Abstract
Recent advances in molecular dynamics (MD) simulation methods and in available computational resources have allowed for more reliable simulations of biological phenomena. From all-atom MD simulations, we are now able to visualize in detail the interactions between antimicrobial peptides (AMPs) and a variety of membrane mimics. This helps us to understand the molecular mechanisms of antimicrobial activity and toxicity. This chapter describes how to set up and conduct molecular dynamics simulations of AMPs and membrane mimics. Details are given for the construction of systems of interest for studying AMPs, which can include simulations of peptides in water, micelles, or lipid bilayers. Explanations of the parameters needed for running a simulation are provided as well.
Keywords: Molecular dynamics, CHARMM, NAMD, micelles, lipid bilayers
1. Introduction
1.1. Why Computer Simulations?
Molecular dynamics (MD) simulations can provide some of the molecular level details necessary to understand the mechanism of action of antimicrobial peptides (AMPs). For many AMPs, it is apparent that the cell membrane is the target. The general interaction between positively charged AMPs and negatively charged bacterial membranes is well understood, as it is the need for hydrophobic content to allow for interactions between the peptide and the membrane core. However, these basic facts are not enough to allow for design of AMPs of therapeutic value. There is no simple, linear correlation between peptide charge or hydrophobicity and antimicrobial activity. Consequently, intuitive design rules for novel AMPs are hard to deduce. The wide variety of sequences and structures of naturally occurring AMPs suggests that there is most certainly not one mechanism of action by which all peptides act.
In this chapter, we discuss the use of MD simulations to study AMPs. The goals in using MD simulations are to both understand the mechanism of action by which these peptides kill bacteria and develop predictive models that can be used to design highly potent, yet non-toxic peptides.
We should note that computer simulations have as steep a learning curve as any sophisticated experimental method. The logistics of running computer simulations can also be challenging, from setting up the system, to choosing numerical integration schemes and statistical ensembles, to managing and analyzing large volumes of data. Nonetheless, as with any sophisticated experimental method, the pleasure of discovery awaits on the other side of a tortuous effort.
1.2. Theory
For a thorough review of the underlying theory of MD simulations, the reader is directed to Allen and Tildesley’s “Computer Simulation of Liquids” (1). MD simulations are based on classical mechanics or, more specifically, on Newton’s second equation of motion. If the force acting on each atom in a system is known, the acceleration can be determined. Using this acceleration, the equations of motion can be integrated, resulting in a trajectory of the positions, velocities, and accelerations of each particle in the system. The force Fi on an atom of mass mi, which is at position ri, is determined from the potential energy of the system, V, as shown in equation [1]. The potential can be used to directly determine the position of a particle as a function of time, as in equation [2].
| [1] |
| [2] |
The equations of motion are solved for each atom in the system. The forces acting on the atoms in the new positions are calculated and the simulation continues for as many time steps as necessary.
Empirical energy functions are used to approximate the force fields for large biological systems composed of many atoms. Some common force fields are AMBER (2), OPLS/AMBER (3), CHARMM (4), and GROMOS (5). We focus here on the CHARMM (Chemistry at HARvard Macromolecular Mechanics) force field, which has been developed over the last 25 years, and is continuously revised to better match new experimental data. There are also several commercially available MD simulation engines for use empowered with the above force fields. We recommend the use of the CHARMM MD simulation engine or NAMD (NAnoscale Molecular Dynamics) (6). A number of CHARMM scripts for the design and analysis of a variety of simulation systems are available on the CHARMM forum (www.charmm.org). However, NAMD is designed to take advantage of advances in parallel computing. This permits longer simulations of larger systems than are possible with CHARMM.
We should note that more recently a graphics user interface has been developed for CHARMM that significantly simplifies the set up of simulation systems and parameters. The software is available at http://www.charmm-gui.org/.
1.2.1. The Potential Function
The potential function is a sum of interaction energies. The value of the potential is determined by summing the bonded terms with the non-bonded potential terms, Vbonded and Vnon-bonded, respectively:
| [3] |
1.2.1.1. Non-bonded Terms
The non-bonded energy terms account for interactions between non-bonded atoms and atoms that are more than three covalent bonds away from each other in a molecule. These are modeled by the van der Waals energy and electrostatic energy, the first and second summation terms in equation [4], respectively.
| [4] |
The calculation of the non-bonded terms in the potential function is the most time-consuming part of the MD simulation. In principle, the interactions between every pair of atoms should be calculated explicitly, meaning that for an N-atom system, N2 calculations would be required. To decrease this number, methods were developed to ignore the interactions between two atoms separated by a distance greater than a specified cutoff distance. For the van der Waals interactions, the potential is “shifted” off over a period ron to roff: Atoms farther from each other than the distance roff are assumed not to interact.
For the electrostatic interactions, cutoff schemes have been shown to be inaccurate when computing electrostatic interactions (7). Instead, the Ewald summation is used, which separates the potential into a slowly decaying long-range component and also a quickly varying short-range component. There are several parameters that need to be optimized when solving the Ewald summation. For large systems, it is best to fix the real sum cut off, Rcutoff, and choose a value for the width of the Gaussians, α, that is large enough to accelerate convergence of the real sum. The value of α determines the relative rate of convergence of the real and reciprocal sums. The accuracy of the sum is independent of α. In practice, when the Ewald sum is used, the Gaussian source distributions centered on point ions are sampled onto a finite grid and the resulting Fourier integral is approximated by a Fourier series, which is evaluated numerically using Fast Fourier Transforms. Complex exponentials in the reciprocal sum are evaluated by β-spline interpolation. Therefore, the grid point spacing and the β-spline order must be optimized. The grid point spacing dictates the number of grids in each direction in the simulation cell onto which the Gaussian charges are interpolated. The β-spline is used to interpolate charges onto the grids. Higher values give more accurate sums but at a greater computational cost. In order for Ewald’s method to be appropriately applied to a simulation, the system must have no net charge.
1.2.1.2. Bonded Terms
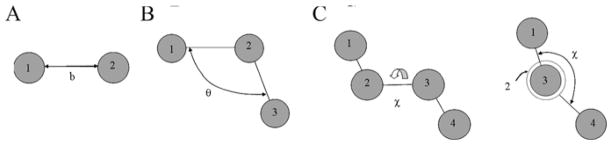
The bonded terms include the bonds, angles, and bond rotations in the molecule (Fig. 17.1) and are computed using equation [5]:
| [5] |
Fig. 17.1.
Bonded interaction motions: (a) bond stretch is the movement between atoms directly bound to each other; (b) angle bend is a function of the relative position of atoms separated by one additional atom (1–3 pairs); (c) rotate-along-bond (dihedrals) is a function of the relative position of atoms separated by two additional atoms (1–4 pairs).
These terms are calculated using approximations for each type of motion. The bond-stretch term is a harmonic potential approximated as a function of displacement from the ideal bond length, b0 for 1,2-pairs of atoms. The force constant, Kb, determines the strength of the bond and is determined by the atoms involved in the bond. The angle-bend term is associated with the movement a bond angle, θ, from its ideal value θ0 and is also represented by a harmonic potential with a force constant, Kθ, which depends on the atoms involved. The rotate-along-bond energy is associated with the torsion angle potential function, which takes into account the steric barriers between 1,4-pairs of atoms (atoms separated by three covalent bonds). This potential expressed as a cosine function.
1.2.1.3. Additional Potentials
There are two additional potential terms in the CHARMM force field. The improper dihedrals term (equation [6]) accounts for out of plane bending, where kω is the force constant and ω –ωo is the out of plane angle. The other component is the Urey–Bradley potential (equation [7]), which accounts for angle bending using 1,3 non-bonded interactions. In this term, kU is the respective force constant and u–uo is the distance between the 1,3 atoms in the harmonic potential:
| [6] |
| [7] |
The CHARMM parameter file contains the parameters for each atom type and bond type. It provides the equilibrium bond lengths and van der Waals radii.
1.2.2. Integration Algorithms
Because there are so many atoms in a MD simulation system and because the energy function is semi-empirical, there is no analytical solution to the equations they formulate. There are many possible integration algorithms. Most are based on Taylor series expansions of the particle position and its derivatives. In CHARMM, the “leapfrog” Verlet algorithm is used. The equations solved in this algorithm are
| [8] |
| [9] |
The velocities are calculated at time t + 1/2 δt. These velocities are then used to calculate positions r at the time t + δt, so that the velocities “leap” over the positions, then the positions “leap” over the velocities (1).
In NAMD, the velocity Verlet algorithm is used. In this method, which minimizes round-off error, the positions, velocities, and accelerations are stored at time t(1). The equations used are
| [10] |
| [11] |
The time step, δt, used with these algorithms is of importance. A smaller time step means slower dynamics. On the other hand, a large time step can cause instability due to large jumps in the conformation negatively impacting the energy of the system. The time step must be much smaller than the fastest characteristic motion within the system. For biological systems, this motion is usually the vibration of hydrogen bonds. A time step of 2 fs (2 × 10−15 s) has been used widely for biological molecules when the SHAKE algorithm is employed to constrain the hydrogen bonds.
1.3. Temperature and Pressure Control
One can show that simply solving Newton’s equations of motion for a system of N particles in volume V conserves the total energy of the system. Indeed any classical mechanical set of equation of motion samples the microcanonical, or NVE ensemble, where N, V, and E are constant.
It is more sensible to simulate biological systems under constant pressure and temperature constraints. The equations of motion need to then be modified appropriately. Temperature is often controlled by using Nose–Hoover–Langevin dynamics. In Langevin dynamics, a stochastic force term is included to introduce the effects of random interactions such as friction between molecules and an occasional high-velocity collision to mimic perturbations that would occur in an experimental system. For constant pressure and temperature simulations in which Langevin dynamics are used to control the temperature, the pressure can be controlled in NAMD with a modified Nose–Hoover method. This method is a combination of the methods described in (8) and (9). The revised equations of motion for this method are given below (equations [12], [13], [14], [15], [16], [17], and [18]), where W is the mass of the piston, τ is the oscillation period, R is the noise on the atoms, and Re is the noise on the piston (10):
| [12] |
| [13] |
| [14] |
| [15] |
| [16] |
| [17] |
| [18] |
1.4. Atomistic Versus Coarse-Grained Simulations
The previous discussion assumes the implementation of fully atomistic simulations. This is the preferred method for the study of antimicrobial peptides, as the ability of the force fields to reproduce structural and conformational changes diminishes with coarse-grained force fields. However, coarse-grained simulations offer a computationally cheaper approach. In these simulations, the atoms are not represented individually, but are grouped into “pseudo-atoms.” This reduction of the system can allow for the simulation of larger systems or for longer timescales.
The level to which the system is reduced can be from the simplest united-atom model, in which the two hydrogens of a methylene group are represented together with the carbon, to groupings of pseudo-atom “beads” that represent two to four methylene groups. As with all-atom simulations, the parameterization for the interactions between beads is empirical. At higher levels of coarse graining, the accuracy of the system behavior is less reliable. For example, simulation of pore formation by the frog AMP magainin has been carried out with a coarse-grained method (11). Simulations of this type might be useful in some applications, but are not the focus of this chapter.
1.5. Implicit Solvation
Implicit solvation (or continuum solvation) is another method for simplifying a fully atomistic simulation. Instead of using fully atomistic “explicit” water molecules, the solvent is represented as a continuous medium, wherein a mean field approach can be used to approximate the behavior of individual solvent molecules. Two common methods use the Poisson–Boltzmann equation or the Generalized Born model. For a more thorough review of the different implicit solvation models, the reader is referred to (12). The Poisson–Boltzmann equation models the electrostatic environment of a solute as a polarizable continuum. In order to further reduce computational expense, more approximations are usually made. For instance, the Generalized Born model is an approximation to the linearized Poisson–Boltzmann equation (13). It is based on modeling the solute as a sphere with different internal dielectric constant than the solvent. Any type of implicit solvation can be used to reduce the number of atoms in the system, but results in the loss of data for the movement of the individual ions and solvent molecules in the system.
1.6. Common Simulation Systems
It has been proposed that AMPs act to kill bacteria through a series of steps: attraction, aggregation, penetration, and lysis (14). Simulations of this process would provide the ultimate level of detail in understanding the mechanism of action of AMPs; however, the timescales on which these processes occur exceed what is currently possible with all-atom MD simulations. Formation of pores has been observed in coarse-grained MD simulations (15, 16), but such simulations do not provide the same resolution as atomistic approaches, and may not accurately reflect system properties and behavior. To accommodate the current limitations on computational power, we can explore various steps of this process individually (Fig. 17.2)
Fig. 17.2.

The three types of systems typically investigated. Peptide in water (with ions) is shown in (a). This type of simulation can be completed in days. View (b) shows a typical peptide–micelle simulation. A typical micelle simulation can be completed in a span of several weeks. A lipid bilayer is shown in (c). With current computational resources, a lipid bilayer simulation takes several months to complete.
1.6.1. Simulations in Water
One of the simplest systems used in MD of AMPs consists of peptides in water. The simulation box is composed of a single peptide, water, counterions (if necessary to achieve charge neutrality), and additional salt if desired. This can be done simply to study the conformational motions of the peptide, or to measure physicochemical properties that might be related to the activity or toxicity of the peptide. When a set of data is available for a large number of related peptides, predictive quantitative structure–activity relationships (QSARs) can be developed. This is an approach widely used in computer-aided drug design (as described in 17 and demonstrated in 18–23). QSARs, obtained through multiple linear regression, correlate properties calculated from the peptide–water simulations to toxicity of the peptide. The hypothesis is that the properties of monomeric peptides can be related to their hemolytic activity and cytotoxicity. Simulations in water require significantly less computational resources than AMPs in environments mimicking lipid membrane, but can nevertheless provide meaningful results.
1.6.2. Simulations in Micelles
Simulations in water can provide a method for rapid screening of new sequences based on properties of single peptides, but in order to obtain information as to how the peptides interact with membrane mimics, and to elucidate the structural or sequence basis for the varying levels of toxicity in several peptide analogues, their interactions with membrane-mimicking micelles can be observed. There are several reasons to use micelles as membrane mimics. Like the lipid bilayers of real membranes, micelles possess a well-defined hydrophobic core and a flexible, hydrophilic interface and are commonly used in place of monolayers or bilayers in experimental methods such as NMR spectroscopy (24–27). Simulations in micelles are much less time and resource consuming than simulations in lipid bilayers, due to their smaller size and faster relaxation times, which have been shown through both direct experiments and simulations to be on the order of 500–1,000 ps for micelles (28–32), in contrast with lipid bilayers, which require tens of nanoseconds (33, 34).
Molecular dynamics studies of micelles have been performed for many types of micelles, including anionic micelles such as those formed by sodium dodecyl sulfate (SDS) (29, 35–38); zwitterionic micelles (e.g., dodecylphosphocholine, DPC) (30, 31, 39–41); and a few mixed composition micelles (42, 43). DPC micelles are considered to be acceptable models of eukaryotic cell membranes, which are generally rich in zwitterionic phospholipids. SDS has been used as a mimic for the negatively charged molecules found in bacterial membranes, because SDS micelles possess an anionic exterior and a hydrophobic interior. The differences of peptide behavior between the SDS micelle and the DPC micelle provide a basis for the study of the activity and toxicity of selected AMPs. Indeed, it is quite possible that AMPs work through different mechanisms to kill bacterial or mammalian cells. In the case of the unstructured indolicidin, for example, the peptide takes on different conformations when interacting with the two types of micelles (44).
1.6.3. Simulations in Lipid Bilayers
The long timescales on which aggregation and penetration of the bilayer by AMPs occur have prohibited the use of fully atomistic MD simulations to study these phenomena. The computational power needed to observe such phenomena using fully atomistic simulations is currently not available. It is possible, however, to study systems for which an initial orientation has been suggested from experimental data. For example, the pore structure of protegrin-1 in a mixed phosphatidylethanolamine (POPE): phosphatidylglycerol (POPG) bilayer was determined from NMR experiments. This was used as a starting configuration for MD simulations of the pore system, allowing a molecular level view of the interactions between the pore and the bilayer system, while eliminating the problems associated with initialization of the system (45).
2. Materials
PDB (Protein Data Bank, http://www.rcsb.org) file for AMP of interest with atomic coordinates.
Coordinate files for micelles or lipids, as needed.
Visualization software, such as VMD or Rasmol.
CHARMM topology and parameter sets (available for free download at http://www.cems.umn.edu/research/kaznessis/antimicrobial.htm.)
Simulation engine (NAMD or CHARMM recommended).
Access to supercomputers or UNIX machines.
3. Methods
3.1. System Construction
The initial configuration of the peptide should be based on an experimentally determined structure (e.g., through X-ray crystallography or NMR), since simulations are not at the point to be able to properly fold a peptide in a reasonable amount of time. The initial peptide coordinate file can be obtained from the Protein Data Bank for many AMPs. The type of simulation will dictate the next steps of system construction.
For simulations carried out in water, typically a large box of water is first constructed. This is often done in CHARMM by copying and translating a box of water. Two pre-made boxes of TIP3P water molecules are available, one containing 125 molecules and the other 216 molecules. The peptide is then placed in the box of water, and the molecules that overlap with the peptide are removed. Counterions and any additional ions should be added at this time. They can be added by replacing water molecules.
For simulations in micelles, coordinates exist for two common types of micelles, DPC and SDS. DPC coordinates can be obtained from the web site of Professor Tuck Wong (http://www.chem.missouri.edu/wong/). SDS coordinates have been published by Alex MacKerell (36). The peptide is can be placed in the center of the micelle with the two centers of mass overlapping. This peptide–micelle complex is then solvated as described in Step 2.
-
A detailed method for building lipid bilayer simulations, with all necessary CHARMM scripts, is available on Benoit Roux’s web site (http://thallium.bsd.uchicago.edu/RouxLab/membrane.html). In this method, the bilayer is built around the peptides, with bad contacts between peptides and lipids or among lipid molecules reduced through a series of rotations and translations. Libraries of lipid conformations are available for a few common lipids, such as dipalmitoylphosphatidylcholine (DPPC). A brief summary of Roux’s method is outlined below:
-
4.1
Use sys1.inp to find the cross-sectional area of the peptide or oligomer. This information is necessary to determine the size of the lipid bilayer that will be needed.
-
4.2
Use sys2.inp to place dummy atoms into preliminary top and bottom leaflets. These atoms are replaced by the head group atoms of the lipids in subsequent steps. The height of the bilayer must be specified based on the length of lipids being used.
-
4.3
Sys3.inp minimizes the dummy atoms.
-
4.4
Sys4.inp requires the library of lipid conformers. In this step, each dummy atom is replaced with a lipid molecule. Each lipid is then systematically translated and rotated to remove bad contacts (such as overlapping atoms).
-
4.5
Use sys5.inp to minimize the system, which at this time is composed of peptide(s) and lipids.
-
4.6
Use sys6.inp and sys7.inp to make a sheet and then a box of water that is of the appropriate size for the system. Sys8.inp is then used to position the water box on each side of the bilayer.
-
4.7
Use sys9.inp to further minimize the system.
-
4.8
Additional minimization scripts (sys10.inp to sys18.inp) can be used.
-
4.1
Create a.psf file for the fully constructed system.
3.2. Running a NAMD Simulation
Convert the prepared coordinate file to a NAMD.pdb file. This can be done using the program crd2pdb, which is available on the NAMD web site (http://www.ks.uiuc.edu/Research/namd/).
Convert the primary structure file (.psf) to a NAMD.psf file using charmm2namd, which is also available on the NAMD web site.
-
Create the simulation script. The following parameters need to be set (see Table 17.1):
-
3.1
Dimensions for the simulation box.
-
3.2
Pressure and temperature at which to run the simulation.
-
3.3
The number of time steps between each output (OutputEnergies, xstFreq, and dcdFreq). They need to be small enough to allow for capture of the details of interest in the simulation but large enough so that the trajectory file does not become too large for storage.
-
3.4
Specify cell basis vectors for periodic boundary conditions. For hexagonal cells, the vectors are [(a, 0, 0), (a/2, , 0), (0, 0, h)], where a is the width of the box (defined as the distance from the center of the simulation to the center of the adjacent image box) and h is the height. For rhombic dodecahedron cells, the vectors are [(a, 0, 0), (0, a, 0), (a/2, a/2, )], where a is the box length (defined as the distance from the center of the simulation to the center of the adjacent image box).
-
3.5
Turn on wrapAll and wrapNearest if using periodic boundary conditions. If wrapAll is specified, when a molecule leaves the simulation box, its coordinates are translated to the other side of the cell when they are output. If wrapNearest is specified, then the coordinates are wrapped to the nearest image to the origin, not the diagonal unit cell centered on the origin.
-
3.6
Turn on rigidbonds so that a 2 fs time step can be used.
-
3.7
Specify the cutoff distance.
-
3.8
Specify nonBondedFreq and fullElectFrequency (the number of steps between calculations of the non-bonded interactions).
-
3.9
Specify the distance for the pair list (pairlistdist).
-
3.10
Specify how often the pair list should be updated (Stepspercycle).
-
3.11
Turn on switching for the van der Waals interactions.
-
3.12
Specify the distance at which the switching function should be activated.
-
3.13
Specify which non-bonded interactions to exclude (typically exclude 1–2 and 1–3 bonded atoms from the non-bonded interactions). Also specify if the 1–4 interactions should be scaled.
-
3.14
Set up the temperature and pressure controllers. Specify the Langevin damping coefficient, target pressure, piston oscillation period, and piston damping coefficient.
-
3.15
Specify the particle mesh Ewald summation parameters. The function needs to be specified “on” and grid sizes must be provided. The grid should be chosen so that that there is approximately one point per Angstrom.
-
3.1
Minimize the system. The default minimization algorithm in NAMD combines conjugate gradient and line search methods. The amount of minimization necessary is dependent on the size of the system and how well the system was built.
Heat the system to the desired temperature. This should be done gradually. The Langevin piston is turned on and given a low temperature set point (usually less than 100 K). Langevin dynamics are run for a few thousand steps and then the temperature is gradually increased. Increasing by 30 K in each cycle to the final temperature set point should allow for the system to remain stable during the heating process.
Run the simulation. The simulation should be carried out until all properties of interest have stabilized.
Table 17.1.
Description of each term to be specified in the NAMD script. Typical values for each term are given as well
| Term | Description | Typical value |
|---|---|---|
| Outputenergies | Number of time steps between energy outputs | 1,000 |
| xstFreq | Number of time steps between to the extended system coordinate file | 1,000 |
| dcdFreq | Number of time steps between writing to the trajectory file | 1,000 |
| wrapAll | Use periodic boundary conditions on all atoms | On |
| wrapNearest | Use nearest image to wrap the coordinates | On |
| rigidbonds | Fixes hydrogen bond lengths to the nominal value | All |
| timestep | Time step in femtoseconds | 2 |
| cutoff | Distance at which non-bonded and bonded interactions are cutoff | 11 |
| nonBondedFreq | Number of time steps between evaluation of the non-bonded interactions | 1 |
| fullElectFrequency | Number of time steps between evaluation of electrostatic interactions | 1 |
| pairlistdist | Distance between pairs for inclusion on the pairlist | 14 |
| stepsPerCycle | Number of time steps between atom reassignments | 20 |
| switching | Use switching for the non-bonded van der Waals interactions | On |
| switchDist | Distance at which the switching function should start to switch | 8 |
| exclude | Which pairs of bonded atoms to exclude | Scaled 1–4 |
| 1–4scaling | Constant factor to modify the electrostatic interactions for the pairs that are modified by exclude | 1 |
| langevin | Use Langevin dynamics | On |
| langevinDamping | Damping coefficient for Langevin dynamics | 10 |
| langevinTemp | Temperature for Langevin calculations | $temp |
| langevinHydrogen | Do not apply Langevin dynamics to hydrogens | No |
| langevinPiston | Use Langevin piston pressure control | On |
| langevinPistonTarget | Target pressure for Langevin piston pressure control | 1.01325 |
| langevinPistonPeriod | Barostat oscillation time in femtoseconds | 200 |
| langevinPistonDecay | Damping timescale in femtoseconds | 10 |
| langevinPistonTemp | Noise temperature | $temp |
4. Notes
There are a number of issues that can arise in the construction of the system. In particular, bad contacts can be built into the system in such a way that they cannot be overcome. In particular, one must be careful about the arrangement of lipid or detergent molecules near aromatic residues. The carbon chains can be placed so that they are inserted through the phenyl ring and will remain there for the duration of the simulation. Similar care must be taken for the position of the carbon chains near constrained loops, such as in β-hairpin peptides.
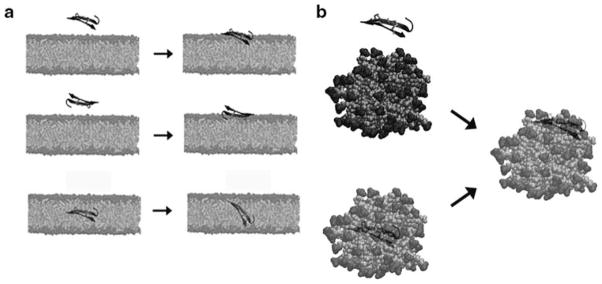
The initial configuration of the system is extremely important. There are still issues in the initialization of peptide–lipid systems because, in general, convergence of peptide–lipid systems is difficult to achieve due to the slow timescales on which the peptides are able to re-orient themselves when binding to the bilayer. Because of this, the proper orientation of the peptides at the start of the simulation is crucial. The need for prior knowledge of the binding conformation, and the restrictions on the accessible timescales, has hindered the predictive ability of lipid bilayer simulations. Specific information about the location of the peptide in a lipid bilayer is rarely available and so one has to assume a starting orientation and position for the peptide. This choice has a significant influence on the behavior of the peptide and on the conformation of the system at the end of the simulation. This problem can be overcome, in part, by using micelles. By starting with the peptide inside the micelle, the peptide “sees” the same surroundings regardless of the initial orientation (Fig. 17.3)
Setup for systems can be done without using CHARMM. Scripts have been written for building systems using VMD (Visual Molecular Dynamics) and are available at the NAMD/VMD web site (http://www.ks.uiuc.edu/Research/vmd/).
The orientation of the simulation box vectors is different in CHARMM than in NAMD. For example, the hexagonal simulation box, recommended for lipid bilayer simulations, is rotated 15 degrees counterclockwise in CHARMM as compared to NAMD. Care must be taken in the start of the simulation of a system that has been built in CHARMM but is being run in NAMD.
The setup of the system should be checked visually at each step using VMD or Rasmol to ensure the proper orientation of molecules relative to each other.
When constructing a lipid bilayer simulation for a pore, care must be taken in the initial stages to prevent the lipids from entering the interior of the pore. MMFP (miscellaneous mean field potential; see mmfp.doc in the CHARMM documentation) constraints are used in sys2.inp in the construction of any lipid bilayer to maintain the proper bilayer thickness. Cylindrical constraints can be used to prevent the lipids from entering the pore.
For simulations of peptides in water or interacting with micelles, the constant pressure–temperature ensemble is recommended. For lipid bilayer simulations, there is some discussion over the proper ensemble (46–48). Although the NPT (constant pressure–constant temperature) ensemble is used in most simulations, problems with the area per lipid can arise for lipid bilayer simulations. This is particularly true for simulations using CHARMM, for which NPzAT ensemble – where the area in the x–y plane is held constant but the z-dimension, normal to the bilayer, is allowed to fluctuate to retain constant pressure – or the NPγT ensemble – where the surface tension is held constant – is recommended. In the simulations that have been presented in the literature, the simulation cell in NPT simulations using CHARMM or NAMD regularly shrinks, reducing the area per lipid. In 2004, Jensen and coworkers simulated a lipid bilayer containing 36 DPPC lipids per leaflet in the NPT ensemble and observed that the x–y dimensions of the cell decreased for the entire 15-ns simulation (49). Images of this system show an unrealistic expansion of the lipid in the z-direction, such that the two leaflets have separated. Feller and Pastor explained the unnatural behavior of lipid bilayers in simulations as being due to the small sizes of the bilayers that are studied (46). Long wavelength undulations of the bilayer are absent in small simulation cells, causing an increase in the surface tension above what would normally be expected from a macroscopic bilayer. The discussion on this topic is still open, but the options to run with constant area or a constant ratio of the x- and y-dimensions are available in NAMD.
For lipid bilayer simulations, the hydration of the lipids and the concentration of ions can have strong effects on the results. When experimental data are available, it should be matched as closely as possible.
Analysis can be done using CHARMM. A number of scripts for analysis of simulations are available in the script archive on the CHARMM Forum (www.charmm.org). Analysis will vary depending on the type of simulation carried out.
All simulations can provide information about the structure of the peptide. The secondary structure can be measured for each time point of the simulation by calculation of the dihedral angles. It is possible to compare the root mean square deviation of the peptide through the simulation to the initial structure or some other chosen structure. The dipole moment of the peptide is easily calculated, as well as the surface area. The area exposed to the solvent can also be easily determined. The shape of the peptide can be measured in terms of eccentricity or the maximum or average dimensions. The moment of inertia and radius of gyration are also easily calculated. The interaction energies between the residues within the peptide or the peptide and water can also be calculated and broken down further into the van der Waals interaction energy and the electrostatic interaction energy. Additionally, though hydrogen bonds are not explicitly modeled in CHARMM, the energy due to hydrogen bond interactions that have been defined by atom type, distance, and angle can be calculated.
Simulations in micelles allow for calculation of peptide properties as well as interaction energies (van der Waals and electrostatic) between peptide and self, peptide and micelle, or peptide and water. Hydrogen bond energy can also be calculated. Micelle shape can be analyzed over the course of the simulation, by monitoring the moments of inertia.
Radial distribution functions with micelle or bilayer core or head groups can provide information about the strength of interaction for each residue and the desired system component.
For bilayer simulations, properties such as thickness, tilt of lipids, order parameters, trans-gauche conformations, hydrogen bonding between head groups, and interactions with ions are often of importance.
Fig. 17.3.
Initialization issues. (a) For lipid bilayer simulations, the initial configuration has a strong bias on the final conformation observed. All three starting configurations should reach the same final conformation, the lower right, which is supported by experimental data. (b) For micelles, the final conformation can be reached with the peptide starting inside or outside the micelle, although again, it is unlikely to observe a peptide rotating while embedded into the micelle, so for simulations with the peptide starting outside of the micelle, the proper “face” must be positioned nearest the micelle surface. Using the conformation with the center of mass of the peptide overlapping the micelle center of mass reduces the bias.
References
- 1.Allen MP, Tildesley DJ. Computer Simulations of Liquids. Oxford: Clarendon Press; 1987. [Google Scholar]
- 2.Pearlman DA, Case DA, Caldwell JW, Ross WS, Cheatham TE, III, DeBolt S, Ferguson D, Seibel G, Kollman P. Amber, a computer program for applying molecular mechanics, normal mode analysis, molecular dynamics and free energy calculations to elucidate the structures and energies of molecules. Comp Phys Commun. 1995;91:1–41. [Google Scholar]
- 3.Jorgensen WL, Tirado-Rives J. The OPLS [optimized potentials for lipid simulations] potential functions for proteins, energy minimizations for crystals of cyclic peptides and crambin. J Am Chem Soc. 1988;110:1657–1666. doi: 10.1021/ja00214a001. [DOI] [PubMed] [Google Scholar]
- 4.Brooks BR, Bruccoleri RE, Olafson BD, States DJ, Swaminathan S, Karplus M. CHARMM: a program for macromolecular energy, minimization, and dynamics simulations. J Comp Chem. 1983;4:187–217. [Google Scholar]
- 5.Berendsen HJC, van der Spoel DL, van Drunen R. GROMACS: a message-passing parallel molecular dynamics implementation. Comp Phys Comm. 1995;91:43–56. [Google Scholar]
- 6.Phillips JC, Braun R, Wang W, Gumbart J, Tajkhorshid E, Villa E, Chipot C, Skeel RD, Kale L, Schulten K. Scalable molecular dynamics with NAMD. J Comput Chem. 2005;26:1781–1802. doi: 10.1002/jcc.20289. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Patra M, Karttunen M, Hyvönen MT, Falck E, Lindqvist P, Vattulainen I. Molecular dynamics simulations of lipid bilayers: major artifacts due to truncating electrostatic interactions. Biophys J. 2003;84:3636–3645. doi: 10.1016/S0006-3495(03)75094-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Martyna GJ, Tobias DJ, Klein ML. Constant pressure molecular dynamics algorithms. J Chem Phys. 1994;101:4177–4189. [Google Scholar]
- 9.Feller SE, Zhang Y, Pastor RW, Brooks BR. Constant pressure molecular dynamics simulation: the Langevin piston method. J Chem Phys. 1995;103:4613–4621. [Google Scholar]
- 10.Bhandarkar M, Brunner R, Chipot C, Dalke A, Dixit S, Grayson P, Gullingsrud J, Gursoy A, Hardy D, Hénin J, Humphrey W, Hurwitz D, Krawetz N, Kumar S, Nelson M, Phillips J, Shinozaki A, Zheng G, Zhu F. NAMD User’s Guide Version 2.6b1. Urbana: Theoretical Biophysics Group, University of Illinois and Beckman Institute; 2005. [Google Scholar]
- 11.Leontiadou H, Mark AE, Marrink SJ. Antimicrobial peptides in action. J Am Chem Soc. 2006;128:12156–12161. doi: 10.1021/ja062927q. [DOI] [PubMed] [Google Scholar]
- 12.Roux B, Simonson T. Implicit solvent models. Biophys Chem. 1999;78:1–20. doi: 10.1016/s0301-4622(98)00226-9. [DOI] [PubMed] [Google Scholar]
- 13.Dominy BN. Parameterization and application of an implicit solvent model for macromolecules. Mol Simulat. 1999;24:259–274. [Google Scholar]
- 14.Brogden KA. Antimicrobial peptides: pore formers or metabolic inhibitors in bacteria? Nat Rev Microbiol. 2005;3:238–250. doi: 10.1038/nrmicro1098. [DOI] [PubMed] [Google Scholar]
- 15.Kandasamy SK, Larson RG. Binding modes of protegrin-1, a beta-strand antimicrobial peptide, in lipid bilayers. Mol Simulat. 2007;33:799–807. [Google Scholar]
- 16.Leontiadou H, Mark AE, Marrink SJ. Ion transport across transmembrane pores. Biophys J. 2007;92:4209–4215. doi: 10.1529/biophysj.106.101295. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Parvu L. QSAR – a piece of drug design. J Cell Mol Med. 2003;7:333–335. doi: 10.1111/j.1582-4934.2003.tb00235.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Jenssen H, Fjell CD, Cherkasov A, Hancock RE. QSAR modeling and computer-aided design of antimicrobial peptides. J Pept Sci. 2008;14:110–114. doi: 10.1002/psc.908. [DOI] [PubMed] [Google Scholar]
- 19.Kaznessis YN, Snow ME, Blankley CJ. Prediction of blood-brain partitioning using Monte Carlo simulations of molecules in water. J Comput Aid Mol Des. 2001;15:697–708. doi: 10.1023/a:1012240703377. [DOI] [PubMed] [Google Scholar]
- 20.Lejon T, Stiberg T, Strøm MB, Svendsen JS. Prediction of antibiotic activity and synthesis of new pentadecapeptides based on lactoferricins. J Pept Sci. 2004;10:329–335. doi: 10.1002/psc.553. [DOI] [PubMed] [Google Scholar]
- 21.Lejon T, Strom MB, Svendsen JS. Antibiotic activity of pentadecapeptides modelled from amino acid descriptors. J Pept Sci. 2001;7:74–81. doi: 10.1002/psc.295. [DOI] [PubMed] [Google Scholar]
- 22.Lejon T, Svendsen JS, Haug BE. Simple parameterization of non-proteinogenic amino acids for QSAR of antibacterial peptides. J Pept Sci. 2002;8:302–306. doi: 10.1002/psc.403. [DOI] [PubMed] [Google Scholar]
- 23.Marrero-Ponce Y, Torrens F, Alvarado YJ, Rotondo R. Bond-based global and local (bond, group and bond-type) quadratic indices and their applications to computer-aided molecular design. 1 QSPR studies of diverse sets of organic chemicals. J Comput Aid Mol Des. 2006;20:685–701. doi: 10.1007/s10822-006-9089-4. [DOI] [PubMed] [Google Scholar]
- 24.Beswick V, Guerois R, Cordier-Ochsenbein F, Coïc YM, Tam HD, Tostain J, Noël JP, Sanson A, Neumann JM. Dodecylphosphocholine micelles as a membrane-like environment: new results from NMR relaxation and paramagnetic relaxation enhancement analysis. Eur Biophys J. 1999;28:48–58. doi: 10.1007/s002490050182. [DOI] [PubMed] [Google Scholar]
- 25.Kallick DA, Tessmer MR, Watts CR, Li CY. The use of dodecylphosphocholine micelles in solution NMR. J Magn Reson B. 1995;109:60–65. doi: 10.1006/jmrb.1995.1146. [DOI] [PubMed] [Google Scholar]
- 26.Kloosterman DA, Goodwin JT, Burton PS, Conradi RA, Stockman BJ, Scahill TA, Blinn JR. An NMR study of conformations of substituted dipeptides in dodecylphosphocholine micelles: implications for drug transport. Biopolymers. 2000;53:396–410. doi: 10.1002/(SICI)1097-0282(20000415)53:5<396::AID-BIP4>3.0.CO;2-5. [DOI] [PubMed] [Google Scholar]
- 27.Lauterwein J, Bösch C, Brown LR, Wüthrich K. Physicochemical studies of the protein-lipid interactions in melittin-containing micelles. Biochim Biophys Acta. 1979;556:244–264. doi: 10.1016/0005-2736(79)90046-4. [DOI] [PubMed] [Google Scholar]
- 28.Fernandez C, Wuthrich K. NMR solution structure determination of membrane proteins reconstituted in detergent micelles. FEBS Lett. 2003;555:144–150. doi: 10.1016/s0014-5793(03)01155-4. [DOI] [PubMed] [Google Scholar]
- 29.Rakitin AR, Pack GR. Molecular dynamics simulations of ionic interactions with dodecyl sulfate micelles. J Phys Chem B. 2004;108:2712–2716. [Google Scholar]
- 30.Wymore T, Gao XF, Wong TC. Molecular dynamics simulation of the structure and dynamics of a dodecylphosphocholine micelle in aqueous solution. J Mol Struct. 1999;485:195–210. [Google Scholar]
- 31.Wong TC, Kamath S. Molecular dynamics simulations of adrenocorticotropin (1–24) peptide in a solvated dodecylphosphocholine (DPC) micelle and in a dimyistoylphosphatidylcholine (DMPC) bilayer. J Biomol Struct Dyn. 2002;20:39–57. doi: 10.1080/07391102.2002.10506821. [DOI] [PubMed] [Google Scholar]
- 32.Tieleman DP, van der Spoel D, Berendsen HJC. Molecular dynamics simulations of dodecylphosphocholine micelles at three different aggregate sizes: micellar structure and lipid chain relaxation. J Phys Chem B. 2000;104:6380–6388. [Google Scholar]
- 33.Shepherd CM, Schaus KA, Vogel HJ, Juffer AH. Molecular dynamics study of peptide-bilayer adsorption. Biophys J. 2001;80:579–596. doi: 10.1016/S0006-3495(01)76039-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Kandasamy SK, Larson RG. Binding and insertion of alpha-helical antimicrobial peptides in POPC bilayers studied by molecular dynamics simulations. Chem Phys Lipids. 2004;132:113–132. doi: 10.1016/j.chemphyslip.2004.09.011. [DOI] [PubMed] [Google Scholar]
- 35.Bruce CD, Berkowitz ML, Perera L, Forbes MDE. Molecular dynamics simulation of sodium dodecyl sulfate micelle in water: micellar structural characteristics and counterion distribution. J Phys Chem B. 2002;106:3788–3793. [Google Scholar]
- 36.MacKerell AD. Molecular dynamics simulation analysis of a sodium dodecyl sulfate micelle in aqueous solution: decreased fluidity of the micelle hydrocarbon interior. J Phys Chem. 1995;99:327–341. [Google Scholar]
- 37.Wymore T, Wong TC. Molecular dynamics study of substance P peptides partitioned in a sodium dodecylsulfate micelle. Biophys J. 1999;76:1213–1227. doi: 10.1016/S0006-3495(99)77285-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Yang ST, Yub Shin SY, Kim YC, Kim Y, Hahm KS, Kim JI. Conformation-dependent antibiotic activity of tritrpticin, a cathelicidin-derived antimicrobial peptide. Biochem Biophys Res Commun. 2002;296:1044–1050. doi: 10.1016/s0006-291x(02)02048-x. [DOI] [PubMed] [Google Scholar]
- 39.Bond PJ, Cuthbertson J, Sansom MS. Simulation studies of the interactions between membrane proteins and detergents. Biochem Soc Trans. 2005;33:910–912. doi: 10.1042/BST20050910. [DOI] [PubMed] [Google Scholar]
- 40.Bond PJ, Cuthbertson JM, Deol SS, Sansom MS. MD simulations of spontaneous membrane protein/detergent micelle formation. J Am Chem Soc. 2004;126:15948–15949. doi: 10.1021/ja044819e. [DOI] [PubMed] [Google Scholar]
- 41.Bond PJ, Sansom MS. Membrane protein dynamics versus environment: simulations of OmpA in a micelle and in a bilayer. J Mol Biol. 2003;329:1035–1053. doi: 10.1016/s0022-2836(03)00408-x. [DOI] [PubMed] [Google Scholar]
- 42.Marrink SJ, Mark AE. Molecular dynamics simulations of mixed micelles modeling human bile. Biochemistry. 2002;41:5375–5382. doi: 10.1021/bi015613i. [DOI] [PubMed] [Google Scholar]
- 43.Wendoloski JJ, Kimatian SJ, Schutt CE, Salemme FR. Molecular dynamics simulation of a phospholipid micelle. Science. 1989;243:636–638. doi: 10.1126/science.2916118. [DOI] [PubMed] [Google Scholar]
- 44.Khandelia H, Kaznessis Y. Cation-pi interactions stabilize the structure of the antimicrobial peptide indolicidin near membranes: molecular dynamics simulations. J Phys Chem B. 2007;111:242–250. doi: 10.1021/jp064776j. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Langham AA, Ahmad AS, Kaznessis YN. On the nature of antimicrobial activity: a model for protegrin-1 pores. J Am Chem Soc. 2008;130:4338–4346. doi: 10.1021/ja0780380. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Feller SE, Pastor RW. On simulating lipid bilayers with an applied surface tension: periodic boundary conditions and undulations. Biophys J. 1996;71:1350–1355. doi: 10.1016/S0006-3495(96)79337-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Jahnig F. What is the surface tension of a lipid bilayer membrane? Biophys J. 1996;71:1348–1349. doi: 10.1016/S0006-3495(96)79336-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Roux B. Commentary: surface tension of biomembranes. Biophys J. 1996;71:1346–1347. doi: 10.1016/S0006-3495(96)79335-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Jensen MO, Mouritsen OG, Peters GH. Simulations of a membrane-anchored peptide: structure, dynamics, and influence on bilayer properties. Biophys J. 2004;86:3556–3575. doi: 10.1529/biophysj.103.029140. [DOI] [PMC free article] [PubMed] [Google Scholar]