Hsp90 stabilizes and prevents degradation of cytoplasmic activation-induced deaminase.
Abstract
Activation-induced deaminase (AID) is the mutator enzyme that initiates somatic hypermutation and isotype switching of the antibody genes in B lymphocytes. Undesired byproducts of AID function are oncogenic mutations. AID expression levels seem to correlate with the extent of its physiological and pathological functions. In this study, we identify AID as a novel Hsp90 (heat shock protein 90 kD) client. We find that cytoplasmic AID is in a dynamic equilibrium regulated by Hsp90. Hsp90 stabilizes cytoplasmic AID, as specific Hsp90 inhibition leads to cytoplasmic polyubiquitination and proteasomal degradation of AID. Consequently, Hsp90 inhibition results in a proportional reduction in antibody gene diversification and off-target mutation. This evolutionarily conserved regulatory mechanism determines the functional steady-state levels of AID in normal B cells and B cell lymphoma lines. Thus, Hsp90 assists AID-mediated antibody diversification by stabilizing AID. Hsp90 inhibition provides the first pharmacological means to down-regulate AID expression and activity, which could be relevant for therapy of some lymphomas and leukemias.
Antibody genes are first rearranged by V(D)J recombination during B lymphocyte development and then further diversified in the periphery after encountering cognate antigen. The latter is achieved by the mechanism of somatic hypermutation (SHM), which introduces random mutations over the exon that encodes the antibody variable region. Coupled to phenotypic selection during the germinal center reaction, SHM results in the overall maturation of the antibody response. SHM is initiated by activation-induced deaminase (AID), which deaminates dC to dU in the Ig loci. Processing of the dU by specific DNA repair enzymes produces the full spectrum of SHM (Di Noia and Neuberger, 2007; Peled et al., 2008). In addition, AID also targets the DNA immediately preceding the constant exons that encode for the different antibody isotypes in the heavy chain locus. Processing of the dU in these switch regions leads to the DNA breaks necessary for class switch recombination (CSR; Stavnezer et al., 2008).
AID being a mutator enzyme sufficient to cause cancer in transgenic models (Okazaki et al., 2003; Pasqualucci et al., 2008), there has been a well-deserved emphasis in studying its regulation. Gene expression regulation is an important step during normal B cell development, with AID being mostly restricted to germinal center B cells (Muramatsu et al., 1999; Crouch et al., 2007). However, AID can normally be expressed outside of the B cell compartment, the exact physiological relevance of which is still unclear, although it may influence the expression of many genes by affecting DNA methylation (Morgan et al., 2004; Macduff et al., 2009; Pauklin et al., 2009; Bhutani et al., 2010; Popp et al., 2010). Importantly, there is ample evidence that AID is expressed in a variety of human lymphomas (Greeve et al., 2003; Pasqualucci et al., 2004) and leukemias (Albesiano et al., 2003; Feldhahn et al., 2007; Klemm et al., 2009; Palacios et al., 2010). In fact, AID plays a role in malignant transformation by initiating DNA double-strand breaks at various non-Ig loci, most prominently c-Myc, which in murine experimental plasmacytoma, and therefore most likely also in human Burkitt’s lymphoma, leads to the hallmark oncogenic c-Myc–IgH chromosomal translocation (Ramiro et al., 2004, 2006; Robbiani et al., 2008, 2009). A role for AID in the etiology of diffuse large B cell lymphoma is also very likely (Pasqualucci et al., 2001, 2004, 2008). In chronic myeloid leukemia (CML), AID mutates the BCR-ABL1 oncogene, leading to resistance to imatinib, the main therapeutic drug (Klemm et al., 2009). Moreover, AID expression in nonlymphoid tumors has also been shown (Endo et al., 2008). Therefore, it is important to understand AID posttranslational regulation, which may differ between normal and transformed cells and have important and varied implications in the several cell types that can express AID.
Multiple mechanisms seem to contribute to restrain AID protein. Subcellular localization is an important step in regulating AID that also impinges on its stability because AID has a significantly shorter half-life in the nucleus than in the cytoplasm (Aoufouchi et al., 2008). In steady-state, the bulk of AID is cytoplasmic as a result of the integration of three mechanisms: nuclear import (Patenaude et al., 2009), nuclear export (Ito et al., 2004; McBride et al., 2004), and cytoplasmic retention (Patenaude et al., 2009). However, it is unknown whether AID stability is regulated in the cytoplasm. Several studies, including the analysis of AID-haploinsufficient mice (Sernández et al., 2008; Takizawa et al., 2008) or mice with altered AID levels resulting from manipulating microRNA regulation (de Yébenes et al., 2008; Dorsett et al., 2008; Teng et al., 2008) or enforcing transgenic overexpression (Robbiani et al., 2009), have suggested that AID protein levels are limiting for and correlate with the efficiency of antibody diversification but also B cell lymphomagenesis. Therefore, any mechanism impinging on the overall AID steady-state levels is important for balancing an efficient humoral immune response with the associated risk of B cell transformation.
We report the physical and functional interaction of AID with the Hsp90 (heat shock protein 90 kD) molecular chaperone pathway. Hsp90 is thought to be more selective of its range of substrates than other chaperones, playing a prominent role in the structural stabilization and functional modulation of many of its client proteins, rather than in their initial folding (Pratt and Toft, 2003; Whitesell and Lindquist, 2005; Pearl and Prodromou, 2006; Picard, 2006; Wandinger et al., 2008). Indeed, the interaction with Hsp90 prevents proteasomal degradation of AID in the cytoplasm, thereby determining the steady-state levels of functional AID.
RESULTS
AID specifically interacts with Hsp90
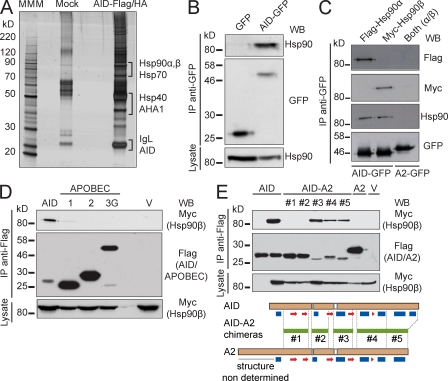
Immunopurification of AID-Flag/HA from cell extracts of Ramos B cells produced a complex but reproducible pattern of copurifying proteins (Fig. 1 A). Of note, we used a stable cell line expressing only 2.5-fold the amount of endogenous AID (Fig. S1 A). After identification of the associated proteins by mass spectrometry, we noticed the presence of several members of the Hsp90 molecular chaperone pathway (Whitesell and Lindquist, 2005), including the two major isoforms of Hsp90 (α and β), the Hsp90 cochaperone AHA-1, Hsp70/Hsc70, and one Hsp40 cochaperone (DnaJa1), as well as several proteasome subunits (Table I). The Hsp90, Hsp70, and Hsp40 proteins are known to cooperate in a multichaperone system (Picard, 2006; Wandinger et al., 2008). Given the importance of Hsp90 in regulating the function of many signal transduction and nucleocytoplasmic shuttling proteins, we decided to further explore this interaction. We confirmed the binding of AID to endogenous Hsp90 by coimmunoprecipitation of AID-GFP from stably expressing Ramos cells (Fig. 1 B). We also confirmed that AID coimmunoprecipitated similarly with tagged versions of Hsp90-α and Hsp90-β (Fig. 1 C). Both isoforms are constitutively expressed in the B cell lines we used, although Hsp90-β is the predominant form in resting primary mouse B cells, with Hsp90-α increasing after cytokine activation (Fig. S1, B and C). These results are in keeping with various studies indicating that mitogenic and cytokine stimuli up-regulate Hsp90-α, whereas Hsp90-β is constitutively expressed (Hansen et al., 1991; Metz et al., 1996; Csermely et al., 1998; Sreedhar et al., 2004). Hsp90-α and Hsp90-β share ∼90% similarity, and although they may have some nonoverlapping roles, for most functions, they are largely equivalent (Csermely et al., 1998; Sreedhar et al., 2004).
Figure 1.
AID interacts with Hsp90. (A) Ramos B cells stably expressing AID-Flag/HA were subjected to consecutive immunoprecipitation with anti-Flag and anti-HA. Precipitated material was eluted with the specific peptides and separated by SDS-PAGE. Proteins were identified by mass spectrometry of tryptic peptides. The proteins relevant to this work are indicated next to the bands from where they were identified. One of two independent experiments is shown. MMM, molecular mass marker. (B) GFP and AID-GFP were immunoprecipitated from extracts of stably expressing Ramos cells and analyzed by Western blot (WB) to detect coimmunoprecipitated endogenous Hsp90. One of three independent experiments is shown. (C) APOBEC2 (A2)-GFP and AID-GFP were immunoprecipitated with anti-GFP from transiently expressing HEK293T cells cotransfected with Flag–Hsp90-α and/or Myc–Hsp90-β. Immunoprecipitates (IP) were probed with anti-Flag and anti-Myc in Western blots. The filters were then probed with anti-Hsp90, which recognizes both isoforms, to verify that the overall Hsp90 level was similar after transfection. Anti-GFP confirmed similar immunoprecipitation of the bait. One of two independent experiments is shown. (D) Lysates from HEK293T cells cotransfected with Myc–Hsp90-β and Flag-tagged versions of AID or the indicated APOBECs or vector alone (V) were immunoprecipitated using anti-Flag and analyzed by Western blot with anti-Myc to verify the presence of Hsp90-β and anti-Flag to confirm the immunoprecipitation of the baits. One of four independent experiments is shown. (E) Lysates from HEK293T cells cotransfected with Myc–Hsp90-β and either vector only (V) or Flag-tagged AID, A2, or AID-A2 chimeras (#1–5, described below in schematic form) were immunoprecipitated with anti-Flag. Immunoprecipitates were analyzed by Western blot using anti-Myc and anti-Flag antibodies. One of three independent experiments is shown. In schematics, horizontal green lines between dashed lines identify the fragments of AID replaced by the homologous region of A2 in each construct. Based on experimental A2 (Prochnow et al., 2007) and predicted AID (Patenaude et al., 2009) secondary structures, blue rectangles indicate α helixes, and red arrows indicate β sheets. Where indicated, aliquots (5%) of the whole cell lysates were probed to control for expression.
Table I.
Proteins copurifying with AID-Flag-HA from Ramos B cells identified by mass spectrometry
| HUGO name | Mascot score | Coverage | Description |
| % | |||
| HSP90AB1 | 2,178 | 44 | Heat shock 90-kD protein 1, β |
| 300 | 8 | ||
| HSP90AA1 | 1,668 | 35 | Heat shock 90-kD protein 1, α |
| 151 | 6 | ||
| HSPA8 | 1,338 | 39 | Heat shock 70-kD protein 8 isoform 1 |
| HSPA6 | 327 | 8 | Heat shock 70-kD protein B |
| AHSA1 | 81 | 3 | AHA-1 activator of heat shock 90-kD protein ATPase homologue 1 |
| 27 | 9 | ||
| DNAJA1 | 212 | 26 | Hsp40 homologue, subfamily A, member 1 |
| PSMD2 | 242 | 14 | Proteasome 26S non-ATPase subunit 2 |
| PSMD1 | 105 | 2 | Proteasome 26S non-ATPase subunit 1 |
| PSMD6 | 105 | 5 | Proteasome 26S non-ATPase subunit 6 |
| 46 | 6 | ||
| PSMC2 | 75 | 3 | Proteasome 26S ATPase subunit 2 |
Proteins with two rows of data were identified in two independent experiments. The gel from one of the experiments is shown in Fig. 1 A. A threshold Mascot score of 50 was used as cut-off, indicating a 95% confidence of being a true identification. For AHSA1, in the second experiment, the mass spectrometry profile was manually examined to confirm the reliability of the observation.
We then characterized the interaction between Hsp90 and AID. Notably, the AID paralogue proteins APOBEC1, APOBEC2 (A2), and APOBEC3G, which share ∼50–60% similarity with AID (Conticello et al., 2005), did not coimmunoprecipitate with Myc–Hsp90-β (Fig. 1 D). This is consistent with a recent study showing that zebra fish A2 interacts with the chaperone Unc45b but not with Hsp90-α (Etard et al., 2010). The region of AID interacting with Hsp90-β could be mapped to the N-terminal half of the molecule by using AID-A2 chimeric proteins (Fig. 1 E). An AID mutant showing impaired oligomerization (Patenaude et al., 2009) was still recognized by Hsp90 (Fig. S1 D). Phosphorylation can modulate client binding to Hsp90 (Dickey et al., 2007), but the known protein kinase A (PKA) sites within the N-terminal region of AID, Thr27, and Ser38, were not essential for the interaction (Fig. S1 D). These results suggest that Hsp90 specifically binds to AID by its N-terminal region in an oligomerization- and phosphorylation-independent fashion.
Hsp90 maintains the steady-state level of AID
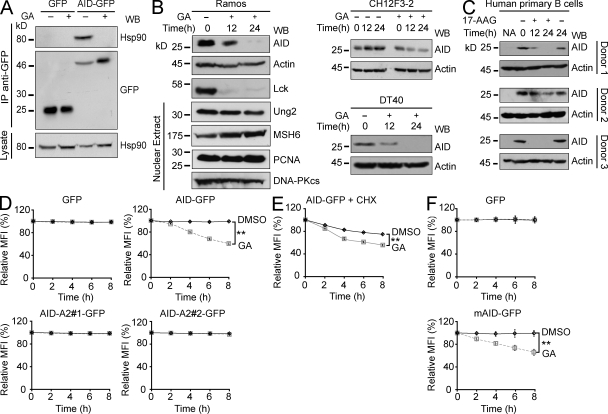
The chaperone activity of Hsp90 depends on an ATP hydrolysis cycle, which is inhibited by geldanamycin (GA) and its derivatives, like 17-allylamino-17-demethoxygeldanamycin (17-AAG; Prodromou et al., 1997; Stebbins et al., 1997; Panaretou et al., 1998; Young and Hartl, 2000). Treating Ramos cells with GA prevented the interaction of AID-GFP with Hsp90, as indicated by the lack of coimmunoprecipitation (Fig. 2 A). More importantly, treatment of human, chicken, and mouse B cell lymphoma lines with GA caused a clear reduction in the levels of endogenous AID at 12 and 24 h. We probed for the known Hsp90 client kinase Lck, which was also reduced, as positive control (Giannini and Bijlmakers, 2004). Other enzymes involved in antibody diversification, including UNG (uracil-DNA N-glycosylase) and MSH6, were not sensitive to Hsp90 inhibition (Fig. 2 B). Finally, endogenous AID in stimulated human primary B cells from multiple donors was also sensitive to Hsp90 inhibition (Fig. 2 C), confirming that the functional interaction between AID and Hsp90 is physiologically relevant.
Figure 2.
Hsp90 actively maintains the steady-state levels of AID. (A) Ramos cells stably expressing AID-GFP or GFP alone were treated with 2 µM GA (+) or DMSO (−) for 2 h. Anti-GFP immunoprecipitates (IP) were fractionated on SDS-PAGE, and blots were probed with anti-Hsp90 and anti-GFP. Aliquots (5%) of the whole cell lysates were probed to control for Hsp90 expression. One of two independent experiments is shown. (B) Human Ramos, mouse CH12F3-2 (pretreated for 16 h with IL-4, TGF-β1, and anti-CD40 to induce AID expression), and chicken DT40 B cell lines were treated with 2 µM GA (+) or DMSO (−) and harvested at the indicated time points after GA. The expression level of the indicated proteins was analyzed by Western blot (WB) in total or nuclear (where indicated) extracts. Identical effect was observed with 17-AAG in all cell lines (not depicted). Representative panels from one of two or three independent experiments (depending on the antibody) are shown. (C) Resting B cells purified from blood of three donors were activated with IL-4 and anti-CD40 and 4 d later treated and analyzed as in B except that the Hsp90 inhibitor 17-AAG (+) was used instead of GA. (D) Ramos cells stably expressing GFP, AID-GFP, or chimeras AID-A2 #1 or #2 were treated in triplicate with 2 µM GA or DMSO. The GFP mean fluorescence intensity (MFI) was monitored by flow cytometry and normalized to t0 = 100%. MFI ± SD is plotted over time. One of three independent experiments is shown (**, P < 0.01). (E) AID-GFP was monitored as in D except that Ramos cells were pretreated with 100 ng/ml CHX for 30 min before Hsp90 inhibition. One of five independent experiments is shown (**, P < 0.01). (F) Purified naive B cells from Aicda−/− mice were activated and retrovirally transduced with mouse AID-GFP or GFP control. Cells were analyzed as in D 2 d after transduction. One of three independent experiments is shown (**, P < 0.01).
To use a more sensitive assay to monitor AID decay at shorter times and to be able to compare AID variants, we established stable Ramos transfectants expressing various AID-GFP constructs from a heterologous promoter. The levels of AID-GFP could thus be monitored over time and accurately quantified by flow cytometry. We confirmed that AID-GFP but not GFP was destabilized upon Hsp90 inhibition in these cell lines with kinetics consistent to that observed for endogenous AID (Fig. 2 D). This indicated a direct action on AID protein rather than on Aicda transcription, which was confirmed by Northern blot of Aicda in Ramos cells (unpublished data). As would be expected, the AID-A2 chimeras that did not interact with Hsp90 were insensitive to GA treatment (Fig. 2 D). Treatments inhibiting or exacerbating PKA activity had no effect on the sensitivity of AID-GFP to GA, further suggesting that these two pathways are not connected (Fig. S2). We then measured the decay kinetics of AID-GFP after pretreating the cells with cycloheximide (CHX) so as to follow the pool of AID that had already been synthesized and not the nascent AID that might be more sensitive to folding requirements. CHX caused the expected decay of AID-GFP (compare control DMSO-treated AID-GFP levels in Fig. 2, D vs. E), which was clearly accelerated by GA (Fig. 2 E), indicating a role for Hsp90 in stabilizing fully synthesized AID. We confirmed that both mouse and human AID-GFP were similarly sensitive to Hsp90 inhibition when expressed in AID-deficient mouse primary splenic B cells (Fig. 2 F and not depicted), thus ruling out any effect of the transformed cell environment of Ramos on our observations. We conclude that functional Hsp90 is necessary to stabilize and maintain the steady-state levels of AID in vivo in primary as well as in transformed cells.
Hsp90 protects cytoplasmic AID from being degraded
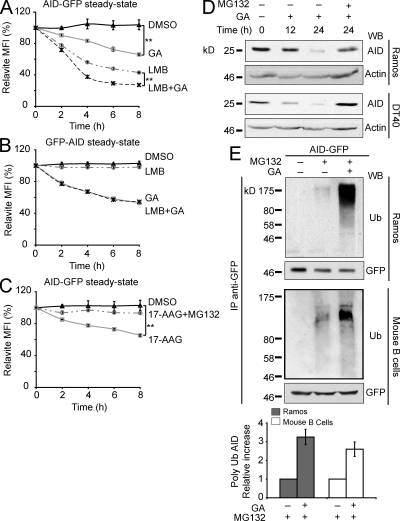
Binding to Hsp90 can regulate protein subcellular localization (DeFranco, 1999; Galigniana et al., 2004). Indeed, our results could be explained by increased AID nuclear import after Hsp90 inhibition and therefore AID destabilization in the nucleus (Aoufouchi et al., 2008). However, we did not observe any changes in AID localization after Hsp90 inhibition, even when combined with a proteasome inhibitor to prevent degradation of nuclear AID (Fig. S3). So, we compared the effects on AID-GFP of inhibiting Hsp90 versus inhibiting nuclear export with leptomycin B (LMB), which enriches AID-GFP in the nucleus (Ito et al., 2004; McBride et al., 2004), in the stable Ramos transfectants. The kinetics of AID-GFP decay after GA or LMB treatment were different (Fig. 3 A). Combined GA and LMB treatment showed an apparently additive effect (Fig. 3 A). Similar results were obtained using DT40 and HeLa cells stably expressing AID-GFP (Fig. S4). The lack of detectable nuclear translocation of AID after Hsp90 inhibition, together with the different decay kinetics after GA and LMB, raised the possibility that AID degradation after each of these treatments happened in different compartments and therefore would involve different pathways. To test this, we used Ramos cells expressing GFP-AID because the N-terminal GFP fusion blocks nuclear import of AID (Patenaude et al., 2009) but not its binding to Hsp90 (Fig. S1 E). Consistently, with its exclusively cytoplasmic localization, GFP-AID did not respond to LMB, but it was still sensitive to GA (Fig. 3 B). Hsp90 clients are in dynamic equilibrium with proteasomal degradation (Pearl and Prodromou, 2006). Indeed, the proteasome inhibitor MG132 prevented the Hsp90 inhibition–induced degradation of AID-GFP as well as of endogenous AID in Ramos and DT40 cells (Fig. 3, C and D; and Fig. S4 A). We obtained identical results using lactacystin, another proteasome inhibitor (unpublished data). A reproducible ∼3.5-fold increase in AID polyubiquitylation was observed in Ramos and primary mouse B cells after combined Hsp90 and proteasome inhibition versus inhibiting only the proteasome (Fig. 3 E). This was not particular to B cells because it was also true for AID-GFP in stably transfected HeLa cells (unpublished data). These experiments show that Hsp90 stabilizes cytoplasmic AID by protecting it from proteasomal degradation.
Figure 3.
Cytoplasmic ubiquitination and proteasomal degradation of AID after Hsp90 inhibition. (A) Ramos cells stably expressing AID-GFP were treated in triplicate with DMSO, 2 µM GA, and/or 50 ng/ml LMB, and the GFP signal was monitored over time by flow cytometry. The MFI normalized to t0 = 100% ± SD is plotted for each treatment (**, P < 0.01). One of five independent experiments is shown. (B) Ramos cells stably expressing GFP-AID, which is completely impaired for nuclear import (Patenaude et al., 2009), were analyzed as in A. One of three independent experiments is shown. (C) Ramos cells stably expressing AID-GFP were pretreated or not for 30 min with 10 µM MG132 before adding DMSO or 2 µM 17-AAG and analyzed as in A (**, P < 0.01). One of five independent experiments is shown. (D) Human Ramos and chicken DT40 B cell lines were treated with 2 µM GA. Where indicated, 10 µM MG132 was added only during the last 12 h of incubation to avoid excessive cell death. Cells were harvested at different time points, lysed, and analyzed by Western blot (WB). One of two independent experiments is shown for each cell line. (E) Ramos B cells stably expressing human AID-GFP or primary mouse B cells transduced with mouse AID-GFP were pretreated with 10 µM MG132 for 30 min before addition or not of 2 µM GA for 5 h. Anti-GFP immunoprecipitates (IP) were analyzed by Western blot using anti-ubiquitin (Ub) and anti-GFP. Polyubiquitinated AID was quantified by densitometry, and the relative means ± SD were plotted for three independent experiments for each cell type.
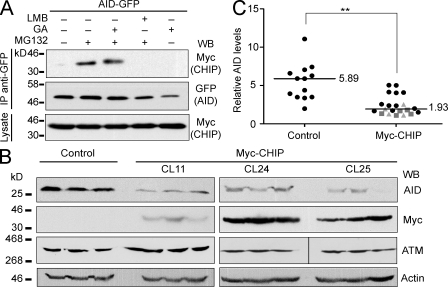
The E3 ubiquitin ligase CHIP (C terminus of Hsc70-interacting protein) is physically associated with Hsp90 and triages many Hsp90 clients (McDonough and Patterson, 2003). Of note, CHIP is constitutively expressed in Ramos and induced upon activation in primary mouse B cells (Fig. S1, B and C). Furthermore, CHIP coimmunoprecipitated with AID-GFP from extracts of stably transfected HeLa cells (Fig. 4 A). The interaction was only apparent when the cells were pretreated with a proteasome inhibitor, which allows the accumulation of this rapid turn over interaction (Li et al., 2004). We reasoned that if Hsp90 was normally stabilizing AID in B cells, the overexpression of CHIP could reduce AID levels by shifting the equilibrium of the pathway from stabilization to degradation. Indeed, all subclones from three independent transfectants of Ramos B cells overexpressing Myc-CHIP showed a significantly reduced steady-state level of AID (Fig. 4, B and C) but not of ATM (ataxia telangiectasia mutated), which is not an Hsp90 client. Altogether, these results indicate that cytoplasmic AID requires constant stabilization by Hsp90 and that altering the balance of this reaction, either by inhibiting Hsp90 or favoring the degradative side of this pathway through CHIP overexpression, leads to greatly diminished AID protein levels through proteasomal degradation in the cytoplasm.
Figure 4.
The Hsp90-associated E3 ubiquitin ligase CHIP can destabilize AID. (A) HeLa cells stably expressing AID-GFP were transfected with Myc-CHIP and 48 h later treated for 5 h with DMSO (−), 2 µM GA, 50 ng/ml LMB, and/or 10 µM MG132 (30-min pretreatment) in the indicated combinations. Anti-GFP immunoprecipitates (IP) were analyzed by Western blot (WB) with anti-GFP and anti-Myc. Aliquots (5%) of the total cell lysates were used to control for Myc-CHIP expression. One of two independent experiments is shown. (B) Endogenous AID was analyzed by Western blot in single cell subclones from untransfected control or three independent Myc-CHIP Ramos transfectants (CL11, CL24, and CL25) after expansion. ATM was used as an Hsp90-independent control, antiactin as loading control, and anti-Myc to confirm the expression of CHIP. Three representative subclones from each transfectant are shown. The vertical black line indicates that intervening lanes have been spliced out. (C) AID protein levels in all control or Myc-CHIP Ramos subclones (distinguished by different symbols) were estimated from nonsaturated Western blots. The signal was normalized to each corresponding actin signal obtained from equivalent exposures and plotted. Median values are indicated (**, P < 0.01).
Inhibition of Hsp90 results in reduced antibody diversification
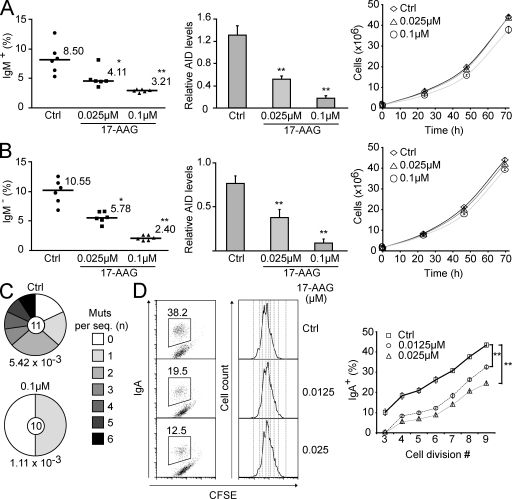
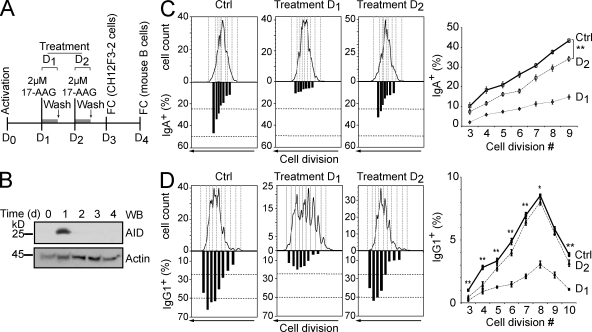
We first used the chicken B cell lymphoma line DT40 to monitor AID-dependent antibody diversification by gene conversion. The frequency of Ig gene conversion is estimated by using a DT40 line with a frameshift mutation in the IgVλ gene that prevents surface IgM expression. Some gene conversion events correct this frameshift, restoring IgM expression. The median percentage of IgM+ cells generated during expansion of several initially IgM− populations is proportional to the rate of Ig gene conversion (Arakawa et al., 2002). The problem is that Hsp90 is essential for eukaryotic cells (Borkovich et al., 1989; Cutforth and Rubin, 1994), which precludes its genetic ablation or complete inhibition. However, we observed that AID decay after Hsp90 inhibition was dose dependent (Fig. S4 C for GA and not depicted for 17-AAG). We used 17-AAG for these assays because we found it to be less toxic than GA for lymphocytes (unpublished data). Low doses of 17-AAG had minimal impact on DT40 cell growth but still caused a robust decrease in AID protein levels (Fig. 5 A). This partial reduction in AID levels was proportional to a reduction in Ig gene conversion (Fig. 5 A). In similar experiments using a DT40 line that diversifies the IgVλ by SHM (Arakawa et al., 2004), we could also confirm a reduction in SHM (this time by monitoring the appearance of sIgM-loss cells from originally sIgM+ populations but also by direct IgVλ sequencing) that was proportional to the decrease in AID protein levels (Fig. 5, B and C). We then analyzed the effect of Hsp90 inhibition on CSR by using the mouse CH12F3-2 cell line, which efficiently switches from IgM to IgA after cytokine stimulation (Nakamura et al., 1996). Because these assays take place over a few days, we used CFSE staining to monitor cell proliferation. Thus, we could compare the efficiency of switching between cells that have undergone the same number of cell divisions, accounting for any cell growth defect that continuous exposure to 17-AAG could cause. There was a clear and dose-dependent reduction in CSR caused by 17-AAG, overall and for each cell division tested (Fig. 5 D). Alternatively, we performed a 12-h treatment with higher doses of 17-AAG, after which the drug was removed (Fig. 6 A). A drastic reduction in CSR to IgA was observed when the CH12F3-2 cells were treated with 17-AAG at day 1 after stimulation, coincident with the time when the peak of AID protein was observed (Fig. 6, B and C). As would be expected, 17-AAG treatment at day 2 had a less but still statistically significant effect on CSR. Importantly, we obtained identical results in switching assays using normal mouse splenic B cells (Fig. 6 D). A drastic decrease of CSR to IgG1 was observed when cells were treated with 17-AAG at day 1 after stimulation. This higher dose of 17-AAG delayed growth of primary B cells, but the effect on CSR was nevertheless very clear when comparing the efficiency of switching per cell division. Again, treating the cells at day 2 after stimulation caused a smaller but statistically significant effect. As expected, treating the cells at day 3 had no effect on the efficiency of CSR measured at day 4 (unpublished data). We conclude that cytoplasmic AID degradation after Hsp90 inhibition has a direct and proportional effect on all antibody diversification mechanisms.
Figure 5.
Reduced antibody diversification in chicken and mouse B cells chronically treated with Hsp90 inhibitors. (A) The rate of Ig gene conversion in DT40 cells was estimated from the proportion of sIgM-gain cells arising from sIgM− populations after 3 wk of expansion in the presence of DMSO (Ctrl) or two different concentrations of 17-AAG. The proportion of sIgM+ cells for each population and the median values are indicated (left). The level of AID protein was quantified by densitometry from nonsaturated Western blots for each population at the end of the experiment and normalized to actin levels. Mean ± SD values for all populations grown in each condition are plotted (middle). The effect of 17-AAG on DT40 growth was monitored by calculating the total number of cells in a culture originating from 105 cells (right). Mean ± SD of triplicate cultures are plotted over time. (B) SHM was monitored in analogous experiments to A except that a sIgM+ ψV− AIDR DT40 cell line, which cannot undergo gene conversion and instead uses SHM to diversify the Ig genes, was used. IgM-loss cells arise as a consequence of SHM with a certain frequency, and so the proportion of sIgM-loss cells arising over time provides an estimate of SHM rate (Arakawa et al., 2004). (C) DNA was extracted from control or 0.1 µM 17-AAG–treated unsorted cultures from B after equal expansion. The IgVλ was PCR amplified and sequenced. The fraction of sequences containing the indicated number of mutations is plotted in the pie chart with the number of sequences analyzed indicated in the center. The calculated mutation frequency (mutations/base pair) is indicated under each pie chart. (D) CH12F3-2 mouse B cells stimulated with IL-4, TGF-β1, and agonist anti-CD40 were cultured with DMSO (Ctrl) or the indicated concentrations of 17-AAG. The cells were stained with CFSE before activation to follow cell divisions. Representative plots of the proportion of IgA+ cells in each population after 3 d (left) and CFSE profiles (middle) are shown. For each cell division, the proportion of sIgA+ cells was calculated, and the results from four independent experiments are summarized in the plot as the mean ± SD values (right). In all panels: *, P < 0.05; **, P < 0.01.
Figure 6.
Acute inhibition of Hsp90 impairs CSR in mouse B cells. (A) Scheme of the experimental strategy fort treatment of CH12F3-2 B cells or primary naive mouse splenic B cells. D, day; FC, flow cytometry. (B) Kinetics of AID protein expression in CH12F3-2 mouse B cells determined by Western blot (WB) at different times after stimulation. One of two independent experiments is shown. (C) CH12F3-2 B cells were stained with CFSE and stimulated with IL-4, TGF-β1, and anti-CD40 to switch to IgA. Either at day 1 or 2 after activation, the cells were treated for 12 h with 2 µM 17-AAG and then returned to normal medium. The proportion of sIgA+ cells per cell division determined by flow cytometry is plotted under each cell division in the corresponding CFSE plot for a representative experiment (left). The mean proportions of sIgA+ cells for each cell division ± SD from four independent experiments are plotted (right; **, P < 0.01). (D) Purified mouse naive splenic B cells were loaded with CFSE and stimulated with IL-4 and LPS to induce switching to IgG1. Either at day 1 or 2 after activation, the cells were treated with 17-AAG and then returned to normal medium. The proportion of sIgG1+ cells per cell division was determined and presented as in C. Data from one representative mouse are shown (left), and pooled data from five mice are plotted (right). To be able to compare all the mice accounting for the inter assay variability, data points were normalized with the percentage of IgG1+ cells in cell division 3 of the control set as 1 (*, P < 0.05; **, P < 0.01). Ctrl, control.
Hsp90 inhibition prevents off-target mutation by AID
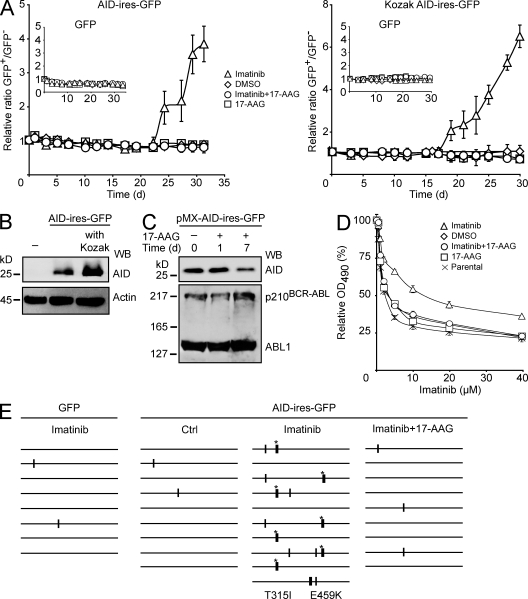
We assayed AID off-target mutation using the recently described role of AID in generating resistance of the oncogenic kinase BCR-ABL1 to the drug imatinib in CML cells (Klemm et al., 2009). We transduced the BCR-ABL1+ K562 cell line with AID-ires-GFP or GFP control retroviruses and, for each construct, prepared mixed populations with uninfected (GFP−) cells at a defined ratio. Mutations in BCR-ABL1 that confer resistance are selected by culturing the cells in the presence of imatinib. This is readily observed by flow cytometry as an increased ratio of GFP+/GFP− cells over time. Indeed, we observed a higher proportion of GFP+ cells within 2–3 wk only in AID-expressing cells that were growing in the presence of imatinib (Fig. 7 A). This was AID dependent as it was not observed in GFP control cultures, and the imatinib-resistant cells appeared earlier in cultures expressing higher levels of AID (Fig. 7, A and B). More relevant to our purpose, the AID-dependent change in the GFP+/GFP− ratio was fully prevented by a very low dose of 17-AAG (Fig. 7 A). Treatment with this dose of 17-AAG visibly reduced AID but not BCR-ABL1 protein levels by day 7 (Fig. 7 C). We confirmed that 17-AAG prevented the AID-mediated increase in imatinib IC50 (Fig. 7 D). We also directly checked for mutations by sequencing BCR-ABL1. An increase in point mutations (many of them known to induce imatinib resistance; Klemm et al., 2009) could be detected in AID-expressing K562 cells growing in imatinib. In contrast, the mutation level in AID-expressing cultures that were jointly treated with imatinib and 17-AAG was indistinguishable from the GFP control (Fig. 7 E and Table II).
Figure 7.
Hsp90 inhibition reduces AID off-target mutations. (A) BCR-ABL1+ K562 cells were transduced with GFP control (insets) or AID-ires-GFP retroviral vectors. Mixed populations of transduced (GFP+) and uninfected (GFP−) cells were cultured in the presence of DMSO, 2 µM imatinib, 2 µM imatinib plus 0.1 µM 17-AAG, or 0.1 µM 17-AAG, and the proportion of GFP+ cells was determined periodically by flow cytometry. Data are plotted as the mean GFP+/GFP− ratio from triplicate populations ± SD, relative to the initial ratio set as 1. Two independent experiments are shown differing only in AID protein level expression from having or not a consensus Kozak sequence before the AID start codon. (B) Western blot (WB) of parental K562 cells (−) and sorted GFP+ populations transduced with pMX-AID-ires-GFP vectors differing only in the presence of the Kozak sequence. AID was detected using anti-AID, and antiactin was used as loading control. (C) Sorted GFP+ cells transduced or not with pMX-KozakAID-ires-GFP vectors as in A were cultured with 0.1 µM 17-AAG for the indicated times. The levels of the indicated proteins were analyzed by Western blot in total extracts using anti-AID and anti-ABL1. One of two independent experiments is shown. (D) GFP+ K562 populations were sorted after expansion from A. Cell viability after culture for 2 d with different concentrations of imatinib was determined using an MTS reduction colorimetric assay. The relative mean OD at 490 nm ± SEM of duplicate wells (untreated cells = 100%) is plotted for each concentration for cell populations transduced with pMXs-KozakAID-ires-GFP that had been treated in A with DMSO, imatinib, imatinib plus 0.1 µM 17-AAG, or 17-AAG. The parental K562 cells were included in this assay. One of two independent experiments is shown. (E) BCR-ABL1 (exon 13 of BCR and exon 9 of ABL1) was PCR amplified from cells expressing GFP control or AID-ires-GFP and expanded under the indicated conditions. Mutations, determined relative to the consensus of all sequences, are indicated on schemes of the ∼700-bp ABL1 region that was directly sequenced from the PCR product. Thin vertical bars represent mutations at A:T bp, whereas thick bars represent mutations at C:G pairs. Mutations previously described to confer imatinib resistance (Branford and Hughes, 2006) are identified by an asterisk and indicated below the sequence stack.
Table II.
BCR-ABL1 mutations in K562 cells
| Construct transduced | Treatment | BCR-ABL1 sequence analysis | Number of clones/total | Amino acid change |
| GFP | Imatinib | Unmutated | 6/8 | No changes |
| 894 CTA to CTG | 1/8 | Silent | ||
| 1062 CTA to CTG | 1/8 | Silent | ||
| AID-ires-GFP | Control | Unmutated | 7/9 | No changes |
| 894 CTA to CTG | 1/9 | Silent | ||
| 1062 CTA to CTG | 1/9 | Silent | ||
| AID-ires-GFP | Imatinib | Unmutated | 2/10 | No changes |
| 894 CTA to CTG | 3/10 | Silent | ||
| 944 ACT to ATT | 4/10 | T315I | ||
| 1062 CTA to CTG | 2/10 | Silent | ||
| 1334/1335 CGT to CTG | 1/10 | R445L | ||
| 1356 CTA to CTG | 2/10 | Silent | ||
| 1375 GAG to AAG | 3/10 | E459K | ||
| AID-ires-GFP | Imatinib + 17-AAG | Unmutated | 6/9 | No changes |
| 894 CTA to CTG | 1/9 | Silent | ||
| 1062 CTA to CTG | 2/9 | Silent |
Identity of the mutations indicated in the diagram in Fig. 7 E. The BCR-ABL1 (exon 13 of BCR and exon 9 of ABL1) was RT-PCR amplified from single cell clones, and a fragment of ∼700 bp in the ABL1 kinase domain was directly sequenced. The number to the left of the mutation indicates the mutated position with respect to the ABL1 open reading frame. The mutated base is underlined within the affected codon. Mutations at C:G pairs and amino acid substitutions previously described to confer clinical imatinib resistance (Branford and Hughes, 2006) are highlighted in bold.
DISCUSSION
We identify and characterize herein the constitutive stabilization of AID by the Hsp90 chaperone pathway. Our results show that Hsp90 largely determines the overall steady-state levels of functional AID protein. Although Hsp90 may also contribute to the biogenesis of AID, its key function appears to be the stabilization of the protein. This is well in accordance with the major role of Hsp90 in protecting the functional competence of many of its clients, beyond simply assisting with their de novo folding (Jakob et al., 1995; Nathan et al., 1997; Whitesell and Lindquist, 2005; Pearl and Prodromou, 2006; Picard, 2006; Wandinger et al., 2008). This is a new mechanism positively regulating AID-mediated antibody diversification that seems evolutionarily conserved, as we find it in chicken, mouse, and human B cells.
The molecular details of AID stabilization in the cytoplasm are likely incomplete because many proteins modulate the Hsp90 pathway (Young et al., 2004; Wandinger et al., 2008). However, our experimental evidence, together with the identification of members of the Hsp90 chaperone pathway that consistently copurified with AID (Table I), suggests a working model in which AID would be regulated similarly to the steroid hormone receptors (Picard, 2006). These receptors first form an early complex with the Hsp70/Hsp40 chaperone/cochaperone, to which Hsp90 and other factors are then recruited. Consecutive cycles of chaperoning are in dynamic equilibrium with proteasomal degradation unless the receptor is stabilized by ligand binding and translocates to the nucleus (Whitesell and Lindquist, 2005; Picard, 2006). Consistent with this model, we found HSPA8, the major constitutively expressed form of Hsp70, among the AID-Flag/HA–interacting partners in Ramos B cells. The same isoform has been found using tagged AID expressed in HEK293 cells (Wu et al., 2005). Hsp40 proteins are the first agents in the Hsp70/Hsp90 chaperone pathway (Kimura et al., 1995; Hernández et al., 2002). We identified and have confirmed the functional interaction of AID with a defined subset of Hsp40 proteins, which will be reported elsewhere (unpublished data). AID stabilization requires the ATP hydrolysis cycle of Hsp90, and the cochaperone AHA-1, which stimulates the ATPase activity of Hsp90 (Panaretou et al., 2002; Lotz et al., 2003), also copurified with AID. As is the case for steroid hormone receptors, we show that cytoplasmic AID exists in a dynamic equilibrium between stabilization and proteasomal degradation. Indeed, inhibiting Hsp90 or overexpressing the Hsp90-associated ubiquitin ligase CHIP (Connell et al., 2001; McDonough and Patterson, 2003) can shift the balance toward AID degradation. Stabilization seems to be the predominant pathway because simultaneous inhibition of the proteasome and Hsp90 leads to the accumulation of much higher levels of polyubiquitinated AID than only proteasome inhibition. However, it remains possible that the destabilizing side of this pathway contributes to limit AID levels, which we are exploring. It is interesting that AID seems to be uniquely dependent on Hsp90 when compared with its paralogue proteins, the APOBEC family. The intrinsic instability of uncomplexed AID probably helps to limit its mutagenic potential. This fits nicely with a study showing that the half-life of AID is highly reduced in the nucleus compared with the cytoplasm (Aoufouchi et al., 2008). We speculate that AID, just as the steroid hormone receptors (Whitesell and Lindquist, 2005; Picard, 2006), might undergo some conformational change to dissociate from Hsp90. An attractive possibility is that oligomerization stabilizes AID, thus emancipating it from Hsp90. In line with this, the Hsp90-interacting domain of AID comprises most of its proposed dimerization interface (Prochnow et al., 2007; Patenaude et al., 2009), and an oligomerization-deficient AID mutant can still bind to Hsp90. We have proposed that dimerization/oligomerization of AID is important for its cytoplasmic retention and nuclear import (Patenaude et al., 2009). However, at variance with the estrogen receptor, Hsp90 does not seem to play a major role in AID cytoplasmic retention. We must hypothesize that AID forms part of another cytoplasmic complex that fulfills this role. This Hsp90-independent fraction of AID might explain the slower kinetics of AID degradation in Ramos cells after Hsp90 inhibition compared with other Hsp90 clients such as Lck (Giannini and Bijlmakers, 2004).
The interaction of AID with Hsp90 has major functional consequences. Hsp90 stabilizes many proteins, and part of the effect observed on antibody diversification after inhibiting Hsp90 could formally be indirect. However, there is a negative dose–response relationship between AID protein levels and Hsp90 inhibition, which correlates with a proportional decrease in antibody diversification. This observation argues for a direct effect. Furthermore, Hsp90 inhibition affected all AID-dependent pathways: SHM, CSR, and Ig gene conversion as well as off-target mutation. All of these pathways are initiated by DNA deamination by AID followed by uracil processing by either UNG or MSH2/MSH6, which were not affected by Hsp90 inhibition. Downstream from there, these pathways diverge (i.e., trans-lesion synthesis for mutations, homologous recombination for gene conversion, and nonhomologous end joining for CSR), so a direct effect on AID is much more likely than an independent effect on each of these pathways. By stabilizing the bulk of AID, the Hsp90 pathway determines the availability of functional AID. Indeed, AID-catalyzed DNA deamination at the Ig loci was directly proportional to the overall level of AID protein remaining after Hsp90 inhibition. This is in agreement with the previously reported AID dose effect on the efficiency of antibody diversification (de Yébenes et al., 2008; Dorsett et al., 2008; Sernández et al., 2008; Takizawa et al., 2008; Teng et al., 2008). Together with our findings, these observations suggest that the ratio of nuclear to cytoplasmic AID is constant. Thus, decreasing the overall level of AID would lead to a proportional decrease in the nuclear fraction and biological activity of AID. Otherwise, if AID were particularly abundant in the nucleus at any stage, a 50% decrease in total AID protein caused by partial Hsp90 inhibition (or by haploinsufficiency) would not necessarily lead to a proportional decrease in antibody diversification. Our work highlights the intimate relationship between the mechanisms of AID subcellular localization and protein stability (Aoufouchi et al., 2008; Patenaude et al., 2009).
Finally, the Hsp90-mediated mechanism stabilizing AID is operative in lymphoma-derived cell lines as well as in cells with ectopic overexpression of AID. The relative Hsp90 dependence of AID in transformed versus normal cells remains to be studied. Nevertheless, our findings offer the first possibility of pharmacologically manipulating AID protein levels to prevent off-target mutation. We provide proof of principle that this is possible in a CML cell line model. A very low dose of Hsp90 inhibitor completely abrogated BCR-ABL1 mutations and imatinib resistance. AID-generated lesions in non-Ig genes are widespread but much less frequent than at the Ig genes (Liu et al., 2008; Robbiani et al., 2009). Indeed, the AID-generated breaks in c-Myc seem to be limiting for the oncogenic c-Myc–IgH chromosomal translocations (Robbiani et al., 2008, 2009). Therefore, it might be possible that very low doses of Hsp90 inhibitors have a disproportionate effect on the frequency of oncogenic lesions versus antibody diversification. It would be worth exploring whether the Hsp90 inhibitors that are currently being tested in the clinic could be useful in treating those cancers in which AID contributes to disease progression (Matsumoto et al., 2007; Pasqualucci et al., 2008; Klemm et al., 2009).
MATERIALS AND METHODS
DNA constructs.
The pEGFP-N3–based (Takara Bio Inc.) expression vectors for human AID-GFP, AID FYRN-GFP, AID-Flag/HA, APOBEC2, and AID-APOBEC2 chimeras have been described previously (Patenaude et al., 2009). Rat APOBEC1 and human APOBEC3G cloned in pEGFP-C3 as well as human AID T27A/T38A, which was subcloned into pEGFP-N3, were gifts from S. Conticello (Istituto Toscano Tumori, Florence, Italy; Conticello et al., 2008). To construct N-terminally Flag-tagged versions of APOBEC1, APOBEC2, and APOBEC3G, enhanced GFP (EGFP) was excised NheI–XhoI from pEGFP-C3 and replaced by the annealed oligonucleotides AO1 and AO2. To construct C-terminally Flag-tagged versions of some of the proteins, EGFP was excised EcoRI–NotI from pEGFP-N3 and replaced by the annealed oligonucleotides OJ215 and OJ216. AID was subcloned as an NheI–NotI fragment under the weaker EF1-α promoter in pEF. AID-APOBEC2 chimeras #1 and #2 were excised from pTrc99a (Patenaude et al., 2009) by partial digestion with NotI and EcoRI and subcloned into the pMXs retroviral vector. Untagged hAID in pMXs-ires-GFP has been described previously (Patenaude et al., 2009). Mouse AID (from R. Harris, University of Minnesota, Minneapolis, MN) was subcloned EcoRI and NotI into pMXs. Flag-human Hsp90-α was inserted as a KpnI–NotI fragment into pcDNA3.1. Myc-human Hsp90-β in pCMV-3Tag2 was a gift of J.-.P Gratton (Institut de Recherches Cliniques de Montréal, Montréal, Québec, Canada). Construct names throughout the manuscript indicate the actual order of the fragments in fusion proteins.
Reagents and antibodies.
Stock aliquots of 2 mM GA, 2 mM 17-AAG, 5 mM H-89, and 25 mM forskolin (LC Laboratories) as well as 50 mM IBMX (3-isobutyl-1-methylxanthine; Sigma-Aldrich) were made in DMSO. Stocks of 5 mM MG132 (EMD) and 25 µg/ml LMB (LC Laboratories) were made in ethanol. CHX (Sigma-Aldrich) was freshly prepared before each experiment 100 mg/ml in ethanol. Stock of 2 mM imatinib (Gleevec; Novartis) in PBS was a gift of T. Moroy and C. Khandanpur (Institut de Recherches Cliniques de Montréal). All drugs were stored at −20°C protected from light. Antibodies and dilutions used were as follows: 1:3,000 anti–EGFP–horseradish peroxidase (HRP; Miltenyi Biotec), 1:3,000 anti–Myc-HRP (Miltenyi Biotec), 1:3,000 anti–Flag-HRP (Sigma-Aldrich), 1:3,000 anti-Hsp90 (sees both isoforms; BD), 1:1,000 anti–Hsp90-α and 1:1,000 anti–Hsp90-β (StressMarq), 1:1,000 anti-AID (Cell Signaling Technology) for human and chicken AID and 1:500 anti-mAID (a gift from F. Alt, Harvard University, Boston, MA) for mouse AID, 1:3,000 antiactin (Sigma-Aldrich), 1:1,000 anti-monoubiquitinated and -polyubiquitinated conjugates antibody (Enzo Life Sciences, Inc.) for endogenous ubiquitin, 1:1,000 mAb anti-CHIP (Sigma-Aldrich), 1:2,000 anti-UNG2 (specific for nuclear isoform; Abcam), 1:2,500 anti-MSH6 (Bethyl Laboratories, Inc.), 1:5,000 anti-PCNA (PC-10; Abcam), 1:1,000 anti–DNA-PK (Santa Cruz Biotechnology, Inc.), 1:1,000 anti-Lck (gift of A. Veillette, Institut de Recherches Cliniques de Montréal), and 1:1,000 anti–c-abl (EMD). Secondary antibodies were used according to the species of the primary antibody: 1:5,000 goat anti–mouse-HRP and 1:10,000 anti–rabbit-HRP (Dako) and 1:5,000 goat anti–rat-HRP (Millipore).
Mice and cell lines.
HeLa cells stably expressing AID-GFP were generated by transfecting with pEF-AID-EGFP and selecting with 2.5 µg/ml puromycin. Ramos cell lines stably expressing GFP, AID-GFP, and AID-Flag/HA have been described previously (Patenaude et al., 2009). Ramos cells expressing Myc-CHIP were generated by transfecting with pcDNA3.1 Myc-CHIP (a gift of L. Petrucelli, Mayo Clinic, Jacksonville, FL) and selecting with G418. Subclones from three independent Myc-CHIP Ramos transfectants and controls were obtained by single cell deposition. Populations of Ramos cells stably expressing chimeras AID-A2 #1 or #2 and DT40 cells stably expressing GFP or AID-GFP as well as the CML cell line K562 (a gift of T. Moroy) stably expressing AID-ires-GFP or GFP control were obtained by retroviral delivery of these genes cloned in pMXs vectors. The supernatant of HEK293T cells cotransfected with pMX and vectors expressing MLV Gag-Pol and VSV-G envelope (3:1:1 ratio) was used to infect 106 cells by spin-infection at 600 g for 1 h at room temperature in the presence of 16 µg/ml polybrene and 20 mM Hepes. Infected cells were sorted to obtain GFP+ homogeneous populations. Primary B cells from Aicda−/− mice (obtained from T. Honjo, Kyoto University, Sakyo-ku, Kyoto, Japan) were prepared and infected as described previously (Patenaude et al., 2009). Experiments using mice followed the guidelines of the Canadian Council on Animal Care and were approved by the Animal Protection Committee at the Institut de Recherches Cliniques de Montréal. Primary human B cells were purified from PBMC obtained by Ficoll gradient centrifugation of voluntary donor blood samples. Resting B cells were isolated using a B cell isolation kit (Miltenyi Biotech) and activated with 5 ng/ml recombinant hIL-4 (PeproTech) and 5 µg/ml recombinant human sCD40L. Work with human samples was performed according to the guidelines of the Institut de Recherches Cliniques de Montréal and Institut National de la Recherche Scientifique–Insitut Armand-Frappier Ethics Committees for Research with Human Samples.
Identification of AID-interacting proteins.
5 × 109 Ramos B cells expressing AID-Flag/HA or empty vector were pelleted, incubated on ice for 10 min, and resuspended in hypotonic buffer I (1 mM Tris-HCl, pH 7.3, 10 mM KCl, 1.5 mM MgCl2, and β-mercaptoethanol). Cells were centrifuged at 2,500 rpm for 10 min at 4°C and lysed by adding hypotonic buffer II (1 mM Tris-HCl, pH 7.3, 10 mM KCl, 1.5 mM MgCl2, 1 mM trichostatin A, 50 µM β-mercaptoethanol, 0.5 mM PMSF, and protease inhibitors [Sigma-Aldrich]). The lysate was centrifuged at 3,900 rpm for 15 min at 4°C, and the supernatant was recentrifuged at 35,000 rpm for 1 h and dialyzed against 20 mM Tris-HCl, pH 7.3, 20% glycerol, 100 mM KCl, 50 µM β-mercaptoethanol, and 0.5 mM PMSF. The dialyzed lysate was incubated with anti-Flag M2 affinity gel (Sigma-Aldrich) overnight at 4°C and then extensively washed and eluted using 3× Flag peptide (Sigma-Aldrich). The eluate was incubated with anti-HA beads (Santa Cruz Biotechnology, Inc.) overnight at 4°C and then washed and eluted using HA peptides (PEP-101P; Covance). Protein was concentrated using StrataClean Resin (Agilent Technologies) before loading on a 4–12% gradient gel (Invitrogen) for SDS-PAGE. The gel was silver stained, each lane was divided into 20 slices, and the slices were submitted for tryptic digestion and peptide identification by mass spectrometry to the Institut de Recherches Cliniques de Montréal Proteomics service using a linear quadrupole IT Orbitrap hybrid mass spectrometer (Thermo Fisher Scientific). Peak generation and protein identification were performed using the MASCOT software package (Perkins et al., 1999).
Coimmunoprecipitation and Western blot.
HEK293T cells cotransfected at a 1:1 ratio with GFP and Myc or Flag-tagged proteins were homogenized in lysis buffer (20 mM Tris, pH 8.0, 137 mM NaCl, 10% glycerol, 2 mM EDTA, 1% Triton X-100, and 20 mM NaF) 48 h after transfection and immunoprecipitated with anti-Flag M2 affinity gel as described previously (Patenaude et al., 2009). Immunoprecipitation of GFP-tagged proteins were performed using the μMACS GFP Isolation kit (Miltenyi Biotech) according to the manufacturer’s instructions. Where indicated, cells were treated with 10 µM MG132 for 30 min and/or 2 µM GA or DMSO for 5 h before lysis. The eluates and lysates were analyzed by Western blot developed with SuperSignal West Pico Chemiluminescent substrate (Thermo Fisher Scientific).
AID stability assays.
The GFP signal of cell lines expressing AID-GFP variants was measured by flow cytometry at various time points after the indicated treatments. Dead cells were excluded by propidium iodide staining. For endogenous AID, 5 × 106 Ramos, DT40, or K562 cells in 5 ml of culture medium were treated with GA or 17-AAG, and aliquots of 1.5–2 × 106 cells were harvested at various time points. Alternatively, 2 × 106 CH12F3-2 cells (a gift of T. Honjo through A. Martin [University of Toronto, Toronto, Ontario, Canada]; Nakamura et al., 1996) were stimulated with 2 ng/ml recombinant human TGF-β1 (R&D Systems), 20 ng/ml recombinant murine IL-4 (PeproTech), and 5 µg/ml functional grade purified anti–mouse CD40 (BD) for 16 h before GA treatment. Human primary B cells at 2 × 106/ml were treated with 2 µM 17-AAG 72 h after activation, and aliquots of 106 cells were harvested at various time points. Cells were washed once with PBS and lysed in SDS-PAGE sample buffer. Lysates were analyzed by Western blotting.
Monitoring antibody diversification.
AID-mediated Ig gene conversion was estimated in DT40 cre1 cells by scoring the frequency of sIgM-gain phenotype, which is directly proportional to the frequency of repair of a frameshift in the IgVλ by gene conversion (Arakawa et al., 2002). DT40 sIgM− cell populations were FACS sorted, and confluent cultures were grown in 24-well plates with Hsp90 inhibitors. This method was favored over using single cell clones because of the effect of Hsp90 inhibition on single cells growth. Cells were grown for 3 wk at 41°C in the presence of the inhibitors, and the surface IgM phenotype was measured by flow cytometry as described previously (Di Noia and Neuberger, 2004). AID-mediated SHM was monitored using the sIgM+ DT40 line ψV− AIDR, in which the IgV pseudogenes have been ablated (gift of H. Arakawa and J.-M. Buerstedde, Institute for Molecular Radiobiology, Neuherberg, Germany; Arakawa et al., 2004). IgM+ cell populations were FACS sorted and expanded for 3 wk in 24-well plates, and the IgM phenotype was analyzed by flow cytometry. Mutations were scored as described previously (Di Noia and Neuberger, 2004) in Vλ sequences PCR amplified from unsorted populations after expansion. To analyze CSR, CH12F3-2 cells were preincubated with CFSE (Invitrogen) according to manufacturer’s instruction before activation with 1 ng/ml TGF-β1, 10 ng/ml recombinant murine IL-4, and 1 µg/ml functional grade purified anti–mouse CD40 (BD). For chronic Hsp90 inhibition, 17-AAG was added 4 h after activation and kept for 3 d. For acute Hsp90 inhibition, 17-AAG was added to the medium for 12 h, and then the cells were washed twice with PBS and resuspended in fresh normal medium. IgA expression was monitored at day 3 after stimulation using PE-conjugated anti–mouse IgA antibody (eBioscience). Resting B cells from AID-deficient mice were purified by MACS CD43 depletion (Miltenyi Biotech) from total splenic lymphocytes (Patenaude et al., 2009). Cells were loaded with CFSE, and 106 cells/well were seeded in 24-well plates with 25 µg/ml LPS (Sigma-Aldrich) and 50 ng/ml mouse IL-4. At different times after activation, 17-AAG was added and washed away 12 h later with PBS, and fresh culture medium was replenished. Isotype switching was analyzed 4 d after activation by flow cytometry after staining with anti–IgG1-biotin (BD), followed by APC-conjugated antibiotin antibody (Miltenyi Biotech) and propidium iodide.
Imatinib resistance assay.
K562 cells stably expressing AID-ires-GFP or GFP control were mixed with the parental cell line at a fixed ratio. The ratio of GFP+ to GFP− cells was measured by flow cytometry every 2 d for populations under the different treatments indicated in the figure (2 µM DMSO, 2 µM imatinib, and 0.1 µM 17-AAG). GFP+ cells were FACS sorted at the end of the experiments for the relevant conditions. Relative imatinib resistance of these populations was determined using Celltiter 96 Aqueous nonradioactive cell proliferation assay (Promega) according to manufacturer’s instruction. For mutation analysis of the BCR-ABL1 gene, single GFP+ CML cells expressing AID or not were sorted at the end of the treatments and expanded to obtain a clone in 96-well plates. RNA was extracted with TRIZOL (Invitrogen), and cDNA synthesis was performed using M-MULV first strand cDNA synthesis kit (New England Biolabs, Inc.). A 1540-bp fragment of the BCR-ABL1 cDNA was PCR amplified using specific primers for BCR (in exon 13) and ABL1 (in exon 9; Klemm et al., 2009) with KOD Hot Start Polymerase (EMD). PCR products were purified and directly sequenced using an ABL1-specific primer.
Statistical analysis.
The unpaired two-tailed Student’s t test was used.
Online supplemental material.
Fig. S1 shows the expression levels of transfected AID in Ramos B cells and endogenous Hsp90 and CHIP in B cells and B cell lines as well as the interaction of Hsp90 with oligomerization-deficient and phospho-null AID mutants and with GFP-AID. Fig. S2 shows that inhibiting or activating PKA in Ramos cells has no effect on the decay of AID-GFP after Hsp90 inhibition. Fig. S3 shows that inhibition of Hsp90 does not lead to relocalization of AID from the cytoplasm to the nucleus. Fig. S4 shows that Hsp90 prevents proteasomal degradation of AID-GFP in DT40 chicken B cells and in HeLa cells and the dose–response curves for AID-GFP levels in Ramos cells treated with GA. Online supplemental material is available at http://www.jem.org/cgi/content/full/jem.20101321/DC1.
Acknowledgments
We thank Dr. J.-P. Gratton for useful discussions and Drs. S. Conticello, D. Muñoz, C.A. Buscaglia, and R. Casellas for comments on the manuscript. We are indebted to H. Yu for helping with AID-Flag/HA purification and to Drs. H. Arakawa, J.-M. Buerstedde, S. Conticello, R. Harris, J.-P. Gratton, T. Moroy, C. Khandanpur, A. Veillette, F. Alt, A. Martin, T. Honjo, and L. Petrucelli for providing reagents. We thank D. Faubert (Institut de Recherches Cliniques de Montréal Proteomics facility) for assistance.
This work was funded by grants from The Cancer Research Society and Canadian Institutes of Health Research (CIHR; MOP-84543) and supported by a Canadian Foundation for Innovation Leaders Opportunity Fund equipment grant to J.M. Di Noia. A. Orthwein was supported in part by a Michel Bélanger fellowship from the Institut de Recherches Cliniques de Montréal, by an Excellence Fellowship from the Department of Microbiology and Immunology, University of Montréal, and by a Cole Foundation Fellowship. A. Lamarre is supported by the Jeanne and J.-Louis Lévesque Chair in Immunovirology of the J.-Louis Lévesque Foundation and by CIHR. J.M. Di Noia and J.C. Young are supported by Canada Research Chairs Tier 2.
The authors have no conflicting financial interests.
Footnotes
Abbreviations used:
- 17-AAG
- 17-allylamino-17-demethoxygeldanamycin
- AID
- activation-induced deaminase
- CHIP
- C terminus of Hsc70-interacting protein
- CHX
- cycloheximide
- CML
- chronic myeloid leukemia
- CSR
- class switch recombination
- EGFP
- enhanced GFP
- GA
- geldanamycin
- HRP
- horseradish peroxidase
- LMB
- leptomycin B
- MFI
- mean fluorescence intensity
- PKA
- protein kinase A
- SHM
- somatic hypermutation
References
- Albesiano E., Messmer B.T., Damle R.N., Allen S.L., Rai K.R., Chiorazzi N. 2003. Activation-induced cytidine deaminase in chronic lymphocytic leukemia B cells: expression as multiple forms in a dynamic, variably sized fraction of the clone. Blood. 102:3333–3339 10.1182/blood-2003-05-1585 [DOI] [PubMed] [Google Scholar]
- Aoufouchi S., Faili A., Zober C., D’Orlando O., Weller S., Weill J.-C., Reynaud C.-A. 2008. Proteasomal degradation restricts the nuclear lifespan of AID. J. Exp. Med. 205:1357–1368 10.1084/jem.20070950 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Arakawa H., Hauschild J., Buerstedde J.-M. 2002. Requirement of the activation-induced deaminase (AID) gene for immunoglobulin gene conversion. Science. 295:1301–1306 10.1126/science.1067308 [DOI] [PubMed] [Google Scholar]
- Arakawa H., Saribasak H., Buerstedde J.-M. 2004. Activation-induced cytidine deaminase initiates immunoglobulin gene conversion and hypermutation by a common intermediate. PLoS Biol. 2:E179 10.1371/journal.pbio.0020179 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bhutani N., Brady J.J., Damian M., Sacco A., Corbel S.Y., Blau H.M. 2010. Reprogramming towards pluripotency requires AID-dependent DNA demethylation. Nature. 463:1042–1047 10.1038/nature08752 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Borkovich K.A., Farrelly F.W., Finkelstein D.B., Taulien J., Lindquist S. 1989. hsp82 is an essential protein that is required in higher concentrations for growth of cells at higher temperatures. Mol. Cell. Biol. 9:3919–3930 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Branford S., Hughes T. 2006. Detection of BCR-ABL mutations and resistance to imatinib mesylate. Methods Mol. Med. 125:93–106 [DOI] [PubMed] [Google Scholar]
- Connell P., Ballinger C.A., Jiang J., Wu Y., Thompson L.J., Höhfeld J., Patterson C. 2001. The co-chaperone CHIP regulates protein triage decisions mediated by heat-shock proteins. Nat. Cell Biol. 3:93–96 10.1038/35050618 [DOI] [PubMed] [Google Scholar]
- Conticello S.G., Thomas C.J., Petersen-Mahrt S.K., Neuberger M.S. 2005. Evolution of the AID/APOBEC family of polynucleotide (deoxy)cytidine deaminases. Mol. Biol. Evol. 22:367–377 10.1093/molbev/msi026 [DOI] [PubMed] [Google Scholar]
- Conticello S.G., Ganesh K., Xue K., Lu M., Rada C., Neuberger M.S. 2008. Interaction between antibody-diversification enzyme AID and spliceosome-associated factor CTNNBL1. Mol. Cell. 31:474–484 10.1016/j.molcel.2008.07.009 [DOI] [PubMed] [Google Scholar]
- Crouch E.E., Li Z., Takizawa M., Fichtner-Feigl S., Gourzi P., Montaño C., Feigenbaum L., Wilson P., Janz S., Papavasiliou F.N., Casellas R. 2007. Regulation of AID expression in the immune response. J. Exp. Med. 204:1145–1156 10.1084/jem.20061952 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Csermely P., Schnaider T., Soti C., Prohászka Z., Nardai G. 1998. The 90-kDa molecular chaperone family: structure, function, and clinical applications. A comprehensive review. Pharmacol. Ther. 79:129–168 10.1016/S0163-7258(98)00013-8 [DOI] [PubMed] [Google Scholar]
- Cutforth T., Rubin G.M. 1994. Mutations in Hsp83 and cdc37 impair signaling by the sevenless receptor tyrosine kinase in Drosophila. Cell. 77:1027–1036 10.1016/0092-8674(94)90442-1 [DOI] [PubMed] [Google Scholar]
- de Yébenes V.G., Belver L., Pisano D.G., González S., Villasante A., Croce C., He L., Ramiro A.R. 2008. miR-181b negatively regulates activation-induced cytidine deaminase in B cells. J. Exp. Med. 205:2199–2206 10.1084/jem.20080579 [DOI] [PMC free article] [PubMed] [Google Scholar]
- DeFranco D.B. 1999. Regulation of steroid receptor subcellular trafficking. Cell Biochem. Biophys. 30:1–24 10.1007/BF02737882 [DOI] [PubMed] [Google Scholar]
- Di Noia J.M., Neuberger M.S. 2004. Immunoglobulin gene conversion in chicken DT40 cells largely proceeds through an abasic site intermediate generated by excision of the uracil produced by AID-mediated deoxycytidine deamination. Eur. J. Immunol. 34:504–508 10.1002/eji.200324631 [DOI] [PubMed] [Google Scholar]
- Di Noia J.M., Neuberger M.S. 2007. Molecular mechanisms of antibody somatic hypermutation. Annu. Rev. Biochem. 76:1–22 10.1146/annurev.biochem.76.061705.090740 [DOI] [PubMed] [Google Scholar]
- Dickey C.A., Kamal A., Lundgren K., Klosak N., Bailey R.M., Dunmore J., Ash P., Shoraka S., Zlatkovic J., Eckman C.B., et al. 2007. The high-affinity HSP90-CHIP complex recognizes and selectively degrades phosphorylated tau client proteins. J. Clin. Invest. 117:648–658 10.1172/JCI29715 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dorsett Y., McBride K.M., Jankovic M., Gazumyan A., Thai T.-H., Robbiani D.F., Di Virgilio M., San-Martin B.R., Heidkamp G., Schwickert T.A., et al. 2008. MicroRNA-155 suppresses activation-induced cytidine deaminase-mediated Myc-Igh translocation. Immunity. 28:630–638 10.1016/j.immuni.2008.04.002 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Endo Y., Marusawa H., Kou T., Nakase H., Fujii S., Fujimori T., Kinoshita K., Honjo T., Chiba T. 2008. Activation-induced cytidine deaminase links between inflammation and the development of colitis-associated colorectal cancers. Gastroenterology. 135:889–898: 898: e1–e3 10.1053/j.gastro.2008.06.091 [DOI] [PubMed] [Google Scholar]
- Etard C., Roostalu U., Strähle U. 2010. Lack of Apobec2-related proteins causes a dystrophic muscle phenotype in zebrafish embryos. J. Cell Biol. 189:527–539 10.1083/jcb.200912125 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Feldhahn N., Henke N., Melchior K., Duy C., Soh B.N., Klein F., von Levetzow G., Giebel B., Li A., Hofmann W.-K., et al. 2007. Activation-induced cytidine deaminase acts as a mutator in BCR-ABL1–transformed acute lymphoblastic leukemia cells. J. Exp. Med. 204:1157–1166 10.1084/jem.20062662 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Galigniana M.D., Harrell J.M., O’Hagen H.M., Ljungman M., Pratt W.B. 2004. Hsp90-binding immunophilins link p53 to dynein during p53 transport to the nucleus. J. Biol. Chem. 279:22483–22489 10.1074/jbc.M402223200 [DOI] [PubMed] [Google Scholar]
- Giannini A., Bijlmakers M.-J. 2004. Regulation of the Src family kinase Lck by Hsp90 and ubiquitination. Mol. Cell. Biol. 24:5667–5676 10.1128/MCB.24.13.5667-5676.2004 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Greeve J., Philipsen A., Krause K., Klapper W., Heidorn K., Castle B.E., Janda J., Marcu K.B., Parwaresch R. 2003. Expression of activation-induced cytidine deaminase in human B-cell non-Hodgkin lymphomas. Blood. 101:3574–3580 10.1182/blood-2002-08-2424 [DOI] [PubMed] [Google Scholar]
- Hansen L.K., Houchins J.P., O’Leary J.J. 1991. Differential regulation of HSC70, HSP70, HSP90 alpha, and HSP90 beta mRNA expression by mitogen activation and heat shock in human lymphocytes. Exp. Cell Res. 192:587–596 10.1016/0014-4827(91)90080-E [DOI] [PubMed] [Google Scholar]
- Hernández M.P., Chadli A., Toft D.O. 2002. HSP40 binding is the first step in the HSP90 chaperoning pathway for the progesterone receptor. J. Biol. Chem. 277:11873–11881 10.1074/jbc.M111445200 [DOI] [PubMed] [Google Scholar]
- Ito S., Nagaoka H., Shinkura R., Begum N., Muramatsu M., Nakata M., Honjo T. 2004. Activation-induced cytidine deaminase shuttles between nucleus and cytoplasm like apolipoprotein B mRNA editing catalytic polypeptide 1. Proc. Natl. Acad. Sci. USA. 101:1975–1980 10.1073/pnas.0307335101 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jakob U., Lilie H., Meyer I., Buchner J. 1995. Transient interaction of Hsp90 with early unfolding intermediates of citrate synthase. Implications for heat shock in vivo. J. Biol. Chem. 270:7288–7294 10.1074/jbc.270.13.7288 [DOI] [PubMed] [Google Scholar]
- Kimura Y., Yahara I., Lindquist S. 1995. Role of the protein chaperone YDJ1 in establishing Hsp90-mediated signal transduction pathways. Science. 268:1362–1365 10.1126/science.7761857 [DOI] [PubMed] [Google Scholar]
- Klemm L., Duy C., Iacobucci I., Kuchen S., von Levetzow G., Feldhahn N., Henke N., Li Z., Hoffmann T.K., Kim Y.M., et al. 2009. The B cell mutator AID promotes B lymphoid blast crisis and drug resistance in chronic myeloid leukemia. Cancer Cell. 16:232–245 10.1016/j.ccr.2009.07.030 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li L., Xin H., Xu X., Huang M., Zhang X., Chen Y., Zhang S., Fu X.-Y., Chang Z. 2004. CHIP mediates degradation of Smad proteins and potentially regulates Smad-induced transcription. Mol. Cell. Biol. 24:856–864 10.1128/MCB.24.2.856-864.2004 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu M., Duke J.L., Richter D.J., Vinuesa C.G., Goodnow C.C., Kleinstein S.H., Schatz D.G. 2008. Two levels of protection for the B cell genome during somatic hypermutation. Nature. 451:841–845 10.1038/nature06547 [DOI] [PubMed] [Google Scholar]
- Lotz G.P., Lin H., Harst A., Obermann W.M.J. 2003. Aha1 binds to the middle domain of Hsp90, contributes to client protein activation, and stimulates the ATPase activity of the molecular chaperone. J. Biol. Chem. 278:17228–17235 10.1074/jbc.M212761200 [DOI] [PubMed] [Google Scholar]
- MacDuff D.A., Demorest Z.L., Harris R.S. 2009. AID can restrict L1 retrotransposition suggesting a dual role in innate and adaptive immunity. Nucleic Acids Res. 37:1854–1867 10.1093/nar/gkp030 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Matsumoto Y., Marusawa H., Kinoshita K., Endo Y., Kou T., Morisawa T., Azuma T., Okazaki I.M., Honjo T., Chiba T. 2007. Helicobacter pylori infection triggers aberrant expression of activation-induced cytidine deaminase in gastric epithelium. Nat. Med. 13:470–476 10.1038/nm1566 [DOI] [PubMed] [Google Scholar]
- McBride K.M., Barreto V., Ramiro A.R., Stavropoulos P., Nussenzweig M.C. 2004. Somatic hypermutation is limited by CRM1-dependent nuclear export of activation-induced deaminase. J. Exp. Med. 199:1235–1244 10.1084/jem.20040373 [DOI] [PMC free article] [PubMed] [Google Scholar]
- McDonough H., Patterson C. 2003. CHIP: a link between the chaperone and proteasome systems. Cell Stress Chaperones. 8:303–308 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Metz K., Ezernieks J., Sebald W., Duschl A. 1996. Interleukin-4 upregulates the heat shock protein Hsp90alpha and enhances transcription of a reporter gene coupled to a single heat shock element. FEBS Lett. 385:25–28 10.1016/0014-5793(96)00341-9 [DOI] [PubMed] [Google Scholar]
- Morgan H.D., Dean W., Coker H.A., Reik W., Petersen-Mahrt S.K. 2004. Activation-induced cytidine deaminase deaminates 5-methylcytosine in DNA and is expressed in pluripotent tissues: implications for epigenetic reprogramming. J. Biol. Chem. 279:52353–52360 10.1074/jbc.M407695200 [DOI] [PubMed] [Google Scholar]
- Muramatsu M., Sankaranand V.S., Anant S., Sugai M., Kinoshita K., Davidson N.O., Honjo T. 1999. Specific expression of activation-induced cytidine deaminase (AID), a novel member of the RNA-editing deaminase family in germinal center B cells. J. Biol. Chem. 274:18470–18476 10.1074/jbc.274.26.18470 [DOI] [PubMed] [Google Scholar]
- Nakamura M., Kondo S., Sugai M., Nazarea M., Imamura S., Honjo T. 1996. High frequency class switching of an IgM+ B lymphoma clone CH12F3 to IgA+ cells. Int. Immunol. 8:193–201 10.1093/intimm/8.2.193 [DOI] [PubMed] [Google Scholar]
- Nathan D.F., Vos M.H., Lindquist S. 1997. In vivo functions of the Saccharomyces cerevisiae Hsp90 chaperone. Proc. Natl. Acad. Sci. USA. 94:12949–12956 10.1073/pnas.94.24.12949 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Okazaki I.M., Hiai H., Kakazu N., Yamada S., Muramatsu M., Kinoshita K., Honjo T. 2003. Constitutive expression of AID leads to tumorigenesis. J. Exp. Med. 197:1173–1181 10.1084/jem.20030275 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Palacios F., Moreno P., Morande P., Abreu C., Correa A., Porro V., Landoni A.I., Gabus R., Giordano M., Dighiero G., et al. 2010. High expression of AID and active class switch recombination might account for a more aggressive disease in unmutated CLL patients: link with an activated microenvironment in CLL disease. Blood. 115:4488–4496 10.1182/blood-2009-12-257758 [DOI] [PubMed] [Google Scholar]
- Panaretou B., Prodromou C., Roe S.M., O’Brien R., Ladbury J.E., Piper P.W., Pearl L.H. 1998. ATP binding and hydrolysis are essential to the function of the Hsp90 molecular chaperone in vivo. EMBO J. 17:4829–4836 10.1093/emboj/17.16.4829 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Panaretou B., Siligardi G., Meyer P., Maloney A., Sullivan J.K., Singh S., Millson S.H., Clarke P.A., Naaby-Hansen S., Stein R., et al. 2002. Activation of the ATPase activity of hsp90 by the stress-regulated cochaperone aha1. Mol. Cell. 10:1307–1318 10.1016/S1097-2765(02)00785-2 [DOI] [PubMed] [Google Scholar]
- Pasqualucci L., Neumeister P., Goossens T., Nanjangud G., Chaganti R.S., Küppers R., Dalla-Favera R. 2001. Hypermutation of multiple proto-oncogenes in B-cell diffuse large-cell lymphomas. Nature. 412:341–346 10.1038/35085588 [DOI] [PubMed] [Google Scholar]
- Pasqualucci L., Guglielmino R., Houldsworth J., Mohr J., Aoufouchi S., Polakiewicz R., Chaganti R.S., Dalla-Favera R. 2004. Expression of the AID protein in normal and neoplastic B cells. Blood. 104:3318–3325 10.1182/blood-2004-04-1558 [DOI] [PubMed] [Google Scholar]
- Pasqualucci L., Bhagat G., Jankovic M., Compagno M., Smith P., Muramatsu M., Honjo T., Morse H.C., III, Nussenzweig M.C., Dalla-Favera R. 2008. AID is required for germinal center-derived lymphomagenesis. Nat. Genet. 40:108–112 10.1038/ng.2007.35 [DOI] [PubMed] [Google Scholar]
- Patenaude A.M., Orthwein A., Hu Y., Campo V.A., Kavli B., Buschiazzo A., Di Noia J.M. 2009. Active nuclear import and cytoplasmic retention of activation-induced deaminase. Nat. Struct. Mol. Biol. 16:517–527 10.1038/nsmb.1598 [DOI] [PubMed] [Google Scholar]
- Pauklin S., Sernández I.V., Bachmann G., Ramiro A.R., Petersen-Mahrt S.K. 2009. Estrogen directly activates AID transcription and function. J. Exp. Med. 206:99–111 10.1084/jem.20080521 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pearl L.H., Prodromou C. 2006. Structure and mechanism of the Hsp90 molecular chaperone machinery. Annu. Rev. Biochem. 75:271–294 10.1146/annurev.biochem.75.103004.142738 [DOI] [PubMed] [Google Scholar]
- Peled J.U., Kuang F.L., Iglesias-Ussel M.D., Roa S., Kalis S.L., Goodman M.F., Scharff M.D. 2008. The biochemistry of somatic hypermutation. Annu. Rev. Immunol. 26:481–511 10.1146/annurev.immunol.26.021607.090236 [DOI] [PubMed] [Google Scholar]
- Perkins D.N., Pappin D.J., Creasy D.M., Cottrell J.S. 1999. Probability-based protein identification by searching sequence databases using mass spectrometry data. Electrophoresis. 20:3551–3567 [DOI] [PubMed] [Google Scholar]
- Picard D. 2006. Chaperoning steroid hormone action. Trends Endocrinol. Metab. 17:229–235 10.1016/j.tem.2006.06.003 [DOI] [PubMed] [Google Scholar]
- Popp C., Dean W., Feng S., Cokus S.J., Andrews S., Pellegrini M., Jacobsen S.E., Reik W. 2010. Genome-wide erasure of DNA methylation in mouse primordial germ cells is affected by AID deficiency. Nature. 463:1101–1105 10.1038/nature08829 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pratt W.B., Toft D.O. 2003. Regulation of signaling protein function and trafficking by the hsp90/hsp70-based chaperone machinery. Exp. Biol. Med. (Maywood). 228:111–133 [DOI] [PubMed] [Google Scholar]
- Prochnow C., Bransteitter R., Klein M.G., Goodman M.F., Chen X.S. 2007. The APOBEC-2 crystal structure and functional implications for the deaminase AID. Nature. 445:447–451 10.1038/nature05492 [DOI] [PubMed] [Google Scholar]
- Prodromou C., Roe S.M., O’Brien R., Ladbury J.E., Piper P.W., Pearl L.H. 1997. Identification and structural characterization of the ATP/ADP-binding site in the Hsp90 molecular chaperone. Cell. 90:65–75 10.1016/S0092-8674(00)80314-1 [DOI] [PubMed] [Google Scholar]
- Ramiro A.R., Jankovic M., Eisenreich T., Difilippantonio S., Chen-Kiang S., Muramatsu M., Honjo T., Nussenzweig A., Nussenzweig M.C. 2004. AID is required for c-myc/IgH chromosome translocations in vivo. Cell. 118:431–438 10.1016/j.cell.2004.08.006 [DOI] [PubMed] [Google Scholar]
- Ramiro A.R., Jankovic M., Callen E., Difilippantonio S., Chen H.-T., McBride K.M., Eisenreich T.R., Chen J., Dickins R.A., Lowe S.W., et al. 2006. Role of genomic instability and p53 in AID-induced c-myc-Igh translocations. Nature. 440:105–109 10.1038/nature04495 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Robbiani D.F., Bothmer A., Callen E., Reina-San-Martin B., Dorsett Y., Difilippantonio S., Bolland D.J., Chen H.T., Corcoran A.E., Nussenzweig A., Nussenzweig M.C. 2008. AID is required for the chromosomal breaks in c-myc that lead to c-myc/IgH translocations. Cell. 135:1028–1038 10.1016/j.cell.2008.09.062 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Robbiani D.F., Bunting S., Feldhahn N., Bothmer A., Camps J., Deroubaix S., McBride K.M., Klein I.A., Stone G., Eisenreich T.R., et al. 2009. AID produces DNA double-strand breaks in non-Ig genes and mature B cell lymphomas with reciprocal chromosome translocations. Mol. Cell. 36:631–641 10.1016/j.molcel.2009.11.007 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sernández I.V., de Yébenes V.G., Dorsett Y., Ramiro A.R. 2008. Haploinsufficiency of activation-induced deaminase for antibody diversification and chromosome translocations both in vitro and in vivo. PLoS One. 3:e3927 10.1371/journal.pone.0003927 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sreedhar A.S., Kalmár E., Csermely P., Shen Y.F. 2004. Hsp90 isoforms: functions, expression and clinical importance. FEBS Lett. 562:11–15 10.1016/S0014-5793(04)00229-7 [DOI] [PubMed] [Google Scholar]
- Stavnezer J., Guikema J.E.J., Schrader C.E. 2008. Mechanism and regulation of class switch recombination. Annu. Rev. Immunol. 26:261–292 10.1146/annurev.immunol.26.021607.090248 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stebbins C.E., Russo A.A., Schneider C., Rosen N., Hartl F.U., Pavletich N.P. 1997. Crystal structure of an Hsp90-geldanamycin complex: targeting of a protein chaperone by an antitumor agent. Cell. 89:239–250 10.1016/S0092-8674(00)80203-2 [DOI] [PubMed] [Google Scholar]
- Takizawa M., Tolarová H., Li Z., Dubois W., Lim S., Callen E., Franco S., Mosaico M., Feigenbaum L., Alt F.W., et al. 2008. AID expression levels determine the extent of cMyc oncogenic translocations and the incidence of B cell tumor development. J. Exp. Med. 205:1949–1957 10.1084/jem.20081007 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Teng G., Hakimpour P., Landgraf P., Rice A., Tuschl T., Casellas R., Papavasiliou F.N. 2008. MicroRNA-155 is a negative regulator of activation-induced cytidine deaminase. Immunity. 28:621–629 10.1016/j.immuni.2008.03.015 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wandinger S.K., Richter K., Buchner J. 2008. The Hsp90 chaperone machinery. J. Biol. Chem. 283:18473–18477 10.1074/jbc.R800007200 [DOI] [PubMed] [Google Scholar]
- Whitesell L., Lindquist S.L. 2005. HSP90 and the chaperoning of cancer. Nat. Rev. Cancer. 5:761–772 10.1038/nrc1716 [DOI] [PubMed] [Google Scholar]
- Wu X., Geraldes P., Platt J.L., Cascalho M. 2005. The double-edged sword of activation-induced cytidine deaminase. J. Immunol. 174:934–941 [DOI] [PubMed] [Google Scholar]
- Young J.C., Hartl F.U. 2000. Polypeptide release by Hsp90 involves ATP hydrolysis and is enhanced by the co-chaperone p23. EMBO J. 19:5930–5940 10.1093/emboj/19.21.5930 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Young J.C., Agashe V.R., Siegers K., Hartl F.U. 2004. Pathways of chaperone-mediated protein folding in the cytosol. Nat. Rev. Mol. Cell Biol. 5:781–791 10.1038/nrm1492 [DOI] [PubMed] [Google Scholar]