Abstract
Objective
To determine whether maternal/fetal SNPs in candidate genes are associated with preterm prelabor rupture of membranes (pPROM).
Study Design
A case-control study was conducted in patients with pPROM (225 mothers and 155 fetuses) and 599 mothers and 628 fetuses with a normal pregnancy; 190 candidate genes and 775 SNPs were studied. Single locus/haplotype association analyses were performed; FDR was used to correct for multiple testing (q*=0.15)].
Results
1) A SNP in TIMP2 in mothers was significantly associated with pPROM(OR=2.12 95% CI [1.47-3.07], p = 0.000068), and this association remained significant after correction for multiple comparisons; 2) Haplotypes for COL4A3 in the mother were associated with pPROM (global p = 0.003); 3) Multilocus analysis identified a three locus model, which included maternal SNPs in COL1A2, DEFA5, and EDN1.
Conclusion
DNA variants in a maternal gene involved in extracellular matrix metabolism doubled the risk of pPROM.
Keywords: Chorioamnionitis, DNA variants, extracellular matrix, genetic association study, genomics, genotype, haplotype, high dimensional biology, MMP, parturition, pPROM, prematurity, SNP
INTRODUCTION
Preterm prelabor rupture of membranes (pPROM) complicates approximately 3 to 4.5% of all pregnancies in the US and it is responsible for about 30% of preterm births.1-15 A genetic predisposition to preterm birth has been suggested16-18 based upon: 1) demonstration of familial aggregation;19-26 2) substantiation with segregation studies;27 3) identification of disease-susceptibility genes;28-30 and 4) racial disparity in rates of pPROM and preterm birth.16;31-54
Genetic factors are known to predispose to pPROM. First, patients with Ehlers-Danlos Syndrome, a rare Mendelian connective tissue disorder with mutations either in collagen genes or genes involved in collagen processing, have a substantial genetic predisposition to preterm delivery preceded by spontaneous rupture of the membranes.55;56 Although Ehlers-Danlos Syndrome is a Mendelian disorder and pPROM is not, the shared aspects of the phenotypes are indicative of related and perhaps common etiology, i.e., genetic predisposition. Further supporting a genetic role with fetal effect is that pregnant women without Ehlers-Danlos Syndrome but with an Ehlers-Danlos Syndrome fetus, present with pPROM more than twice as often (50%) than in affected women (20%) with or without an affected fetus.56 Similarly, patients with Marfan syndrome, a disorder involving mutations of the fibrillin-1 gene leading to abnormalities of collagen structure and hyaluronic acid synthesis, have a 6% rate of pPROM,57 which is higher than the general population, supporting the view that a genetic factor that predisposes to Marfan plays a role in the risk for pPROM.58 These two syndromes only explain a small fraction of pPROM risk, but their existence demonstrates the principle of a genetic role in pPROM.
Polymorphisms in several genes have been studied in pPROM.59-100 Some genes include matrix metalloproteinase genes (MMP1, MMP8 and MMP9) and SERPINH1 [heat-shock protein 47 (Hsp47)], all of which affect extracellular matrix protein degradation in fetal membranes. Variants in these genes could be associated with membrane weakening and rupture, although a direct functional link has yet to be established. Functional studies have also demonstrated a role for some of the variants in the expression and activity of these molecules involved in extracellular matrix metabolism.61-63;76;86;91;92;94
The objective of this genetic association study was to determine if either maternal or fetal carriage of DNA variants predispose to pPROM. Seven hundred seventy five single nucleotide polymorphisms (SNPs) from 190 candidate genes that have been implicated in the mechanisms of disease responsible for spontaneous preterm labor, pPROM, small-for-gestational age (SGA), and preeclampsia, were analyzed. The study was conducted in a Hispanic population at a single site from Chile and with extreme care to phenotypic characterization.
MATERIALS AND METHODS
Study Design
This was a case-control study that included patients with pPROM and their neonates (mothers: 225 and fetuses: 155) who delivered preterm (21-36 weeks of gestation) as well as controls and their neonates (mothers: 599 and fetuses: 628). A patient was considered to have preterm PROM if she met the following criteria: 1) gestational age below 37 weeks; 2) a history of leaking of fluid reported by the mother; 3) sterile speculum examination demonstrating pooling of fluid and a nitrazine test which was positive; 4) a ferning test was considered confirmatory but not necessary for the diagnosis of preterm PROM; and 5) the term “prelabor” rupture of membranes was used in our manuscript to indicate that the leaking of fluid was required to have occurred at least one hour prior to the onset of regular contractions. These criteria have been used by other investigators.5 The control group included women who delivered a neonate of appropriate weight for gestational age101 at term (37–42 weeks of gestation) without complications of pregnancy including preterm labor with term delivery, preeclampsia, eclampsia, HELLP syndrome, term PROM, SGA, large-for-gestational age neonates, fetal demise, placental abruption, placenta previa, or chorioamnionitis. Clinical chorioamnionitis was diagnosed according to the criteria proposed by Gibbs et al.102 including maternal temperature of ≥37.8°C and two or more of the following criteria: uterine tenderness, malodorous vaginal discharge, maternal leukocytosis (≥15000 cells/mm3), maternal tachycardia (>100 beats/min) and fetal tachycardia (>160 beats/min).
Patients of Hispanic origin were recruited at the Sotero del Rio Hospital, in Puente Alto, Chile. All eligible mothers were enrolled in a research protocol, which requested permission to collect DNA from the mother and her neonate for research purposes. The exclusion criteria, beside those explained above for controls, included: 1) known major fetal chromosomal and/or structural anomalies; 2) multiple pregnancy; 3) serious medical illness (chronic renal failure, congestive heart failure, connective tissue disorders, etc.); 4) refusal to provide written informed consent; and 5) a clinical emergency, which prevented counseling of the patient about participation in the study, such as fetal distress or maternal hemorrhage. A blood sample was obtained from the mother at the time of enrollment in the protocol, and from the umbilical cord (fetal blood) after delivery. Demographic and clinical characteristics of the mothers were obtained from a data collection form administered by trained medical and paramedical personnel. The collection of samples and their utilization for research purposes was approved by the Institutional Review Boards of the Sotero del Rio Hospital, Santiago, Chile (an affiliate of the Pontificia Catholic University of Santiago, Chile), and the Eunice Kennedy Shriver National Institute of Child Health and Human Development, NIH, DHHS.
Genotyping
Candidate genes were selected for analysis based on biological plausibility for a role in pPROM and other pregnancy complications including spontaneous preterm labor with intact membranes, SGA, and preeclampsia. Genes involved in processes such as the control of the immune response (pattern recognition receptors, cytokines, chemokines and their respective receptors), uteroplacental ischemia, or angiogenesis were considered appropriate candidates for this study. A complete list of the 190 genes and all SNPs genotyped are included in the supplemental materials (Supplemental Table 1).
SNP discovery within the candidate genes was performed by DNA sequencing at Genaissance Pharmaceuticals, Inc. (New Haven, CT, USA) using its Index Repository that includes a total of 93 subjects with Native American, Hispanic/Latino, European, Asian, and African-American ancestry.103 The protocol for this has been previously described.30 SNPs selected for genotyping were intended to capture at least 90% of the haplotypic diversity of each gene covering variation in the coding regions,104 100 bases at each end of the introns, 1000 bases upstream of the start codon, and 100 bases downstream of the stop codon.
Template DNA for genotyping was obtained by whole-genome amplification105 of genomic DNA106 isolated from blood using an automated DNA isolation protocol (BioRobot 9604, Qiagen, Valencia, CA, USA). Genotyping was carried out using the MassARRAY TM System (Sequenom, Inc., San Diego, CA, USA) at the high-throughput genotyping facility at Genaissance. Each genotyping assay involved PCR amplification from template DNA in a target region defined by specific primers for the respective polymorphic sites, purification of the amplicon, annealing of the indicated extension primer to one strand of the amplicon adjacent to the polymorphic site, extending the primer by one nucleotide using the MassEXTEND TM reaction (Sequenom, Inc., San Diego, CA, USA), and detection of the allele-specific extension product by mass spectrometry.107
Quality Control
SNPs were verified for Mendelian consistency and genotyping efficiency of both SNPs and samples as described elsewhere.30 Briefly, we considered the number of Mendelian inconsistencies between mother and fetus to identify potential relationship errors (e.g. sample mix-ups or mislabeling). In the case of multiple inconsistencies in a given pair, the pair was excluded from further analysis (10 pairs in controls and 5 pairs in cases). Tests for deviations from HWE were performed for mothers and fetuses separately and again separately for diagnostic subgroups. Because it is currently unclear how to unequivocally distinguish between deviations from HWE due to genotyping error, and deviations from HWE due to biological causes, such as location at or near a disease susceptibility locus, we noted SNPs that deviate from HWE, but we did not remove them from the analysis.108-113 If necessary, we could follow-up these observations with additional testing. Therefore, in the case of deviations from HWE we tagged the SNPs but proceeded with the analyses.
Finally, we tested for population stratification in cases and controls using STRUCTURE,114 which indicated that case and control Chilean samples both cluster with HapMap European samples (data not shown).
Statistical analysis
Continuous demographic and clinical characteristics of cases and controls [gestational age, birth weight, maternal age, and body mass index (BMI)] were tested for normality using Shapiro-Wilks test. All measurements deviated significantly from normality; therefore, Mann-Whitney two-sample rank sum tests were used for case-control comparisons. χ2 tests were used to test for differences in parity, Apgar scores at 1 and 5 minutes, smoking, and differences in fetal gender between cases and controls. Stata 10.0 statistical software (StataCorp, College Station, TX, USA) was used for all analyses.
Single locus tests of association
Statistical tests for single locus association and for deviations from HWE were calculated using PLINK software.115 Statistical significance for deviations from HWE in cases and controls was determined using Fisher’s exact test. Single locus tests of association were performed with logistic regression using an additive genotypic model where the minor allele was coded as the risk allele. Standard summary statistics, odds ratios (OR) and confidence intervals (CI) were reported for these tests of association. Prior to performing single locus and haplotype analyses, rare SNPs in our data set (allele frequency less than 0.01) were removed (21 SNPs in maternal samples and 44 in fetal samples) as were redundant SNPs (those in strong linkage disequilibrium (LD). LD based SNP pruning was performed using PLINK software, with a cutoff of r2 = 0.8. Fifty-two maternal SNPs and 59 fetal SNPs were removed because they were in LD with other SNPs in the data set. Of the 775 SNPs that passed quality control, we analyzed 702 maternal and 665 fetal SNPs. We also excluded a small number of X chromosome SNPs (seven total) for fetal data, as neonates included in the study were both male and female, and power is greatly reduced in the male and female samples analyzed separately. These criteria accounted for the difference in the number of SNPs tested in mothers and fetuses.
Multiple testing corrections
A false discovery rate (FDR) correction was performed to adjust for multiple comparisons using a q* of 0.15 in single locus tests of association in maternal and fetal analyses separately.116 The q* indicates the expected proportion of results that are identified as interesting that are actually false. This is in contrast to α (typically set to 0.05), which indicates the probability of obtaining even one false positive result among all tests for which the null hypothesis is rejected. FDR is used to measure global error, that is, the expected number of false rejections of the null hypothesis among the total number of rejections. The critical significance level was calculated by ranking the results by p values and then multiplying this rank by q* divided by the total number of tests using the step-up approach of Benjamini and Hochberg.117 The threshold q* = 0.15 is deliberately generous, for the purposes of discovery, in which false acceptance of the null is more problematic than false rejection.
Haplotype tests of association
Haplotype analyses were performed on genes with at least one significantly associated SNP (p < 0.01) and at least two SNPs in the same gene. Haplotype frequencies, as well as haplotype-based association analyses for pPROM with two- and three-marker sliding windows, were calculated using PLINK software. Only haplotypes that had a frequency of ≥ 0.05 were analyzed, and only SNPs that had less than 5% missing data were used. The strongest associated haplotype windows are reported and those that demonstrated marginal significance with an omnibus test (p ≤ 0.05) were analyzed for haplotype-specific effects. We present the calculation of OR for each haplotype (using the most common haplotype as referent), as well as determination of case and control haplotype frequencies. Standard summary statistics for pairwise LD were calculated using Haploview.118;119 Haplotype blocks were assigned using the confidence interval algorithm created by Gabriel et al.120
Histologic chorioamnionitis analysis
A systematic histologic examination of all placentas available was performed based on diagnostic criteria previously described.121 All statistically significant single locus and haplotype associations were further analyzed for allele and haplotype differences between patients with pPROM with histologic chorioamnionitis alone or with funisitis (n=85) and term controls (delivered >37 weeks without histologic chorioamnionitis or funisitis, n=488). The purpose of these analyses was to further evaluate whether histologic chorioamnionitis was driving the observed associations.
Multi-locus analysis
Exploratory multi-locus analyses were performed using Multifactor Dimensionality Reduction (MDR) to identify interactions among maternal, fetal, and maternal/fetal SNPs. MDR has been previously described by Ritchie et al.122 and is available as open source software at www.epistasis.org. Briefly, MDR is a non-parametric (does not assume any statistical model) and model free (no assumption mode of genetic inheritance) unique tool for identifying gene-gene interactions. MDR collapses all of the genetic data into two categories (high and low risk) by comparing all single locus and all multi-locus combinations, and then categorizing each genotype into either high-risk or low-risk on the basis of the ratio of cases to controls that have that genotype. MDR ultimately selects one genetic model, either single or multi-locus, that most successfully predicts phenotype or disease status. Analyses were performed: 1) separately for maternal and fetal data (tag SNPs only); and 2) combined for available maternal and fetal paired DNA samples. In our case, we analyzed 672 fetal and 702 maternal SNPs for a total of 1374 in the combined analysis. The different number in maternal and fetal samples was due to different QC results for the two, and the fact that we did not want to remove possible interactions among genes in mothers and fetuses. Data were analyzed for two- and three-way interactions with 10-fold cross-validation and average balanced accuracy as the metrics for evaluating a model.123 Several filtering steps and parameters were explored and are described on Supplemental Table 2. The MDR algorithm was implemented with the full array of tag SNPs as well as after filtering, using the Tuned ReliefF (TuRF) approach as described in detail by Moore and White.124 TuRF is a modification of ReliefF. Briefly, ReliefF is a method that estimates the quality of attributes (e.g. SNPs) through a nearest neighbor algorithm that selects neighbors from the same and different classes based on the values of the SNPs (in this case genotypes).125 TuRF is a modification of ReliefF method that systematically removes SNPs that poorly differentiate cases and controls.124 The motivation behind this algorithm is that the ReliefF estimates of the true associating SNPs will improve as the non-associating SNPs are removed from the dataset. In addition, SNPs were filtered based on results of the single SNP analyses and only SNPs that had a marginal p value of ≤ 0.1 were included, or only those with a p value < 0.05 were analyzed separately. Permutation testing with 1,000 permutations was used to determine statistical significance of all MDR models, addressing potential multiple testing issues.
MDR as described above is ideal for a balanced data set where the number of cases and controls are the same or close to the same. However, computational methods have been developed since the initial development of MDR to test for prediction accuracies in an imbalanced data set, such as ours.123 The method, termed balanced accuracy, corrects for imbalanced data by taking an average of the sensitivity and specificity and is defined as the arithmetic mean of sensitivity and specificity. We tested for balanced accuracy in this manuscript.
Bioinformatics Tools
The SNPper (http://snpper.chip.org) database using dbSNP Build 125 was used to determine marker positions (bp), marker function, and identify amino acid changes.
Pathway analysis
To examine whether the SNPs found to be putatively associated with pPROM mapped to different biological networks and disease functions, an exploratory analysis was performed using Ingenuity Pathway Analyses (IPA) (Ingenuity Systems, Inc., Redwood City, CA, USA).126-129 The genes with variants that were associated with pPROM (p<0.05) were entered into IPA analysis and were termed “focus genes.” The IPA measured associations of these molecules with other molecules, their network interactions, and biological functions stored in its knowledge base. The knowledge base is scientist-curated and encompasses relationships between proteins, genes, cells, tissues, xenobiotics, and diseases. Our focus genes served as seeds for the IPA algorithm, which models functional networks by identifying interconnected molecules, including molecules not among the focus genes from the IPA knowledge base. The software illustrates the networks graphically, and calculates a score for each network, which represents the approximate “fit” between the eligible focus molecules and each network. The network score is based on the hypergeometric distribution and is reported as the -log (Fisher’s exact test result). The IPA software was used to calculate the most significant biological processes associated with each network modeled by IPA. The top functions for a network were ascertained in IPA using the right-tailed Fisher’s exact test for over-representation of network molecules in a given process.
RESULTS
Table 1 displays the clinical and demographic characteristics of the study population. Women with pPROM had a lower median gestational age at delivery, a lower birth weight, and different distributions of 1st minute Apgar score, 5th minute Apgar score and BMI. These differences were expected (except BMI) by the design of the study. The cases had more male than female newborns. Analyses were adjusted for potential confounders (BMI and fetal gender) in all single locus tests of association.
Table 1.
Demographic and clinical characteristics of the study population
| Variable | Cases (n = 225*) |
Controls (n = 599*) |
p-value | ||
|---|---|---|---|---|---|
| Median (25th-75th) |
Mean (SD) |
Median (25th-75th) |
Mean (SD) |
||
| Parity (number of previous pregnancies) | 1 [0-2] | 1.39 (1) | 1 [0-1] | 0.91 (1) | <0.0001 |
| Maternal age (years) | 27 [21-34] | 28 (8) | 24 [20-30] | 25 (6) | <0.0001 |
| BMI | 24 [22-27] | 25 (5) | 24 [22-26] | 24 (4) | 0.018 |
| Smoking | 15% | 14% | 0.910 | ||
| Clinical chorioamnionitis | 11% | 0% | - | ||
| Gestational age at delivery (weeks) | 32 [28-34] | 31 (4) | 40 [39-41] | 40 (1) | <0.0001 |
| Birth weight (grams) | 1730 [1200-2200] |
1676 (642) | 3440 [3230-3650] |
3449 (287) | <0.0001 |
| Fetal gender (% male) | 61% | 51% | 0.015 | ||
| 1st Minute Apgar score | 8 [4-9] | 6 (3) | 9 [9-9] | 8 (1) | <0.0001 |
| 5th Minute Apgar score | 9 [7-9] | 7 (3) | 9 [9-9] | 9 (0.3) | <0.0001 |
BMI: body mass index; SD: standard deviation
Maternal samples: 225 cases and 599 controls; fetal samples: 155 cases and 628 controls
Single Locus Tests of Association
Summary information for the SNPs with the most significant associations (p < 0.01) with pPROM in maternal and fetal DNA is provided on Table 2 (Table 3 for unadjusted analyses). There was one significant deviation from HWE in maternal controls at Prostaglandin E receptor 1, subtype EP1 (PTGER1) SNP rs3745459 (p = 9 × 10−5) that also significantly deviated from HWE in cases (p = 4 × 10−5). There were also significant deviations from HWE in controls in the corticotropin-releasing hormone receptor-1 gene (CRHR1), SNP rs28364026 (in fetal samples p = 3 × 10−8; in maternal samples p = 4 × 10−6), but not in cases. Therefore, although these two SNPs were analyzed and were found to associate with pPROM, these results should be interpreted with caution.
Table 2.
Gene summary information strongest associations (p < 0.01)
| Population | Gene Name | Gene Code | rs# | Chromosome | Position (bp) | Function |
|---|---|---|---|---|---|---|
| Maternal | Tissue inhibitor of metalloproteinase 2 | TIMP2 | rs2277698 | 17 | 74378612 | Coding Exon (S101S) |
| Angiogenin, ribonuclease, RNase family 5 | ANG | rs11701 | 14 | 20231893 | Intron | |
| Toll-like receptor 1 | TLR1 | rs3923647 | 4 | 38475934 | Coding Exon (H305L) | |
| Nitric oxide synthase 3 (endothelial cell) | NOS3 | rs3730305 | 7 | 1.5E+08 | Intron | |
| Alpha 3 type IV collagen isoform 5 precursor | COL4A3 | rs1882435 | 2 | 2.28E+08 | Intron | |
| Prostaglandin E receptor 1, subtype EP1 | PTGER1 | rs3745459 | 19 | 14445317 | Coding Exon (A272A) | |
|
| ||||||
| Fetal | Chemokine (C-C motif) receptor 2 | CCR2 | rs3749461 | 3 | 46370317 | Promoter |
| Matrix metalloproteinase 19 isoform rasi-1 | MMP19 | rs1056784 | 12 | 54519580 | Coding Exon (P245S) | |
| Corticotropin releasing hormone receptor 1 | CRHR1 | rs28364026 | 17 | 41268075 | Promoter | |
| Collagen, type IV, alpha 3 | COL4A3 | rs1882435 | 2 | 2.28E+08 | Intron | |
|
CD55 molecule, decay accelerating factor for
complement |
CD55/DAF | rs10746462 | 1 | 2.06E+08 | Intron | |
| Defensin, beta 1 preproprotein | DEFB1 | rs5743418 | 8 | 6722970 | Promoter | |
| Lipase C precursor | LIPC | rs6080 | 15 | 56625225 | Intron | |
| Insulin-like growth factor 1 receptor | IGF1R | rs3743262 | 15 | 97282996 | Coding Exon (T766T) | |
| Plasminogen activator, tissue type isoform 3 | PLAT | rs8178750 | 8 | 42164028 | Intron | |
| Tumor necrosis factor alpha | TNF | rs1800610 | 6 | 31651806 | Intron | |
| Lymphotoxin alpha precursor | LTA | rs1041981 | 6 | 31648763 | Coding Exon (T60N) | |
Table 3.
Single locus tests of association (p < 0.01) unadjusted for covariates
| Population | Gene Code | rs# | Minor Allele |
Minor Allele Frequency |
OR | 95% CI | P-Value | ||
|---|---|---|---|---|---|---|---|---|---|
| Cases | Controls | Lower | Upper | ||||||
|
Maternal (Cases n = 225; Controls n = 599) |
REN | rs8192282 | A | 0.13 | 0.08 | 1.68 | 1.18 | 2.39 | 0.004 |
| COL4A3 | rs1882435 | A | 0.33 | 0.26 | 1.4 | 1.1 | 1.78 | 0.006 | |
| TLR1 | rs3923647 | T | 0.06 | 0.02 | 2.59 | 1.47 | 4.58 | 0.001 | |
| CSPG2 | rs2287926 | A | 0.14 | 0.09 | 1.57 | 1.12 | 2.22 | 0.009 | |
| RNASE4 | rs11701 | G | 0.13 | 0.18 | 0.64 | 0.47 | 0.89 | 0.008 | |
| IGF1R | rs3743262 | T | 0.2 | 0.14 | 1.58 | 1.18 | 2.12 | 0.002 | |
| TIMP2 | rs2277698 | A | 0.13 | 0.07 | 1.88 | 1.32 | 2.69 | 0.0005 | |
|
| |||||||||
|
Fetal (Cases n = 155; Controls n = 628) |
F5 | rs6019 | C | 0.07 | 0.04 | 1.99 | 1.18 | 3.33 | 0.0096 |
| CD55 | rs10746462 | A | 0.31 | 0.24 | 1.5 | 1.13 | 1.99 | 0.005 | |
| COL4A3 | rs1882435 | A | 0.35 | 0.26 | 1.51 | 1.16 | 1.97 | 0.002 | |
| CCR2 | rs3749461 | G | 0.07 | 0.03 | 2.24 | 1.26 | 3.97 | 0.006 | |
| FGF1 | rs34003 | G | 0.3 | 0.4 | 0.67 | 0.51 | 0.88 | 0.004 | |
| IL18BP | rs5743658 | C | 0.05 | 0.02 | 2.59 | 1.32 | 5.09 | 0.006 | |
| MMP19 | rs1056784 | T | 0.02 | 0 | 7.16 | 2.07 | 24.79 | 0.002 | |
| IMP5 | rs283640261 | A | 0.08 | 0.15 | 0.57 | 0.38 | 0.85 | 0.007 | |
The most significant association in maternal DNA, after adjusting for fetal gender and BMI, was at a synonymous coding SNP (S101S) in tissue inhibitor of metalloproteinase 2 (TIMP2) rs2277698 (OR = 2.12 [95% CI 1.47-3.07], p = 6.8 × 10−6) (Table 4). The minor allele frequency of this SNP (A) was 0.13 in cases and 0.07 in controls. The most significant association observed in fetal DNA was in a SNP in the chemokine (C-C motif) receptor 2 (CCR2) promoter region, rs3749461 (OR = 2.62 [95% CI 1.44-4.75], p = 0.002). The minor allele frequency for this SNP (G) was 0.07 in cases and 0.03 in controls. Only the association with maternal SNP rs2277698 remained statistically significant after correction for multiple testing using FDR.
Table 4.
Single locus tests of association (p < 0.01) adjusted for fetal sex and BMI
| Population | Gene Code | rs# | Minor Allele |
Minor Allele Frequency |
OR2 | 95% CI3 | p-value | ||
|---|---|---|---|---|---|---|---|---|---|
| Cases | Controls | Lower | Upper | ||||||
|
Maternal (Cases n = 225; Controls n = 599) |
TIMP2 | rs2277698 | A | 0.13 | 0.07 | 2.12 | 1.47 | 3.07 | 0.000068* |
| ANG | rs11701 | G | 0.13 | 0.18 | 0.58 | 0.41 | 0.83 | 0.003 | |
| TLR1 | rs3923647 | T | 0.06 | 0.02 | 2.40 | 1.30 | 4.43 | 0.005 | |
| NOS3 | rs3730305 | A | 0.07 | 0.04 | 1.91 | 1.21 | 3.01 | 0.005 | |
| COL4A3 | rs1882435 | A | 0.33 | 0.26 | 1.42 | 1.10 | 1.83 | 0.007 | |
| PTGER1 | rs37454591 | T | 0.05 | 0.02 | 2.00 | 1.20 | 3.34 | 0.008 | |
|
| |||||||||
|
Fetal (Cases n = 155; Controls n = 628) |
CCR2 | rs3749461 | G | 0.07 | 0.03 | 2.62 | 1.44 | 4.75 | 0.002 |
| MMP19 | rs1056784 | T | 0.02 | 0.00 | 6.81 | 1.95 | 23.83 | 0.003 | |
| CRHR1 | rs283640261 | A | 0.08 | 0.15 | 0.52 | 0.33 | 0.80 | 0.003 | |
| COL4A3 | rs1882435 | A | 0.35 | 0.26 | 1.51 | 1.14 | 1.99 | 0.004 | |
| CD55/DAF | rs10746462 | A | 0.31 | 0.24 | 1.52 | 1.13 | 2.04 | 0.006 | |
| DEFB1 | rs5743418 | T | 0.04 | 0.02 | 2.84 | 1.33 | 6.07 | 0.007 | |
| LIPC | rs6080 | A | 0.04 | 0.02 | 2.74 | 1.31 | 5.73 | 0.007 | |
| IGF1R | rs3743262 | T | 0.20 | 0.14 | 1.62 | 1.14 | 2.30 | 0.008 | |
| PLAT | rs8178750 | T | 0.05 | 0.09 | 0.42 | 0.22 | 0.80 | 0.008 | |
| TNF | rs1800610 | A | 0.25 | 0.32 | 0.67 | 0.50 | 0.91 | 0.009 | |
| LTA | rs1041981 | A | 0.34 | 0.27 | 1.47 | 1.10 | 1.97 | 0.0097 | |
These SNPs are deviated from HWE.
OR is the Odds Ratio for the additive genotypic model.
95% CI is the 95% confidence interval of the Odds Ratio.
Significant after FDR correction.
Additional SNPs that were associated with pPROM at a p value < 0.05 are presented in Table 5. Although we have not emphasized these findings in the present report, in some instances they represent associations in genes reported that may lend support to previous findings or may be additional SNPs in genes reported with a p value < 0.01. Such findings strengthen the likelihood of an association because it will be based on multiple SNPs for the same gene (e.g. NOS3 in mothers, collagen genes in mothers, and MMP19 in the fetus).
Table 5.
Single SNP associations with pPROM (unadjusted p < 0.05)
| A. Maternal DNA | ||||||
|---|---|---|---|---|---|---|
| Gene | SNP | A1 | OR | L95 | U95 | p |
| TIMP2 | rs2277698 | A | 2.122 | 1.466 | 3.073 | 6.79E-05 |
| ANG | rs11701 | G | 0.5805 | 0.4076 | 0.8268 | 0.002579 |
| TLR1 | rs3923647 | T | 2.399 | 1.301 | 4.426 | 0.005087 |
| NOS3 | rs3730305 | A | 1.909 | 1.211 | 3.01 | 0.005372 |
| COL4A3 | rs1882435 | A | 1.42 | 1.101 | 1.832 | 0.007009 |
| PTGER1 | rs3745459 | T | 2.001 | 1.199 | 3.341 | 0.007988 |
| CSF1 | rs1058885 | C | 1.426 | 1.086 | 1.873 | 0.01072 |
| IL6R | rs8192282 | A | 1.637 | 1.121 | 2.391 | 0.01081 |
| VWF | rs1800377 | A | 0.4161 | 0.2094 | 0.8271 | 0.01236 |
| COL4A4 | rs12475686 | T | 1.361 | 1.066 | 1.738 | 0.01349 |
| IGF1 | rs5742620 | A | 2.361 | 1.194 | 4.667 | 0.01353 |
| REN | rs3730103 | G | 1.87 | 1.128 | 3.101 | 0.01521 |
| CSF1 | rs333970 | C | 1.343 | 1.05 | 1.717 | 0.01877 |
| ACE | rs4311 | T | 1.342 | 1.05 | 1.717 | 0.01899 |
| MMP10 | rs486055 | A | 1.837 | 1.105 | 3.054 | 0.01912 |
| LPL | rs270 | A | 0.6478 | 0.4501 | 0.9323 | 0.01943 |
| IL1A | rs3783550 | C | 1.324 | 1.046 | 1.676 | 0.01946 |
| TNFRSF1B | rs5746051 | G | 0.66 | 0.4654 | 0.936 | 0.01975 |
| IGF1R | rs3743262 | T | 1.447 | 1.058 | 1.978 | 0.02072 |
| NOS2A | rs2779248 | G | 1.359 | 1.045 | 1.768 | 0.02193 |
| CSPG2 | rs2287926 | A | 1.519 | 1.058 | 2.179 | 0.02331 |
| COL1A1 | rs17639446 | G | 1.667 | 1.072 | 2.592 | 0.02331 |
| TNR | rs1385540 | T | 1.367 | 1.023 | 1.827 | 0.03462 |
| F3 | rs610277 | C | 2.15 | 1.057 | 4.376 | 0.03467 |
| IL1A | rs17561 | T | 0.7351 | 0.5516 | 0.9796 | 0.03569 |
| ACE | rs4354 | T | 0.2752 | 0.08158 | 0.9286 | 0.03758 |
| TBXAS1 | GNSC_53711588 | T | 3.073 | 1.048 | 9.012 | 0.04087 |
| MMP1 | rs470132 | T | 1.387 | 1.013 | 1.898 | 0.04103 |
| IFNGR2 | rs9808753 | G | 1.412 | 1.013 | 1.967 | 0.04152 |
| PLAT | rs8178750 | T | 0.6275 | 0.4008 | 0.9824 | 0.04157 |
| MMP10 | rs17860949 | T | 1.471 | 1.014 | 2.132 | 0.04191 |
| FGF4 | rs3740640 | G | 1.636 | 1.017 | 2.633 | 0.0425 |
| LIPC | GNSC_16324977 | A | 1.893 | 1.007 | 3.556 | 0.04736 |
| NOS3 | rs1800782 | T | 1.664 | 1.005 | 2.756 | 0.04792 |
| B. Fetal DNA | ||||||
|---|---|---|---|---|---|---|
| Gene | SNP | A1 | OR | L95 | U95 | p |
| CCR2 | 46370317 | G | 2.62 | 1.444 | 4.753 | 0.001529 |
| MMP19 | 54519580 | T | 6.807 | 1.945 | 23.83 | 0.002696 |
| CRHR1 | 41268075 | A | 0.5161 | 0.3314 | 0.8036 | 0.003411 |
| COL4A3 | 227810996 | A | 1.51 | 1.144 | 1.994 | 0.003652 |
| DAF | 205577171 | A | 1.517 | 1.13 | 2.038 | 0.005597 |
| DEFB1 | 6722970 | T | 2.84 | 1.33 | 6.065 | 0.007014 |
| LIPC | 56625225 | A | 2.739 | 1.309 | 5.732 | 0.00749 |
| IGF1R | 97282996 | T | 1.616 | 1.135 | 2.302 | 0.007757 |
| PLAT | 42164028 | T | 0.4245 | 0.2244 | 0.8029 | 0.008413 |
| TNF | 31651806 | A | 0.6721 | 0.4981 | 0.907 | 0.009364 |
| LTA | 31648763 | A | 1.472 | 1.098 | 1.974 | 0.009728 |
| PROS1 | 95129086 | A | 1.445 | 1.087 | 1.922 | 0.01135 |
| SERPINE1 | 100567623 | T | 4.524 | 1.402 | 14.6 | 0.01157 |
| GNB3 | 6820171 | A | 2.005 | 1.166 | 3.447 | 0.01192 |
| COL4A1 | 109659786 | G | 1.598 | 1.107 | 2.307 | 0.01231 |
| PLAUR | 48851659 | G | 2.075 | 1.166 | 3.693 | 0.01301 |
| COL4A4 | 227681867 | T | 1.415 | 1.076 | 1.862 | 0.01314 |
| FGF1 | 141955251 | G | 0.7177 | 0.5443 | 0.9465 | 0.01879 |
| IGF2R | 160443699 | T | 0.4062 | 0.1912 | 0.8632 | 0.01915 |
| IL5RA | 3093142 | A | 1.53 | 1.07 | 2.188 | 0.01968 |
| COL5A2 | 189683203 | C | 1.422 | 1.056 | 1.915 | 0.02058 |
| TLR2 | 154844859 | C | 1.881 | 1.088 | 3.254 | 0.02377 |
| IL18BP | 71387372 | C | 2.223 | 1.077 | 4.586 | 0.03069 |
| MMP16 | 89409417 | A | 1.521 | 1.036 | 2.235 | 0.03247 |
| MMP19 | 54521518 | T | 0.4382 | 0.2047 | 0.9377 | 0.03354 |
| CSF1 | 110267989 | C | 1.387 | 1.021 | 1.884 | 0.03618 |
| PTGS1 | 124173328 | T | 0.2857 | 0.0868 | 0.9405 | 0.03932 |
| IL12RB1 | 18031384 | T | 1.433 | 1.016 | 2.023 | 0.0406 |
| APOC3 | 116206884 | A | 1.347 | 1.009 | 1.797 | 0.04324 |
| IGF1 | 101398994 | C | 0.5903 | 0.3539 | 0.9845 | 0.04339 |
| AGT | 228912600 | T | 0.6469 | 0.4224 | 0.9906 | 0.04512 |
| FLT1 | 27910114 | A | 0.7273 | 0.5323 | 0.9937 | 0.04551 |
| FN1 | 215951895 | C | 0.7467 | 0.5605 | 0.9947 | 0.04589 |
| F5 | 167808137 | C | 1.744 | 1.006 | 3.024 | 0.04763 |
| VWF | 6043694 | A | 0.4215 | 0.1792 | 0.9916 | 0.04779 |
Haplotype Tests of Association
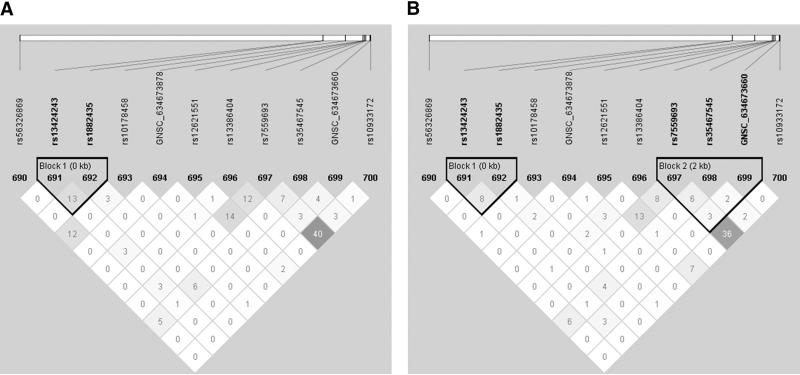
Haplotype analyses of genes with at least one significant SNP (p < 0.05) and two SNPs in the gene identified one gene, Alpha 3 type IV collagen isoform precursor (COL4A3), in maternal DNA samples that was associated with risk for pPROM (Table 6). The haplotype included markers rs1882435-rs10178458-GNSC_634673878 (global p = 0.003). This haplotype had rs1882435, a SNP that was associated with the risk of pPROM (Table 4) (p = 0.007) where the (A) allele is the risk allele. Upon examining the individual haplotypes, it was clear that all statistically significant haplotypes contained the rs1882435 risk allele, although the effect size is greater and the p value is much less for the haplotype than for the single SNP results. Examination of the LD plot for COL4A3 (Figure 1) demonstrated that these three markers were in overall weak LD (r2 ≤ 0.03), further supporting a true haplotype effect.
Table 6.
Haplotype sliding windows association results unadjusted for covariates (p < 0.05)
| Population | Gene Code | SNP rs# | Haplotype | Frequency | OR | 95% CI | p-value | ||
|---|---|---|---|---|---|---|---|---|---|
| Cases | Controls | Lower | Upper | ||||||
| Maternal | COL4A3 | rs1882435-rs10178458-GNSC_634673878 | Global p | 0.003 | |||||
| CCT (Referent) | 0.59 | 0.66 | |||||||
| ACT | 0.32 | 0.23 | 1.55 | 1.20 | 1.99 | 0.0004 | |||
| CTT | 0.09 | 0.11 | 0.92 | 0.62 | 1.36 | 0.680 | |||
Figure 1. Haploview plots of genes identified in analyses of haplotype tests of association in maternal samples.
LD plots were generated in Haploview and are presented for: A) COL4A3 cases r2; and B) COL4A3 controls r2. Within each triangle is presented the pairwise correlation coefficient (r2) LD plots white, (r2 = 0), shades of grey, (0 < r2 < 1), black, (r2 = 1).
Histologic chorioamnionitis
Sub-analyses of all statistically significant single locus and haplotype associations for differences between cases with histologic chorioamnionitis and controls (Tables 7 and 8) demonstrated a decrease in the OR for the maternal SNP in TIMP2, rs2277698, with the OR dropping from 2.12 to 1.22, as well as a loss of statistical significance for maternal samples (Table 7). The OR for CCR2 SNP rs3749461 increased from 2.62 to 3.41 and remained statistically significant (Table 7). The COL4A3 haplotype rs1882435-rs10178458-GNSC_634673878 that associated in maternal samples was not statistically different between cases with histologic chorioamnionitis and controls (Table 8).
Table 7.
Histologic chorioamnionitis analysis of statistically significant associations (p < 0.01, see Table 5)
| Population | Gene Code | rs# | OR | 95% CI | p-value | |
|---|---|---|---|---|---|---|
| Lower | Upper | |||||
| Maternal (Cases n = 78; Controls n = 452) |
COL4A3 | rs1882435 | 1.20 | 0.82 | 1.77 | 0.352 |
| TLR1 | rs3923647 | 2.14 | 0.93 | 4.91 | 0.074 | |
| NOS3 | rs3730305 | 2.54 | 1.42 | 4.52 | 0.002 | |
| ANG | rs11701 | 0.49 | 0.28 | 0.86 | 0.013 | |
| TIMP2 | rs2277698 | 1.22 | 0.67 | 2.23 | 0.517 | |
| PTGER1 | rs3745459 | 1.48 | 0.62 | 3.55 | 0.382 | |
|
|
||||||
| Fetal (Cases n = 57; Controls n = 469) |
CCR2 | rs3749461 | 3.41 | 1.49 | 7.81 | 0.004 |
| CD55/DAF | rs10746462 | 1.90 | 1.24 | 2.93 | 0.003 | |
| COL4A3 | rs1882435 | 1.65 | 1.09 | 2.50 | 0.019 | |
| LTA | rs1041981 | 1.77 | 1.14 | 2.75 | 0.012 | |
| TNF | rs1800610 | 0.79 | 0.50 | 1.24 | 0.304 | |
| DEFB1 | rs5743418 | 1.09 | 0.26 | 4.51 | 0.907 | |
| PLAT | rs8178750 | 0.43 | 0.16 | 1.18 | 0.101 | |
| MMP19 | rs1056784 | 7.67 | 1.47 | 39.98 | 0.016 | |
| LIPC | rs6080 | 2.48 | 0.92 | 6.71 | 0.073 | |
| IGF1R | rs3743262 | 1.47 | 0.87 | 2.50 | 0.153 | |
| CRHR1 | rs28364026 | 0.57 | 0.30 | 1.08 | 0.083 | |
Table 8.
Histologic chorioamnionitis analyses of statistically significant haplotype associations (p<0.05)
| Population | Gene Code | SNP rs# | Haplotype | Frequency | OR | 95% CI | p-value | ||
|---|---|---|---|---|---|---|---|---|---|
| Cases | Controls | Lower | Upper | ||||||
| Maternal | COL4A3 | rs1882435-rs10178458-GNSC_634673878 | Global p | 0.211 | |||||
| CCT (Referent) | 0.63 | 0.66 | - | - | - | - | |||
| ACT | 0.29 | 0.23 | 1.32 | 0.87 | 1.97 | 0.161 | |||
| CTT | 0.08 | 0.11 | 0.74 | 0.36 | 1.41 | 0.349 | |||
MDR analysis
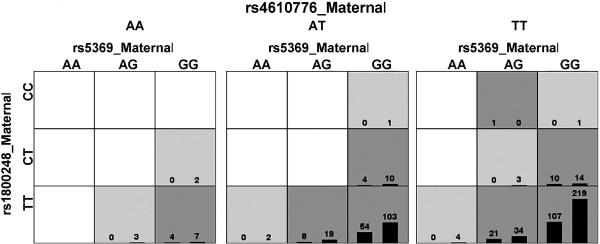
Exploratory MDR analyses were performed using different filtering approaches (Table 9). The only model with a p < 0.05 and a high cross validation consistency (10 of 10) was found in analyses of SNPs filtered using TuRF in the combined fetal and maternal analyses (Table 9C, Figure 2). This model included rs5369_maternal rs1800248_maternal rs4610776_maternal and had a testing balanced accuracy of 0.60 (p = 0.047), with a cross validation consistency of 10/10. The SNP, rs5369, is a synonymous substitution in exon 3 in endothelin 1 (EDN1), the SNP, rs1800248, also encodes a synonymous substitution in exon 47 in collagen type I alpha 2 (COL1A2) and SNP, rs4610776, is 5′ to the transcribed part of the defensin alpha 5 gene (DEFA5).
Table 9.
Summary of MDR analyses
| A. Preterm PROM Maternal with tagged SNPs | ||||
|---|---|---|---|---|
| Model | Training Balance Accuracy |
Testing Balance Accuracy |
Cross Validation Consistency |
p-value |
| All SNPs | ||||
| rs11701 | 0.5700 | 0.5199 | 3/10 | 0.8263 |
| rs2301339 rs5445 | 0.6265 | 0.5839 | 5/10 | 0.1062 |
| rs28763986 rs6083 rs1800774 | 0.6896 | 0.4652 | 1/10 | 0.5703 |
| Turf Option - 10 SNPs | ||||
| rs11701 | 0.5677 | 0.5315 | 6/10 | 0.5779 |
| rs2069849 rs11701 | 0.6103 | 0.5621 | 5/10 | 0.3066 |
| rs1385540 rs454078 rs2301339 | 0.6430 | 0.5743 | 4/10 | 0.2292 |
| Genotypic p-value cutoff - 0.05 | ||||
| rs11701 | 0.5700 | 0.5119 | 3/10 | 0.8263 |
| rs2071307 rs1254600 | 0.6129 | 0.5393 | 3/10 | 0.5234 |
| rs352140 rs2479426 rs2293117 | 0.6673 | 0.5732 | 6/10 | 0.2350 |
| Genotypic p-value cutoff - 0.10 | ||||
| rs11701 | 0.5674 | 0.5350 | 5/10 | 0.5360 |
| rs17876029 rs11701 | 0.6057 | 0.5273 | 2/10 | 0.6554 |
| rs8192282 rs8178610 rs2479426 | 0.6521 | 0.5842 | 4/10 | 0.1513 |
| B. Preterm PROM Fetal with tagged SNPs | ||||
|---|---|---|---|---|
| Model | Training Balance Accuracy |
Testing Balance Accuracy |
Cross Validation Consistency |
p-value |
| All SNPs | ||||
| rs34003 | 0.5848 | 0.5729 | 9/10 | 0.2881 |
| rs2069762 rs2301339 | 0.6410 | 0.5687 | 5/10 | 0.3097 |
| rs2069762 rs1041981 rs2252070 | 0.7042 | 0.5016 | 1/10 | 0.9820 |
| Turf Option - 10 SNPs | ||||
| rs25645 | 0.5639 | 0.5286 | 7/10 | 0.6870 |
| rs11541998 rs25645 | 0.6086 | 0.5270 | 4/10 | 0.7071 |
| rs11764718 rs25645 rs3746190 | 0.6633 | 0.5596 | 6/10 | 0.4020 |
| Genotypic p-value cutoff - 0.05 | ||||
| rs2020920 | 0.5312 | 0.4934 | 4/10 | 0.8692 |
| rs3917727 rs2071538 | 0.5740 | 0.5217 | 8/10 | 0.7487 |
| rs3917727 rs2071538 rs16940668 | 0.6071 | 0.5336 | 10/10 | 0.6173 |
| Genotypic p-value cutoff - 0.10 | ||||
| rs2069762 | 0.5559 | 0.5431 | 10/10 | 0.4278 |
| rs2069762 rs1799962 | 0.5876 | 0.5615 | 4/10 | 0.2881 |
| rs5990 rs2069762 rs2071538 | 0.6441 | 0.5607 | 7/10 | 0.3909 |
| C. Preterm PROM Maternal-Fetal Combined with tagged SNPs | ||||
|---|---|---|---|---|
| Model | Training Balance Accuracy |
Testing Balance Accuracy |
Cross Validation Consistency |
p-value |
| All SNPs | ||||
| rs2071538_2 | 0.6837 | 0.6636 | 4/10 | 0.0002 |
| rs6750027_2 rs549908 | 0.7134 | 0.6586 | 2/10 | 0.0010 |
| GNSC_634673660_2 rs549908 rs1077835_2 | 0.7543 | 0.6817 | 2/10 | 0.0015 |
| Turf Option - 10 SNPs | ||||
| rs5445 | 0.5536 | 0.5521 | 8/10 | 0.2572 |
| rs5369 rs1800248 | 0.5925 | 0.5791 | 10/10 | 0.1061 |
| rs5369 rs1800248 rs4610776 | 0.6211 | 0.5995 | 10/10 | 0.0465 |
| Genotypic p-value cutoff - 0.05 | ||||
| rs5743418_2 | 0.6825 | 0.6363 | 5/10 | 0.0013 |
| rs1058885 rs645114_2 | 0.7088 | 0.6686 | 4/10 | 0.0008 |
| rs1882435_2 rs4311 rs4251883_2 | 0.7408 | 0.6984 | 3/10 | 0.0003 |
| Genotypic p-value cutoff - 0.10 | ||||
| rs5743418_2 | 0.6817 | 0.6434 | 4/10 | 0.0008 |
| rs1058885 rs645114_2 | 0.7088 | 0.6686 | 4/10 | 0.0008 |
| rs1058885 rs1882435_2 rs6909681_2 | 0.7474 | 0.6778 | 4/10 | 0.0012 |
Bold indicates a statistically significant interaction (permutation p < 0.05).
Combined analysis consisted of matching fetal to maternal individuals and adding the SNPs to the analysis (i.e., instead of an individual having 672 SNPs this would increase to 1374). Any result with a “_2” indicates the genotypes were fetal. Any maternal or fetal genotypes without matches were removed from the combined analysis.
Figure 2. MDR results for maternal-fetal analyses.
MDR model for a three-way interaction involving maternal SNPs rs4610776 (DEFA5) and rs5359 (EDN1) and rs1800248 (COL1A2). Each panel represents a three locus genotype; the genotype for each SNP is labeled on the figure. Each large square (3 × 3 box) represents a different genotype for rs4610776 (AA on left, AT in the middle, and TT on the right). Within each square, each row of cells delineates rs1800248 genotypes (top CC, middle CT and bottom TT) and each column the rs35369 genotypes. Therefore, each small cell describes a single and unique three locus genotype. Within each cell are two bars that represent the number of cases with this genotype (left hand bar) and number of controls (right hand bar). Each multilocus cell is denoted as “high risk” (dark gray) or “low risk” (light gray) for spontaneous preterm labor/delivery with intact membranes. Empty cells are shown in white. Risk status is determined by the ratio of cases to controls adjusted by the number of cases and controls studied. The testing average balanced accuracy is 60% (p-value = 0.047) with a cross-validation consistency of 10 out of 10.
Pathway analysis
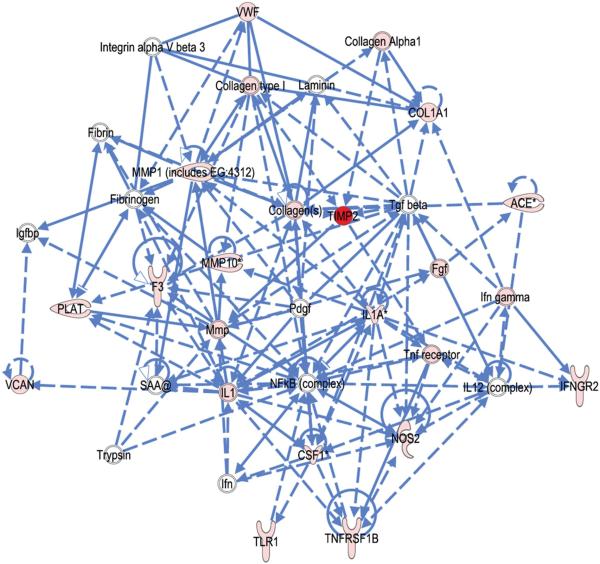
To discover novel networks of interacting molecules, the IPA was seeded with SNPs meeting the criteria of p < 0.05. In mothers, the IPA network algorithm discovered that the focus molecules were significantly interconnected in four networks (scoring 3 to 36, which corresponds to p=10−3 to p=10−36) (Supplemental Table 3A). These networks were joined together by a few molecules, namely TIMP2 among our input genes (in two of four networks) as well as several network partners derived from the IPA database, which included MMPs and a wide representation of extracellular matrix proteins. The top ranked network is illustrated in Figure 3. IPA identified regulatory interactions involving our “focus SNPs” (pink and red in Figure 3) that incorporated other molecules of interest in pPROM. The IPA algorithm identified the top functions of this network as “organismal injury and abnormalities”, “connective tissue disorders” and “inflammatory disease.” Notably, “connective tissue disorders” or “connective tissue development” pathways were identified in three of the four top ranking networks, all of which include multiple inflammatory and extracellular matrix metabolism related genes, supporting the involvement of these genes in pPROM.
Figure 3. Connection map for the first ranked network generated by IPA from maternal focus gene input.
The biomarkers passing the p < 0.05 significance threshold (focus molecules, depicted as pink or red) were entered into the IPA software for an unsupervised functional analysis to discern regulatory networks involving these molecules. The asterisk indicates that there was more than one SNP probe for the gene tested and the most significant value was placed into the analysis. Solid lines show direct interaction (binding/physical contact); dashed line, indirect interaction supported by the literature but possibly involving one or more intermediate molecules that have not been investigated definitively. Molecular interactions involving only binding are connected with a solid line (no arrowhead) since directionality cannot be inferred.
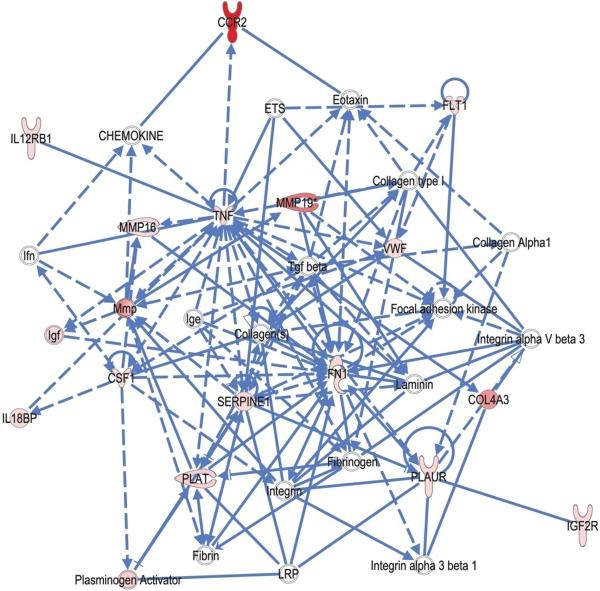
Using the fetal genes as input, the IPA network algorithm discovered that “fetal focus genes” are highly interconnected. It modeled these into four networks (scoring 3 to 35, corresponding to p=10−3 to p=10−35) (Supplemental Table 3B; Figure 4). Similar to the maternal model, the top-ranking network identified “organismal injury and abnormalities” as one of the top disease function which contain collagen type IV, MMPs, pro-inflammatory cytokines/chemokines-related molecules (including TNF alpha, CSF-1, CCR2, IL12 receptor beta 1, IL18 binding protein). Notably, other pathways identified by IPA contained a substantial number of extracellular matrix proteins such as collagens, MMPs, and related molecules (e.g. plasminogen activator). Other top functions included infection mechanisms and cell death that have previously been implicated in pPROM.
Figure 4. Connection map for the first ranked network generated by IPA from fetal focus gene input.
The biomarkers passing the p < 0.05 significance threshold (focus molecules, depicted as pink or red) were entered into the IPA software for an unsupervised functional analysis to discern regulatory networks involving these molecules. The asterisk indicates that there was more than one SNP probe for the gene tested and the most significant value was placed into the analysis. Solid lines show direct interaction (binding/physical contact); dashed line, indirect interaction supported by the literature but possibly involving one or more intermediate molecules that have not been investigated definitively. Molecular interactions involving only binding are connected with a solid line (no arrowhead) since directionality cannot be inferred.
COMMENT
Principal findings of the study
We report the results of a relatively large carefully phenotyped genetic association study of women with pPROM in a homogeneous Hispanic population. This genetic association study of maternal and fetal candidate genes identified DNA variants that predispose to pPROM leading to preterm delivery. The main observations were: 1) A SNP in TIMP2 in mothers was significantly associated with this phenotype; 2) Haplotypes for COL4A3 in the mother were associated with pPROM; 3) Multilocus analysis identified a three locus model, which included maternal SNPs in COL1A2, DEFA5, as well as EDN1; and 4) Pathway analysis suggests that maternal and fetal genes involved in the regulation of extracellular matrix metabolism and inflammation are involved in the biological processes that predispose to pPROM. Taken together, these findings support the hypothesis that genetic variation plays a significant role in predisposition to pPROM, and that this involves DNA variants in genes that participate in the inflammatory response and extra cellular matrix metabolism.
Single locus analysis for mothers
The observed association between TIMP2 and pPROM is novel and lends support to the view that the genetic control of extracellular matrix metabolism is an important factor predisposing to pPROM. This result is consistent with our initial argument regarding the relationship between Ehlers-Danlos Syndrome and risk of pPROM as well as with previously demonstrated imbalance between MMPs and TIMPs in the amniotic fluid of women with pPROM in the presence or absence of intra-amniotic infection.130-134
These observations are consistent with in vitro studies in which microbial products added to fetal membrane explants generated an imbalance between MMPs and TIMPs, tilting the balance towards matrix degradation.135-141 TIMP2 plays an important role in regulating the activities of matrix degrading enzymes. MMP1, MMP8 and MMP9 have been implicated in the mechanisms responsible for membrane rupture. Indeed, the amniotic fluid concentrations of all these enzymes are increased in patients with pPROM (with and without intra-amniotic infection/inflammation).130-134;142-148 Inasmuch as TIMP2 can modulate the activities of MMPs, the association of a DNA variant in TIMP2 with pPROM is of considerable interest. MMP2 is a constitutive enzyme, while MMP9 is inducible.149 Both have been found in amniotic fluid and the concentrations of both zymogen and inhibitor free active forms of MMP9 are elevated in the amniotic fluid of women with pPROM.133;144;150-153 We have previously reported that amniotic fluid TIMP2 concentrations are lower in women with spontaneous labor (term and preterm), with intact or ruptured membranes, regardless of the microbial status of the amniotic cavity, than in women not in labor.133 A decrease in TIMP2 amniotic fluid concentration is thought to favor MMP activity promoting extracellular matrix degradation, which has been associated with labor.152 In a parallel genetic association study of women with preterm labor with intact membranes, we found a significant association between the same TIMP2 SNP and this phenotype (in press). Therefore, there is consistency in the finding of an association between the carriage of this particular DNA variant in TIMP2 and spontaneous preterm labor/delivery, regardless of membrane status. The SNP associated with preterm PROM in TIMP2 is located in an exon; however, there is no evidence at this time that this SNP is functional. In other words, that it changes the protein level.
Haplotype analyses identified novel genes predisposing to pPROM
Maternal haplotype analyses revealed that haplotypes in COL4A3 were associated with pPROM. One particular haplotype (ACT) was associated with a 55% increased risk of pPROM (See Table 6). Collagen IV is a major component of the basement membrane of the amnion, chorion, and the uterine cervix. The degradation of collagen type IV is important for parturition. MMP2 and MMP9 specifically cleave collagen type IV; and TIMP2, where we observed the most significant single locus SNP association with pPROM, is a regulator of the activity for these enzymes. Therefore, the findings of haplotype analysis for collagen IV and the single locus association in the mother supports the relationship between structural proteins of the extracellular matrix (collagen IV) and a regulator of its degradation (TIMP2), lending substantial biological plausibility to both associations.
Histologic chorioamnionitis
The sub-analysis of cases with histologic chorioamnionitis was informative because several associations were either weakened or completely disappeared when compared to the entire data set. This may mean that, in this subset, the major association is not driven by infection but by other biological processes that are independent of this. For example, the major association with TIMP2 in maternal DNA changes from highly significant in the entire data set to not significant in the histologic chorioamnionitis subset. Such changes may reflect variation in gene by environment interactions for this and other genes. Thus, the data are suggestive that most of the associations are not motivated by histologic chorioamnionitis. However, we recognize the need to be cautious in this interpretation because the sample size in the subset was substantially less than in the entire dataset, thereby reducing power.
Multi-locus analyses
Preterm PROM is syndromic in nature,154;155 and multiple mechanisms of disease are likely to be involved.2;156-173 To address the complexity of the genetic predisposition to this phenotype,174;175 we performed exploratory multi-locus analyses using MDR to explicitly address the potential role of interactions among genes (maternal, fetal, and maternal-fetal).176 The results of these analyses indicate that three maternal genes, COL1A2, DEFA5, and EDN1, may interact to modify the risk for pPROM. These genes are involved in collagen metabolism,58;177 susceptibility to bacterial infection178 and uterine contractility,179-181 respectively. Taken together, these findings may support the hypothesis that genetic epistasis between three major components of the common pathway of parturition (uterine contractility, cervical ripening, and membrane rupture) affect risk of pPROM.
Collagen I is a fibrillar protein which, together with type III collagen, are the major structural proteins present in the chorio-amniotic membranes and confer tensile strength to the membranes. Collagen I is also an important structural protein in the uterine cervix,182 and may play a role in the process of cervical remodeling during pregnancy.183;184 DNA variants in the collagen I gene may alter the predisposition to pPROM by altering this structural protein in the reproductive tract; specifically, membranes and the cervix. Cervical insufficiency has been recognized as a cause of pPROM, and this would link maternal collagen I (structure and degradation) with rupture of membranes.
Concentrations of vaginal defensins are elevated in the presence of bacterial vaginosis,185;186 a condition characterized by a gene-environment interaction in the etiology of preterm birth.73;187 We have previously reported that DEFA5 is expressed by endocervical cells,188 and this protein has been found in vaginal fluid, but expression has also been found in the stratified squamous epithelium of the vagina and ectocervix.189 This antimicrobial peptide is also detectable in cervico-vaginal lavage fluid.190 The highest concentrations in this fluid occur during the secretory phase of the menstrual cycle, indicating that it may be under progesterone control.189 Defensin 5 has been implicated in the control of microbial proliferation in the lower genital tract and in preventing ascending intrauterine infection. Therefore, it is possible that DNA variants in this gene may modify the susceptibility to infection, and therefore, pPROM.
Finally, we note that the multi-locus analyses identified a SNP in EDN1 (endothelin 1) as contributing to gene-to-gene interaction predisposing to pPROM. We have previously reported that amniotic fluid EDN1 concentrations are elevated in the presence of intra-amniotic infection.191 This molecule induces smooth muscle contraction and is an uterotonic agent.179-181 Importantly, Margarit et al.192 reported that women destined to develop pPROM have higher amniotic fluid concentrations of EDN1 in the midtrimester than those who do not have pPROM.
An integrated view of these findings is that three components of the common pathway of parturition (uterine contractility, cervical ripening, and membrane rupture) can be modified simultaneously by the genes identified in multi-locus analyses. Moreover, the finding that DNA variants in DEFA5 may also contribute to risk, link alterations in the host defense mechanisms in the lower genital tract and activation of the common pathway of parturition.
Pathway analysis
There is an increasing realization of the importance of pathways in the etiology of complex phenotypes. We utilized IPA to examine the contribution of genetic variants in determining networks and disease functions. As with the results presented above, the findings of IPA support the hypothesis that genes involved in extracellular matrix metabolism and inflammation pathways are associated with pPROM. Such findings are also consistent with a large body of literature supporting this view. Because the IPA knowledgebase has extensive coverage of molecular mechanisms across broad domains of biology and pathology, and it constructs the network models based on > 1 million known molecular interactions, the network partners discovered by our IPA analysis could represent novel pPROM biomarker candidates.
Strengths and limitations of the study
The strengths of our study include a well-defined phenotype (pPROM) and a homogeneous population. This is the largest study to examine the genetic predisposition to pPROM in Hispanics. Moreover, the study includes both maternal and fetal DNA and a relatively large number of genes and DNA variants. The number of DNA variants selected was estimated to cover 90% of the exonic and proximal DNA variation in the candidate genes. Importantly, we identified that maternal DNA variants contributed to modify the risk. Limitations of these types of studies are that confirmation of the findings is required and that we have not examined the effect of environmental factors that are known to play a role in the risk of pPROM. In addition, functional studies are needed to assess the precise physiological implications of the DNA variants identified in this study. Although previous studies have found associations between fetal DNA variants and pPROM, we did not find any significant association in fetal DNA that passed correction for multiple testing. This may represent false negative results. Also, we did not study the identical variants found in previous studies because our genotyping platform was not appropriate for these variants. Finally, the findings observed in this Hispanic population may not be representative of other ethnic groups. Further studies are required to replicate our findings and those of others, as well as to identify if fetal DNA variants may play a significant role in the predisposition to pPROM. Moreover, the role of maternal-fetal interactions and incompatibility needs to be explored. It is possible that differences in DNA variants between the maternal and fetal genome predispose to adverse pregnancy outcome.193-195
Conclusion
This genetic association study of candidate genes involved in adverse pregnancy outcome revealed that maternal DNA variants are associated with pPROM.
Supplementary Material
Acknowledgments
Acknowledgment: This research was supported, in part, by the Perinatology Research Branch, Division of Intramural Research, Eunice Kennedy Shriver National Institute of Child Health and Human Development, NIH, DHHS.
Footnotes
Condensation: A genetic association study identifies DNA variants in the fetus and mother that may predispose to preterm prelabor rupture of membranes.
Reference List
- 1.Garite TJ, Freeman RK, Linzey EM, Braly P. The use of amniocentesis in patients with premature rupture of membranes. Obstet.Gynecol. 1979;54:226–30. [PubMed] [Google Scholar]
- 2.Taylor J, Garite TJ. Premature rupture of membranes before fetal viability. Obstet.Gynecol. 1984;64:615–20. [PubMed] [Google Scholar]
- 3.Gravett MG, Eschenbach DA. Possible role of Ureaplasma urealyticum in preterm premature rupture of the fetal membranes. Pediatr.Infect.Dis. 1986;5:S253–S257. doi: 10.1097/00006454-198611010-00010. [DOI] [PubMed] [Google Scholar]
- 4.Mercer BM, Arheart KL. Antimicrobial therapy in expectant management of preterm premature rupture of the membranes. Lancet. 1995;346:1271–79. doi: 10.1016/s0140-6736(95)91868-x. [DOI] [PubMed] [Google Scholar]
- 5.Mercer BM, Miodovnik M, Thurnau GR, Goldenberg RL, Das AF, Ramsey RD, et al. Antibiotic therapy for reduction of infant morbidity after preterm premature rupture of the membranes. A randomized controlled trial. National Institute of Child Health and Human Development Maternal-Fetal Medicine Units Network. JAMA. 1997;278:989–95. [PubMed] [Google Scholar]
- 6.Parry S, Strauss JF., III Premature rupture of the fetal membranes. N.Engl.J Med. 1998;338:663–70. doi: 10.1056/NEJM199803053381006. [DOI] [PubMed] [Google Scholar]
- 7.Mercer BM, Goldenberg RL, Meis PJ, Moawad AH, Shellhaas C, Das A, et al. The Preterm Prediction Study: prediction of preterm premature rupture of membranes through clinical findings and ancillary testing. The National Institute of Child Health and Human Development Maternal-Fetal Medicine Units Network. Am J Obstet Gynecol. 2000;183:738–45. doi: 10.1067/mob.2000.106766. [DOI] [PubMed] [Google Scholar]
- 8.Garite TJ. Management of premature rupture of membranes. Clin.Perinatol. 2001;28:837–47. doi: 10.1016/s0095-5108(03)00081-2. [DOI] [PubMed] [Google Scholar]
- 9.Kenyon SL, Taylor DJ, Tarnow-Mordi W. Broad-spectrum antibiotics for preterm, prelabour rupture of fetal membranes: the ORACLE I randomised trial. ORACLE Collaborative Group. Lancet. 2001;357:979–88. doi: 10.1016/s0140-6736(00)04233-1. [DOI] [PubMed] [Google Scholar]
- 10.Kenyon S, Taylor DJ, Tarnow-Mordi WO. ORACLE--antibiotics for preterm prelabour rupture of the membranes: short-term and long-term outcomes. Acta Paediatr.Suppl. 2002;91:12–15. doi: 10.1111/j.1651-2227.2002.tb00153.x. [DOI] [PubMed] [Google Scholar]
- 11.Mercer BM. Preterm premature rupture of the membranes: current approaches to evaluation and management. Obstet.Gynecol.Clin.North Am. 2005;32:411–28. doi: 10.1016/j.ogc.2005.03.003. [DOI] [PubMed] [Google Scholar]
- 12.Santolaya-Forgas J, Romero R, Espinoza J, Erez O, Friel AL, Kusanovic JP, et al. Prelabor rupture of membranes. In: Reece EA, Hobbins JC, editors. Clinical Obstetrics: The Fetus and the Mother. Blackwell Publishing; 2007. pp. 1130–88. [Google Scholar]
- 13.Goldenberg RL, Culhane JF, Iams JD, Romero R. Epidemiology and causes of preterm birth. Lancet. 2008;371:75–84. doi: 10.1016/S0140-6736(08)60074-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Kenyon S, Pike K, Jones DR, Brocklehurst P, Marlow N, Salt A, et al. Childhood outcomes after prescription of antibiotics to pregnant women with preterm rupture of the membranes: 7-year follow-up of the ORACLE I trial. Lancet. 2008;372:1310–18. doi: 10.1016/S0140-6736(08)61202-7. [DOI] [PubMed] [Google Scholar]
- 15.Waters TP, Mercer BM. The management of preterm premature rupture of the membranes near the limit of fetal viability. Am J Obstet.Gynecol. 2009;201:230–40. doi: 10.1016/j.ajog.2009.06.049. [DOI] [PubMed] [Google Scholar]
- 16.DeFranco E, Teramo K, Muglia L. Genetic influences on preterm birth. Semin.Reprod.Med. 2007;25:40–51. doi: 10.1055/s-2006-956774. [DOI] [PubMed] [Google Scholar]
- 17.Plunkett J, Muglia LJ. Genetic contributions to preterm birth: implications from epidemiological and genetic association studies. Ann.Med. 2008;40:167–95. doi: 10.1080/07853890701806181. [DOI] [PubMed] [Google Scholar]
- 18.Himes KP, Simhan HN. Genetic susceptibility to infection-mediated preterm birth. Infect.Dis.Clin.North Am. 2008;22:741–53. vii. doi: 10.1016/j.idc.2008.05.004. [DOI] [PubMed] [Google Scholar]
- 19.Porter TF, Fraser AM, Hunter CY, Ward RH, Varner MW. The risk of preterm birth across generations. Obstet.Gynecol. 1997;90:63–67. doi: 10.1016/S0029-7844(97)00215-9. [DOI] [PubMed] [Google Scholar]
- 20.Winkvist A, Mogren I, Hogberg U. Familial patterns in birth characteristics: impact on individual and population risks. Int.J.Epidemiol. 1998;27:248–54. doi: 10.1093/ije/27.2.248. [DOI] [PubMed] [Google Scholar]
- 21.Clausson B, Lichtenstein P, Cnattingius S. Genetic influence on birthweight and gestational length determined by studies in offspring of twins. BJOG. 2000;107:375–81. doi: 10.1111/j.1471-0528.2000.tb13234.x. [DOI] [PubMed] [Google Scholar]
- 22.Treloar SA, Macones GA, Mitchell LE, Martin NG. Genetic influences on premature parturition in an Australian twin sample. Twin.Res. 2000;3:80–82. doi: 10.1375/136905200320565526. [DOI] [PubMed] [Google Scholar]
- 23.Ward K, Argyle V, Meade M, Nelson L. The heritability of preterm delivery. Obstet.Gynecol. 2005;106:1235–39. doi: 10.1097/01.AOG.0000189091.35982.85. [DOI] [PubMed] [Google Scholar]
- 24.Plunkett J, Borecki I, Morgan T, Stamilio D, Muglia LJ. Population-based estimate of sibling risk for preterm birth, preterm premature rupture of membranes, placental abruption and pre-eclampsia. BMC.Genet. 2008;9:44. doi: 10.1186/1471-2156-9-44. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Kistka ZA, DeFranco EA, Ligthart L, Willemsen G, Plunkett J, Muglia LJ, et al. Heritability of parturition timing: an extended twin design analysis. Am J Obstet.Gynecol. 2008;199:43–45. doi: 10.1016/j.ajog.2007.12.014. [DOI] [PubMed] [Google Scholar]
- 26.Boyd HA, Poulsen G, Wohlfahrt J, Murray JC, Feenstra B, Melbye M. Maternal contributions to preterm delivery. Am J Epidemiol. 2009;170:1358–64. doi: 10.1093/aje/kwp324. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Plunkett J, Feitosa MF, Trusgnich M, Wangler MF, Palomar L, Kistka ZA, et al. Mother’s genome or maternally-inherited genes acting in the fetus influence gestational age in familial preterm birth. Hum.Hered. 2009;68:209–19. doi: 10.1159/000224641. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Menon R, Merialdi M, Betran AP, Dolan S, Jiang L, Fortunato SJ, et al. Analysis of association between maternal tumor necrosis factor-alpha promoter polymorphism (−308), tumor necrosis factor concentration, and preterm birth. Am J Obstet.Gynecol. 2006;195:1240–48. doi: 10.1016/j.ajog.2006.05.037. [DOI] [PubMed] [Google Scholar]
- 29.Velez DR, Fortunato SJ, Williams SM, Menon R. Interleukin-6 (IL-6) and receptor (IL6-R) gene haplotypes associate with amniotic fluid protein concentrations in preterm birth. Hum.Mol Genet. 2008;17:1619–30. doi: 10.1093/hmg/ddn049. [DOI] [PubMed] [Google Scholar]
- 30.Romero R, Velez DR, Kusanovic JP, Hassan SS, Mazaki-Tovi S, Vaisbuch E, Kim CJ, Chaiworapongsa T, Pearce B, Friel L, Bartlett J, Anant MK, Salisbury BA, Vovis GF, Lee MS, Gomez R, Behnke E, Oyarzun E, Tromp G, Williams SM, Menon R. Identification of fetal and maternal single nucleotide polymorphisms in candidate genes that predispose to spontaneous preterm labor with intact membranes. Am J Obstet.Gynecol. 2010 doi: 10.1016/j.ajog.2010.03.026. Ref Type: In Press. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Savitz DA, Blackmore CA, Thorp JM. Epidemiologic characteristics of preterm delivery: etiologic heterogeneity. Am J Obstet.Gynecol. 1991;164:467–71. doi: 10.1016/s0002-9378(11)80001-3. [DOI] [PubMed] [Google Scholar]
- 32.Zhang J, Savitz DA. Preterm birth subtypes among blacks and whites. Epidemiology. 1992;3:428–33. doi: 10.1097/00001648-199209000-00008. [DOI] [PubMed] [Google Scholar]
- 33.Blackmore CA, Savitz DA, Edwards LJ, Harlow SD, Bowes WA., Jr. Racial differences in the patterns of preterm delivery in central North Carolina, USA. Paediatr.Perinat.Epidemiol. 1995;9:281–95. doi: 10.1111/j.1365-3016.1995.tb00144.x. [DOI] [PubMed] [Google Scholar]
- 34.Berkowitz GS, Blackmore-Prince C, Lapinski RH, Savitz DA. Risk factors for preterm birth subtypes. Epidemiology. 1998;9:279–85. [PubMed] [Google Scholar]
- 35.Nguyen DP, Genc M, Vardhana S, Babula O, Onderdonk A, Witkin SS. Ethnic differences of polymorphisms in cytokine and innate immune system genes in pregnant women. Obstet.Gynecol. 2004;104:293–300. doi: 10.1097/01.AOG.0000133486.85400.5e. [DOI] [PubMed] [Google Scholar]
- 36.Fiscella K. Race, genes and preterm delivery. J Natl.Med.Assoc. 2005;97:1516–26. [PMC free article] [PubMed] [Google Scholar]
- 37.Menon R, Velez DR, Thorsen P, Vogel I, Jacobsson B, Williams SM, et al. Ethnic differences in key candidate genes for spontaneous preterm birth: TNF-alpha and its receptors. Hum.Hered. 2006;62:107–18. doi: 10.1159/000096301. [DOI] [PubMed] [Google Scholar]
- 38.Menon R, Merialdi M, Lombardi SJ, Fortunato SJ. Differences in the placental membrane cytokine response: a possible explanation for the racial disparity in preterm birth. Am.J.Reprod.Immunol. 2006;56:112–18. doi: 10.1111/j.1600-0897.2006.00394.x. [DOI] [PubMed] [Google Scholar]
- 39.Kistka ZA, Palomar L, Lee KA, Boslaugh SE, Wangler MF, Cole FS, et al. Racial disparity in the frequency of recurrence of preterm birth. Am J Obstet.Gynecol. 2007;196:131–36. doi: 10.1016/j.ajog.2006.06.093. [DOI] [PubMed] [Google Scholar]
- 40.Menon R, Williams SM, Fortunato SJ. Amniotic fluid interleukin-1beta and interleukin-8 concentrations: racial disparity in preterm birth. Reprod.Sci. 2007;14:253–59. doi: 10.1177/1933719107301336. [DOI] [PubMed] [Google Scholar]
- 41.Palomar L, DeFranco EA, Lee KA, Allsworth JE, Muglia LJ. Paternal race is a risk factor for preterm birth. Am J Obstet.Gynecol. 2007;197:152–57. doi: 10.1016/j.ajog.2007.03.035. [DOI] [PubMed] [Google Scholar]
- 42.Velez DR, Menon R, Thorsen P, Jiang L, Simhan H, Morgan N, et al. Ethnic differences in interleukin 6 (IL-6) and IL6 receptor genes in spontaneous preterm birth and effects on amniotic fluid protein levels. Ann.Hum.Genet. 2007;71:586–600. doi: 10.1111/j.1469-1809.2007.00352.x. [DOI] [PubMed] [Google Scholar]
- 43.Fortunato SJ, Menon R, Velez DR, Thorsen P, Williams SM. Racial disparity in maternal-fetal genetic epistasis in spontaneous preterm birth. Am.J.Obstet.Gynecol. 2008;198:666–69. doi: 10.1016/j.ajog.2008.02.003. [DOI] [PubMed] [Google Scholar]
- 44.Menon R, Velez DR, Morgan N, Lombardi SJ, Fortunato SJ, Williams SM. Genetic regulation of amniotic fluid TNF-alpha and soluble TNF receptor concentrations affected by race and preterm birth. Hum.Genet. 2008;124:243–53. doi: 10.1007/s00439-008-0547-z. [DOI] [PubMed] [Google Scholar]
- 45.Shen TT, DeFranco EA, Stamilio DM, Chang JJ, Muglia LJ. A population-based study of race-specific risk for preterm premature rupture of membranes. Am J Obstet.Gynecol. 2008;199:373–77. doi: 10.1016/j.ajog.2008.05.011. [DOI] [PubMed] [Google Scholar]
- 46.Simhan HN, Krohn MA. Paternal race and preterm birth. Am J Obstet.Gynecol. 2008;198:644–46. doi: 10.1016/j.ajog.2007.11.046. [DOI] [PubMed] [Google Scholar]
- 47.Simhan HN, Bodnar LM, Krohn MA. Paternal race and bacterial vaginosis during the first trimester of pregnancy. Am J Obstet.Gynecol. 2008;198:196–4. doi: 10.1016/j.ajog.2007.09.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Ryckman KK, Williams SM, Krohn MA, Simhan HN. Racial differences in cervical cytokine concentrations between pregnant women with and without bacterial vaginosis. J Reprod.Immunol. 2008;78:166–71. doi: 10.1016/j.jri.2008.01.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Alexander GR, Wingate MS, Bader D, Kogan MD. The increasing racial disparity in infant mortality rates: composition and contributors to recent US trends. Am J Obstet.Gynecol. 2008;198:51–59. doi: 10.1016/j.ajog.2007.06.006. [DOI] [PubMed] [Google Scholar]
- 50.Menon R, Camargo MC, Thorsen P, Lombardi SJ, Fortunato SJ. Amniotic fluid interleukin-6 increase is an indicator of spontaneous preterm birth in white but not black Americans. Am J Obstet.Gynecol. 2008;198:77. doi: 10.1016/j.ajog.2007.06.071. [DOI] [PubMed] [Google Scholar]
- 51.Menon R, Thorsen P, Vogel I, Jacobsson B, Morgan N, Jiang L, et al. Racial disparity in amniotic fluid concentrations of tumor necrosis factor (TNF)- alpha and soluble TNF receptors in spontaneous preterm birth. Am J Obstet.Gynecol. 2008;198:533–10. doi: 10.1016/j.ajog.2007.11.025. [DOI] [PubMed] [Google Scholar]
- 52.Menon R, Pearce B, Velez DR, Merialdi M, Williams SM, Fortunato SJ, et al. Racial disparity in pathophysiologic pathways of preterm birth based on genetic variants. Reprod.Biol.Endocrinol. 2009;7:62. doi: 10.1186/1477-7827-7-62. 62. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Velez DR, Fortunato S, Thorsen P, Lombardi SJ, Williams SM, Menon R. Spontaneous preterm birth in African Americans is associated with infection and inflammatory response gene variants. Am J Obstet Gynecol. 2009;200:209–27. doi: 10.1016/j.ajog.2008.08.051. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Misra DP, Caldwell C, Young AA, Jr., Abelson S. Do fathers matter? Paternal contributions to birth outcomes and racial disparities. Am J Obstet.Gynecol. 2010;202:99–100. doi: 10.1016/j.ajog.2009.11.031. [DOI] [PubMed] [Google Scholar]
- 55.Barabas AP. Ehlers-Danlos syndrome associated with prematurity and premature rupture of fetal membranes; possible increase in incidence. BMJ. 1966;2:682–84. doi: 10.1136/bmj.2.5515.682. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Lind J, Wallenburg HC. Pregnancy and the Ehlers-Danlos syndrome: a retrospective study in a Dutch population. Acta Obstet Gynecol Scand. 2002;81:293–300. doi: 10.1034/j.1600-0412.2002.810403.x. [DOI] [PubMed] [Google Scholar]
- 57.Meijboom LJ, Drenthen W, Pieper PG, Groenink M, van der Post JA, Timmermans J, et al. Obstetric complications in Marfan syndrome. Int.J Cardiol. 2006;110:53–59. doi: 10.1016/j.ijcard.2005.07.017. [DOI] [PubMed] [Google Scholar]
- 58.Anum EA, Hill LD, Pandya A, Strauss JF., III Connective tissue and related disorders and preterm birth: clues to genes contributing to prematurity. Placenta. 2009;30:207–15. doi: 10.1016/j.placenta.2008.12.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Dizon-Townson DS, Major H, Varner M, Ward K. A promoter mutation that increases transcription of the tumor necrosis factor-alpha gene is not associated with preterm delivery. Am.J.Obstet.Gynecol. 1997;177:810–13. doi: 10.1016/s0002-9378(97)70273-4. [DOI] [PubMed] [Google Scholar]
- 60.Roberts AK, Monzon-Bordonaba F, Van Deerlin PG, Holder J, Macones GA, Morgan MA, et al. Association of polymorphism within the promoter of the tumor necrosis factor alpha gene with increased risk of preterm premature rupture of the fetal membranes. Am J Obstet Gynecol. 1999;180:1297–302. doi: 10.1016/s0002-9378(99)70632-0. [DOI] [PubMed] [Google Scholar]
- 61.Ferrand PE, Parry S, Sammel M, Macones GA, Kuivaniemi H, Romero R, et al. A polymorphism in the matrix metalloproteinase-9 promoter is associated with increased risk of preterm premature rupture of membranes in African Americans. Mol.Hum.Reprod. 2002;8:494–501. doi: 10.1093/molehr/8.5.494. [DOI] [PubMed] [Google Scholar]
- 62.Ferrand PE, Fujimoto T, Chennathukuzhi V, Parry S, Macones GA, Sammel M, et al. The CARD15 2936insC mutation and TLR4 896 A>G polymorphism in African Americans and risk of preterm premature rupture of membranes (PPROM) Mol.Hum.Reprod. 2002;8:1031–34. doi: 10.1093/molehr/8.11.1031. [DOI] [PubMed] [Google Scholar]
- 63.Fujimoto T, Parry S, Urbanek M, Sammel M, Macones G, Kuivaniemi H, et al. A single nucleotide polymorphism in the matrix metalloproteinase-1 (MMP-1) promoter influences amnion cell MMP-1 expression and risk for preterm premature rupture of the fetal membranes. J.Biol.Chem. 2002;277:6296–302. doi: 10.1074/jbc.M107865200. [DOI] [PubMed] [Google Scholar]
- 64.Genc MR, Gerber S, Nesin M, Witkin SS. Polymorphism in the interleukin-1 gene complex and spontaneous preterm delivery. Am.J.Obstet.Gynecol. 2002;187:157–63. doi: 10.1067/mob.2002.122407. [DOI] [PubMed] [Google Scholar]
- 65.Lorenz E, Hallman M, Marttila R, Haataja R, Schwartz DA. Association between the Asp299Gly polymorphisms in the Toll-like receptor 4 and premature births in the Finnish population. Pediatr.Res. 2002;52:373–76. doi: 10.1203/00006450-200209000-00011. [DOI] [PubMed] [Google Scholar]
- 66.Kalish RB, Vardhana S, Gupta M, Chasen ST, Perni SC, Witkin SS. Interleukin-1 receptor antagonist gene polymorphism and multifetal pregnancy outcome. Am.J.Obstet.Gynecol. 2003;189:911–14. doi: 10.1067/s0002-9378(03)00770-1. [DOI] [PubMed] [Google Scholar]
- 67.Witkin SS, Vardhana S, Yih M, Doh K, Bongiovanni AM, Gerber S. Polymorphism in intron 2 of the fetal interleukin-1 receptor antagonist genotype influences midtrimester amniotic fluid concentrations of interleukin-1beta and interleukin-1 receptor antagonist and pregnancy outcome. Am J Obstet Gynecol. 2003;189:1413–17. doi: 10.1067/s0002-9378(03)00630-6. [DOI] [PubMed] [Google Scholar]
- 68.Annells MF, Hart PH, Mullighan CG, Heatley SL, Robinson JS, Bardy P, et al. Interleukins-1, -4, -6, -10, tumor necrosis factor, transforming growth factor-beta, FAS, and mannose-binding protein C gene polymorphisms in Australian women: Risk of preterm birth. Am.J.Obstet.Gynecol. 2004;191:2056–67. doi: 10.1016/j.ajog.2004.04.021. [DOI] [PubMed] [Google Scholar]
- 69.Doh K, Sziller I, Vardhana S, Kovacs E, Papp Z, Witkin SS. Beta2-adrenergic receptor gene polymorphisms and pregnancy outcome. J.Perinat.Med. 2004;32:413–17. doi: 10.1515/JPM.2004.138. [DOI] [PubMed] [Google Scholar]
- 70.Genc MR, Onderdonk AB, Vardhana S, Delaney ML, Norwitz ER, Tuomala RE, et al. Polymorphism in intron 2 of the interleukin-1 receptor antagonist gene, local midtrimester cytokine response to vaginal flora, and subsequent preterm birth. Am.J.Obstet.Gynecol. 2004;191:1324–30. doi: 10.1016/j.ajog.2004.05.074. [DOI] [PubMed] [Google Scholar]
- 71.Kalish RB, Vardhana S, Gupta M, Perni SC, Chasen ST, Witkin SS. Polymorphisms in the tumor necrosis factor-alpha gene at position −308 and the inducible 70 kd heat shock protein gene at position +1267 in multifetal pregnancies and preterm premature rupture of fetal membranes. Am.J.Obstet.Gynecol. 2004;191:1368–74. doi: 10.1016/j.ajog.2004.07.007. [DOI] [PubMed] [Google Scholar]
- 72.Kalish RB, Vardhana S, Gupta M, Perni SC, Witkin SS. Interleukin-4 and -10 gene polymorphisms and spontaneous preterm birth in multifetal gestations. Am.J.Obstet.Gynecol. 2004;190:702–06. doi: 10.1016/j.ajog.2003.09.066. [DOI] [PubMed] [Google Scholar]
- 73.Macones GA, Parry S, Elkousy M, Clothier B, Ural SH, Strauss JF., III A polymorphism in the promoter region of TNF and bacterial vaginosis: preliminary evidence of gene-environment interaction in the etiology of spontaneous preterm birth. Am.J.Obstet.Gynecol. 2004;190:1504–08. doi: 10.1016/j.ajog.2004.01.001. [DOI] [PubMed] [Google Scholar]
- 74.Moore S, Ide M, Randhawa M, Walker JJ, Reid JG, Simpson NA. An investigation into the association among preterm birth, cytokine gene polymorphisms and periodontal disease. BJOG. 2004;111:125–32. doi: 10.1046/j.1471-0528.2003.00024.x-i1. [DOI] [PubMed] [Google Scholar]
- 75.Valdez LL, Quintero A, Garcia E, Olivares N, Celis A, Rivas F, Jr., et al. Thrombophilic polymorphisms in preterm delivery. Blood Cells Mol Dis. 2004;33:51–56. doi: 10.1016/j.bcmd.2004.04.011. [DOI] [PubMed] [Google Scholar]
- 76.Wang H, Parry S, Macones G, Sammel MD, Ferrand PE, Kuivaniemi H, et al. Functionally significant SNP MMP8 promoter haplotypes and preterm premature rupture of membranes (PPROM) Hum.Mol.Genet. 2004;13:2659–69. doi: 10.1093/hmg/ddh287. [DOI] [PubMed] [Google Scholar]
- 77.Crider KS, Whitehead N, Buus RM. Genetic variation associated with preterm birth: a HuGE review. Genet.Med. 2005;7:593–604. doi: 10.1097/01.gim.0000187223.69947.db. [DOI] [PubMed] [Google Scholar]
- 78.Engel SA, Erichsen HC, Savitz DA, Thorp J, Chanock SJ, Olshan AF. Risk of spontaneous preterm birth is associated with common proinflammatory cytokine polymorphisms. Epidemiology. 2005;16:469–77. doi: 10.1097/01.ede.0000164539.09250.31. [DOI] [PubMed] [Google Scholar]
- 79.Fuks A, Parton LA, Polavarapu S, Netta D, Strassberg S, Godi I, et al. Polymorphism of Fas and Fas ligand in preterm premature rupture of membranes in singleton pregnancies. Am.J.Obstet.Gynecol. 2005;193:1132–36. doi: 10.1016/j.ajog.2005.05.082. [DOI] [PubMed] [Google Scholar]
- 80.Kalish RB, Nguyen DP, Vardhana S, Gupta M, Perni SC, Witkin SS. A single nucleotide A>G polymorphism at position −670 in the Fas gene promoter: relationship to preterm premature rupture of fetal membranes in multifetal pregnancies. Am.J.Obstet.Gynecol. 2005;192:208–12. doi: 10.1016/j.ajog.2004.06.106. [DOI] [PubMed] [Google Scholar]
- 81.Perni SC, Vardhana S, Kalish R, Chasen S, Witkin SS. Clara cell protein 16 concentration in mid-trimester amniotic fluid: association with fetal gender, fetal G>A +38 CC16 gene polymorphism and pregnancy outcome. J Reprod.Immunol. 2005;68:85–90. doi: 10.1016/j.jri.2005.08.001. [DOI] [PubMed] [Google Scholar]
- 82.Kalish RB, Vardhana S, Normand NJ, Gupta M, Witkin SS. Association of a maternal CD14 -159 gene polymorphism with preterm premature rupture of membranes and spontaneous preterm birth in multi-fetal pregnancies. J.Reprod.Immunol. 2006;70:109–17. doi: 10.1016/j.jri.2005.12.002. [DOI] [PubMed] [Google Scholar]
- 83.Murtha AP, Nieves A, Hauser ER, Swamy GK, Yonish BA, Sinclair TR, et al. Association of maternal IL-1 receptor antagonist intron 2 gene polymorphism and preterm birth. Am.J.Obstet.Gynecol. 2006;195:1249–53. doi: 10.1016/j.ajog.2006.09.002. [DOI] [PubMed] [Google Scholar]
- 84.Engel SM, Olshan AF, Siega-Riz AM, Savitz DA, Chanock SJ. Polymorphisms in folate metabolizing genes and risk for spontaneous preterm and small-for-gestational age birth. Am.J.Obstet.Gynecol. 2006;195:1231–11. doi: 10.1016/j.ajog.2006.07.024. [DOI] [PubMed] [Google Scholar]
- 85.Erichsen HC, Engel SA, Eck PK, Welch R, Yeager M, Levine M, et al. Genetic variation in the sodium-dependent vitamin C transporters, SLC23A1, and SLC23A2 and risk for preterm delivery. Am.J.Epidemiol. 2006;163:245–54. doi: 10.1093/aje/kwj035. [DOI] [PubMed] [Google Scholar]
- 86.Wang H, Parry S, Macones G, Sammel MD, Kuivaniemi H, Tromp G, et al. A functional SNP in the promoter of the SERPINH1 gene increases risk of preterm premature rupture of membranes in African Americans. Proc.Natl.Acad.Sci.U.S.A. 2006;103:13463–67. doi: 10.1073/pnas.0603676103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 87.Valdez-Velazquez LL, Quintero-Ramos A, Perez SA, Mendoza-Carrera F, Montoya-Fuentes H, Rivas F, Jr., et al. Genetic polymorphisms of the renin-angiotensin system in preterm delivery and premature rupture of membranes. J Renin.Angiotensin.Aldosterone.Syst. 2007;8:160–68. doi: 10.3317/jraas.2007.026. [DOI] [PubMed] [Google Scholar]
- 88.Grisaru-Granovsky S, Tevet A, Bar-Shavit R, Salah Z, Elstein D, Samueloff A, et al. Association study of protease activated receptor 1 gene polymorphisms and adverse pregnancy outcomes: results of a pilot study in Israel. Am.J.Med.Genet.A. 2007;143A:2557–63. doi: 10.1002/ajmg.a.31985. [DOI] [PubMed] [Google Scholar]
- 89.Chaves JH, Babayan A, Bezerra CM, Linhares IM, Witkin SS. Maternal and neonatal interleukin-1 receptor antagonist genotype and pregnancy outcome in a population with a high rate of pre-term birth. Am J Reprod.Immunol. 2008;60:312–17. doi: 10.1111/j.1600-0897.2008.00625.x. [DOI] [PubMed] [Google Scholar]
- 90.Rey G, Skowronek F, Alciaturi J, Alonso J, Bertoni B, Sapiro R. Toll receptor 4 Asp299Gly polymorphism and its association with preterm birth and premature rupture of membranes in a South American population. Mol Hum.Reprod. 2008;14:555–59. doi: 10.1093/molehr/gan049. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 91.Wang H, Ogawa M, Wood JR, Bartolomei MS, Sammel MD, Kusanovic JP, et al. Genetic and epigenetic mechanisms combine to control MMP1 expression and its association with preterm premature rupture of membranes. Hum.Mol.Genet. 2008;17:1087–96. doi: 10.1093/hmg/ddm381. [DOI] [PubMed] [Google Scholar]
- 92.Wang H, Sammel MD, Tromp G, Gotsch F, Halder I, Shriver MD, et al. A 12-bp deletion in the 5′-flanking region of the SERPINH1 gene affects promoter activity and protects against preterm premature rupture of membranes in African Americans. Hum.Mutat. 2008;29:332. doi: 10.1002/humu.9522. [DOI] [PubMed] [Google Scholar]
- 93.Cho JK, Kim YH, Park IY, Shin JC, Oh MK, Park SJ, et al. Polymorphism of haptoglobin in patients with premature rupture of membrane. Yonsei Med.J. 2009;50:132–36. doi: 10.3349/ymj.2009.50.1.132. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 94.Ferrell G, Lu M, Stoddard P, Sammel MD, Romero R, Strauss JF, III, et al. A single nucleotide polymorphism in the promoter of the LOXL1 gene and its relationship to pelvic organ prolapse and preterm premature rupture of membranes. Reprod.Sci. 2009;16:438–46. doi: 10.1177/1933719108330567. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 95.Kalinka J, Bitner A. Selected cytokine gene polymorphisms and the risk of preterm delivery in the population of Polish women. Ginekol.Pol. 2009;80:111–17. [PubMed] [Google Scholar]
- 96.Lukaszewski T, Barlik M, Seremak-Mrozikiewicz A, Kurzawinska G, Mrozikiewicz PM, Sieroszewski P, et al. Polymorphism in the genes of Toll-like receptors type 2 and type 4 (TLR-2 and TLR-4) and the risk of premature rupture of the membranes--preliminary study. Ginekol.Pol. 2009;80:914–19. [PubMed] [Google Scholar]
- 97.Salminen A, Paananen R, Karjalainen MK, Tuohimaa A, Luukkonen A, Ojaniemi M, et al. Genetic association of SP-C with duration of preterm premature rupture of fetal membranes and expression in gestational tissues. Ann.Med. 2009;41:629–42. doi: 10.1080/07853890903186176. [DOI] [PubMed] [Google Scholar]
- 98.Kramer MS, Kahn SR, Rozen R, Evans R, Platt RW, et al. Vasculopathic and thrombophilic risk factors for spontaneous preterm birth. Int.J.Epidemiol. 2009;38:715–23. doi: 10.1093/ije/dyp167. [DOI] [PubMed] [Google Scholar]
- 99.Sata F, Toya S, Yamada H, Suzuki K, Saijo Y, Yamazaki A, et al. Proinflammatory cytokine polymorphisms and the risk of preterm birth and low birthweight in a Japanese population. Mol Hum.Reprod. 2009;15:121–30. doi: 10.1093/molehr/gan078. [DOI] [PubMed] [Google Scholar]
- 100.Speer EM, Gentile DA, Zeevi A, Pillage G, Huo D, Skoner DP. Role of single nucleotide polymorphisms of cytokine genes in spontaneous preterm delivery. Hum.Immunol. 2006;67:915–23. doi: 10.1016/j.humimm.2006.08.291. [DOI] [PubMed] [Google Scholar]
- 101.Gonzalez RP, Gomez RM, Castro RS, Nien JK, Merino PO, Etchegaray AB, et al. A national birth weight distribution curve according to gestational age in Chile from 1993 to 2000. Rev.Med.Chil. 2004;132:1155–65. doi: 10.4067/s0034-98872004001000001. [DOI] [PubMed] [Google Scholar]
- 102.Gibbs RS, Blanco JD, St Clair PJ, Castaneda YS. Quantitative bacteriology of amniotic fluid from women with clinical intraamniotic infection at term. J Infect.Dis. 1982;145:1–8. doi: 10.1093/infdis/145.1.1. [DOI] [PubMed] [Google Scholar]
- 103.Stephens JC, Schneider JA, Tanguay DA, Choi J, Acharya T, Stanley SE, et al. Haplotype variation and linkage disequilibrium in 313 human genes. Science. 2001;293:489–93. doi: 10.1126/science.1059431. [DOI] [PubMed] [Google Scholar]
- 104.Judson R, Salisbury B, Schneider J, Windemuth A, Stephens JC. How many SNPs does a genome-wide haplotype map require? Pharmacogenomics. 2002;3:379–91. doi: 10.1517/14622416.3.3.379. [DOI] [PubMed] [Google Scholar]
- 105.Kuivaniemi H, Yoon S, Shibamura H, Skunca M, Vongpunsawad S, Tromp G. Primer-extension preamplified DNA is a reliable template for genotyping. Clin.Chem. 2002;48:1601–04. [PubMed] [Google Scholar]
- 106.Romero R, Kuivaniemi H, Tromp G, Olson J. The design, execution, and interpretation of genetic association studies to decipher complex diseases. Am.J.Obstet.Gynecol. 2002;187:1299–312. doi: 10.1067/mob.2002.128319. [DOI] [PubMed] [Google Scholar]
- 107.Winkelmann BR, Hoffmann MM, Nauck M, Kumar AM, Nandabalan K, Judson RS, et al. Haplotypes of the cholesteryl ester transfer protein gene predict lipid-modifying response to statin therapy. Pharmacogenomics.J. 2003;3:284–96. doi: 10.1038/sj.tpj.6500195. [DOI] [PubMed] [Google Scholar]
- 108.Lee WC. Detecting population stratification using a panel of single nucleotide polymorphisms. Int.J.Epidemiol. 2003;32:1120. doi: 10.1093/ije/dyg301. [DOI] [PubMed] [Google Scholar]
- 109.Feder JN, Gnirke A, Thomas W, Tsuchihashi Z, Ruddy DA, Basava A, et al. A novel MHC class I-like gene is mutated in patients with hereditary haemochromatosis. Nat.Genet. 1996;13:399–408. doi: 10.1038/ng0896-399. [DOI] [PubMed] [Google Scholar]
- 110.Nielsen DM, Ehm MG, Weir BS. Detecting marker-disease association by testing for Hardy-Weinberg disequilibrium at a marker locus. Am.J.Hum.Genet. 1998;63:1531–40. doi: 10.1086/302114. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 111.Wittke-Thompson JK, Pluzhnikov A, Cox NJ. Rational inferences about departures from Hardy-Weinberg equilibrium. Am.J.Hum.Genet. 2005;76:967–86. doi: 10.1086/430507. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 112.Trikalinos TA, Salanti G, Khoury MJ, Ioannidis JP. Impact of violations and deviations in Hardy-Weinberg equilibrium on postulated gene-disease associations. Am.J.Epidemiol. 2006;163:300–09. doi: 10.1093/aje/kwj046. [DOI] [PubMed] [Google Scholar]
- 113.Ryckman KK, Jiang L, Li C, Bartlett J, Haines JL, Williams SM. A prevalence-based association test for case-control studies. Genet.Epidemiol. 2008;32:600–05. doi: 10.1002/gepi.20342. [DOI] [PubMed] [Google Scholar]
- 114.Pritchard JK, Stephens M, Donnelly P. Inference of population structure using multilocus genotype data. Genetics. 2000;155:945–59. doi: 10.1093/genetics/155.2.945. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 115.Purcell S, Neale B, Todd-Brown K, Thomas L, Ferreira MA, Bender D, et al. PLINK: a tool set for whole-genome association and population-based linkage analyses. Am.J.Hum.Genet. 2007;81:559–75. doi: 10.1086/519795. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 116.Benjamini Y, Drai D, Elmer G, Kafkafi N, Golani I. Controlling the false discovery rate in behavior genetics research. Behav.Brain Res. 2001;125:279–84. doi: 10.1016/s0166-4328(01)00297-2. [DOI] [PubMed] [Google Scholar]
- 117.Benjamini Y, Hochberg Y. Controlling the false discovery rate: a practical and powerful approach to multiple testing. J Roy Statist Soc Ser B. 1995;57:289–300. [Google Scholar]
- 118.Devlin B, Risch N. A comparison of linkage disequilibrium measures for fine-scale mapping. Genomics. 1995;29:311–22. doi: 10.1006/geno.1995.9003. [DOI] [PubMed] [Google Scholar]
- 119.Barrett JC, Fry B, Maller J, Daly MJ. Haploview: analysis and visualization of LD and haplotype maps. Bioinformatics. 2005;21:263–65. doi: 10.1093/bioinformatics/bth457. [DOI] [PubMed] [Google Scholar]
- 120.Gabriel SB, Schaffner SF, Nguyen H, Moore JM, Roy J, Blumenstiel B, et al. The structure of haplotype blocks in the human genome. Science. 2002;296:2225–29. doi: 10.1126/science.1069424. [DOI] [PubMed] [Google Scholar]
- 121.Redline RW, Heller D, Keating S, Kingdom J. Placental diagnostic criteria and clinical correlation--a workshop report. Placenta. 2005;26(Suppl A):S114–S117. doi: 10.1016/j.placenta.2005.02.009. [DOI] [PubMed] [Google Scholar]
- 122.Ritchie MD, Hahn LW, Roodi N, Bailey LR, Dupont WD, Parl FF, et al. Multifactor-dimensionality reduction reveals high-order interactions among estrogen-metabolism genes in sporadic breast cancer. Am.J.Hum.Genet. 2001;69:138–47. doi: 10.1086/321276. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 123.Velez DR, White BC, Motsinger AA, Bush WS, Ritchie MD, Williams SM, et al. A balanced accuracy function for epistasis modeling in imbalanced datasets using multifactor dimensionality reduction. Genet.Epidemiol. 2007;31:306–15. doi: 10.1002/gepi.20211. [DOI] [PubMed] [Google Scholar]
- 124.Moore JH, White BC. Tuning ReliefF for Genome-Wide Genetic Analysis. In: Marchiori E, Moore JH, Rajapakse JC, editors. Evolutionary Computation, Machine Learning and Data Mining in Bioinformatics. Springer Berlin; Heidelberg: 2007. pp. 166–75. [Google Scholar]
- 125.Kira K, Rendell LA. A Practical Approach to Feature Selection. In: Sleeman D, Edwards P, editors. Proceedings of the Ninth International Workshop on Machine Learning; Aberdeen: Morgan-Kaufman; 1992. pp. 249–56. [Google Scholar]
- 126.Calvano SE, Xiao W, Richards DR, Felciano RM, Baker HV, Cho RJ, et al. A network-based analysis of systemic inflammation in humans. Nature. 2005;437:1032–37. doi: 10.1038/nature03985. [DOI] [PubMed] [Google Scholar]
- 127.Kuijl C, Savage ND, Marsman M, Tuin AW, Janssen L, Egan DA, et al. Intracellular bacterial growth is controlled by a kinase network around PKB/AKT1. Nature. 2007;450:725–30. doi: 10.1038/nature06345. [DOI] [PubMed] [Google Scholar]
- 128.Loza MJ, McCall CE, Li L, Isaacs WB, Xu J, Chang BL. Assembly of inflammation-related genes for pathway-focused genetic analysis. PLoS One. 2007;2:e1035. doi: 10.1371/journal.pone.0001035. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 129.Winn VD, Haimov-Kochman R, Paquet AC, Yang YJ, Madhusudhan MS, Gormley M, et al. Gene expression profiling of the human maternal-fetal interface reveals dramatic changes between midgestation and term. Endocrinology. 2007;148:1059–79. doi: 10.1210/en.2006-0683. [DOI] [PubMed] [Google Scholar]
- 130.Vadillo-Ortega F, Hernandez A, Gonzalez-Avila G, Bermejo L, Iwata K, Strauss JF., III Increased matrix metalloproteinase activity and reduced tissue inhibitor of metalloproteinases-1 levels in amniotic fluids from pregnancies complicated by premature rupture of membranes. Am.J.Obstet.Gynecol. 1996;174:1371–76. doi: 10.1016/s0002-9378(96)70687-7. [DOI] [PubMed] [Google Scholar]
- 131.Fortunato SJ, Menon R, Lombardi SJ. MMP/TIMP imbalance in amniotic fluid during PROM: an indirect support for endogenous pathway to membrane rupture. J Perinat.Med. 1999;27:362–68. doi: 10.1515/JPM.1999.049. [DOI] [PubMed] [Google Scholar]
- 132.Fortunato SJ, Menon R. Distinct molecular events suggest different pathways for preterm labor and premature rupture of membranes. Am.J.Obstet.Gynecol. 2001;184:1399–405. doi: 10.1067/mob.2001.115122. [DOI] [PubMed] [Google Scholar]
- 133.Maymon E, Romero R, Pacora P, Gomez R, Mazor M, Edwin S, et al. A role for the 72 kDa gelatinase (MMP-2) and its inhibitor (TIMP-2) in human parturition, premature rupture of membranes and intraamniotic infection. J.Perinat.Med. 2001;29:308–16. doi: 10.1515/JPM.2001.044. [DOI] [PubMed] [Google Scholar]
- 134.Vadillo-Ortega F, Sadowsky DW, Haluska GJ, Hernandez-Guerrero C, Guevara-Silva R, Gravett MG, et al. Identification of matrix metalloproteinase-9 in amniotic fluid and amniochorion in spontaneous labor and after experimental intrauterine infection or interleukin-1 beta infusion in pregnant rhesus monkeys. Am J Obstet.Gynecol. 2002;186:128–38. doi: 10.1067/mob.2002.118916. [DOI] [PubMed] [Google Scholar]
- 135.Fortunato SJ, Menon R, Swan K, Baricos W. Expression of matrix degrading enzymes and tissue inhibitors of metalloproteinases (TIMP) in human fetal membranes. 15th annual meeting of the Society of Perinatal Obstetricians Atlanta. Am.J.Obstet.Gynecol. 1995;170 [Google Scholar]
- 136.Fortunato SJ, Menon R, Lombardi SJ. Collagenolytic enzymes (gelatinases) and their inhibitors in human amniochorionic membrane. Am J Obstet.Gynecol. 1997;177:731–41. doi: 10.1016/s0002-9378(97)70260-6. [DOI] [PubMed] [Google Scholar]
- 137.Fortunato SJ, Menon R, Lombardi SJ. Expression of a progelatinase activator (MT1-MMP) in human fetal membranes. Am J Reprod.Immunol. 1998;39:316–22. doi: 10.1111/j.1600-0897.1998.tb00524.x. [DOI] [PubMed] [Google Scholar]
- 138.Fortunato SJ, Menon R, Lombardi SJ. Stromelysins in placental membranes and amniotic fluid with premature rupture of membranes. Obstet.Gynecol. 1999;94:435–40. doi: 10.1016/s0029-7844(99)00336-1. [DOI] [PubMed] [Google Scholar]
- 139.Fortunato SJ, Menon R, Lombardi SJ. Amniochorion gelatinase-gelatinase inhibitor imbalance in vitro: a possible infectious pathway to rupture. Obstet.Gynecol. 2000;95:240–44. doi: 10.1016/s0029-7844(99)00503-7. [DOI] [PubMed] [Google Scholar]
- 140.Zaga-Clavellina V, Merchant-Larios H, Garcia-Lopez G, Maida-Claros R, Vadillo-Ortega F. Differential secretion of matrix metalloproteinase-2 and -9 after selective infection with group B streptococci in human fetal membranes. J Soc.Gynecol.Investig. 2006;13:271–79. doi: 10.1016/j.jsgi.2006.02.006. [DOI] [PubMed] [Google Scholar]
- 141.Garcia-Lopez G, Vadillo-Ortega F, Merchant-Larios H, Maida-Claros R, Osorio M, Soriano-Becerril D, et al. Evidence of in vitro differential secretion of 72 and 92 kDa type IV collagenases after selective exposure to lipopolysaccharide in human fetal membranes. Mol.Hum.Reprod. 2007;13:409–18. doi: 10.1093/molehr/gam025. [DOI] [PubMed] [Google Scholar]
- 142.Maymon E, Romero R, Pacora P, Gervasi MT, Bianco K, Ghezzi F, et al. Evidence for the participation of interstitial collagenase (matrix metalloproteinase 1) in preterm premature rupture of membranes. Am J.Obstet.Gynecol. 2000;183:914–20. doi: 10.1067/mob.2000.108879. [DOI] [PubMed] [Google Scholar]
- 143.Maymon E, Romero R, Pacora P, Gomez R, Athayde N, Edwin S, et al. Human neutrophil collagenase (matrix metalloproteinase 8) in parturition, premature rupture of the membranes, and intrauterine infection. Am J.Obstet.Gynecol. 2000;183:94–99. doi: 10.1067/mob.2000.105344. [DOI] [PubMed] [Google Scholar]
- 144.Maymon E, Romero R, Pacora P, Gervasi MT, Gomez R, Edwin SS, et al. Evidence of in vivo differential bioavailability of the active forms of matrix metalloproteinases 9 and 2 in parturition, spontaneous rupture of membranes, and intra-amniotic infection. Am.J.Obstet.Gynecol. 2000;183:887–94. doi: 10.1067/mob.2000.108878. [DOI] [PubMed] [Google Scholar]
- 145.Maymon E, Romero R, Pacora P, Gervasi MT, Edwin SS, Gomez R, et al. Matrilysin (matrix metalloproteinase 7) in parturition, premature rupture of membranes, and intrauterine infection. Am J.Obstet.Gynecol. 2000;182:1545–53. doi: 10.1067/mob.2000.107652. [DOI] [PubMed] [Google Scholar]
- 146.Maymon E, Romero R, Chaiworapongsa T, Kim JC, Berman S, Gomez R, et al. Value of amniotic fluid neutrophil collagenase concentrations in preterm premature rupture of membranes. Am J.Obstet.Gynecol. 2001;185:1143–48. doi: 10.1067/mob.2001.118166. [DOI] [PubMed] [Google Scholar]
- 147.Angus SR, Segel SY, Hsu CD, Locksmith GJ, Clark P, Sammel MD, et al. Amniotic fluid matrix metalloproteinase-8 indicates intra-amniotic infection. Am J Obstet.Gynecol. 2001;185:1232–38. doi: 10.1067/mob.2001.118654. [DOI] [PubMed] [Google Scholar]
- 148.Fortunato SJ, LaFleur B, Menon R. Collagenase-3 (MMP-13) in fetal membranes and amniotic fluid during pregnancy. Am J.Reprod.Immunol. 2003;49:120–25. doi: 10.1034/j.1600-0897.2003.00012.x. [DOI] [PubMed] [Google Scholar]
- 149.Fortunato SJ, Menon R, Bryant C, Lombardi SJ. Programmed cell death (apoptosis) as a possible pathway to metalloproteinase activation and fetal membrane degradation in premature rupture of membranes. Am J.Obstet.Gynecol. 2000;182:1468–76. doi: 10.1067/mob.2000.107330. [DOI] [PubMed] [Google Scholar]
- 150.Athayde N, Edwin SS, Romero R, Gomez R, Maymon E, Pacora P, et al. A role for matrix metalloproteinase-9 in spontaneous rupture of the fetal membranes. Am J.Obstet.Gynecol. 1998;179:1248–53. doi: 10.1016/s0002-9378(98)70141-3. [DOI] [PubMed] [Google Scholar]
- 151.Athayde N, Romero R, Gomez R, Maymon E, Pacora P, Mazor M, et al. Matrix metalloproteinases-9 in preterm and term human parturition. J.Matern.Fetal Med. 1999;8:213–19. doi: 10.1002/(SICI)1520-6661(199909/10)8:5<213::AID-MFM3>3.0.CO;2-R. [DOI] [PubMed] [Google Scholar]
- 152.Riley SC, Leask R, Chard T, Wathen NC, Calder AA, Howe DC. Secretion of matrix metalloproteinase-2, matrix metalloproteinase-9 and tissue inhibitor of metalloproteinases into the intrauterine compartments during early pregnancy. Mol.Hum.Reprod. 1999;5:376–81. doi: 10.1093/molehr/5.4.376. [DOI] [PubMed] [Google Scholar]
- 153.Lavee M, Goldman S, niel-Spiegel E, Shalev E. Matrix metalloproteinase-2 is elevated in midtrimester amniotic fluid prior to the development of preeclampsia. Reprod.Biol.Endocrinol. 2009;7:85. doi: 10.1186/1477-7827-7-85. 85. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 154.Di Renzo GC. The great obstetrical syndromes. J.Matern.Fetal Neonatal Med. 2009;22:633–35. doi: 10.1080/14767050902866804. [DOI] [PubMed] [Google Scholar]
- 155.Romero R. Prenatal medicine: the child is the father of the man. J.Matern.Fetal Neonatal Med. 2009;22:636–39. doi: 10.1080/14767050902784171. 1996. [DOI] [PubMed] [Google Scholar]
- 156.Garite TJ. Premature rupture of the membranes: the enigma of the obstetrician. Am J Obstet.Gynecol. 1985;151:1001–05. doi: 10.1016/0002-9378(85)90369-2. [DOI] [PubMed] [Google Scholar]
- 157.Romero R, Quintero R, Oyarzun E, Wu YK, Sabo V, Mazor M, et al. Intraamniotic infection and the onset of labor in preterm premature rupture of the membranes. Am J Obstet.Gynecol. 1988;159:661–66. doi: 10.1016/s0002-9378(88)80030-9. [DOI] [PubMed] [Google Scholar]
- 158.Eschenbach DA. Intrauterine infection and premature membrane rupture. Curr.Opin.Obstet.Gynecol. 1989;1:23–26. [PubMed] [Google Scholar]
- 159.Gibbs RS. Premature rupture of the membranes: intraamniotic infection. Pediatr.Infect.Dis.J. 1990;9:776. [PubMed] [Google Scholar]
- 160.Asrat T, Nageotte MP, Garite TJ, Gocke SE, Dorchester W. Gram stain results from amniocentesis in patients with preterm premature rupture of membranes--comparison of maternal and fetal characteristics. Am J Obstet.Gynecol. 1990;163:887–89. doi: 10.1016/0002-9378(90)91089-u. [DOI] [PubMed] [Google Scholar]
- 161.Arias F, Rodriquez L, Rayne SC, Kraus FT. Maternal placental vasculopathy and infection: two distinct subgroups among patients with preterm labor and preterm ruptured membranes. Am.J.Obstet.Gynecol. 1993;168:585–91. doi: 10.1016/0002-9378(93)90499-9. [DOI] [PubMed] [Google Scholar]
- 162.Mercer BM, Arheart KL. Antibiotic therapy for preterm premature rupture of the membranes. Semin.Perinatol. 1996;20:426–38. doi: 10.1016/s0146-0005(96)80010-3. [DOI] [PubMed] [Google Scholar]
- 163.Arias F, Victoria A, Cho K, Kraus F. Placental histology and clinical characteristics of patients with preterm premature rupture of membranes. Obstet Gynecol. 1997;89:265–71. doi: 10.1016/S0029-7844(96)00451-6. [DOI] [PubMed] [Google Scholar]
- 164.Lovett SM, Weiss JD, Diogo MJ, Williams PT, Garite TJ. A prospective, double-blind, randomized, controlled clinical trial of ampicillin-sulbactam for preterm premature rupture of membranes in women receiving antenatal corticosteroid therapy. Am J Obstet Gynecol. 1997;176:1030–38. doi: 10.1016/s0002-9378(97)70398-3. [DOI] [PubMed] [Google Scholar]
- 165.Belady PH, Farkouh LJ, Gibbs RS. Intra-amniotic infection and premature rupture of the membranes. Clin.Perinatol. 1997;24:43–57. [PubMed] [Google Scholar]
- 166.Bendon RW, Faye-Petersen O, Pavlova Z, Qureshi F, Mercer B, Miodovnik M, et al. Fetal membrane histology in preterm premature rupture of membranes: comparison to controls, and between antibiotic and placebo treatment. The National Institute of Child Health and Human Development Maternal Fetal Medicine Units Network, Bethesda, MD, USA. Pediatr.Dev.Pathol. 1999;2:552–58. doi: 10.1007/s100249900161. [DOI] [PubMed] [Google Scholar]
- 167.Goncalves LF, Chaiworapongsa T, Romero R. Intrauterine infection and prematurity. Ment.Retard.Dev.Disabil.Res.Rev. 2002;8:3–13. doi: 10.1002/mrdd.10008. [DOI] [PubMed] [Google Scholar]
- 168.Chaiworapongsa T, Espinoza J, Yoshimatsu J, Kim YM, Bujold E, Edwin S, et al. Activation of coagulation system in preterm labor and preterm premature rupture of membranes. J Matern.Fetal Neonatal Med. 2002;11:368–73. doi: 10.1080/jmf.11.6.368.373. [DOI] [PubMed] [Google Scholar]
- 169.Siega-Riz AM, Promislow JH, Savitz DA, Thorp JM, Jr., McDonald T. Vitamin C intake and the risk of preterm delivery. Am J Obstet.Gynecol. 2003;189:519–25. doi: 10.1067/s0002-9378(03)00363-6. [DOI] [PubMed] [Google Scholar]
- 170.Simhan HN, Caritis SN, Krohn MA, Hillier SL. The vaginal inflammatory milieu and the risk of early premature preterm rupture of membranes. Am.J Obstet Gynecol. 2005;192:213–18. doi: 10.1016/j.ajog.2004.07.021. [DOI] [PubMed] [Google Scholar]
- 171.Casanueva E, Ripoll C, Tolentino M, Morales RM, Pfeffer F, Vilchis P, et al. Vitamin C supplementation to prevent premature rupture of the chorioamniotic membranes: a randomized trial. Am.J Clin.Nutr. 2005;81:859–63. doi: 10.1093/ajcn/81.4.859. [DOI] [PubMed] [Google Scholar]
- 172.Erez O, Espinoza J, Chaiworapongsa T, Gotsch F, Kusanovic JP, Than NG, et al. A link between a hemostatic disorder and preterm PROM: a role for tissue factor and tissue factor pathway inhibitor. J Matern.Fetal Neonatal Med. 2008;21:732–44. doi: 10.1080/14767050802361807. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 173.Savasan ZA, Romero R, Chaiworapongsa T, Kusanovic JP, Kim SK, Mazaki-Tovi S, et al. Evidence in support of a role for anti-angiogenic factors in preterm prelabor rupture of membranes. J Matern.Fetal Neonatal Med. 2010 doi: 10.3109/14767050903440471. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 174.Pennell CE, Jacobsson B, Williams SM, Buus RM, Muglia LJ, Dolan SM, et al. Genetic epidemiological studies of preterm birth: Guidelines for research. Am.J.Obstet.Gynecol. 2006 doi: 10.1016/j.ajog.2006.03.109. [DOI] [PubMed] [Google Scholar]
- 175.Biggio J, Christiaens I, Katz M, Menon R, Merialdi M, Morken NH, et al. A call for an international consortium on the genetics of preterm birth. Am J Obstet.Gynecol. 2008;199:95–97. doi: 10.1016/j.ajog.2008.06.012. [DOI] [PubMed] [Google Scholar]
- 176.Li S, Lu Q, Fu W, Romero R, Cui Y. A regularized regression approach for dissecting genetic conflicts that increase disease risk in pregnancy. Stat.Appl.Genet.Mol.Biol. 2009;8 doi: 10.2202/1544-6115.1474. Article. [DOI] [PubMed] [Google Scholar]
- 177.Ramirez F, Tanaka S, Bou-Gharios G. Transcriptional regulation of the human alpha2(I) collagen gene (COL1A2), an informative model system to study fibrotic diseases. Matrix Biol. 2006;25:365–72. doi: 10.1016/j.matbio.2006.05.002. [DOI] [PubMed] [Google Scholar]
- 178.Porter EM, van DE, Valore EV, Ganz T. Broad-spectrum antimicrobial activity of human intestinal defensin 5. Infect.Immun. 1997;65:2396–401. doi: 10.1128/iai.65.6.2396-2401.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 179.Word RA, Kamm KE, Casey ML. Contractile effects of prostaglandins, oxytocin, and endothelin-1 in human myometrium in vitro: refractoriness of myometrial tissue of pregnant women to prostaglandins E2 and F2 alpha. J Clin.Endocrinol.Metab. 1992;75:1027–32. doi: 10.1210/jcem.75.4.1400867. [DOI] [PubMed] [Google Scholar]
- 180.Dallot E, Pouchelet M, Gouhier N, Cabrol D, Ferre F, Breuiller-Fouche M. Contraction of cultured human uterine smooth muscle cells after stimulation with endothelin-1. Biol.Reprod. 2003;68:937–42. doi: 10.1095/biolreprod.102.008367. [DOI] [PubMed] [Google Scholar]
- 181.Di Liberto G, Dallot E, Eude-Le P,I, Cabrol D, Ferre F, Breuiller-Fouche M. A critical role for PKC zeta in endothelin-1-induced uterine contractions at the end of pregnancy. Am J Physiol Cell Physiol. 2003;285:C599–C607. doi: 10.1152/ajpcell.00040.2003. [DOI] [PubMed] [Google Scholar]
- 182.DANFORTH DN, BUCKINGHAM JC, RODDICK JW., Jr. Connective tissue changes incident to cervical effacement. Am J Obstet.Gynecol. 1960;80:939–45. doi: 10.1016/0002-9378(60)90472-5. [DOI] [PubMed] [Google Scholar]
- 183.Uldbjerg N, Ekman G, Malmstrom A, Olsson K, Ulmsten U. Ripening of the human uterine cervix related to changes in collagen, glycosaminoglycans, and collagenolytic activity. Am J Obstet.Gynecol. 1983;147:662–66. doi: 10.1016/0002-9378(83)90446-5. [DOI] [PubMed] [Google Scholar]
- 184.Iwahashi M, Muragaki Y, Ooshima A, Umesaki N. Decreased type I collagen expression in human uterine cervix during pregnancy. J Clin.Endocrinol.Metab. 2003;88:2231–35. doi: 10.1210/jc.2002-021213. [DOI] [PubMed] [Google Scholar]
- 185.Balu RB, Savitz DA, Ananth CV, Hartmann KE, Miller WC, Thorp JM, et al. Bacterial vaginosis and vaginal fluid defensins during pregnancy. Am J Obstet.Gynecol. 2002;187:1267–71. doi: 10.1067/mob.2002.126989. [DOI] [PubMed] [Google Scholar]
- 186.Balu RB, Savitz DA, Ananth CV, Hartmann KE, Miller WC, Thorp JM, et al. Bacterial vaginosis, vaginal fluid neutrophil defensins, and preterm birth. Obstet.Gynecol. 2003;101:862–68. doi: 10.1016/s0029-7844(03)00042-5. [DOI] [PubMed] [Google Scholar]
- 187.Romero R, Chaiworapongsa T, Kuivaniemi H, Tromp G. Bacterial vaginosis, the inflammatory response and the risk of preterm birth: a role for genetic epidemiology in the prevention of preterm birth. Am.J Obstet Gynecol. 2004;190:1509–19. doi: 10.1016/j.ajog.2004.01.002. [DOI] [PubMed] [Google Scholar]
- 188.Svinarich DM, Wolf NA, Gomez R, Gonik B, Romero R. Detection of human defensin 5 in reproductive tissues. Am J Obstet Gynecol. 1997;176:470–75. doi: 10.1016/s0002-9378(97)70517-9. [DOI] [PubMed] [Google Scholar]
- 189.Quayle AJ, Porter EM, Nussbaum AA, Wang YM, Brabec C, Yip KP, et al. Gene expression, immunolocalization, and secretion of human defensin-5 in human female reproductive tract. Am J Pathol. 1998;152:1247–58. [PMC free article] [PubMed] [Google Scholar]
- 190.Fan SR, Liu XP, Liao QP. Human defensins and cytokines in vaginal lavage fluid of women with bacterial vaginosis. Int J Gynaecol.Obstet. 2008;103:50–54. doi: 10.1016/j.ijgo.2008.05.020. [DOI] [PubMed] [Google Scholar]
- 191.Romero R, Avila C, Edwin SS, Mitchell MD. Endothelin-1,2 levels are increased in the amniotic fluid of women with preterm labor and microbial invasion of the amniotic cavity. Am J Obstet.Gynecol. 1992;166:95–99. doi: 10.1016/0002-9378(92)91837-z. [DOI] [PubMed] [Google Scholar]
- 192.Margarit L, Griffiths AN, Tsapanos V, Tsakas S, Decavalas G. Amniotic fluid endothelin levels and the incidence of premature rupture of membranes. Int.J Gynaecol.Obstet. 2006;93:18–21. doi: 10.1016/j.ijgo.2005.12.013. [DOI] [PubMed] [Google Scholar]
- 193.Cui Y, Fu W, Sun K, Romero R, Wu R. Mapping Nucleotide Sequences that Encode Complex Binary Disease Traits with HapMap. Curr.Genomics. 2007;8:307–22. doi: 10.2174/138920207782446188. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 194.Goddard KA, Tromp G, Romero R, Olson JM, Lu Q, Xu Z, et al. Candidate-gene association study of mothers with pre-eclampsia, and their infants, analyzing 775 SNPs in 190 genes. Hum.Hered. 2007;63:1–16. doi: 10.1159/000097926. [DOI] [PubMed] [Google Scholar]
- 195.Cui Y, Kang G, Sun K, Qian M, Romero R, Fu W. Gene-centric genomewide association study via entropy. Genetics. 2008;179:637–50. doi: 10.1534/genetics.107.082370. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.