Abstract
Intestinal epithelia are maintained by intestinal stem cells (ISCs) that divide to replace dying absorptive and secretory cells that make up this tissue. Lineage labeling studies, both in vertebrates and Drosophila, have revealed the relationships between ISCs and their progeny. In addition, a number of signaling pathways involved in ISC proliferation and differentiation have been identified. Further studies will clarify the signals originating from the ISC niche and determine the processes that control the number and uniform distribution of niches throughout the epithelium.
Introduction and context
The intestine is a tissue that undergoes extensive turnover as epithelial cells shed continuously into the lumen. Although much of what is known about intestinal stem cells (ISCs) has come from studies of the mammalian intestine [1], the recent discovery of Drosophila ISCs [2,3] has revealed a remarkable conservation in the function of ISCs and in the molecular pathways that regulate their self-renewal, division, and differentiation [1]. By building on the strengths of both mammalian and Drosophila systems, investigators can now clarify the precise mechanisms underlying ISC behavior.
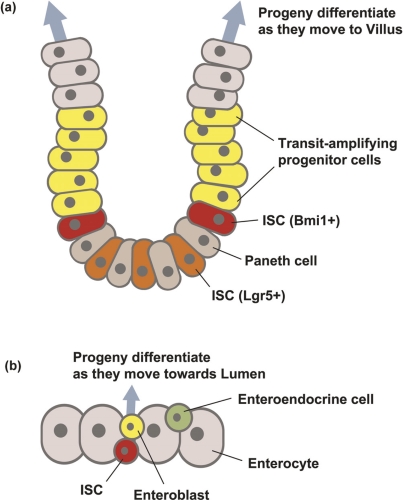
Mammalian ISCs are found within the folds of the monostratified intestinal epithelium, inside regions called crypts, which are located at the base of each intestinal villus [1,4] (Figure 1a). Each mammalian crypt is thought to house several stem cells, and multiple crypts contribute to each villus. A single ISC produces a lineage of cells stretching from the crypt upward into the villus, where it intermingles with adjacent ISC lineages. Two populations of ISCs - a fast-dividing group, marked by the wingless int-1 (Wnt) pathway target gene Lgr5 (leucine-rich repeat-containing G protein-coupled receptor 5), and a slow-dividing group, marked by the DNA remodeling protein Bmi1 (Bmi1 polycomb ring finger oncogene) - are thought to replace cells during homeostasis and regeneration, respectively [4]. Clonal lineage-tracing experiments in mice have shown that all of the different cells of the intestine can be produced by either a single multipotent Lgr5(+) ISC or a Bmi1(+) ISC [5-7]. Ongoing research is addressing the relationships between these fast- and slow-dividing populations in order to understand how these ISCs respond to maintain a constant number of absorptive and secretory intestinal cells in the digestive tract [4].
Figure 1. Intestinal stem cells (ISCs) in mammals and Drosophila.
In these schematics of cross-sections of intestinal epithelia, the ISCs are located basally, adjacent to the surrounding muscle or mesenchyme, and the lumen is located at the top. (a) The mammalian crypt houses both Bmi1+ (red) and Lgr5+ (orange) ISC populations at its base. Clones of cells arising from each ISC intermingle within the epithelium. After ISC division, one daughter will differentiate into a transit-amplifying progenitor (yellow), which will divide as it moves upward from the crypt and toward the villus. Differentiation of the amplified progeny continues as cells leave the crypt to take up residence in the villi. (b) Drosophila ISCs are also located basally (red). An ISC clone will contain all of the cell types present in the gut epithelium. Similar to mammalian ISCs, Drosophila ISCs divide and one daughter undergoes differentiation into an enteroblast (yellow). Unlike mammalian ISCs, transit-amplifying progenitors are not produced and the enteroblast differentiates directly into an enteroendocrine cell (green) or an enterocyte (beige). Bmi1, Bmi1 polycomb ring finger oncogene; Lgr5, leucine-rich repeat-containing G protein-coupled receptor 5.
Lineage-tracing experiments in Drosophila have similarly revealed the presence of clones arising from ISCs [3] (Figure 1b). As in mice, such clones contain all of the different cells of the intestine; however, Drosophila clones are more discrete; only one stem cell is associated with each clone, and these appear to remain more or less separated from one another. This may be due to structural differences as there are no crypts in the Drosophila gut epithelium nor are there villi. Despite these superficial differences, clones from both Drosophila and mice persist for long periods of time, indicating that a self-renewing stem cell is the clone founder. Indeed, the similarity between these systems goes beyond the cellular level because in both organisms ISC proliferation and differentiation are regulated by Wnt and Notch signaling, respectively [1,2,8,9].
Clones containing multiple cell types demonstrate that both Drosophila and mammalian ISCs produce different types of daughter cells. This has been well documented in the fly, in which one ISC founder produces an ISC daughter, which self-renews, and an enteroblast (Eb) daughter, which differentiates directly into an enteroendocrine or enterocyte cell [3]. Thus, ISC clones in Drosophila are thought to contain only one ISC and a mixture of other gut cell types. It is not yet clear whether the division of ISCs is asymmetric, as in dividing neuroblasts [10], or whether it is initially symmetric but then one daughter differentiates according to its position, as occurs in germline stem cells (GSCs) [11]. One candidate protein that determines cell division symmetry, the Notch signaling inhibitor Numb [10], has been tested and does not appear to play a role in ISCs [12]. Further studies will be needed to clarify whether ISCs divide symmetrically or asymmetrically.
The divisions of mammalian ISCs are likely to be similar to those in Drosophila, albeit producing a transit-amplifying progenitor rather than an undividing Eb. Human ISCs have also been shown to undergo monoclonal conversion or niche succession, the expansion of a single ISC to overtake the entire crypt, and fission, the division of one crypt into two - meaning that these cells possess the ability to divide symmetrically to produce two ISC daughters [13,14]. In the mouse, the loss of adenomatous polyposis coli [15], a negative regulator of the Wnt pathway, or PTEN (phosphatase and tensin homolog), a member of the PI3K-Akt (phosphatidylinositol 3-kinase-Akt) signaling pathway that negatively regulates the Wnt pathway in ISCs [16], increases crypt fission. These studies suggest that there are mechanisms whereby mammalian ISCs can undergo expansion by symmetric division and a subsequent duplication of the crypt itself. Although to date there have been no reports that Drosophila ISCs divide symmetrically, it is possible that ISC clone expansion could occur in Drosophila ISCs also or in their Eb progeny.
Major recent advances
In both Drosophila and mammals, ISCs produce a clone of cells containing one or more ISCs, progenitors, and differentiated cells. In both cases, ISCs lie at the origin of what can be thought of as a proliferative unit, which is a region (in the tissue) where the production of differentiated intestinal cells occurs. The even distribution of Drosophila clones or mammalian crypts throughout the gut epithelium suggests that some mechanism determines the uniformity of such proliferative units in this tissue. In addition, it is not clear what governs the localization of mammalian ISCs to the base of crypts [6,7] or the basal localization of Drosophila ISCs in their pseudostratified epithelium [3].
The answer to these questions seems to lie in the concept of a stem cell niche - a locale (within a tissue) where stem cells reside - because it contains the correct concentrations of cell signaling ligands that are taken up by stem cells to activate proliferation and/or inactivate differentiation processes [17]. Without their niches, stem cells would be lost, as has been aptly demonstrated in GSCs [11,18]. In GSCs, the niche is large enough to support only two or three GSCs and this explains both the position and the upper limit of the GSC pool in the germline. Similarly, an understanding of the number and position of ISC proliferative units will emerge from the study of the localization and distribution of niches in the midgut.
A recent report addressed the establishment of ISC clones in the developing midgut. During Drosophila development, adult midgut progenitors (AMPs) originate from the embryonic endoderm, disperse, and divide to expand the midgut during the larval stage [19]. Surrounding visceral muscle cells secrete ligands to activate epidermal growth factor signaling in AMPs, causing them to divide to fill the developing tissue. This close relationship between the gut epithelium and its surrounding muscle niche continues into adulthood, during which the surrounding muscle serves as the source of Wg (wingless) and Upd (unpaired) ligands in order to activate Wnt and JAK/STAT (janus kinase/signal transducer and activator of transcription) signaling in ISCs, respectively [9,20]. The dependence of ISCs on signals emanating from the surrounding muscle cells explains in part why ISCs are located basally in the epithelium.
The relationship of the AMPs to bona fide ISCs is unclear as only a subset of these AMPs becomes ISCs in the adult. A provocative report has shed light on this question by showing that during early development, the first AMP division is asymmetric in order to generate an AMP daughter and a peripheral cell (PC) daughter [21]. This division is crucial because subsequently the PC daughter becomes the niche, whose short-range secretion of bone morphogenetic protein (BMP) ligand is necessary to prevent the AMPs from differentiating. AMPs that lose contact with the PC differentiate, presumably because of loss of BMP signaling, and thus ISCs are formed in an ISC/PC ratio of 1:1. The notion of surrounding cells signaling to stem cells is a common theme. During adulthood, the progeny of ISCs again signal to ISCs, this time through the JAK/STAT pathway during homeostasis [20,22,23], to activate these cells during regeneration [24]. These data reveal the complexity of the ISC niche, where both the surrounding muscle cells and the progeny of ISCs themselves cooperate to maintain or regulate the ISCs at that position.
In mammals, the niche is less well defined, and the Wnt, Notch, and BMP signaling pathway ligands are known to regulate ISCs, from either the surrounding mesenchymal tissue or the adjacent paneth cells in the crypt [17,25]. It was found recently that single isolated ISCs produce crypt-like structures ex situ [26], suggesting that, like the Drosophila AMP-PC niche, cells comprising the mammalian ISC niche can be produced by ISCs themselves. These results may also mean that, unlike Drosophila ISCs, mammalian ISCs do not require signals from surrounding non-gut cells.
Future directions
ISCs and the niche are interrelated: the disruption of signaling in the niche results in rapid differentiation and ISC loss. There are many unanswered questions about the biology of ISCs and their niche:
1. Signals originating from the ISC niche. It is clear that the Wnt, Notch, BMP, and JAK/STAT signaling pathways are involved in the regulation of ISC division and in the differentiation of their progeny. Other pathways may operate in these cells as well and future research will seek to understand the source of these signals and how they are integrated in the ISC to produce a response. In particular, the role of adhesion molecules, such as integrins that play a role in maintaining GSCs, remains to be characterized.
2. Mechanisms underlying the number of ISCs. Why ISC number sometimes increases and results in an expansion of ISC proliferative units complete with their own niches is unclear. Whereas, to date, Drosophila ISCs have not been shown to undergo expansionary symmetric divisions, mammalian ISCs have been; ISCs can overtake the entire crypt niche, and crypts can expand by fission or budding. It will be interesting to see how these duplications involve the ISC niche. For example, does niche expansion occur before or after ISC expansion?
3. Processes that control the number and uniform distribution of ISC niches throughout the epithelium. Although ISCs seem to produce their own niches during development, it is unclear why some AMPs produce PC niche cells but others do not. The means by which ISCs remain associated with their niche cells is also not yet clear; adhesion proteins may be important both in keeping ISCs associated with their niche and perhaps in transducing contact-dependent signals from niche cells to ISCs. It is likely that the contact of ISCs with the surrounding muscle, or mesenchymal tissue, or their progeny underlies the answers to this question.
Just as the niche is intertwined with ISCs, these questions are intertwined with one another. One thing is clear: ISC biologists can now take advantage of both mammalian and Drosophila systems to address these questions.
Acknowledgments
This work was supported by Human Frontier Science Program (PK), the National Institutes of Health, and the Howard Hughes Medical Institute (NP).
Abbreviations
- AMP
adult midgut progenitor
- Bmi1
Bmi1 polycomb ring finger oncogene
- BMP
bone morphogenetic protein
- Eb
enteroblast
- GSC
germline stem cell
- ISC
intestinal stem cell
- JAK/STAT
janus kinase/signal transducer and activator of transcription
- Lgr5
leucine-rich repeat-containing G protein-coupled receptor 5
- PC
peripheral cell
- Wnt
wingless int-1
Competing Interests
The authors declare that they have no competing interests.
The electronic version of this article is the complete one and can be found at: http://f1000.com/reports/b/2/73
References
- 1.Casali A, Batlle E. Intestinal stem cells in mammals and Drosophila. Cell Stem Cell. 2009;4:124–7. doi: 10.1016/j.stem.2009.01.009. [DOI] [PubMed] [Google Scholar]
- 2.Micchelli CA, Perrimon N. Evidence that stem cells reside in the adult Drosophila midgut epithelium. Nature. 2006;439:475–9. doi: 10.1038/nature04371. [DOI] [PubMed] [Google Scholar]; F1000 Factor 3.0 RecommendedEvaluated by Carl Thummel 03 Feb 2006
- 3.Ohlstein B, Spradling A. The adult Drosophila posterior midgut is maintained by pluripotent stem cells. Nature. 2006;439:470–4. doi: 10.1038/nature04333. [DOI] [PubMed] [Google Scholar]; F1000 Factor 6.4 Must ReadEvaluated by Alejandro Sanchez-Alvarado 20 Jan 2006, Carl Thummel 03 Feb 2006
- 4.Li L, Clevers H. Coexistence of quiescent and active adult stem cells in mammals. Science. 2010;327:542–5. doi: 10.1126/science.1180794. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Bjerknes M, Cheng H. Clonal analysis of mouse intestinal epithelial progenitors. Gastroenterology. 1999;116:7–14. doi: 10.1016/S0016-5085(99)70222-2. [DOI] [PubMed] [Google Scholar]
- 6.Barker N, van Es JH, Kuipers J, Kujala P, van den Born M, Cozijnsen M, Haegebarth A, Korving J, Begthel H, Peters PJ, Clevers H. Identification of stem cells in small intestine and colon by marker gene Lgr5. Nature. 2007;449:1003–7. doi: 10.1038/nature06196. [DOI] [PubMed] [Google Scholar]; F1000 Factor 10.9 ExceptionalEvaluated by Salvador Aznar-Benitah 26 Oct 2007, Matthew John Smalley 01 Nov 2007, Maarten van Lohuizen 12 Nov 2007, Amy Wagers 06 Dec 2007, Leanne Jones 21 Dec 2007, Mariann Bienz 10 Jan 2008, John Lazo 06 Feb 2009
- 7.Sangiorgi E, Capecchi MR. Bmi1 is expressed in vivo in intestinal stem cells. Nat Genet. 2008;40:915–20. doi: 10.1038/ng.165. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Ohlstein B, Spradling A. Multipotent Drosophila intestinal stem cells specify daughter cell fates by differential notch signaling. Science. 2007;315:988–92. doi: 10.1126/science.1136606. [DOI] [PubMed] [Google Scholar]; F1000 Factor 4.8 Must ReadEvaluated by Carl Thummel 01 Mar 2007, Stephen Doxsey 12 Jun 2007
- 9.Lin G, Xu N, Xi R. Paracrine Wingless signalling controls self-renewal of Drosophila intestinal stem cells. Nature. 2008;455:1119–23. doi: 10.1038/nature07329. [DOI] [PubMed] [Google Scholar]
- 10.Neumuller RA, Knoblich JA. Dividing cellular asymmetry: asymmetric cell division and its implications for stem cells and cancer. Genes Dev. 2009;23:2675–99. doi: 10.1101/gad.1850809. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Fuller MT, Spradling AC. Male and female Drosophila germline stem cells: two versions of immortality. Science. 2007;316:402–4. doi: 10.1126/science.1140861. [DOI] [PubMed] [Google Scholar]
- 12.Bardin AJ, Perdigoto CN, Southall TD, Brand AH, Schweisguth F. Transcriptional control of stem cell maintenance in the Drosophila intestine. Development. 2010;137:705–14. doi: 10.1242/dev.039404. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.McDonald SA, Preston SL, Greaves LC, Leedham SJ, Lovell MA, Jankowski JA, Turnbull DM, Wright NA. Clonal expansion in the human gut: mitochondrial DNA mutations show us the way. Cell Cycle. 2006;5:808–11. doi: 10.4161/cc.5.8.2641. [DOI] [PubMed] [Google Scholar]
- 14.Gutierrez-Gonzalez L, Deheragoda M, Elia G, Leedham SJ, Shankar A, Imber C, Jankowski JA, Turnbull DM, Novelli M, Wright NA, McDonald SA. Analysis of the clonal architecture of the human small intestinal epithelium establishes a common stem cell for all lineages and reveals a mechanism for the fixation and spread of mutations. J Pathol. 2009;217:489–96. doi: 10.1002/path.2502. [DOI] [PubMed] [Google Scholar]
- 15.Wasan HS, Park HS, Liu KC, Mandir NK, Winnett A, Sasieni P, Bodmer WF, Goodlad RA, Wright NA. APC in the regulation of intestinal crypt fission. J Pathol. 1998;185:246–55. doi: 10.1002/(SICI)1096-9896(199807)185:3 246::AID-PATH90 3.0.CO;2-8. [DOI] [PubMed] [Google Scholar]
- 16.He XC, Yin T, Grindley JC, Tian Q, Sato T, Tao WA, Dirisina R, Porter-Westpfahl KS, Hembree M, Johnson T, Wiedemann LM, Barrett TA, Hood L, Wu H, Li L. PTEN-deficient intestinal stem cells initiate intestinal polyposis. Nat Genet. 2007;39:189–98. doi: 10.1038/ng1928. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Voog J, Jones DL. Stem cells and the niche: a dynamic duo. Cell Stem Cell. 2010;6:103–15. doi: 10.1016/j.stem.2010.01.011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Xie T, Spradling AC. A niche maintaining germ line stem cells in the Drosophila ovary. Science. 2000;290:328–30. doi: 10.1126/science.290.5490.328. [DOI] [PubMed] [Google Scholar]; F1000 Factor 3.0 RecommendedEvaluated by Gordon Fishell 02 Nov 2001
- 19.Jiang H, Edgar BA. EGFR signaling regulates the proliferation of Drosophila adult midgut progenitors. Development. 2009;136:483–93. doi: 10.1242/dev.026955. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Lin G, Xu N, Xi R. Paracrine unpaired signaling through the JAK/STAT pathway controls self-renewal and lineage differentiation of Drosophila intestinal stem cells. J Mol Cell Biol. 2010;2:37–49. doi: 10.1093/jmcb/mjp028. [DOI] [PubMed] [Google Scholar]
- 21.Mathur D, Bost A, Driver I, Ohlstein B. A transient niche regulates the specification of Drosophila intestinal stem cells. Science. 2010;327:210–3. doi: 10.1126/science.1181958. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Beebe K, Lee WC, Micchelli CA. JAK/STAT signaling coordinates stem cell proliferation and multilineage differentiation in the Drosophila intestinal stem cell lineage. Dev Biol. 2010;338:28–37. doi: 10.1016/j.ydbio.2009.10.045. [DOI] [PubMed] [Google Scholar]
- 23.Liu W, Singh SR, Hou SX. JAK-STAT is restrained by Notch to control cell proliferation of the Drosophila intestinal stem cells. J Cell Biochem. 2010;109:992–9. doi: 10.1002/jcb.22482. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Jiang H, Patel PH, Kohlmaier A, Grenley MO, McEwen DG, Edgar BA. Cytokine/Jak/Stat signaling mediates regeneration and homeostasis in the Drosophila midgut. Cell. 2009;137:1343–55. doi: 10.1016/j.cell.2009.05.014. [DOI] [PMC free article] [PubMed] [Google Scholar]; F1000 Factor 8.6 ExceptionalEvaluated by Yukiko Yamashita 02 Jul 2009, Asma Nusrat 17 Jul 2009, Erika Matunis 21 Jul 2009, Ken Irvine 06 Aug 2009, Laurence Rahme 21 Aug 2009
- 25.Greco V, Guo S. Compartmentalized organization: a common and required feature of stem cell niches? Development. 2010;137:1586–94. doi: 10.1242/dev.041103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Sato T, Vries RG, Snippert HJ, van de Wetering M, Barker N, Stange DE, van Es JH, Abo A, Kujala P, Peters PJ, Clevers H. Single Lgr5 stem cells build crypt-villus structures in vitro without a mesenchymal niche. Nature. 2009;459:262–5. doi: 10.1038/nature07935. [DOI] [PubMed] [Google Scholar]; F1000 Factor 8.1 ExceptionalEvaluated by Bruce Morgan 01 Apr 2009, Bill Lowry 03 Apr 2009, Philip Maini 06 Oct 2009