IkBβ forms a complex with the NF-κB subunits RelA and c-Rel that inhibits the transcription of IL-1β and other genes. Mice lacking IkBβ are protected against LPS-induced shock.
Abstract
Inhibitor of κB (IκB) β (IκBβ) represents one of the major primary regulators of NF-κB in mammals. In contrast to the defined regulatory interplay between NF-κB and IκBα, much less is known about the biological function of IκBβ. To elucidate the physiological role of IκBβ in NF-κB signaling in vivo, we generated IκBβ-deficient mice. These animals proved to be highly refractory to LPS-induced lethality, accompanied by a strong reduction in sepsis-associated cytokine production. In response to LPS, IκBβ is recruited to the IL-1β promoter forming a complex with the NF-κB subunits RelA/c-Rel required for IL-1β transcription. Further transcriptome analysis of LPS-stimulated wild-type and IκBβ-deficient BM-derived macrophages revealed several other genes with known regulatory functions in innate immunity arguing that a subset of NF-κB target genes is under control of IκBβ. Collectively, these findings provide an essential proinflammatory role for IκBβ in vivo, and establish a critical function for IκBβ as a transcriptional coactivator under inflammatory conditions.
NF-κB plays an important role in the regulation of diverse biological processes such as development, immune and inflammatory responses, and apoptosis (Baldwin, 1996; Gilmore, 2006; Ghosh and Hayden, 2008). Through its ubiquitous appearance, NF-κB is involved in regulation of a wide range of genes, such as genes encoding cytokines, adhesion molecules, cytokine receptors, immunoregulatory molecules, and antiapoptotic proteins. In mammals, the NF-κB transcription factor family includes five members: p50/NF-κB1, p52/NF-κB2, RelA/p65, c-Rel, and RelB (Ghosh and Karin, 2002; Ghosh and Hayden, 2008). These polypeptide subunits form homo- and heterodimers that are sequestered through stable association with inhibitor of κB (IκB) proteins in the cytoplasm of resting cells. Activators of the NF-κB pathway, such as cytokines, growth factors, and bacterial and viral products, strongly enhance the activity of the IκB kinase complex (IKK). IKK phosphorylates the IκB inhibitor proteins, leading to their rapid proteasomal degradation (Karin and Ben-Neriah, 2000; Ghosh and Hayden, 2008). After degradation of IκB, NF-κB dimers are able to enter the nucleus, bind specifically to DNA, and modulate transcription of various target genes.
Because the initial discovery of the IκB proteins as the cytoplasmic inhibitors of NF-κB, considerable effort has been given to understand regulation and modes of action (Baeuerle and Baltimore, 1988). IκBα and IκBβ are the major signal-responsive isoforms within the IκB family that also includes IκBε, IκBγ, p100, p105, Bcl-3, and the newly described IκBζ (Yamamoto et al., 2004; Hoffmann and Baltimore, 2006). Although IκBα and IκBβ show many common structural features, they exhibit functional differences (Thompson et al., 1995; Tran et al., 1997). IκBα is rapidly degraded upon stimulation, followed by immediate NF-κB–dependent resynthesis. Newly synthesized IκBα enters the nucleus and removes NF-κB complexes from the DNA to export them back to the cytoplasm (Sun et al., 1993; Klement et al., 1996; Hoffmann et al., 2002). In contrast, IκBβ is degraded much more slowly, and its resynthesis is not regulated by NF-κB. Depending on the cell type and stimulus, IκBβ undergoes persistent degradation, contributing to constitutive NF-κB activation (Thompson et al., 1995; Bourke et al., 2000). Furthermore, it has been shown that IκBα–NF-κB complexes undergo cytoplasmic to nuclear shuttling in resting cells, whereas IκBβ–NF-κB complexes commonly stay in the cytoplasm (Tran et al., 1997; Huang and Miyamoto, 2001; Malek et al., 2001; Ghosh and Karin, 2002). A hypophosphorylated form of IκBβ has been shown to reside in the nucleus of certain cell types upon stimulation. Nuclear IκBβ is capable of forming a complex with DNA–NF-κB dimers, but is unable to dislocate NF-κB from the DNA, thereby prolonging NF-κB activity (Suyang et al., 1996; DeLuca et al., 1999).
Accumulating evidence points to a broader nuclear function of the IκB protein family (Bates and Miyamoto, 2004). Certain IκB protein family members associate specifically with definite NF-κB proteins, acting as transcription coactivators at distinct genes. Thus, IκBα cooperates with RelA/p65 in the regulation of the Notch-target gene hes1 after stimulation with TNF (Aguilera et al., 2004). IκBζ associates specifically with p50 to the NF-κB–binding site of the IL-6 promoter (Yamamoto et al., 2004).
Less is known regarding the function of IκBβ in vivo. To analyze the physiological function of IκBβ, we generated IκBβ-deficient (IκBβ−/−) mice. We demonstrate that IκBβ−/− mice are highly resistant to LPS-induced septic shock. LPS resistance is caused by impaired cytokine expression in IκBβ−/− mice. Using the IL-1β gene as a model of IκBβ regulated NF-κB target genes, we demonstrate that IκBβ is essential for IL-1β production upon LPS. In addition, we show that the transcription of IL-1β depends on a positively acting p65–c-Rel–IκBβ complex.
RESULTS
Generation and immunological phenotype of IκBβ−/− mice
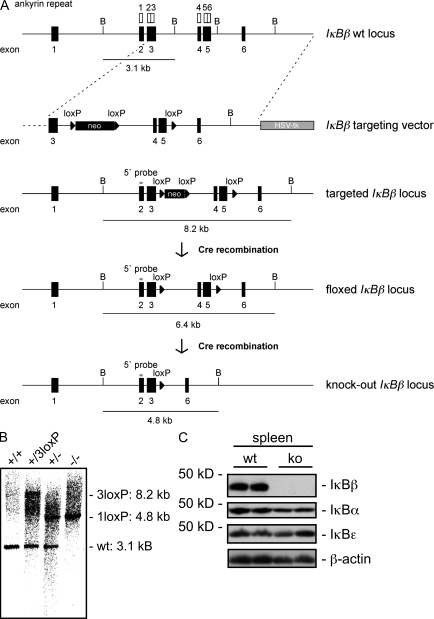
To elucidate the physiological role of IκBβ, we generated IκBβ−/− mice using targeted gene disruption (as described in the Materials and methods section; Fig. 1 A). Exons 4 and 5 of the IκBβ gene, which code for the ankyrin repeats 4–6 that are essential for the function of IκB proteins and the binding to NF-κB, were deleted (Inoue et al., 1992). Southern blot analysis of the genomic tail DNA of F2 mice demonstrated a complete deletion of the IκBβ alleles (Fig. 1 B). Western blot analysis of whole spleen extracts indicated that IκBβ expression was completely abolished in IκBβ−/− mice with no change in the expression of IκBα and IκBε (Fig. 1 C).
Figure 1.
Disruption of the IκBβ gene. (A) Schematic structure of WT IκBβ locus. Ankyrin repeats of IκBβ encoded by exons 2–5 are indicated. Furthermore, the targeting vector, the targeted IκBβ locus, the floxed IκBβ locus, and the IκBβ knock-out locus, generated by Cre recombination-mediated deletion of exons 4 and 5 are shown. Solid boxes represent exons, and lines represent introns. Neo, loxP-flanked PGK-neomycin cassette; HSV-tk, HSV-thymidine kinase gene; B, BamHI site. The length of BamHI-generated restriction fragments detected by Southern blotting with a 5′ flanking probe is indicated. Location of the 5′ flanking probe in exon 2 is shown. (B) Southern blot analysis of genomic DNA from targeted ES cells (+/3loxP), WT (+/+) mice, IκBβ+/−, and IκBβ−/− F2 mice. (C) Immunoblot analysis of IκBβ, IκBα, and IκBε in whole-cell extract of WT and IκBβ−/− (ko) spleens. The membrane was stripped and probed for β-actin to ensure equal protein loading.
IκBβ−/− mice born with the expected Mendelian frequency were viable and showed no distinct abnormalities in appearance. Flow cytometric analysis of isolated spleen cells from WT and IκBβ−/− mice demonstrated an increase in marginal zone B cells and a reduction of naive B cells (Fig. S1, A and B). Furthermore, increased memory T cell population in the spleen of IκBβ−/− mice was observed (Fig. S1 C) and analyses of BM exhibited an increase in BM-derived macrophages (BMDMs) in IκBβ−/− mice (Fig. S1 D).
IκBβ−/− mice are highly resistant to LPS-induced septic shock
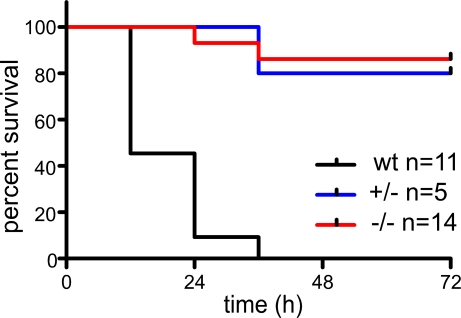
Previous studies demonstrated that LPS stimulation leads to IκBβ degradation and persistent NF-κB activity. To elucidate the role of IκBβ during endotoxic shock, we analyzed the LPS responsiveness in IκBβ−/− mice (Thompson et al., 1995). Mice were intraperitoneally injected with a high dose of LPS (30 mg/kg), and survival was monitored (Fig. 2). IκBβ−/− mice demonstrate a remarkable resistance to the lethal effect of LPS (log-rank test IκBβ−/− versus WT, P < 0.0001) in contrast to WT animals that showed a 100% lethality within 36 h after LPS injection. Although surviving IκBβ+/− and IκBβ−/− mice showed signs of LPS-induced shock in the first hours, they completely recovered after 72 h, arguing for a significant protection against LPS-induced septic shock. The phenotype of the heterozygous mice suggests that both IκBβ alleles are required for the full LPS response in vivo.
Figure 2.
Survival of IκBβ−/− mice after high-dose LPS challenge. Survival curves of IκBβ−/−, IκBβ+/−, and WT mice after the injection of LPS (30 mg/kg). Kaplan-Meier analysis demonstrated a significant difference in survival between IκBβ−/− and WT (log-rank test IκBβ−/− versus WT; P < 0.0001). Data are from three separate experiments and the number of mice in each group is indicated.
Deficiency of IκBβ reduces the biosynthesis of the proinflammatory cytokines TNF, IL-1β, and IL-6
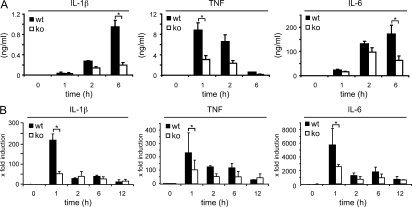
LPS induces rapid production of inflammatory cytokines in vivo, leading to multiorgan failure of the host (Morrison and Ryan, 1987). To determine whether increased resistance to endotoxic shock in IκBβ−/− mice was caused by decreased proinflammatory cytokines, we measured serum concentration of inflammatory cytokines TNF, IL-1β, and IL-6 after LPS challenge. As expected, the serum levels of these cytokines were significantly increased upon LPS treatment in WT mice (Fig. 3 A). In contrast, only a moderate increase of the serum concentrations of TNF, IL-1β, and IL-6 was observed in IκBβ−/− mice. Furthermore, mRNA levels of TNF, IL-1β, and IL-6 in the liver after LPS injection remained significantly lower in IκBβ−/− mice (Fig. 3 B). These data suggest that IκBβ is essential for in vivo production of inflammatory cytokines during LPS-induced septic shock.
Figure 3.
Deficiency in IκBβ reduces LPS-triggered production of sepsis inducing cytokines. (A) Serum levels of IL-1β, TNF, and IL-6 in IκBβ−/− and control mice after LPS injection (30 mg/kg). TNF, IL-1β, and IL-6 were measured in serum collected from tail vain 0, 1, 2, and 6 h after injection of LPS (Student’s t test; *, P < 0.001 versus controls). (B) Liver IL-1β, TNF, and IL-6 mRNA expression levels after LPS challenge (30 mg/kg). At the indicated time points after LPS injection (30 mg/kg), total RNA from whole livers were prepared and mRNA levels were quantified using real-time PCR analysis (Student’s t test; *, P < 0.05 versus controls). For each time point, four animals per strain were examined in two independent experiments.
IκBβ deficiency modify IL-1β cytokine production in LPS-stimulated BMDMs
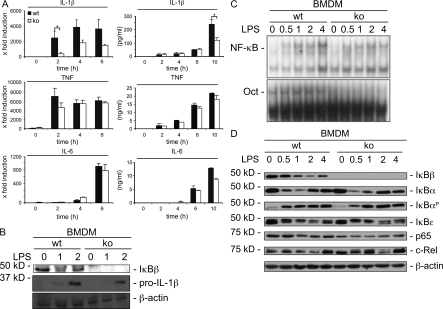
As macrophages represent the major source of inflammatory cytokine production during sepsis, we analyzed cytokine expression in WT and IκBβ−/− BMDMs. In contrast to WT BMDMs that showed increased TNF, IL-6, and IL-1β mRNA expression and protein secretion upon LPS stimulation, endotoxin treatment led to impaired IL-1β mRNA induction, pro–IL-1β expression, and IL-1β secretion in IκBβ−/− BMDMs (Fig. 4, A and B). Although no significant changes in either TNF mRNA induction or secretion were observed after LPS treatment, IL-6 secretion was affected only at later time points in IκBβ−/− BMDMs (Fig. 4 A), suggesting specific regulation of IL-1β transcription by IκBβ in BMDMs. In addition to IL-1β, we observed in transcriptome profiles of LPS-stimulated WT and IκBβ−/− BMDMs that several other genes that are known to be important for the regulation of innate immunity are LPS induced in an IκBβ-dependent manner (Table S1).
Figure 4.
Knockout of IκBβ exhibit reduced IL-1β expression in macrophages (BMDMs). (A) WT and IκBβ−/− BMDM (three animals for each group) were stimulated with LPS (1 µg/ml) as indicated. Total RNA was prepared and IL-1β, TNF, and IL-6 mRNA levels were quantified using real-time PCR analysis (left; Student’s t test; *, P < 0.01 versus control). IL-1β, IL-6, and TNF cytokine secretion in response to LPS was determined by ELISA (right; Student’s t test; *, P < 0.01 versus control). Results are shown as the mean of two independent experiments. (B) BMDMs from WT and IκBβ−/− mice were stimulated with LPS (1 µg/ml) for indicated time points or were left as an untreated control. Western blot detects IκBβ and pro–IL-1β expression. Similar results were obtained from two additional experiments. β-actin was used as loading control. (C) EMSA using a radiolabeled probe containing an NF-κB–binding site. BMDMs were LPS-treated (100 ng/ml) as indicated and nuclear extracts were analyzed by EMSA. An Oct-consensus oligonucleotide was used to control equal protein input. Three independent experiments revealed similar results. (D) BMDMs isolated from WT and IκBβ−/− mice were stimulated with LPS (100 ng/ml) and analyzed by Western blot. Expression levels of indicated NF-κB proteins and IκB members were determined at the indicated time points. One out of three independent experiments is shown.
Binding of LPS to Toll-like receptor 4 (TLR4) activates NF-κB through IKK in a MyD88-dependent manner. To test for altered signaling kinetics and DNA binding in IκBβ−/− BMDMs, we performed electrophoretic mobility shift assays (EMSAs). Once normalized to Oct1 DNA binding, LPS-induced NF-κB signaling kinetics were indistinguishable (Fig. 4 C). No differences in the protein expression level of NF-κB1 or NF-κB2 were evident in BMDMs of IκBβ−/− mice (unpublished data). RelA/p65 expression was decreased and c-Rel expression increased in BMDMs of IκBβ−/− mice (Fig. 4 D). IκBα was phosphorylated and both, IκBα and IκBε, were degraded with the same kinetics upon LPS treatment in WT and IκBβ−/− mice (Fig. 4 D). To determine whether the defect of IκBβ−/− macrophages that produce IL-1β was specific to LPS, we stimulated cells with TNF and other TLR ligands. The IκBβ deficiency did not influence IL-1β secretion induced by TNF, CpgA (TLR9 agonist), and CpgB (TLR9 agonist) in BMDMs (Fig. S2). Although IL-1β secretion was decreased in polyI:C (TLR3 agonist) and Pam3CysSK4 (TLR2 agonist)-treated BMDMs, this reduction was not statistically significant (Fig. S2). Collectively, these data argue that IκBβ functions specifically in certain NF-κB pathways.
Influence of IκBβ small interfering RNA (siRNA) on IL-1β transcriptional regulation
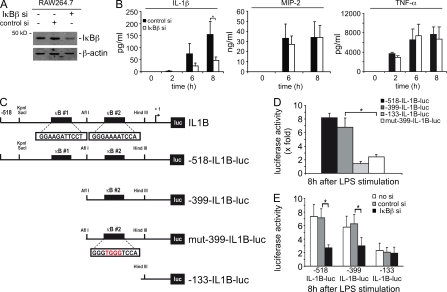
To dissect the function of IκBβ in IL-1β expression in further detail in vitro, we used RNA interference in the macrophage cell line RAW264.7. Transfection of RAW264.7 macrophages with an IκBβ-specific siRNA led to a pronounced reduction in the IκBβ protein expression (Fig. 5 A). To test the IκBβ function in IL-1β secretion, we stimulated IκBβ siRNA-transfected RAW264.7 macrophages with LPS and measured IL-1β secretion over time (Fig. 5 B). Whereas control siRNA-transfected RAW264.7 cells secreted IL-1β, IL-1β production was impaired in IκBβ siRNA-transfected cells (Fig. 5 B). In contrast, no change in LPS-induced secretion of other NF-κB target genes MIP-2 and TNF were observed in IκBβ siRNA-transfected RAW264.7 macrophages, demonstrating specific interaction of IκBβ and IL-1β transcription (Fig. 5 B; Kim et al., 2003).
Figure 5.
IκBβ knock down results a significant reduction in IL-1β cytokine expression and secretion. (A) Silencing of IκBβ expression in RAW264.7 macrophages using siRNA. RAW264.7 macrophages were transfected with control or IκBβ-specific siRNAs or were left as an untransfected control. After 48 h, Western blot detected IκBβ expression. β-actin was used as loading control. (B) Measurement of IL-1β, MIP-2, and TNF cytokine secretion in control and IκBβ siRNA-transfected RAW264.7 cells. 48 h after transfection, cells were treated with LPS (100 ng/ml) and cytokine secretion was measured by ELISA. Data were obtained from three independent experiments performed in triplicate, and the results are presented as mean and SEM. (C) Schematic maps of different luciferase reporter genes containing IL-1β promoter luciferase reporter gene constructs. Three different luc-constructs encoding IL-1β promoter sequence starting from −518, −399, and −133, respectively, were used in transient transfection assays in RAW264.7 macrophages. The IL-1β–Luc construct IL-1B-518 includes two functional κB sites, whereas the IL-1B-399 contains only one functional κB site. No κB sites are present in the −133 IL-1β–Luc construct. In the mut-399-IL1B-luc construct, a mutation was introduced into the κB site by site-directed mutagenesis, as indicated. (D) RAW264.7 macrophages were transfected with 1 µg of the indicated IL-1β promoter luciferase reporter gene constructs. 48 h after the transfection, cells were stimulated with LPS (100 ng/ml) for 8 h and luciferase activity was determined. Data were obtained from three independent experiments performed in triplicate, and the results are presented as mean and SEM. (E) RAW264.7 macrophages were cotransfected using 1 µg of the indicated IL-1β promoter luciferase reporter gene constructs, a control siRNA, an IκBβ-specific siRNA, or they were left as an untransfected control. 48 h after the transfection, cells were stimulated with LPS (100 ng/ml) for 8 h and luciferase activity was determined. Data were obtained from three independent experiments performed in triplicate, and the results are presented as mean and SEM.
To further elucidate IκBβ-dependent regulation of the IL-1β promoter, we transfected several IL-1β reporter gene constructs into RAW264.7 macrophages (Fig. 5 C). The −518 bp IL-1β reporter gene, harboring two functional κB binding sites, showed an eightfold increase activity 8 h after stimulation with LPS in RAW264.7 macrophages (Cogswell et al., 1994; Fig. 5 D). Deleting the distal NF-κB–binding site (−399IL-1B) did not impair inducibility, whereas mutation or deletion of the proximal κB site significantly decreased LPS induction. These data suggest that the proximal κB site contributes to LPS-induction of the IL-1β promoter in RAW264.7 macrophages. However, we cannot completely exclude the contribution of the distal κB site.
To investigate whether IκBβ is essential for LPS-mediated induction of the IL-1β promoter, RAW267.6 macrophages were cotransfected with IκBβ siRNA and IL-1β reporter gene constructs. As shown in Fig. 5 E, knockdown of IκBβ clearly reduced IL-1β promoter induction after LPS stimulation, again pointing to a coactivator function of IκBβ toward IL-1β transcription.
Recruitment of IκBβ to the IL-1β promoter in complex with NF-κB p65–c-Rel
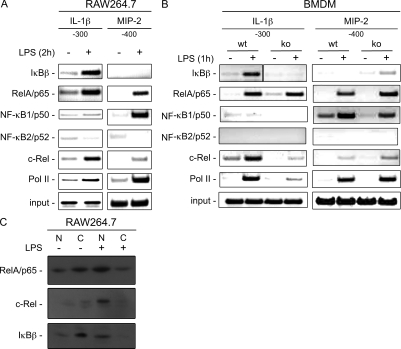
To test whether IκBβ directly binds to the IL-1β promoter, we performed chromatin immunoprecipitation (ChIP) assays. ChIP analysis using RAW264.7 macrophages demonstrated recruitment of IκBβ to the κB site of IL-1β promoter upon stimulation with LPS (Fig. 6 A). In contrast, IκBβ was not recruited to NF-κB–binding site in the MIP-2 gene promoter, suggesting specific regulation of the IL-1β promoter by IκBβ (Widmer et al., 1993). To characterize the activation complex in more detail, we investigated p65, p50, p52, and c-Rel binding. In addition to IκBβ, we found recruitment of the p65–c-Rel complex to the IL-1β promoter after LPS stimulation in RAW264.7 macrophages, suggesting that a p65–c-Rel dimer, assembled by IκBβ, induces IL-1β transcription after LPS treatment. In contrast, a classical NF-κB (p50/p65) dimer was recruited to the MIP-2 promoter upon LPS treatment and no binding of IκBβ was observed, demonstrating specificity for the IL-1β promoter (Fig. 6 A). Functionality of c-Rel for LPS-induced IL-1β transcription was also demonstrated using RNA interference in RAW264.7 macrophages (Fig. S3). To further demonstrate the influence of IκBβ for dimer formation at the IL-1β promoter at the genetic level, we used ChIP assays in IκBβ−/− BMDMs. Binding of IκBβ to the IL-1β promoter upon LPS treatment was confirmed in BMDMs (Fig. 6 B). Whereas, c-Rel binds to the IL-1β promoter in WT macrophages, reduced binding was observed in IκBβ−/− BMDMs (Fig. 6 B). A significant lower binding of RNA polymerase II to the IL-1β promoter indicated decreased transcriptional activity in IκBβ−/− cells after LPS treatment (Fig. 6 B). On the other hand, complex formation and recruitment of RNA polymerase II after LPS treatment was not changed at the MIP-2 promoter gene in IκBβ−/− BMDMs, confirming specificity for the IL-1β promoter (Fig. 6 B). Consistent with a nuclear function of IκBβ, we observed nuclear accumulation of IκBβ in LPS-treated BMDMs (Fig. S4). Previous studies demonstrated the interaction of IκBβ with c-Rel–p65 complexes in stimulated WEHI 231 cells in the nucleus (Phillips and Ghosh, 1997). To investigate direct interaction of the IκBβ–p65–c-Rel complex with DNA, we performed pulldown assays in Raw264.7 macrophages using biotinylated κB oligonucleotides corresponding to the proximal κB binding site of the IL-1β promoter. In nuclear extracts an IκBβ–p65–c-Rel trimer was found bound to the proximal κB binding site of the IL-1β promoter after LPS treatment (Fig. 6 C). No binding of c-Rel was detected in unstimulated cells, indicating that LPS-signaling induces molecular changes, like p65–c-Rel dimer formation or conformational changes of c-Rel, which are needed to detect c-Rel binding to the κB oligonucleotide in the assay used. Altogether, these data suggest that IκBβ is recruited to the IL-1β promoter after LPS treatment and is needed for NF-κB complex formation and transcriptional activation.
Figure 6.
Recruitment of IκBβ to the IL-1β promoter in complex with NF-κB p65/c-Rel. (A) ChIP in RAW264.7 macrophages. RAW264.7 macrophages were stimulated for 2 h with LPS (100 ng/ml). Chromatin was immunoprecipitated with IκBβ-, RelA/p65-, NF-κB1/p50–, NF-κB2/p52–, c-Rel-, and RNA-polymerase II–specific antibodies or IgG as a negative control. Precipitated DNA or 10% of the chromatin input was amplified with gene-specific primers for IL-1β or MIP-2 promoters. Three independent experiments revealed similar results. (B) ChIP in BMDMs. After stimulation with LPS (100 ng/ml) for 1 h, chromatin was immunoprecipitated with IκBβ-, RelA/p65-, NF-κB1/p50–, NF-κB2/p52–, c-Rel-, and RNA-polymerase II–specific antibodies or IgG as a negative control. Precipitated DNA or 10% of the chromatin input was amplified with gene-specific primers for IL-1β or MIP-2 promoters. Three independent experiments revealed similar results. (C) Biotin-streptavidin pulldown assay with a κB oligonucleotide, corresponding to the proximal κB site of the IL-1β promoter. RAW264.7 macrophages were stimulated with LPS (100 ng/ml) for 2 h. Nuclear and cytoplasmic extracts were incubated with the κB oligonucleotide and pulled down with streptavidin-agarose. Western blot detected RelA/p65, c-Rel, and IκBβ. One out of three independent experiments is shown.
DISCUSSION
Several reports of mice with targeted disruptions of IκB family members demonstrated that the different proteins play distinct biological roles. In contrast to IκBα, much less is known regarding to the in vivo function of IκBβ. In this study, we analyzed IκBβ−/− mice and demonstrate a novel function of IκBβ in the whole organism. IκBβ−/− mice share none of the hallmarks compared with IκBα−/− mice (Beg et al., 1995; Klement et al., 1996). Similar to IκBε−/− mice, IκBβ−/− mice survive to adulthood and show no overt abnormalities (Mémet et al., 1999). However, our results demonstrate that IκBβ is essential in regulating innate immunity in a LPS model of septic shock.
Bacterial infection can induce a systemic response characterized by multiple organ failure and high mortality rate. LPS, a major integral structural component of the outer membrane of Gram-negative bacteria, is a potent initiator of inflammation and endotoxin shock. LPS activates macrophages to produce cytokines, such as IL-1β, TNF, and IL-6, which serve as critical mediators of septic shock (Morrison and Ryan, 1987). Excessive production of these cytokines leads to capillary leakage, vascular hemorrhage, tissue destruction, and subsequent lethal organ failure. Thus, the expression of proinflammatory cytokines like IL-1β, TNF, and IL-6 needs to be tightly regulated during an inflammatory response. We now demonstrate that IκBβ is a critical regulator of LPS-induced septic shock. IκBβ deficiency confers LPS resistance in vivo, which is caused by the impaired secretion of the proinflammatory cytokines IL-1β, TNF, and IL-6. In BMDM, IL-1β was determined as a specific molecular IκBβ target, whereas the activation of the TNF and IL-6 genes remained unaffected after LPS treatment in this particular cellular model. Because IL-1β−/− mice are not protected from high-dose, LPS-induced septic shock, other IκBβ targets have to contribute to the observed LPS resistance (Fantuzzi et al., 1996). The importance of the IL-1 system for high-dose, LPS-induced septic shock is reflected by the LPS resistance of the IL-1β converting enzyme–deficient mice, known to have neither detectable serum levels of IL-1β nor IL-1α upon LPS challenge (Li et al., 1995). Interestingly, in microarray analysis of LPS-treated BMDMs, IL-1α induction after LPS stimulation was impaired in IκBβ−/− BMDMs (Table S1). However, Glaccum et al. (1997) reported that IL-1R−/− mice, which are refractory to both IL-1α and IL-1β signaling, are not resistant to LPS-induced septic shock, indicating that additional genes must contribute to the resistance in IκBβ−/− mice to LPS-induced lethality. The observation that several genes with important functions in innate immunity such as the chemokine (C-X-C motif) ligand 1 (Cxcl1), suppressor of cytokine signaling 3 (Socs3), interleukin 12p40 (Il12b) or others induced by LPS in BMDMs in an IκBβ-dependent manner, points to the possibility that a complex IκBβ-controlled genetic network mediates the LPS resistance observed in IκBβ−/− mice. Analysis of the IκBβ-dependent genes (Table S1) using Genomatix Pathway System software revealed a significant enrichment of genes controlled by the canonical IL-1–IKK–NF-κB signaling pathway (P < 0.01) and MyD88 response genes (P < 0.001), arguing that a subset of NF-κB– and MyD88-regulated genes is regulated by IκBβ (unpublished data). Furthermore, we observed a discrepancy between LPS-induced secretion of the proinflammatory cytokines in the BMDM model and LPS-induced cytokine expression measured in liver and serum. In BMDMs, only IL-1β secretion was impaired because of IκBβ deficiency, whereas the LPS-induced expression of IL-1β, TNF, and IL-6 was dependent on IκBβ in vivo. The liver is important for the initiation of defense mechanisms and the initiation of multiorgan failure during sepsis. LPS has been shown to activate hepatic Kupffer cells to synthesize and secrete inflammatory cytokines such as IL-1β, TNF, and IL-6 (Koo et al., 1999). Therefore, we cannot exclude that a different set of genes controlled by IκBβ in response to LPS in Kupfer cells, including IL-1β, TNF, and IL-6, are responsible for the LPS-resistance observed in IκBβ−/− mice in vivo. Thus, tissue and cell type specificities have to be considered in this context.
LPS is sensed by TLR4. Signaling via TLR4 activates a TRIF-dependent pathway of the induction of IFN-β and IFN-inducible genes in a MyD88-dependent pathway leading to activation of a NF-κB–dependent genetic program (Beutler, 2004; Beutler, 2009). The mechanisms by which LPS induces septic shock is related to its ability to activate NF-κB. For example, the highly LPS susceptible secretory leukoprotease inhibitor–deficient mice are characterized by an increased NF-κB signaling magnitude, and the LPS-resistant poly ADP-ribose polymerase-1–deficient mice demonstrate a distinct impaired NF-κB activation (Oliver et al., 1999; Nakamura et al., 2003). IκBβ is thought to control late-phase NF-κB activation (Hoffmann et al., 2002). This IκBβ activity was not observed in LPS-stimulated BMDMs because LPS stimulation results in similar NF-κB activation kinetics in WT and IκBβ−/− cells. Because IκBε is present in IκBβ−/− cells and degraded with the same kinetics as in WT BMDMs, IκBε may compensate for the IκBβ loss.
We detected a gene-specific activator function of IκBβ during the early LPS-induced NF-κB response. As a model of IκBβ-dependent transcriptional regulation, we focused onto the control of the IL-1β promoter because induction of this gene by LPS mostly depends on IκBβ, revealing a ten fold decreased inducibility in WT compared with IκBβ−/− BMDMs (Table S1). We observed a direct recruitment of IκBβ to the κB-binding site of the IL-1β promoter in complex with p65/c-Rel. Interestingly, in BMDMs of IκBβ−/− mice the lack of IκBβ binding to the IL-1β promoter leads to the loss of c-Rel recruitment as well as reduced binding of RNA polymerase II, indicating reduced transcriptional activation. Therefore, the remaining p65/RelA homodimers are not sufficient to activate IL-1β transcription, suggesting that IκBβ is indispensable in formation of a transcriptional active p65–c-Rel complex at the IL-1β promoter. This is in line with recent observations, demonstrating that each NF-κB dimer supports a different amount of transcriptional activation at a specific gene promoter and that the IL-1β gene-promoter is most responsive to p65/RelA and c-Rel in vitro (Algarté et al., 1999; Hiscott et al., 1993; Lin et al., 1995; Saccani et al., 2003).
As previously characterized, a stable complex of IκBβ and NF-κB p65/c-Rel can be found in the nucleus of WEHI 231 cells (Phillips and Ghosh, 1997). Using an oligonucleotide with specific κB-binding sequence, we can also identify a NF-κB p65–c-Rel–IκBβ complex in the nucleus of RAW264.7 macrophages after treatment with LPS, suggesting that IκBβ exists in the nucleus of macrophages, as found constitutively in WEHI 231 cells and LPS-stimulated 70/Z3 cells (Phillips and Ghosh, 1997; Suyang et al., 1996). Consistently, nuclear translocation of IκBβ was observed in LPS-stimulated BMDMs. Because IκBβ is clearly needed to recruit c-Rel to the IL-1β promoter, we suggest a more active role for IκBβ in gene transcription than a sole chaperone function.
In addition to the inhibitory function of the IκB protein family in resting cells, promoter-specific functions are becoming more evident. LPS signaling induces expression of IκBζ in macrophages, which is important for the induction of a subset of LPS-induced genes, like IL-6, by forming a promoter-bound p50–p65–IκBζ complex. Interestingly, LPS induction of IL-6 in macrophages was not shown to be IκBβ dependent (Yamamoto et al., 2004). Together with our data, which demonstrates specificity of IκBβ toward certain NF-κB– and MyD88-regulated promoters and recruitment of a p65/c-Rel dimer to the IL-1β promoter, we propose that the IκB proteins function to confer selectivity in NF-κB dimer usage, and therefore in signaling specificity. In addition to IκBβ and IκBζ, IκBα was shown to repress the hes1 promoter by direct binding (Aguilera et al., 2004). Furthermore, it was demonstrated that IκBα interacts with corepressors, like SMRT and N-Cor and different histone deacetylases (Aguilera et al., 2004). Whether IκBβ interacts with the epigenetic machinery is unknown in the moment and awaits further experiments.
Together, we now provide genetic evidence that IκBβ is essential for resistance toward LPS induced septic-shock. At the molecular level, IκBβ binds to a subset of NF-κB–dependent promoters and activates a subset of LPS-induced genes, like the IL-1β gene. This establishes IκBβ as an essential coactivator for gene transcription in vivo.
MATERIALS AND METHODS
Targeted disruption of the IκBβ gene.
The 8.2-kb genomic clone containing exons 3–6 of IκBβ was isolated from a genomic 129/Sv λ-DASHII-phage library. In the targeting vector, a loxP-flanked PGK-neomycin (neo)-cassette was introduced into intron 3. An additional loxP site was cloned into the AvrII site in intron 5. Thereby the BamHI sites in intron 3 and intron 5 were destroyed. The HSV-thymidine kinase gene was inserted 3.5 kb downstream of the neo-cassette. Embryonic stem cells (line E 14.1) were electroporated with the linearized vector (10 µg) and selected with G418 (300 µg/ml; Biochrom) and gancyclovir (2 µM; Sigma-Aldrich). Resistent clones were screened for homologous recombination by PCR. Positive clones were verified by Southern blot analysis using an external 5′ flanking probe (exon 2) and a neo probe. Correctly targeted ES cells were electroporated with pi-Cre-Plasmid (10 µg), to remove the neo-cassette, and selected with neomycin. Neomycin-sensitive clones were analyzed by Southern blot and PCR analyses to validate the correct deletion of the neo-cassette. These clones were aggregated to C57BL/6 morulae, and resulting chimeric mice were crossed with Deleter-Cre-mice to generate IκBβ knockout mice (Schwenk et al., 1995). Disruption of the IκBβ gene was verified by Southern blot und PCR analyses of tail DNA. Homozygous offspring were obtained at the predicted frequency by interbreeding heterozygous mice. The null phenotype created by mutation of the IκBβ gene was confirmed by Western blot analysis of spleen extracts. For LPS injection, IκBβ−/− mice were backcrossed at least 7 times to the C57BL/6 background.
Reagents.
CpG 1826 B-type, CpG 2216 A-type, PAM3CSK4, and PolyIC were purchased from Sigma-Aldrich.
Systemic challenge of WT and IκBβ−/− mice.
WT C57BL/6 (littermate controls or purchased from Charles River Laboratories), IκBβ+/− litters, and IκBβ−/− mice were injected i.p. with LPS 30 mg/kg (L-2630, strain 0111:B4; Sigma-Aldrich). Mice were monitored over 72 h for signs of sepsis and lethality. Blood was taken from the tail vein 0, 1, 2, and 6 h after LPS injection to investigate serum levels of IL-1β, TNF, and IL-6 using ELISA. All animal studies were conducted in compliance with European guidelines for the care and use of laboratory animals and were approved by the local authorities (Regierung von Oberbayern).
Immunocytochemistry.
For immunodetection of IκBβ and RelA/p65 in WT and IκBβ−/− LPS or untreated BMDMs, cells were fixed in 4% formaldehyde (Sigma-Aldrich), permeabilized with 0.3% Triton-X (Sigma-Aldrich), and stained with IκBβ- and RelA/p65-specific primary antibodies (Santa Cruz Biotechnology, Inc.), followed by a FITC-labeled secondary antibody (Invitrogen). Cells were then counterstained with DAPI (Vector Laboratories) to identify nuclei and subjected to fluorescence microscopy (Axiovert 200 M; Carl Zeiss, Inc.). Emitted fluorescence was collected on a color charge-coupled device camera system (AxioCam MRc; Carl Zeiss, Inc.). High-resolution images were captured and analyzed using AxioVision 4.3 software (Carl Zeiss, Inc.).
ChIP.
ChIP assays were performed as previously described (Fritsche et al., 2009; Schneider et al., 2006, 2010). An equal amount of chromatin (50–100 µg) was used for each precipitation. The following antibodies were used: IκB-β, RelA/p65, c-Rel, p50, p52, RNA-Polymerase II, and control IgG, all from Santa Cruz Biotechnology. One twentieth of the precipitated chromatin was used for each PCR reaction. To ensure linearity, 28 to 38 cycles were performed, and one representative result is shown. Sequences of the promoter specific primers are as follows: IL-1β promoter (−347/−151): sense, 5′-TCCCTGGAAATCAAGGGGTGG-3′, antisense 5′-TCTGGGTGTGCATCTACGTGCC-3′; MIP-2 promoter (-433/-138): sense 5′-CAACAGTGTACTTACGCAGACG-3′, antisense 5′-CTAGCTGCCTGCCTCATTCTAC-3′.
Quantitative real-time RT-PCR.
Total RNA was isolated from liver, BMDMs, or RAW264.7 macrophages was isolated using the RNeasy kit (Qiagen) following the manufacturer’s instructions. Quantitative mRNA analyses were performed as previously described using real-time PCR analysis (TaqMan, PE Applied Biosystems; Schneider et al., 2006; Fritsche et al., 2009). Sequences of the primers are: IL-1β, forward 5′-CTCAATGGACAGAATATCAACCAACA-3′ and reverse 5′-ACAGGACAGGTATAGATTCTTTCCTTTG-3′; IL-6, forward 5′-TCGGAGGCTTAATTACACATGTTCT-3′ and reverse IL-6 R 5′-GCATCATCGTTGTTCATACAATCA-3′; TNF, forward 5′-ATGAGAAGTTCCCAAATGGCC-3′ and reverse 5′-TCCACTTGGTGGTTCGCTACG-3′; Cyclophilin, forward 5′-ATGGTCAACCCCACCGTGT-3′ and reverse 5′-TTCTGCTGTCTTTGGAACTTTGTC-3′.
Determination of cytokine secretion.
Cytokine levels in blood sera and culture supernatants of IL-1β, TNF, and IL-6 were determined using commercially available ELISA kits, according to the manufacturer’s instructions (R&D Systems).
Southern and Western blot analysis.
10 µg of genomic tail DNA was digested with BamH I, yielding 4.8 and 3.1 kb fragments for IκBβ−/− and IκBβ WT alleles, respectively. DNA was separated on agarose gels and transferred to nitrocellulose membrane (GeneScreen Plus; DuPont). Hybridization was performed using hybridization buffer (1 M NaCl, 100 mg/ml dextran sulfate, 1% SDS, and 50 µg/ml salmon sperm DNA) and a Rediprime random primer labeling kit (GE Healthcare) with P32 α-dCTP–labeled probes.
Whole-cell lysates were prepared and Western blots and immunoprecipitations were done as previously described (Fritsche et al., 2009; Schneider et al., 2006, 2010). The following antibodies were used: IκB-β, p65, p50, p52, c-Rel, IκB-α, phospho-IκBα, IκB-ε (Santa Cruz Biotechnology, Inc.), IL-1β (R&D Systems), and β-actin (Sigma-Aldrich). Proteins were detected by Odyssey Infrared Imaging System (Licor).
Cell isolation and culture.
For generating BMDMs, mice were killed by cervical dislocation under ether anesthesia and BM was flushed from humerus, femur, and tibia of 6–8 mice. Cells were collected and washed as previously described (Ohashi et al., 2000). Pluznik medium containing RPMI 1640, 5% heat-inactivated horse serum (PAA), 15% FCS (PAA), 15% culture supernatant from M-CSF producing L929 cells (DSM ACC), and 1% P/S was used for cell culturing. After 6 d, the enriched macrophages were used for experiments as indicated.
Flow cytometry.
Fluorescence staining of isolated mouse splenocytes was performed as described previously (Rad et al., 2006). The following antibodies were used: PE-conjugated anti-IgD (SouthernBiotech); biotinylated anti-CD3 (Caltag Laboratories); PE-conjugated anti-Terr-119, PE-conjugated anti-CD45RB, PE-conjugated anti-CD19, APC-conjugated anti-CD62L, FITC-conjugated anti-CD23, FITC-conjugated anti-CD21, biotinylated anti-IgM, APC-conjugated anti-CD11b, biotinylated anti-CD3 (BD). Streptavidin-PerCP was from BD. Fluorescence was analyzed by using a FACSCalibur (BD) flow cytometer and CellQuest software (BD).
Biotin-streptavidin pulldown assay.
Assays were performed as previously described (Schild et al., 2009; Schneider et al., 2010). Approximately 7 × 107 RAW264.7 macrophages were used for each time point. Nuclear and cytosolic extract were prepared by using nuclear extraction kit (Active Motif) according to the manufacturer’s instructions. The following 5′ biotin-labeled oligonucleotide, corresponding to the positions −261 to −270 of the IL-1β promoter, was used: 5′-ACCCCAGGAAAACCCAATATTT-3′.
IL-1β promoter reporter gene assay and mutagenesis.
To determine IL-1β promoter activity the −518-IL1B-luc, −399-IL1B-luc, and −199-IL1B-luc luciferase reporter gene constructs were used. Point mutations within the −399-IL-1B-luc plasmid were generated using the QuikChange Site-Directed Mutagenesis kit (Stratagene) in conjugation with the following oligonucleotides: 5′-CATTCTTCTAACGTGTTGGAATCCACTATTATGTGGAC-3′ and 5′-GTCCACATTAAACTGGATTCCAACACGTTAGAAGAATC-3′. Mutated residues are shown in Fig. 5 C. Mutations were confirmed by sequencing. 1 µg of each reporter gene constructs was transfected using Oligofectamine (Invitrogen). Luciferase activity was normalized to protein concentration and analyzed as previously described (Reichert et al., 2007).
Assay with siRNA specific to IκBβ and c-Rel.
RAW264.7 macrophages were transfected with siRNA duplex (Ambion) specific for mouse IκBβ or with scramble duplex in a concentration of 200 nM with Oligofectamine (Invitrogen) according to the manufacturer’s instructions. The following siRNAs were used: IκBβ, 5′-GACUGGAGGCUACAACUAG-3′; c-Rel, 5′-AUAGCAUGUUGACAUCAGACAUACU-3′; control siRNA, 5′-CAGUCGCGUUUGCGACUGG-3′.
EMSA.
EMSAs were performed as previously described (Arkan et al., 2001) using NF-κB (5′-AGTTGAGGGGACTTTCCCAGGC-3′ and 3′-TCAACTCCCCTGAAAGGGTCCG-5′) and Oct-1 (5′-TGTCGAATGCAAATCACTAGAA-3′ and 3′-ACAGCTTACGTTTAGTGATCTT-5′) oligonucleotides.
Gene expression profiling.
Gene expression profiling was performed as previously described (Reichert et al., 2007; Fritsche et al., 2009). Duplicates of total RNA were prepared using RNeasy kit (Qiagen). Labeled cRNA was produced and hybridized onto the Affymetrix GeneChip Mouse Genome 430 2.0 Array set according to Affymetrix standard protocols. Expression data were analyzed using Microarray Suite 5.0. Genes induced at least fivefold in WT BMDMs 2 h after LPS stimulation (100 ng/ml) and who’s induction is reduce to <55% in IκBβ−/− compared with WT BMDMs are presented in Table S1.
Statistical analysis.
Unless otherwise indicated, all data were obtained from at least three independent experiments performed in triplicate and the results are presented as mean and standard error of the mean (SEM). To demonstrate statistical significance a two-tailed Student's t test or Kaplan-Meier with a log-rank test was used. Statistical significance was assigned to P < 0.05.
Online supplemental material.
Fig. S1 shows that IκBβ mice−/− demonstrate an increase of splenic marginal zone B cells and memory T cells and an enforced differentiation of macrophages within the BM. Fig. S2 shows IL-1β secretion in response to NF-κB activators in IκBβ−/− BMDMs. Fig. S3 shows that c-Rel knockdown results a significant reduction in IL-1β cytokine expression and secretion. Fig. S4 shows that LPS induces the nuclear translocation of IκBβ in BMDMs. Table S1 shows genes activated by LPS in BMDM in an IκBβ-dependent way. Online supplemental material is available at http://www.jem.org/cgi/content/full/jem.20100864/DC1.
Acknowledgments
This work was founded by DFG SFB456 (to GS and RMS), DFG AR 710/2-1 (to MCA) and by Deutsche Krebshilfe grant 107977 (to MCA).
The authors have no conflicting financial interests.
Footnotes
Abbreviations used:
- BMDM
- BM-derived macrophage
- ChIP
- chromatin immunoprecipitation
- EMSA
- electrophoretic mobility shift assay
- IκB
- inhibitor of κB
- IKK
- IκB kinase
- siRNA
- small interfering RNA
- TLR
- Toll-like receptor
References
- Aguilera C., Hoya-Arias R., Haegeman G., Espinosa L., Bigas A. 2004. Recruitment of IkappaBalpha to the hes1 promoter is associated with transcriptional repression. Proc. Natl. Acad. Sci. USA. 101:16537–16542 10.1073/pnas.0404429101 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Algarté M., Kwon H., Génin P., Hiscott J. 1999. Identification by in vivo genomic footprinting of a transcriptional switch containing NF-kappaB and Sp1 that regulates the IkappaBalpha promoter. Mol. Cell. Biol. 19:6140–6153 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Arkan M.C., Leonarduzzi G., Biasi F., Başağa H., Poli G. 2001. Physiological amounts of ascorbate potentiate phorbol ester-induced nuclear-binding of AP-1 transcription factor in cells of macrophagic lineage. Free Radic. Biol. Med. 31:374–382 10.1016/S0891-5849(01)00601-3 [DOI] [PubMed] [Google Scholar]
- Baeuerle P.A., Baltimore D. 1988. I kappa B: a specific inhibitor of the NF-kappa B transcription factor. Science. 242:540–546 10.1126/science.3140380 [DOI] [PubMed] [Google Scholar]
- Baldwin A.S., Jr 1996. The NF-kappa B and I kappa B proteins: new discoveries and insights. Annu. Rev. Immunol. 14:649–683 10.1146/annurev.immunol.14.1.649 [DOI] [PubMed] [Google Scholar]
- Bates P.W., Miyamoto S. 2004. Expanded nuclear roles for IkappaBs. Sci. STKE. 2004:pe48 10.1126/stke.2542004pe48 [DOI] [PubMed] [Google Scholar]
- Beg A.A., Sha W.C., Bronson R.T., Baltimore D. 1995. Constitutive NF-kappa B activation, enhanced granulopoiesis, and neonatal lethality in I kappa B alpha-deficient mice. Genes Dev. 9:2736–2746 10.1101/gad.9.22.2736 [DOI] [PubMed] [Google Scholar]
- Beutler B. 2004. Inferences, questions and possibilities in Toll-like receptor signalling. Nature. 430:257–263 10.1038/nature02761 [DOI] [PubMed] [Google Scholar]
- Beutler B.A. 2009. TLRs and innate immunity. Blood. 113:1399–1407 10.1182/blood-2008-07-019307 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bourke E., Kennedy E.J., Moynagh P.N. 2000. Loss of Ikappa B-beta is associated with prolonged NF-kappa B activity in human glial cells. J. Biol. Chem. 275:39996–40002 10.1074/jbc.M007693200 [DOI] [PubMed] [Google Scholar]
- Cogswell J.P., Godlevski M.M., Wisely G.B., Clay W.C., Leesnitzer L.M., Ways J.P., Gray J.G. 1994. NF-kappa B regulates IL-1 beta transcription through a consensus NF-kappa B binding site and a nonconsensus CRE-like site. J. Immunol. 153:712–723 [PubMed] [Google Scholar]
- DeLuca C., Petropoulos L., Zmeureanu D., Hiscott J. 1999. Nuclear IkappaBbeta maintains persistent NF-kappaB activation in HIV-1-infected myeloid cells. J. Biol. Chem. 274:13010–13016 10.1074/jbc.274.19.13010 [DOI] [PubMed] [Google Scholar]
- Fantuzzi G., Zheng H., Faggioni R., Benigni F., Ghezzi P., Sipe J.D., Shaw A.R., Dinarello C.A. 1996. Effect of endotoxin in IL-1 beta-deficient mice. J. Immunol. 157:291–296 [PubMed] [Google Scholar]
- Fritsche P., Seidler B., Schüler S., Schnieke A., Göttlicher M., Schmid R.M., Saur D., Schneider G. 2009. HDAC2 mediates therapeutic resistance of pancreatic cancer cells via the BH3-only protein NOXA. Gut. 58:1399–1409 10.1136/gut.2009.180711 [DOI] [PubMed] [Google Scholar]
- Ghosh S., Hayden M.S. 2008. New regulators of NF-kappaB in inflammation. Nat. Rev. Immunol. 8:837–848 10.1038/nri2423 [DOI] [PubMed] [Google Scholar]
- Ghosh S., Karin M. 2002. Missing pieces in the NF-kappaB puzzle. Cell. 109(Suppl):S81–S96 10.1016/S0092-8674(02)00703-1 [DOI] [PubMed] [Google Scholar]
- Gilmore T.D. 2006. Introduction to NF-kappaB: players, pathways, perspectives. Oncogene. 25:6680–6684 10.1038/sj.onc.1209954 [DOI] [PubMed] [Google Scholar]
- Glaccum M.B., Stocking K.L., Charrier K., Smith J.L., Willis C.R., Maliszewski C., Livingston D.J., Peschon J.J., Morrissey P.J. 1997. Phenotypic and functional characterization of mice that lack the type I receptor for IL-1. J. Immunol. 159:3364–3371 [PubMed] [Google Scholar]
- Hiscott J., Marois J., Garoufalis J., D’Addario M., Roulston A., Kwan I., Pepin N., Lacoste J., Nguyen H., Bensi G., et al. 1993. Characterization of a functional NF-kappa B site in the human interleukin 1 beta promoter: evidence for a positive autoregulatory loop. Mol. Cell. Biol. 13:6231–6240 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hoffmann A., Baltimore D. 2006. Circuitry of nuclear factor kappaB signaling. Immunol. Rev. 210:171–186 10.1111/j.0105-2896.2006.00375.x [DOI] [PubMed] [Google Scholar]
- Hoffmann A., Levchenko A., Scott M.L., Baltimore D. 2002. The IkappaB-NF-kappaB signaling module: temporal control and selective gene activation. Science. 298:1241–1245 10.1126/science.1071914 [DOI] [PubMed] [Google Scholar]
- Huang T.T., Miyamoto S. 2001. Postrepression activation of NF-kappaB requires the amino-terminal nuclear export signal specific to IkappaBalpha. Mol. Cell. Biol. 21:4737–4747 10.1128/MCB.21.14.4737-4747.2001 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Inoue J., Kerr L.D., Rashid D., Davis N., Bose H.R., Jr, Verma I.M. 1992. Direct association of pp40/I kappa B beta with rel/NF-kappa B transcription factors: role of ankyrin repeats in the inhibition of DNA binding activity. Proc. Natl. Acad. Sci. USA. 89:4333–4337 10.1073/pnas.89.10.4333 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Karin M., Ben-Neriah Y. 2000. Phosphorylation meets ubiquitination: the control of NF-[kappa]B activity. Annu. Rev. Immunol. 18:621–663 10.1146/annurev.immunol.18.1.621 [DOI] [PubMed] [Google Scholar]
- Kim D.S., Han J.H., Kwon H.J. 2003. NF-kappaB and c-Jun-dependent regulation of macrophage inflammatory protein-2 gene expression in response to lipopolysaccharide in RAW 264.7 cells. Mol. Immunol. 40:633–643 10.1016/j.molimm.2003.07.001 [DOI] [PubMed] [Google Scholar]
- Klement J.F., Rice N.R., Car B.D., Abbondanzo S.J., Powers G.D., Bhatt P.H., Chen C.H., Rosen C.A., Stewart C.L. 1996. IkappaBalpha deficiency results in a sustained NF-kappaB response and severe widespread dermatitis in mice. Mol. Cell. Biol. 16:2341–2349 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Koo D.J., Chaudry I.H., Wang P. 1999. Kupffer cells are responsible for producing inflammatory cytokines and hepatocellular dysfunction during early sepsis. J. Surg. Res. 83:151–157 10.1006/jsre.1999.5584 [DOI] [PubMed] [Google Scholar]
- Li P., Allen H., Banerjee S., Franklin S., Herzog L., Johnston C., McDowell J., Paskind M., Rodman L., Salfeld J., et al. 1995. Mice deficient in IL-1 beta-converting enzyme are defective in production of mature IL-1 beta and resistant to endotoxic shock. Cell. 80:401–411 10.1016/0092-8674(95)90490-5 [DOI] [PubMed] [Google Scholar]
- Lin R., Gewert D., Hiscott J. 1995. Differential transcriptional activation in vitro by NF-kappa B/Rel proteins. J. Biol. Chem. 270:3123–3131 10.1074/jbc.270.7.3123 [DOI] [PubMed] [Google Scholar]
- Malek S., Chen Y., Huxford T., Ghosh G. 2001. IkappaBbeta, but not IkappaBalpha, functions as a classical cytoplasmic inhibitor of NF-kappaB dimers by masking both NF-kappaB nuclear localization sequences in resting cells. J. Biol. Chem. 276:45225–45235 10.1074/jbc.M105865200 [DOI] [PubMed] [Google Scholar]
- Mémet S., Laouini D., Epinat J.C., Whiteside S.T., Goudeau B., Philpott D., Kayal S., Sansonetti P.J., Berche P., Kanellopoulos J., Israël A. 1999. IkappaBepsilon-deficient mice: reduction of one T cell precursor subspecies and enhanced Ig isotype switching and cytokine synthesis. J. Immunol. 163:5994–6005 [PubMed] [Google Scholar]
- Morrison D.C., Ryan J.L. 1987. Endotoxins and disease mechanisms. Annu. Rev. Med. 38:417–432 [DOI] [PubMed] [Google Scholar]
- Nakamura A., Mori Y., Hagiwara K., Suzuki T., Sakakibara T., Kikuchi T., Igarashi T., Ebina M., Abe T., Miyazaki J., et al. 2003. Increased susceptibility to LPS-induced endotoxin shock in secretory leukoprotease inhibitor (SLPI)-deficient mice. J. Exp. Med. 197:669–674 10.1084/jem.20021824 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ohashi K., Burkart V., Flohé S., Kolb H. 2000. Cutting edge: heat shock protein 60 is a putative endogenous ligand of the toll-like receptor-4 complex. J. Immunol. 164:558–561 [DOI] [PubMed] [Google Scholar]
- Oliver F.J., Ménissier-de Murcia J., Nacci C., Decker P., Andriantsitohaina R., Muller S., de la Rubia G., Stoclet J.C., de Murcia G. 1999. Resistance to endotoxic shock as a consequence of defective NF-kappaB activation in poly (ADP-ribose) polymerase-1 deficient mice. EMBO J. 18:4446–4454 10.1093/emboj/18.16.4446 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Phillips R.J., Ghosh S. 1997. Regulation of IkappaB beta in WEHI 231 mature B cells. Mol. Cell. Biol. 17:4390–4396 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rad R., Brenner L., Bauer S., Schwendy S., Layland L., da Costa C.P., Reindl W., Dossumbekova A., Friedrich M., Saur D., et al. 2006. CD25+/Foxp3+ T cells regulate gastric inflammation and Helicobacter pylori colonization in vivo. Gastroenterology. 131:525–537 10.1053/j.gastro.2006.05.001 [DOI] [PubMed] [Google Scholar]
- Reichert M., Saur D., Hamacher R., Schmid R.M., Schneider G. 2007. Phosphoinositide-3-kinase signaling controls S-phase kinase-associated protein 2 transcription via E2F1 in pancreatic ductal adenocarcinoma cells. Cancer Res. 67:4149–4156 10.1158/0008-5472.CAN-06-4484 [DOI] [PubMed] [Google Scholar]
- Saccani S., Pantano S., Natoli G. 2003. Modulation of NF-kappaB activity by exchange of dimers. Mol. Cell. 11:1563–1574 10.1016/S1097-2765(03)00227-2 [DOI] [PubMed] [Google Scholar]
- Schild C., Wirth M., Reichert M., Schmid R.M., Saur D., Schneider G. 2009. PI3K signaling maintains c-myc expression to regulate transcription of E2F1 in pancreatic cancer cells. Mol. Carcinog. 48:1149–1158 10.1002/mc.20569 [DOI] [PubMed] [Google Scholar]
- Schneider G., Saur D., Siveke J.T., Fritsch R., Greten F.R., Schmid R.M. 2006. IKKalpha controls p52/RelB at the skp2 gene promoter to regulate G1- to S-phase progression. EMBO J. 25:3801–3812 10.1038/sj.emboj.7601259 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schneider G., Henrich A., Greiner G., Wolf V., Lovas A., Wieczorek M., Wagner T., Reichardt S., von Werder A., Schmid R.M., et al. 2010. Cross talk between stimulated NF-kappaB and the tumor suppressor p53. Oncogene. 29:2795–2806 10.1038/onc.2010.46 [DOI] [PubMed] [Google Scholar]
- Schwenk F., Baron U., Rajewsky K. 1995. A cre-transgenic mouse strain for the ubiquitous deletion of loxP-flanked gene segments including deletion in germ cells. Nucleic Acids Res. 23:5080–5081 10.1093/nar/23.24.5080 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sun S.C., Ganchi P.A., Ballard D.W., Greene W.C. 1993. NF-kappa B controls expression of inhibitor I kappa B alpha: evidence for an inducible autoregulatory pathway. Science. 259:1912–1915 10.1126/science.8096091 [DOI] [PubMed] [Google Scholar]
- Suyang H., Phillips R., Douglas I., Ghosh S. 1996. Role of unphosphorylated, newly synthesized I kappa B beta in persistent activation of NF-kappa B. Mol. Cell. Biol. 16:5444–5449 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Thompson J.E., Phillips R.J., Erdjument-Bromage H., Tempst P., Ghosh S. 1995. I kappa B-beta regulates the persistent response in a biphasic activation of NF-kappa B. Cell. 80:573–582 10.1016/0092-8674(95)90511-1 [DOI] [PubMed] [Google Scholar]
- Tran K., Merika M., Thanos D. 1997. Distinct functional properties of IkappaB alpha and IkappaB beta. Mol. Cell. Biol. 17:5386–5399 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Widmer U., Manogue K.R., Cerami A., Sherry B. 1993. Genomic cloning and promoter analysis of macrophage inflammatory protein (MIP)-2, MIP-1 alpha, and MIP-1 beta, members of the chemokine superfamily of proinflammatory cytokines. J. Immunol. 150:4996–5012 [PubMed] [Google Scholar]
- Yamamoto M., Yamazaki S., Uematsu S., Sato S., Hemmi H., Hoshino K., Kaisho T., Kuwata H., Takeuchi O., Takeshige K., et al. 2004. Regulation of Toll/IL-1-receptor-mediated gene expression by the inducible nuclear protein IkappaBzeta. Nature. 430:218–222 10.1038/nature02738 [DOI] [PubMed] [Google Scholar]