Abstract
There are several different types of self-incompatibility in different flowering plant species, and there has recently been progress in understanding their molecular genetics by using combined molecular and evolutionary approaches. Questions include the mechanism of self-incompatibility (both the nature of the proteins encoded by the genes and whether incompatibility systems all have separate genes for the pollen and pistil recognition proteins, which is the focus of this mini-review) and whether these systems involve chromosome regions with suppressed recombination and, if so, the size of these regions.
Introduction and context
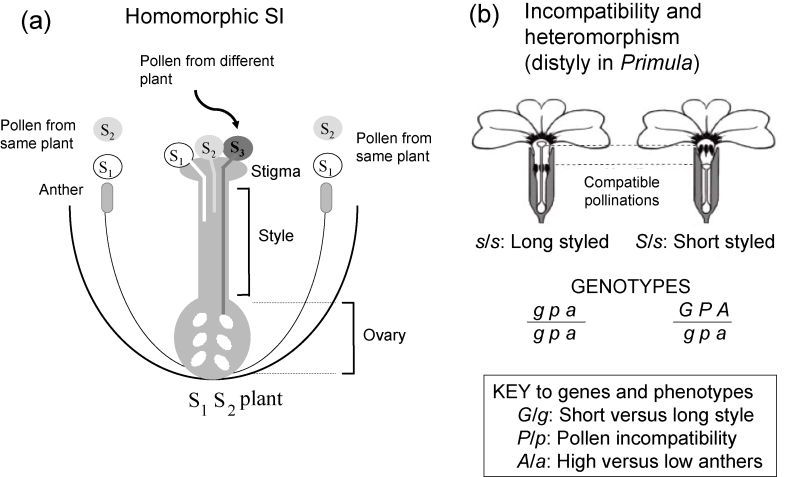
Self-incompatibility (SI) systems in flowering plants are either homomorphic, with many different incompatibility types (Figure 1a) whose flowers are indistinguishable, or heteromorphic, with only two or three incompatibility types that also have different positions of flower parts (often called heterostyled plants). The incompatibility types of plants with homomorphic incompatibility can be determined only by testing the compatibility of different individuals, whereas in heterostyled plants, the flower morphology usually indicates the incompatibility type [1]. Heterostyly is an interesting example of a genetically controlled polymorphism for flower morphology. In distyly, half of the plants have flowers with long styles and anthers deep inside the flowers, whereas the other half have the reciprocal arrangement (Figure 1b); an incompatibility system ensures that long- and short-styled plants are each compatible only with the other morph.
Figure 1. Flower parts and S genotypes.
(a) Homomorphic self-incompatibility (SI). Gametophytic control of pollen incompatibility types is shown; the haploid pollen grains express the allele they carry. This system is known in Solanaceae, Papaveraceae, Rosaceae, and Antirrhinum species (in an unrelated angiosperm family, Plantaginaceae). In other families, pollen specificities are controlled by the genotype of the diploid anther tissue (sporophytic system). This is known in Brassicaceae and in Ipomoea, in the family Convolvulaceae (e.g., [32]). (b) Heteromorphic SI. Long- and short-styled primrose flowers (showing the pollinations that are compatible) from [29-31] and the three genes hypothesised to control style length, pollen incompatibility type, and anther position.
For many years, the mechanisms of these types of recognition systems remained unknown. Genetic studies showed that SI systems do not involve self-recognition (cross-incompatibility occurs between different individuals that have the same incompatibility type) but rather are genetically determined chemical recognition systems. These studies also showed that there are different types of SI in different flowering plant families. Although genetic studies raised interesting questions in both homomorphic and heteromorphic systems, progress stalled until molecular studies became possible.
Now, molecular genetic work and molecular evolutionary approaches in different systems have revealed the recognition genes and the detailed mechanisms involved in the recognition event in several homomorphic SI systems [2,3], including the steps downstream of these events [4,5]. Similar progress should be possible in other self-incompatible plants, including those with heteromorphic systems, in which no S genes have yet been identified. Once genes are identified, it becomes possible to study how self-recognition works (i.e., the mechanism or mechanisms rejecting incompatible pollen).
Early genetic studies of incompatibility in Nicotiana [6] and Oenothera [7], with homomorphic systems, discovered multiple alleles at a single genetic locus (the incompatibility or self-incompatibility locus [S-locus]), and multi-allele S-loci are now known in several other plant families. In many self-incompatible species, the incompatibility type of the haploid pollen grains is controlled by the allele carried (gametophytic control; Figure 1a); the pistil cells are diploid, and both alleles are expressed – the plant rejects pollen carrying either of its own alleles. With allele numbers as high as 50 or more, it was very surprising that, when sequences of the S-locus genome region of some of these species were obtained, separate genes were found for the recognition proteins expressed by pollen grains and pistils (i.e., the genetic S-locus includes at least two incompatibility genes: one for the pistil incompatibility protein and one or – as explained below – perhaps more than one gene controlling the pollen incompatibility types). With two or more genes, correct combinations of alleles must be maintained to avoid combinations that allow self-compatibility, and obviously very close genetic linkage between the pistil and pollen genes will prevent such disadvantageous combinations. However, it is mystifying how new combinations (alleles with new incompatibility types) can ever arise given that at least two genes must change appropriately – this is a puzzle that has not yet been solved.
Major recent advances
Homomorphic systems
A recent study adds Papaver rhoeas to the species in which two component genes have been identified. Sixteen years after the pistil gene was characterised [8], the pollen gene of the S1 type was identified only 457 base pairs away [9]; the distance might be different in alleles of other incompatibility types as variation in the physical arrangement of genes is known in other systems, including self-incompatible Arabidopsis species, such as Arabidopsis lyrata [10]. Sequences of alleles with three different incompatibility types were as different as those of the P. rhoeas pistil incompatibility gene (with mean non-synonymous differences per site around 30-40% and extremely high synonymous site differences), and each sequence was found only in plants with a particular incompatibility type. This supports long-term maintenance of different pairs of alleles at the pollen and pistil loci (reviewed in [11]), consistent with in vitro tests [9] providing evidence that the gene encodes the pollen recognition protein. Although trans-membrane motifs are recognisable, the sequence suggests a new kind of recognition protein.
In the family Brassicaceae, self-incompatible Brassica species and A. lyrata and its close relative Arabidopsis halleri have an incompatibility system that involves a protein kinase stigma protein that recognises a pollen surface protein [10,12]. The two proteins are encoded by adjacent genes (respectively, called SRK and SCR [or SP11]). The amino acid sequences of both genes are highly diverged when alleles of plants with different incompatibility types are compared [10,13-15]. Pollen incompatibility types are controlled by the diploid genotype of the donor plant (sporophytic control), and in addition to the advances in understanding the mechanism of SI, the control of the incompatibility type expressed by pollen is now understood [16,17].
Breakdown of SI is almost as interesting as its mechanism and maintenance, and a recent study advances the understanding of the evolution of self-compatibility in Arabidopsis thaliana [18]. Both the pollen and pistil genes are still present in A. thaliana [10]. Different A. thaliana individuals have different sequences of these genes [19], recognisably corresponding to sequences of types known in A. lyrata [20], so self-compatibility clearly did not evolve simply by a loss-of-function mutation (in one S gene) that spread throughout the species (as this would have led to sequence uniformity in the species). However, the new paper [18] found that 95% of European A. thaliana have one sequence type, A, in which SCR is disrupted by a 213-base pair inversion without actual loss of any of its sequence (relative to the intact version in the rarer sequence type, C, and the corresponding A. halleri sequence). Transgenic tests showed that stigmas of A. thaliana plants with intact type A SRK sequences (found in several A strains) specifically reject pollen of A. halleri plants carrying this type of SCR allele; that is, the kinases of these strains probably function normally. Furthermore, transgenic A. thaliana plants with such type A alleles acquire self-compatibility in experiments when the SCR inversion was re-inverted, suggesting that this is their sole defect.
These results greatly clarify the loss of SI in this plant. European A. thaliana clearly evolved via a mutation inactivating SCR function, and the mutations inactivating most type A SRK sequences probably occurred later and spread in populations after the SRK gene had become functionless in the absence of active pollen ligand. A. thaliana plants from elsewhere most likely acquired self-compatibility through independent mutations, probably also affecting S-locus genes [21]. Plants with type B alleles have several mutations and rearrangements in the S-locus region, but it is difficult to determine which was the initial one [21]. Satisfyingly, loss of SI through a pollen mutation is predicted to be the commonest type of breakdown [22]. However, more cases of breakdown of SI need to be studied before it will be clear whether there is really a general trend for pollen mutations to be the cause.
Heterostyled (heteromomorphic) systems
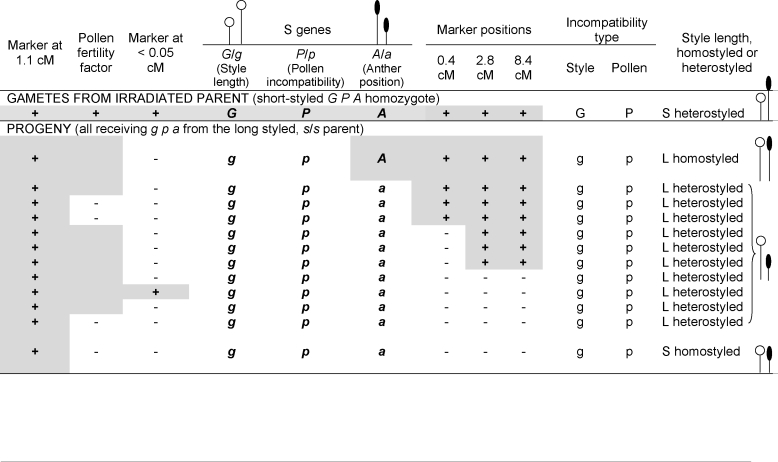
Like homomorphic systems, distyly is thought to be controlled by tightly linked incompatibility genes, but unlike homomorphic systems, several genes are believed to control different aspects of the flower morphology as well as the incompatibility type. This ‘supergene’ hypothesis for the distyly S-locus is now supported by deletion mutants in plants of Turnera subulata [23]. The new work used markers in loci previously shown to be closely genetically linked to the S-locus. The parent plants were chosen so that mutants with loss of the dominant allele at any of the loci causing short-styled flowers could be detected by their flower phenotypes. Among these deletion mutants, most appeared to be normal long-styled (including their incompatibility) and two were ‘homostyled’. All of them had lost markers, and all genotypes except one fit a simple multiple-gene model (Figure 2). Overall, the results reject the possibility, which previously could not be excluded, that alleles at a single heterostyly gene somehow control all three flower characters.
Figure 2. Phenotypic effects of deletions in Turnera subulata, detected using genetic markers.
The presence (+ and grey boxes when the data are observed and grey boxes without text when the presence of a gene is inferred) or absence (-, white boxes) of five genetic markers present in a T. subulata parent plant is shown. Two markers are on the left of the self-incompatibility locus (S-locus), and three are on the right (at the indicated genetic map distances). The parent plant was short-styled and homozygous for the dominant alleles at the S-locus genes (G, P, and A; Figure 1b) [23]. This parent plant was irradiated to produce deletions in pollen gametes and crossed with a long-styled plant (carrying only the recessive alleles at the S-locus: genotype gpa) that also lacked the markers. Deletions detected as changed flower phenotypes in some progeny of the irradiated parent were checked for loss of one or more markers (to ensure that the changed flower phenotype is due to a change in the S-locus region). The incompatibility reactions of the deletion progeny are also listed. Stigma incompatibility types were tested by pollination tests with pollen from long-styled and short-styled plants (loss of the G gene should change the stigma reaction from the short-styled type to the long-styled type, rejecting pollen from long-styled donors and accepting pollen from short-styled ones – denoted in the figure by ‘g’). Similarly, loss of the P gene was tested on stigmas of both style types, by using the pollen of the eight non-sterile progeny. The results overall show that the A gene must lie to one side of G and P. One short homostyled plant (at the bottom of the figure) does not fit the inference from the markers, assuming a single deletion. This phenotype requires changing the anther position from the high position of the short-styled parent (i.e., an A → a deletion) and also its pollen incompatibility type to accept pollen from long-styled donors (i.e., a deletion changing P → p). In other words, its S-locus genotype under the three-gene model should be Gpa, requiring deletions on both sides of the G gene.
Future directions
As candidate pollen as well as pistil genes are discovered in more species, it seems increasingly likely that SI always involves recognition reactions between proteins encoded by different genes (‘lock-and-key systems’, including fungal incompatibility systems involving pheromones and receptors [24,25], which are not homologous to any known plant S-gene products). To date, the very different SI systems in all of the unrelated flowering plant taxa in which the molecular basis of SI is understood (Solanaceae, Papaveraceae, Rosaceae, and Antirrhinum, all with gametophytic control of pollen incompatibility, and Brassicaceae with a sporophytic system) have such systems with two or more genes. However, many self-incompatible plants remain unstudied at the molecular level, and single-gene systems may yet be found. With recently developed methods for high-throughput genetic mapping [26], it may soon become possible to discover the S-locus regions in plants in other taxa with gametophytic systems, such as Oenothera organensis [7] and in grasses, which have two incompatibility loci, both as yet undiscovered (e.g., [27,28]).
Genetic maps should lead to tests of whether the S-locus regions have suppressed recombination (as expected if alleles at two or more loci must be kept in the correct, incompatible combinations). Molecular evolutionary studies to estimate recombination rates in S genes and other nearby genes should allow estimates of how many loci are included in any non-recombining region. It should also be possible to discover whether suppressed recombination has evolved around these genes in a chromosome region where recombination occurs versus S-locus regions evolving in regions where recombination is suppressed (e.g., the large pericentromere regions of many plants); the Hordeum bulbosum S-locus appears to be in a pericentromere region, as was previously found in some other plants [28].
If large regions, containing many genes, are maintained with only a subset of all possible combinations of alleles, the S-locus regions may resemble sex-determining regions of sex chromosomes, such as Y chromosomes, most of which do not recombine with the X, and this has resulted in Y-linked genes becoming fixed for sequence variants (leading to sequence divergence from their X-linked homologues) and also for mutant alleles. In the distylous system of T. subulata, most of the deletions identified were transmitted poorly to progeny when used as male parents in crosses, suggesting that they cause defects in pollen function and hinting that the region may contain many genes and may thus resemble a sex-determining region. It may soon be possible to get direct evidence on this point in T. subulata and also in primroses (Primula vulgaris; Figure 1b), the classical species for studying distyly [29-31].
However, discovering an S-locus region may not allow the component genes to be identified. If they are located within a large non-recombining region, it will be difficult to determine which of the genes in S-locus regions are the S genes [28]. As mentioned above, analysis of nucleotide diversity is necessary to complement molecular genetic studies. Using the predicted high number of sequence differences between different S alleles [11] will, however, identify the recognition genes only in recombining genome regions. Without recombination, the whole region will have high inter-allele sequence divergence if the alleles have been maintained polymorphic for a long time, just as non-recombining Y- and X-linked alleles are highly diverged, and the differences will include synonymous as well as amino acid differences [14]. Recombination rate estimates may help illuminate the puzzlingly low sequence divergence between the putative pollen gene alleles in Antirrhinum and the presence of multiple highly variable putative pollen gene copies in apple and pear species [11].
The generation of new S alleles in homomorphic systems remains a mystery. Currently, ideas for how new functional combinations of pollen and pistil alleles might be generated in a species are theoretical [3,22], and the empirical results still shed little light on this puzzle.
Abbreviations
- S-locus
self-incompatibility locus
- SCR
Sex combs reduced
- SI
self-incompatibility
- SP11
S-locus protein 11
- SRK
S-locus receptor kinase
Competing Interests
The author declares that she has no competing interests.
The electronic version of this article is the complete one and can be found at: http://f1000.com/reports/b/2/68
References
- 1.Barrett SCH. Heterostylous genetic polymorphisms: model systems for evolutionary analysis. In: Barrett SCH, editor. Evolution and Function of Heterostyly. Heidelberg, Germany: Springer-Verlag; 1992. pp. 1–29. [Google Scholar]
- 2.Boggs N, Dwyer K, Nasrallah ME, Nasrallah JB. In vivo detection of residues required for ligand-selective activation of the S-locus receptor in Arabidopsis. Curr Biol. 2009;19:786–91. doi: 10.1016/j.cub.2009.03.037. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Chookajorn T, Kachroo A, Ripoll DR, Clark AG, Nasrallah JB. Specificity determinants and diversification of the Brassica self-incompatibility pollen ligand. Proc Natl Acad Sci U S A. 2003;101:911–7. doi: 10.1073/pnas.2637116100. [DOI] [PMC free article] [PubMed] [Google Scholar]; F1000 Factor 6.0 Must ReadEvaluated by Thierry Gaude 15 Jan 2004
- 4.Hua ZH, Fields A, Kao TH. Biochemical models for S-RNase-based self-incompatibility. Mol Plant. 2008;1:575–85. doi: 10.1093/mp/ssn032. [DOI] [PubMed] [Google Scholar]
- 5.Haasen K, Goring D. The recognition and rejection of self-incompatible pollen in the Brassicaceae. Bot Stud. 2010;51:1–6. [Google Scholar]
- 6.East EM, Mangelsdorf AJ. A new interpretation of the hereditary behaviour of self-sterile plants. Proc Natl Acad Sci U S A. 1925;11:166–71. doi: 10.1073/pnas.11.2.166. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Emerson S. A preliminary survey of the Oenothera organensis population. Genetics. 1939;24:524–37. doi: 10.1093/genetics/24.4.524. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Foote HCC, Ride JP, Franklin-Tong VE, Walker EA, Lawrence MJ, Franklin FCH. Cloning and expression of a distinctive class of self-incompatibility (S) gene from Papaver rhoeas L. Proc Natl Acad Sci U S A. 1994;91:2265–9. doi: 10.1073/pnas.91.6.2265. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Wheeler MJ, de Graaf BH, Hadjiosif N, Perry RM, Poulter NS, Osman K, Vatovec S, Harper A, Franklin FC, Franklin-Tong VE. Identification of the pollen self-incompatibility determinant in Papaver rhoeas. Nature. 2009;459:992–5. doi: 10.1038/nature08027. [DOI] [PMC free article] [PubMed] [Google Scholar]; F1000 Factor 4.8 Must ReadEvaluated by David Smyth 03 Jul 2009, Deborah Charlesworth 09 Jul 2009
- 10.Kusaba M, Dwyer K, Hendershot J, Vrebalov J, Nasrallah JB, Nasrallah ME. Self-incompatibility in the genus Arabidopsis: characterization of the S locus in the outcrossing A. lyrata and its autogamous relative, A. thaliana. Plant Cell. 2001;13:627–43. doi: 10.1105/tpc.13.3.627. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Newbigin E, Paape T, Kohn JR. RNase-based self-incompatibility: puzzled by pollen S. Plant Cell. 2008;20:2286–92. doi: 10.1105/tpc.108.060327. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Takayama S, Shiba H, Iwano M, Shimosato H, Che F-S, Kai N, Suzuki G, Hinata K, Isogai A. The pollen determinant of self-incompatibility in Brassica campestris. Proc Natl Acad Sci U S A. 2000;97:1920–5. doi: 10.1073/pnas.040556397. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Watanabe M, Ito A, Takada Y, Ninomiya C, Kakizaki T, Takahata Y, Hatakeyama K, Hinata K, Suzuki G, Takasaki T, Satta Y, Shiba H, Takayama S, Isogai A. Highly divergent sequences of the pollen self-incompatibility (S) gene in class-I S haplotypes of Brassica campestris (syn. rapa) L. FEBS Lett. 2000;473:139–44. doi: 10.1016/S0014-5793(00)01514-3. [DOI] [PubMed] [Google Scholar]
- 14.Charlesworth D, Bartolomé C, Schierup MH, Mable BK. Haplotype structure of the stigmatic self-incompatibility gene in natural populations of Arabidopsis lyrata. Mol Biol Evol. 2003;20:1741–53. doi: 10.1093/molbev/msg170. [DOI] [PubMed] [Google Scholar]
- 15.Sato T, Nishio T, Kimura R, Kusaba M, Suzuki G, Hatakeyama K, Ockendon D, Satta Y. Coevolution of the S-locus genes SRK, SLG and SP11/SCR in Brassica oleracea and B. rapa. Genetics. 2002;162:931–40. doi: 10.1093/genetics/162.2.931. [DOI] [PMC free article] [PubMed] [Google Scholar]; F1000 Factor 3.0 RecommendedEvaluated by Peter Langridge 27 May 2003
- 16.Shiba H, Iwano M, Entani T, Ishimoto K, Shimosato H, Che FS, Satta Y, Ito A, Takada Y, Watanabe M, Isogai A, Takayama S. The dominance of alleles controlling self-incompatibility in Brassica pollen is regulated at the RNA level. Plant Cell. 2002;14:491–504. doi: 10.1105/tpc.010378. [DOI] [PMC free article] [PubMed] [Google Scholar]; F1000 Factor 6.0 Must ReadEvaluated by Thierry Gaude 13 Mar 2002
- 17.Kusaba M, Tung CW, Nasrallah ME, Nasrallah JB. Monoallelic expression and dominance interactions in anthers of self-incompatible Arabidopsis lyrata. Plant Physiol. 2002;128:17–20. doi: 10.1104/pp.010790. [DOI] [PMC free article] [PubMed] [Google Scholar]; F1000 Factor 3.0 RecommendedEvaluated by Deborah Charlesworth 06 Feb 2002
- 18.Tsuchimatsu T, Suwabe K, Shimizu-Inatsugi R, Isokawa S, Pavlidis P, Stadler T, Suzuki G, Takayama S, Watanabe M, Shimizu KK. Evolution of self-compatibility in Arabidopsis by a mutation in the male specificity gene. Nature. 2010;464:1342–6. doi: 10.1038/nature08927. [DOI] [PubMed] [Google Scholar]
- 19.Nasrallah ME, Liu P, Sherman-Broyles S, Boggs NA, Nasrallah JB. Natural variation in expression of self-incompatibility in Arabidopsis thaliana: implications for the evolution of selfing. Proc Natl Acad Sci U S A. 2004;101:16070–4. doi: 10.1073/pnas.0406970101. [DOI] [PMC free article] [PubMed] [Google Scholar]; F1000 Factor 6.0 Must ReadEvaluated by Deborah Charlesworth 29 Oct 2004
- 20.Bechsgaard JS, Castric V, Charlesworth D, Vekemans X, Schierup MH. The transition to self-compatibility in Arabidopsis thaliana and evolution within S-haplotypes over 10 Myr. Mol Biol Evol. 2006;23:1741–50. doi: 10.1093/molbev/msl042. [DOI] [PubMed] [Google Scholar]
- 21.Boggs N, Nasrallah JB, Nasrallah ME. Independent S-locus mutations caused self-fertility in Arabidopsis thaliana. PLoS Genet. 2009;5:e1000426. doi: 10.1371/journal.pgen.1000426. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Uyenoyama MK, Zhang Y, Newbigin E. On the origin of self-incompatibility haplotypes: transition through self-compatible intermediates. Genetics. 2001;157:1805–17. doi: 10.1093/genetics/157.4.1805. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Labonne J, Tamari F, Shore J. Characterization of X-ray-generated floral mutants carrying deletions at the S-locus of distylous Turnera subulata. Heredity. 2010;105:235–43. doi: 10.1038/hdy.2010.39. [DOI] [PubMed] [Google Scholar]; F1000 Factor 3.0 RecommendedEvaluated by Deborah Charlesworth 10 May 2010
- 24.Casselton LA. Molecular recognition in fungal mating. Endeavour. 1997;21:159–63. doi: 10.1016/S0160-9327(97)01057-0. [DOI] [PubMed] [Google Scholar]
- 25.Casselton LA. Molecular genetics of mating recognition in basidiomycete fungi. Microbiol Mol Biol Rev. 1998;62:55–70. doi: 10.1128/mmbr.62.1.55-70.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Baird NA, Etter PD, Atwood TS, Currey MC, Shiver AL, Lewis ZA, Selker EU, Cresko WA, Johnson EA. Rapid SNP discovery and genetic mapping using sequenced RAD markers. PLoS One. 2008;3:e3376. doi: 10.1371/journal.pone.0003376. [DOI] [PMC free article] [PubMed] [Google Scholar]; F1000 Factor 6.4 Must ReadEvaluated by Tony Long 23 Dec 2008, Matthew Hahn 05 Aug 2009
- 27.Shinozuka H, Cogan N, Smith K, Spangenberg G, Forster J. Fine-scale comparative genetic and physical mapping supports map-based cloning strategies for the self-incompatibility loci of perennial ryegrass (Lolium perenne L.) Plant Mol Biol. 2009;72:343–55. doi: 10.1007/s11103-009-9574-y. [DOI] [PubMed] [Google Scholar]
- 28.Kakeda K, Ibuki T, Suzuki J, Tadano H, Kurita Y, Hanai Y, Kowyama Y. Molecular and genetic characterization of the S locus in Hordeum bulbosum L., a wild self-incompatible species related to cultivated barley. Mol Genet Genomics. 2008;280:509–19. doi: 10.1007/s00438-008-0383-9. [DOI] [PubMed] [Google Scholar]
- 29.Darwin CR. The Different Forms of Flowers on Plants of the Same Species. London: John Murray; 1877. [Google Scholar]
- 30.Haldane JBS. Two new allelomorphs for heterostyly in Primula. Am Nat. 1933;67:559–60. doi: 10.1086/280515. [DOI] [Google Scholar]
- 31.Li J, Webster M, Furuya M, Gilmartin P. Identification and characterization of pin and thrum alleles of two genes that co-segregate with the Primula S locus. Plant J. 2007;51:18–31. doi: 10.1111/j.1365-313X.2007.03125.x. [DOI] [PubMed] [Google Scholar]
- 32.Tomita RN, Suzuki G, Yoshida K, Yano Y, Tsuchiya T, Kakeda K, Mukai Y, Kowyama Y. Molecular characterization of a 313-kb genomic region containing the self-incompatibility locus of Ipomoea trifida, a diploid relative of sweet potato. Breed Sci. 2004;52:165–75. doi: 10.1270/jsbbs.54.165. [DOI] [Google Scholar]