are quite complex and regulated by a number of different factors. We found that eye receptor genes are up-regulated in virgins relative to all other groups. These results are interesting because virgins are still attempting to take flights while mated queens have decreased their number of attempts and instrumentally inseminated queens have ceased trying to fly. Once queens have completed the mating process, they remain in the hive to lay eggs and never fly again (except possibly to swarm the following year). It is possible that following mating, the expression levels of these genes decrease because they are no longer required for a queen to function properly within the colony. Furthermore, oxidoreductases were expressed at higher levels in the naturally mated group relative to all other groups (Table S2). Oxidoreductases are known to be important in increasing lifespan and immunity (Lai et al. 2007; McGraw et al. 2004). These results suggest that perhaps something about the II process is not recapitulating some aspects of natural mating that triggers an increase in oxidoreductases, which could help to explain why inseminated queens are often less fecund and have a shorter lifespan that naturally mated queens (but see Cobey 2007).
In addition to the effect of II, there was also an effect of exposure to semen on brain expression patterns. 308 genes were differentially expressed between saline- and semen-inseminated queens (p<0.05), and principal components and clustering analyses also revealed an effect of semen-exposure on brain gene expression. The PCA revealed that about 35% of the variation in brain gene expression was associated with the effects of semen exposure. K-means clustering also identified two clusters of transcripts associated with semen exposure. However, results from another study indicate that there is no significant effect of semen exposure on ovary development or pheromone production (Kocher et al. 2009), at least in the early stages, again suggesting that brain transcriptional changes are uncoupled, or at least operating at a different timescale, from post-mating physiological changes.
Overall, we found a significant degree of overlap between this study and previous studies of transcriptional changes associated with mating in Drosophila melanogaster, but not in Anopheles gambiae (Kocher et al. 2008; Lawniczak and Begun 2004; Mack et al. 2006; McGraw et al. 2008; 2004; Rogers et al. 2008). Notably, there was a significant overlap between the early (2-day) post-mating changes and late (5-day, egg-laying) post-mating changes in honey bees, suggesting that many of the transcriptional changes that we observe can remain stable throughout the transition from flight-related behavior to egg-laying behavior. Furthermore, our results overlapped significantly with results from several studies conducted in Drosophila (Lawniczak and Begun 2004; Mack et al. 2006; McGraw et al. 2008; 2004), suggesting that some of the post-mating transcriptional machinery may be conserved across species. Finally, we have identified several genes that are associated with post-mating changes in females in both Apis mellifera and Drosophila melanogaster. These genes are associated with defense response, antioxidant activity, and oxidoreductase activity. Expression levels of these genes were characterized in the brains of honey bee queens, but in the whole-bodies of Drosophila. While the magnitude, direction, and location of these transcriptional changes are not necessarily the same, we have nevertheless identified a set of genes that are associated with post-mating changes in females that appear to be conserved across insect orders. These results suggest that some aspects of mating appear to be conserved across multiple insect orders, and they are consistent with the ideas that there is a tradeoff between fecundity and lifespan (Stearns 1992) and that mating induces changes in the immune system (Lawniczak et al. 2007).
We also compared our results from this study to previously published gene expression and linkage mapping data for various traits in honey bees, including: QMP exposure (Grozinger et al. 2003b), nursing/foraging behavior (Whitfield et al. 2003), methoprene treatment (Whitfield et al. 2006), worker ovary activation (Grozinger et al. 2007), and pollen hoarding (Hunt et al. 2007). Genes associated with the early post-mating transition also appear to be associated with low retinue response in workers (Kocher et al. in review). The functional basis for this overlap is unclear. We captured transcriptional variation in the brains of queens that were in the very early stages of ovary development. It could be that low-responding workers may share similar expression profiles because these individuals are also primed for ovary development in the absence of a queen. Furthermore, there was significantly less overlap than expected between genes associated with mating and genes associated with worker division of labor (especially nursing behavior) and genes regulated by queen pheromone, which also regulates worker division of labor. This suggests that the genes associated with early post-mating changes in queens may be quite distinct from those associated with worker division of labor. Interestingly, genes that are down-regulated by methoprene in workers also appear to be regulated by the mating process in queens. Methoprene is a juvenile hormone (JH) analog, and JH is known to play a very important role in ovarian development and mating in insects, though its role in honey bee reproduction is unclear (Robinson and Vargo 1997).
In summary, mating in honey bee queens induces rapid and permanent changes in behavior, physiology, and gene expression. In these early post-mating stages, there are no significant differences in flight behavior, pheromone production or ovary activation between saline and semen inseminated queens (Kocher et al. 2009), suggesting that insemination volume, not substance, is the main factor that triggers these post-mating changes. However, we did find that 308 genes were expressed at significantly different levels in the brains of semen and saline inseminated queens, suggesting that the insemination substance does effect brain gene expression, and may also have subtle effects on behavior and physiology. Furthermore, there are dramatic differences between instrumentally inseminated and naturally mated queens in terms of behavior, physiology (including ovary activation, pheromone profiles and Vg levels), and brain gene expression, suggesting that improvements can be made to the instrumental insemination procedure to more closely mimic natural mating. Finally, some of these transcriptional changes we observed appear to be consistent across insect orders, and suggest that mating induces changes in genes associated with immune response, antioxidant activity, and oxidoreductase activity.
Experimental Procedures
Field Methods
The queens (Apis mellifera carnica) were reared at the NCSU Bee Research Facility in Raleigh, NC as detailed in Kocher et al. (2008). The source colony used for this experiment was headed by a queen (Glenn Apiaries, Fallbrook, CA) instrumentally inseminated with semen from a single drone. Because male bees (drones) are haploid, the average coefficient of relatedness (G) among offspring of such an instrumentally inseminated queen is 0.75. Prior to emergence as adults, queen cells were placed into colonies with approximately 1,000 workers per colony 2 days before expected emergence. 2 days following emergence, surviving queens were randomly assigned to a group (virgin, saline-inseminated, semen-inseminated, or naturally mated), marked with paint, and returned to their respective colonies. Colony entrances were modified with a plexiglas-covered runway, and queen excluders were placed at the entrance so that individuals attempting to fly could be observed (see Tarpy and Page 2000). On the 7th day following their expected emergence, queens in the II groups were anesthetized with carbon dioxide for 4.0 minutes during the II procedure. These queens were inseminated with either 10.0 μl of saline (Williams and Harbo 1982) or 10.0 μl of semen collected from brother drones using a Schley Model II insemination device and a calibrated Harbo syringe. The semen was collected into a glass capillary tube from approximately 100 drones from a common colony the same morning as the inseminations, expelled into a glass vial, and thoroughly mixed with a sterile metal spatula prior to recollection with the insemination device (see Seeley and Tarpy 2007). For queens > 4 days after emergence, the colony entrances were monitored from 2–6pm daily and any attempts to fly were recorded. Queens in the naturally mated group were allowed to take multiple orientation flights, but only one mating flight: once a queen returned to the colony with the mating sign, she was no longer allowed to fly and was confined to the colony. Two days following mating or instrumental insemination, queens were collected on dry ice and stored at −80°C for processing. Virgin queens were collected on the same day as instrumentally inseminated queens. Since there was variability in the timing the queens naturally mated, there was a 1–2 day variation in the collection of these queens compared to the other three groups. Prior to dissection, queen heads were removed and partially lyophilized to facilitate dissection (Grozinger et al. 2003a), after which their brains were dissected out on dry ice and stored for future processing. In total, there were 18 virgin queens, 12 saline-inseminated queens, 16 semen-inseminated queens, and 14 naturally mated queens.
Quantification of Vitellogenin RNA levels by quantitative real-time-PCR (qRT-PCR)
Fat body samples from individual queens were extracted using an RNeasy RNA extraction kit (Qiagen, Valencia, CA). cDNA was synthesized from 200ng RNA using the SYBRGreen Master Mix (Applied Biosystems, Foster City, CA). qRT-PCR was performed with an ABI Prism 7900 sequence detector and the SYBR green detection method (Applied Biosystems). The housekeeping gene, eIFS-8 was used as a control (Grozinger 2003; Whitfield 2003). 6 queens from each group were used for this analysis. For each sample, triplicate qRT-PCR reactions were performed and averaged. Quantification was performed using a standard curve generated using genomic DNA. A negative control (cDNA reaction without RT-enzyme) and dissociation curve was used as well. Groups were normalized to one sample for ease of graphical representation, and a nonparametric Wilcoxon rank sums was used in JMP 7.0 (SAS, Cary, NC)to determine significance. The primer sequences were developed in PrimerExpress (Applied Biosystems) as follows.
Vitellogenin
Forward:5′TTGACCAAGACAAGCGGAACT 3′
Reverse: 5′AAGGTTCGAATTAACGATGAAAGC 3′
eIFS-8
Forward: 5′TGAGTGTCTGCTATGGATTGCAA3′
Reverse: 5′CGTGGAGTGTTATCGTAAGTAGCAA 3′
Microarray extractions and hybridizations
4 individuals from each group were selected for microarray analysis, for a total of 16 microarrays. RNA was extracted from brains using PicoPure RNA Isolation kits (Arcturus, CA). 200 ng RNA was amplified using MessageAmp II aRNA Amplification kits (Ambion, Austin, TX). 2μg of amplified RNA was labeled with Cy3 or Cy5 dye using Kreatech labeling kits (Applied Biosystems, Salt Lake City, UT). 60pmol each of two sets of labeled probe were then hybridized to the whole-genome oligonucleotide arrays supplied from the laboratory of Dr. Gene Robinson (University of Illinois, Urbana-Champaign). Brains were hybridized using a repeated loop design with dye-swaps incorporated. Arrays were then scanned using the Axon Genepix 4000B scanner (Molecular Devices, Sunnyvale, CA) using GENEPIX software (Agilent Technologies, Santa Clara, CA). Each array was visually inspected, and spots were manually adjusted to ensure proper annotation of each transcript.
Microarray Data Analysis
Spots with intensity less than 300 (the mean background intensity of these arrays) in two or more arrays were removed from the analysis, and spots with less than 10 observations remaining were also excluded from the data set. Expression data was log-transformed and normalized using a mixed-model ANOVA (proc MIXED, SAS, Cary, NC) with the following model: Y = μ + dye + array + block + dye*array + array*block + ε, where Y is expression, dye is a fixed effect, and array, block, and their interactions are random effects. Pre- and post-normalization, Cy3/Cy5 plots and MA plots were generated and visually inspected to ensure that each array was high quality. Detection of significance for differential expression on residuals was performed using a mixed-model ANOVA with the model: Y = μ + behavior + dye + array + dye*array + ε, where y is the residual from the previous model, behavior and dye are fixed effects, and array and dye*array are random effects. P-values were corrected for multiple testing using a false discovery rate < 0.001 (proc MULTTEST, SAS). Contrast statements were employed to identify transcripts (out of the list of 631) that were differentially expressed between different mating groups. Because these transcripts survived an FDR threshold of <0.001, we were confident that they were significantly associated with the mating process in queens, and as such we did not feel it was necessary to correct for multiple testing a second time. Hierarchical and k-means clustering was performed in Genesis (Sturn et al. 2002). Approximate-unbiased p-values and bootstrap values were obtained for sample clustering using the pvclust package (Suzuki and Shimodaira 2006) in R version 2.5.0 (method=“cor”, hclust=“ward”, n=1000). Principal components analysis was performed in JMP 7.0 (SAS, Cary, NC). Gene Ontology analysis was conducted in DAVID (Dennis et al. 2003; Huang da et al. 2009). Microarray data is available through ArrayExpress with accession number E-MEXP-2205.
Comparative Analysis
The significant genes from the brain expression dataset were compared to several published transcriptional datasets (Grozinger et al. 2007; Grozinger et al. 2003b; Kocher et al. 2008; Lawniczak and Begun 2004; Mack et al. 2006; McGraw et al. 2008; 2004; Rogers et al. 2008; Whitfield et al. 2006; 2003). For previously published honey bee studies, all comparisons were conducted using the honey bee predicted gene set. Orthologous genes were determined using the BLASTP algorithm to identify the best Drosophila match to the predicted honey bee genes. More information about the microarrays and the annotation can be found at the honey bee oligonucleotide array website(http://www.biotech.uiuc.edu/centers/Keck/Functional_genomics/HoneyBeeOligo.htm). The significance of overlap was determined using a two-tailed Fisher Exact test using the number of genes that were common to both studies. For graphical purposes, the deviation of observed overlap from expected overlap was converted into a z-score.
Supplementary Material
Acknowledgments
We would like to thank the Grozinger and Tarpy labs for their comments on this manuscript, as well as J. Keller, J. Summers, W. Hensey, D. Hopkins, and K. Hutcherson for their assistance in the field. This work is supported by a USDA-NRI grant to CMG and DRT (2006-35607-16625), as well as an NIH Training Grant to SDK.
References
- Amdam GV, Csondes A, Fondrk MK, Page RE., Jr Complex social behaviour derived from maternal reproductive traits. Nature. 2006;439:76–8. doi: 10.1038/nature04340. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cobey SW. Comparison studies of instrumentally inseminated and naturally mated honey bee queens and factors affecting their performance. Apidologie. 2007;38:390–410. [Google Scholar]
- Dennis G, Jr, Sherman BT, Hosack DA, Yang J, Gao W, Lane HC, Lempicki RA. DAVID: Database for Annotation, Visualization, and Integrated Discovery. Genome Biology. 2003;4:3. [PubMed] [Google Scholar]
- Engels W, Gonçalves LS, Engles E. Effects of carbon dioxide on vitellogenin metabolism in unmated queen honeybees. Journal of Apicultural Research. 1976;15:3–10. [Google Scholar]
- Engels W, Imperatriz-Fonseca VL. Caste development, reproductive strategies and control of fertility in honey bees and stingless bees. In: Engels W, editor. Social Insects: An Evolutionary Approach to Caste and Reproduction. Springer-Verlag; Heidelberg, Germany: 1990. pp. 167–230. [Google Scholar]
- Grozinger CM, Fan Y, Hoover SE, Winston ML. Genome-wide analysis reveals differences in brain gene expression patterns associated with caste and reproductive status in honey bees (Apis mellifera) Molecular Ecology. 2007;16:4837–48. doi: 10.1111/j.1365-294X.2007.03545.x. [DOI] [PubMed] [Google Scholar]
- Grozinger CM, Sharabash NM, Whitfield CW, Robinson GE. Pheromone-mediated gene expression in the honey bee brain. Proceedings of the National Academy of Sciences USA. 2003a;100:14519–14525. doi: 10.1073/pnas.2335884100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Grozinger CM, Sharabash NM, Whitfield CW, Robinson GE. Pheromone-mediated gene expression in the honey bee brain. Proceedings of the National Academy of Sciences USA. 2003b;100(Suppl 2):14519–25. doi: 10.1073/pnas.2335884100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Harris JW, Woodring J, Harbo JR. Effects of carbon dioxide on levels of biogenic amines in the brains of queenless worker and virgin queen honey bees (Apis mellifera) Journal of Apicultural Research. 1996;35:69–78. [Google Scholar]
- Haydak MH. The queen honeybee. 1949. The queen honeybee; pp. 68–94. [Google Scholar]
- Huang da W, Sherman BT, Lempicki RA. Systematic and integrative analysis of large gene lists using DAVID bioinformatics resources. Nature Protocols. 2009;4:44–57. doi: 10.1038/nprot.2008.211. [DOI] [PubMed] [Google Scholar]
- Hunt GJ, Amdam GV, Schlipalius D, Emore C, Sardesai N, Williams CE, Rueppell O, Guzman-Novoa E, Arechavaleta-Velasco M, Chandra S, Fondrk MK, Beye M, Page RE., Jr Behavioral genomics of honeybee foraging and nest defense. Naturwissenschaften. 2007;94:247–67. doi: 10.1007/s00114-006-0183-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kaftanoglu O, Peng YS. Effects of insemination on the initiation of oviposition in the queen honey bee. Journal of Apicultural Research. 1982;21:3–6. [Google Scholar]
- Keeling CI, Slessor KN, Higo HA, Winston ML. New components of the honey bee (Apis mellifera L.) queen retinue pheromone. Proceedings of the National Academy of Sciences USA. 2003;100:4486–91. doi: 10.1073/pnas.0836984100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kocher SD, Ayroles JA, Stone EA, Grozinger CM. Individual variation in pheromone response correlates with reproductive traits and brain gene expression in honey bees. doi: 10.1371/journal.pone.0009116. in review. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kocher SD, Richard FJ, Tarpy DR, Grozinger CM. Genomic analysis of post-mating changes in the honey bee queen (Apis mellifera) BMC Genomics. 2008;9:232. doi: 10.1186/1471-2164-9-232. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kocher SD, Richard FJ, Tarpy DR, Grozinger CM. Queen reproductive state modulates pheromone production and queen-worker interactions in honey bees. Behavioral Ecology. 2009;20(5):1007–1014. doi: 10.1093/beheco/arp090. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lai CQ, Parnell LD, Lyman RF, Ordovas JA, Mackay TFC. Candidate genes affecting Drosophila life span identified by integrating microarray gene expression analysis and QTL mapping. Mechanisms of Ageing and Development. 2007;128:237–249. doi: 10.1016/j.mad.2006.12.003. [DOI] [PubMed] [Google Scholar]
- Laidlaw JHH, Page RE., Jr . Queen rearing and bee breeding. Wicwas; Cheshire, CT: 1997. [Google Scholar]
- Lawniczak MKN, Barnes AI, Linklater JR, Boone JM, Wigby S, Chapman T. Mating and immunity in invertebrates. Trends in Ecology & Evolution. 2007;22:48–55. doi: 10.1016/j.tree.2006.09.012. [DOI] [PubMed] [Google Scholar]
- Lawniczak MKN, Begun DJ. A genome-wide analysis of courting and mating responses in Drosophila melanogaster females. Genome. 2004;47:900–910. doi: 10.1139/g04-050. [DOI] [PubMed] [Google Scholar]
- Lodesani M, Vecchi MA. A study on the development of the ovaries of queen bees (Apis mellifera ligustica) instrumentally and naturally mated. Apicoltore Moderno. 1996;87:65–70. [Google Scholar]
- Mack PD, Kapelnikov A, Heifetz Y, Bender M. Mating-responsive genes in reproductive tissues of female Drosophila melanogaster. Proceedings of the National Academy of Sciences USA. 2006;103:10358–63. doi: 10.1073/pnas.0604046103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mackensen O. Effect of carbon dioxide on initial oviposition of artificially inseminated and virgin queen bees. Journal of Economic Entomology. 1947;40:344–349. doi: 10.1093/jee/40.3.344. [DOI] [PubMed] [Google Scholar]
- McGraw LA, Clark AG, Wolfner MF. Post-mating gene expression profiles of female Drosophila melanogaster in response to time and to four male accessory gland proteins. Genetics. 2008;179:1395–408. doi: 10.1534/genetics.108.086934. [DOI] [PMC free article] [PubMed] [Google Scholar]
- McGraw LA, Gibson G, Clark AG, Wolfner MF. Genes regulated by mating, sperm or seminal proteins in mated female Drosophila melanogaster. Current Biology. 2004;14:1509–1514. doi: 10.1016/j.cub.2004.08.028. [DOI] [PubMed] [Google Scholar]
- Nijhout HF. Abdominal stretch reception in Dipetalogaster-Maximus (Hemiptera, Reduviidae) Journal of Insect Physiology. 1984;30:629–633. [Google Scholar]
- Plettner E, Otis GW, Wimalaratne PDC, Winston ML, Slessor KN, Pankiw T, Punchihewa PWK. Species-and caste-determined mandibular gland signals in honeybees (Apis) Journal of Chemical Ecology. 1997;23:363–377. [Google Scholar]
- Plettner E, Slessor KN, Winston ML, Robinson GE, Page RE., Jr Mandibular gland components and ovarian development as measures of caste differentiation in the honey bee (Apis mellifera L.) Journal of Insect Physiology. 1993;39:235–240. [Google Scholar]
- Roberts WC. Multiple mating of queen bees proved by progeny and flight tests. Gleanings in Bee Culture. 1944;72:255–260. [Google Scholar]
- Robinson GE, Vargo EL. Juvenile hormone in adult eusocial Hymenoptera: gonadotropin and behavioral pacemaker. Archives of Insect Biochemistry and Physiology. 1997;35:559–83. doi: 10.1002/(SICI)1520-6327(1997)35:4<559::AID-ARCH13>3.0.CO;2-9. [DOI] [PubMed] [Google Scholar]
- Rogers DW, Mancini E, Whitten MM, Baldini F, Thailayil J, della Torre A, Levashina E, Catteruccia F. Molecular Bases of Post-Mating Behaviour in Anopheles Gambiae. American Journal of Tropical Medicine and Hygiene. 2008;79:314–314. [Google Scholar]
- Schluns H, Moritz RFA, Neumann P, Kryger P, Koeniger G. Multiple nuptial flights, sperm transfer and the evolution of extreme polyandry in honey bee queens. Animal Behavior. 2005;70:125–131. [Google Scholar]
- Seeley TD, Tarpy DR. Queen promiscuity lowers disease within honeybee colonies. Proceedings of Biological Sciences. 2007;274:67–72. doi: 10.1098/rspb.2006.3702. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stearns SC. The Evolution of Life Histories. Oxford University Press; New York: 1992. [Google Scholar]
- Sturn A, Quackenbush J, Trajanoski Z. Genesis: cluster analysis of microarray data. Bioinformatics. 2002;18:207–208. doi: 10.1093/bioinformatics/18.1.207. [DOI] [PubMed] [Google Scholar]
- Sugawara T. Stretch reception in bursa copulatrix of the butterfly, Pieris rapae crucivora, and its role in behaviour. Journal of Comparative Physiology. 1979;130:191–199. [Google Scholar]
- Suzuki R, Shimodaira H. Pvclust: an R package for assessing the uncertainty in hierarchical clustering. Bioinformatics. 2006;22:1540–2. doi: 10.1093/bioinformatics/btl117. [DOI] [PubMed] [Google Scholar]
- Tanaka ED, Hartfelder K. The initial stages of oogenesis and their relation to differential fertility in the honey bee (Apis mellifera) castes. Arthropod Structure & Development. 2004;33:431–442. doi: 10.1016/j.asd.2004.06.006. [DOI] [PubMed] [Google Scholar]
- Tarpy DR. PhD. University of California; Davis: 2000. Behavioral and evolutionary mechanisms of polyandry in honey bees (Apis mellifera) [Google Scholar]
- Tarpy DR, Nielsen R, Nielsen DI. A scientific note on the revised estimates of effective paternity frequency in Apis. Insectes Sociaux. 2004;51:203–204. [Google Scholar]
- Tarpy DR, Page RE. No behavioral control over mating frequency in queen honey bees (Apis mellifera L.): implications for the evolution of extreme polyandry. American Naturalist. 2000;155:820–827. doi: 10.1086/303358. [DOI] [PubMed] [Google Scholar]
- Tarpy DR, Page RE., Jr The curious promiscuity of queen honey bees (Apis mellifera): evolutionary and behavioral mechanisms. Annales Zoologici Fennici. 2001;38:255–265. [Google Scholar]
- Valle D. Vitellogenesis in Insects and Other Groups -a Review. Memorias Do Instituto Oswaldo Cruz. 1993;88:1–26. doi: 10.1590/s0074-02761993000100005. [DOI] [PubMed] [Google Scholar]
- Whitfield CW, Ben-Shahar Y, Brillet C, Leoncini I, Crauser D, Leconte Y, Rodriguez-Zas S, Robinson GE. Genomic dissection of behavioral maturation in the honey bee. Proceedings of the National Academy of Sciences USA. 2006;103:16068–75. doi: 10.1073/pnas.0606909103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Whitfield CW, Cziko AM, Robinson GE. Gene expression profiles in the brain predict behavior in individual honey bees. Science. 2003;302:296–299. doi: 10.1126/science.1086807. [DOI] [PubMed] [Google Scholar]
- Williams J, Harbo JR. Bioassays for diluents of honey bee semen. Annual Entomological Society of America. 1982;75:457–459. [Google Scholar]
- Winston ML. The Biology of the Honey Bee. Harvard University Press; Cambridge, MA: 1991. [Google Scholar]
- Wolfner MF. Tokens of love: functions and regulation of Drosophila male accessory gland products. Insect Biochemistry and Molecular Biology. 1997;27:179–192. doi: 10.1016/s0965-1748(96)00084-7. [DOI] [PubMed] [Google Scholar]
- Wolfner MF. The gifts that keep on giving: physiological functions and evolutionary dynamics of male seminal proteins in Drosophila. Heredity. 2002;88:85–93. doi: 10.1038/sj.hdy.6800017. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.