Abstract
p21-activated kinases (PAKs) act downstream of Rho-family GTPase and are linked to steps in both cancer initiation and progression. There are six mammalian PAK isoforms that are divided into two groups, and for different reasons both groups are attractive targets for cancer therapy. We describe the background and recent development of a PAK inhibitor, PF-3758309, which exhibits relatively good selectivity and high potency for PAKs. Experiments using PF-3758309 confirm that inhibiting PAK is a beneficial strategy to combat some tumors, and this activity is likely related to modulation of both cell proliferation and survival. The genetic loss of NF2 (neurofibromatosis type 2) leading to increased cell proliferation through a Ras-Rac-PAK pathway may represent a good test system to analyze this new PAK inhibitor.
Introduction and context
Rho-family GTPases signal through a variety of well-studied ‘effector’ protein kinases (reviewed in [1]). The p21-activated kinases (PAKs) contribute to cell cycle progression and the plasticity of the actin cytoskeleton and thus can contribute to a number of steps of cancer initiation and progression [2]. The mammalian PAK kinases fall into two distinct subgroups based on distinct catalytic and regulatory domain homology (Figure 1). Mammalian PAK1, PAK2, and PAK3 comprise the better-understood group I PAKs that are activated by Rac1 and Cdc42 and can be purified with a Rac1 and the guanine nucleotide exchange factor (GEF) termed PIX (PAK-interacting exchange factor) [3]. In contrast, PAK4, PAK5, and PAK6 (group II) are not activated by Cdc42 binding to the CRIB (Cdc42/Rac interaction and binding) region [4].
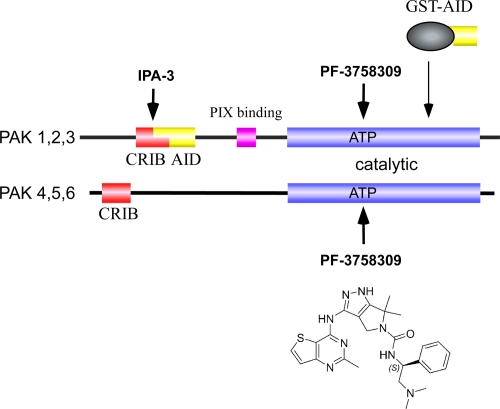
Figure 1. Schematic diagram of the regions targeted by various p21-activated kinase (PAK) inhibitors.
Both group I and group II PAKs contain related catalytic and Cdc42/Rac interaction and binding (CRIB) domains. PAK1, PAK2, and PAK3 are inhibited in trans via an auto-inhibitory domain (AID), which when expressed as a glutathione S-transferase (GST) fusion protein can effectively inhibit the kinase [16]. The PAK1-, PAK2-, and PAK3-binding partner PIX (PAK-interacting exchange factor) interacts via a highly unusual SH3-interacting region [3]. The small-molecule inhibitor IPA-3 (p21-activated kinase inhibitor 3) is a symmetric dimer joined by a disulphide bond [19] which targets the CRIB region. The structure of PF-3758309 is shown for reference; PF-3758309 binds the ATP-binding pocket of PAKs as revealed by the X-ray structure of the PAK4-PF-3758309 complex [21].
PAK1 is highly expressed in the brain and promotes synaptic plasticity [5]. It is notable that both PAK1 and the PIX-associated GIT1 protein [6] co-localize with huntingtin inclusions and that constitutively active PAK1 enhances the huntingtin aggregation [7]. The potential role of PAKs in cancer progression was recently reviewed [8]. The group 1 PAKs may be oncogenic only in specific backgrounds such as with loss of NF2 (neurofibromatosis type 2) [9]. Inhibition of PAK could be beneficial in immune systems in which PAK1-/- macrophages have an attenuated ERK (extracellular signal-regulated kinase) response [10]. There is general evidence for misregulated PAK4 in human tumor cell lines [11] and PAK5 somatic mutations as a cancer ‘driver’ [12]. The PAK4 locus is present on an amplicon associated with colorectal and pancreatic cancers [13,14].
Experiments involving upregulation or downregulation of PAK kinase activity are the basis for most of our current understanding of the PAK kinases. It is therefore unfortunate that a widely used activation loop, phospho-mimetic mutant PAK1 (T423E), is actually not active in vivo [15].
Previous strategies to inhibit PAKs
Aside from siRNA (short interfering RNA)-mediated knockdown of one or multiple PAK isoforms, most credible information regarding PAK function has been gleaned through the use of the PAK1 auto-inhibitory domain (AID), which exhibits a dissociation constant (Ki) of approximately 90 nM toward PAK1 activation in vitro [16]. The glutathione S-transferase (GST) AID does exert effects independent of PAK1 inhibition [17]: some rationale for this off-target effect of PAK1 AID comes from the observation that it avidly binds fragile-X proteins [18]. Fortunately, the PAK2 AID does not have this unwanted side effect as a genetically encoded GST fusion protein (Zhao Z-S, Manser E, unpublished observations). There has been significant interest in development of drugs against PAKs as therapeutic agents, and these would greatly aid in elucidating the biology of PAKs.
Major recent advances
Inhibitor of group I PAKs
The only reported PAK inhibitor for this group in which selectivity has been assessed against a wide spectrum of kinases is IPA-3 (p21-activated kinase inhibitor 3). This small-molecule PAK inhibitor was developed from a screen designed to detect an inhibition of Cdc42-mediated PAK1 activation rather than kinase activity [19]. This allosteric inhibitor of PAK kinase activation does not target the ATP-binding pocket but rather the regulatory interface where Cdc42 or Rac1 binds PAK (Figure 1). The mechanism of action of IPA-3 involves binding and covalent attachment to the PAK1 regulatory domain [20], but this reaction is reversible. Since the compound contains a disulphide bond, it is inactivated inside cells, and this makes it unsuitable for therapeutic use.
Inhibitor of group II PAKs
A recent groundbreaking paper characterizes an orally administrable compound developed by Pfizer Inc. (New York, NY, USA), termed PF-3758309 [21], which exhibits potent binding and inhibition (Ki of ~5-15 nM) of PAKs in an ATP-competitive manner (Figure 1). PF-3758309 was profiled for its growth-inhibitory activity in a panel of 92 tumor cell lines, half of which exhibited IC50 (inhibitory concentration for 50% of maximal effect) values of less than 10 nM. Pathway profiling of this inhibitor has suggested links to apoptosis and the cytoskeleton as expected for PAK4 [22]. In all, this paper gives support to the notion of PAKs as therapeutic targets in cancer. However, PF-3758309 acts on both group I and II PAKs as well as AMPK (AMP-dependent kinase) and RSK (ribosomal S6 kinase) at nanomolar concentrations. The in vivo inhibition of GEF-H1 phosphorylation at Ser-810 indicates PAK4 or PAK1 inhibition (or both) [23]. Measured in vitro, the Ki toward PAK4 is approximately 19 nM whereas the value quoted for PAK2 is 10 times higher (~190 nM) and that for PAK1 is approximately 14 nM. Given that the active sites of the group I PAK kinases are essentially identical, this is unexpected.
Future directions
The data on PF-3758309 as described [21] indicate that this drug is an excellent inhibitor of both group I and II PAKs. The two PAK classes can have somewhat different substrate specificities in vitro [24], although thus far we are not aware of any that are validated in vivo. Comparing the effects of IPA-3 versus PF-3758309 would allow one to differentiate between these targets. With respect to genuine isoform-specific PAK inhibitors, targeting the kinase domain would seem to be problematic. Since PAK1 knockout mice have only minor abnormalities in the central nervous system and immune systems [5,25], compensation in any case between PAK1 and PAK2 is likely in most tissues. The complication with comparing knockout models with inhibitors is that PAKs have kinase-independent activities [26].
At present, PAK-oriented therapy seems obvious for oncology in which PAKs contribute to progression and persistence of the disease; the xenograft models in mice suggest that PF-3758309 does not impact animal survival over the period of the experiments [21]. The potential of PAK inhibitors such as PF-3758309 for treatment of NF2 patients is quite exciting. NF2 is usually diagnosed in young adults by tinnitus or imbalance and is characterized by bilateral vestibular schwannomas growing on the eighth cranial nerve. The NF2 gene product termed merlin is thought to be activated by PAK or PKA (protein kinase A) phosphorylation at Ser-518. Merlin is proposed to negatively regulate PAK1 [27], and thus primary schwannoma samples derived from NF2 patients have elevated PAK1 activity [9]. IPA-3 blocks inappropriate PAK2 activation and associated PIX-mediated events in schwannoma cells [28].
Acknowledgments
This work was supported by the Agency for Science, Technology and Research (A*STAR, Singapore).
Abbreviations
- AID
auto-inhibitory domain
- GEF
guanine nucleotide exchange factor
- GST
glutathione S-transferase
- IPA-3
p21-activated kinase inhibitor 3
- Ki
dissociation constant of the enzyme-inhibitor complex
- NF2
neurofibromatosis type 2
- PAK
p21-activated kinase
- PIX
PAK-interacting exchange factor
Competing Interests
The authors declare that they have no competing interests. The Small G-Protein Signalling and Kinases (sGSK) Group has no direct affiliation with GlaxoSmithKline (Uxbridge, Middlesex, UK).
The electronic version of this article is the complete one and can be found at: http://f1000.com/reports/b/2/70
References
- 1.Zhao ZS, Manser E. PAK and other Rho-associated kinases--effectors with surprisingly diverse mechanisms of regulation. Biochem J. 2005;386:201–14. doi: 10.1042/BJ20041638. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Arias-Romero LE, Chernoff J. A tale of two Paks. Biol Cell. 2008;100:97–108. doi: 10.1042/BC20070109. [DOI] [PubMed] [Google Scholar]
- 3.Manser E, Loo TH, Koh CG, Zhao ZS, Chen XQ, Tan L, Tan I, Leung T, Lim L. PAK kinases are directly coupled to the PIX family of nucleotide exchange factors. Mol Cell. 1998;1:183–92. doi: 10.1016/S1097-2765(00)80019-2. [DOI] [PubMed] [Google Scholar]
- 4.Wells CM, Jones GE. The emerging importance of group II PAKs. Biochem J. 2010;425:465–73. doi: 10.1042/BJ20091173. [DOI] [PubMed] [Google Scholar]
- 5.Asrar S, Meng Y, Zhou Z, Todorovski Z, Huang WW, Jia Z. Regulation of hippocampal long-term potentiation by p21-activated protein kinase 1 (PAK1) Neuropharmacology. 2009;56:73–80. doi: 10.1016/j.neuropharm.2008.06.055. [DOI] [PubMed] [Google Scholar]
- 6.Goehler H, Lalowski M, Stelzl U, Waelter S, Stroedicke M, Worm U, Droege A, Lindenberg KS, Knoblich M, Haenig C, Herbst M, Suopanki J, Scherzinger E, Abraham C, Bauer B, Hasenbank R, Fritzsche A, Ludewig AH, Büssow K, Coleman SH, Gutekunst CA, Landwehrmeyer BG, Lehrach H, Wanker EE. A protein interaction network links GIT1, an enhancer of huntingtin aggregation, to Huntington's disease. Mol Cell. 2004;15:853–65. doi: 10.1016/j.molcel.2004.09.016. [DOI] [PubMed] [Google Scholar]
- 7.Luo S, Mizuta H, Rubinsztein DC. p21-activated kinase 1 promotes soluble mutant huntingtin self-interaction and enhances toxicity. Hum Mol Genet. 2008;17:895–905. doi: 10.1093/hmg/ddm362. [DOI] [PubMed] [Google Scholar]
- 8.Dummler B, Ohshiro K, Kumar R, Field J. Pak protein kinases and their role in cancer. Cancer Metastasis Rev. 2009;28:51–63. doi: 10.1007/s10555-008-9168-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Yi C, Wilker EW, Yaffe MB, Stemmer-Rachamimov A, Kissil JL. Validation of the p21-activated kinases as targets for inhibition in neurofibromatosis type 2. Cancer Res. 2008;68:7932–7. doi: 10.1158/0008-5472.CAN-08-0866. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Smith SD, Jaffer ZM, Chernoff J, Ridley AJ. PAK1-mediated activation of ERK1/2 regulates lamellipodial dynamics. J Cell Sci. 2008;121:3729–36. doi: 10.1242/jcs.027680. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Callow MG, Clairvoyant F, Zhu S, Schryver B, Whyte DB, Bischoff JR, Jallal B, Smeal T. Requirement for PAK4 in the anchorage-independent growth of human cancer cell lines. J Biol Chem. 2002;277:550–8. doi: 10.1074/jbc.M105732200. [DOI] [PubMed] [Google Scholar]
- 12.Greenman C, Stephens P, Smith R, Dalgliesh GL, Hunter C, Bignell G, Davies H, Teague J, Butler A, Stevens C, Edkins S, O’Meara S, Vastrik I, Schmidt EE, Avis T, Barthorpe S, Bhamra G, Buck G, Choudhury B, Clements J, Cole J, Dicks E, Forbes S, Gray K, Halliday K, Harrison R, Hills K, Hinton J, Jenkinson A, Jones D, et al. Patterns of somatic mutation in human cancer genomes. Nature. 2007;446:153–8. doi: 10.1038/nature05610. [DOI] [PMC free article] [PubMed] [Google Scholar]; F1000 Factor 10.1 ExceptionalEvaluated by Channing Der 14 Mar 2007, Leonardo Pardo 20 Mar 2007, Anthony Means 28 Mar 2007, Michael Cole 19 Apr 2007
- 13.Chen S, Auletta T, Dovirak O, Hutter C, Kuntz K, El-ftesi S, Kendall J, Han H, Von Hoff DD, Ashfaq R, Maitra A, Iacobuzio-Donahue CA, Hruban RH, Lucito R. Copy number alterations in pancreatic cancer identify recurrent PAK4 amplification. Cancer Biol Ther. 2008;7:1793–802. doi: 10.4161/cbt.7.11.6840. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Kimmelman AC, Hezel AF, Aguirre AJ, Zheng H, Paik JH, Ying H, Chu GC, Zhang JX, Sahin E, Yeo G, Ponugoti A, Nabioullin R, Deroo S, Yang S, Wang X, McGrath JP, Protopopova M, Ivanova E, Zhang J, Feng B, Tsao MS, Redston M, Protopopov A, Xiao Y, Futreal PA, Hahn WC, Klimstra DS, Chin L, DePinho RA. Genomic alterations link Rho family of GTPases to the highly invasive phenotype of pancreas cancer. Proc Natl Acad Sci U S A. 2008;105:19372–7. doi: 10.1073/pnas.0809966105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Ng Y-W, Raghunathan D, Chan PM, Baskaran Y, Smith DJ, Lee C-H, Verma C, Manser E. Why an A-loop phospho-mimetic fails to activate PAK1: understanding an inaccessible kinase state by molecular dynamics simulations. Structure. 2010;18:879–90. doi: 10.1016/j.str.2010.04.011. [DOI] [PubMed] [Google Scholar]
- 16.Zhao ZS, Manser E, Chen XQ, Chong C, Leung T, Lim L. A conserved negative regulatory region in alphaPAK: inhibition of PAK kinases reveals their morphological roles downstream of Cdc42 and Rac1. Mol Cell Biol. 1998;18:2153–63. doi: 10.1128/mcb.18.4.2153. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Thullberg M, Gad A, Beeser A, Chernoff J, Stromblad S. The kinase-inhibitory domain of p21-activated kinase 1 (PAK1) inhibits cell cycle progression independent of PAK1 kinase activity. Oncogene. 2007;26:1820–8. doi: 10.1038/sj.onc.1209983. [DOI] [PubMed] [Google Scholar]
- 18.Say E, Tay HG, Zhao ZS, Baskaran Y, Li R, Lim L, Manser E. A functional requirement for PAK1 binding to the KH(2) domain of the fragile X protein-related FXR1. Mol Cell. 2010;38:236–49. doi: 10.1016/j.molcel.2010.04.004. [DOI] [PubMed] [Google Scholar]
- 19.Deacon SW, Beeser A, Fukui JA, Rennefahrt UE, Myers C, Chernoff J, Peterson JR. An isoform-selective, small-molecule inhibitor targets the autoregulatory mechanism of p21-activated kinase. Chem Biol. 2008;15:322–31. doi: 10.1016/j.chembiol.2008.03.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Viaud J, Peterson JR. An allosteric kinase inhibitor binds the p21-activated kinase autoregulatory domain covalently. Mol Cancer Ther. 2009;8:2559–65. doi: 10.1158/1535-7163.MCT-09-0102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Murray BW, Guo C, Piraino J, Westwick JK, Zhang C, Lamerdin J, Dagostino E, Knighton D, Loi CM, Zager M, Kraynov E, Popoff I, Christensen JG, Martinez R, Kephart SE, Marakovits J, Karlicek S, Bergqvist S, Smeal T. Small-molecule p21-activated kinase inhibitor PF-3758309 is a potent inhibitor of oncogenic signaling and tumor growth. Proc Natl Acad Sci U S A. 2010;107:9446–51. doi: 10.1073/pnas.0911863107. [DOI] [PMC free article] [PubMed] [Google Scholar]; F1000 Factor 6.0 Must ReadEvaluated by Ed Manser 03 Jun 2010
- 22.Eswaran J, Soundararajan M, Knapp S. Targeting group II PAKs in cancer and metastasis. Cancer Metastasis Rev. 2009;28:209–17. doi: 10.1007/s10555-008-9181-4. [DOI] [PubMed] [Google Scholar]
- 23.Zenke FT, Krendel M, DerMardirossian C, King CC, Bohl BP, Bokoch GM. p21-activated kinase 1 phosphorylates and regulates 14-3-3 binding to GEF-H1, a microtubule-localized Rho exchange factor. J Biol Chem. 2004;279:18392–400. doi: 10.1074/jbc.M400084200. [DOI] [PubMed] [Google Scholar]
- 24.Rennefahrt UE, Deacon SW, Parker SA, Devarajan K, Beeser A, Chernoff J, Knapp S, Turk BE, Peterson JR. Specificity profiling of Pak kinases allows identification of novel phosphorylation sites. J Biol Chem. 2007;282:15667–78. doi: 10.1074/jbc.M700253200. [DOI] [PubMed] [Google Scholar]
- 25.Allen JD, Jaffer ZM, Park SJ, Burgin S, Hofmann C, Sells MA, Chen S, Derr-Yellin E, Michels EG, McDaniel A, Bessler WK, Ingram DA, Atkinson SJ, Travers JB, Chernoff J, Clapp DW. p21-activated kinase regulates mast cell degranulation via effects on calcium mobilization and cytoskeletal dynamics. Blood. 2009;113:2695–705. doi: 10.1182/blood-2008-06-160861. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Gnesutta N, Minden A. Death receptor-induced activation of initiator caspase 8 is antagonized by serine/threonine kinase PAK4. Mol Cell Biol. 2003;23:7838–48. doi: 10.1128/MCB.23.21.7838-7848.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Xiao GH, Beeser A, Chernoff J, Testa JR. p21-activated kinase links Rac/Cdc42 signaling to merlin. J Biol Chem. 2002;277:883–6. doi: 10.1074/jbc.C100553200. [DOI] [PubMed] [Google Scholar]
- 28.Flaiz C, Chernoff J, Ammoun S, Peterson JR, Hanemann CO. PAK kinase regulates Rac GTPase and is a potential target in human schwannomas. Exp Neurol. 2009;218:137–44. doi: 10.1016/j.expneurol.2009.04.019. [DOI] [PMC free article] [PubMed] [Google Scholar]